Abstract
Background and Aims The ability to fix atmospheric nitrogen is thought to play an important role in the invasion success of legumes. Interactions between legumes and nitrogen-fixing bacteria (rhizobia) span a continuum of specialization, and promiscuous legumes are thought to have higher chances of forming effective symbioses in novel ranges. Using Australian Acacia species in South Africa, it was hypothesized that widespread and highly invasive species will be more generalist in their rhizobial symbiotic requirements and more effective in fixing atmospheric nitrogen compared with localized and less invasive species.
Methods To test these hypotheses, eight localized and 11 widespread acacias were examined using next-generation sequencing data for the nodulation gene, nodC, to compare the identity, species richness, diversity and compositional similarity of rhizobia associated with these acacias. Stable isotope analysis was also used to determine levels of nitrogen obtained from the atmosphere via symbiotic nitrogen fixation.
Key Results No differences were found in richness, diversity and community composition between localized and widespread acacias. Similarly, widespread and localized acacias did not differ in their ability to fix atmospheric nitrogen. However, for some species by site comparisons, significant differences in δ15N isotopic signatures were found, indicating differential symbiotic effectiveness between these species at specific localities.
Conclusions Overall, the results support recent findings that root nodule rhizobial diversity and community composition do not differ between acacias that vary in their invasiveness. Differential invasiveness of acacias in South Africa is probably linked to attributes such as differences in propagule pressure, reasons for (e.g. forestry vs. ornamental) and extent of, plantings in the country.
Keywords: Acacia, invasive, mutualism, nitrogen fixation effectiveness, rhizobia, root nodule, symbiotic promiscuity
INTRODUCTION
The ability to establish mutualistic interactions is thought to be an important determinant of colonization success and spread of non-native plants (Traveset and Richardson, 2014). These interactions result in higher fitness as afforded by increased nutrition (e.g. biological nitrogen fixation), reproduction (e.g. pollination) and spread (e.g. seed dispersal). Like most biological interactions, plant mutualisms fall along a continuum of specialization, with plants at one end of the spectrum capable of forming mutualisms with a wide range of partners (i.e. exhibiting high levels of symbiotic promiscuity or generalism), but at the other end only associating with one or a few partners (i.e. specialization) (Bascompte, 2009). Promiscuity may allow plants to utilize potential mutualists found in their new ranges more easily (e.g. Aizen et al., 2012; Heleno et al., 2013). Intuitively then, promiscuity on the part of either plant or potential mutualists should enhance the colonization probability of introduced plants that are often unaccompanied by their own mutualists (Parker et al., 2006; Stanton-Geddes and Anderson, 2011; Wandrag et al., 2013).
The establishment success of plants, specifically invasive species, has been increasingly linked to their interactions with mutualistic soil microbes (Inderjit and Cahill, 2015; Vestergård et al., 2015). For example, most legumes form symbioses with bacteria called rhizobia, resulting in biological nitrogen fixation, a physiological adaptation that has been linked to their success as invasive species (Rodríguez-Echeverría et al., 2009, 2011). Legumes benefit from the acquisition of fixed atmospheric nitrogen from rhizobia in specialized structures called root nodules, while simultaneously providing rhizobia with carbon resources (Franche et al., 2009). The formation of root nodules involves complex signalling pathways between plants and bacteria (Stacey, 2007). For example, various rhizobial nodulation genes (nod genes) respond to plant root exudates (typically flavonoids) by producing nodulating factors (so-called Nod factors), leading to root nodule formation (Hopkins and Hüner, 2009). Nod genes, which are important determinants of specialization in legume–rhizobium interactions (Spaink, 2000), are located on symbiotic plasmids or highly mobile ‘symbiotic islands’, which can be transferred between different bacterial species, and even genera, through conjugation (Ding and Hynes, 2009). This means that bacteria with the same identity based on core genes (e.g. 16S rDNA) might not be able to nodulate the same legume species if they carry different symbiotic genes.
Nodulation per se (i.e. establishment of an interaction) does not always translate into effective symbiosis benefiting the plant, as a single plant individual can be colonized by multiple strains of bacteria differing in nitrogen-fixing effectiveness (Mårtensson et al., 1989; Kiers et al., 2006). Different strains of the same rhizobial species can differ in their effectiveness (Dwivedi et al., 2015), even in association with the same host legume species (Thrall et al., 2000, 2011; Klock et al., 2015). Like other mutualisms, legume–rhizobium interactions are susceptible to cheating strategies (Franche et al., 2009; Klock et al., 2015), whereby the less effective ‘cheater’ strains act as free riders providing limited or no benefits to the host plant (Franche et al., 2009; Barrett et al., 2015), for example certain strains of Bradyrhizobuim japonicum and Rhizobium meliloti (Amarger, 1981; Singleton and Stockinger, 1983; Kiers et al., 2003). Moreover, contrary to the widely held view that individual nodules typically comprise a single strain of rhizobia, it is now known that single nodules can, in some instances, harbour multiple strains (Denison, 2000; Kiers et al., 2006; Checcucci et al., 2016), including non-rhizobial endophytes whose functions are not yet fully understood (Hoque et al., 2011; Birnbaum et al., 2016). Thus, cheating behaviour should theoretically be possible within individual nodules (Checcucci et al., 2016) and may therefore impact on overall symbiotic effectiveness. To counter the effects of cheating, some legumes have acquired the ability to select for more ‘co-operative’ rhizobia depending on their nitrogen-fixing effectiveness (Kiers et al., 2003).
Congeneric legumes that differ in their levels of invasiveness are excellent study systems to investigate how changes in diversity, composition and effectiveness of root nodule-associated rhizobial communities impact plant invasion. For example, trees in the genus Acacia Mill. have been extensively transported around the globe to regions outside their native Australian ranges for various reasons (e.g. forestry, fuel or ornamental) (Kull and Rangan, 2008; Carruthers et al., 2011; Kull et al., 2011). Globally acacias differ in invasiveness and their introduction histories (Richardson et al., 2011; Rodríguez-Echeverría et al., 2011). For example, in South Africa, widespread acacias are considered to be some of the country’s most damaging invasive species (e.g. A. dealbata, A. decurrens and A. mearnsii; Le Maitre et al., 2011, 2015), while others are restricted to a single locality and are found in relatively low abundance (e.g. A. paradoxa; Zenni et al., 2009). Differences in invasiveness of acacias have been attributed to varying propagule pressure and residence times (Richardson and Rejmánek, 2011). Differential invasiveness may, however, also reflect differences in the effectiveness of their mutualistic relationships with rhizobia. Globally, highly invasive acacias appear to be promiscuous rhizobial hosts, predominantly nodulated by various Bradyrhizobium strains (Rodríguez-Echeverría et al., 2007, 2011; Crisóstomo et al., 2013; Le Roux et al., 2016). In some instances, these rhizobia have been co-introduced with acacias into their new ranges (e.g. Rodríguez-Echeverría, 2010; Crisóstomo et al., 2013; Ndlovu et al., 2013) while in other instances they appear to form associations with novel rhizobia (e.g. Ndlovu et al., 2013). Co-introduction means that introduced plants may not be limited by their ability to find compatible rhizobia in their new ranges, which might also be less effective. There is some evidence suggesting that invasive acacias are generally more promiscuous than naturalized or non-invasive acacias (Klock et al., 2015), but such generalism appears to be constrained by geographical scale (Klock et al., 2016).
Here, using next-generation sequencing data for the nodulation gene, nodC, obtained from root nodule communities from various introduced acacias that differ in their degree of invasiveness in South Africa, together with stable isotope analysis of nitrogen, we tested the hypothesis that mutualist promiscuity enhances symbiotic effectiveness and that promiscuity is linked to invasion success. Specifically, we hypothesized that widespread acacias (i.e. successful invaders) will show higher levels of symbiotic promiscuity and effectiveness, compared with localized acacias (those that have not spread extensively). We tested three predictions arising from these hypotheses (1) that widespread acacias should associate with more diverse rhizobial communities than co-occurring localized acacias; (2) that widespread acacias should exploit a rhizobial community compositionally different from that of localized species; and (3) that widespread acacias should form associations with more effective rhizobial strains and will therefore accumulate more fixed atmospheric nitrogen.
