Abstract
Background and Aims Arachnitis uniflora is a mycoheterotrophic plant that exploits arbuscular mycorrhizal fungi of neighbouring plants. We tested A. uniflora's specificity towards fungi across its large latitudinal range, as well as the role of historical events and current environmental, geographical and altitudinal variables on fungal genetic diversity.
Methods Arachnitis uniflora mycorrhizas were sampled at 25 sites. Fungal phylogenetic relationships were reconstructed, genetic diversity was calculated and the main divergent lineages were dated. Phylogeographical analysis was performed with the main fungal clade. Fungal diversity correlations with environmental factors were investigated.
Key Results Glomeraceae fungi dominated, with a main clade that likely originated in the Upper Cretaceous and diversified in the Miocene. Two other arbuscular mycorrhizal fungal families not previously known to be targeted by A. uniflora were detected rarely and appear to be facultative associations. High genetic diversity, found in Bolivia and both northern and southern Patagonia, was correlated with temperature, rainfall and soil features.
Conclusions Fungal genetic diversity and its distribution can be explained by the ancient evolutionary history of the target fungi and by micro-scale environmental conditions with a geographical mosaic pattern.
Keywords: Arbuscular mycorrhizal fungi, Andean–Patagonian forest, Arachnitis uniflora, genetic diversity, mycoheterotrophy, phylogeography
INTRODUCTION
The most common mycorrhizal symbiosis occurs between arbuscular mycorrhizal fungi (AMF) and plants; in fact, ∼70–90 % of land plant species form mycorrhizas with members of the fungal phylum Glomeromycota (Parniske, 2008; Smith and Read, 2008; Merckx et al., 2012). The relationship between AMF and green plants is interpreted as mutualistic, and it is generally assumed that there is bidirectional transfer of nutrients. In this kind of relationship the fungi are obligate partners, because they cannot complete their life cycle without plants. In contrast, arbuscular mycorrhizal green plants are autotrophic and mostly capable of development in the absence of AMF colonization; therefore, plants are considered facultative symbionts in this relationship (Smith and Read, 2008). A third partner, such as an achlorophyllous plant, can interfere with the arbuscular mycorrhizal mutualism; this additional partner takes advantage of that mycorrhizal relationship by becoming a ‘cheater’, which invades a mutualism between two other organisms. Thus, a tripartite symbiosis arises where an obligate cheater plant becomes part of, and depends on the fate of, the mutualism it targets (Bidartondo, 2005; Smith and Read, 2008; Waterman et al., 2013). Achlorophyllous plants sustained by fungi are referred to as mycoheterotrophs (Leake, 1994). Mycoheterotrophy shows multiple independent origins, and is present in ten families of angiosperms, seven of which are monocots (Merckx et al., 2013a, b).
The biochemical and structural mechanisms of mycoheterotrophic plants to avoid recognition and/or suppress defences in their fungal hosts (sensuMerckx et al., 2009) while they sequester fungal carbon are unclear; however, the evolution and maintenance of these pathways may also generate selective pressures towards specialization (Futuyma and Moreno, 1988; Waterman et al., 2013). Furthermore, the dependence of a mycoheterotrophic plant on fungi for its establishment, survival and/or diversification agrees with the observation that some mycoheterotrophic associations are extremely specific (Bidartondo and Bruns, 2002; Bidartondo et al., 2002; Taylor et al., 2002; Leake, 2004). On the other hand, generalists have greater potential to adapt to new environments, providing more opportunities for speciation and a reduced risk of extinction (Zayed et al., 2005). The development of molecular tools shows that not all mycoheterotrophs are extreme specialists; indeed, some mycoheterotrophic plants have the ability to associate with more than one fungal family. Generalist mycoheterotrophic relationships have been found in Sciaphila ledermannii (Triuridaceae) which associates with Acaulosporaceae, Gigasporaceae and Glomeraceae, in Campylosiphon congestus (Burmanniaceae) and Gymnosiphon capitatus (Burmanniaceae) with Acaulosporaceae and Glomeraceae, and in Voyria species (Gentianaceae) with Gigasporaceae and Glomeraceae (Bidartondo et al., 2002; Franke et al., 2006; Merckx et al., 2010, 2012; Courty et al., 2011). Nonetheless, Glomeraceae is the most common family targeted by mycoheterotrophic plants (Merckx et al., 2012), possibly due to its ability to contact roots quickly and to produce an extensive mycelium inside the roots (Hart and Reader, 2002).
There is substantial evidence showing that nutrient availability, dispersal limitation, host plant communities and other environmental factors affect AMF distribution, abundance, root colonization, hyphal development and spore germination (Camargo-Ricalde, 2002; Smith and Read, 2008; Kivlin et al., 2011). In mycoheterotrophic relationships, it is difficult to separate the requirements of each partner, but in general the host fungi delimit the habitat the achlorophyllous plant can occupy (McKendrick et al., 2000; Waterman et al., 2013).
Environmental influence upon the biota is not restricted to the present day. In the last several million years, glaciations and associated climate change, tectonic events, volcanism, palaeobasins, seashore shifts and marine introgressions have dramatically altered the landscape, e.g. in South America (Ortiz-Jaureguizar and Cladera, 2006; Ramos and Ghiglione, 2008; Martínez and Kutschker, 2011; Ponce et al., 2011; Sérsic et al., 2011), with concomitant effects on the patterns of distribution and diversification in the biota.
Here, we studied Arachnitis uniflora from the completely mycoheterotrophic monocot family Corsiaceae (Fig. 1, inset). This species grows principally in dense and shaded Andean–Patagonian temperate forests of Argentina and Chile, in sub-humid and humid tropical Andean forests in Bolivia, and in the treeless Malvinas–Falkland Islands (Dimitri, 1972; Cribb et al., 1995; Ibisch et al., 1996). Bidartondo et al. (2002) showed a specific association with AMF belonging to Glomus group A (currently placed within Glomeraceae) in three populations of A. uniflora. It is unclear whether fungal diversity varies as the number of populations increases, especially when including distant areas, as well as different floristic regions. Though evidence indicates that specialization towards a fungal species does not preclude a wide distribution (e.g. the orchid Eulophia zollingeri; Ogura-Tsujita and Yukawa, 2008), the extensive geographical range of A. uniflora suggests that fungal variation may await discovery due to historical factors and/or adaptation to environmental conditions. To test these expectations, we used fungal DNA sequence data to identify mycorrhizal fungi associated with individuals of A. uniflora across nearly all of its geographical range (with the exception of the Malvinas–Falkland Islands). We tested the influence on fungal diversity of current environmental, geographical and altitudinal variables, as well as the role of historical events through molecular dating of fungal clades.
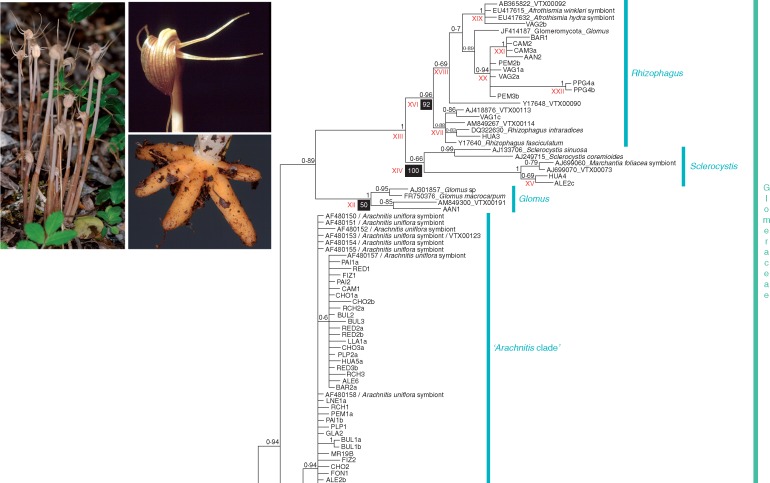
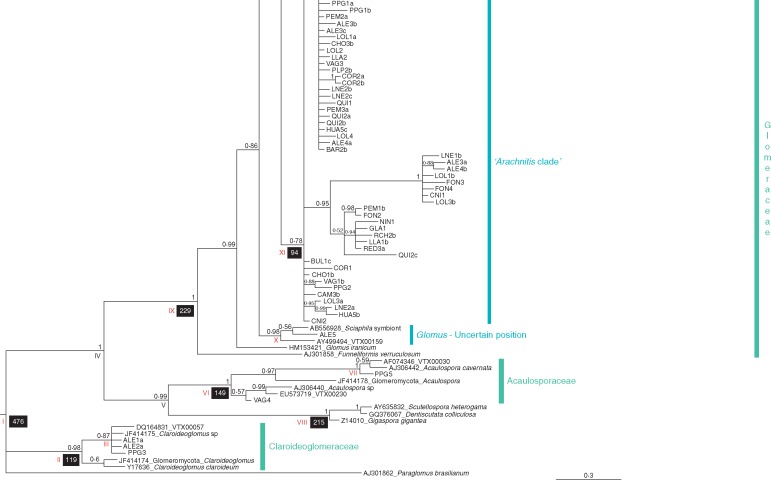
>Fig. 1.
Bayes„ian in„fer„ence tree based on the mid„dle frag„ment (604 pb) of the nu„clear 18S rDNA gene se„quences of the glomeromycotan in„di„vid„uals found in Arachnitis uniflora roots and other fun„gal lin„eages used as iden„ti„fiers, which are described in Supplementary Data Table S1. Terminal nodes denoted with„out GenBank ac„ces„sion num„bers cor„res„pond to fungi found in this work. Node sup„port is shown as Bayes„ian pos„ter„ior prob„abil„ity (BPP) over each node. Nodes with BPP <0·5 were collapsed to polytomies. Black boxes under nodes in„di„cate main di„ver„gence times (in Mya); all Roman nu„merals cor„res„pond to the node ages shown in Table 2, to„gether with their 95 % HPD inter„vals. (Inset) Image of A. uniflora plants and de„tails of flower and roots.
