Abstract
Background Among the environmental limitations that affect plant growth, viruses cause major crop losses worldwide and represent serious threats to food security. Significant advances in the field of plant–virus interactions have led to an expansion of potential strategies for genetically engineered resistance in crops during recent years. Nevertheless, the evolution of viral virulence represents a constant challenge in agriculture that has led to a continuing interest in the molecular mechanisms of plant–virus interactions that affect disease or resistance.
Scope and Conclusion This review summarizes the molecular mechanisms of the antiviral immune system in plants and the latest breakthroughs reported in plant defence against viruses. Particular attention is given to the immune receptors and transduction pathways in antiviral innate immunity. Plants counteract viral infection with a sophisticated innate immune system that resembles the non-viral pathogenic system, which is broadly divided into pathogen-associated molecular pattern (PAMP)-triggered immunity and effector-triggered immunity. An additional recently uncovered virus-specific defence mechanism relies on host translation suppression mediated by a transmembrane immune receptor. In all cases, the recognition of the virus by the plant during infection is central for the activation of these innate defences, and, conversely, the detection of host plants enables the virus to activate virulence strategies. Plants also circumvent viral infection through RNA interference mechanisms by utilizing small RNAs, which are often suppressed by co-evolving virus suppressors. Additionally, plants defend themselves against viruses through hormone-mediated defences and activation of the ubiquitin–26S proteasome system (UPS), which alternatively impairs and facilitates viral infection. Therefore, plant defence and virulence strategies co-evolve and co-exist; hence, disease development is largely dependent on the extent and rate at which these opposing signals emerge in host and non-host interactions. A deeper understanding of plant antiviral immunity may facilitate innovative biotechnological, genetic and breeding approaches for crop protection and improvement.
Keywords: Antiviral immunity, antiviral immune receptors, PAMP-triggered immunity, effector-triggered immunity, NSP-interacting kinase, NIK-mediated translation suppression, antiviral RNA silencing, hormone-mediated defence, proteasome degradation, NBS-LRR resistance protein, receptor-like kinase, LRR-RLK
INTRODUCTION
As obligate parasites with limited viral genome-encoded functions, plant viruses extensively use the host intracellular machinery for replication of their genomes, expression of viral genes and establishment of infection. As a consequence, they interact profoundly with the host during their biological cycle. In contrast to animal viruses, which use host surface receptors and endocytic activities to invade host cells, plant viruses are delivered into the cells by insect vectors or through opportunistic mechanical wounds. Once inside the cells, the viral particles, which minimally consist of nucleic acids encapsulated by the coat protein or capsid, are disassembled to release the viral genome and to initiate the infectious cycle, which includes expression and replication of the viral genome, cell to cell and long-distance movement of the viral particles and/or viral genome and vector-mediated transmission to new hosts. The extensive interactions between plant viruses and their hosts during infection lead to the physiological disorders responsible for plant diseases, which represent major constraints to agricultural productivity worldwide.
Plants employ multiple defence mechanisms to restrict viral replication and movement, such as gene silencing, immune receptor signalling, hormone-mediated defence, protein degradation and regulation of metabolism (Incarbone and Dunoyer, 2013). In virus–plant interactions, one of the major mechanisms for plant antiviral immunity relies on RNA silencing, which is often suppressed by co-evolving viral suppressors, thus enhancing viral pathogenicity in susceptible hosts. In addition, plants use nucleotide-binding leucine-rich repeat (NB-LRR) domain-containing resistance proteins, which recognize viral effectors and activate effector-triggered immunity (ETI) in a defence mechanism similar to that employed in non-viral infections (Mandadi and Scholthof, 2013). Plants have also been found to use innate pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) to limit viral infection (Kørner et al., 2013). More recently, a transmembrane immune receptor, which is structurally similar to co-receptor-like kinases involved in PTI, has been shown to activate host translation suppression to fight DNA viruses, a newly discovered mechanism for antiviral defences in plants (Zorzatto et al., 2015).
Viral infections can also lead to hormonal disruption, which manifests as simultaneous induction of many antagonistic hormones and triggering of defence responses (Alazem and Lin, 2015). Virus–host interactions can aberrantly regulate phytohormone pathways, leading to disease development and hormone-mediated defensive responses. Plants employ the ubiquitin–proteasome pathway (UPS) as an antiviral defence strategy and, concomitantly, viruses have been reported to exploit the UPS to induce, inhibit or modify ubiquitin (Ub)-related host proteins (Alcaide-Loridan and Jupin, 2012). In this review, we summarize recent reports on host–virus interactions, highlighting mechanisms adopted by plants to overcome viral infections in a continuous coevolutionary race for dominance. A major focus is antiviral immune receptors and their signal transduction pathways.
PLANT INNATE IMMUNE SYSTEM: DETECTION AND SIGNALLING IN ANTIVIRAL DEFENCES
Effector-triggered immunity: intracellular immune receptor R (resistance protein) in virus–plant interactions
The plant innate immune pathway employs a two-level detection system, which involves plasma membrane-localized and intracellular immune receptors, to activate defences against invaders (Dodds and Rathjen, 2010; Zipfel, 2014). In the first level of defence, PTI is mediated by surface-localized pattern recognition receptors (PRRs), which detect and recognize PAMPs (Böhm et al., 2014; Macho and Zipfel, 2014). The second level, ETI, involves intracellular immune receptors, designated as resistance proteins (R), which recognize – directly or indirectly – virulence effectors secreted by the pathogens into the host intracellular environment, thereby activating a defence response (Jones and Dangl, 2006) (Fig. 1).
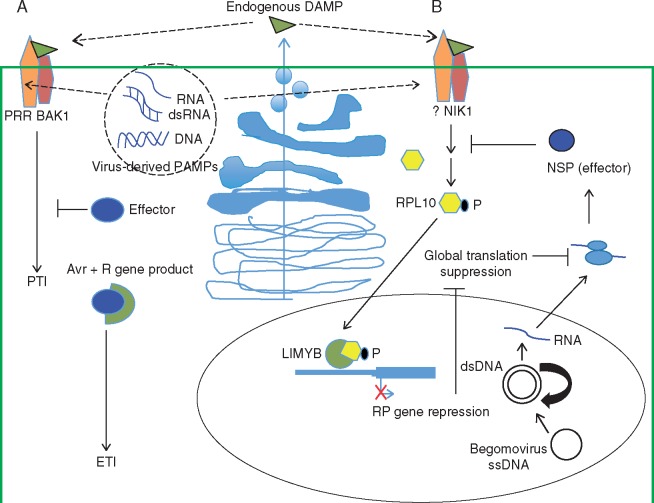
Fig. 1.
Antiviral innate immunity in plants. (A) PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) in virus–host interactions. During viral infection, the replication and expression of the viral genome lead to the accumulation of virus-derived nucleic acids with features of pathogen-associated molecular patterns (PAMPs), which may be recognized by host pattern recognition receptors (PRRs) that, in turn, heterooligomerize with co-receptors, such as BAK1 and BKK1, to trigger PTI. Alternatively, PTI may be activated upon PRR recognition of damage-associated molecular patterns (DAMPs), which are induced by infection and delivered to the apoplast by the host cells via the secretory apparatus. In a successful infection, expression of the viral genome results in accumulation of virus effectors to suppress PTI, leading to disease. In resistant genotypes, however, the resistance genes specifically recognize, directly or indirectly, the viral effectors, called avirulence (Avr) factors, activating ETI and conferring resistance. (B) The translational control arm of the NIK1-mediated signalling in antiviral innate immunity. Virus infection-induced oligomerization of NIK1 promotes transphosphorylation at the crucial Thr474, activating the kinase. Alternatively, NIK1 interacts with an unknown ligand-binding LRR-RLK in a stimulus-dependent manner. Although viral infection triggers NIK1-mediated antiviral signalling, the molecular basis of this elicitation is unknown and may be either intracellular virus-derived nucleic acid PAMPs or endogenous DAMPs released in the apoplasts by the host cells. Upon activation, NIK1 indirectly mediates the RPL10 phosphorylation, promoting its translocation to the nucleus, where it interacts with LIMYB to down-regulate the expression of translation-related genes. Therefore, the propagation of the antiviral signal culminates with suppression of host global protein synthesis, which also impairs translation of viral mRNA, as a defence mechanism. In begomovirus–host compatible interactions, the binding of begomovirus NSP to the NIK1 kinase domain (A-loop) inhibits autophosphorylation at Thr474, thereby preventing receptor kinase activation and RPL10 phosphorylation, overcoming this layer of defence. The viral single-stranded DNA replicates via double-stranded DNA intermediates that are transcribed in the nucleus of plant-infected cells. NSP binds to the nascent viral DNA and facilitates its movement to the cytoplasm and acts in concert with the classical movement protein MP to transport the viral DNA to the adjacent, uninfected cells.
The tobacco N gene was the first-identified R gene, which confers resistance against the Tobacco mosaic virus (TMV) (Whitham et al., 1994). Since then, many R genes involved in antiviral resistance in plants have been identified (Gururani et al., 2012; Mandadi and Scholtor, 2013), such as Sw-5 for Tomato spotted wilt virus (TSWV) in tomato (Brommonschenkel et al., 2000), Rx1 and Rx2 for Potato virus X (PVX) in potato (Bendahmane et al., 1999, 2000), RTM1 and RTM2 for Tobacco etch virus (TEV), RCY1 for Cucumber mosaic virus (CMV) in arabidopsis (Chisholm et al., 2000; Whitham et al., 2000; Takahashi et al., 2001) and the I locus for Bean common mosaic virus (Vallejo et al., 2006). A majority of the known R proteins belong either to the coiled-coil (CC)-NB-LRR or Toll/interleukin-1 receptor (TIR)-NBS-LRR class (Zhu et al., 2013; for a further review, see Gururani et al., 2012).
