Abstract
Background and Aims Olive is considered a native plant of the eastern side of the Mediterranean basin, from where it should have spread westward along the Mediterranean shores, while little is known about its diffusion in the eastern direction.
Methods Genetic diversity levels and population genetic structure of a wide set of olive ecotypes and varieties collected from several provinces of Iran, representing a high percentage of the entire olive resources present in the area, was screened with 49 chloroplast and ten nuclear simple sequence repeat markers, and coupled with archaeo-botanical and historical data on Mediterranean olive varieties. Approximate Bayesian Computation was applied to define the demographic history of olives including Iranian germplasm, and species distribution modelling was performed to understand the impact of the Late Quaternary on olive distribution.
Key Results The results of the present study demonstrated that: (1) the climatic conditions of the last glacial maximum had an important role on the actual olive distribution, (2) all Iranian olive samples had the same maternal inheritance as Mediterranean cultivars, and (3) the nuclear gene flow from the Mediterranean basin to the Iranian plateau was almost absent, as well as the contribution of subspecies cuspidata to the diversity of Iranian olives.
Conclusions Based on this evidence, a new scenario for the origin and distribution of this important fruit crop has been traced. The evaluation of olive trees growing in the eastern part of the Levant highlighted a new perspective on the spread and distribution of olive, suggesting two routes of olive differentiation, one westward, spreading along the Mediterranean basin, and another moving towards the east and reaching the Iranian plateau before its domestication.
Keywords: Olea europaea subsp. europaea var. europaea, centre of origin, chloroplast markers, SSR markers, ecotypes, genetic differentiation, population genetics
INTRODUCTION
The Near East area and neighbouring regions are recognized as the primary centre where the first cultivation took place, and where the colonization of the Mediterranean region for many crop and animal species started (Zohary and Hopf, 2000; Petit et al., 2005; Rodriguez-Sanchez et al., 2009; Lovell et al., 2010; Linares, 2011; Myles et al., 2011; Zeder, 2011; Kaniewski et al., 2012; Mayol et al., 2015).
The ancient distribution and the process of domestication of olive (Olea europaea L. subsp. europaea, var. europaea) and its relationships with local wild populations and ancestral varieties are still openly debated issues. In the last two decades, numerous phylogeographical and phylogenetic analyses have been carried out to clarify the diffusion of olive along the Mediterranean basin, by using nuclear, plastidial and mitochondrial markers (Besnard et al., 2002, 2007, 2009; Contento et al., 2002; Breton et al., 2006) and applying archaeological, historical and ecological data (Terral, 1996; Terral et al., 2004, 2005; Kaniewski et al., 2012; Zohary et al., 2012). According to Besnard et al. (2013a), the north-eastern Levant (Syrian–Turkish border) can be identified as the primary centre of olive domestication, assuming the westward route of olive diffusion, even if the central part of the Mediterranean basin has been recently assumed as a local centre of domestication (Diez et al., 2015).
However, first evidence on the presence of olives with unknown origin in the eastern side of the Levant area provided elements for new considerations on their origin and diffusion. In fact, studies performed on olive trees presently growing in Iran, often under extreme climate and soil conditions and with unknown cultivation status, have highlighted their clear genetic differentiation from Mediterranean cultivars (Noormohammadi et al., 2014; Mousavi et al., 2014), indicating a possible founder effect from an unknown centre of origin (Hosseini-Mazinani et al., 2014).
The presence of Olea europaea has been documented by archaeo-botanical and pollen evidence from the lower Palaeolithic, onward along the Fertile Crescent area (including ancient Mesopotamia and the eastern coast of the Mediterranean, the actual Near- and Middle-East countries), which hosted the earliest human civilizations and the wild progenitors of the most important crops and domesticated animals in early agriculture (Niklewski and van Zeist, 1970; Noy et al., 1973; van Zeist and Bakker-Heeres, 1984; Darmon, 1987; Liphschitz et al., 1991; Kislev et al., 1992; Willcox, 1996; Diamond, 2002; van Zeist and Bottema, 2009; Lovell et al., 2010; Zohary et al., 2012).
Olive distribution, in addition to being under the influence of human activities, has also been influenced by the effects of climate change, as for naturally spread wild olives (Besnard et al., 2013b). The frequency, direction and timing of Quaternary climate change in Iran and western Asia as a whole are sparsely documented, but the dominance of herbaceous over tree pollen indicates lower precipitation rates before and during the Last Glacial Maximum (LGM, 22 000 years BP) than today, while during the early Holocene rainfall levels were increased (Kehl, 2009).
Based on this background, the present work aimed to verify: (1) how human factors and climate events may have affected the present distribution pattern of olive, and (2) if, when and how olive may have moved in two opposite directions (eastward and westward) from the putative centre of origin.
A series of climate scenarios, that are likely to have modelled the distribution of the species, was reconstructed, to evaluate the relative importance of current and past climatic conditions on the observed patterns of genetic variation. To investigate the demographic history of O. europaea, a dual approach was applied: an assignment test (to determine the probability of an individual originating from a gene pool) and an Approximate Bayesian Computation (ABC) analysis (Beaumont et al., 2002), to select the most likely scenario shaping genetic diversity and to set an approximate time frame for the inferred history, as already reported for other cultivated plant species (Tamaki et al., 2016; Parat et al., 2016).
MATERIALS AND METHODS
Plant material and DNA extraction
Apart from recently planted olive groves, olive in Iran occurs as isolated trees or groups of a few plants growing in natural or semi-natural ecosystems, in areas populated by wild relatives of other fruit crops, such as pistachio, almond and pomegranate. Some trees grow near or within archaeological, historical or sacred sites, and benefit from the respect and attention of local people (Supplementary Data Fig. S1). We included in the present study all samples previously analysed by Hosseini-Mazinani et al. (2014), together with new olive samples, to keep as much variability still present in Iran as possible. Cultivated varieties imported from abroad were excluded from this set. Some individuals were characterized by large bushes or by multiple or single trees, reaching considerable trunk diameter and fruit size (Hosseini-Mazinani and Torkzaban, 2013). In each population, from 30 to 100 % of individuals were sampled (1–12 samples, depending on the size of the target population), for a total of 132 olives, representative of 34 sites and 15 provinces, and 20 cultivated varieties with local origin (Supplementary Data Table S1, Figs S1 and S2). Table S1 also reports climatic data, coordinates and altitudes of the sampling sites (www.irimo.ir). According to the previous work (Hosseini-Mazinani et al., 2014), and given their unknown origin, these olives are hereafter termed olive ecotypes. Samples deriving from three south-eastern Iranian provinces, previously distinguished as subspecies cuspidata (Hosseini-Mazinani et al., 2014), were also included in the analysis, together with new samples derived from the same areas, for a total of 37 specimens.
Total DNA was extracted from leaves by using a GeneElute Plant Genomic DNA Miniprep Kit (Sigma-Aldrich), following the manufacturer’s instructions.
To establish the relationships with Mediterranean cultivated olives, simple sequence repeat (SSR) data for 273 Mediterranean olive cultivars, derived from the CNR-IBBR olive molecular database and from previously published data (Baldoni et al., 2009; Trujillo et al., 2014; Mariotti et al., 2016), were also included in the analysis (Supplementary Data Table S2).
Chloroplast and nuclear markers
To detect maternal inheritance of the Iranian samples, chloroplast genotyping was performed by using 29 chloroplast (cp)SSRs and indels and 20 single nucleotide polymorphisms (SNPs), the latter through SNaPshot technique (Mariotti et al., 2010; Hosseini-Mazinani et al., 2014). Furthermore, potentially polymorphic regions (Besnard et al., 2011) were analysed by direct sequencing using primers listed in Supplementary Data Table S3. Data obtained from this analysis were compared with those of Mediterranean cultivars previously published (Mariotti et al., 2010; Besnard et al., 2013a).
In the present work, ten SSR markers were applied, representing the best ranked loci over 77 evaluated microsatellite regions, recommended for olive genotyping due to their highest discrimination power, independent segregation and single locus inheritance (Baldoni et al., 2009). PCR amplifications were performed following the previously published method (Hosseini-Mazinani et al., 2014). Output data were analysed by GeneMapper 3.7 (Applied Biosystems-Hitachi).
Frequency analysis and genetic differentiation of whole samples
Frequency analyses were performed on a total of 462 genotypes, including 152 Iranian samples (132 ecotypes and 20 local varieties), 273 Mediterranean varieties and 37 Olea europaea subsp. cuspidata samples.
FreeNA (Chapuis and Estoup, 2007) was used to estimate the presence of null alleles, with the ENA correction allowing us to refine differentiation by excluding null alleles. Allelic richness (AR) and gene diversity (GD) were calculated using Fstat (Goudet, 2002). Intra- and inter-population genetic statistics [number of alleles (Na), effective number of alleles (Ne), observed (Ho) and expected (He) heterozygosities and fixation index (F)] were calculated using GenAlEx 6.5 (Peakall and Smouse, 2013), as well as the principal coordinates analysis (PCoA), to visualize the genetic distances among accessions.