MATERIALS AND METHODS
Study species
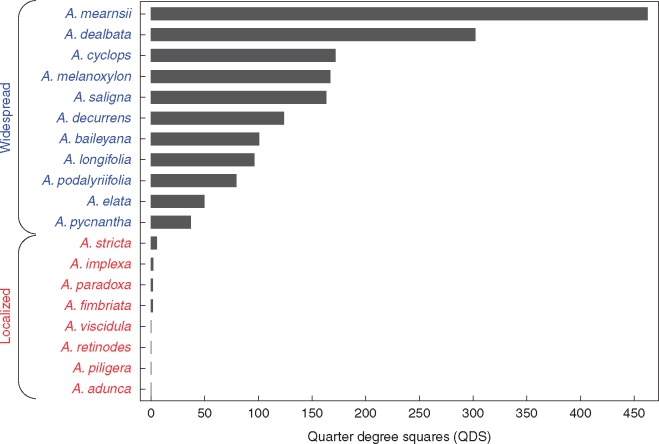
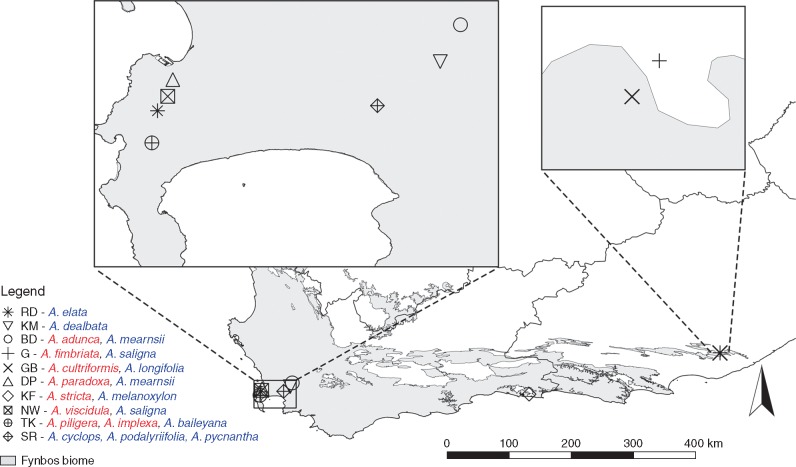
We sampled rhizobial communities from populations of all known naturalized Australian Acacia species found in South Africa (with the exception of A. decurrens), i.e. 19 species in total. Wild populations were recently discovered of two species, A. cultriformis and A. piligera (J. R. U. Wilson, pers. comm.), not previously recorded in the South African Plant Invaders Atlas (SAPIA) database (Henderson, 1998), but which are suspected to have been introduced long ago to the country (Poynton, 2009), and were included here. To classify taxa according to status, i.e. as widespread or localized, we extracted all distribution records from the SAPIA database (Henderson, 1998). These were used to determine the number of quarter degree squares (QDS: 15 min × 15 min grids cells in the WGS84 geodetic datum) occupied by each species (Fig. 1). We then used the natural break in QDS occurrences to classify species as widespread (QDS ≥ 38) or localized (QDS < 38). This approach identified eight localized species and 11 widespread species (Supplementary Data Table S1). Importantly, pairs of localized and widespread species were sampled at each sampling site, with the geographic location of sites determined by the distribution of localized species (Fig. 2). This site-level paired sampling design allowed us to investigate differences in rhizobial communities associated with localized and widespread Acacia species at the local/site scale, thus controlling for strong spatial compositional turnover often found in microbial communities (Slabbert et al., 2010). For five widespread taxa (distributed over three sites), we could not find populations co-occurring with a localized species and thus they were sampled alone. These species were not included in paired comparisons, but were included in the visualization of bacterial community composition between species.
Fig. 1.
Occurrence records of Australian Acacia species extracted from the South African Plant Invaders Atlas (SAPIA) database. Following the natural break in the occurrence records, all species present in ≥ 38 quarter degree squares were considered widespread, while the remaining taxa were considered localized.
Fig. 2.
Map illustrating site locations in South Africa and Acacia species sampled at each site during this study. Colours indicate the status of the species (red, localized; blue, widespread). Acacias from sites RD, KM and SR were not used in paired bacterial diversity and community comparisons of localized and widespread species, but were included in the PCoA plot in order to investigate how their bacterial communities relate to other acacias.
Root nodule collections, DNA isolation and next-generation sequencing
Root nodules were collected from July to November 2015 by excavation of acacia saplings and removal of nodules from roots. Between five and ten root nodules per individual plant were collected from at least five individuals per species at each site. The individuals were at least 5 m apart from each in order to prevent potential sampling of co-infected plants. Root nodules were kept on silica until needed for DNA extraction. For DNA extraction, we pooled five root nodules per individual plant and five replicate plants for each species at each site. Pooled desiccated root nodules for each plant were tissue-lysed to create a homogenous mixture for DNA extraction. DNA was extracted from tissue-lysed root nodules using the DNeasy® Plant Mini Kit (Qiagen, supplied by White Head Scientific, Cape Town, South Africa) following the manufacturer’s protocol. Samples were nanodropped to determine DNA quality and concentration.
The nodulation gene, nodC, was amplified for nodule-extracted DNA using the primers nodCF12F (5′-CCG GAT AGG MTG GKB CCR TA-3′) and nodCRI2R (5′-GTG CAC AAS GCR TAD RCC TTC AH-3′), and with sample-specific barcodes in the forward primer. This barcode has been successfully applied across rhizobia in both the Alpha- and Betaproteobacteria (Le Roux et al., 2016). Amplification was done using a 30 cycle PCR and the HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA, USA) under the following PCR conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s, 53 °C for 40 s and 72 °C for 1 min, followed by a final elongation at 72 °C for 5 min. After amplification, PCR products were checked on a 2 % agarose gel to determine the success of amplification and the relative intensity of bands. Multiple PCR samples (each sample representing the contents of five nodules from an individual plant) were barcoded first and then pooled together in equal proportions based on their molecular weight and DNA concentrations. Pooled PCR samples were purified using calibrated Ampure XP beads (Agencourt Bioscience Corporation, MA, USA) and used to prepare DNA libraries by following the Illumina TruSeq DNA library preparation protocol. Sequencing was performed at the Molecular Research LP next-generation sequencing service (www.mrdnalab.com, Shallowater, TX, USA) on an Illumina MiSeq instrument following the manufacturer’s guidelines. Our approach of utilizing next-generation sequencing techniques has the advantage of including multiple rhizobial operational taxonomic units (OTUs) per nodule (Checcucci et al., 2016), since conventional methods of culturing are not able to detect all of these endophytes (Birnbaum et al., 2016).