The Rx gene from potato, which encodes an NBS-LRR-type protein with a CC domain at the N-terminus (CC-NBS-LRR), may be the best-characterized resistance gene in plant–virus interactions (Bendahmane et al., 1999). The Rx N-terminal CC domain interacts intramolecularly with the Rx NB-LRR region and intermolecularly with the Rx cofactor RanGAP2 (Ran GTPase-activating protein 2) (Rairdan et al., 2008; Tameling et al., 2010). The C-terminus of the LRR domain is also thought to be involved in the specific recognition of the viral effector, which is functionally represented by the coat protein (CP), although a direct interaction between the CP and Rx has not been demonstrated (Bendahmane et al., 1995; Candresse et al., 2010; Dangl and Jones, 2001; Farnham and Baulcombe, 2006). The current mechanistic model for Rx function predicts that Rx is activated upon recognition of the Ran GTPase-mediated interaction with the CP.
Tobacco N protein represents a well-characterized example of the TIR-NBS-LRR class of R proteins in plant–virus interactions. The N resistance protein directly interacts with the helicase domain of the TMV replicase to trigger resistance (Ueda et al., 2006). Full resistance to TMV, however, depends on the N receptor-interacting protein 1 (NRIP1), which is recruited from the cytoplasm to the cytosol and nucleus to interact directly with both the N resistance protein and TMV replicase (Caplan et al., 2008). In both Rx-mediated resistance and N-mediated resistance, the R protein is activated in the cytoplasm, but full functionality of the Rx and N resistance proteins depends on their nucleocytoplasmic distribution. The R signalling cascade in plant–virus interactions consists of rapid activation of mitogen-activated protein kinases (MAPKs) and the involvement of molecular chaperone complexes controlling R protein stabilization and destabilization (Kadota and Shirasu, 2012; Hoser et al., 2013).
Generally, in plant–pathogen interactions, the immune responses downstream of R protein activation are associated with reactive oxygen species (ROS) production, calcium ion influx, MAPK activation, salicylic acid (SA) accumulation and massive transcriptional reprogramming, including the induction of genes associated with defence responses. Frequently, as in the case of N-mediated resistance, R protein activation also leads to the induction of a hypersensitive response (HR), which is often associated with programmed cell death of the infected and adjacent cells, confining the pathogen to the local site of infection. Concomitantly with the induction of the local defence response, R protein activation also activates defence signalling at distal tissues of infection, referred to as systemic acquired resistance (SAR), a defence mechanism shared by both Rx-mediated resistance and N-mediated resistance and induced by SA accumulation. More detailed information on SA signalling in defence is discussed in the hormone-mediated defence section.
Well-characterized exceptions to the NBS-LRR configuration of R proteins include the non-NBS-LRR-encoding RTM genes, which confer dominant resistance to TEV, Lettuce mosaic potyvirus (LMV) and Plum pox potyvirus (PPV) (Cosson et al., 2012), and the tomato Tm-1 gene, which encodes a protein with a TIM-barrel-like structure and confers dominant resistance to TMV. The Tm-1-encoded product interacts directly with the viral replicase, impairing viral genome replication (Ishibashi and Ishikawa, 2013).
Recessive resistance
In addition to dominant R genes, recessive R genes have also been reported, and most of them confer resistance against viruses (Kang et al., 2005). A compatible virus–host interaction leading to systemic infection requires replication of the virus genome in addition to cell to cell and long-distance movement through the plant vascular system. Disruption of any of these processes results in incompatible interactions, which is often mediated by host resistance factors. The recessive gene-encoded products are involved in compatibility functions; they are not immune receptors and are not associated with the ETI but rather act as essential factors required for the virus to complete its biological cycle. Therefore, many plant natural resistance genes have been mapped to mutations of essential host factors for virus infection. Examples of recessive resistance genes include eukaryotic translation initiation factors, such as eIF4E and eIF4G, which play an essential role in successful infection by potyviruses, bymoviruses, cucumoviruses, ipomoviruses, sobemoviruses, carmoviruses and waikiviruses, and thereby resistance is conferred by eIF4E and eIF4G loss-of-function mutations or modification of their gene products (Revers and Nicaise, 2014).
Antiviral immune receptors in PAMP-triggered immunity
The first layer of innate immunity is immediately activated upon host detection of highly conserved structural motifs exclusively expressed by pathogens, known as PAMPs, or endogenous danger signals released by the host during a wound or pathogenic attack known as damage-associated molecular patterns (DAMPs), which function as elicitors (Macho and Zipfel, 2014). The recognition of different PAMPs or DAMPs by specific cell surface sensors, designated PRRs, activates a sophisticated defence signalling cascade which inhibits a broad spectrum of potential pathogens, including bacteria, viruses, fungi and oomycetes. In plants, the PRRs are represented by receptor-like kinases (RLKs) and receptor-like proteins (RLPs) located at the cell surface. Both RLKs and RLPs often require a co-receptor to form an active complex to initiate signalling. The best-characterized co-receptor in PTI is the BRASSINOSTEROID INSENSITIVE1 (BRI1)-associated kinase 1, BAK1, which forms active signalling complexes with both RLKs and RLPs after PAMP detection by PRRs (Liebrand et al., 2014; Postma et al., 2016). BAK1 belongs to the LRR-RLK family and has an N-terminal extracellular LRR domain, which is structurally similar to mammalian Toll-like receptor (TLR) immune sensors, a transmembrane segment and an intracellular kinase domain. BAK1 heterodimerizes with several LRR-RLK immune sensors, including FLS2 (flagellin receptor), EFR (bacterial elongation factor-Tu receptor) and PEPR1 (damage-associated peptide 1 receptor), and is functionally required in immunity and signalling triggered by multiple bacterial PAMPs. The BAK1 positive regulation in plant immunity involves phosphorylation reactions between the BAK1 co-receptor and the corresponding PRR.
In the case of viral pathogens, the innate immune system has been primarily described in mammalian cells, which often detects specific biochemical features that are exclusive to the viral nucleic acid genome. Viral genomes exist as single- or double-stranded RNA or DNA and can be monopartite or partitioned into two or more segments. In mammalian cells, the TLRs comprise a large family of nucleic acid-sensing PRRs, which have relevant roles in antiviral defence. TLRs are similar to LRR-RLKs; they are single, membrane-spanning receptors with an LRR extracellular domain. Different members of the TLR family recognize different biochemical features present in viral, but not in host, nucleic acids, such as single-stranded RNA without a 5′ cap, double-stranded RNA (dsRNA) or unmethylated DNA. Specific recognition also relies on the opportunistic subcellular localization of TLRs and the viral genome in host cells. Although specific PRRs for viral recognition have not yet been found in plants, accumulated data indicate that plant PTI signalling inhibits viral infection similarly to non-viral pathogens. In fact, plant–virus interactions induce a complex set of typical PTI responses, including ROS production, ion fluxes, SA accumulation, defence gene activation, such as PR-1, and callose deposition (for a review, see Nicaise, 2014). In addition, upstream and downstream components of the PTI signalling pathway have been shown to play a role in antiviral defence. The functions of the PTI co-receptors BAK1 and BKK1 (BAK1-like kinase 1) are required to build an effective defence against RNA viruses in arabidopsis (Yang et al., 2010; Kørner et al., 2013), and MAPK4, a negative regulator of plant PTI signalling, suppresses soybean defence against Bean pod mottle virus (BPMV; Liu et al., 2011). Furthermore, the pre-activation of PTI by the elicitor chitosan, through interaction with chitin-binding PRRs, has also been shown to be effective against viruses (Iriti and Varoni, 2014). Finally, according to the zigzag evolutionary model of plant innate immunity (Jones and Dangl, 2006), the involvement and activation of ETI in plant–virus interactions is conceptually associated with successful PTI inhibition by a viral effector, further substantiating the argument that an antiviral PTI mechanism operates in plants as well (Fig. 1). Given the mode of virus delivery into plant cells and the obligatory conservative nature of PAMPs, which is not a property of rapidly evolving plant virus proteins, the molecular nature of the virus signatures recognized by plant PTI is very probably similar to those presented by mammalian viruses during infection. Therefore, the discovery of plant antiviral PRRs is expected to accelerate the characterization of nucleic acid-sensing PRRs and/or DAMP-sensing PRRs in plants.
Immune receptor-mediated suppression of translation: a new paradigm for antiviral defences in plants
NIK1 as an antiviral immune receptor.
The immune receptor NIK1 [nuclear shuttle protein (NSP)-interacting kinase 1], a RLK family member, has a remarkable role in the defence response against geminiviruses (Fontes et al., 2004). Although NIK1 shows structural similarities to BAK1, the mechanism for NIK-mediated antiviral defence is completely different from classical BAK1-mediated PTI (Machado et al., 2015).
NIKs (NIK1, NIK2 and NIK3) were first identified as targets of the NSP from Begomovirus, the largest genus of the Geminiviridae family (Fontes et. al., 2004). The NSP–NIK interaction is conserved among begomovirus NSPs and NIK homologues from different hosts (Mariano et al., 2004). NIK homologues from arabidopsis, tomato and soybean interact with NSPs from Cabbage leaf curl virus (CaLCuV) and from tomato-infecting begomoviruses, such as Tomato golden mosaic virus (TGMV), Tomato crinkle leaf yellow virus (TCrLYV) and Tomato yellow spot virus (ToYSV) (Fontes et al., 2004; Mariano et al., 2004; Sakamoto et al., 2012). These interactions suppress the NIK kinase activity and prevent the activation of the antiviral signal transduction pathway, creating a suitable environment for begomovirus infection (Santos et al., 2009, 2010). Consistent with a role for NIK in antiviral defence, loss-of-function nik1, nik2 and nik3 mutants showed enhanced susceptibility to CaLCuV infection (Fontes et al., 2004; Rocha et al., 2008; Santos et al., 2009). In addition, overexpression of NIK1 delays viral infection and attenuates symptom development in tomato (Carvalho et al., 2008). Finally, mutations in the activation loop (A-loop) of NIK1 that prevent its autophosphorylation also compromise the capacity of NIK1 to elicit a response against begomoviruses (Santos et al., 2009).