MATERIALS AND METHODS
Plant species and sampling
Arachnitis uniflora Phil. grows underground and surfaces only during flowering and fruiting, when a shoot ∼6–40 cm tall is formed, ending in a single zygomorphic flower (Fig. 1, inset). The root system consists of a star-like cluster of about ten tuberous roots that store starch and oil and are densely colonized internally by mycorrhizal fungi (Dimitri, 1972; Ibisch et al., 1996; Domínguez and Sérsic, 2004; Domínguez et al., 2005) (Fig. 1, inset). It inhabits shaded forests, where it co-exists with Austrocedrus chilensis (Cupressaceae), Nothofagus spp. (Nothofagaceae), Osmorhiza chilensis (Apiaceae), Araucaria araucana (Araucariaceae), Chusquea culeou (Poaceae), Luma apiculata (Myrtaceae) and Lomatia hirsuta (Proteaceae).
One hundred and twenty-three A. uniflora were collected from 24 sites along the Andean–Patagonian forests from Argentina and Chile and the Bolivian forests to cover the geographical range of the species (Table 1). The whole root system was removed, washed and immediately stored in 2 % CTAB buffer, and preserved at −20 °C until further processing. All voucher specimens were deposited in the CORD herbarium. All nucleotide sequences from the LAV site (Verde Lake, Villa La Angostura) were obtained by Bidartondo et al. (2002) and were retrieved from GenBank.
Table 1.
Collection sites and codes (A, Argentina; B, Bolivia; C, Chile), coordinates, altitude, genetic diversity indices (for the obtained Glomeromycota sequences and exclusively for Arachnitis clade), and haplotypes found in each site, for fungi associated with Arachnitis uniflora roots
| Site ID | Site | Latitude | Longitude | Altitude | π in Glomeromycota | H in the Arachnitis clade | Haplotypes |
|---|---|---|---|---|---|---|---|
| VAG | Valle Grande (B) | −18·572 | −64·043 | 2387 | 0·042 | 1·000 | H1, H11 |
| PPG | Pampa Grande (B) | −18·677 | −63·917 | 2378 | 0·062 | 0·667 | H1, H11 |
| COR | Corel (C) | −35·535 | −71·197 | 487·3 | 0·009 | 0·667 | H10, H14 |
| QUI | Quillón (C) | −36·668 | −72·460 | 41·2 | 0·012 | 0·500 | H1, H16 |
| HUA | Hualpén (C) | −36·805 | −73·172 | 17·7 | 0·037 | 1·000 | H1–H2, H17 |
| AAN | Alto Antuco (C) | −37·369 | −71·697 | 749 | 0·054 | – | – |
| CNI | Ñielol Hill (C) | −38·724 | −72·589 | 217·6 | 0·025 | 1·000 | H3, H8 |
| LLA | Llancalil (C) | −39·236 | −71·636 | 333 | 0·011 | 1·000 | H1–H2, H6 |
| RCH | Ruca Choroy Lake (A) | −39·246 | −71·189 | 1241·9 | 0·010 | 1·000 | H1–H2, H6, H20 |
| LNE | Los Nevados (C) | −39·327 | −71·834 | 400 | 0·016 | 0·900 | H1, H3, H17–H18 |
| PLP | La Peña (C) | −39·507 | −72·582 | 153·9 | 0·003 | 0·667 | H1–H2 |
| LOL | Lolog Lake (A) | −40·683 | −71·373 | 936·9 | 0·019 | 0·867 | H1, H3, H13, H17 |
| LAVd | Verde Lake, Villa La Angostura (A) | −40·777 | −71·659 | 813 | 0·000 | 0·000 | H1 |
| BAR | San Carlos de Bariloche (A) | −41·053 | −71·541 | 822·3 | 0·015 | 0·476 | H1–H2 |
| PEM | Perito Moreno Hill, El Bolsón (A) | −41·857 | −71·537 | 593·2 | 0·027 | 0·500 | H1, H4 |
| CHO | Cholila (A) | −42·461 | −71·610 | 580 | 0·006 | 0·733 | H1–H2, H8 |
| ALE | Los Alerces National Park (A) | −42·887 | −71·608 | 526·2 | 0·040 | 0·857 | H1–H2, H12, H15, H19 |
| NIN | Los Niños Lagoon (A) | −44·006 | −71·490 | 1026·3 | – | – | H6 |
| FON | Fontana Lake (A) | −44·890 | −71·527 | 927·8 | 0·024 | 0·833 | H1, H3–H4 |
| FIZ | Fitz Roy, El Chaltén (A) | −49·268 | −72·950 | 693·2 | 0·005 | 1·000 | H1–H2 |
| GLA | Los Glaciares National Park (A) | −50·484 | −72·875 | 403 | 0·010 | 1·000 | H1, H6 |
| PAI | Torres del Paine National Park (C) | −51·092 | −73·198 | 258·2 | 0·002 | 0·667 | H1, H2 |
| BUL | Fuerte Bulnes (C) | −53·610 | −70·945 | 403·9 | 0·006 | 0·800 | H2, H7–H8 |
| CAM | Puerto Cameron (C) | −53·750 | −70·095 | 116·4 | 0·034 | 1·000 | H8–H9 |
| RED | Redonda Island (A) | −54·863 | −68·479 | 402·2 | 0·007 | 0·700 | H2, H5–H6 |
H, haplotype diversity; π, nucleotide diversity.
Only sequences obtained from GenBank (LAV site).
Fungal DNA extraction, amplification and sequencing
Total DNA from one thin section of one root of each A. uniflora individual was extracted according to Gardes and Bruns (1993) with a purification step using GeneClean (QBioGene). The whole fungal 18S ribosomal rDNA gene was amplified (JumpStart, Sigma), using the specific primers NS1 (White et al., 1990) and EF3 (Smit et al., 1999). The PCR procedure was 2 min at 94 °C; 34 cycles of 30 s at 94 °C, 30 s at 53 °C and 1 min 30 s at 72 °C; and final extension for 7 min at 72 °C. Because initial DNA sequence screening produced some electropherograms of multiple overlapping copies, cloning was performed using TOPO TA kits (Invitrogen, UK). At least four putative positive colonies from each amplification product were used. The cycling scheme was 7 min at 94 °C; 25 cycles of 30 s at 94 °C, 30 s at 53 °C and 1 min 30 s at 72 °C; and final extension for 5 min at 72 °C. The primers used were NS1/NS3, NS3/NS5 and NS5/EF3 (White et al., 1990), covering the whole 18S gene. Each fragment was sequenced (BigDye, Applied Biosystems) with a 3730 Genetic Analyzer (Applied Biosystems). Two different matrices were constructed, one including the sequences of the 18S (1514 bp) and another containing only the fragment with the highest number of polymorphic sites (S). This fragment (604 bp) corresponds to the middle part of the 18S amplified with the NS3/NS5 primers. Sequences were identified by BLAST, and non-glomeromycotan DNA sequences were discarded. Additionally, the sequences were also identified against taxa in the MaarjAM database of Glomeromycota (Öpik et al., 2010).
Phylogenetic analysis
The fungal sequences obtained from A. uniflora roots were assigned by BLAST to virtual taxa (VTX) in the MaarjAM database; those VTX with ≥ 97 % sequence similarity were added to the matrices (ten sequences). Eight A. uniflora Glomus group A sequences, classified as Glomeraceae by Schüßler and Walker (2010), obtained by Bidartondo et al. (2002), were retrieved from GenBank. Finally, 23 sequences of other genera of Glomeraceae cited in Krüger et al. (2011) and Redecker et al. (2013) were included. Paraglomus (Paraglomeraceae) was selected as outgroup. The complete dataset is shown in Table S1.
Phylogenetic relationships among sequences were reconstructed by Bayesian inference (BI). The GTR + I + G model of DNA evolution was selected under the Akaike information criterion (AIC) as implemented in MrModeltest 2.2 (Nylander, 2004). Analysis was performed in MrBayes 3.1 (Ronquist and Huelsenbeck, 2003) and consisted of two independent runs of 1 ×107 generations with four chains (three heated and one cold), sampling every 100 cycles; the first 10 % of the sampled trees (corresponding to the burn-in period) were discarded. Correlation among runs was evaluated in Tracer v.1.6 (Rambaut et al., 2014), considering the effective sample size on each parameter (ESSs; Supplementary Data Table S2). All these procedures were implemented on both matrices (complete and middle part of the 18S gene).
Molecular dating
Estimation of the divergence time in the different clades of AMF associated with A. uniflora was conducted using three calibration points. The split between Ascomycota, Basidiomycota and Glomeromycota was set to 595 Mya and the crown node of Glomeromycota was constrained to 460 Mya (Redecker et al., 2000). The calibration point to Gigasporaceae was 240 Mya (Padovan et al., 2005). Because the selected priors were located outside the Glomeraceae, it was necessary to add sequences from Ascomycota, Basidiomycota and representatives of each Glomeromycota family to the alignment used in the above analysis. These sequences were retrieved from GenBank, following Merckx and Bidartondo (2008) and are shown in Table S1. Divergence times were estimated using a Bayesian approach and implementing a relaxed molecular clock model with BEAST v.1.6 (Drummond and Rambaut, 2007). The substitution model was GTR with a Gamma site heterogeneity model with four categories following the MrModeltest result, the clock model was set as an uncorrelated log-normal relaxed model, and the birth–death process was selected as the speciation model. The Monte Carlo Markov chain was set to run for 2 × 107 generations, sampling every 1000 cycles; the first 10 % of the sampled trees (corresponding to the burn-in period) were discarded. Correlation among runs was evaluated in Tracer v.1.6 considering the ESSs values on each parameter (Table S2).
Genetic diversity and structure
Nucleotide diversity (π; Nei, 1987) and number of AMF taxa (Glomeromycota families and Glomeraceae genera; see Results section) were calculated at each site, while those sequences belonging to the main AMF clade associated with A. uniflora roots (see Results section) were selected to calculate haplotype diversity (H; Nei, 1987). Indexes were calculated in DnaSP 5.10 (Rozas et al., 2003). The distributions of these AMF diversities across sites were plotted using the point-to-grid statistical analysis tool in Diva-GIS 7.5.0 (Hijmans et al., 2005). This plotting was conducted with a grid size of 0·3×0·3° (33·3 × 33·3 km at the equator) and a circular neighbourhood option of 1·5° (166·5 km).
