Abstract
Heritable changes in gene expression are important contributors to phenotypic differences within and between species and are caused by mutations in cis-regulatory elements and trans-regulatory factors. Although previous work has suggested that cis-regulatory differences preferentially accumulate with time, technical restrictions to closely related species and limited comparisons have made this observation difficult to test. To address this problem, we used allele-specific RNA-seq data from Saccharomyces species and hybrids to expand both the evolutionary timescale and number of species in which the evolution of regulatory divergence has been investigated. We find that as sequence divergence increases, cis-regulatory differences do indeed become the dominant type of regulatory difference between species, ultimately becoming a better predictor of expression divergence than trans-regulatory divergence. When both cis- and trans-regulatory differences accumulate for the same gene, they more often have effects in opposite directions than in the same direction, indicating widespread compensatory changes underlying the evolution of gene expression. The frequency of compensatory changes within and between species and the magnitude of effect for the underlying cis- and trans-regulatory differences suggests that compensatory changes accumulate primarily due to selection against divergence in gene expression as a result of weak stabilizing selection on gene expression levels. These results show that cis-regulatory differences and compensatory changes in regulation play increasingly important roles in the evolution of gene expression as time increases.
Keywords: cis, trans, compensatory, yeast
Introduction
Heritable changes in gene expression are important contributors to phenotypic differences within and between species and are caused by mutations in cis-regulatory elements (e.g., promoter and enhancers) and trans-regulatory factors (e.g., transcription factors and noncoding RNAs) (Carroll 2008; Stern and Orgogozo 2008). The net contributions of cis- and trans-regulatory differences to total expression differences can be estimated by comparing the relative expression of two parental strains or species to the relative allelic expression of F1 hybrids made by crossing these strains or species (Cowles et al. 2002; Wittkopp et al. 2004). Such studies suggest that trans-regulatory differences often make larger contributions to gene expression differences than cis-regulatory differences within species (Wittkopp et al. 2004; Wang et al. 2007; Sung et al. 2009; Zhang and Borevitz 2009; Emerson et al. 2010; Bell et al. 2013; Schaefke et al. 2013; Suvorov et al. 2013; Coolon et al. 2014; Chen et al. 2015). This observation is typically attributed to the greater mutational target size of trans-regulatory mutations relative to cis-regulatory mutations (Wittkopp et al. 2004; Gruber et al. 2012; Metzger et al. 2016).
Despite this difference in mutational input, cis-regulatory differences can make substantial contributions to gene expression differences within species (Goncalves et al. 2012; Davidson and Balakrishnan 2016). In addition, cis-regulatory differences make either similar (Landry et al. 2005; Tirosh et al. 2009; McManus et al. 2010; Shi et al. 2012; Coolon et al. 2014; Lemmon et al. 2014; Guerrero et al. 2016) or greater (Zhuang and Adams 2007; Wittkopp et al. 2008a; Graze et al. 2009; Tirosh et al. 2009; Shi et al. 2012; Mack et al. 2016) contributions to expression differences than trans-regulatory differences between species, suggesting that cis-regulatory differences preferentially accumulate over time (Wittkopp et al. 2008a; Emerson and Li 2010; Gordon and Ruvinsky 2012; Coolon et al. 2014). This accumulation of cis-regulatory differences is likely due to a combination of purifying selection against trans-regulatory changes (Denver et al. 2005; Prud’homme et al. 2007) and positive selection for cis-regulatory changes (Lemos et al. 2008; Emerson et al. 2010; McManus et al. 2010; Llopart 2012; Schaefke et al. 2013; Coolon et al. 2014; Coolon et al. 2015).
Studies of regulatory divergence have also revealed that cis- and trans-regulatory differences often influence expression of the same gene (Wittkopp et al. 2004; Landry et al. 2005; Zhuang and Adams 2007; Wittkopp et al. 2008a; Graze et al. 2009; Tirosh et al. 2009; Zhang and Borevitz 2009; Emerson et al. 2010; McManus et al. 2010; Goncalves et al. 2012; Shi et al. 2012; Bell et al. 2013; Schaefke et al. 2013; Suvorov et al. 2013; Coolon et al. 2014; Lemmon et al. 2014; Chen et al. 2015; Davidson and Balakrishnan 2016; Guerrero et al. 2016; Mack et al. 2016). When regulatory differences affect gene expression in the same direction, they reinforce one another, resulting in a change in gene expression more extreme than either individual regulatory change. However, when regulatory changes have opposite effects on gene expression, they may compensate for one another, resulting in gene expression divergence less extreme than the individual regulatory changes. Interestingly, cis- and trans-regulatory differences for the same gene are more often observed in opposite directions than reinforcing changes, a pattern that is thought to be caused by natural selection (Landry et al. 2005; Goncalves et al. 2012; Bell et al. 2013; Schaefke et al. 2013; Coolon et al. 2014; Chen et al. 2015; Davidson and Balakrishnan 2016; Mack et al. 2016).