Bioinformatics
We used mothur version 1.36.1 (Schloss et al., 2009) to perform downstream analyses of raw MiSeq sequence data. Briefly, we removed all DNA sequences that had low quality scores (<25 % quality) and which had more than two differences to the barcode primer sequences. We screened out all sequences that had any ambiguous bases and a maximum of six homopolymers. We optimized the minimum and maximum lengths of sequences by choosing those sequences that started and ended in positions that were occupied by 90 % of all sequences, resulting in sequences of between 333 and 336 bp long. Because individual root nodules can comprise multiple strains of rhizobia (Checcucci et al., 2016) we sub-sampled 1000 sequences from each replicate (i.e. five pooled nodules from an individual plant), yielding a total of 5000 sequences for every 25 nodules from each species. For one species, A. cyclops, only 3000 sequences were obtained as only three replicates yielded good quality sequencing data. This approach also allowed us to account for the usually strong correlation between the number of observed OTUs and the number of sequences obtained, thus reducing the biases these measures introduce to alpha and beta diversity measures (Schloss et al., 2011). Note that although ‘bacterial strain’ commonly refers to a group of genetically identical individuals and ‘OTU’ to a certain level of genetic similarity, we use these terms interchangeably here. From these sub-samples, we removed all chimeric sequences independent of a reference database using the uchime algorithm (in mothur) (Edgar et al., 2011) and the template as self, i.e. de novo removal. Since no reference database is available for nodC, we computed pairwise sequence similarities with the Needleman–Wunsch algorithm and then clustered the sequences into root nodule rhizobial OTUs (RNR OTUs) at 97 % DNA sequence similarity with the nearest neighbour algorithm. We then removed singleton and doubleton OTUs (i.e. OTUs with only one or two reads across all 98 000 nodC sequences). The resulting RNR OTU table (individual acacia tree by RNR OTU count matrix) had many instances of extremely low RNR OTU abundances (e.g. < 5 DNA sequence reads across all 98 replicates – 0·1 % of total sequences), which probably represent data generation errors. We removed these extremely low abundance OTUs from the data set in two ways. First, after inspection of a cumulative sequence contribution curve (Supplementary Data Fig. S1), we only retained RNR OTUs representing the majority of sequences across all replicates for further downstream analyses. This approach identified 25 RNR OTUs that accounted for 99·1 % of all DNA sequence reads. Next, all cells in the OTU × replicate matrix that contained <1 % of sequences for that replicate were converted to zero. To determine the taxonomic affinity of OTUs, we blasted their associated sequences against the NCBI’s GenBank database (http://blast.ncbi.nlm.nih.gov/Blast).
Phylogeny
Representative DNA sequences for each RNR OTU were aligned, and a Bayesian inference phylogeny reconstructed using Mr Bayes v 3.2 (Ronquist and Huelsenbeck, 2003). Sequence data from the genus Mesorhizobium were included as outgroup taxa. jModelTest (Posada, 2008) and the Akaike information criterion (Akaike, 1973) were used to determine the best fit model for our data. The Bayesian model was run for 4 million iterations sampling every 1000th generation, and a consensus tree was built, discarding the first 25 % of trees as burn-in specifying the GTR + G substitution model. Posterior probabilities (PPs) were calculated using a majority rule consensus method to assess tree topology support.
Root nodule rhizobial community diversity
We aggregated RNR OTUs across replicates to determine diversity at the population level, i.e. we summed all replicates for each species per site that was sampled. In order to determine whether our sampling effort was adequate, we performed rarefaction analyses in mothur version 1.36.1 (Schloss et al., 2009). From the aggregated RNR OTU matrix, we calculated species richness (S; total number of RNR OTUs per species, giving equal weight to both abundant and rare RNR OTUs), Shannon diversity (H; diversity measure that takes into account the abundance differences between dominant and rare RNR OTUs), inverse Simpson diversity (Si; diversity measure that weights the abundance of dominant RNR OTUs higher than rare ones) and evenness (J; which measures how equally the abundances of RNR OTUs are spread in the sample) (Hill, 1973; Chao et al., 2014). We calculated evenness as the natural logarithm of H/S (Hill, 1973). Diversity indices were calculated using the R package vegan (version 2.3-3; Oksanen et al., 2016) and the function ‘renyi’. Together, these three metrics account for the influence of both common and rare RNR OTUs. We also calculated Faith’s phylogenetic diversity (PD; Faith, 1992) based on the retrieved RNR OTU phylogeny using the package picante (Kembel et al., 2010). This measure includes phylogenetic relatedness when calculating diversity for a sample of various abundances.
In order to investigate differences between localized and widespread species, we conducted paired t-tests to compare the various metrics described above across pairs of species sampled at each site.
Root nodule rhizobial community composition
To visualize acacia species-associated root nodule community compositions, we ran a principal co-ordinates analysis (PCoA) based on a Bray–Curtis dissimilarity matrix, calculated with the function ‘vegdist’ in the R package vegan (Oksanen et al., 2016). We did this for all acacia species that were sampled and not just for co-occurring localized and widespread species. We then used a permutation multivariate analysis of variance (PERMANOVA) (Anderson, 2001) with 9999 permutations in PRIMER v7 (Clarke et al., 2014) to test for the influence of status (widespread vs. localized) on differences in RNR OTU composition; we used site as a random factor and status as a fixed factor. To determine whether localized and widespread species were significantly over- or underdispersed in terms of their group centroids (i.e. multivariate homogeneity of group dispersions/variances), we used the function ‘betadisper’ in the vegan package with 9999 permutations. We also wanted to see whether acacias within sites are associating with compositionally more similar rhizobial communities compared with acacias from other sites. For this, we made a design matrix with zeros (0) coding for within-site distances and ones (1) coding for between-site distances (Rundle and Jackson, 1996). We then used a Mantel test, also in package vegan, with 9999 permutations to test the correlation between the Bray–Curtis dissimilarity matrix and the design matrix. Finally, in order to visualize the OTU abundances, we created a heat map with four abundance categories (excluding zero): 10–<100, 100–<1000, 1000–<2500 and ≥2500 using the package gplots (version 2.17.0) and function ‘heatmap.2’ (Warnes et al., 2015).
Stable isotope analysis
The 15N/14N ratio (δ15N) of plant leaves/shoots has been used as a proxy for biological nitrogen fixation (BNF) (Hobbie et al., 1998; Rodríguez-Echeverría et al., 2009; Lötter et al., 2014a, b). This is because nitrogenase preferentially incorporates the lighter isotope of N2 gas (Sra et al., 2004; Unkovich, 2013), and has the effect of diluting the overall isotopic signature, leading to decreased δ15N values. Thus, smaller values of δ are indicative of added N from biological nitrogen fixation (Rodríguez-Echeverría et al., 2009; Lötter et al., 2014a, b). Conversely, soil-available nitrogen generally has a higher value of 15N compared with the atmosphere (Unkovich et al., 2008), leading to increased values of δ15N in plants exploiting soil-derived nitrogen as the primary source of nitrogen. Thus, to infer whether widespread and localized acacias differed in their abilities to obtain nitrogen from the atmosphere, and therefore the effectiveness of their rhizobial mutualists, we used stable isotope analyses (Environmental Isotope Laboratory, iThemba Labs, WITS University, Johannesburg). The first 3–5 fully expanded leaves were collected from the same individuals from which root nodules were collected. All leaves were immediately oven-dried for 1 week at 45 °C to prevent the formation of mould after collection. Dried leaf samples were crushed into a fine powder and nitrogen isotopic analyses carried out using a Flash HT Plus integrated via a ConFlo IV system with a Delta V Plus Isotope Ratio Mass Spectrometer (Thermo Scientific, Bremen, Germany). Samples were combusted at 1020 °C and the nitrogen isotope values corrected against an in-house standard (Merck Gel δ15N = +6·80).
Isotope values were expressed in parts per thousand (‰) following Lötter et al. (2014a, b) and Rodríguez-Echeverría et al. (2009):
where δ15N is the heavy N isotope, and R is the ratio of heavier to light isotopes (15N/14N) for the sample and standard (being atmospheric nitrogen), respectively. δ15N values were used in t-tests for each site, comparing each widespread with co-occurring localized species pair. For site TK, which had two localized species, we conducted a pairwise t-test with Bonferroni correction.
All statistical analyses [paired t-tests, analyses of variance (ANOVAs) and PCoA] were performed in the R programming language (version 3.2.2) (R Core Team, 2016) with functions from the base package.