A haplotype network was built using the median-joining algorithm implemented in Network 5.0 (Bandelt et al., 1999); for this analysis only the sequences grouped in the main AMF clade were selected and autapomorphies and gaps were not considered. Five ambiguous connections (loops) were resolved using the three criteria postulated by the coalescent theory (Crandall and Templeton, 1993).
Patterns of genetic diversity and structure were estimated by genetic landscape shape interpolation analyses using the program Alleles In Space 1.0 (Miller, 2005). This procedure allows the graphical representation of inter-individual genetic distance to detect the location of putative barriers or contact zones with dissimilar gene composition. The analysis was carried out with a grid size of 50×50 and a distance weight value of a = 1. The analysed DNA sequences were those belonging to the main clade of AMF detected, and distributed along Andean-Patagonian range.
The spatial structure of the genetic diversity of the AMF belonging to the main clade associated with A. uniflora was analysed across the distribution range using Bayesian inference implemented in Geneland v.4.0.0 (Guillot et al., 2005). Five independent replicate runs were performed, with the number of populations ranging between 1 and 25, assuming a correlated allelic frequency model, and a spatial model without uncertainty on coordinates. Each run consisted of 8 × 106 iterations, with a thinning interval of 1000 and a burn-in phase of 800 iterations. Given the complete consistency in the most probable number of populations among runs, the run with the highest posterior probability value was selected, with which membership maps with a 50 × 50 pixel spatial domain were created, according to Guillot et al., 2005.
AMF diversity and environmental conditions
The geographical coordinates and altitude of each site were recorded with a GPS. Using the WorldClim database (Hijmans et al., 2005), bioclimatic variables were obtained for each locality at a spatial resolution of 1 km2. From the 19 bioclimatic variables were selected those with biological relevance, trying to avoid correlated variables: annual temperature (°C) and precipitation (mm) averages, temperature and precipitation of the warmest, coldest, driest and wettest quarters (i.e. 3-month seasons; Supplementary Data Table S3). To characterize edaphic features, one soil sample of ∼500 g was removed from the ground surface to a depth of 15 cm at each location. For each sample, pH, percentages of organic carbon (C), nitrogen (N), sand (Sd) and silt (St), and concentrations of phosphorus (P, ppm) and potassium (K, meq 100 g−1) were determined by the soil laboratory at the Facultad de Ciencias Agrarias (Universidad Nacional de Córdoba). The relationships between AMF nucleotide diversity (Table 1) and environmental factors (Table S3) were tested using linear regressions with Infostat v.2014 (Di Rienzo et al., 2014).
RESULTS
Extracted DNA of 123 A. uniflora individual roots revealed Glomeromycota sequences in 69 different plants. Sequences of four clones per root retrieved a total of 104 Glomeromycota sequences belonging to 24 sites along the distribution range. Non-Glomeromycota fungi (e.g. Pezizales in Ascomycota and Agaricales in Basidiomycota, principally) were not considered in this study because previous morphological and molecular studies performed on roots of A. uniflora never revealed the presence of ectomycorrhizal fungi (Bidartondo et al., 2002; Domínguez and Sérsic, 2004; Domínguez et al., 2009).
Phylogenetic analysis
Bayesian phylogenetic relationships were reconstructed with the 104 glomeromycotan DNA sequences, with the addition of the sequences obtained from the GenBank and MaarjAM databases, summarizing 145 sequences (Table S1). Trees were generated with both length matrices; the topologies of both trees were similar (Fig. 1 and Supplementary Data Appendix S1); however, stronger support for each node was found for the tree constructed with the middle part of the 18S gene than for the tree obtained from the entire gene. Thus, further analyses were carried out using only the middle portion.
The selected Bayesian phylogenetic tree (Fig. 1) revealed the presence of fungal symbionts of A. uniflora belonging to three well-supported glomeromycotan families, sensuSchüßler and Walker (2010): Glomeraceae [Bayesian posterior probability (BPP) =1·00], Acaulosporaceae (BPP =1·00) and Claroideoglomeraceae (BPP =0·98). The Glomeraceae clade was well supported and retrieved the largest portion of the analysed sequences (95·5 %), where three genera were recognized: Glomus (paraphyletic), Sclerocystis (BPP = 0·66) and Rhizophagus (BPP = 0·96); however, 84·1 % of these sequences were clustered in an unresolved Glomeraceae group (hereafter termed the ‘Arachnitis clade’; BPP = 0·78) identified as virtual taxon VTX00123 (Arachnitis uniflora symbiont, from Bidartondo et al., 2002) in the MaarjAM database.
The Glomeraceae family clade (Fig. 1) shows an early division separating the genus Funneliformis from the remaining genera, which form a large, well-supported clade (BPP = 0·99). Most remaining lineages cluster in two clades (BPP = 0·86), a small one formed by a Glomus of uncertain position (BPP = 0·98) and containing one fungus from A. uniflora (ALE5) and a large clade with two sub-clades, the Arachnitis clade, which is unresolved and contains most A. uniflora fungi; and a second clade (BPP = 0·89) that also divides in two, one clade containing Sclerocystis (BPP = 0·66) and Rhizophagus (BPP = 0·96), and the other with Glomus (BPP = 1·00) and one fungal sequence from A. uniflora (AAN1) roots. The Sclerocystis and Rhizophagus clades contain two and 13 sequences from A. uniflora, respectively. This tree topology suggests that Glomus would be paraphyletic.
Outside the Glomeraceae, the families Claroideoglomeraceae and Acaulosporaceae constituted two well supported clades, containing three and two individuals from A. uniflora, respectively (Fig. 1). Acaulosporaceae was located within a bigger clade, grouping Diversisporales GenBank sequences (BPP = 0·99). Acaulosporaceae representatives were restricted to the Bolivian forest sites (VAG4 and PPG5). Claroideoglomeraceae fungi were found in one Bolivian site (PPG3) and in two different plants at ALE (ALE1a and ALE2a; Fig. 1, Table 1).
Only two plant individuals presented two fungal families in the same root (Glomeraceae and Claroideoglomeraceae) at ALE, but we detected up to three different AMF clades per root at ALE, CAM, PEM and VAG, among which the Arachnitis clade was always present. Within each site, ALE and PPG showed four different Glomeromycota clades, HUA and VAG showed three AMF clades and AAN, BAR, CAM and PEM showed two clades; in the remaining sites only the dominant AMF group, the Arachnitis clade, was present. Only at AAN did we find no Arachnitis clade representatives (Fig. 1, Table 1).
Genetic diversity and structure
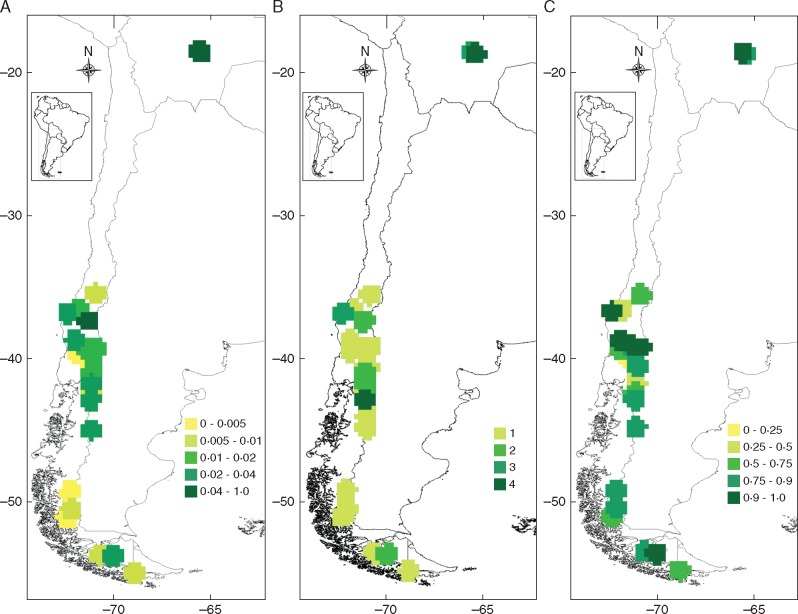
Considering the Glomeromycota detected in A. uniflora roots, the nucleotide diversity index (π) showed the highest values for Bolivian and AAN sites, with π = 0·0617 in PPG, π = 0·0545 in AAN and π = 0·0416 in VAG (Table 1). The lowest values were found in LAV (π = 0). It was not possible to calculate π in NIN because just one fungal sequence was found. In general, AMF genetic diversity tended to increase towards lower latitudes (Fig. 2A); high diversity was detected in Bolivia and the northernmost sites of Chile. However, a cline pattern is not clear due to medium to high genetic diversity detected at the southernmost latitudes. A similar pattern was found when considering the number of different AMF taxa (Acaulosporaceae, Claroideoglomeraceae, Glomus, Sclerocystis, Rhizophagus and Arachnitis clade) at each site (Fig. 2B).
Fig. 2.
Geographical distribution patterns of genetic diversity. (A) nucleotide diversity of all Arachnitis uniflora-associated fungal tax at each site. (B) Number of glomeromycotan clades per site. (C) Haplotype diversity using only those sequences belonging to the Arachnitis clade (Fig. 1). Colour scales indicate genetic diversity values based on a 33·3 × 33·3 km grid cell.
Spatial structuring of the genetic diversity of the Arachnitis clade evidenced three population clusters (Fig. 3A), according to the BPP. The COR site formed an exclusive cluster (Fig. 3B; BPP = 0·45); Bolivia grouped together with almost all Patagonian sites (i.e. north of COR towards PAI; Fig. 3C; BPP = 0·34); with the exception of the three sites associated with Tierra del Fuego, which formed the last genetic group (Fig. 3D; BPP = 0·4).
Fig. 3.