The SSR data were further analysed by STRUCTURE 2.3.4 software (Pritchard et al., 2009), running 100 replicate Monte Carlo Markov chains (MCMCs) with a burn-in period of 100 000 followed by a sampling period of 100 000 for 500 000 iterations, applied for each K. The range of possible number of clusters (K) was from 1 to 10, considering independent alleles and admixture of individuals. Bayesian analysis divided sampled individuals into a number of K clusters and the most likely value of K was estimated using ΔK (Evanno et al., 2005), performed by STRUCTURE Harvester (Earl and von Holdt, 2012). GenAlEx 6.5 was used to estimate pairwise population matrices of Dest, Gst statistics and Fst pairwise distance with 999 permutations, in order to detect the differentiation between the STRUCTURE groups.
To evaluate the assignment of individuals and groups within the populations found by STRUCTURE, three different statistical criteria were applied, as implemented in GeneClass 2 software (Piry et al., 2004): Cavalli-Sforza and Edwards’ (1967) Chord distance, the likelihood frequency-based method of Paetkau et al. (1995) (extended by Piry et al., 2004), and a Bayesian method developed by Baudouin and Lebrun (2001). Monte Carlo resampling of 10 000 individuals was used to compute the probability of the multilocus genotype of each individual being encountered in a given reference population following Paetkau et al. (2004). Furthermore, to establish the contribution of Iranian Olea europaea subsp. cuspidata to the present genetic variability of Iranian olives, shared and private alleles within and between the cuspidata samples, Mediterranean varieties, Iranian ecotypes and cultivars were assessed by GenAlEx 6.5, followed by a GeneClass 2 assignment, applying the described criteria.
Genetic diversity and differentiation within Iran
On the Iranian olive trees (cultivars and ecotypes), to assign the relationships within and between different sample sets, molecular evolutionary analyses were performed by Darwin v.5 software (Perrier and Jacquemoud-Collet, 2006), using the weighted neighbour-joining method, running 10 000 bootstrap replications. STRUCTURE 2.3.4 software was run using the same parameters previously reported. The range of possible number of clusters (K) was set from 1 to 34 (number of populations based on geographical sampling sites), with independent alleles and admixture model. STRUCTURE Harvester identified the most likely K values. The estimation of pairwise population matrices was assessed by GenAlEx.
Demographic history inferred by ABC
To trace the demographic history of olive, a two-step ABC procedure (Beaumont et al., 2002), implemented in DIYABC v 2.1.0 (Cornuet et al., 2014), was performed. The entire data set (A) (425 genotypes, excluding cuspidata samples) was first analysed to draft the history of the hierarchical genetic structure identified by STRUCTURE analysis (see Results), then ABC analysis was focused only on the Iranian individuals (B).
(A) We based DIYABC analysis on the genetic clustering results of 425 individuals (excluding cuspidata samples), although for simplicity we considered the Iranian samples as a single group. The three groups were: Western Mediterranean cultivars (WM), Central-Eastern Mediterranean cultivars (CEM), and Iranian cultivars and ecotypes (IR). We examined three simple population demography scenarios (Supplementary Data Fig. S3), in which, t# refers to timescale (scaled by generation time) and N# refers to effective population size of the corresponding population (WM, CEM, IR, the branch connecting to CEM between t1 and t2, or the ancestral population) during each time (i.e. 0 – t1 and t1 – t2). Furthermore, IR was set to be traced back to an ancestral population (as requested by DIYABC), since it was the population with the highest level of differentiation and with a high level of genetic diversity. Nevertheless, the level of diversity was high also in WM, so we decided to perform a preliminary test to ascertain which one of the two most differentiated populations (WM or IR) coalesced to the ancestral population. The models, in which IR was set to trace back to the ancestral population, obtained good support, and therefore all successive analyses included this condition. The three scenarios are as follows (Fig. S3A): Scenario 1 (hierarchical split model), testing a colonization event from the middle-east region, WM merged with CEM at t1, and subsequently CEM merged with IR at t2. N1, N2 and N3 were the effective population sizes of WM, CEM and IR, respectively, Na was the effective population size of the ancestral population, and Nb was the effective population size of CEM at t1 and t2; Scenario 2 (simple split model), in which all three populations diverged at the same time, t2; and Scenario 3 (isolation with admixture model), testing the possibility that the two most differentiated groups, WM and IR, generated CEM by admixture at t1, and then WM merged with IR at t2.
(B) The STRUCTURE analysis within Iranian olive ecotypes and varieties indicated the presence of a clear genetic structure with four clusters (see Results), and therefore we defined four populations: Pop1, Pop2, Pop3 and Pop4. In the four scenarios examined (Fig. S3B), Pop2 was set to be the most ancient population, since it was the population with the highest level of diversity. Furthermore, we decided to put Pop4 merging with Pop3 or Pop1. The four scenarios are as follows (Fig. S3B): Scenario 1 (hierarchical split model 1), in which we assumed that Pop3 merged with Pop4 at t1, Pop4 merged with Pop1 at t2, and Pop1 merged with Pop2 at t3; Scenario 2 (hierarchical split model 2), which was similar to scenario 1 but assuming that Pop4 merged with Pop3 at t1, and that Pop3 merged with Pop1 at t2; Scenario 3 (hierarchical split model 3), the simplest one, in which Pop4 merged with Pop3 at t1, and Pop1 and Pop3 merged with Pop2 at the same time (t3); and Scenario 4 (hierarchical split model 4), similar to scenario 3 but assuming that Pop4 merged with Pop1 at t1. Considering that in natural conditions an olive seedling can start flowering after 15–20 years or earlier and may remain productive for at least 30–50 years (despite the long lifespan of olive, in natural conditions its competitiveness with other tree species is low), the estimation of generation time in olive is complicated. In accordance with other authors (Besnard et al., 2014; Diez et al., 2015), 25 years was considered as the average generation time.
DIYABC-Mutation model, summary statistics and model checking
For all simulations, we used the Generalized Stepwise Mutation model (GSM; Estoup et al., 2002), with single nucleotide indels (SNIs). To obtain better posterior distribution, we performed initial runs to explore the parameter space, thus changing the default prior values (Supplementary Data Tables S4 and S5). The minimum and maximum priors for SSR mutation rate were set at 5 × 10−4–5 × 10−3 (Tables S4 and S5). Mean expected heterozygosity (He) and number of alleles (Na) were used as summary statistics for single populations and He, Na, and Fst for population pairs. A million simulations were performed for each scenario and the most likely scenario was evaluated by comparing posterior probabilities, using logistic regression. Goodness-of-fit was assessed for each scenario by model checking using the principal component analysis (PCA) approach implemented in DIYABC, which measures the discrepancy between simulated and real data. Finally, each scenario was used in a simulation of 1000 datasets to estimate Type I and Type II errors by using ‘Evaluate the confidence in scenario choice’ option in DIYABC. Type I error for the most likely scenario X was estimated as the probability at which scenario X is rejected when it is the true scenario. Type II error for the most likely scenario X was computed as the probability of choosing the scenario X when it was not the true scenario.
Species distribution modelling and climatic data
Species distribution modelling (SDM), based on climatic information from the database of WorldClim - Global Climate Data (Hijmans et al., 2005), was performed to define the shape of possible distribution throughout the late Quaternary: the last interglacial period (LIG; 120 000–140 000 years BP), the LGM (21 000 years BP), Mid-Holocene (MH; 6000 years BP) and Pre-present (1950–2000 years BP). The maximum entropy model, implemented in a MAXENT 3.3.3 algorithm (Phillips et al., 2006), was used. Random null distributions were built to test for the significance of our SDM. For this test, we built a new SDM using 34 occurrence points for Olea europaea Iranian ecotypes (Table S1). Predictions were also generated for the WM and CEM gene pools, to examine whether genetic divergences among them were environmentally induced. The former distribution of O. europaea by projecting our model on to LGM, MH, LIG and Pre-present conditions using 19 bioclimatic variables data was predicted (Supplementary Data Table S6). Predicted distributions during the LGM and MH were generated by downloading both the community climate system model (CCSM) and the model for interdisciplinary research on climate (MIROC). The modelled distributions were generated with 70 % of the points (training data) and cross-validated with 30 % of the remaining localities (test data), averaged over 10 runs and all other parameters were set to default. The performance of models was assessed using the area under the receiver operating characteristic curve (AUC; Hanley and McNeil, 1982). We used the suitability threshold to derive projected presence/absence distribution from logistic outputs. To show the effect of 19 bioclimatic variables from 140 000 years BP (LIG) up to Pre-present conditions on the three groups individuated by Bayesian analyses (WM, CEM and IR), PCA was performed by PAST software v. 3.10 (Hammer et al., 2001).
RESULTS
Chloroplast variation analysis
All Iranian samples and reference cultivars shared the most common E1 lineage (chlorotype E1.1), shown by 90 % of the Mediterranean cultivated varieties (Besnard et al., 2013a). The only exceptions were 37 genotypes of subsp. cuspidata, clearly belonging to lineage C, which is typical of the south Asian populations of cuspidata (Besnard et al., 2007, 2011), and distinguished into two different chlorotypes, sublineages C1 and C2, as previously reported by Hosseini-Mazinani et al. (2014) (Supplementary Data Table S7).
Genetic diversity and differentiation between Mediterranean and Iranian samples
Diversity data include Mediterranean cultivars and Iranian ecotypes, with the exclusion of the cuspidata samples, considering them belonging to the different subspecies.
SSR null allele values, which ranged from 0·006 to 0·044, were considered negligible values by Chapuis and Estoup (2007). The only exceptions were loci DCA16 and GAPU103A, for which values were moderate: 0·060 and 0·061, respectively. Iranian genotypes showed the highest values for frequency indices, such as Ne, GD, He and F, whereas Ho was higher in the Mediterranean samples (Supplementary Data Table S8).