Mechanisms of NIK1 activation.
As a single-pass transmembrane receptor kinase, NIK is expected to dimerize or multimerize with itself and/or co-receptors to promote transphosphorylation and subsequent activation of the kinase. However, there is a complete lack of information on the critical early event that triggers NIK1 signalling and transduction, which culminates with suppression of host global translation as an antiviral response. Recently, a comparison between the transcriptomes induced by begomovirus infection and expression of a constitutively activated NIK1 receptor revealed that begomovirus infection is the activating stimulus of NIK1-mediated defence, although the molecular basis for this elicitation is still unknown. By comparison with the mechanism of mammalian antiviral immune receptor activation, one can predict that unique biochemical features of the begomovirus genome function as possible ligands that trigger or stabilize NIK dimerization or multimerization with a co-receptor. Begomoviruses are single-stranded DNA viruses, which replicate via double-stranded DNA intermediates in the nuclei of infected cells. The divergent transcription units of the viral genome result in single-stranded transcripts and double-stranded overlapping RNAs as possible sources for specific nucleic acid ligands. In mammals, the cytoplasmic receptor PKR (protein kinase receptor), which is activated by dsRNA molecules of > 40 bp, mediates global translation suppression by phosphorylating eIF2α on Ser51 as an antiviral response (Jackson et al., 2010). Alternatively or additionally, NIK1 activation may depend on host molecular signatures (DAMPs) released in the apoplast in response to viral infection.
Activation of many kinases requires phosphorylation of the activation segment (A-loop) that is defined by the region delimited by two conserved tripeptide motifs, DFG and APE (Nolen et al., 2004). This region is highly conserved among members of the LRR-RLK II subfamily and other members of the extended LRR-RLK family. The phosphorylation status of the activation segment has been shown to dictate NIK1 kinase activity (Carvalho et al., 2008; Fontes et al., 2004; Santos et al., 2009). NIK1 is phosphorylated in vitro at the conserved positions Thr474 and Thr469, and mutations in the A-loop compromise its autophosphorylation capacity (Santos et al., 2009). Replacement of Thr474 with alanine strongly inhibits the autophosphorylation activity and the capacity of NIK1 to elicit a defence response, whereas replacement of Thr474 with a phosphomimetic aspartate residue increases autophosphorylation activity and results in constitutive activation of a NIK1 mutant receptor that it is no longer inhibited by begomovirus NSP (Santos et al., 2009). These results indicate that phosphorylation at the essential Thr474 residue in the A-loop constitutes a key regulatory mechanism for NIK activation.
Although replacement of the essential Thr474 residue with the aspartate residue bypasses the NSP inhibitory effect on kinase activity, it does not impair NSP binding to an 80 amino acid stretch (positions 422–502) of NIK that encompasses the putative active site for Ser/Thr kinases (sub-domain VIb–HrDvKssNxLLD) and the activation loop (sub-domain VII–DFGAk/rx, plus sub-domain VIII–GtxGyiaPEY; Fontes et al., 2004). These results suggest that the NSP inhibitor acts upstream of the phosphorylation at position 474.
While phosphorylation at Thr474 is linked to an activation loop-dependent mechanism for NIK function, phosphorylation of Thr469 appears to have an autoinhibitory role (Santos et al., 2009). Replacing Thr469 with alanine relieves repression and enhances substrate phosphorylation. Furthermore, mutation at Thr469 does not inhibit autophosphorylation activity or impair the capacity of the mutant protein to elicit a defence response and to redirect the downstream component RPL10 to the nucleus. It has been proposed that autophosphorylation of Thr469 within the NIK1 A-loop allows the kinase to control the sustained signalling more efficiently. Whether this inhibitory mechanism allows NIK1 to phosphorylate pathway components differentially remains to be determined.
Downstream components of the NIK-mediated antiviral response.
A ribosomal protein, RPL10, identified as a binding partner for NIKs, acts as a downstream effector of the NIK-mediated antiviral response. Arabidopsis rpl10 mutants showed enhanced susceptibility to geminivirus infection, recapitulating the nik1 phenotype (Rocha et al., 2008). Ectopic expression of NIK1 or a hyperactive NIK1 mutant led to relocation of phosphorylated RPL10A from the cytosol to the nuclei (Carvalho et al., 2008). In addition, an inactive NIK1 mutant failed to redirect the protein to the nuclei of co-transfected cells, while a mutant RPL10A defective for NIK1 phosphorylation is not redirected to the nucleus and does not mount a defence response against begomoviruses. These data suggest that the nucleocytoplasmic shuttling of RPL10 is regulated by phosphorylation and is dependent on the kinase activity of NIK1, classifying RPL10 as a downstream effector of NIK1-mediated signalling.
Although RPL10 binds to NIK1 in vitro and in vivo, it is not efficiently phosphorylated by NIK1 in vitro and may not serve as a direct NIK1 substrate in vivo. Nevertheless, the nucleocytoplasmic shuttling of RPL10 is regulated by phosphorylation and is dependent on the kinase activity of NIK1. In fact, NIK1 does not relocate a phosphorylation-deficient mutant of RPL10 to the nucleus (Carvalho et al., 2008). Furthermore, the gain-of-function T474D mutant is more effective at redirecting RPL10 to the nucleus, and inactive mutants of NIK1 fail to alter the cytosolic localization of RPL10 (Santos et al., 2009). Mutations in the A-loop similarly affect the capacity of NIK1 to elicit an antiviral response and to mediate the phosphorylation-dependent nuclear relocalization of RPL10.
In order to gain new insights into the molecular mechanisms of NIK1 in antiviral immunity, arabidopsis transgenic lines harbouring the gain-of-function mutant T474D on a nik1 knockout background were analysed for gene expression (Zorzatto et al., 2015). The constitutive activation of NIK-mediated signalling resulted in the down-regulation of translation-related genes and the suppression of global translation, decreasing the loading of host mRNAs in actively translating polysomes (Zorzatto et al., 2015). In begomovirus-infected lines, the association of viral mRNA with actively translating polysomes was lower in T474D lines than in the wild type, indicating that the begomovirus is not capable of sustaining high levels of viral mRNA translation when global host translation is impaired. Accordingly, the transgenic lines ectopically expressing T474D displayed enhanced resistance to begomovirus, demonstrating that suppression of global protein synthesis may effectively protect plant cells against DNA viruses.
Further analyses detected LIMYB, an RPL10-interacting MYB domain-containing transcriptional factor, as another downstream component of the NIK1-mediated antiviral pathway (Zorzatto et al., 2015). LIMYB binds to and acts in concert with RPL10 to repress fully the expression of ribosomal gene expression. LIMYB overexpression represses ribosomal protein (RP) genes at the transcriptional level, resulting in protein synthesis inhibition, decreased viral mRNA association with polysome fractions and enhanced tolerance to the begomovirus CaLCuV. In contrast, loss of LIMYB function releases repression of RP genes and recapitulates the enhanced susceptibility phenotype of the nik1 null alleles. T474D also downregulates the expression of the same sub-set of LIMYB-regulated RP genes but requires LIMYB to repress RP gene expression. Therefore, LIMYB is a downstream transcriptional repressor in the NIK1-mediated pathway, which links NIK1 activation to the downregulation of translational machinery-related genes, thereby suppressing global host translation as an antiviral immunity strategy in plants.
NIK1-mediated translation suppression may act as a conserved antiviral mechanism in begomovirus–host interactions. Tomato T474D transgenic lines were tolerant to the tomato-infecting begomoviruses ToYSV and ToSRV (Tomato severe rugose virus), which display highly divergent genomic sequences and hence are only distantly related within the group of tomato-infecting begomoviruses (Brustolini et al., 2015). As in arabidopsis-infected T474D lines, overexpression of T474D in tomato represses RP genes, suppresses global protein synthesis and decreases viral mRNA association with the polysome fractions (Brustolini et al., 2015). Therefore, the enhanced tolerance to tomato-infecting begomovirus displayed by the T474D-expressing lines is associated with the translational control branch of the NIK-mediated antiviral responses. These observations underscore the potential of a sustained NIK-mediated defence pathway to confer broad-spectrum tolerance to begomoviruses in distinct plant species. However, whether NIK-mediated suppression of global translation functions against plant RNA viruses it is still a matter of debate.
Mechanistic model for the NIK1-mediated antiviral signalling pathway.
Since the discovery of NIKs, several features of the NIK1-mediated antiviral signalling and its interaction with the begomovirus NSP have been elucidated (Fig. 1). We now know that the transmembrane receptor NIK1, a serine/threonine kinase transducer, is activated by viral infection to trigger a defence response against the virus itself, although the molecular basis for this elicitation remains unknown. Based on common features of the LRR-RLKII family, we propose that the extracellular domain of NIK undergoes oligomerization with itself or with an unidentified ligand-dependent LRR-RLK receptor following viral infection. The ligand may be DAMPs delivered into the apoplast by the secretory apparatus upon detection of viral infection. Alternatively, NIK1 may recognize virus-derived nucleic acids as PAMPs that promote oligomerization of the antiviral immune receptor. Regulation of NIK kinase activity depends on a conformational change of the A-loop induced by phosphorylation of Thr474. Activated NIK regulates the nucleocytoplasmic trafficking of RPL10, which in turn interacts with the transcriptional repressor LIMYB to downregulate RP genes, leading to suppression of host and viral mRNA translation, thereby linking the antiviral response to receptor activation.