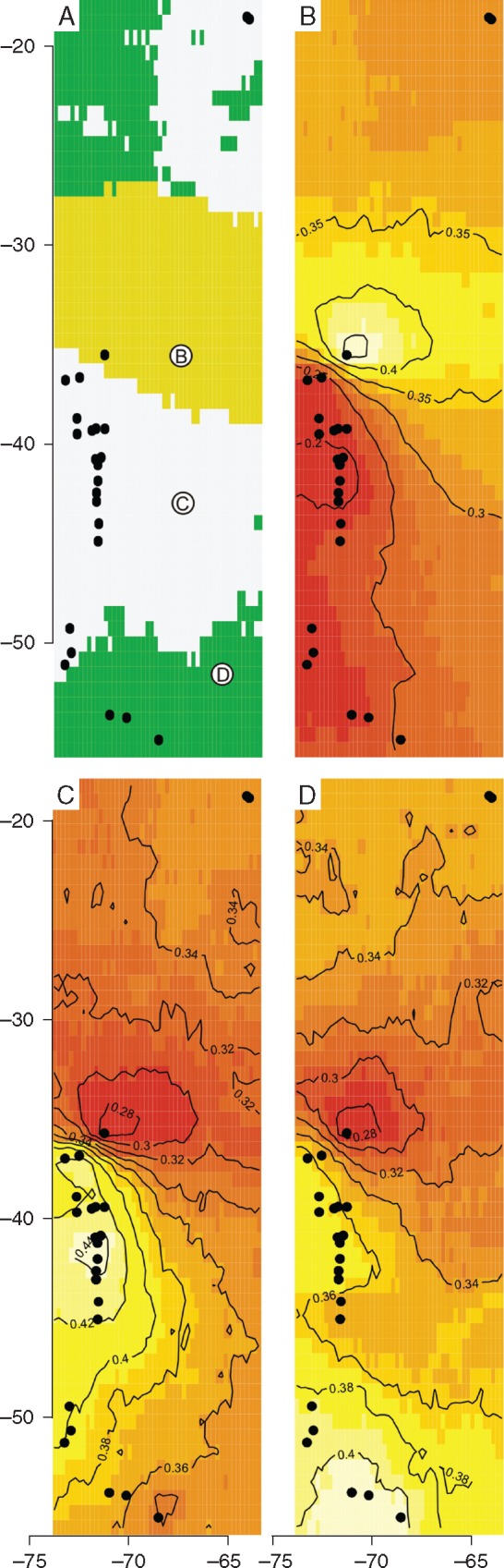
Bayesian clustering of the AMF belonging to the Arachnitis clade. (A) Most probable population membership arrangement. Each colour indicates a cluster. (B–D) Maps of each cluster selected by Geneland; colours indicate the Bayesian posterior probability (BPP), white representing the maximum value and red the lowest. Black dots show each studied site. (B) COR group. (C) Most Patagonian sites and Bolivian representatives. (D) Southernmost Patagonian sites, associated with Tierra del Fuego.
Genetic diversity and phylogeography of the Arachnitis clade
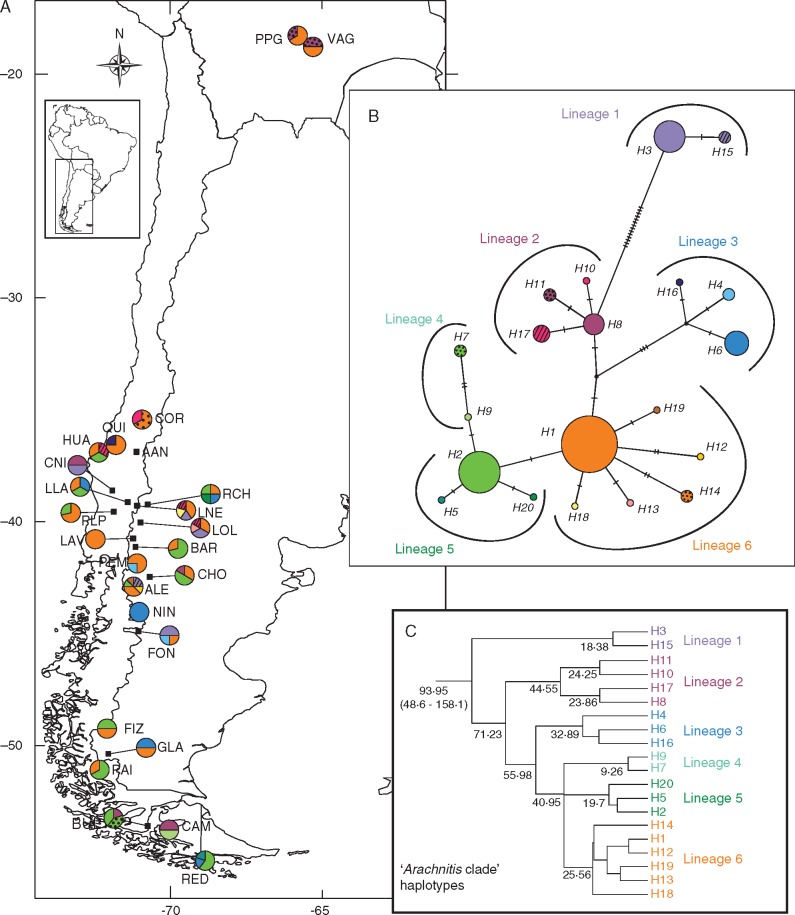
As the Arachnitis clade (i.e. VTX00123) included most DNA sequences and covered nearly the whole geographical range of the plant species, it was used for phylogeographical analyses. Haplotype diversity showed no spatial structure; eight populations showed the highest haplotype diversity values (H = 1; Table 1, Fig. 2C), while the lowest values ranging from H = 0·69 to H = 0. The NIN and AAN sites were not included in the diversity analysis because of scarcity of sequences. The haplotype network displayed a total of 20 haplotypes. Haplotypes H1 and H2 were the most frequent and widespread (Fig. 4); H1 was distributed from the Bolivian sites to PAI, one of the southernmost continental sites, while H2, although absent in Bolivia, extended along the Patagonian forest to Tierra del Fuego. Haplotypes H6 and H8 were also widespread, but less frequent. Considering the mutational steps, H3 and its derivative H15 were the most divergent, separated from H8 by 14 and 15 mutational steps, respectively; these two haplotypes were found between latitudes 39° and 45°S. Haplotypes H4, H6 and H16 formed a group that originated from a non-sampled or extinct ancestor (median vector) and diverged from the rest by three mutational steps. All other haplotypes differed in one or two mutational steps from each other. The most geographically isolated Bolivian sites contained, besides H1, an exclusive haplotype (H11). Haplotypes 5, 7, 9,10 and 12–20 were exclusive to single populations (Table 1). With the exception of NIN and LAV, all populations were polymorphic, with ALE having five haplotypes, the highest number.
Fig. 4.
Geographical distribution and genealogical relationship of the nuclear 18S rDNA haplotypes found in fungi associated with Arachnitis uniflora roots and particularly those belonging to the Arachnitis clade. Colour correspondence exists between panels. (A) Haplotype distribution. The pie charts reflect the frequency of occurrence of each haplotype in each site. Site codes are referenced in Table 1. (B) Haplotype network. Each short line on the bar between haplotypes represents one mutational step. Black dots represent median vectors and are represented as non-sampled or extinct ancestors. (C) Haplotype lineages found within the Arachnitis clade and their respective divergence times (over each node).
Divergence times among haplotypes from the Arachnitis clade were estimated and the phylogenetic tree obtained gave the same network structure, with six main haplotype lineages (Fig. 4C). The origin of this diversification dated from the Upper Cretaceous [93·95 Mya; 95 % highest posterior density (HPD) interval 48·6–158·1 Mya], while the occurrence of the first lineages ranged from 71·2 to 40·9 Mya during the Palaeocene–Eocene, principally, and the last divergences of the current haplotypes occurred during the Miocene (24·2–9·3 Mya). Other important clades of the AMF associated with A. uniflora were e.g. Glomeraceae (229·1 Mya; 95 % HPD interval 145·3–315·2 Mya), Claroideoglomeraceae (119·4 Mya; 95 % HPD interval 37·8–241 Mya) and Acaulosporaceae (145 Mya; 95 % HPD interval 59·4–250·1 Mya), and are shown in Fig. 1 and Table 2.
Table 2.