PCoA separated genotypes into three main groups: Iranian, Eastern-Central Mediterranean and western Mediterranean (Fig. 1), with coordinates explaining 8·89 and 6·69 % of the total variance.
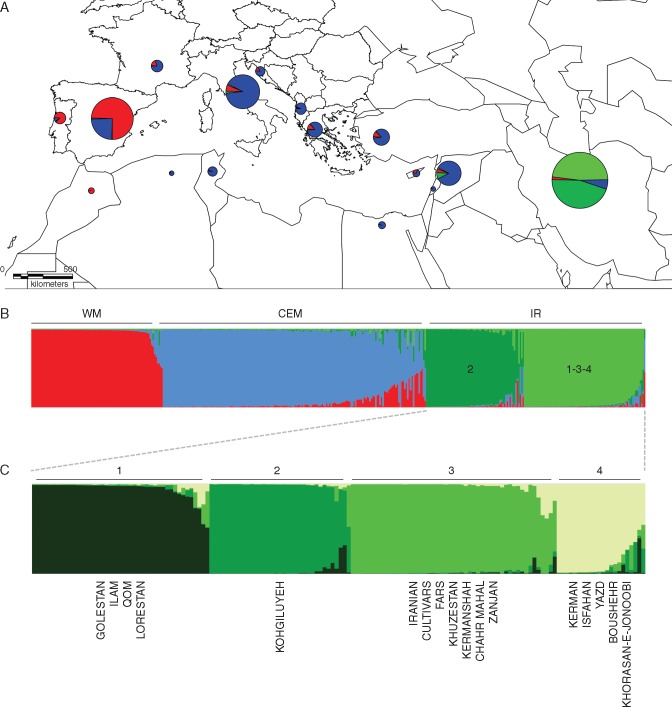
Fig. 1.
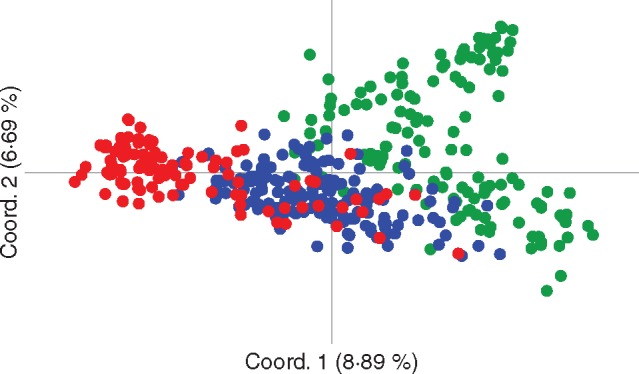
Differentiation among Iranian and Mediterranean olives based on the variation of SSR markers, based on principal coordinates analysis (PCoA) of the olive genotypes across the Mediterranean and Iranian distribution range. Red = Western Mediterranean cultivars; blue = Central-Eastern Mediterranean cultivars; green = Iranian genotypes and cultivars.
When STRUCTURE analysis was performed, the number of most likely sub-populations (K) peaked at K = 4. The four groups were clearly separated on the basis of their geographical pattern (Fig. 2A): the Mediterranean cultivars clustered into Western (WM) and Central-Eastern Mediterranean (CEM) groups, as previously shown by PCoA, whereas the Iranian samples were split into two other groups (Fig. 2B).
Fig. 2.
(A) Geographical distribution of different gene pools detected through Mediterranean and Iran using Structure software. Size of circles refers to the number of samples. (B) Four populations detected based on the entire set of samples. Red: Western Mediterranean (WM) cultivars including Morocco, Spain and Portugal; blue: Central-Eastern Mediterranean (CEM) cultivars including Algeria, Tunisia, France, Italy, Croatia, Albania, Greece, Turkey, Egypt, Cyprus, Lebanon and Syria; dark and light green: Iranian cultivars and ecotypes (IR). (C) Iranian samples split into four populations (1–4).
To study the genetic differentiation between the three geographical areas (WM, CEM and IR), we considered the Iranian groups together, whereas the genetic variability within Iranian samples is described in a separate section of this work. Different parameters and pairwise population distances were calculated to determine differentiation between the three mentioned groups (Table 1). Pairwise population values decreased toward the west, suggesting an important genetic differentiation between eastern and western groups (Table 1B).
Table 1.
Diversity (A) and differentiation (B) estimates for the three populations (groups) obtained by STRUCTURE analysis (below the diagonal)
| (A) | Na | Ho | He | F |
|---|---|---|---|---|
| WM | 10·90 | 0·806 | 0·72 | –0·179 |
| CEM | 17·70 | 0·857 | 0·83 | –0·038 |
| IR | 18·30 | 0·749 | 0·84 | 0·111 |
| (B) | WM | CEM | IR | |
| Dest | WM | 0·000 | 0·001 | 0·001 |
| CEM | 0·175 | 0·000 | 0·001 | |
| IR | 0·422 | 0·299 | 0·000 | |
| G′st(Hed) | WM | 0·000 | 0·002 | 0·002 |
| CEM | 0·196 | 0·000 | 0·002 | |
| IR | 0·454 | 0·319 | 0·000 | |
| Fst | WM | 0·000 | 0·001 | 0·001 |
| CEM | 0·027 | 0·000 | 0·001 | |
| IR | 0·058 | 0·030 | 0·000 |
Fst = Inbreeding coefficient within subpopulations, relative to total = genetic differentiation among populations, Fst = (Ht−Hs)/ Ht; G′st(Hed) = Hedrick’s standardized G′st, further corrected for bias when the numbers of populations is small; Dest = Jost’s estimate of differentiation. Values for Dest, G′st(Hed) and Fst values are below the diagonal. Probability P (rand > = data) based on 999 permutations is shown above the diagonal.
The assignment analysis indicated the proportions of individuals derived from their source population (Table 2). WM, representing the largest population (179 samples), showed the highest values, while the lowest was the one of IR. The limitation of genetic sharing among populations derived from their geographical pattern. For all calculated methods, the CEM population showed considerable migration to WM, ranging between 57·78 % (Distance) to 62·03 % (Bayesian). The Iranian population (IR) did not originate from the other populations, while the migration from Iran to CEM was 4·13–7·56 % and to WM 5·19–8·81 %.
Table 2.
Percentage of assigned individuals performed by GENECLASS2
| Distance |
Frequency |
Bayesian |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| WM | CEM | IR | WM | CEM | IR | WM | CEM | IR | |
| WM | 66·53 | 57·78 | 8·81 | 57·56 | 59·74 | 5·19 | 59·02 | 62·03 | 6·26 |
| CEM | 1·81 | 53·95 | 7·56 | 1·34 | 50·33 | 4·45 | 1·30 | 51·40 | 4·13 |
| IR | 0·02 | 1·37 | 43·08 | 0·04 | 1·62 | 45·88 | 0·01 | 1·37 | 45·81 |
Three groups of populations (WM, CEM and IR), as identified by population structure analyses (considering the two Iranian populations as a single group), were compared. Exclusion of individuals is based in three different criteria: the Cavalli–Sforza and Edwards distance, the frequency-based method of Paetkau et al. (1995) and the Bayesian approach of Baudouin and Lebrun (2001).
Contribution of subspecies cuspidata
When considering the entire pool of samples, including subsp. cuspidata specimens, of 255 total alleles found in all analysed accessions, 23 were private to cuspidata samples, 35 to Mediterranean varieties and 32 to the Iranian olives. When only the Iranian olives were compared with cuspidata, 93 common alleles were found. Mediterranean cultivars and cuspidata shared 88 alleles, whereas 93 and 42 alleles were private to Mediterranean and to cuspidata, respectively. Population differentiation values derived by Dest, G′st(Hed) and Fst pairwise matrices (Table 3A), calculated between Mediterranean cultivars, Iranian olives and Iranian subsp. cuspidata samples, showed the highest values between cuspidata and Mediterranean varieties, followed by those between cuspidata and the Iranian olives. The lowest values were between Iranian and Mediterranean samples. The individual assignment analysis suggested no migration among Iranian individuals, cuspidata and Mediterranean cultivars (Table 3B).
Table 3.
(A) Pairwise population matrices of Dest, Hedrick's standardized G'st (G'st(Hed)) and Fst. (B) Percentage of assigned individuals performed by GENECLASS2. Three groups of populations
| (A) | Med cvs | IR | cusp | |
|---|---|---|---|---|
| Dest | Mediterranean cultivars | 0·000 | 0·001 | 0·001 |
| Iranian olives | 0·321 | 0·000 | 0·001 | |
| subsp. cuspidata | 0·634* | 0·590 | 0·000 | |
| G'st(Hed) | Mediterranean cultivars | 0·000 | 0·002 | 0·002 |
| Iranian olives | 0·343 | 0·000 | 0·002 | |
| subsp. cuspidata | 0·658 | 0·612 | 0·000 | |
| Fst | Mediterranean cultivars | 0·000 | 0·001 | 0·001 |
| Iranian olives | 0·036 | 0·000 | 0·001 | |
| subsp. cuspidata | 0·072 | 0·059 | 0·000 |
| (B) | Distance |
Frequency |
Bayesian |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Med cvs | IR | cusp | Med cvs | IR | cusp | Med cvs | IR | cusp | |
| Med cvs | 52·06 | 45·84 | 0·01** | 55·26 | 7·24 | 0·16 | 53·19 | 4·96 | 0·06 |
| IR | 0·98 | 4·51 | 0·05 | 1·22 | 43·12 | 0·49 | 0·90 | 46·00 | 0·21 |
| cusp | 0·00 | 0·05 | 27·91 | 0·00 | 0·00 | 34·29 | 0·00 | 0·02 | 40·85 |
In bold type are the highest distance values. Probability P (rand >= data) based on 999 permutations.