RESULTS
Root nodule rhizobial community diversity
From the 98 000 nodC sequences selected for downstream analyses, 2480 (2·5 %) were removed as chimeric. After clustering at 97 % sequence similarity, a further 1256 sequences representing singleton or doubleton RNR OTUs were removed. A total of 170 RNR OTUs were recovered from the remaining 94 264 DNA sequences. Not surprisingly, the first 25 RNR OTUs accounted for 99·1 % (93444 DNA reads) (see Fig. S1), all of which are representative of the genus Bradyrhizobium. Rarefaction analyses showed nodule bacterial taxon accumulation curves to reach asymptotes in all instances (Fig. S2), indicating that sampling was representative after removal of singletons/doubletons and low frequency interactions (i.e. representing <0·1 % of sequences for an individual tree).
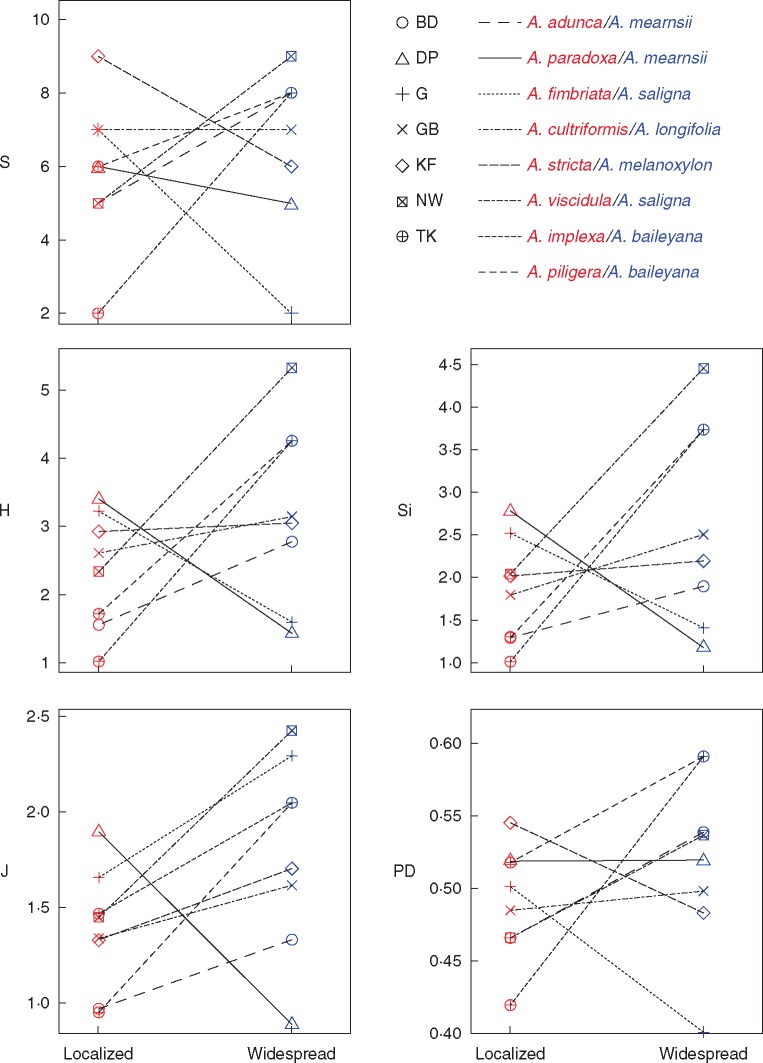
The RNR OTU richness, Shannon diversity, inverse Simpson diversity, evenness or phylogenetic diversity did not differ significantly between pairs of co-occurring widespread/ localized acacias (Fig. 3; Table 1; Supplementary Data Table S2). While the overall diversity of nodule communities did not differ significantly between widespread and localized acacias across sites (paired t-tests in Table 1), the trend was towards higher bacterial diversity associated with widespread acacias for all metrics (Table 1; Fig. 3). For six of the eight sampled widespread/localized acacia pairs, the widespread species housed more diverse bacterial communities (Fig. 3).
Fig. 3.
Diversity metrics (S, richness; H, Shannon diversity; Si, inverse Simpson diversity; J, evenness; PD, Faith’s phylogenetic distance) for co-occurring widespread and localized acacia species pairs (lines) at different sites (symbols).
Table 1.
Summary statistics for paired t-tests of RNR OTUs for co-occurring localized and widespread Acacia species pairs for richness (S), Shannon diversity (H), inverse Simpson diversity (Si), evenness (J) and phylogenetic diversity (PD)
| Metric | t | d.f. | P-value |
|---|---|---|---|
| S | –0·893 | 7 | 0·402 |
| H | –1·960 | 7 | 0·091 |
| Si | –1·791 | 7 | 0·117 |
| J | –1·593 | 7 | 0·155 |
| PD | –0·994 | 7 | 0·353 |
Root nodule rhizobial community composition
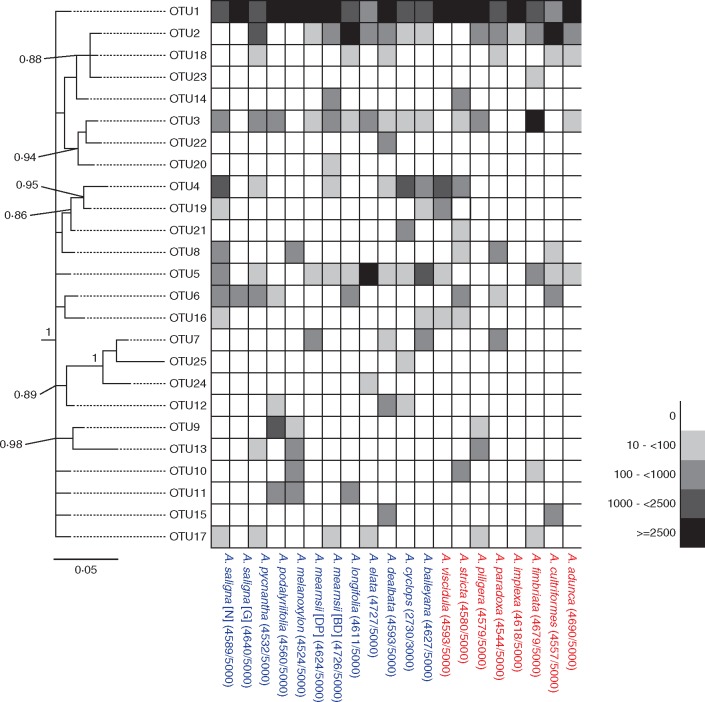
The first two components of the PCoA explained 62 % of the variation in bacterial community composition (PC1, 39 %; PC2, 23 %). There were no consistent compositional differences in the communities of bacteria with which widespread and localized acacias are associated (F-value = 0·31, P = 0·865). There was substantial variation in the composition of bacterial communities associated with both widespread and localized acacias (Fig. 4), and the dispersion of communities did not vary with status (F-value = 0·288, P = 0·601). Finally, while there was a tendency for compositional differences within sites (i.e. between widespread and localized acacia pairs) to be lower than differences between acacia species across sites (r2 = 0·0087, P = 0·0597) this trend was not significant. Interestingly, one ubiquitous RNR OTU (OTU1) associated with all acacias, although to various extents (Fig. 5), while three OTUs were unique to widespread species (OTU20, 24 and 25) and one OTU was unique to a localized species (OTU23, associated with Acacia fimbriata).
Fig. 4.
Principal co-ordinates analysis (PCoA) plot of Acacia RNR OTU community associations across all sites. A PERMANOVA model, with site as a random factor, indicated no significant difference in community composition between localized and widespread acacias.
Fig. 5.
Heat map indicating RNR OTU abundance and associations with each Acacia species (blue, widespread; red, localized). Numbers in parentheses next to species names indicate total number of sequence reads used after quality filtering. The left-hand side RNR OTU tree represents the retrieved Bayesian topology based on nodC DNA barcodes. Tree topology support is indicated as posterior probabilities at nodes, and the bar represents number of substitutions per site.