Divergence ages and their confidence intervals (95 % HPD) obtained in the dating of AMF associated with A. uniflora
| Lineage nodes | Median age | 95 % HPD interval |
|---|---|---|
| Clades not shown in Fig. 1 | ||
| Ascomycota–Basidiomycota–Glomeromycota divergence | 595·01 | 521–667·41 |
| Ascomycota | 216·44 | 55·12–404·42 |
| Basidiomycota | 286·53 | 131·14–488·87 |
| Diversisporales | 309·75 | 245·25–381·73 |
| Glomeromycota clades shown in Fig. 1 | ||
| I, Glomeromycota | 476·04 | 420·64–527·5 |
| II, Claroideoglomeraceae | 119·39 | 37·81–241 |
| III | 75·62 | 26·11–153·94 |
| IV | 335·01 | 268·21–406·05 |
| V | 279·64 | 221–346·53 |
| VI, Acaulosporaceae | 148·99 | 59·43–250·14 |
| VII | 52·08 | 17·53–107·53 |
| VIII, Gigasporacea | 215·00 | 177·12–252·73 |
| IX, Glomeraceae | 229·14 | 145·33–315·18 |
| X, Glomus; uncertain position | 41·67 | 7·15–105·4 |
| XI, Arachnitis clade | 93·95 | 48·57–158·08 |
| XII, Glomus | 49·8 | 13·61–105·64 |
| XIII | 134·56 | 72·08–214·44 |
| XIV, Sclerocystis | 100·16 | 40·04–173·77 |
| XV | 16·78 | 1·54–51 |
| XVI, Rhizophagus | 91·66 | 44·86–151·32 |
| XVII | 46·95 | 14·48–94·38 |
| XVIII | 69·44 | 32·46–118 |
| XIX | 36·97 | 8·32–82·16 |
| XX | 44·75 | 18·88–82·69 |
| XXI | 15·1 | 3·23–34·59 |
| XXII | 2·66 | 0–14·27 |
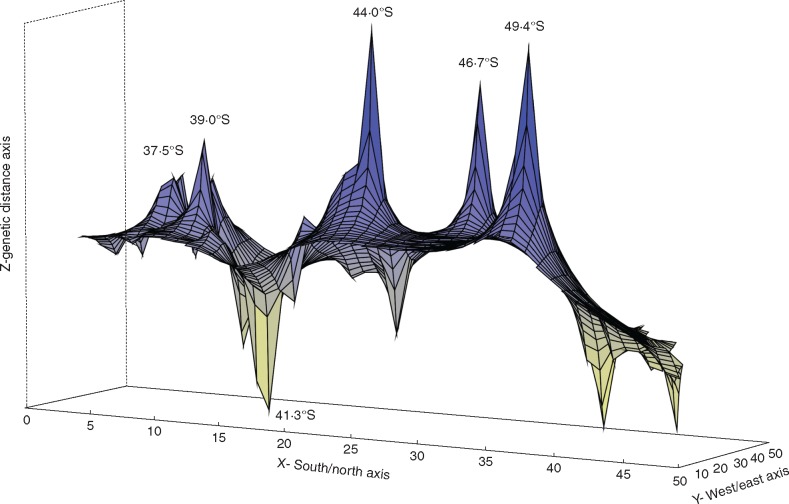
Genetic landscape shape interpolation analyses (Fig. 5) produced surface plots that show two major genetic discontinuities, indicating probable contact areas at 44° around the NIN and FON sites, and another at latitude 49·4°S, between FIZ and GLA, both delimiting a northern and a southern region in the Andean–Patagonian forest. The northern region was in general characterized by the presence of haplotypes derived from H1 (lineage 6; H12, H13, H14, H18 and H19) and haplotypes derived from H8 (lineage 2; H10 and H17 only situated in this region). The most divergent haplotypes, H3 and H15 (lineage 1), were found only between 37·5° and 46·7°S breaks. Although lineage 3 (H4, H6 and H16) was widespread, its haplotypes were principally distributed in this region, with H4 and H16 only present here. The southern region was dominated by haplotypes derived from H2 (lineages 4 and 5; H5, H7, H9 and H20), being present H1, too. Other smaller peaks also appear at 37·5°, 39° and 46·7°S. Depressions indicate more homogeneous genetic zones, occurring principally around 41·3°S and southwards from 49·4°S.
Fig. 5.
Genetic landscape shape interpolation analysis using a 50×50 grid size and a distance weighting parameter of a = 1. The x- and y-axes correspond to geographic locations; the z-axis shows genetic distances. Positive peaks show genetic discontinuities or possible barriers to gene flow, and are referenced with latitude coordinates. The analysis was conducted with fungal sequences belonging to the Arachnitis clade (Fig. 1).
AMF diversity and environmental conditions
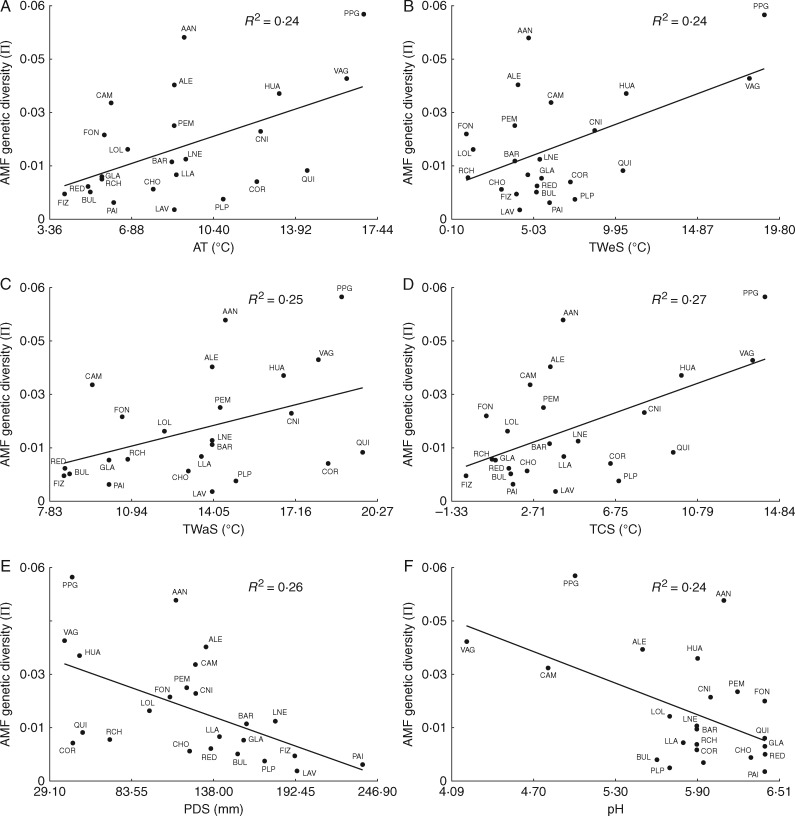
Environmental conditions were highly variable across the sampled sites. For example, annual temperature ranged from 4 to 16·8 °C and rainfall ·and from 665 to 2143 mm (Table S3). Temperature of the coldest season and precipitation in the warmest season were the most variable bioclimatic factors, with coefficients of variation (CVs) of 89·6 and 62·5, respectively. In relation to edaphic traits, P concentration was the most variable, ranging from 0·4 to 114·7 ppm (CV =156·2). Linear regressions performed to contrast fungal nucleotide diversity with environmental factors showed some significant associations. Within variables related to temperature, annual temperature (AT) (R2 = 0·24; P = 0·0092), mean temperature of the wettest season (TWeS) (R2=0·24; P=0·009), mean temperature of the warmest season (TWaS) (R2 = 0·15; P = 0·036) and mean temperature of the coldest season (TCS) (R2=0·27; P=0·0058) were positively and significantly associated with AMF diversity (Fig. 6A–D). One variable related to precipitation (PDS, R2 = 0·26, P=0·0064) and another to edaphic traits (pH, R2 = 0·24, P = 0·0105) were significantly negatively correlated with AMF genetic diversity (Fig. 6E, F). Only significant regressions are shown in Fig. 6.
Fig. 6.
Relationship between nucleotide diversity of Arachnitis uniflora fungi and environmental factors at each site, listed in Table 1. Only significant regressions are shown. (A) Annual temperature. (B) Mean temperature during the wettest season. (C) Mean temperature during the warmest season. (D) Mean temperature during the coldest season. (E) Precipitation during the driest season. (F) Soil pH.
DISCUSSION
Arbuscular mycorrhizal fungi in A. uniflora roots
Nearly 95 % of the Glomeromycota sequences from A. uniflora roots belong to Glomeraceae, supporting conclusions from Bidartondo et al. (2002), who showed that all A. uniflora samples were associated with only one AMF lineage, Glomus group A. Glomeraceae is the most common AMF in mycoheterotrophic plants (Merckx et al., 2012), and A. uniflora is not an exception. However, our study revealed the existence of two additional families associated with A. uniflora roots (Acaulosporaceae and Claroideoglomeraceae). The present study and other recent works provide evidence that families other than Glomeraceae can be associated with mycoheterotrophic plants. Acaulosporaceae representatives, as well as the Glomeraceae genera Rhizophagus, Sclerocyctis and Glomus, were also recorded in other mycoheterotrophic species (Russell and Bulman, 2005; Franke et al., 2006; Merckx and Bidartondo, 2008; Merckx et al., 2010, 2012; Courty et al., 2011; Yamato et al., 2011). However, Claroideoglomeraceae had not been reported before in mycoheterotrophic plants (Franke et al., 2006; Merckx et al., 2012). This is the first study to address the genetic diversity of fungi associated with a mycoheterotrophic plant across its geographical range. In addition to the clade representing most common A. uniflora symbionts, the high geographical coverage of this study may have made it possible to recover AMF taxa that associate more rarely with this plant species. These latter may be less effective co-colonizers and/or could represent taxa specific to a certain habitat or geographical region within the range of A. uniflora.
The topology of the phylogenetic tree (Fig. 1) is in general congruent with previously published Glomeromycota phylogenetic trees (i.e. Krüger et al., 2011; Redecker et al., 2013). Compared with these, topological differences occurred mainly in the positions of Claroideoglomeraceae (sister family of Glomeraceae) and Funneliformis (sister genus of Glomus). It is important to note that the topology recovered in the tree constructed with the entire 18S sequence (Appendix S1) was similar to the previously cited phylogenies, but node supports were in general lower, with several non-monophyletic groups. In Fig. 1Glomus appears to be polyphyletic; in fact, this was already observed by Schüßler and Walker (2010), who categorized some Glomus individuals and/or clades as ‘species of uncertain position’. Although in these kinds of analysis high phylogenetic resolution is always desirable, our main intention was to test how diverse were the AMF associated with A. uniflora.
In the present study, the Bolivian sites showed the highest AMF taxa and genetic diversity, containing representatives of three glomeromycotan families and with Acaulosporaceae exclusively present at those sites. Noticeably, Claroideoglomeraceae was found disjunct in one Bolivian site and in ALE in the Andean–Patagonian region.
As mentioned above, the family Glomeraceae, and essentially the Arachnitis clade, are the most common AMF found in A. uniflora. This confirms that this plant depends on representatives of Glomeraceae (or the Arachnitis clade) for its establishment and/or survival. The other fungal families present more rarely in A. uniflora could represent ‘facultative’ mycobionts, as suggested by Franke et al. (2006), or incipient host shifts according to the geographical mosaic theory of coevolution (Thompson, 2005). The latter postulates that variation in species assemblages, in this case that of AMF, would lead to local adaptations, e.g. host shifts. It is important to remark that here we only addressed the AMF part of this symbiosis. With respect to the colonizing ability of the different AMF that makes shifts possible, Hart and Reader (2002) noted that members of Glomeraceae usually contacted roots quickly and produced a more extensive mycelium inside the roots than in soil, while members of the Acaulosporaceae contacted roots more slowly and established a much less extensive mycelium in either roots or soil (Chagnon et al., 2013).