In bold type are the percentage of subsp. cuspidata assignment.
Med cvs, Mediterranean cultivars; IR, Iranian ecotypes and cultivars; cusp, subspecies cuspidata.
Genetic diversity and differentiation within Iranian olives
Within the Iranian olives (excluding cuspidata trees), Ho was lower than He at all loci except EMO90 and GAPU101, and F values showed positive and significant deviations from zero for most loci (mean value = 0·11), indicating a high level of inbreeding within Iranian samples (Supplementary Data Table S9).
Neighbour-joining analysis on the Iranian ecotypes and cultivars (Supplementary Data Fig. S4) showed that Koghiluyeh olive trees clustered completely apart, while all other samples grouped together (Ilam, Golestan, Boushehr, Yazd and Isfahan provinces). The high bootstrap values (cut off under 50 %) confirmed the reliability of the results. When the Bayesian clustering analysis was performed only on the Iranian samples, the best log-likelihood value highlighted the presence of four genetic groups: Pop1 comprised samples from Golestan, Ilam, Lorestan and Qom provinces, Pop2 was exclusively represented by Kohgiluyeh samples, Pop3 included Zanjan, Khuzestan, Chahar Mahal, Kermanshah and Fars ecotypes and 16 out of 20 main Iranian cultivars, and Pop4 contained five provinces from the centre, south and east of Iran and three other varieties, with some admixture with Pop2 and Pop3 (Fig. 2C). Between the groups identified by STRUCTURE, the highest genetic differentiation was between Pop2 and Pop3 (Supplementary Data Table S10).
Demographic history of olive varieties
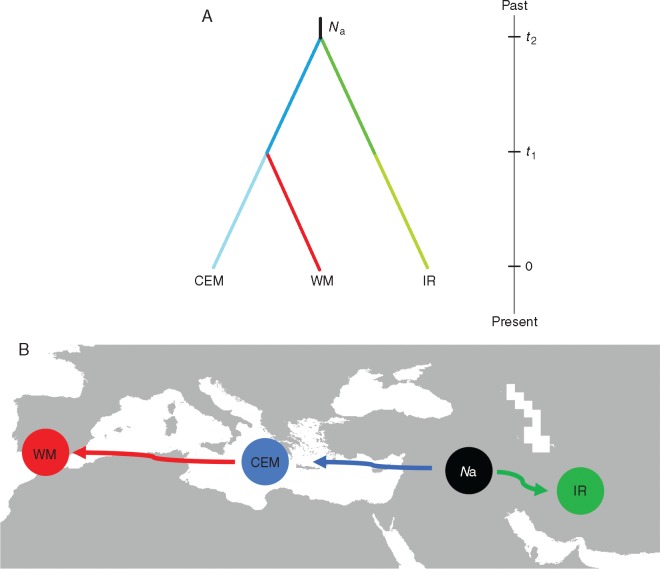
The highest posterior probability defined by DIYABC analysis was found for Scenario 1 (hierarchical split model) (Fig. 3A, B), and its value [0·9204, 95 % confidence interval (CI): 0·9101–0·9307] was higher than for scenarios 2 (0·0008) and 3 (0·0788) (Table 4). This result suggests that the divergence of IR from a hypothetical ancestral population occurred before the divergence of CEM and WM. The median values of effective population sizes for this scenario were 1·78, 2·69, 9·01, 1·35 and 3·56 for N1 (WM), N2 (CEM), N3 (IR), Na (ancestral population) and Nb (the branch conducting to CEM between t1 and t2), respectively (Supplementary Data Table S11). The results showed that the effective population size of the ancestral population (Na) was approximately 6·6 times lower than that of IR, suggesting an expansion event at time t2. At time t1, a further expansion event generated population CEM, increasing the effective population size by around 7·5 times with respect to its size at t2. Furthermore, at t1 a bottleneck event gave rise to WM, reducing the effective population size around twofold. The median values of the divergence times t1 and t2 were 648 (95 % CI: 170–983) and 5190 (95 % CI: 1440–9510) generations ago, respectively (Table S11). Assuming 25 years as the generation time, divergence times scaled to 16 200 years ago (95 % CI: 4250–24 575) and 129 750 years ago (95 % CI: 36 000–237 750) for t1 and t2, respectively. The posterior probability, summary statistics (Supplementary Data Table S12) and PCoA results (Supplementary Data Fig. S5) suggested that scenario 1 provides a good fit to the data. The type I error rate was 0·2120, and the average type II error rate was 0·0945. The most probable scenario is reported in Fig. 3A.
Fig. 3.
(A) Demographic scenarios revealed from the ABC analyses, considering three gene pools (WM, CEM, IR): Na: ancestral effective population size; t1–t2, divergence times for the depicted events of domestication. (B) Hypothetical routes of ‘colonization’ of domesticated olive from the centre of domestication to the Mediterranean area and towards the east, up to Iran.
Table 4.
Posterior probability of each tested demographic scenario and its 95 % confidence interval based on the logistic estimate according to DIYABC
| Scenario | Posterior probability 95 % CI (minimum–maximum) |
|---|---|
| (A) Analysis of the Mediterranean cultivars | |
| Scenario 1 | 0·9204 (0·9101–0·9307) |
| Scenario 2 | 0·0008 (0·0000–0·1196) |
| Scenario 3 | 0·0788 (0·0000–0·1977) |
| (B) Analysis of the Iranian samples | |
| Scenario 1 | 0·2279 (0·1979–0·2579) |
| Scenario 2 | 0·2126 (0·1819–0·2434) |
| Scenario 3 | 0·4057 (0·3646–0·4469) |
| Scenario 4 | 0·1538 (0·0831–0·2245) |
Demographic history of the Iranian samples
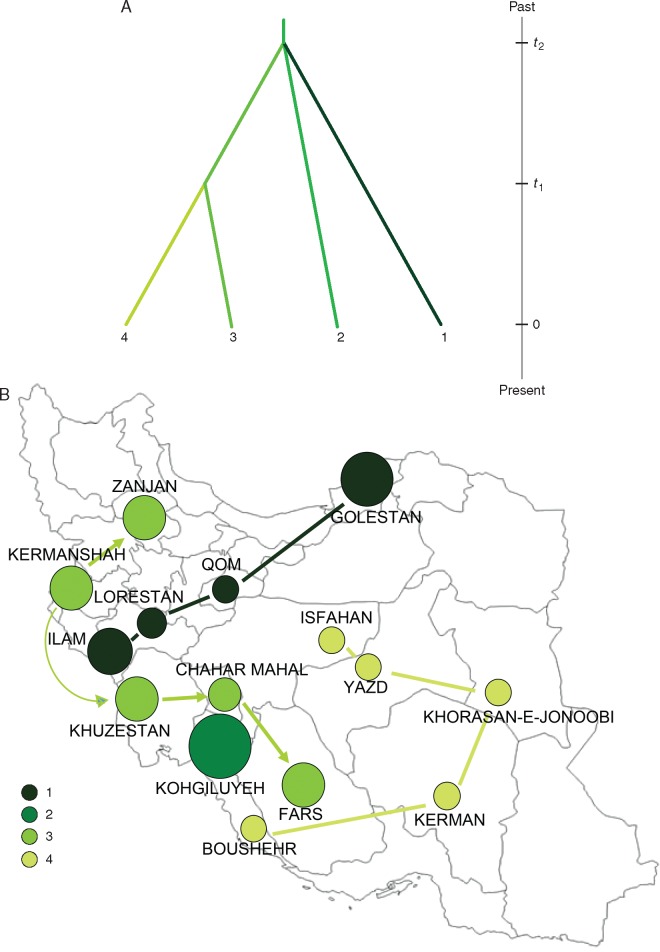
Scenario 3 (hierarchical split model 3) was the demographic model with the highest posterior probability (Fig. 4A). Its value was not very high (0·4057, 95 % CI: 0·3646–0·4469), but its CI did not overlap with those of the other scenarios (Supplementary Data Table S13). The median values of the effective population sizes were 3420, 1800, 3280 and 4530 for Pop1, Pop2, Pop3 and Pop4, respectively (Supplementary Data Table S14). This result showed that the effective population size of Pop2 was at least 1·8 times lower than those of Pop1 and Pop3, suggesting an expansion event at time t2 (Fig. 4B). At time t1, another expansion event resulted in the divergence of Pop3 and Pop4, which increased their effective population size 1·4-fold. The median values of the divergence times, t1 and t2, were 2170 (95 % CI: 653–4640) and 4080 (95 % CI: 1360–13 100) generations ago, respectively (Table S14). Divergence times scaled to 54 250 years ago (95 % CI: 16 325–116 000) and 102 000 years ago (95 % CI: 34 000–327 500) for t1 and t2, respectively. The posterior probability, summary statistics (Table S14) and PCA results (Supplementary Data Fig. S6) suggest that scenario 3 provides a good fit to the data (Fig. 4A). The type I error rate was 0·384 and the average type II error rate was 0·156.