Nuclear shuttle protein prevents activation of the pathway by binding to the NIK kinase domain and sterically interfering with phosphorylation of Thr474 in the A-loop. As a consequence, phosphorylation of RPL10 is impaired, and the RP is trapped in the cytoplasm during begomovirus infection. NSP inhibition of NIK1 prevents activation of the NIK-mediated signalling pathway, resulting in an intracellular environment that is more favourable for viral proliferation and spread. The viral single-stranded DNA replicates via double-stranded DNA intermediates that are transcribed in the nucleus of plant-infected cells (Hanley-Bowdoin et al., 2013). NSP binds to the nascent viral DNA and facilitates its movement to the cytoplasm, acting in concert with the classical movement protein MP to transport the viral DNA to the adjacent, uninfected cells.
RNA SILENCING MACHINERY: AN ADAPTIVE ANTIVIRAL IMMUNITY MECHANISM
The RNA silencing pathway or RNA interference (RNAi) is a well-established natural antiviral defence mechanism in plants, in which the viruses are both inducers and targets of RNA silencing (Wang et al., 2010; Szittya et al., 2013). To inhibit RNA silencing, well-adapted plant viruses are known to encode silencing-suppressor proteins, which can counteract the host silencing-based antiviral process (Wieczorek and Obrepalska-Steplowska, 2015). In this review, we summarize the conceptual advances in the antiviral RNAi mechanism and the evolving virulence strategies to overcome this adaptive plant defence (Fig. 2). For more detailed information, a collection of excellent, updated reviews describing antiviral RNA silencing mechanisms and suppressors is available (Carbonell and Carrington, 2015; Csorba et al, 2015; Zhang et al., 2015).
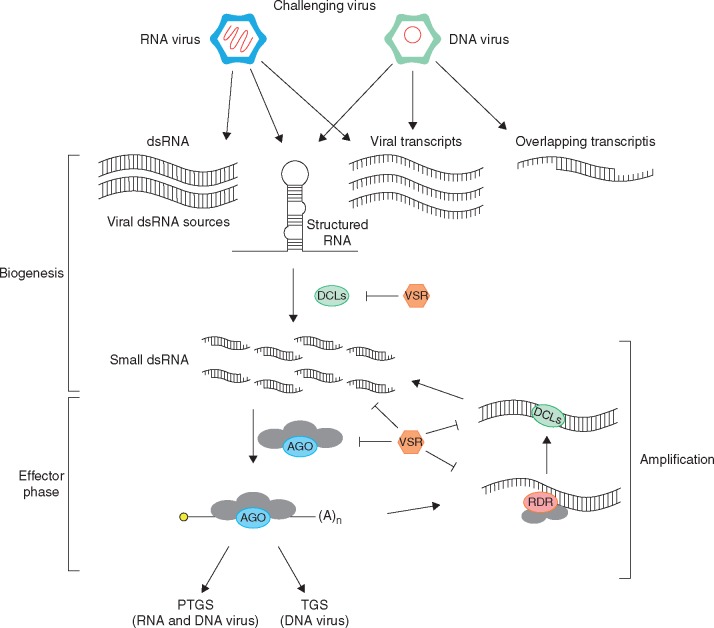
Fig. 2.
Adaptive antiviral immunity in plants: general model of antiviral RNA silencing and its suppression by viral suppressors of RNA silencing (VSRs). The Silencing response is triggered by viral dsRNA molecules (vsRNA, ds-siRNA, 21, 22 rr 24 nt) from different sources, which are produced by Dicer-like proteins (DCLs). These vsRNAs are subsequently loaded into Argonaute (AGO)-containing silencing complexes. In post-transcriptional gene silencing (PTGS), viral RNA is targeted by the RNA-induced silencing complex (RISC) for degradation or translational repression, while the RNA-induced transcriptional silencing complex (RITS) causes histone and/or DNA methylation, leading to transcriptional gene silencing (TGS). The effector phase can also result in the amplification of silencing response through the action of RNA-dependent RNA polymerase (RDR) proteins, which produce more dsRNA substrates for DCL processing. VSRs can target multiple steps of the RNA silencing pathway, defeating host antiviral mechanisms by interfering in dicing, vsRNA loading, AGO activation and amplification.
The diversity in RNAi mechanisms relies mainly on the existence of multiple copies of AGO (Argonaute), RNA-dependent RNA polymerase (RDR), DRB (double-stranded RNA binding) and DCL (Dicer-like) genes, which probably result from gene duplication followed by specialization (Parent et al., 2015; Zhang et al., 2015). DCL2 and DCL4 have a crucial role in antiviral defence (Deleris et al. 2006; Qu et al., 2008; Garcia-Ruiz et al., 2010). Arabidopsis plants containing loss-of-function mutations within the Dicer-like 2 (DCL2), Argonaute 2 (AGO2) and HEN1 RNA methyltransferase were more susceptible to Turnip crinkle virus (TCV) infection (Zhang et al., 2012). Arabidopsis dlc4 mutants inoculated with TCV lacking P38 (silencing suppressor) exhibited large primary lesions, but viral systemic movement was compromised. However, viral infection was fully established in dcl2–dcl4 double mutants (Deleris et al., 2006). Recently, Andika et al. (2015) demonstrated the differential requirement for the DCL4 and DCL2 proteins in the inhibition of intracellular and systemic infection, respectively, by PVX in arabidopsis, which highlights the host’s ability to fight against both local and systemic viral infection.
Although DCL3 has a minor role against RNA viruses, it is crucial against DNA viruses (Qu et al., 2008; Csorba et al., 2015). Arabidopsis dcl3 mutants are unable to recover from geminivirus infection, while remission was observed in wild-type, dcl2 and dcl4 plants (Raja et al., 2014). Plants employ RNA-directed DNA methylation (RdDM) as an epigenetic defence against geminiviruses (Raja et al., 2008, 2014; Ruiz-Ferrer and Voinnet, 2009; Hanley-Bowdoin et al., 2013). Arabidopsis methylation-deficient mutants are hypersusceptible to geminivirus infection. Additionally, cytosine methylation levels are significantly reduced in viral DNA isolated from methylation-deficient mutants (Raja et al., 2008). DCLs interact with DRBs to produce small RNAs. The DRB3 protein functions with Dicer-like 3 (DCL3) and Argonaute 4 (AGO4) in methylation-mediated antiviral defence (Raja et al., 2014). In turn, some DNA viruses can suppress silencing by interfering with the methyl cycle. A silencing suppressor from begomovirus, AC2, inhibits host adenosine kinase (ADK) activity, which is required for RNA silencing (Wang et al., 2003, 2005). The AL2-mediated silencing suppression was followed by reduced cytosine methylation (Buchmann et al., 2009). The betasatellite-encoded protein, βC1, from Tomato yellow leaf curls China virus (TYLCCNV) also targets the methyl cycle through inhibition of S-adenosylhomocysteine hydrolase (SAHH) activity (Yang et al., 2011). Geminiviruses also employ an alternative mechanism to interfere with the host DNA methylation machinery during the infection by reducing the transcript levels of Methyltransferase 1 (MET1) and Chromomethylase 3 (CMT3), key enzymes of the plant methylation cycle (Rodriguez-Negrete et al., 2013). The replicase-associated protein (Rep) is responsible for the repression of MET1 and CMT3, and another viral protein, C4, has an auxiliary role in MET1 down-regulation (Rodriguez-Negrete et al., 2013).
The AGO proteins are essential in antiviral defence against both RNA and DNA viruses. AGO1, AGO2, AGO4, AGO5, AGO7 and AGO10 have been shown to display antiviral activity in arabidopsis, while AGO1 and AGO18 play antiviral defence roles in rice (reviewed in Carbonell and Carrington, 2015). RDR activities contribute to the amplification of antiviral activity. RDR1 and RDR6 play an essential role in the amplification of virus-derived small interfering RNAs (siRNAs; Wang et al., 2010). The biogenesis of Tobacco rattle virus (TRV)-derived siRNAs involves the combined activity of RDR1, RDR2 and RDR6 (Donaire et al., 2008). DCL4 and RDR1 are major contributors to biogenesis of Turnip mosaic virus (TuMV)-derived siRNAs, although a full antiviral defence also requires DCL2 and RDR6 (Gacia-Ruiz et al., 2010). OsRDR6 knockdown transgenic rice show hypersusceptibility to Rice stripe virus (RSV). These phenotypes are associated with increased accumulation of RSV genomic RNA and reduced RSV-derived siRNA accumulation compared with the wild-type plants (Jiang et al., 2012). Hong et al. (2015) also reported an increase in susceptibility to Rice dwarf phytoreovirus (RDV) in OsRDR6 downregulated rice followed by a reduction in the RDV vsiRNA levels. However, overexpression of OsRDR6 had no effect on RDV infection.
Many viral suppressor proteins can target multiple steps of the RNA silencing pathway to defeat host antiviral mechanisms. One strategy used by viral suppressors is impairment of viral siRNA biogenesis by inhibiting DCL proteins and/or the activity of cofactors, sequestering dsRNA/siRNA or promoting AGO protein destabilization prior to RISC assembly (reviewed in Csorba et al., 2015). The p22 suppressor of Tomato chlorosis virus (ToCV) binds long dsRNAs in vitro, preventing them from being cleaved by an RNase III-type Dicer homologue, which might block the silencing process by interfering with the generation of siRNAs (Landeo-Rios et al., 2015). The silencing suppressor of Lettuce necrotic yellows virus (LNYV), phosphoprotein P, targets multiple proteins involved in the RNA silencing pathway, including those involved in the RISC complex and dsRNA amplification. LNYV P impairs RNA silencing through inhibition of micro RNA (miRNA)-guided AGO1 cleavage and translational repression and also compromises RDR6/SGS3-dependent amplification of silencing (Mann et al., 2016). One of the best-characterized suppressors of antiviral RNA silencing is the potyviral helper component proteinase (HCPro), which plays multiple roles in the suppression of vsiRNA biogenesis, such as ds–siRNA binding, HEN1 binding, blocking HEN1 methyltransferase activity, blocking primary siRNA biogenesis by RAV2 interaction and downregulating RDR6 (Zhang et al., 2015). A recent study suggested two mechanisms by which HCPro exerts its RNA silencing suppressor functions (Ivanov et al., 2016). HCPro may block siRNA methylation of HEN1 via inhibition of S-adenosyl-l-methionine synthase (SAMS) and SAHH, two key enzymes of the methionine cycle. HCPro may also attenuate viral RNA translational repression through association with AGO1 and ribosomes.