The mode of inheritance of a mutation, whether it is recessive, dominant, additive, or exhibits over or underdominance, can influence the probability that the mutation will fix in a population. For changes in gene regulation, the mode of inheritance and the mechanism of regulatory divergence are often related. In particular, cis-regulatory differences are more likely to be additive than trans-regulatory differences whereas trans-regulatory differences are more likely to be dominant or recessive than cis-regulatory differences. In addition, genes with compensatory changes are often over or underdominant (Landry et al. 2007; Lemos et al. 2008; McManus et al. 2010; Gruber et al. 2012).
Despite these patterns of regulatory evolution being observed in many studies, they are not universal. For example, the relative contribution of cis-regulatory differences to total expression differences varies amongst taxa and several recent papers have failed to observe a general increase in the relative frequency of cis-regulatory differences over time (Coolon et al. 2014; Lemmon et al. 2014; Guerrero et al. 2016). In addition, the inability to make F1 hybrids between distantly related species means that the patterns of regulatory evolution described to date only reflect observations from closely related species.
Here, we use genome-wide expression data from several Saccharomyces species to determine how cis- and trans-regulatory differences contribute to the evolution of gene expression over increasing evolutionary distance. We use RNA-seq data collected within (Schaefke et al. 2013) and between (Schraiber et al. 2013) species with sequence divergence between 1% and 40%, as well as intra and interspecific hybrids made by crossing these strains and species. This level of sequence divergence is roughly equivalent to that observed between humans and chickens (Dujon 2006; Hittinger 2013) and substantially increases the evolutionary distance upon which the evolution of cis- and trans-regulation has been interrogated, allowing long term trends in the evolution of gene regulation to be identified.
Materials and Methods
Estimates of gene expression divergence for flies and mammals were taken from Coolon et al. (2014), which used data from Brawand et al. (2011) for mammals and from McManus et al. (2010), Meisel et al. (2012), and Suvorov et al. (2013) for flies. Estimates of sequence divergence (substitutions per site) were taken from the literature for flies (Lin et al. 2008), mammals (Prasad et al. 2008), and yeast (Scannell et al. 2011). For the yeast species, these estimates were calculated using a molecular clock approach that accounts for rate variation across taxa using ∼100 genes (Scannell et al. 2011). For flies and mammals these estimates were from 4-fold degenerate sites within coding regions. In all cases, estimates of sequence divergence were derived using maximum likelihood approaches that account for the possibility of multiple substitutions at the same site.
Estimates of regulatory divergence within species used allele-specific expression data from S. cerevisiae strains BY4741 (BY) and RM11 (RM) and their F1 hybrid (Schaefke et al. 2013). These data showed good agreement with previous data from the same cross (Spearman’s rho > 0.87 for both cocultures and hybrids) (Emerson et al. 2010). However, the coverage of this previous experiment was substantially lower and only the data from Schaefke et al. 2013 were used. Relative allelic expression within the F1 hybrids suggested the presence of either aneuploidy or a large copy number variant on both chromosomes II and XIII and these regions were removed from further analysis in all comparisons. No evidence of these variants was observed in the Emerson et al. 2010 data. Estimates of regulatory divergence between species used allele-specific expression data from S. cerevisiae (YHL068), S. paradoxus (CBS 432), S. mikatae (IFO 1815), S. bayanus (CBS 7001), and F1 hybrids between S. cerevisiae and each of the other three Saccharomyces species (Schraiber et al. 2013). In total, eight comparisons were made: four total expression comparisons between parental strains or species, and four allele-specific expression comparisons in F1 hybrids. For each comparison, read counts at each gene from two independent biological replicates were combined. Because differences in total expression influences the power to detect regulatory changes, total counts were down-sampled to the minimum value observed across comparisons for each gene using Fisher’s noncentral hypergeometric distribution as implemented in the BiasedUrn R package (Fog 2015). As a result of this down-sampling, each gene in each comparison has the same number of allele-specific reads, thus removing differences in power across samples (Coolon et al. 2014). Genes with <20 total reads in any comparison were removed and only the set of genes with data across all comparisons used. In total, 3133 genes were retained (supplementary additional file 2, Supplementary Material online).
For each of the four strain/species comparisons, differences in total expression were estimated as the log2 ratio of counts in the parental strains or species for each gene (log2(P1/P2), where P1 and P2 are the allele specific reads from strain/species 1 and 2, respectively). Likewise, the log2 ratio of allele-specific counts within the F1 hybrids were used to estimate cis-regulatory differences for each gene (log2(H1/H2), where H1 and H2 are the allele specific reads from the hybrid). Estimates of trans-regulatory differences for each gene were taken as the difference between total parental expression differences and hybrid cis-regulatory differences (log2(P1/P2) − log2(H1/H2)).