Fig. 4.
(A) Demographic scenarios reconstructed by the ABC analyses for the Iranian genotypes, considering four populations (1, 2, 3 and 4): Na: ancestral effective population size; t1–t2, divergence times for the depicted events. (B) Geographic occurrence of the four populations along the Iranian provinces, according to the distribution of sampling sites.
Impact of Quaternary glaciation on olive distribution modelling
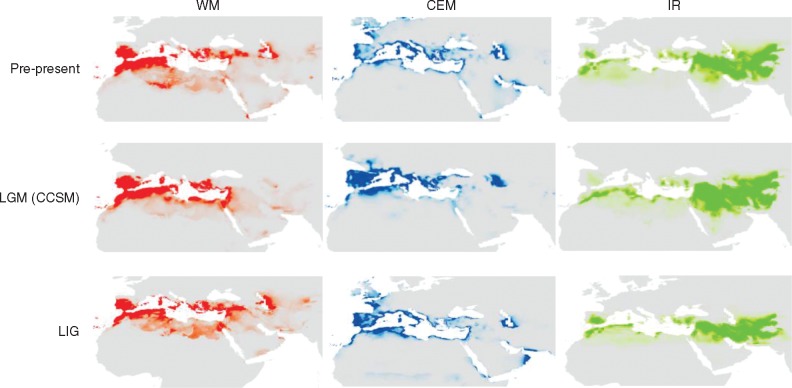
Averaged AUC values for the replicate runs were > 0·991 for all distribution models, supporting their predictive power. The two models of MIROC and CCSM provided comparable species distributions. Furthermore, MH (6000 years BP) and present condition showed a similar pattern (Supplementary Data Fig. S7). MAXENT demonstrated in general a different distribution of O. europaea between the eastern cluster and the others, the IR cluster covering the western and eastern part of the Fertile Crescent and reaching the extreme west of the Mediterranean basin (Fig. 5). For all three groups (IR, CEM and WM), the distribution of O. europaea for the LIG (140 000 years BP) was more similar to the present condition than to the LGM (22 000 years BP). The CCSM model predicted that in WM the species distribution at the LGM was smaller than for the Pre-present and LIG, but higher for CEM and IR. Furthermore, 22 000 years BP the distribution area of population IR was spreading up to the centre of the Arabian Peninsula. To evaluate the effects of 19 bioclimatic variables on species distribution in different periods, PCA was performed based on jackknife results (Supplementary Data Fig. S8). Bio18 and 19 (precipitation of the warmest and coldest quarter, respectively) significantly separated the IR (LGM and present) from WM and CEM (LIG, LGM and present), while IR (LIG) differentiated by Bio03 and 09 (isothermality and mean temperature of the driest quarter, respectively) was in general closer to WM and CEM (Fig. S8).
Fig. 5.
MAXENT predicted suitability for WM, CEM and IR gene pools of olive during three time periods: LIG, last interglacial (120 000–140 000 years BP); LGM-CCSM, last glacial maximum (22 000 years BP), employing the Community Climate System Model (CCSM), and Pre-present (1950–2000). Red (WM), blue (CEM) and green (IR) colours indicate higher probabilities of suitable climatic conditions.
DISCUSSION
In this study, we reshaped the distribution of Olea europaea subsp. europaea var. europaea, investigating an enlarged set of samples, from the Mediterranean basin to the Iranian plateau, through the analysis of molecular and past/present climatic data, under the framework of archaeo-botanical data.
We highlighted that the genetic differentiation between the two olive gene pools occurred long before domestication, spreading westward, reaching the Mediterranean area, and eastward, settling down in the Iranian plateau. We also found that the distribution area of the main Mediterranean olive lineage spread not only in the Near East countries (north-eastern Levant) (Besnard et al., 2013a) but also in the Middle East, including the Fertile Crescent and the Iranian plateau. The presence of olive in Iran probably started from a local centre of diffusion placed in the south-west area of the country and dated back about 100 000 years ago, considering 25 years for each generation, but this time could be increased for a plant that remains productive for hundreds of years. Archaeo-botanical evidence showed that olive was widespread in the country 5800 years BP (Van Zeist, 2008), and its cultivation and use have been described since 3000 years ago (Morgan, 2015; Djamali et al., 2016).
Demographic history of olive
Bayesian analyses, dividing olive genotypes into Iranian (IR), Western (WM) and Central-Eastern Mediterranean (CEM) groups, highlighted a high genetic differentiation between the Iranian samples and the Mediterranean cultivars, showing a clear separation among them, as previously hypothesized (Noormohammadi et al., 2014; Mousavi et al., 2014; Hosseini-Mazinani et al., 2014). The reduction of Mediterranean cultivar populations from three well-differentiated gene pools (western, central and eastern Mediterranean), as reported by many authors (Sarri et al., 2006; Haouane et al., 2011; Belaj et al., 2012), into two (CEM and WM, in the present work) was probably due to the strong effect of the Iranian group on total differentiation, not analysed in the previous studies.
Through ABC simulations, previously applied in some cultivated plant species including olive (Diez et al., 2015; Tamaki et al., 2016; Parat et al., 2016), we observed an ancient expansion event from a common ancestor, giving rise to a migration wave from the east to the west, followed by a more recent separation of the Mediterranean varieties into two gene pools at time t1, about 13 000 years ago. The most probable scenario supported that the Iranian population diverged and expanded from an ancestral gene pool around 130 000 years BP, even if DIYABC did not model continuous gene flow at each generation (Tsuda et al., 2015) and the examined species has a generation time that is difficult to determine, which could have led to an under-estimation of the divergence time. Taking into account the lower confidential interval limit (36 000 years ago), the divergence time t2 is still well before the LGM (22 000 years BP). At that time, the ABC results suggested a westward expansion of olives from the Middle-Eastern territories, as hypothesized for other crop and forest trees (Myles et al., 2011; Zeder, 2011; Mayol et al., 2015). An ancient separation of IR and WM was also supported by the almost total absence of migration events from WM to IR and vice versa, as suggested by the assignment test analysis.
An ancient separation of Iranian and Mediterranean gene pools was affirmed (5000 generations), followed by the separation between CEM and WM populations (600 generations).
How climate conditions may have shaped the present distribution of olive
The SDM analysis revealed the divergence of the central and western part of the Mediterranean from the Near and Middle East. PCA, based on 19 bioclimatic variables, described mean temperatures of the driest quarter as factors acting on olive distribution during the LIG (140 000 years BP). Since the LGM, along the MH (6 000 years BP) up to the present, precipitation (during warmest and coldest quarters) was the most important climatic factor determining separation of WM and CEM from Eastern (Iran) olive clusters. Several studies reported the joint influence of isolation by distance and environmental adaptation to promote genetic divergence of plant populations (Lee and Mitchell-Olds, 2011; Temunovic et al., 2012; Mosca et al., 2014). Thus, the role of climate-driven adaptation clearly supports the divergence of Eastern and Western olive groups before olive domestication.
The presence of olive along the Iranian area is documented since 38 000 years BP
According to Hole (1998), agriculture spread to Iran from the south-west of the Fertile Crescent, and latest excavations in the foothills of the Zagros Mountains have provided new and substantial evidence that the eastern region of the Fertile Crescent was an independent centre of plant and animal domestication (Matthews et al., 2010; Darabi et al., 2011; Niknami and Nikzad, 2012). The first report of Olea pollen in the Fertile Crescent was from Lake Zarivar (Kurdestan province, west of Iran), where Olea pollen grains were found in sampled sediments at different depths, corresponding to pollen-assemblage (p-a) zone 3 (38 000–15 000 cal years BP). In the Zarivar area, scattered tree stands were still present, mainly Pistacia, Acer and Quercus, to a lesser extent in subzone 3a. The p-a zone 4 (15 000–12 000 cal years BP) was related to the Lateglacial period, which was unfavourable for the growth of forest trees and characterized by drought resulting from high temperatures, while p-a zones 5–7 (12 000–5900 cal. years BP) was a period with a progressive expansion of trees (van Zeist and Bottema, 1977). Furthermore, van Zeist (2008) affirmed that the Olea pollen grains of zone 7 could belong to cultivated olive. Before that period, a local exploitation of food resources, including olive, was reported based on pollen evidence of Cerealia, Olea and Malva collected at the site of Zawi-Chemi (Iraq, about 4 km south of the Shanidar cave, near the Iranian border), associated with a cultural sequence dating back to 10 870 years BP (Al-Ameri et al., 2011).
At the same time, in the western part of the Iranian plateau, nomad populations cultivated semi-wild orchards and non-irrigated vines, figs and almonds at high altitudes (1500–2000 m a.s.l.) (Amanolahi, 1986; Potts, 2014). Palynological investigations documented the presence of Olea pollen grains dated to 4300 years BP (Djamali et al., 2009) in south-western Iran (Fars Province), corresponding to an increase of human activities in this province (Alizadeh, 2006).
Moreover, charcoal fragments of Olea sp. retrieved in several contexts of Konar Sandal (Jiroft, Kerman province), from the Bronze Age to the Iron Age, were interpreted as representing cultivated trees (Mashkour et al., 2013), or as O. europaea subsp. cuspidata, the latter still growing in the south-eastern part of Iran (Hosseini-Mazinani et al., 2014). The presence of olive cultivation has been continuous in Iran since 3200 years BP, because of construction of an intensive hydraulic network, mainly related to innovative agricultural systems, and increased at about 2700 years BP, with a prominent peak between 2500 and 2100 years BP (Djamali et al., 2010, 2016).