Over the past decades, significant advances have been made in the current understanding of the role of RNA silencing in plant antiviral immunity responses. Concomitantly, diverse mechanisms employed by viruses to avoid silencing-mediated resistance have been unravelled, most of them through silencing suppressor activities. Additionally, there are reports that plants have evolved specific defences against RNA silencing suppression. Collectively, these findings provide new insight into the molecular mechanisms mediating plant–virus interactions, and they concomitantly highlight a complex and lasting arms race between pathogens and their hosts.
HORMONE-MEDIATED ANTIVIRAL DEFENCES
Plant hormones play important roles in intercellular and systemic signalling systems, regulating developmental processes and plant responses to a wide range of biotic and abiotic stresses (Bari and Jones, 2009). In susceptible hosts, plant viruses often manipulate biochemical events and molecular interactions required for their replication and movement, leading to misregulation and disruption of hormone signalling (Alazem and Lin, 2015).
Salicylic acid is a key component of the plant response to pathogens and is involved in the establishment of local and systemic resistance (Vlot et al., 2009; Pieterse et al., 2012). The role of SA in viral defence was initially reported in the interaction between the TMV and the tobacco N resistance gene (Gaffney et al., 1993; Jovel et al., 2011). Tobacco transgenic lines deficient in SA accumulation were defective in their ability to induce SAR against TMV and inefficiently restricted virus movement (Gaffney et al., 1993). The SA pathway is typically activated by both DNA and RNA viruses (Whitham et al., 2006; Ascencio-Ibanez et al., 2008). Arabidopsis cpr1 (constitutive expresser of PR genes) mutants, in which SA-mediated SAR is constitutively activated, were less susceptible to CaLCuV infection (Bowling et al., 1994). Additionally, the arabidopsis mutant lsb1 (less susceptible to BSCTV 1) showed impairment in Beet severe curly top virus (BSCTV) DNA replication and reduced infectivity (Chen et al., 2010). Previous studies showed that upregulation of LSB1/GDU3 affects geminivirus infection by activating the SA pathway (Chen et al., 2010).
The SA defence response is also triggered by Potato virus Y (PVY) and Tomato ringspot virus (ToRSV) (Jovel et al., 2011; Baebler et al., 2014). The lack of SA accumulation in the NahG potato plants (transgenic lines deficient in SA accumulation) causes unrestricted viral spreading and consequent disease symptoms (Baebler et al, 2014). Transcriptomic analysis confirmed the central role of SA in inducing the Ny-1-mediated responses and showed that the absence of SA leads to significant changes at the gene expression level, including a delay in activation of defence genes. In a similar manner, SA-dependent mechanisms were implicated in the restriction of ToRSV spread in tobacco. Lesion size and viral systemic spread were reduced with SA pre-treatment but enhanced in NahG transgenic lines deficient in SA accumulation, (Jovel et al., 2011). The eds5 (enhanced disease susceptibility 5) mutation and the NahG transgene partially defeated the resistance of Col-24-C to Cucumber mosaic virus strain-Y (CM-Y) (Takahashi et al., 2004).
Plum pox virus (PPV) replication is restricted to inoculated leaves in tobacco plants, but the virus is able to infect P1/HC-Pro-expressing plants systemically (Alamillo et al., 2006). Interestingly, PPV was also able to move systemically in NahG-expressing tobacco plants. Further analysis revealed reduced accumulation of viral-derived small RNAs in the NahG transgenic plants and enhanced expression of SA-mediated defence transcripts, such as those of pathogenesis-related (PR) proteins PR-1 and PR-2, alternative oxidase-1 and the putative RNA-dependent RNA polymerase NtRDR1, in response to PPV infection, suggesting that SA might act as an enhancer of RNA silencing in tobacco. SA treatments also induced resistance against TMV and activated the RNA silencing-related genes DCL1, DCL2, RDR1 and RDR2 in tomato plants (Campos et al., 2014).
The role of jasmonic acid (JA) signalling in virus defence is controversial. Genes involved in the JA pathway are generally suppressed during geminivirus infection (Ascencio-Ibanez et al., 2008). The viral pathogenesis factor βC1 from TYLCCNV attenuates expression of several JA-responsive genes (Yang et al., 2008). In contrast, multiple genes related to JA signalling were upregulated in transgenic tobacco plants expressing the viral silencing suppressor AC2 derived from African cassava mosaic virus (Soitamo et al., 2012). The AC2 protein also interacts with CSN5a, a COP9 signalosome component, interfering with the derubylation activity of the CSN complex and disturbing several cellular processes, including jasmonate responses (Lozano-Duran et al., 2011). Exogenous jasmonate treatment of A. thaliana plants disrupts geminivirus infection, suggesting that the suppression of the jasmonate response might be crucial for infection (Lozano-Duran et al., 2011). In contrast, exogenously applied methyl jasmonate (MeJA) reduced local resistance to TMV and permitted systemic viral movement in Nicotiana tabacum (tobacco) cultivars while the silencing of CORONATINE-INSENSITIVE 1 (COI1), a JA receptor, reduced viral accumulation in a tobacco cultivar possessing the N gene, as did that of allene oxide synthase, a JA biosynthetic enzyme (Oka et al., 2013).
Brassinosteroids (BRs) have also been identified as a plant defence inducer against viruses (Nakashita et al., 2003). Wild-type tobacco treated with brassinolide (BL) exhibited enhanced resistance to TMV. BL-treated tobacco plants did not show SA accumulation or induction of PR gene expression, suggesting that BL-induced resistance is distinct from SAR (Nakashita et al., 2003). Geminiviruses also interact with the BR signalling pathway. Viral C4 (or AC4 in some viruses) interacts with BRASSINOSTEROID-INSENSITIVE 2 (BIN2), which is a negative regulator of BR signalling (Piroux et al., 2007). Although the functional relevance of this interaction remains to be investigated, ectopic expression of the BCTV C4 protein in A. thaliana drastically alters plant development, possibly through the disruption of multiple hormonal pathways (Mills-Lujan and Deom, 2010). A BR receptor, the LRR-RLK brassinosteroid insensitive-1 (BRI1), and PRRs interact with the co-receptor BAK1 in a ligand-dependent manner. BAK1 was also found to be essential for plant basal immunity during compatible interactions with RNA viruses. For example, TCV, ORMV and TMV accumulated to higher levels in the bak1-4 and bak1-5 mutants than in wild-type plants (Korner et al., 2013).
Previous studies showed that the ethylene (ET) pathway might play an important role in antiviral defence (Fischer and Dröge-Laser, 2004; Love et al., 2005, 2007). Overexpression of NtERF5, an ET-responsive transcription factor, conferred enhanced resistance to TMV infection, showing reduced size of local HR lesions and impaired systemic spreading of the virus (Fischer and Dröge-Laser, 2004). Mutations in ET signalling were also reported to alter plant susceptibility to viruses. Two arabidopsis ET signalling mutants, etr1 and ein2, showed reduced susceptibility to Cauliflower mosaic virus (CaMV) infection (Love et al., 2005, 2007). The transcription factor WRKY8, which mediates the ET signalling pathway, is involved in the response against TMV-cg (crucifer-infecting Tobacco mosaic virus) (Chen et al., 2013). In wrky8 mutants, several ET-synthesized or responsive transcription factors, such as ACS6 and ERF104, were more strongly induced in TMV-cg systemically infected leaves. Functional analysis using mutants showed that the acs6, erf104 and ein2 mutants had reduced accumulation of TMV-cg RNA in systemically infected leaves compared with the wild type, indicating an important role for ET in anti-TMV-cg defence. The ET signalling pathway was also correlated with TuMV-initiated suppression of defence responses and enhanced aphid reproduction in plants (Casteel et al., 2015). Transgenic expression of Nia-Pro (nuclear inclusion a-protease domain) in arabidopsis alters ethylene responses and suppresses aphid-induced callose formation in an ET-dependent manner.
Abscisic acid (ABA) plays a key role in modulating plant responses to different biotic and abiotic stresses. Although the involvement of ABA in biotic stress has been studied extensively, the roles of ABA in viral replication and movement are not well characterized (Alazem et al., 2014, 2015). Previous studies suggested virus-induced changes in ABA metabolism during infection (Whenham et al., 1986; Fraser and Whenham, 1989). Tomato plants harbouring the Tm-1 gene for resistance to TMV contain higher concentrations of ABA than susceptible plants (Fraser and Whenham, 1989). Exogenous applications of ABA reduced the systemic accumulation of TMV-cg. Mutations in ABA deficient 1, ABA deficient 2, ABA deficient 3 or abi4 accelerated systemic TMV-cg accumulation in arabidopsis (Chen et al., 2013). ABA2 has also been shown to play a role in the accumulation of Bamboo mosaic potexvirus (BaMV) and CMV (Alazem et al., 2014). Mutants downstream of ABA2 (aao3, abi1-1, abi3-1 and abi4-1) were susceptible to BaMV. The aba2-1 mutant showed decreased accumulation of BaMV (+)RNA, (–)RNA and coat protein, with the most dramatic effect being observed for (–)RNA. ABA is also involved in the increase in callose deposition on plasmodesmata by inhibiting β-1,3-glucanase transcription, which may restrict cell to cell movement of the virus and enhance resistance (Beffa et al., 1996; Rezzonico et al., 1998; Mauch-Mani and Mauch, 2005).