Geographical structure of genetic diversity
Considering the geographical distribution patterns of the whole genetic diversity indices (Fig. 2), it was possible to recognize several areas of high diversity that can be hypothesized as glacial refugia for AMF associated with A. uniflora, in concordance with sites of high genetic diversity of A. uniflora (M. Renny et al., unpubl. res.). The northernmost sites in Bolivia showed high fungal genetic diversity due principally to the presence of fungi of three families and, though sharing H1 with the remaining sites, showed an exclusive haplotype (H11), probably as a response of fragmentation and isolation processes. Across the Patagonian sites, another five putative refugia were identified, such as in the Chilean coastal mountain range at 36·8°S (HUA), also proposed by Sérsic et al. (2011), in the Central Depression of Chile at 38·7°S (CNI), where Vergara et al. (2014) found a refugial population of Nothofagus obliqua and Acosta et al. (2014) found one Nothofagus population with an exclusive haplotype within the same place. It is noteworthy that Nothofagus is a common co-occurring genus with A. uniflora along its Andean–Patagonian range, and specifically N. dombeyii was reported to be associated with A. uniflora (Bidartondo et al., 2002). Other refugial areas were located in a longitudinal zone along the western (LNE) and eastern (RCH and LOL) flanks of the Andes between 39·3°S and 40·7°S, at 42·5°– 43°S (CHO and ALE), in concordance with refugia proposed in broad analyses by Sérsic et al. (2011) and Souto et al. (2015), and in specific studies by Premoli et al. (2000), Marchelli and Gallo (2004) and Cosacov et al. (2010); while the HUA refuge was proposed only by Sérsic et al. (2011). The southernmost refuge was proposed at 53·7°S (CAM), in agreement with Jakob et al. (2009), Tremetsberger et al. (2009) and Souto et al. (2015). It is remarkable that this last refuge in CAM reinforces the idea of local persistence of the forest in southern latitudes (Premoli et al., 2000). Notably, the appearance of putative co-colonizers or ‘facultative’ fungi coincides with sites VAG, PPG, ALE, HUA and CAM, and zones around these, proposed here as refuges.
Twenty-four of the 25 analysed sites were represented by individuals belonging to the Arachnitis clade; this large geographical representation allowed a detailed phylogeographical analysis with this clade. The pattern achieved with the haplotype network and geographical distribution of haplotypes showed no clear geographical structure, evidenced by the wide distribution of the most frequent haplotypes, although it is possible to distinguish two diversification areas: the northern area had a prevalence of haplotypes derived from H1, while the southern area contained mostly haplotypes derived from H2. This is in concordance with the clusters obtained by the statistical test implemented in Geneland.
Centred on the two most frequent and widespread haplotypes (H1 and H2), the network (Fig. 4B) showed a star-like topology – a pattern suggesting a rapid expansion of an ancestral haplotype over a large geographical area (Avise, 2000). Moreover, the four most abundant haplotypes (H1, H2, H6 and H8) were shared in almost all populations, indicating ancient fluid relationships among populations. Thus, geographical distances seem not to be relevant barriers to haplotype connection, as was already reported by Davison et al. (2015); indeed, Bolivian populations shared one out of two haplotypes with the Patagonian forests.
Two main breaks detected with the landscape analyses (interpreted as meeting zones between long-term divergent genetic sources; Fig. 5) at latitudes 44°S and 49·4°S, between sites FON and FIZ, were consistent with the clusters previously mentioned, defining the northern and southern areas. These breaks coincide with a region where no A. uniflora records in herbaria or other databases exist; however, we cannot dismiss the possibility that this mainly underground species grows in the area. The presence of a peak at latitude 46·7°S in the middle of the gap suggests that the lack of collections from this area may have affected the surface plot results (Fig. 5). Minor breaks were detected for latitudes 37·5° and 39°S between HUA and CNI, and around LLA, respectively. All latitudinal breaks detected are in agreement with barriers proposed by Sérsic et al. (2011) for the Patagonian Andes, except for 46·7°, which was found by Mathiasen and Premoli (2010).
In general, refugia and breaks are correlated with climatic changes associated with Pleistocene glaciations (Sérsic et al., 2011 and references therein), though the molecular dating of haplotype divergences would also show concordance with previous events in the Patagonian Andes. Thus, it was possible to identify an ancient diversification origin within the Arachnitis clade during the Upper Cretaceous (93·95 Mya; 95 % HPD 48·6–158·1 Mya). As Corsiaceae diversified more recently, during the Eocene (53–36 Mya; Mennes et al., 2015), it would suggest that these ancient AMF diversifications must have occurred in association with green plants of the community. Fungal diversification and distribution would have enough time to be shaped by successive historic events, beginning in the Cretaceous and continuing until the Pleistocene climate changes. Along the same lines, a study performed on Nothofagus (Acosta and Premoli, 2010; Acosta et al., 2014) provides five genetic discontinuities (at 37·5°, 39°, 42° 40°, 46° and 50°S) in agreement with those found here, which were explained as a result of marine ingressions during the Oligocene–Miocene. Finally, the last fungal Arachnitis clade haplotype divergences, which occurred during the Miocene, could have occurred in association with diversification events within the Corsiaceae ancestors of A. uniflora.
Environmental drivers of AMF genetic diversity
Of the three studied environmental factors, temperature (TCS, AT, TWeS and TWaS) was strongly and positively associated with AMF genetic diversity (Fig. 6A–D). Rainfall and edaphic features were poorly correlated with AMF diversity, and were represented only by PDS (from the precipitation variables; Fig. 6E) and pH values (from the edaphic variables; Fig. 6F); both significantly and negatively correlated with genetic diversity. There is knowledge about the influence of several factors – temperature, soil nutrients and characteristics, latitudinal range, and others – on the distribution, richness and fitness of AMF (i.e. Green et al., 1976; Koske, 1987; Porter et al., 1987; Johnson et al., 1991; Allen et al., 1995; Lekberg et al., 2007; Kivlin et al., 2011; Davison et al., 2015; Öpik and Davison, 2016). Indeed other factors, like the surrounding community or particular AMF species' requirements, can alter the diversity patterns in fungi (Camargo-Ricalde, 2002; Landis et al., 2004; Kivlin et al., 2011).
In line with this close relationship between temperature pattern and fungal diversity in A. uniflora roots, Pirozynski (1968) suggested that temperature is the major factor determining the distribution and occurrence of fungi in general. Moreover, Green et al. (1976) argued that pH and temperature similarly drive AMF distribution, in agreement with our results (Fig. 6E, F). Temperature has a stimulating effect on AMF (e.g. Schenck et al., 1975; Koske, 1987; Hu et al., 2013) by increasing hyphal P uptake and transport (Gavito et al., 2003). The results here suggest that genetic diversity of AMF positively correlates with pH, as observed in VAG, CAM, PPG and ALE (Fig. 6F, Table S3), which is in agreement with previous findings (Anderson and Liberta, 1992; Coughlan et al., 2000).
Finally, increasing diversity of fungi associated with A. uniflora was correlated with sites with low precipitation during the driest season. In the Andean–Patagonian forests, this period coincides with the spring–summer time. In general, AMF colonization peaks are reported in spring and summer seasons (Sigüenza et al., 1996; Lugo et al., 2003; Bohrer et al., 2004) associated with water stress (Kennedy et al., 2002). This driest time was correlated too with the highest spore density periods for different ecosystems (i.e. Mago and Mukerji, 1994; Carvalho et al., 2001; Lugo and Cabello, 2002; Escudero and Mendoza, 2005; Sivakumar, 2013), in line with our findings.
Concluding remarks
The non-photosynthetic mycorrhizal plant A. uniflora can associate with fungi of three glomeromycotan families, among which Claroideoglomeraceae had never before been found in mycoheterotrophic plants. Glomeraceae fungi of three genera are by far the most common and widespread fungi that nourish this plant species. The Arachnitis clade, the main group of AMF with A. uniflora, experienced several and successive climatic and geological events that moulded their distribution and diversity. In line with these processes, it is possible that fungi rarely found in this plant may have resulted from facultative associations during adverse environment conditions in several time periods.
In this tripartite relationship, AMF and their chlorophyllous hosts have far more control than mycoheterotrophic plants, considering the huge differences in biomass and because mycoheterotrophic plants depend on AMF and their green hosts, but not vice versa. We found that temperature is the most important factor accompanying the distribution of AMF genetic diversity; however, there was no geographical pattern to temperature. Rather, environment acts at the micro-scale, where some determining factors (e.g. temperature, pH and dry periods acting together or combined differently) create geographical mosaics with particular conditions that allow A. uniflora to increase its fungal diversity.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: sequences included in the phylogenetic analysis. Table S2: ESS values from Bayesian analyses obtained with Tracer. Table S3: collection sites and IDs, and environmental variables (temperature, precipitation and soil traits) of the sampled sites for fungi associated with A. uniflora roots. Appendix S1: Bayesian inference tree based on the complete (1514 pb) nuclear 18S rDNA gene sequences of the glomeromycotan individuals found in A. uniflora roots and other fungal lineages used as identifiers, which are described in Table S1. Terminal nodes denoted without GenBank accession numbers correspond to fungi found in this work. Node supports are shown as Bayesian posterior probability (BPP) values at each node. Nodes with BPP < 0·5 were collapsed to polytomies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Strelin, A. A. Cocucci, A. Cosacov, M. Baranzelli, G. Ferreiro and F. Sazatornil for assistance with field work; the National Parks Administration (APN, Argentina) and National Forest Corporation (CONAF, Chile) for sampling permissions; L. Martinez-Suz and B. Atkinson for assistance in the laboratory; and G. Grilli for help with sequence data analysis. M.R. acknowledges a Bentham-Moxon Trust grant and a Science and Technical Secretary of National University of Córdoba (SECyT-UNC) fellowship. We thank J. Geml and another anonymous reviewer for comments and suggestions, which improved the manuscript. M.C.A., A.N.S. and N.C. acknowledge the National Research Council of Argentina (CONICET) as researchers and M.R. as doctoral fellowship holders.