A major role in the pattern of distribution of olive should be attributed to the traditional system of water supply for irrigation, such as the qanat and dam structures, peculiar to the Iranian territory (Motiee et al., 2006; Djamali et al., 2016) and which has been destroyed deliberately or due to catastrophic events.
There is also evidence that ancient olive cultivation in Iran could have been related to the production of table olives, as reported in Chinese literature (Hirth, 1910; Laufer, 1919), especially during the maximum expansion of olive cultivation, about 1000 years BP. The large fruits of many olive samples included in our analysis confirm their suitability as table olive varieties (Mousavi et al., 2014), and the few remnants of olive trees still present in Iran are compatible with a high technological level of olive production reached in the past.
The evidence of large or very large fruits for most Iranian ecotypes (Hosseini-Mazinani and Torkzaban, 2013; Mousavi et al., 2014), a character clearly subjected to selection during the process of domestication (Alcantara and Rey, 2003), corroborated the evidence that Iranian ecotypes do not represent a wild form of olive (Olea europaea subsp. europaea var. sylvestris, named oleasters), as naturally occurring along most of the Mediterranean shores but, more likely, they represent the vestiges of ancient orchards, abandoned for hundreds of years and returned to wild conditions, or the seed dissemination of palaeo-varieties that survived in spite of environmental constraints.
Genetic differentiation of Iranian olives
The unique mother lineage, observed by chloroplast analysis, indicated the Iranian samples as sister populations of most cultivated Mediterranean olive varieties. The actual genetic differentiation between the two gene pools could be related to genetic drift and founder effects during natural or human colonization (Bagnoli et al., 2009). Moreover, even if Iranian olive ecotypes and subsp. cuspidata coexisted in the same environments, the contribution of cuspidata to the genetic diversity of the Iranian cultivars and ecotypes must be excluded, following the assignment test and diversity indices.
Environmental conditions certainly played a crucial role in the distribution of olive during the last millennia in Iran, as have historical and geo-political issues. The genetic structure revealed that ecotypes from the western province of Ilam were genetically grouped with those growing along the south bank of the Caspian Sea, up to Golestan province. This linear axis, from west to north-east of the country, overlaps the migration routes of Nomads and transhumant shepherds, who played a major role in crop domestication around 10 000 years BP (Koocheki and Gliessman, 2005; Potts, 2014). Another Iranian olive group was placed on the sidelines of the Zagros Mountains, which represent the north–south geo-climatic barrier, limiting the possibility of olive migration toward the east and promoting its north–south diffusion. Almost all main Iranian olive cultivars were assigned to this population, and diffused up to the Zanjan province where olive cultivation has been described for thousands of years (Djamali et al., 2010; Tofighian and Khademi Nadooshan, 2011). Within the fourth genetic cluster, the monumental olives of Kerman province and Fosoon from Khorasan-e-Jonoobi (105 trees registered as natural heritage by the Iranian Cultural Heritage Organization in 2011) were included, as well as Yazd and Isfahan trees, related to ancient worship rituals, as reported by Al-Tabari (1999), who described several ‘fire temples’ during the Sasanian dynasty, with gardens including thousands of olive trees at least 1500 years BP. Interestingly, ABC analyses grouped all Kohgiluyeh samples into a separate population, probably representing the remnant of the most ancient olives grown in Iran. Kohgiluyeh province was a suitable place for olive growth since the Holocene (5500 years ago). In fact, climatic conditions in Iran in that period were similar to the present, with high precipitation/evaporation ratio because of low evaporation resulting from low summer temperatures (Ferrigno, 1991).
The evaluation of a large set of Iranian olives has provided a new perspective on the spread of olive before and during its domestication, strictly related to climatic variables and human activities. The genetic pattern of differentiation between the Mediterranean basin and Middle East countries supports the hypothesis of an east–west isolation mediated by climatic conditions, with a prominent peak during Quaternary glaciations, and linked in particular to precipitation and evaporation (Kehl, 2009; Mikolajewicz, 2011). The structure of variation, coupled to palaeo-climatic and historical data, suggests an enlarged centre of olive domestication eastward, up to western Iran. A secondary differentiation might have occurred through crossings with local oleasters (Baldoni et al., 2006; Belaj et al., 2007) or with other Olea europaea subspecies, giving rise to the current genetic variability of the Mediterranean.
Multidisciplinary studies, including archaeological, genetic and climatic aspects, performed on the genetic resources of Middle Eastern countries could provide new scenarios for the origin not only of cultivated olive, but also of many other plant crop and livestock species.
AUTHOR CONTRIBUTIONS
This work was part of S.M.’s PhD dissertation (supervised by K.A. and M.H.). M.H., S.M., L.B. and R.M. conceived the project and sampling design. M.H., B.T. and S.M. contributed to collection of genotypes. R.M., S.M. and N.C. performed all molecular work and genotype scoring. L.C. provided all archaeo-botanical and historical information. S.M., R.M., F.B., S.P., G.G.V. and L.B. analysed and interpreted the genotypic data. All authors wrote the article and contributed edits and comments.
SUPPLEMENTARY INFORMATION
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: (a) map indicating the sites of sample collection, classified as: wild environment (blue); archaeological areas (pink); burial areas (purple). (b) map of the archaeological and historical sites cited in the text. Figure S2: (a) ancient building close to Malekshahi olive sample (Ilam); (b) an olive ecotype in its natural habitat (Kohgiluyeh); (c) an ancient building near the Ordib olive sample (Isfahan); (d) Lemesk tree sample growing within a burial area (Golestan); (e) olive trees on an artificial hill of archaeological interest; (f) olive tree close to a sacred site (Golestan). Figure S3: demographic models tested with Approximate Bayesian Computation (ABC) considering Mediterranean (A) and Iranian cultivars (B) of O. europaea. Figure S4: neighbour-joining unrooted tree obtained from the SSR profiles of all Iranian ecotypes by Darwin software. Figure S5: principal component analysis of summary statistics based on 10 000 simulated datasets in DIYABC (scenario 1) of O. europaea Mediterranean cultivars. Figure S6: principal component analysis of summary statistics based on 10 000 simulated datasets in DIYABC (scenario 1) of O. europaea Iranian cultivars. Figure S7: MAXENT predicted suitability for IR, CEM and WM gene pools of Olea europaea during four time periods: LIG, last interglacial (120 000–140 000 years BP); LGM-CCSM and LGM-MIROC, last glacial maximum (22 000 years BP); and MH, Mid-Holocene (6000 years BP) employing two different palaeoclimate layers, the Community Climate System Model (CCSM) and the Model for Interdisciplinary Research on Climate (MIROC); Pre, present conditions (1950–2000). Figure S8: principal component analysis based on Jackknife results for 19 bioclimatic variables for the three geographical areas (WM, CEM, IR) from the LIG up to Pre-present condition. Table S1: list of the olive samples from Iran analysed in the study. Table S2: list of Mediterranean varieties and Iranian main cultivars, based on country of diffusion and source of plant material. Table S3: list of primers used to amplify and detect cpSNP and cpSSR polymorphisms. Table S4: prior distributions in DIYABC analysis of the Mediterranean cultivars. Table S5: prior distributions in DIYABC analysis of the Iranian cultivars. Table S6: nineteen bioclimatic descriptors based on BIOCLIM data. Table S7: chloroplast genotyping results, based on 29 length polymorphisms and 20 cpSNPs. Table S8: genetic diversity within Iranian olives and within Mediterranean countries. Number of alleles (N), number of effective alleles (Ne), genetic diversity (GD), observed heterozygosity (Ho), expected heterozygosity (He) and fixation index (F) are reported. Table S9: genetic indices for each SSR locus within Iranian cultivars and genotypes: observed heterozygosity (Ho), expected heterozygosity (He) and fixation index (F). Table S10: pairwaise genetic distance of four Iranian populations detected by Structure analysis, Dest, G′st Hedrick’s standardized (Hed) and Fst. Table S11: parameter estimates for the best demographic scenario based on Approximate Bayesian Computation (ABC) of the Mediterranean cultivars of O. europaea. Table S12: model checking in Approximate Bayesian Computation (ABC) of O. europaea Mediterranean cultivars. Table S13: model checking in Approximate Bayesian Computation (ABC) of O. europaea Iranian ecotypes. Table S14: parameter estimates for the best demographic scenario based on Approximate Bayesian Computation (ABC) of the Iranian ecotypes of O. europaea.
Supplementary Material
ACKNOWLEDGMENTS
This research was performed within the international collaboration between CNR-IBBR (Italy), NIGEB (Iran) and Tarbiat Modares University (TMU), which provided facilities and financial support. Partial support was derived from the BeFOre project–European Union’s Horizon 2020 Research and Innovation Programme, G.A.N. 645595. The Iranian Ministry of Agriculture (Olive Research Office) contributed to collecting samples from different parts of the Iran. We thank Mr Mahmoud Haghighi for financial support of the propagation and conservation of the Iranian ecotypes and to Dr Martina Rossi for helping during the analysis and discussions.