Stable isotope analysis
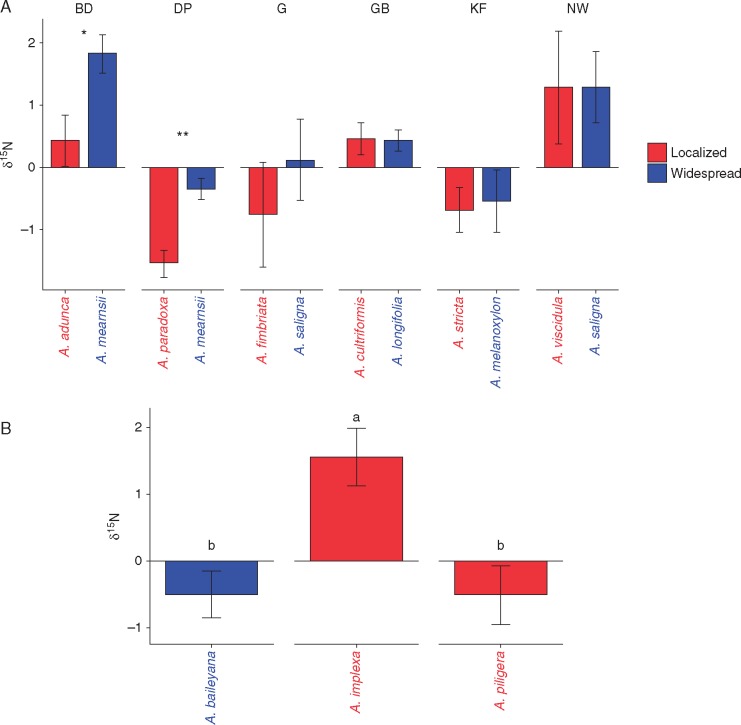
For two pairs of widespread and localized acacias there were significant differences in δ15N values, and in both cases the localized species had lower values (A. adunca, P < 0·05; and A. paradoxa, P < 0·01, Table S3), indicating that it had received more nitrogen from the atmosphere through fixation (Fig. 6A) (Rodríguez-Echeverría et al., 2009; Lötter et al., 2014a, b). A mixed trend was observed at site TK, where one of the localized species (A. implexa) differed significantly (P < 0·05) from the widespread species (A. baileyana), and also from the other co-occurring localized species (A. piligera). Thus, here the two localized species differed significantly in their BNF abilities. Apart from these sites, overall there were no differences in BNF between localized and widespread species. It is interesting to note that at sites DP, G and KF there seems to be an increased uptake of atmospheric nitrogen by acacias. One possibility for this might be lower level of soil nitrogen available for use by these acacias, or that these might represent cases of more effective mutualisms.
Fig. 6.
Variation in stable nitrogen (N) isotopic signatures for widespread and localized Acacia species (mean ± s.e.) for (A) sites which had pairs of co-occurring widespread and localized species; and (B) site TK which had two localized and one widespread species. For (A) significance is indicated as: *P < 0·05, **P < 0·01; for (B) the same letter above bars indicates that the treatments are not significantly different from each other (P ≤ 0·05, pairwise t-test with Bonferroni correction). Positive values indicate a higher sample abundance of 15N compared with atmosphere, while negative values indicate a lower abundance of 15N compared with the reference value.
DISCUSSION
We found no support for our predictions that mutualist promiscuity or effectiveness influences invasiveness of acacias in South Africa. Counter to expectations under our initial hypothesis, rhizobial communities of widespread and localized acacias do not exhibit consistent diversity differences. Furthermore, analyses of RNR community composition between widespread and localized species indicate no consistent preference for particular community assemblages between these two groups. These findings are in agreement with Klock et al. (2016) who found rhizobial richness and community composition to be similar for Australian acacias that have differential invasiveness in California. In the native Australian ranges of acacias, narrowly distributed species also have similar levels of rhizobial associations with extremely widespread species (Murray et al., 2001), i.e. range-restricted species do not show greater levels of specialization. Interestingly, such patterns have been demonstrated for other legume species, for example non-native Trifolium species that associate with equally diverse rhizobial communities in their introduced (New Zealand) and native (UK) ranges (McGinn et al., 2016). Although there seems to be a general paucity of data regarding the rhizobial symbionts nodulating acacias in their native Australian environment, there seems to be little difference in the promiscuity of invasive and non-invasive acacias (Rodríguez-Echeverría et al., 2011). For example, based on 16S rRNA genetic data, A. dealbata, A. longifolia, A. mearnsii, A. melanoxylon and A. saligna are able to nodulate with both Bradyrhizobium and Rhizobium strains (Lawrie, 1983; Barnet et al., 1985; Marsudi et al., 1999; Lafay and Burdon, 2001; Yates et al., 2004), while A. pycnantha nodulates with both Bradyrhizobium and Burkholderia strains, suggesting that these acacias are all generalist in their symbiotic requirements.
Similar to native Australian regions (Burdon et al., 1999; Stepkowski et al., 2012) and other introduced ranges (Weir et al., 2004; Rodríguez-Echeverría et al., 2011; Crisóstomo et al., 2013; Ndlovu et al., 2013), we found naturalized acacias in South Africa to be primarily associated with slow-growing rhizobia from the genus Bradyrhizobium. Bradyrhizobium is a cosmopolitan genus and is associated with a diverse range of legumes globally (Weir et al., 2004; Andam and Parker, 2008; Birnbaum et al., 2012). Without data from the native ranges, and the short sequence reads employed here, it is difficult to elucidate whether rhizobial symbionts have been co-introduced with acacias to South Africa. Such co-introductions are a common phenomenon among woody invaders and their microbial mutualists (Nuñez and Dickie, 2014). It is conceivable that the high propagule pressure underlying many acacia introductions to South Africa (Poynton, 2009) may have facilitated co-introductions of compatible rhizobia, as has been demonstrated for A. pycnantha in South Africa (Ndlovu et al., 2013). Co-invasions of acacias and their associated rhizobia have been found for A. longifolia and A. saligna in Europe (Rodríguez-Echeverría, 2010; Crisóstomo et al., 2013) and for A. longifolia in New Zealand (Weir et al., 2004).
Recent studies have found native legumes of South Africa’s Greater Cape Floristic Region to nodulate predominantly with bacterial symbionts other than Bradyrhizobium [e.g. Burkholderia and Mezorhizobium strains (Kock, 2004; Lemaire et al., 2015)], while invasive acacias preferentially nodulate with Bradyrhizobium (Ndlovu et al., 2013; Le Roux et al., 2016). A similar pattern was observed for acacias in New Zealand where they are nodulated only by Bradyrhizobium strains, while native legumes associate predominantly with strains of Mesorhizobium (Weir et al., 2004). Therefore, considering the general rarity of Bradyrhizobium in the Cape, and their prominent association with Australian acacias in this study, it is unlikely that they represent mutualists with which native legumes frequently nodulate. An intriguing possibility for the ubiquitous nature of Bradyrhizobium in association with all acacias studied to date, and specifically with regards to localized species included in the current study, is that soil bacterial communities have been transformed to such an extent by the widespread species that their strains now dominate the below-ground environment, i.e. facilitated proliferation of certain strains upon acacia invasion. Indeed, a recent study found that rhizosphere soils of invasive A. dealbata in South Africa are significantly enriched for Bradyrhizobium strains compared with soils where the species is absent (C. Kamutando et al., unpubl. res.). Such transformations of soil community composition could therefore potentially create opportunities for localized species to encounter, and associate with rhizobial strains utilized by widespread acacias. Overall we found no evidence that a lack of generalism characterizes localized acacias in South Africa. Instead, we found that both localized and widespread acacias associate equally well with a range of bacterial OTUs and thus appear not to be constrained by locating compatible rhizobia. This is in agreement with observations from California where non-native acacias differing in invasiveness showed similar levels of symbiotic promiscuity and performances when grown in non-native soils (Klock et al., 2016). Although it is possible that certain strains have been co-introduced with some of the acacias studied here, the cosmopolitan status of Bradyrhizobium makes conclusions regarding this difficult. To determine conclusively whether co-introductions of rhizobia occurred with acacias to South Africa would require a phylogeographic study of Australian and South African rhizobia (e.g. Ndlovu et al., 2013).