Following previous work, the extent of gene expression conservation was estimated by correlating read counts (P1 vs. P2) for each gene using Spearman’s rho (Brawand et al. 2011; Meisel et al. 2012; Coolon et al. 2014). One minus Spearman’s rho was then used to estimate gene expression divergence. An identical approach was used to estimate cis-regulatory divergence by correlating allele specific reads from hybrids (H1 vs. H2). To estimate trans-regulatory divergence, trans-regulatory differences were first used to estimate the number of reads expected due to trans-regulatory changes (T1 and T2). This was done by assuming that the log2 ratio of these counts was equal to the trans-regulatory difference (i.e., log2(T1/T2) = log2(P1/P2) − log2(H1/H2)) and that the total number of reads was the same as the parental and hybrids comparisons (i.e., T1 + T2 = P1 + P2 = H1 + H2). From these equations, the values of T1 and T2 were determined for each gene and trans-regulatory divergence calculated from the correlation of T1 and T2.
To estimate expression and regulatory divergence using a metric independent of correlations, we estimated D from Meisel et al. (2012). This metric is the absolute value of the difference in expression scaled by half the sum of expression. For total expression divergence, we used the difference in allele specific counts between the parental strains and species. For cis-regulatory divergence, we used the difference in allele specific counts in hybrids. For trans-regulatory divergence, we used the difference in the estimated trans-regulatory allele specific counts.
To determine the dominant regulatory mechanism by which expression diverged for each comparison, Spearman’s rho was used to correlate the estimates of total expression divergence with the estimates of cis- and trans-regulatory divergence. To calculate the percent of regulatory divergence due to cis-regulatory differences at each gene within each comparison, absolute cis-regulatory differences were divided by the sum of absolute cis-regulatory differences plus absolute trans-regulatory differences (|c|/(|c|+|P-c|), where c is the difference in allele-specific expression within a hybrid, or cis-regulatory divergence, and P is the difference in expression between two strain or species, or Parental expression difference). The percent of regulatory divergence due to cis-regulatory divergence was compared at different sequence divergence amounts using a Wilcoxon rank sum test. To determine the change in magnitude of expression and regulatory differences with sequence divergence, the median absolute difference in total expression, cis-, and trans-regulation for each comparison were used.
To test for differences in the frequency of regulatory change, we categorized each gene within each comparison using a series of statistical tests. To test for significant differences in total expression, allele-specific counts between parental strains or species were compared using a binomial exact test for each gene. To test for significant cis-regulatory differences, allele-specific counts from F1 hybrids were compared using a binomial exact test for each gene. To detect significant trans-regulatory differences, Fisher’s exact test was used to compare the ratio of allele-specific counts in the parental strains or species with the ratio of allele-specific counts in the F1 hybrids for each gene. The frequency of regulatory changes was determined from the criteria listed in supplementary additional file 1, table S1, Supplementary Material online. Because genes that would have been categorized as ambiguous in previous work typically had low read counts, they were combined with the conserved category (McManus et al. 2010; Coolon et al. 2014). For all tests, a false discovery rate (FDR) corrected P value of 0.01 was used (Benjamini and Hochberg 1995).
To test for differences in the mode of inheritance, the total read count of both alleles was used. Differences in power between strains and species was removed by down-sampling across all comparisons using Fisher’s noncentral hypergeometric distribution within the R package, biased Urn (Fog 2015). Any gene in which the average number of reads across samples was below 20 was removed (supplementary additional file 3, Supplementary Material online). A binomial exact test was then used to compare total expression between parental strains or species as well as between each parental strain or species and their F1 hybrid. Each gene was classified into one of four categories based on these statistical tests (supplementary additional file 1, table S2, Supplementary Material online). An FDR corrected P value cutoff of 0.01 was used to determine statistical significance. Enrichment for the overlap between regulatory and inheritance categories was calculated using chi-square tests. For each test, a 2 × 2 contingency table was created by collapsing all combinations of categories but one. Each combination of regulatory and inheritance categories was tested individually.
Intraspecific data from (Schaefke et al. 2013) can be accessed at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lzeddgeiamgkkfk&acc=GSE46838 (last accessed Feburary 27, 2017) and http://www.ncbi.nlm.nih.gov/bioproject/PRJNA194385 (last accessed Feburary 27, 2017). Interspecific data from (Schraiber et al. 2013) can be accessed at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38875 (last accessed Feburary 27, 2017). A graphical depiction of how the metrics used are related is included in supplementary additional file 1, figure S4, Supplementary Material online. In all cases, 99% confidence intervals were determined by bootstrapping 10,000 times and P values determined using 10,000 permutations. Processed data used for supporting the conclusions of this article are included in supplementary additional files 2 and 3, Supplementary Material online. The code used for analysis is included in supplementary additional files 4 and 5, Supplementary Material online. All calculations were done using R 3.1.3 (R Core Team 2013).