LITERATURE CITED
- Al-Ameri TK, Jasim SY, Al-Khafaji AJS.. 2011. Middle paleolithic to neolithic cultural history of North Iraq. Arab Journal of Geosciences 4: 945–972. [Google Scholar]
- Alcantara JM, Rey PJ.. 2003. Conflicting selection pressures on seed size: evolutionary ecology of fruit size in a bird‐dispersed tree, Olea europaea. Journal of Evolutionary Biology 16: 1168–1176. [DOI] [PubMed] [Google Scholar]
- Alizadeh A. 2006. The origins of state organizations in prehistoric highland Fars, Southern Iran In: Excavations at Tall-e Bakun. Chicago: Oriental Institute Publications. [Google Scholar]
- Al-Tabari ADM. 1999. The Sasanids, the Byzantines, the Lakhmids, and Yemen, Vol. 5, Bosworth C.E. (trans.), Near Eastern Studies. New York: State University of New York Press. [Google Scholar]
- Amanolahi S. 1986. The ecological adaptation of the Lutfi herder-horticulturists of south Iran. Human Ecology 14: 355–360. [Google Scholar]
- Bagnoli F, Vendramin GG, Buonamici A, et al. 2009. Is Cupressus sempervirens native in Italy? An answer from genetic and palaeobotanical data. Molecular Ecology 18: 2276–2286. [DOI] [PubMed] [Google Scholar]
- Baldoni L, Tosti N, Ricciolini C, et al. 2006. Genetic structure of wild and cultivated olives in the central Mediterranean basin. Annals of Botany 98: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni L, Cultrera NGM, Mariotti R, et al. 2009. A consensus list of microsatellite markers for olive genotyping. Molecular Breeding 24: 213–231. [Google Scholar]
- Baudouin L, Lebrun P.. 2001. An operational Bayesian approach for the identification of sexually reproduced cross fertilized populations using molecular markers. Acta Horticulturae 546: 81–93. [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ.. 2002. Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaj A, Muñoz-Diez C, Baldoni L, Porceddu A, Barranco D, Satovic Z.. 2007. Genetic diversity and population structure of wild olives from the north-western Mediterranean assessed by SSR markers. Annals of Botany 100: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaj A, del Carmen, Dominguez-García M, Atienza SG, et al. 2012. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genetics & Genomes 8: 365–378. [Google Scholar]
- Besnard G, Khadari B, Baradat P, Berville A.. 2002. Olea europaea (Oleaceae) phylogeography based on chloroplast DNA polymorphism. Theoretical and Applied Genetics 104: 1353–1361. [DOI] [PubMed] [Google Scholar]
- Besnard G, Rubio de Casas R, Vargas P.. 2007. Plastid and nuclear DNA polymorphism reveals historical processes of isolation and reticulation in the olive tree complex (Olea europaea). Journal of Biogeography 34: 736–752. [Google Scholar]
- Besnard G, de Casas RR, Christin PA, Vargas P.. 2009. Phylogenetics of Olea (Oleaceae) based on plastid and nuclear ribosomal DNA sequences: tertiary climatic shifts and lineage differentiation times. Annals of Botany 104: 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Hernandez P, Khadari B, Dorado G, Savolainen V.. 2011. Genomic profiling of plastid DNA variation in the Mediterranean olive tree. BMC Plant Biology 11: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Khadari B, Navascues M, et al. 2013a. The complex history of the olive tree: from Late Quaternary diversification of Mediterranean lineages to primary domestication in the northern Levant. Proceedings of the Royal Society B: Biological Sciences 280: 20122833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, El Bakkali A, Haouane H, Baali-Cherif D, Moukhli A., Khadari B.. 2013b. Population genetics of Mediterranean and Saharan olives: geographic patterns of differentiation and evidence for early generations of admixture. Annals of Botany 112: 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Dupuy J, Larter M, Cuneo P, Cooke D, Chikhi L.. 2014. History of the invasive African olive tree in Australia and Hawaii: evidence for sequential bottlenecks and hybridization with the Mediterranean olive. Evolutionary Applications 7: 195–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C, Tersac M, Berville A.. 2006. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. Journal of Biogeography 33: 1916–1928. [Google Scholar]
- Cavalli-Sforza LL, Edwards AW.. 1967. Phylogenetic analysis. Models and estimation procedures. American Journal of Human Genetics 19: 233. [PMC free article] [PubMed] [Google Scholar]
- Chapuis MP, Estoup A.. 2007. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 24: 621–631. [DOI] [PubMed] [Google Scholar]
- Contento A, Ceccarelli M, Gelati M, Maggini F, Baldoni L, Cionini P.. 2002. Diversity of Olea genotypes and the origin of cultivated olives. Theoretical and Applied Genetics 104: 1229–1238. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Pudlo P, Veyssier J, et al. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30: 1187–1189. [DOI] [PubMed] [Google Scholar]
- Darabi H, Naseri R, Young R, Fazeli H.. 2011. The absolute chronology of East Chia Sabz: a Pre-Pottery Neolithic site in Western Iran In: Budja M, ed. 18th Neolithic Seminar Documenta Praehistorica 38: 255–365. [Google Scholar]
- Darmon F. 1987. Analyses polliniques de trois sites natoufiens (ancien, récent, final) dans la région de Salibiya-Fazaël. Paléorient 13: 121–129. [Google Scholar]
- Diamond J. 2002. Evolution, consequences and future of plant and animal domestication. Nature 418: 700–707. [DOI] [PubMed] [Google Scholar]
- Diez CM, Trujillo I, Martinez-Urdiroz N, et al. 2015. Olive domestication and diversification in the Mediterranean Basin. New Phytologist 206: 436–447. [DOI] [PubMed] [Google Scholar]
- Djamali M, De Beaulieu JL, Miller NF, et al. 2009. Vegetation history of the SE section of the Zagros Mountains during the last five millennia; a pollen record from the Maharlou Lake, Fars Province, Iran. Vegetation History and Archaeobotany 18: 123–136. [Google Scholar]
- Djamali M, Miller NF, Ramezani E, et al. 2010. Notes on arboricultural and agricultural practices in ancient Iran based on new pollen data. Paléorient 36: 175–188. [Google Scholar]
- Djamali M, Jones MD, Migliore J, et al. 2016. Olive cultivation in the heart of the Persian Achaemenid Empire: new insights into agricultural practices and environmental changes reflected in a late Holocene pollen record from Lake Parishan, SW Iran. Vegetation History and Archaeobotany 25: 255–269. [Google Scholar]
- Earl DA, von Holdt BM.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- Estoup A, Jarne P, Cornuet JM.. 2002. Homoplasy and mutation model at microsatellite loci and their consequence for population genetics analysis. Molecular Ecology 11: 1591–1604. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Ferrigno JG. 1991. Glaciers of Iran. Glaciers of the Middle East and Africa: Satellite Image Atlas of Glaciers of the World, pp. G31–G47. [Google Scholar]
- Goudet J. 2002. FSTAT: a program to estimate and test gene diversities and fixation indices. v. 2.9.3.2 (Online). http://www.unil.ch/izea/softwares/fstat.html.
- Hammer O, Harper DAT, Ryan PD.. 2001. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4: 9. [Google Scholar]
- Hanley JA, McNeil BJ.. 1982. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143: 29–36. [DOI] [PubMed] [Google Scholar]
- Haouane H, El Bakkali A, Moukhli A, et al. 2011. Genetic structure and core collection of the World Olive Germplasm Bank of Marrakech: towards the optimised management and use of Mediterranean olive genetic resources. Genetica 139:1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A.. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Hirth F. 1913. The mystery of Fu-lin. Journal of the American Oriental Society 1: 193–208. [Google Scholar]
- Hole F. 1998. The spread of agriculture into the eastern arc of the Fertile Crescent: food for the herders In: Damania AB, Valkoun J, Willcox G, Qualset CO, eds. The Origins of Agriculture and Crop Domestication: The Harlan Symposium. FAO, ICARDA, IPGRI, pp. 83–92. [Google Scholar]
- Hosseini-Mazinani M, Torkzaban B.. 2013. Iranian Olive Catalogue. Tehran: National Institute of Genetic Engineering and Biotechnology press. [Google Scholar]
- Hosseini-Mazinani M, Mariotti R, Torkzaban B, et al. 2014. High genetic diversity detected in olives beyond the boundaries of the Mediterranean Sea. PLoS One 9: e93146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniewski D, Van Campo E, Boiy T, et al. 2012. Primary domestication and early uses of the emblematic olive tree: palaeobotanical, historical and molecular evidence from the Middle East. Biological Reviews 87: 885–899. [DOI] [PubMed] [Google Scholar]
- Kehl M. 2009. Quaternary climate change in Iran the state of knowledge. Erdkunde 63: 1–17. [Google Scholar]
- Kislev ME, Nadel D, Carmi I.. 1992. Epipalaeolithic (19 000 BP) cereal and fruit diet at Ohalo II, Sea of Galilee, Israel. Review of Palaeobotany and Palynology 73: 161–166. [Google Scholar]
- Koocheki A, Gliessman SR.. 2005. Pastoral nomadism, a sustainable system for grazing land management in arid areas. Journal of Sustainable Agriculture 25: 113–131. [Google Scholar]
- Laufer B. 1919. Sino-Iranica. Chinese contributions to the history of civilization in ancient Iran, with special reference to the history of cultivated plants and products. Anthropological Series XV, No. 3 Chicago.