Viral infections may also disturb auxin, cytokinin and gibberellin signalling pathways. The replicase protein of TMV interacts with the related Aux/IAA proteins in arabidopsis and tomato, leading to modifications in auxin-mediated gene regulation and disease development (Padmanabhan et al., 2005, 2008). The geminivirus South African cassava mosaic virus [ZA:99] activated expression of auxin-inducible genes in arabidopsis (Pierce and Rey, 2013). In a similar manner, the geminivirus AC2/AL2 protein interacts with an ADK in arabidopsis, leading to increased expression of primary cytokinin-responsive genes (Baliji et al., 2010). Gibberellic acid may have a defence role against biotrophic or necrotrophic pathogens via modulation of the balance between SA- and JA/ET-mediated signalling pathways (Robert-Seilaniantz et al., 2007; Alazem and Lin, 2015). The P2 protein of the Rice dwarf virus (RDV) interacts with ent-kaurene oxidase in vivo, a key factor in the biosynthesis of gibberellins, leading to a dwarf phenotype in rice, which was rescued after exogenous application of GA3 (Zhu et al., 2005).
It is quite clear that plant hormones play a critical role in many aspects of plant biology, including development and pathogen defence. During viral infection, symptoms and viral accumulation have been correlated with disturbance in phytohormone levels. Despite advances in the knowledge of hormone-mediated antiviral functions, there are still many questions to be answered, including how cross-talk between hormone pathways modulates the host defence response to impair viral infection.
PROTEASOME DEGRADATION
The UPS plays a central role in a wide range of fundamental plant processes, including degradation and functional modification of cellular proteins, and signalling in response to abiotic and biotic stimuli (Sadanandom et al., 2012; Luo, 2016). In the context of virus–plant interactions, the UPS is targeted by many viruses to maintain suitable levels of viral proteins and to achieve a successful infection. However, the UPS also acts as a host defence mechanism to eliminate viral components (Alcaide-Loridan and Jupin, 2012). Several interactions between viral proteins and components of the ubiquitin and ubiquitin-like protein pathways have been reported. The helper component proteases (HcPro) of Lettuce mosaic virus (LMV) and PVY were reported to interact directly with different subunits of the 20S proteasome (Jin et al., 2007; Dielen et al., 2011). The HcPro from Papaya ringspot virus (PRSV) interacts with the papaya homologue of arabidopsis PAA (a1 subunit of the 20S proteasome), and inhibition of the proteasome increased the accumulation of PRSV in papaya and accelerated development of symptoms and viral RNA accumulation (Sahana et al., 2012). Transgenic tobacco expressing the geminivirus protein βC1 displayed a reduction in polyubiquitination activity, probably due to interaction between βC1 from Cotton leaf curl Multan virus (CLCuMV) and the host ubiquitin-conjugating (UBC) enzyme, SlUBC3, leading to disruption of the UPS (Eini et al., 2009). Some viruses depend on interactions with the ubiquitin pathway to achieve a successful infection. The geminivirus BSCTV encodes the protein C2, a transcriptional activator, which binds to S-adenosylmethionine decarboxylase 1 (SAMDC1) (Zhang et al., 2011). This study suggests that BSCTV C2 attenuates the 26S proteasome-mediated degradation of SAMDC1 to establish a hypomethylated environment to facilitate viral accumulation. The Turnip yellow mosaic virus (TYMV) RNA-dependent RNA polymerase (66K) is degraded by the UPS in infected cells, which compromises viral infectivity (Camborde et al., 2010). The virus, in turn, makes use of a viral deubiquitinating enzyme (DUB) to stabilize RdRp and contribute positively to infection (Chenon et al., 2012).
Plant viruses also use UPS processes to promote virulence. The downregulation of RPM9, a 26S proteasome subunit, inhibits the systemic spread of TMV and TUMV in Nicotiana benthamiana (Jin et al., 2006). The viral replication protein (Rep) from geminiviruses binds to host SUMO-conjugating enzyme 1 (SCE1), which is required for viral infection (Castillo et al., 2004; Sanchez-Duran et al., 2011). Geminiviruses also interfere with the activity of the COP9 signalosome complex through interaction of the viral protein C2 and the host CSN5 protein, compromising many cellular processes regulated by the CUL1-based SCF ubiquitin E3 ligases (Lozano-Duran et al, 2011). In the case of the tombusvirus Tomato bushy stunt virus (TBSV), a Cdc34p E2 UBC enzyme has been identified to interact with the TBSV p33 replication protein, promoting its ubiquitination (Li et al., 2008). Downregulation of Cdc34p compromises tombusvirus replicase activity (Li et al., 2008).
Geminiviruses alter the cell cycle of infected host cells to create a suitable environment for viral replication (Hanley-Bowdoin et al., 2013). The expression of the pathogenicity factor C4 from Beet severe curly top virus (BSCTV) affects the cell cycle in arabidopsis, leading to abnormal cell divisions, and induces a host RING finger protein (RKP), which targets cyclin kinase inhibitors for proteasomal degradation. Mutations in RKP reduced the susceptibility to BSCTV and impaired BSCTV replication in plant cells (Lai et al., 2009). Some viruses may also induce host protein degradation to defeat the RNA silencing pathway. The polerovirus silencing suppressor P0 from Beet western yellows virus (BWYV) targets AGO1 for degradation in a still unknown proteasome-insensitive mechanism (Baumberger et al., 2007).
In summary, the UPS involvement in plant defence mechanisms occurs at different levels, from ubiquitin to the 26S proteasome (Dielen et al., 2011). Viruses hijack the host UPS to control the quality of their own proteins and to enhance effectiveness. Concomitantly, plants use this pathway as another layer of resistance, mainly targeting viral proteins for degradation. Over the past decades, many reports have revealed a complex network involving host UPS components and viral proteins from several different groups of plant viruses, which suggests that perturbation of the Ub pathway might be a conserved mechanism in virus–host interactions.
CONCLUSIONS
In response to viral infection, plants activate a multilayered defence response, including immune receptor signalling, RNA silencing, hormone-mediated defence pathways and protein degradation. Viruses, however, can subvert the plant’s defence signalling by suppressing the host immune system and/or manipulating the host defence signalling network to their own benefit by affecting hormone signalling or the proteasome degradation pathway. Therefore, a better understanding of plant–virus interaction dynamics is crucial if we are to use the plant immune system rationally and more effectively to control viral infections.
In spite of the significant advances in our knowledge of the antiviral immunity in plants made in the last decade, several questions about the dynamics between the virulence strategy of the viruses and the plant immune system remain. For example, we do not know the identities of the virus-derived PAMPs or plant-derived DAMPs that induce antiviral PTI and the viral effectors that suppress it. Furthermore, antiviral PRRs have not been identified. A better understanding of the repertoire of virus effectors (Avr factor) and the NBS-LRR host targets (R proteins) and their mode of action in activating ETI and/or suppressing PTI will help to define the evolutionary pressure acting upon the host and viruses and determine how to deploy the immune system towards a more efficient control of virus infection. We also need to define the NIK1-mediated suppression of translation as a general or virus-specific antiviral strategy in plants. So far, a sustained NIK1 pathway has been shown to be effective against begomoviruses, one of the largest groups of plant DNA viruses, which cannot circumvent the regulatory mechanism of host translation. Although plant RNA viruses have developed a variety of non-canonical mechanisms to translate their RNAs and overcome the regulatory mechanism of host translation, they interact tightly with the host protein synthesis machinery such that host translation initiation factor-encoded genes can function as recessive resistance genes. Furthermore, the translational repression activity of the effector AGO has been recently demonstrated to play a role in the antiviral RNA silencing mechanism. These examples support the argument that hindering the translation of viral mRNA (globally or specifically) is a promising avenue for virus control. Nevertheless, an emerging theme in the plant immunity scenario is that RNA silencing is connected to the other plant defence layers controlling and co-ordinating protein-based innate immunity and SAR into a more robust defence. Therefore, strategies for integration of different plant defence layers (innate immunity, SAR and RNAi) in a co-ordinated manner are expected to ensure a robust and more durable defence response against plant viruses for crop protection.
ACKNOWLEDGEMENTS
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico [573600/2008-2 and 447578/2014-6 to E.P.B.F.] and the Fundação de Amparo à Pesquisa do Estado de Minas Gerais [CBB-APQ-03175-13 and CBB-APQ-01491-14 to E.P.B.F.]. I.P.C. was supported by a CAPES graduate fellowship.