Results
Gene Expression and Regulatory Divergence Reach a Plateau As Sequence Divergence Increases
To determine how gene expression and regulation evolve, we used allele specific RNA-seq data from within and between several Saccharomyces yeast species. For short evolutionary time scales, we used two S. cerevisiae strains, BY and RM (0.0028 substitutions per site) (Schaefke et al. 2013; Maclean et al. 2016). For sequentially longer evolutionary time scales, we compared S. cerevisiae (Sc) to S. paradoxus (Sp, 0.30 substitutions per site), S. mikatae (Sm, 0.48 substitutions per site), and S. bayanus (Sb, 0.90 substitutions per site) (Scannell et al. 2011; Schraiber et al. 2013). We quantified genome-wide divergence in gene expression and regulation using Spearman’s rho. Because all correlations are sensitive to the dynamic range of expression, we used a common set of genes expressed in all strains and species, down sampling the counts for each gene to be equivalent across all comparisons.
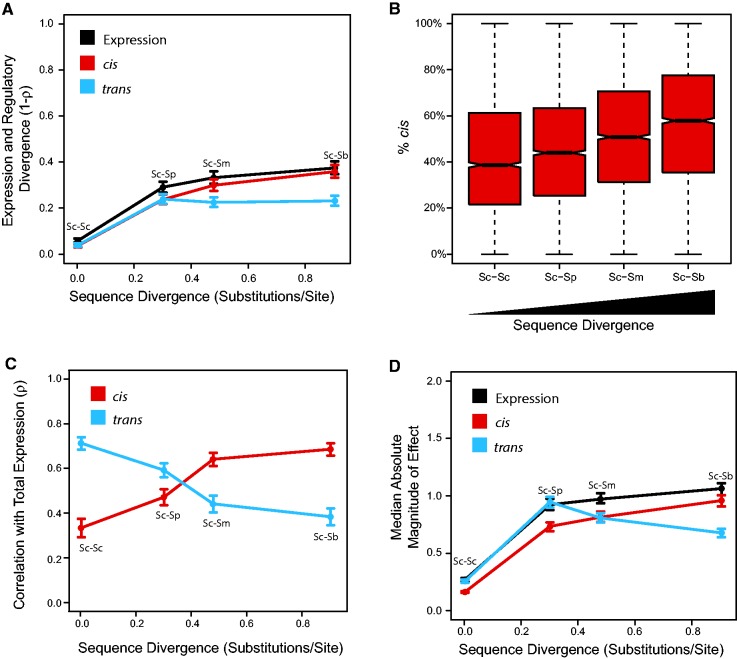
We found that as sequence divergence increased, gene expression divergence increased significantly (fig. 1A, black; P < 0.01 for all comparisons, permutation test). However, the rate of expression divergence slowed with time, suggesting a plateau in the extent of gene expression divergence. Using allele specific expression from F1 hybrids (Sc × Sc, Sc × Sp, Sc × Sm, and Sc × Sb) to measure cis-regulatory differences, we observed a similar pattern (fig. 1A, red; P < 1 × 10−5 for all comparisons, permutation test). We next compared total expression differences to cis-regulatory differences to estimate trans-regulatory differences, using these differences to calculate trans-regulatory divergence. While trans-regulatory divergence was lowest within species (fig. 1A, blue; P < 1 × 10−5 for all comparisons of within versus between species, permutation test), all between species comparisons were not significantly different, suggesting that trans-regulatory divergence had reached a plateau (P > 0.3 for all comparisons, permutation test). In addition, while cis- and trans-regulatory divergence were not significantly different within species, or for the closest between species comparison, trans-regulatory divergence was significantly lower than cis-regulatory divergence for the two greatest sequence divergence comparisons (P < 1 × 10−5 for both comparisons, permutation test). Using a metric of gene expression divergence not based on correlations produced similar patterns (supplementary additional file 1, fig. S1, Supplementary Material online) (Meisel et al. 2012). These results suggest that both regulatory and expression divergence plateau as sequence divergence increases, albeit at different rates.
Fig. 1.—
cis-regulatory differences become the dominant form of regulatory divergence between species. (A) Total expression and regulatory divergence over time. Divergence in gene expression and regulation were calculated as one minus Spearman’s rho. Sequence divergence was calculated from 4-fold degenerate sites within coding regions. Error bars are 99% CI from bootstrap analysis. (B) Percent of total regulatory divergence due to cis-regulatory differences. Notches in box plots represent 95% CI for the median. (C) Correlation between total expression divergence and cis-regulatory divergence or trans-regulatory divergence. Error bars are 99% CI from bootstrap analysis. (D) Median absolute magnitude of difference in total expression and regulation. Error bars are 99% CI from bootstrap analysis. Black: Total expression, Red: cis-regulatory, Blue; trans-regulatory. Sc: S. cerevisiae. Sp: S. paradoxus. Sm: S. mikatae. Sb: S. bayanus.