The abundance of compatible rhizobia in soils seems to be an important factor for determining growth responses of acacias (Thrall et al., 2007a) as well as their invasiveness (Parker, 2001). It therefore seems that the successful establishment and spread of acacias in new environments might be dependent on the density of compatible rhizobia (Thrall et al., 2007a, 2008). This means that the first arriving acacias would induce a local proliferation of their preferred symbionts and that later arriving species might end up acquiring symbionts that might not necessarily have been the preferred genotypes had such species been the first to have colonized the site. This also means that it is possible that certain species might not have been capable of colonizing a site if the initial species did not cause a local proliferation of compatible symbionts.
The similarity in rhizobial diversity found between localized and widespread acacias in this study may reflect similar levels of host promiscuity and invasive potential, and therefore that localized species have not yet reached their full potential ranges in South Africa. However, this is unlikely to be the case as most localized species included here have never been recorded as invasive (Richardson and Rejmánek, 2011, but see Zenni et al., 2009; Kaplan et al., 2012).
Rhizobial diversity and community structure are important factors influencing the productivity of acacias, but increased diversity alone does not always translate into higher plant fitness (Barrett et al., 2015). That is in part because different rhizobial strains may differ in their symbiotic effectiveness (Franche et al., 2009; Klock et al., 2015). We found no consistent differences in rhizobial community composition between localized and widespread acacias (also see Klock et al., 2016) and this may reflect high-frequency associations of all acacias with one or a few OTUs of rhizobia. For example, RNR OTU1 made up the vast majority of the associations with most acacias (average read count per associated acacia species = 2522) albeit at different frequencies (Fig. 5), and may represent one of the most effective and preferred acacia strains. In New Zealand, Bever et al. (2013) found extensive variation in symbiotic effectiveness of acacia-associated rhizobial strains; with some strains broadly effective across all species tested, some strains varying in effectiveness depending on host species, and some strains being relatively ineffective across all acacias tested. At site level, we only found two instances where symbiotic effectiveness differed between widespread and localized acacias, albeit in the opposite direction from that which we hypothesized. Widespread A. mearnsii showed significantly less accumulated atmospheric nitrogen compared with co-occurring localized species [A. adunca (site BD) and A. paradoxa (site DP)] (Fig. 6). This could indicate that A. mearnsii is less reliant on atmospheric nitrogen and is better able to utilize already scarce nutrients, or that it had less effective associations than co-occurring localized species. Interestingly, in both these instances, all three species shared OTU1 at high frequencies (Fig. 5). This suggests that shared OTUs between different acacias may indeed, dependent on host species, differ in their symbiotic effectiveness. Such differential effectiveness of a single rhizobium strain on different hosts/genotypes is well documented (e.g. Checcucci et al., 2016). Overall, however, it appears that nitrogen fixation effectiveness is similar for widespread and localized acacias in South Africa.
Our sampling design might suffer from the fact that inferences are restricted to local scales, since shifts in the distribution of rhizobial genotypes across the landscape is likely and the capacity to utilize different partners across a landscape may be critical for the success of widespread legumes. Thus, it is not clear whether these patterns will hold at landscape levels. Also, there could be many additional factors that might influence the invasive capacity of acacias in South Africa. First, propagule pressure and introduction history have been implicated as being important factors determining invasiveness (Wilson et al., 2007). Widespread and localized species have shared similar residence times in South Africa (t = –1·1771, d.f. = 13, P > 0·05, Table S4), but do differ substantially in propagule pressure and number of introductions (t = –2·6819, d.f. = 14, P < 0·05; data from Poynton, 2009), with widespread species often characterized by extremely high propagule pressure. Then, soil microbes other than rhizobia might influence plant performance, including pathogens and other mutualistic organisms (Thrall et al., 2007b), as well as the abiotic conditions of soils (Klock et al., 2016). Although our data suggest that rhizobial diversity and community composition do not translate into differential mutualistic effectiveness for acacias and therefore their varying degrees of invasiveness in South Africa, there are a wide variety of local environmental factors that may influence the relative amount of symbiotic nitrogen gained. The inherent ability of plants to obtain nitrogen and other important nutrients from the soil, the nature of the recipient environment and its conditions (e.g. nutrient status), the ubiquitous nature of some cosmopolitan strains of compatible rhizobia, together with the potential for co-introduction of these strains, indicate that multiple factors could aid acacias to become successful invaders. The potential modification of whole soil bacterial communities through facilitated proliferation of invasive plants means that, in terms of restoration, soil microbiome composition should be considered a major impact by invasive plants. Restoration may need to realign soil microbial communities to native plant communities in order to maximize restoration potential (Harris, 2009). Future work should address the effectiveness of these mutualistic associations on finer and more precise scales and under common garden conditions, for example by inoculating various acacias with specific rhizobial strains and calculating symbiotic responses using fitness correlates such as growth kinetics (Thrall et al., 2011; Klock et al., 2015), in conjunction with isotope studies.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: locality details of all Acacia species included in this study. Table S2: richness, diversity, evenness and phylogenetic diversity metrics for all species. Table S3: stable δ15N isotope values for the various Acacia species used in this study. Table S4: introduction histories of the Acacia species analysed in this study. Figure S1: cumulative distribution of sequence reads across identified OTUs showing that 25 OTUs account for the majority of sequences (99·1 %). Figure S2: rarefaction curves for all acacia species.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Angel Valverde for help with NGS bioinformatics. J.L.R. acknowledges support from the National Research Foundation (grant no. 93591) and the Oppenheimer Memorial Trust. C.H. acknowledges support from the National Research Foundation (grant nos 81825 and 76912) and the Australian Research Council (Discovery Project DP150103017). The authors declare that there is no conflict of interest arising from this publication.