- Lee CR, Mitchell-Olds T.. 2011. Quantifying effects of environmental and geographical factors on patterns of genetic differentiation. Molecular Ecology 20: 4631–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JC. 2011. Biogeography and evolution of Abies (Pinaceae) in the Mediterranean Basin: the roles of long-term climatic change and glacial refugia. Journal of Biogeography 38: 619–630. [Google Scholar]
- Liphschitz N, Gophna R, Hartman M, Biger G.. 1991. The beginning of olive (Olea europaea) cultivation in the old world: a reassessment. Journal of Archaeological Science 18: 441–453. [Google Scholar]
- Lovell JL, Meadows J, Jacobsen GE.. 2010. Upland olive domestication in the Chalcolithic Period: new 14C determination from el-Khawarij (Ajlun), Jordan. Radiocarbon 52: 364–371. [Google Scholar]
- Mariotti R, Cultrera NGM, Díez CM, Baldoni L, Rubini A.. 2010. Identification of new polymorphic regions and differentiation of cultivated olives (Olea europaea L.) through plastome sequence comparison. BMC Plant Biology 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti R, Cultrera NGM, Mousavi S, et al. 2016. Development, evaluation, and validation of new EST-SSR markers in olive (Olea europaea L.). Tree Genetics and Genomes 12: 120. [Google Scholar]
- Mashkour M, Tengberg M, Shirazi Z, Madjidzadeh Y.. 2013. Bio-archaeological studies at Konar Sandal, Halil Rud Basin, Southeastern Iran. Environmental Archaeology 18: 222–246. [Google Scholar]
- Matthews R, Mohammadifar Y, Matthews W, Motarjem A.. 2010. Investigating the Early Neolithic of Western Iran: the Central Zagros Archaeological Project (CZAP). Antiquity 084: 323. [Google Scholar]
- Mayol M, Riba M, González-Martínez SC, et al. 2015. Adapting through glacial cycles: insights from a long-lived tree (Taxus baccata). New Phytologist 208: 973–986. [DOI] [PubMed] [Google Scholar]
- Mikolajewicz U. 2011. Modeling mediterranean ocean climate of the last glacial maximum. Climate of the Past 7: 161–180. [Google Scholar]
- Morgan J. 2015. The book of pears: the definitive history and guide to over 500 varieties. White River Junction, VT: Chelsea Green Publishing, pp. 27–48. [Google Scholar]
- Mosca M, Gonzalez-Martınez SC, Neale DB.. 2014. Environmental versus geographical determinants of genetic structure in two subalpine conifers. New Phytologist 201: 180–192. [DOI] [PubMed] [Google Scholar]
- Motiee H, Mcbean E, Semsar A, Gharabaghi B, Ghomashchi V.. 2006. Assessment of the contributions of traditional qanats in sustainable water resources management. Int J Water Resour D 22: 575–88. [Google Scholar]
- Mousavi S, Hosseini-Mazinani M, Arzani K, et al. 2014. Molecular and morphological characterization of Golestan (Iran) olive ecotypes provides evidence for the presence of promising genotypes. Genetic Resources and Crop Evolution 61: 775–785. [Google Scholar]
- Myles S, Boyko AR, Owens CL, et al. 2011. Genetic structure and domestication history of the grape. Proceedings of the National Academy of Sciences 108: 3530–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklewski JW, van Zeist W.. 1970. A late Quaternary pollen diagram from northwestern Syria. Acta Botanica Neerlandica 19: 737–754. [Google Scholar]
- Niknami KA, Nikzad M.. 2012. New evidence of the Neolithic period in West Central Zagros: the Sarfirouzabad-Mahidasht Region, Iran. Documenta Praehistorica 39: 453–458. [Google Scholar]
- Noormohammadi Z, Trujillo I, Belaj A, Ataei S, Hosseini-Mazinan M.. 2014. Genetic structure of Iranian olive cultivars and their relationship with Mediterranean’s cultivars revealed by SSR markers. Scientia Horticulturae 178: 175–183. [Google Scholar]
- Noy T, Legge AJ, Higgs ES.. 1973. Recent excavations at Nahal Oren, Israel. Proceedings of the Prehistoric Society 39: 75–99. [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C.. 1995. Microsatellite analysis of population structure in Canadian polar bears. Molecular Ecology 4: 347–354. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A.. 2004. Genetic assignment methods for the direct, real‐time estimation of migration rate: a simulation‐based exploration of accuracy and power. Molecular Ecology 13: 55–65. [DOI] [PubMed] [Google Scholar]
- Parat F, Schwertfirm G, Rudolph U, et al. 2016. Geography and end use drive the diversification of worldwide winter rye populations. Molecular Ecology 25: 500–14. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE.. 2013. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier X, Jacquemoud-Collet JP.. 2006. DARwin software. http://darwin.cirad.fr/darwin.
- Petit RJ, Hampe A, Cheddadi R.. 2005. Climate changes and tree phylogeography in the Mediterranean. Taxon 54: 877–885. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE.. 2006. Maximum entropy modeling of species geographic distributions. Ecological Modelling 190: 231–259. [Google Scholar]
- Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A.. 2004. GENECLASS2: a software for genetic assignment and first-generation migrant detection. Journal of Heredity 95: 536–539. [DOI] [PubMed] [Google Scholar]
- Potts DT. 2014. Nomadism in Iran from antiquity to the Modern Era. New York: Oxford University Press. [Google Scholar]
- Pritchard J, Wen X, Falush D.. 2009. Documentation for structure software: version 2.3. http://pritch.bsd.uchicago.edu/structure.html.
- Rodriguez-Sanchez F, Guzman B, Valido A, Vargas P, Arroyo J.. 2009. Late Neogene history of the laurel tree (Laurus L., Lauraceae) based on phylogeographical analyses of Mediterranean and Macaronesian populations. Journal of Biogeography 36: 1270–1281. [Google Scholar]
- Sarri V, Baldoni L, Porceddu A, et al. 2006. Microsatellite markers are powerful tools for discriminating among olive cultivars and assigning them to geographically defined populations. Genome 49: 1606–1615. [DOI] [PubMed] [Google Scholar]
- Tamaki I, Kuze T, Hirota K, Mizuno M.. 2016. Genetic variation and population demography of the landrace population of Camellia sinensis in Kasuga, Gifu Prefecture, Japan. Genetic Resources and Crop Evolution 1: 1–9. [Google Scholar]
- Temunovic M, Franjic J, Satovic Z, Grgurev M, Frascaria-Lacoste N, Fernandez-Manjarres JF.. 2012. Environmental heterogeneity explains the genetic structure of continental and Mediterranean populations of Fraxinus angustifolia Vahl. PLoS One 7: e42764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terral JF. 1996. Wild and cultivate olive (Olea europaea L.): a new approach to an old problem using inorganic analyses of modern wood and archaeological charcoal. Review of Palaeobotany and Palynology 91: 383–397. [Google Scholar]
- Terral JF, Alonso N, Chatti N, et al. 2004. Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. Journal of Biogeography 31: 63–77. [Google Scholar]
- Terral JF, Badal E, Heinz C. et al. 2005. Paleoecologie de l’olivier et paléoclimats au Quaternaire récent en Méditerranée nord-occidentale: la mémoire du bois. Archéo-Plantes 2: 5–28. [Google Scholar]
- Tofighian H, Khademi Nadooshan F.. 2011. Sasanians in the Persian Gulf according to archaeological data www.Iranchambersociety.com.
- Trujillo I, Ojeda MA, Urdiroz NM, et al. 2014. Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) using SSR and morphological markers. Tree Genetics and Genomes 10: 141–155. [Google Scholar]
- Tsuda Y, Nakao K, Ide Y, Tsumura Y.. 2015. The population demography of Betula maximowicziana, a cool‐temperate tree species in Japan, in relation to the last glacial period: its admixture‐like genetic structure is the result of simple population splitting not admixing. Molecular Ecology 24: 1403–1418. [DOI] [PubMed] [Google Scholar]
- van Zeist W. 2008. Late Pleistocene and Holocene vegetation at Zeribar. Diatom Monographs 8: 53–104. [Google Scholar]
- van Zeist W, Bakker-Heeres JAH.. 1984. Archaeobotanical studies in the Levant, 2. Neolithic and Halaf levels and Ras Shamra. Palaeohistoria 26: 151–170. [Google Scholar]
- van Zeist W, Bottema S.. 1977. Palynological investigations in western Iran. Palaeohistoria Bussum 19: 19–85. [Google Scholar]
- van Zeist W, Bottema S.. 2009. A palynological study of the Acheulian site of Gesher Benot Ya’aqov, Israel. Vegetation History and Archaeobotany 18: 105–121. [Google Scholar]
- Willcox G. 1996. Evidence for plant exploitation and vegetation history from three Early Neolithic pre-pottery sites on the Euphrates (Syria). Vegetation History and Archaeobotany 5: 143–152. [Google Scholar]
- Zeder MA. 2011. The origins of agriculture in the Near East. Current Anthropology 52(S4): S221–S235. [Google Scholar]
- Zohary D, Hopf M.. 2000. Domestication of plants in the Old World: the origin and spread of cultivated plants in West Asia. Europe, and the Nile Valley. New York: Oxford University Press. [Google Scholar]
- Zohary D, Hopf M, Weiss E.. 2012. Domestication of plants in the old world, 4th edn.Oxford: Oxford University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.