cis-Regulatory Differences Become the Dominant Mechanism of Regulatory Change As Sequence Divergence Increases
Differences in the plateaus of cis- and trans-regulatory divergence suggest differences in the rates of accumulation of cis- and trans-regulatory differences, and thus changes in the relative contributions of cis- and trans-regulatory differences, over time. To estimate how the proportion of total regulatory changes varies with sequence divergence, we calculated the percent of total regulatory change due to cis-regulatory differences (%cis). We found that the median percentage of regulatory divergence due to cis-regulatory differences increased significantly as sequence divergence increased, ranging from approximately 40% within species to nearly 60% between Sc and Sb (P < 8 × 10−6 for all comparisons, Wilcoxon rank sum test, fig. 1B). Consistent with this result, trans-regulatory divergence correlated best with total expression divergence within species, whereas cis-regulatory divergence became the better predictor of gene expression divergence as sequence divergence increased (fig. 1C).
Changes in the relative contribution of cis- and trans-regulatory differences to total regulatory divergence can be caused by changes in the magnitude of cis- and trans-regulatory differences. We thus estimated the absolute difference in expression and regulation for each gene for each comparison. For total expression, the median difference was lowest between Sc strains (P < 1 × 10−5 for all comparisons, permutation test) and largest between Sc and Sb (fig. 1D, black; P < 1 × 10−5, permutation test). Similarly, the median absolute cis-regulatory difference increased significantly as sequence divergence increased (fig. 1D, red; P < 1 × 10−5 for all comparisons, permutation test). By contrast, while the median absolute trans-regulatory difference was higher between species than within species (P < 1 × 10−5 for comparing within Sc to Sc vs. Sp, permutation test), trans-regulatory differences between species were significantly smaller at greater sequence divergence levels (fig. 1D, blue; P < 1 × 10−5 for all comparisons, permutation test).
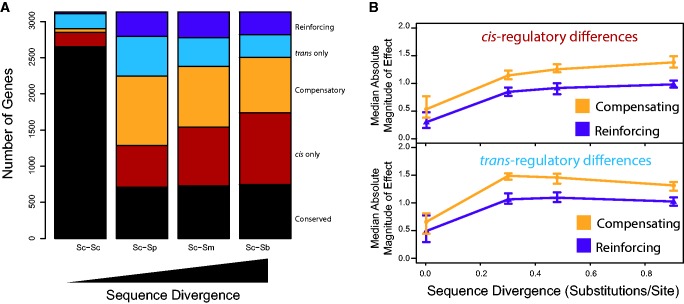
Changes in the relative contribution of cis- and trans-regulatory differences to total regulatory divergence can also be caused by difference in the frequency of cis- and trans-regulatory differences. We thus estimated the frequency of regulatory changes by classifying individual genes within each comparison into one of five regulatory divergence classes based on statistical support: cis-regulatory only, trans-regulatory only, reinforcing (cis- and trans- differences in the same direction), compensating (cis- and trans-differences in opposite directions), and conserved (none of the above). These classifications are similar to those used in previous work (McManus et al. 2010; Coolon et al. 2014), but combine genes with significant cis- and trans-regulatory differences in opposite directions into a single compensatory class instead of distinguishing between partial (cis × trans in previous work) and full compensation (compensatory in previous work) (Landry et al. 2005; Schaefke et al. 2013). As a consequence of this classification, the statistical power to detect compensatory and reinforcing changes is equivalent.
Using this classification scheme, we found that the fraction of genes with a conserved pattern of expression was highest within species (BY vs. RM, 85%, P < 1x10−15, chi-square test, fig. 2A) and relatively constant between species (∼23%, P = 0.63, chi-square test). The fraction of genes with only cis-regulatory differences increased with sequence divergence, going from approximately 6% within species to over 30% between Sc and Sb (P < 7 × 10−7 for all comparisons, chi-square test). This latter result is consistent with the increased role of cis-regulatory differences in total expression divergence as sequence divergence increases. By contrast, the number of genes with only trans-regulatory differences increased initially from within to between species (∼7–17%, P < 1 × 10−15, chi-square test), but then decreased significantly as sequence divergence increased (P < 0.001 for all comparisons, chi-square test). As a consequence, approximately 10% of genes had only a trans-regulatory difference in expression at the longest evolutionary distances considered. Similarly, the proportion of genes with any trans-regulatory change (trans-only, compensatory, and reinforcing) decreased with increasing sequence divergence between species (P < 8 × 10−7 for all comparisons, chi-square test). Thus, while the frequency of cis-regulatory differences increases as sequence divergence increases, the frequency of trans-regulatory differences decreases between species as sequence divergence increases.