LITERATURE CITED
- Aizen MA, Sabatino M, Tylianakis JM.. 2012. Specialization and rarity predict nonrandom loss of interactions from mutualist networks. Science 335: 1486–1489. [DOI] [PubMed] [Google Scholar]
- Akaike H. 1973. Akaike Information theory as an extension of the maximum likelihood principle In: Petrov BN, Csaki F, eds. Second international symposium on information theory. Budapest: Akademiai Kiado, 267–281. [Google Scholar]
- Amarger N. 1981. Competition for nodule formation between effective and ineffective strains of Rhizobium meliloti. Soil Biology and Biochemistry 13: 475–480. [Google Scholar]
- Andam CP, Parker MA.. 2008. Origins of Bradyrhizobium nodule symbionts from two legume trees in the Philippines. Journal of Biogeography 35: 1030–1039. [Google Scholar]
- Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecology 26: 32–46. [Google Scholar]
- Barnet YM, Catt PC, Hearne DH.. 1985. Biological nitrogen fixation and root-nodule bacteria (Rhizobium sp. and Bradyrhizobium sp.) in two rehabilitating sand dune areas planted with Acacia spp. Australian Journal of Botany 33: 595–610. [Google Scholar]
- Barrett LG, Bever JD, Bissett A, Thrall PH.. 2015. Partner diversity and identity impacts on plant productivity in Acacia–rhizobial interactions. Journal of Ecology 103: 130–142. [Google Scholar]
- Bascompte J. 2009. Mutualistic networks. Frontiers in Ecology and the Environment 7: 429–436. [Google Scholar]
- Bever JD, Broadhurst LM, Thrall PH.. 2013. Microbial phylotype composition and diversity predicts plant productivity and plant-soil feedbacks. Ecology Letters 16: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum C, Barrett LG, Thrall PH, Leishman MR.. 2012. Mutualisms are not constraining cross-continental invasion success of Acacia species within Australia. Diversity and Distributions 18: 962–976. [Google Scholar]
- Birnbaum C, Bissett A, Thrall PH, Leishman MR.. 2016. Nitrogen-fixing bacterial communities in invasive legume nodules and associated soils are similar across introduced and native range populations in Australia. Journal of Biogeography 43: 1631–1644. [Google Scholar]
- Burdon JJ, Gibson AH, Searle SD, Woods MJ, Brockwell J.. 1999. Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian Acacia: within-species interactions. Journal of Applied Ecology 37: 398–408. [Google Scholar]
- Carruthers J, Robin L, Hattingh JP, Kull CA, Rangan H, van Wilgen BW.. 2011. A native at home and abroad: the history, politics, ethics and aesthetics of acacias. Diversity and Distributions 17: 810–821. [Google Scholar]
- Chao A, Gotelli NJ, Hsieh TC, et al. 2014. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecological Monographs 84: 45–67. [Google Scholar]
- Checcucci A, Azzarello E, Bazzicalupo M, et al. 2016. Mixed nodule infection in Sinorhizobium meliloti–Medicago sativa symbiosis suggest the presence of cheating behavior. Frontiers in Plant Science 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Gorley RN, Somerfield PJ, Warwick RM.. 2014. . Change in marine communities: an approach to statistical analysis and interpretation, Version 7, 3rd edn.Plymouth: PRIMER-E. [Google Scholar]
- Crisóstomo JA, Rodríguez-Echeverría S, Freitas H.. 2013. Co-introduction of exotic rhizobia to the rhizosphere of the invasive legume Acacia saligna, an intercontinental study. Applied Soil Ecology 64: 118–126. [Google Scholar]
- Denison RF. 2000. Legume sanctions and the evolution of symbiotic cooperation by rhizobia. American Naturalist 156: 567–576. [DOI] [PubMed] [Google Scholar]
- Ding H, Hynes MF.. 2009. Plasmid transfer systems in the rhizobia. Canadian Journal of Microbiology 55: 917–927. [DOI] [PubMed] [Google Scholar]
- Dwivedi SL, Sahrawat KL, Upadhyaya HD, et al. 2015. Advances in host plant and rhizobium genomics to enhance symbiotic nitrogen fixation in grain legumes. Amsterdam: Elsevier Ltd. [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R.. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biological Conservation 61: 1–10. [Google Scholar]
- Franche C, Lindström K, Elmerich C.. 2009. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant and Soil 321: 35–59. [Google Scholar]
- Harris J. 2009. Soil microbial communities and restoration ecology: facilitators or followers? Science 325: 573–574. [DOI] [PubMed] [Google Scholar]
- Heleno RH, Olesen JM, Nogales M, Vargas P, Traveset A.. 2013. Seed dispersal networks in the Galápagos and the consequences of alien plant invasions. Proceedings of the Royal Society B: Biological Sciences 280: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L. 1998. Southern African Plant Invaders Atlas (SAPIA). Applied Plant Sciences 12: 31–32. [Google Scholar]
- Hill MO. 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54: 427–432. [Google Scholar]
- Hobbie EA, MacKo SA, Shugart HH.. 1998. Patterns in N dynamics and N isotopes during primary succession in Glacier Bay, Alaska. Chemical Geology 152: 3–11. [Google Scholar]
- Hopkins WG, Hüner NP.. 2009. Introduction to plant physiology. Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
- Hoque MS, Broadhurst LM, Thrall PH.. 2011. Genetic characterization of root-nodule bacteria associated with Acacia salicina and A. stenophylla (Mimosaceae) across south-eastern Australia. International Journal of Systematic and Evolutionary Microbiology 61: 299–309. [DOI] [PubMed] [Google Scholar]
- Inderjit Cahill JF. 2015. Linkages of plant–soil feedbacks and underlying invasion mechanisms. AoB PLANTS 7: pii: plv022. doi:10.1093/aobpla/plv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan H, Van Zyl HWF, Le Roux JJ, Richardson DM, Wilson JRU.. 2012. Distribution and management of Acacia implexa (Benth.) in South Africa: a suitable target for eradication? South African Journal of Botany 83: 23–35. [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, et al. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Rousseau RA, West SA, Denison RF.. 2003. Host sanctions and the legume–rhizobium mutualism. Nature 425: 78–81. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Rousseau RA, Denison RF.. 2006. Measured sanctions: legume hosts detect quantitative variation in rhizobium cooperation and punish accordingly. Evolutionary Ecology Research 8: 1077–1086. [Google Scholar]
- Klock MM, Barrett LG, Thrall PH, Harms KE.. 2015. Host promiscuity in symbiont associations can influence exotic legume establishment and colonization of novel ranges. Diversity and Distributions 21: 1193–1203. [Google Scholar]
- Klock MM, Barrett LG, Thrall PH, Harms KE.. 2016. Differential plant invasiveness is not always driven by host promiscuity with bacterial symbionts. AoB PLANTS 8: pii: plw060. doi:10.1093/aobpla/plw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock MM. 2004. Diversity of root nodulating bacteria associated with Cyclopia species. PhD Thesis, University of Pretoria.
- Kull CA, Rangan H.. 2008. Acacia exchanges: wattles, thorn trees, and the study of plant movements. Geoforum 39: 1258–1272. [Google Scholar]
- Kull CA, Shackleton CM, Cunningham PJ, et al. 2011. Adoption, use and perception of Australian acacias around the world. Diversity and Distributions 17: 822–836. [Google Scholar]
- Lafay B, Burdon JJ.. 2001. Small-subunit rRNA genotyping of rhizobia nodulating Australian Acacia spp. Applied and Environmental Microbiology 67: 396–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie AC. 1983. Relationships among rhizobia from native Australian legumes. Applied and Environmental Microbiology 45: 1822–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire B, Dlodlo O, Chimphango S, et al. 2015. Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiology Ecology 91: 1–17. [DOI] [PubMed] [Google Scholar]
- Lötter D, Van Garderen EA, Tadross M, Valentine AJ.. 2014a. Seasonal variation in the nitrogen nutrition and carbon assimilation in wild and cultivated Aspalathus linearis (rooibos tea). Australian Journal of Botany 62: 65–73. [Google Scholar]
- Lötter D, Valentine AJ, Archer Van Garderen E, Tadross M.. 2014b. Physiological responses of a fynbos legume, Aspalathus linearis to drought stress. South African Journal of Botany 94: 218–223. [Google Scholar]
- Le Maitre DC, Gaertner M, Marchante E, et al. 2011. Impacts of invasive Australian acacias: implications for management and restoration. Diversity and Distributions 17: 1015–1029. [Google Scholar]
- Le Maitre DC, Gush MB, Dzikiti S.. 2015. Impacts of invading alien plant species on water flows at stand and catchment scales. AoB PLANTS 7: pii: plv043. doi:10.1093/aobpla/plv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux JJ, Mavengere NR, Ellis AG.. 2016. The structure of legume–rhizobium interaction networks and their response to tree invasions. AoB PLANTS 8: pii: plw038. doi:10.1093/aobpla/plw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsudi NDS, Glenn AR, Dilworth MJ.. 1999. Identification and characterization of fast- and slow-growing root nodule bacteria from South-Western Australian soils able to nodulate Acacia saligna. Soil Biology and Biochemistry 31: 1229–1238. [Google Scholar]
- Mårtensson AM, Brutti L, Ljunggren H.. 1989. Competition between strains of Bradyrhizobium japonicum for nodulation of soybeans at different nitrogen fertilizer levels. Plant and Soil 117: 219–225. [Google Scholar]
- McGinn KJ, van der Putten WH, Duncan RP, Shelby N, Weser C, Hulme PE.. 2016. Trifolium species associate with a similar richness of soil-borne mutualists in their introduced and natives. Journal of Biogeography 43: 944–954. [Google Scholar]
- Murray BR, Thrall PH, Woods MJ.. 2001. Acacia species and rhizobial interactions: implications for restoration of native vegetation. Ecological Management and Restoration 2: 213–219. [Google Scholar]
- Ndlovu J, Richardson DM, Wilson JRU, Le Roux JJ.. 2013. Co-invasion of South African ecosystems by an Australian legume and its rhizobial symbionts. Journal of Biogeography 40: 1240–1251. [Google Scholar]
- Nuñez MA, Dickie IA.. 2014. Invasive belowground mutualists of woody plants. Biological Invasions 16: 645–661. [Google Scholar]
- Oksanen JF, Blanchet FG, Kindt R, et al. 2016. vegan: community ecology package. R package version 2.3-3.