LITERATURE CITED
- Acosta MC, Premoli AC.. 2010. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Molecular Phylogenetics and Evolution 54: 235–242. [DOI] [PubMed] [Google Scholar]
- Acosta MC, Mathiasen P, Premoli AC.. 2014. Retracing the evolutionary history of Nothofagus in its geo-climatic context: new developments in the emerging field of phylogeology. Geobiology 12: 497–510. [DOI] [PubMed] [Google Scholar]
- Allen EB, Allen MF, Helm DJ, Trappe JM, Molina R, Rincon E.. 1995. Patterns and regulation of mycorrhizal plant and fungal diversity. Plant and Soil 170: 47–62. [Google Scholar]
- Anderson RC, Liberta AE.. 1992. Influence of supplemental inorganic nutrients on growth, survivorship, and mycorrhizal relationships of Schizachyrium scoparium (Poaceae) grown in fumigated and unfumigated soil. American Journal of Botany 79: 406–414. [Google Scholar]
- Avise JC. 2000. Phylogeography: the history and formation of species. Cambridge: Harvard University Press. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bidartondo MI. 2005. The evolutionary ecology of myco-heterotrophy. New Phytologist 167: 335–352. [DOI] [PubMed] [Google Scholar]
- Bidartondo MI, Bruns TD.. 2002. Fine‐level mycorrhizal specificity in the Monotropoideae (Ericaceae): specificity for fungal species groups. Molecular Ecology 11: 557–569. [DOI] [PubMed] [Google Scholar]
- Bidartondo MI, Redecker D, Hijri I, et al. 2002. Epiparasitic plants specialized on arbuscular mycorrhizal fungi. Nature 419: 389–392. [DOI] [PubMed] [Google Scholar]
- Bohrer KE, Friese CF, Amon JP.. 2004. Seasonal dynamics of arbuscular mycorrhizal fungi in differing wetland habitats. Mycorrhiza 14: 329–337. [DOI] [PubMed] [Google Scholar]
- Camargo-Ricalde SL. 2002. Dispersal, distribution and establishment of arbuscular mycorrhizal fungi: a review. Boletín de la Sociedad Botánica de México 71: 33–44. [Google Scholar]
- Carvalho LM, Caçador I, Martins-Loução M.. 2001. Temporal and spatial variation of arbuscular mycorrhizas in salt marsh plants of the Tagus estuary (Portugal). Mycorrhiza 11: 303–309. [DOI] [PubMed] [Google Scholar]
- Chagnon PL, Bradley RL, Maherali H, Klironomos JN.. 2013. A trait-based framework to understand life history of mycorrhizal fungi. Trends in Plant Science 18: 484–491. [DOI] [PubMed] [Google Scholar]
- Cosacov A, Sérsic AN, Sosa V, Johnson LA, Cocucci AA.. 2010. Molecular evidence of ice-age refugia in the Patagonia steppe and post-glacial colonisation of the Andes slopes: insights from the endemic species Calceolaria polyrhiza (Calceolariaceae). Journal of Biogeography 37: 1463–1477. [Google Scholar]
- Coughlan AP, Dalpé Y, Lapointe L, Piché Y.. 2000. Soil pH-induced changes in root colonization, diversity, and reproduction of symbiotic arbuscular mycorrhizal fungi from healthy and declining maple forests. Canadian Journal of Forest Research 30: 1543–1554. [Google Scholar]
- Courty P, Walder F, Boller T, et al. 2011. C and N metabolism in mycorrhizal networks and mycoheterotrophic plants of tropical forests: a stable isotope analysis. Plant Physiology 156: 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall KA, Templeton AR.. 1993. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribb PJ, Wilkin P, Clements M.. 1995. Corsiaceae: a new family for the Falklands Islands. Kew Bulletin 50: 171–172. [Google Scholar]
- Davison J, Moora M, Öpik M, et al. 2015. Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349: 970–973. [DOI] [PubMed] [Google Scholar]
- Dimitri MJ. 1972. Una nueva especie del género Arachnitis Phil. (Corsiaceae). Revista Facultad de Agronomía de la Universidad Nacional de La Plata 48: 37–45. [Google Scholar]
- Domínguez LS, Sérsic A.. 2004. The southernmost myco-heterotrophic plant, Arachnitis uniflora: root morphology and anatomy. Mycologia 96: 1143–1151. [DOI] [PubMed] [Google Scholar]
- Domínguez L, Sérsic A, Melville L, Peterson RL.. 2005. “Prepackaged symbioses” – propagules on roots of the epiparasitic plant Arachnitis uniflora Phil. New Phytologist 169: 191–198. [DOI] [PubMed] [Google Scholar]
- Domínguez LS, Melville L, Sérsic A, Faccio A, Peterson RL. 2009. The mycoheterotroph, Arachnitis uniflora, has a unique association with arbuscular mycorrhizal fungi. Botany 87: 1198–1208. [Google Scholar]
- Drummond AJ, Rambaut A.. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero V, Mendoza R.. 2005. Seasonal variation of arbuscular mycorrhizal fungi in temperate grasslands along a wide hydrologic gradient. Mycorrhiza 15: 291–299. [DOI] [PubMed] [Google Scholar]
- Franke T, Beenken L, Döring M, Kocyan A, Agerer R.. 2006. Arbuscular mycorrhizal fungi of the Glomus-group A lineage (Glomerales; Glomeromycota) detected in myco-heterotrophic plants from tropical Africa. Mycological Progress 5: 24–31. [Google Scholar]
- Futuyma DJ, Moreno G.. 1988. The evolution of ecological specialization. Annual Review of Ecology and Systematics 19: 207–233. [Google Scholar]
- Gardes M, Bruns TD.. 1993. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Gavito ME, Schweiger P, Jakobsen I.. 2003. P uptake by arbuscular mycorrhizal hyphae: effect of soil temperature and atmospheric CO2 enrichment. Global Change Biology 9: 106–116. [Google Scholar]
- Green NE, Graham SO, Schenck NC.. 1976. The influence of pH on the germination of vesicular-arbuscular mycorrhizal spores. Mycologia 68: 929–934. [PubMed] [Google Scholar]
- Guillot G, Mortier F, Estoup A.. 2005. Geneland: a program for landscape genetics. Molecular Ecology Notes 5: 712–715. [Google Scholar]
- Hart MM, Reader RJ.. 2002. Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytologist 153: 335–344. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A.. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Hu Y, Rillig MC, Xiang D, Hao Z, Chen B.. 2013. Changes of AM fungal abundance along environmental gradients in the arid and semi-arid grasslands of northern China. PLoS One 8: e57593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibisch PL, Neinhuis C, Rojas PN.. 1996. On the biology, biogeography, and taxonomy of Arachnitis Phil. nom. cons. (Corsiaceae) in respect to a new record from Bolivia. Willdenowia 26: 321–332. [Google Scholar]
- Jakob SS, Martinez–Meyer E, Blattner FR.. 2009. Phylogeographic analyses and paleodistribution modeling indicate Pleistocene in situ survival of Hordeum species (Poaceae) in southern Patagonia without genetic or spatial restriction. Molecular Biology and Evolution 26: 907–923. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Zak DR, Tilman D, Pfleger FL.. 1991. Dynamics of vesicular-arbuscular mycorrhizae during old field succession. Oecologia 86: 349–358. [DOI] [PubMed] [Google Scholar]
- Kennedy LJ, Tiller RL, Stutz JC.. 2002. Associations between arbuscular mycorrhizal fungi and Sporobolus wrightii in riparian habitats in arid South-west North America. Journal of Arid Environments 50: 459–475. [Google Scholar]
- Kivlin SN, Hawkes CV, Treseder KK.. 2011. Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biology and Biochemistry 43: 2294–2303. [Google Scholar]
- Koske RE. 1987. Distribution of VA mycorrhizal fungi along a latitudinal temperature gradient. Mycologia 79: 55–68. [Google Scholar]
- Krüger M, Krüger C, Walker C, Stockinger H, Schüßler A.. 2011. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytologist 193: 970–984. [DOI] [PubMed] [Google Scholar]
- Landis FC, Gargas A, Givnish TJ.. 2004. Relationships among arbuscular mycorrhizal fungi, vascular plants and environmental conditions in oak savannas. New Phytologist 164: 493–504. [Google Scholar]
- Leake JR. 1994. The biology of myco‐heterotrophic (‘saprophytic') plants. New Phytologist 127: 171–216. [DOI] [PubMed] [Google Scholar]
- Leake JR. 2004. Myco-heterotroph/epiparasitic plant interactions with ectomycorrhizal and arbuscular mycorrhizal fungi. Current Opinion in Plant Biology 7: 422–428. [DOI] [PubMed] [Google Scholar]
- Lekberg Y, Koide RT, Rohr JR, Aldrich-Wolfe L, Morton JB.. 2007. Role of niche restrictions and dispersal in the composition of arbuscular mycorrhizal fungal communities. Journal of Ecology 95: 95–105. [Google Scholar]
- Lugo MA, Cabello MN.. 2002. Native arbuscular mycorrhizal fungi (AMF) from mountain grassland (Córdoba, Argentina) I: seasonal variation of fungal spore diversity. Mycologia 94: 579–586. [DOI] [PubMed] [Google Scholar]
- Lugo MA, Maza MEG, Cabello MN.. 2003. Arbuscular mycorrhizal fungi in a mountain grassland II: seasonal variation of colonization studied, along with its relation to grazing and metabolic host type. Mycologia 95: 407–415. [PubMed] [Google Scholar]
- Mago P, Mukerji KG.. 1994. Vesicular arbuscular mycorrhizae in Lamiaceae: I. Seasonal variation in some members. Phytomorphology 44: 83–88. [Google Scholar]
- Marchelli P, Gallo LA.. 2004. The combined role of glaciations and hybridization in shaping the distribution of genetic variation in a Patagonian southern beech. Journal of Biogeography 31: 451–460. [Google Scholar]
- Martínez OA, Kutschker A.. 2011. The ‘Rodados Patagónicos’ (Patagonian shingle formation) of eastern Patagonia: environmental conditions of gravel sedimentation. Biological Journal of the Linnean Society 103: 336–345. [Google Scholar]