Fig. 2.—
Compensatory changes in regulation are common between species. (A) Number of genes within each regulatory divergence category versus sequence divergence. Genes were classified into one of five categories based on statistical support: Black, conserved. Red, cis-regulatory difference only. Orange, compensatory. Blue, trans-regulatory difference only. Purple, reinforcing. (B) Median absolute magnitude of effect for the cis-regulatory (top) and trans-regulatory (bottom) components of genes with a compensatory mode of regulatory divergence (orange) or reinforcing mode of regulatory divergence (purple) versus sequence divergence. Error bars are 99% CI from bootstrap analysis.
Compensatory Changes in Gene Regulation Are Common
Within species, the vast majority of genes with regulatory differences had either a cis-regulatory difference (41%) or a trans-regulatory difference (43%), and only rarely had both (16%) (fig. 2A). By contrast, between species, genes with both cis- and trans-regulatory differences were a greater fraction of all genes (∼35–40%) than were genes with only cis-regulatory differences (∼18–32%) or only trans-regulatory differences (∼10–17%), indicating that differences in regulation accumulate at multiple levels between species. Amongst genes showing both cis- and trans-regulatory differences, we observed that compensatory differences were ∼2–3 times more common than reinforcing differences. This abundance of compensatory changes means that between species, ∼25–30% of all genes show evidence of compensatory changes and that compensatory changes are either the largest or second largest regulatory divergence category (behind cis-regulatory only). Thus, compensatory changes are a major mechanism by which gene expression evolves between species.
Because neutral evolution is predicted to create equal numbers of reinforcing and compensatory differences, the greater frequency of compensatory differences relative to reinforcing differences is thought to reflect natural selection preferentially fixing beneficial compensatory mutations (Landry et al. 2005; McManus et al. 2010; Goncalves et al. 2012; Mack et al. 2016). However, an overabundance of compensatory changes could also arise from selection acting against reinforcing changes due to stabilizing selection on gene expression levels (Lemos et al. 2005; Schraiber et al. 2013; Hodgins-Davis et al. 2015) or from opposing biases in the direction of cis- and trans-regulatory mutations (Metzger et al. 2016), rather than from selection for compensatory changes.
To distinguish amongst these possibilities, we first compared the relative frequency of compensatory and reinforcing changes within and between species. If compensatory changes are primarily fixed by positive selection, their frequency relative to reinforcing changes should increase as sequence divergence increases. However, we found that the relative frequency of compensatory and reinforcing differences was not significantly different across comparisons (∼68–74% compensatory, fig. 2A, P = 0.13, chi-square test), suggesting that positive selection for compensatory changes is not the dominant mechanism responsible for creating the greater frequency of compensatory changes.
Reinforcing changes in gene expression are expected to have a greater effect on, and move expression further from its optimum than, compensatory changes. In the presence of stabilizing selection, large changes from the optimal level of expression are deleterious and reinforcing changes are thus expected to be eliminated more often by stabilizing selection than compensatory changes. As a consequence, in the presence of stabilizing selection, the cis- and trans-regulatory changes contributing to a reinforcing change are expected to be smaller than the cis- and trans-regulatory changes contributing to compensatory changes. By contrast, if the overabundance of compensatory changes is due to opposing biases in the direction of cis- and trans-regulatory changes, no difference in the magnitude of regulatory differences is expected between reinforcing and compensatory changes. Consistent with widespread stabilizing selection on gene expression levels, we found that genes with a compensatory pattern of regulatory divergence have a 1.4-fold (1.2–1.6, 95% CI) greater cis-regulatory difference and a 1.3-fold (1.2–1.5, 95% CI) greater trans-regulatory difference than genes with a reinforcing pattern of regulatory divergence between species (fig. 2B, P < 1 × 10−5 for both cis- and trans-regulatory effects, bootstrap). These results suggest that selection against reinforcing changes due to stabilizing selection on gene expression levels is the primary mechanism responsible for the preferential accumulation of compensatory changes between species.
Mode of Inheritance Is Closely Tied to the Mechanism of Regulatory Divergence
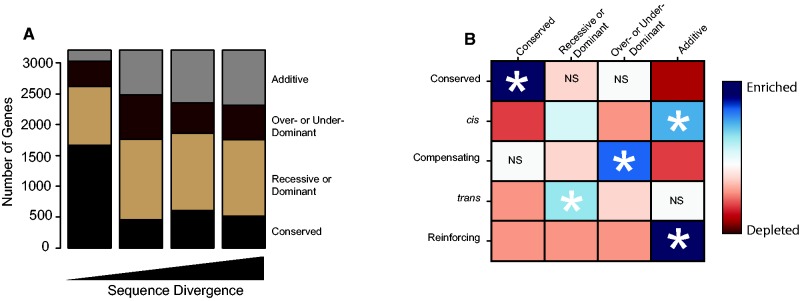
To determine how inheritance patterns change as sequence divergence increases, we divided all genes into four inheritance categories for each comparison based on statistical tests: conserved (no total expression difference between the hybrid and either parent), recessive/dominant (total expression difference between the hybrid and one parent only), additive (total expression difference between the hybrid and both parents, with hybrid expression intermediate to that of the parents), and over/underdominant or misexpressed (total expression difference between the hybrid and both parents, with hybrid expression more extreme than both parents). Consistent with expression and regulatory divergence between species, we found that the number of conserved genes was higher within than between species (fig. 3A, P < 1 × 10−15, chi-square test). In addition, we found that the number of genes with an additive mode of inheritance increased with divergence (P = 4 × 10−11, chi-square test). By contrast, the recessive/dominant and over/underdominant categories showed more complicated patterns that were not consistent as sequence divergence increased.