LITERATURE CITED
- Alazem M, Lin KY, Lin NS.. 2014. The abscisic acid pathway has multifaceted effects on the accumulation of bamboo mosaic virus. Molecular Plant-Microbe Interactions 27: 177–189. [DOI] [PubMed] [Google Scholar]
- Alazem M, Lin NS.. 2015. Roles of plant hormones in the regulation of host–virus interactions. Molecular Plant Pathology 16: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide-Loridan C, Jupin I.. 2012. Ubiquitin and plant viruses, let’s play together! Plant Physiology 160: 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamillo JM, Saenz P, Garcia JA.. 2006. Salicylic acid-mediated and RNA-silencing defence mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. The Plant Journal 48: 217–227. [DOI] [PubMed] [Google Scholar]
- Andika IB, Maruyama K, Sun L, Kondo H., Tamada T, Suzuki N.. 2015. Different Dicer-like protein components required for intracellular and systemic antiviral silencing in Arabidopsis thaliana. Plant Signaling and Behavior 10: e1039214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibanez JT, Sozzani R, Lee TJ, et al. 2008. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiology 148: 436–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler S, Witek K, Petek M, et al. 2014. Salicylic acid is an indispensable component of the Ny-1 resistance gene-mediated response against Potato virus Y infection in potato. Journal of Experimental Botany 65: 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliji S, Lacatus G, Sunter G.. 2010. The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 402: 238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JDG.. 2009. Role of plant hormones in plant defence responses. Plant Molecular Biology 69: 473–488. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC.. 2007. The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Current Biology 17: 1609–1614. [DOI] [PubMed] [Google Scholar]
- Beffa RS, Hofer RM, Thomas M, Meins F Jr.. 1996. Decreased susceptibility to virus disease of β-1,3-glucanase-deficient plants generated by antisense transformation. The Plant Cell 8: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Kohm BA, Dedi C, Baulcombe DC.. 1995. The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. The Plant Journal 8: 933–941. [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC.. 1999. The Rx gene from potato controls separate virus resistance and cell death responses. The Plant Cell 11: 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Querci M, Kanyuka K, Baulcombe DC.. 2000. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. The Plant Journal 21: 73–81. [DOI] [PubMed] [Google Scholar]
- Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T.. 2014. Immune receptor complexes at the plant cell surface. Current Opinion in Plant Biology 20: 7–54. [DOI] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O.. 2014. The diversity, biogenesis and activities of endogenous silencing small RNAs in Arabidopsis. Annual Review Plant Biology 65: 473–503. [DOI] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X.. 1994. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. The Plant Cell 6: 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brommonschenkel SH, Frary A, Frary A, Tanksley SD.. 2000. The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Molecular Plant-Microbe Interactions 13: 1130–1138. [DOI] [PubMed] [Google Scholar]
- Brustolini OJB, Machado JPB, Condori-Apfata JA, et al. 2015. Sustained NIK-mediated antiviral signalling confers broadspectrum tolerance to begomoviruses in cultivated plants. Plant Biotechnology Journal 13: 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM.. 2009. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. Journal of Virology 83: 5005–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camborde L, Planchais S, Tournier V, et al. 2010. The ubiquitin–proteasome system regulates the accumulation of Turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. The Plant Cell 22: 3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos L, Granell P, Tarraga S, et al. 2014. Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant Physiology and Biochemistry 77: 35–43. [DOI] [PubMed] [Google Scholar]
- Candresse T, Marais A, Faure C, Dubrana MP, Gombert J, Bendahmane A.. 2010. Multiple coat protein mutations abolish recognition of Pepino mosaic potexvirus (PepMV) by the potato Rx resistance gene in transgenic tomatoes. Molecular Plant-Microbe Interactions 23: 376–383. [DOI] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP.. 2008. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Carrington JC.. 2015. Antiviral roles of plant ARGONAUTES. Current Opinion in Plant Biology 27: 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Santos AA, Pires SR, et al. 2008. Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PLoS Pathogens 4: e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel C, De Alwis M, Bak A, Dong H, Steven A, Jander G.. 2015. Disruption of ethylene responses by Turnip mosaic virus mediates suppression of plant defense against the aphidvector, Myzus persicae. Plant Physiology 169: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AG, Kong LJ, Hanley-Bowdoin L, Bejarano ER.. 2004. Interaction between a geminivirus replication protein and the plant sumoylation system. Journal of Virology 78: 2758–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang Z, Teng K, et al. 2010. Upregulation of LSB1/GDU3 affects geminivirus infection by activating the salicylic acid pathway. The Plant Journal 62: 12–23. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang L, Li D, Wang F, Yu D. 2013. WRKY8 transcription fact functions in the TMV-cg defense response by mediating both abscisic acid a ethylene signaling in Arabidopsis. Proceedings of the Nationtal Academy of Sciences. USA 110: E1963–E1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenon M, Camborde L, Cheminant S, Jupin I.. 2012. A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase andaffects viral infectivity. EMBO Journal 31: 741–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Mahajan SK, Whitham SA, Yamamoto ML, Carrington JC.. 2000. Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proceedings of the Nationtal Academy of Sciences, USA 97: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Schurdi-Levraud V, Le QH, et al. 2012. The RTM resistance to potyviruses in Arabidopsis thaliana: natural variation of the RTM genes and evidence for the implication of additional genes. PLoS One 7: e39169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Kontra L, Burgyan J.. 2015. Viral silencing suppressors: tools forged to fine-tune host–pathogen coexistence. Virology 479–480: 85–103. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG.. 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O.. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71. [DOI] [PubMed] [Google Scholar]
- Dielen AS, Sassaki FT, Walter J, Michon T, Menard G.. 2011. The 20S proteasome alpha-5 subunit of Arabidopsis thaliana carries an RNase activity and interacts in planta with the lettuce mosaic potyvirus HcPro protein. Molecular Plant Pathology 12: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP.. 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Review Genetics 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Donaire L, Barajas D, Martinez-Garcia B, Martinez-Priego L, Pagan I, Llave C.. 2008. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. Journal of Virology 82: 5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eini O, Dogra S, Selth LA, Dry IB, et al. 2009. Interaction with a host ubiquitin-conjugating enzyme is required for the pathogenicity of a geminiviral DNA beta satellite. Molecular Plant Microbe-Interactions 22: 737–746. [DOI] [PubMed] [Google Scholar]
- Farnham G, Baulcombe DC.. 2006. Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proceedings of the National Academy of Sciences, USA 103: 18828–18833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Dröge-Laser W.. 2004. Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Molecular Plant-Microbe Interactions 17: 1162–1171. [DOI] [PubMed] [Google Scholar]
- Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J.. 2004. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes and Development 18: 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser RSS, Whenham RJ.. 1989. Abscisic-acid metabolism in tomato plantsinfected with tobacco mosaic-virus – relationships with growth, symptoms and the Tm-1 gene for Tmv resistance. Physiological and Molecular Plant Pathology 34: 215–226. [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, et al. 1993. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756. [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, et al. 2010. Arabidopsis RNA-dependent RNA polymerases and dicer- like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. The Plant Cell 22: 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, Park SW.. 2012. Plant disease resistance genes: current status and future directions. Physiological and Molecular Plant Pathology 78: 51–65. [Google Scholar]
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S.. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nature Reviews Microbiology 11: 777–788. [DOI] [PubMed] [Google Scholar]
- Hong W, Qian D, Sun R, et al. 2015. OsRDR6 plays role in host defense against double-stranded RNA virus, Rice Dwarf Phytoreovirus. Scientific Reports 5: 11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoser R, Zurczak M, Lichocka M, et al. 2013. Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytologist 200: 158–171. [DOI] [PubMed] [Google Scholar]
- Iriti M, Varoni EM.. 2014. Chitosan-induced antiviral activity and innate immunity in plants. Environmental Science and Pollution Research 22: 2935–2944. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Ishikawa M.. 2013. The resistance protein Tm-1 inhibits formation of a Tomato mosaic virus replication protein–host membrane protein complex. Journal of Virology, 87: 7933–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov KI, Eskelin K, Basic M, et al. 2016. Molecular insights into the function of the viral RNA silencing suppressor HCPro. The Plant Journal 85: 30–45. [DOI] [PubMed] [Google Scholar]
- Incarbone M, Dunoyer P.. 2013. RNA silencing and its suppression: novel insights from in planta analyses. Trends in Plant Science 18: 382–392. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV.. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Reviews Molecular Cell Biology 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Qian D, Zheng H, et al. 2012. RNA-dependent RNA polymerase 6 of rice (Oryza sativa) plays role in host defense against negative-strand RNA virus, Rice stripe virus. Virus Research 163: 512–519. [DOI] [PubMed] [Google Scholar]
- Jin H, Li S, Villegas A Jr.. 2006. Down-regulation of the 26S proteasome subunit RPN9 inhibits viral systemic transport and alters plant vascular development. Plant Physiology 142: 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ma D, Dong J, et al. 2007. HC-Pro protein of potato virus Y can interact with three Arabidopsis 20S proteasome subunits in planta. Journal of Virology 81: 12881–12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL.. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jovel J, Walker M, Sanfaçon H.. 2011. Salicylic acid-dependent restriction of tomato ringspot virus spread in tobacco is accompanied by a hypersensitive response, local RNA silencing, and moderate systemic resistance. Molecular Plant-Microbe Interactions 24: 706–718. [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K.. 2012. The HSP90 complex of plants. Biochim Biophys Acta 1823: 689–697. [DOI] [PubMed] [Google Scholar]
- Kang BC, Yeam I, Jahn MM.. 2005. Genetics of plant virus resistance. Annual Review of Phytopathology 43: 581–562. [DOI] [PubMed] [Google Scholar]
- Kørner CJ, Klauser D, Niehl A, et al. 2013. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Molecular Plant-Microbe Interactions 26: 1271–1280. [DOI] [PubMed] [Google Scholar]
- Lai J, Chen H, Teng K, et al. 2009. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. The Plant Journal 57: 905–917. [DOI] [PubMed] [Google Scholar]
- Landeo-Rios Y, Navas-Castillo J, Moriones E, Canizare MC.. 2015. The p22 RNA silencing suppressor of the crinivirus Tomato chlorosis virus preferentially binds long dsRNAs preventing them from cleavage. Virology 488: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD.. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. Journal of Virology 82: 6911–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TWH, van den Burg HA, Joosten MHAJ.. 2014. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends in Plant Science 19: 123–132. [DOI] [PubMed] [Google Scholar]