- Mathiasen P, Premoli AC.. 2010. Out in the cold: genetic variation of Nothofagus pumilio (Nothofagaceae) provides evidence for latitudinally distinct evolutionary histories in austral South America. Molecular Ecology 19: 371–385. [DOI] [PubMed] [Google Scholar]
- McKendrick SL, Leake JR, Read DJ.. 2000. Symbiotic germination and development of myco-heterotrophic plants in nature: transfer of carbon from ectomycorrhizal Salix repens and Betula pendula to the orchid Corallorhiza trifida through shared hyphal connections. New Phytologist 145: 539–548. [DOI] [PubMed] [Google Scholar]
- Mennes CB, Lam VKY, Rudall PJ, et al. 2015. Ancient Gondwana break-up explains the distribution of the mycoheterotrophic family Corsiaceae (Liliales). Journal of Biogeography 42: 1123–1136. [Google Scholar]
- Merckx V, Bidartondo MI.. 2008. Breakdown and delayed cospeciation in the arbuscular mycorrhizal mutualism. Proceedings of the Royal Society B. Biological Sciences 275: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx V, Bidartondo MI, Hynson NA.. 2009. Myco-heterotrophy: when fungi host plants. Annals of Botany 104: 1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx V, Stöckel M, Fleischmann A, Bruns TD, Gebauer G.. 2010. 15N and 13C natural abundance of two mycoheterotrophic and a putative partially mycoheterotrophic species associated with arbuscular mycorrhizal fungi. New Phytologist 188: 590–596. [DOI] [PubMed] [Google Scholar]
- Merckx V, Janssens SB, Hynson NA, Specht CD, Bruns TD, Smets EF.. 2012. Mycoheterotrophic interactions are not limited to a narrow phylogenetic range of arbuscular mycorrhizal fungi. Molecular Ecology 21: 1524–1532. [DOI] [PubMed] [Google Scholar]
- Merckx V,, Freudenstein JV,, Kissling J, et al. 2013a. Taxonomy and classification In: Merckx V, ed. Mycoheterotrophy. The biology of plants living on fungi. London: Springer, 19–101. [Google Scholar]
- Merckx V, Mennes CB, Peay KG, Geml J.. 2013b. Evolution and Diversification In: Merckx V, ed. Mycoheterotrophy. The biology of plants living on fungi. London: Springer, 215–244. [Google Scholar]
- Miller MP. 2005. Alleles in space: computer software for the joint analysis of interindividual spatial and genetic information. Journal of Heredity 96: 722–724. [DOI] [PubMed] [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- Nylander JAA. 2004. MrModeltest v.2. Program distributed by the author. Sweden: Uppsala University; (March 2016). [Google Scholar]
- Ogura-Tsujita Y, Yukawa T.. 2008. Epipactis helleborine shows strong mycorrhizal preference towards ectomycorrhizal fungi with contrasting geographic distributions in Japan. Mycorrhiza 18: 331–338. [DOI] [PubMed] [Google Scholar]
- Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, et al. 2010. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytologist 188: 223–241. [DOI] [PubMed] [Google Scholar]
- Öpik M, Davison J.. 2016. Uniting species- and community-oriented approaches to understand arbuscular mycorrhizal fungal diversity. Fungal Ecology 24: 106–113. [Google Scholar]
- Ortiz-Jaureguizar E, Cladera GA.. 2006. Palaeoenvironmental evolution of southern South America during the Cenozoic. Journal of Arid Environments 66: 498–532. [Google Scholar]
- Padovan ACB, Sanson GFO, Brunstein A, Briones MRS.. 2005. Fungi evolution revisited: application of the penalized likelihood method to a Bayesian fungal phylogeny provides a new perspective on phylogenetic relationships and divergence dates of Ascomycota groups. Journal of Molecular Evolution 60: 726–735. [DOI] [PubMed] [Google Scholar]
- Parniske M. 2008. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nature Reviews Microbiology 6: 763–775. [DOI] [PubMed] [Google Scholar]
- Pirozynski KA. 1968. Geographical distribution of fungi. In: Ainsworth GC, Sussman AS, eds. The Fungi New York: Academic Press, 487–504. [Google Scholar]
- Ponce JF, Rabassa J, Coronato A, Borromei AM.. 2011. Palaeogeographical evolution of the Atlantic coast of Pampa and Patagonia from the last glacial maximum to the Middle Holocene. Biological Journal of the Linnean Society 103: 363–379. [Google Scholar]
- Porter WM, Robson AD, Abott LK.. 1987. Field survey of the distribution of vesicular-arbuscular mycorrhizal fungi in relation to soil pH. Journal of Applied Ecology 24: 659–662. [Google Scholar]
- Premoli A, Kitzberger T, Veblen T.. 2000. Isozyme variation and recent biogeographical history of the long-lived conifer Fitzroya cupressoides. Journal of Biogeography 27: 251–260. [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ.. 2014. Tracer v1.6 http://beast.bio.ed.ac.uk/Tracer (January 2017).
- Ramos VA, Ghiglione MC.. 2008. Tectonic evolution of the Patagonian Andes In: Rabassa J, ed. The late Cenozoic of Patagonia and Tierra del Fuego. Developments in quaternary sciences 11. Oxford: Elsevier, 57–71. [Google Scholar]
- Redecker D, Kodner R, Graham LE.. 2000. Glomalean fungi from the Ordovician. Science 289: 1920–1921. [DOI] [PubMed] [Google Scholar]
- Redecker D, Schüßler A, Stockinger H, Stürmer SL, Morton JB, Walker C.. 2013. An evidence-based consensus for the classification of arbuscular mycorrhizal fungi (Glomeromycota). Mycorrhiza 23: 515–531. [DOI] [PubMed] [Google Scholar]
- Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW.. 2014. InfoStat versión 2014. Córdoba, Argentina: Grupo InfoStat, Universidad Nacional de Córdoba; http://www.infostat.com.ar. [Google Scholar]
- Ronquist F, Huelsenbeck J.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R.. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Russell J, Bulman S.. 2005. The liverwort Marchantia foliacea forms a specialized symbiosis with arbuscular mycorrhizal fungi in the genus Glomus. New Phytologist 165: 567–579. [DOI] [PubMed] [Google Scholar]
- Schenck NC, Graham SO, Green NE.. 1975. Temperature and light effect on contamination and spore germination of vesicular arbuscular mycorrhizal fungi. Mycologia 67: 1189–1192. [PubMed] [Google Scholar]
- Schüßler A, Walker C.. 2010. The Glomeromycota. A species list with new families and new genera Gloucester, in libraries at The Royal Botanic Garden Edinburgh, The Royal Botanic Garden Kew, Botanische Staatssammlung Munich and Oregon State University. http://www.amd-phylogeny.com.
- Sérsic AN, Cosacov A, Cocucci AA, et al. 2011. Emerging phylogeographical patterns of plants and terrestrial vertebrates from Patagonia. Biological Journal of the Linnean Society 103: 475–494. [Google Scholar]
- Sigüenza C, Espejel I, Allen EB.. 1996. Seasonality of mycorrhizae in coastal sand dunes of Baja California. Mycorrhiza 6: 151–157. [Google Scholar]
- Sivakumar N. 2013. Effect of edaphic factors and seasonal variation on spore density and root colonization of arbuscular mycorrhizal fungi in sugarcane fields. Annals of Microbiology 63: 151–160. [Google Scholar]
- Smit E, Leeflang P, Glandorf B, van Elsas JD, Wernars K.. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Applied and Environmental Microbiology 65: 2614–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read D.. 2008. Mycorrhizal symbiosis, 3rd edn New York: Elsevier Academic Press. [Google Scholar]
- Souto CP, Mathiasen P, Acosta MC, et al. 2015. Identifying genetic hotspots by mapping molecular diversity of widespread trees: when commonness matters. Journal of Heredity 106: 537–545. [DOI] [PubMed] [Google Scholar]
- Taylor DL, Bruns TD, Leake JR, Read D.. 2002. Mycorrhizal specificity and function in myco-heterotrophic plants In: Sanders I, van der Hijden M, eds. Mycorrhizal ecology. Ecological studies, Vol. 157 Berlin: Springer, 375–413. [Google Scholar]
- Thompson JN. 2005. The geographic mosaic of coevolution. Chicago: University of Chicago Press. [Google Scholar]
- Tremetsberger K, Urtubey E, Terrab A, et al. 2009. Pleistocene refugia and polytopic replacement of diploids by tetraploids in the Patagonian and Subantarctic plant Hypochaeris incana (Asteraceae, Cichorieae). Molecular Ecology 18: 3668–3682. [DOI] [PubMed] [Google Scholar]
- Vergara R, Gitzendanner MA, Soltis DE, Soltis PS.. 2014. Population genetic structure, genetic diversity, and natural history of the South American species of Nothofagus subgenus Lophozonia (Nothofagaceae) inferred from nuclear microsatellite data. Ecology and Evolution 4: 2450–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman RJ, Klooster MR, Hentrich H, Bidartondo MI.. 2013. Species interactions of mycoheterotrophic plants: specialization and its potential consequences In: Merckx V, ed. Mycoheterotrophy. The biology of plants living on fungi. London: Springer, 267–296. [Google Scholar]
- White TJ,, Bruns T,, Lee SB,, Taylor JW.. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR protocols. New York: Academic Press, 315–322. [Google Scholar]
- Yamato M, Yagame T, Shimomura N, et al. 2011. Specific arbuscular mycorrhizal fungi associated with non-photosynthetic Petrosavia sakuraii (Petrosaviaceae). Mycorrhiza 21: 631–639. [DOI] [PubMed] [Google Scholar]
- Zayed A, Packer L, Grixti JC, Ruz L, Owen RE, Toro H.. 2005. Increased genetic differentiation in a specialist versus a generalist bee: implications for conservation. Conservation Genetics 6: 1017–1026. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.