- Parker MA. 2001. Mutualism as a constraint on invasion success for legumes and rhizobia. Diversity and Distributions 7: 125–136. [Google Scholar]
- Parker MA, Malek W, Parker IM.. 2006. Growth of an invasive legume is symbiont limited in newly occupied habitats. Diversity and Distributions 12: 563–571. [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Poynton RJ. 2009. Tree planting in southern Africa. Vol. 3 Other genera. Pretoria: Department of Agriculture, Forestry and Fisheries. [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Richardson DM, Carruthers J, Hui C, et al. 2011. Human-mediated introductions of Australian acacias – a global experiment in biogeography. Diversity and Distributions 17: 771–787. [Google Scholar]
- Richardson DM, Rejmánek M.. 2011. Trees and shrubs as invasive alien species – a global review. Diversity and Distributions 17: 788–809. [Google Scholar]
- Rodríguez-Echeverría S. 2010. Rhizobial hitchhikers from Down Under: invasional meltdown in a plant–bacteria mutualism? Journal of Biogeography 37: 1611–1622. [Google Scholar]
- Rodríguez-Echeverría S, Crisóstomo JA, Freitas H.. 2007. Genetic diversity of rhizobia associated with Acacia longifolia in two stages of invasion of coastal sand dunes. Applied and Environmental Microbiology 73: 5066–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Echeverría S, Crisóstomo JA, Nabais C, Freitas H.. 2009. Belowground mutualists and the invasive ability of Acacia longifolia in coastal dunes of Portugal. Biological Invasions 11: 651–661. [Google Scholar]
- Rodríguez-Echeverría S, Le Roux JJ, Crisóstomo JA, Ndlovu J.. 2011. Jack-of-all-trades and master of many? How does associated rhizobial diversity influence the colonization success of Australian Acacia species? Diversity and Distributions 17: 946–957. [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Jackson DA.. 1996. Spatial and temporal variation in littoral-zone fish communities: a new statistical approach. Canadian Journal of Fisheries and Aquatic Sciences 53: 2167–2176. [Google Scholar]
- Schloss PD, Gevers D, Westcott SL.. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6: e27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton PW, Stockinger KR.. 1983. Compensation against ineffective nodulation in soybean. Crop Science 23: 69–72. [Google Scholar]
- Slabbert E, Kongor RY, Esler KJ, Jacobs K.. 2010. Microbial diversity and community structure in Fynbos soil. Molecular Ecology 19: 1031–1041. [DOI] [PubMed] [Google Scholar]
- Spaink HP. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annual Review of Microbiology 54: 257–288. [DOI] [PubMed] [Google Scholar]
- Sra AK, Hu Y, Martin GE, Snow DD, Ribbe MW, Kohen A.. 2004. Competitive 15N kinetic isotope effects of nitrogenase-catalyzed dinitrogen reduction. Journal of the American Chemical Society 126: 12768–12769. [DOI] [PubMed] [Google Scholar]
- Stacey G. 2007. The Rhizobium–legume nitrogen-fixing symbiosis In: Bothe H, Ferguson SJ, Newton WE, eds. Biology of the nitrogen cycle. Amsterdam: Elsevier Inc, 147–163. [Google Scholar]
- Stanton-Geddes J, Anderson CG.. 2011. Does a facultative mutualism limit species range expansion? Oecologia 167: 149–155. [DOI] [PubMed] [Google Scholar]
- Stepkowski T, Watkin E, McInnes A, Gurda D, Gracz J, Steenkamp ET.. 2012. Distinct Bradyrhizbium communities nodulate legumes native to temperate and tropical monsoon Australia. Molecular Phylogenetics and Evolution 63: 265–277. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Burdon JJ, Woods MJ.. 2000. Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian legumes: interactions within and between genera. Journal of Applied Ecology 37: 52–65. [Google Scholar]
- Thrall PH, Slattery JF, Broadhurst LM, Bickford S.. 2007a. Geographic patterns of symbiont abundance and adaptation in native Australian Acacia–rhizobia interactions. Journal of Ecology 95: 1110–1122. [Google Scholar]
- Thrall PH, Hochberg ME, Burdon JJ, Bever JD.. 2007b. Coevolution of symbiotic mutualists and parasites in a community context. Trends in Ecology and Evolution 22: 120–126. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Bever JD, Slattery JF.. 2008. Rhizobial mediation of Acacia adaptation to soil salinity: evidence of underlying trade-offs and tests of expected patterns. Journal of Ecology 96: 746–755. [Google Scholar]
- Thrall PH, Laine A-L, Broadhurst LM, Bagnall DJ, Brockwell J.. 2011. Symbiotic effectiveness of rhizobial mutualists varies in interactions with native Australian legume genera. PLoS One 6: e23545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traveset A, Richardson DM.. 2014. Mutualistic interactions and biological invasions. Annual Review of Ecology, Evolution, and Systematics 45: 89–113. [Google Scholar]
- Unkovich M. 2013. Isotope discrimination provides new insight into biological nitrogen fixation. New Phytologist 198: 643–646. [DOI] [PubMed] [Google Scholar]
- Unkovich M, Herridge D, Peoples M, et al. 2008. Measuring plant-associated nitrogen fixation in agricultural systems. Canberra, Australia: Australian Centre for International Agricultural Research. [Google Scholar]
- Vestergård M, Rønn R, Ekelund F.. 2015. Above–belowground interactions govern the course and impact of biological invasions. AoB PLANTS 7: pii: plv025. doi:10.1093/aobpla/plv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandrag EM, Sheppard A, Duncan RP, Hulme PE.. 2013. Reduced availability of rhizobia limits the performance but not invasiveness of introduced Acacia. Journal of Ecology 101: 1103–1113. [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, et al. 2015. gplots: various R programming tools for plotting data. R package version 2.17.0.
- Weir BS, Turner SJ, Silvester WB, Park D-C, Young JM.. 2004. Unexpectedly diverse Mesorhizobium strains and Rhizobium leguminosarum nodulate native legume genera of New Zealand, while introduced legume weeds are nodulated by Bradyrhizobium species. Applied and Environmental Microbiology 70: 5980–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JRU, Richardson DM, Rouget M, et al. 2007. Residence time and potential range: crucial considerations in modelling plant invasion. Diversity and Distributions 13: 11–12. [Google Scholar]
- Yates RJ, Howieson JG, Nandasena KG, O’Hara GW.. 2004. Root-nodule bacteria from indigenous legumes in the north-west of Western Australia and their interaction with exotic legumes. Soil Biology and Biochemistry 36: 1319–1329. [Google Scholar]
- Zenni RD, Wilson JRU, Le Roux JJ, Richardson DM.. 2009. Evaluating the invasiveness of Acacia paradoxa in South Africa. South African Journal of Botany 75: 485–496. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.