- Liu JZ, Horstman HD, Braun E, et al. 2011. Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiology 157: 1363–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love AJ, Yun BW, Laval V, Loake GJ, Milner JJ.. 2005. Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense-signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiology 139: 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love AJ, Laval V, Geri C, et al. 2007. Components of Arabidopsis defense- and ethylene-signaling pathways regulate susceptibility to Cauliflower mosaic virus by restricting long-distance movement. Molecular Plant-Microbe Interactions 20: 659–670. [DOI] [PubMed] [Google Scholar]
- Lozano-Duran R, Rosas-Diaz T, Luna AP, Bejarano ER.. 2011. Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLos One 6: e22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H. 2016. Interplay between the virus and the ubiquitin–proteasome system: molecular mechanism of viral pathogenesis. Current Opinion in Virology 17: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado JPB, Brustolini OJB, Mendes GC, Santos AA, Fontes EPB.. 2015. NIK1, a host factor specialized in antiviral defense or a novel general regulator of plant immunity? Bioessays 37: 1236–1242. [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C.. 2014. Plant PRRs and the activation of innate immune signaling. Molecular Cell 54: 263–272. [DOI] [PubMed] [Google Scholar]
- Mandadi KK, Scholthof K-BG.. 2013. Plant immune responses against viruses: how does a virus cause disease? The Plant Cell 25: 1489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann KS, Johnson KN, Carroll BJ, Dietzgen RG.. 2016. Cytorhabdovirus P protein suppresses RISC-mediated cleavage and RNA silencing amplification in planta. Virology 490: 27–40. [DOI] [PubMed] [Google Scholar]
- Mariano AC, Andrade MO, Santos AA, et al. 2004. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology 318: 24–31. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F.. 2005. The role of abscisic acid in plant–pathogen interactions. Current Opinion in Plant Biology 8: 409–414. [DOI] [PubMed] [Google Scholar]
- Mills-Lujan K, Deom CM.. 2010. Geminivirus C4 protein alters Arabidopsis development. Protoplasma 239: 95–110. [DOI] [PubMed] [Google Scholar]
- Nakashita H, Yasuda M, Nitta T, et al. 2003. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. The Plant Journal 33: 887–898. [DOI] [PubMed] [Google Scholar]
- Nicaise V. 2014. Crop immunity against viruses: outcomes and future challenges. Frontiers in Plant Science 5: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G.. 2004. Regulation of protein kinases: controlling activity through activation segment conformation. Molecular Cell 15: 661–675. [DOI] [PubMed] [Google Scholar]
- Oka K, Kobayashi M, Mitsuhara I, Seo S.. 2013. Jasmonic acid negatively regulates resistance to Tobacco mosaic virus in tobacco. Plant and Cell Physiology 54: 1999–2010. [DOI] [PubMed] [Google Scholar]
- Padmanabhan MS, Goregaoker SP, Golem S, Shiferaw H, Culver JN.. 2005. Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. Journal of Virology 79: 2549–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan MS, Kramer SR, Wang X, Culver JN.. 2008. Tobacco mosaic virus replicase–auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. Journal of Virology 82: 2477–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent JS, Bouteiller N, Elmayan T, Vaucheret H.. 2015. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. The Plant Journal 81: 223–232. [DOI] [PubMed] [Google Scholar]
- Pierce EJ, Rey ME.. 2013. Assessing global transcriptome changes in response to South African cassava mosaic virus [ZA99] infection in susceptible Arabidopsis thaliana. PLos One 8: e67534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, van der Does D, Zamioudis C, Leon-Reyes A, van Wees SCM.. 2012. Hormonal modulation of plant immunity. Annual Reviews of Cell and Developmental Biology 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Piroux N, Saunders K, Page A, Stanley J.. 2007. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKη, a component of the brassinosteroid signalling pathway. Virology 362: 428–440. [DOI] [PubMed] [Google Scholar]
- Postma J, Liebrand TW, Bi G, et al. 2016. Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytologist 210: 627–642. [DOI] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ.. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviralRNA silencing pathway negatively regulated by DCL1. Proceedings of the Nationtal Academy of Sciences, USA 105: 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P.. 2008. The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. The Plant Cell 20: 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja P, Sanville BC, Buchmann RC, Bisaro DM.. 2008. Viral genome methylation as an epigenetic defense against geminiviruses. Journal of Virology 82: 8997–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja P, Jackel JN, Li S, Heard IM, Bisaro DM.. 2014. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. Journal of Virology 88: 2611–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezzonico E, Flury N, Meins F Jr, Beffa R.. 1998. Transcriptional down-regulation by abscisic acid of pathogenesis-related beta-1,3-glucanase genes in tobacco cell cultures. Plant Physiology 117: 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers F, Nicaise V.. 2014. Plant resistance to infection by viruses. eLS doi: 10.1002/9780470015902.a0000757.pub3. [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JD.. 2007. Pathological hormone imbalances. Current Opinion in Plant Biology 10: 372–379. [DOI] [PubMed] [Google Scholar]
- Rocha CS, Santos AA, Machado JPB, Fontes EPB.. 2008. The ribosomal protein L10/QM-like protein is a component of the NIK mediated antiviral signaling. Virology 380: 165–169. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Negrete E, Lozano-Durán R, Piedra-Aguilera A, Cruzado L, Bejarano ER, Castillo AG.. 2013. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytologyst 199: 464–475. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O.. 2009. Roles of plant small RNAs in biotic stress responses. Annual Review of Plant Biology 60: 485–510. [DOI] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S.. 2012. The ubiquitin–proteasome system: central modifier of plant signalling. New Phytologist 196: 13–28. [DOI] [PubMed] [Google Scholar]
- Sahana N, Kaur H, Basavaraj, et al. 2012. Inhibition of the host proteasome facilitates papaya ringspot virus accumulation and proteosomal catalytic activity is modulated by viral factor HcPro. PLoS One 7: e52546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Deguchi M, Brustolini OJB, et al. 2012. The tomato RLK superfamily: phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC Plant Biology 12: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Durán MA, Dallas MB, Ascencio-Ibañez JT, et al. 2011. Interaction between geminivirus replication protein and the SUMO-conjugating enzyme is required for viral infection. Journal of Virology 85: 9789–9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AA, Carvalho CM, Florentino LH, et al. 2009. Conserved threonine residues within the A-loop of the receptor NIK differentially regulate the kinase function required for antiviral signaling. PLoS One 4: e5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AA, Lopes KVG, Apfata JAC, Fontes EPB.. 2010. NSP interacting kinase, NIK: a transducer of plant defence signalling. Journal of Experimental Botany 61: 3839–3845. [DOI] [PubMed] [Google Scholar]
- Soitamo AJ, Jada B, Lehto K.. 2012. Expression of geminiviral AC2 RNA silencing suppressor changes sugar and jasmonate responsive gene expression in transgenic tobacco plants. BMC Plant Biology 12: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Burgyan J.. 2013. RNA interference-mediated intrinsic antiviral immunity in plants. Current Topics in Microbiology and Immunology 371: 153–181. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Suzuki M, Natsuaki K, Shigyo T, Hino K, et al. 2001. Mapping the virus and host genes involved in the resistance response in cucumber mosaic virus infected Arabidopsis thaliana. Plant and Cell Physiology 42: 340–347. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kanayama Y, Zheng MS, et al. 2004. Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant and Cell Physiology 45: 803–809. [DOI] [PubMed] [Google Scholar]
- Tameling WI, Nooijen C, Ludwig N, et al. 2010. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. The Plant Cell 22: 4176–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yamaguchi Y, Sano H.. 2006. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Molecular Biology 61: 31–45. [DOI] [PubMed] [Google Scholar]
- Vallejos CE, Astua-Monge G, Jones V, et al. 2006. Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics 172: 1229–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF.. 2009. Salicylic acid, a multifaceted hormone to combat disease. Annual Review Phytopathology 47: 177–206. [DOI] [PubMed] [Google Scholar]
- Wang H, Hao L, Shung CY, Sunter G, Bisaro DM.. 2003. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. The Plant Cell 15: 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM.. 2005. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. Journal of Virology 79: 7410–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, et al. 2010. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proceedings of the Nationtal Academy of Sciences, USA 107: 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whenham RJ, Fraser RSS, Brown LP, Payne JA.. 1986. Tobacco Mosaic Virus-induced increase in abscisic-acid concentration in tobacco leaves – intracellular location in light and dark-green areas, and relationship to symptom development. Planta 168: 592–598. [DOI] [PubMed] [Google Scholar]
- Witham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B.. 1994. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115. [DOI] [PubMed] [Google Scholar]
- Whitham SA, Anderberg RJ, Chisholm ST, Carrington JC.. 2000. Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. The Plant Cell 12: 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham SA, Yang C, Goodin MM.. 2006. Global impact: elucidating plant responses to viral infection. Molecular Plant-Microbe Interaction 19: 1207–1215. [DOI] [PubMed] [Google Scholar]
- Wieczorek P, Obrepalska-Steplowska A.. 2015. Suppress to survive – implication of plant viruses in PTGS. Plant Molecular Biology Report 33: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gou X, He K, Xi D, et al. 2010. BAK1 and BKK1 in Arabidopsis thaliana confer reduced susceptibility to turnip crinkle virus. European Journal of Plant Pathology 127: 149–156. [Google Scholar]
- Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua N.. 2008. βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes and Development 22: 2564–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xie Y, Raja P, et al. 2011. Suppression of methylation-mediated transcriptional gene silencing by betaC1–SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathogens 7: e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wu Z, Li Y, Wu J.. 2015. Biogenesis, function, and applications of virus-derived small RNAs in plants. Frontiers in Microbiology 6: 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang X, Singh J, Li D, Qu F.. 2012. Temperature-dependent survival of turnip crinkle virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires dcl2,AGO2,and HEN1. Journal of Virology 86: 6847–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen H, Huang X, et al. 2011. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. The Plant Cell 23: 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SF, Gao F, Cao XS, et al. 2005. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiology 139: 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Jeong RD, Lim GH, et al. 2013. Double-stranded RNA-binding protein 4 is required for resistance signaling against viral and bacterial pathogens. Cell Reports 4: 1168–1184. [DOI] [PubMed] [Google Scholar]
- Zipfel C. 2014. Plant pattern-recognition receptors. Trends in Immunology 35: 345–351. [DOI] [PubMed] [Google Scholar]
- Zorzatto C, Machado JPB, Lopes KVG, et al. 2015. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 520: 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]