Fig. 3.—
Mode of inheritance and mechanism of regulatory divergence are closely related. (A) Number of genes within each mode of inheritance category versus sequence divergence. Genes were classified into one of four categories based on statistical support: Black, conserved. Light brown, dominant/recessive. Dark brown, misexpressed. Gray, additive. (B) Comparison of regulatory divergence categories and modes of inheritance categories. Mode of inheritance categories are on the top. Intersections show magnitude of enrichment (blue) and depletion (red) of genes within each combination of regulatory and inheritance categories. Asterisks mark the strongest enrichment for each regulatory divergence category. Statistical significance of enrichment and depletion is shown by the brightness of each box, with NS signifying nonsignificant effects and darker colors corresponding to lower P values. All categories not marked by NS are significant at less than a Bonferroni corrected P value of 0.0005.
To determine whether associations between regulatory divergence categories and mode of inheritance categories are stable as gene regulation evolves, we compared the observed number of genes within each combination of regulatory divergence and mode of inheritance categories to the expected number in the absence of an association. We found clear correspondences between regulatory divergence class and mode of inheritance class such that each regulatory divergence class was highly enriched for a single mode of inheritance (fig. 3B). For example, genes with conserved patterns of regulatory divergence were enriched for genes with conserved inheritance and depleted for genes with additive modes of inheritance. In addition, genes with cis-regulatory differences had largely additive effects. Interestingly, genes with a reinforced pattern of divergence were also enriched for additive effects, however, the cause of this association is unclear. In contrast to cis-regulatory and reinforcing differences, trans-regulatory differences were enriched for being either dominant or recessive in one strain/species relative to another; which specific allele was dominant and which allele was recessive was largely dependent on the specific gene and cross. Finally, we observed that compensatory changes were enriched for over and underdominance, consistent with transgressive expression due to opposing effects of regulatory mutations in different genetic backgrounds that are exposed within hybrids (Landry et al. 2005; McManus et al. 2010). Overall, we found highly congruent patterns across comparisons that were independent of sequence divergence (supplementary additional file 1, fig. S2, Supplementary Material online). These results suggest that the mode of inheritance and mechanism of regulatory divergence are highly correlated, are established early during evolution, and are largely maintained over time.
Discussion
This study provides the first systematic look at the evolution of cis- and trans-regulatory changes over a long range of evolutionary distances. We find that 1) cis-regulatory differences play an ever-greater role in gene expression divergence relative to trans-regulatory differences as sequence divergence increases; 2) compensatory changes in regulation are a common mechanism underlying the evolution of gene expression; and 3) the evolution of gene expression and regulation follows consistent patterns across taxa.
cis-Regulatory Divergence Becomes the Better Predictor of Expression As Sequence Divergence Increases
Two of the clearest patterns we observe as sequence divergence increases are an increase in the proportion of regulatory divergence due to cis-regulatory differences and an increase in the importance of cis-regulatory divergence in explaining total regulatory divergence. These patterns are consistent with predictions about the evolution of cis-regulatory sequences (Wray et al. 2003; Wittkopp 2005; Wray 2007), as well as data from early studies of regulatory divergence between species (Zhuang and Adams 2007; Wittkopp et al. 2008a; Graze et al. 2009; Tirosh et al. 2009). However, several recent studies have failed to consistently find this pattern, suggesting a more complicated relationship between cis-regulatory divergence and evolutionary distance (Coolon et al. 2014; Lemmon et al. 2014; Guerrero et al. 2016). One possible source of this complication is unique demographic differences in the species compared. For example, comparisons of regulatory divergence between three Drosophila species was potentially influenced by the unique evolutionary and demographic history of D. sechellia (Coolon et al. 2014). Likewise, both Solanum pennellii (tomato, Guerrero et al. 2016) and Zea mays (corn, Lemmon et al. 2014) are domesticated plants and have therefore experienced unique evolutionary histories which may result in differences in the contributions of cis- and trans-regulatory differences relative to undomesticated taxa. In the case of the Saccharomyces yeast studied here, the lineage most likely to have a unique evolutionary history is that of S. cerevisiae due to its domestication and widespread use in industrial and agricultural fermentation processes (Hittinger 2013; Liti 2015). However, all comparisons are made with S. cerevisiae and any aspects of its evolutionary history that are unusual are shared amongst all comparisons made in this study and thus unlikely to be responsible for the observed patterns.
Another reason a relationship between evolutionary distance and cis-regulatory divergence may be observed here and not in other studies is that the yeast species studied are more distantly related than species used in previous work, resulting in a stronger and easier to detect pattern. Interestingly, cis-regulatory divergence, like gene expression divergence, reaches a plateau with increasing sequence divergence. As a consequence, cis-regulatory divergence is most similar for the three between species comparisons and the within species comparison appears to be the outlier. Thus, if the relative excess of trans-regulatory differences within species compared with between species is dependent on the system studied, then until sufficient sequence divergence has occurred, the relative proportions of cis- and trans-regulatory divergence between different taxonomic groups should not be directly compared. This interpretation would account for the variable contributions of cis-regulatory differences observed between species in different systems while still being consistent with a preferential accumulation of cis-regulatory differences that is observed within the same system. By comparing strains that differ in their evolutionary relatedness, but are still the same species, future work should be able to determine the extent to which cis-regulatory divergence depends on divergence time versus the speciation process.
Compensatory Changes in Regulation Are a Major Mechanism Underlying the Evolution of Gene Expression
Perhaps the most unexpected aspect of the data is the plateau in both expression and regulatory divergence with increasing sequence divergence. In particular, the low amount of trans-regulatory divergence relative to cis-regulatory and total expression divergence, as well as the comparatively low sequence divergence levels at which the plateau occurs, have not previously been observed. There are at least three, nonmutually exclusive, possible explanations for the pattern of trans-regulatory divergence. First, trans-regulatory divergence may simply reflect greater rates of trans-regulatory divergence between S. paradoxus and S. cerevisiae relative to the comparisons between S. cerevisiae and S. mikatae or S. bayanus. Such a difference could arise due to differences in the action of natural selection or differences in the frequency and magnitude of trans-regulatory mutations along specific branches. Initial analysis of the interspecific data using techniques to identify changes in selective pressure within genes from a common pathway identified acceleration of trans-regulatory divergence specifically within the S. paradoxus branch (Schraiber et al. 2013). However, because all comparisons of regulatory divergence are made to a common reference species (S. cerevisiae), it is difficult to polarize changes in regulation and identify the individual branches in which they were most likely to occur. As a consequence, lineage-specific changes are both difficult to show and difficult to rule out given the current data. One potential resolution to this problem would be to measure regulatory divergence for all pairwise crosses between multiple species to identify the branches on which regulatory changes likely occurred and obtain independent measurements of regulatory divergence for species with similar levels of sequence divergence.
Second, the methodology used to identify cis- and trans-regulatory differences can potentially misinterpret combined cis- and trans-regulatory changes as cis-only regulatory changes. For example, if the trans-regulatory elements of two species do not interact equally with the two cis-regulatory alleles within a hybrid, then the trans-regulatory changes can be incorrectly added to the cis-regulatory differences (Wittkopp et al. 2008b; Takahasi et al. 2011). In the extreme case, where the trans-regulatory factors from each species only interact with their cognate cis-regulatory sequences, no trans-regulatory differences would be observed using our method. Such regulatory incompatibilities may increase with sequence divergence, potentially resulting in an increase in the proportion of cis-regulatory divergence and an apparent decrease in the occurrence of trans-regulatory divergence with increasing sequence divergence. Although recent work has highlighted several instances of trans-regulatory rewiring (Voordeckers et al. 2015), these case studies are rare and have been documented primarily between much more distantly related species than studied here. For species which can readily hybridize in the wild, such as those used in the current work (Hittinger 2013), it seems unlikely that the majority of both species regulatory networks would no longer interact with one another. In addition, an increase in the incompatibility of cis- and trans-regulatory elements at larger sequence divergences should result in a decrease in the frequency of trans-regulatory divergence across all regulatory divergence categories. However, the category with the largest decrease in frequency is for trans-regulatory only differences, with compensatory changes showing a much smaller decrease, and reinforcing changes showing no difference in frequency regardless of sequence divergence. Thus, while regulatory incompatibilities between cis- and trans-regulatory elements may contribute to the observed pattern, they seem unlikely to explain the entire relationship between trans-regulatory divergence and sequence divergence.
Finally, because evolution is fundamentally a tug of war between forces that change and forces that maintain organismal form and function, ample opportunities exist for compensatory changes. The employed methodology is intended to detect the net contribution of cis- and trans-regulatory differences to expression differences and is not intended to determine the exact number of regulatory changes that have occurred. Thus, if two trans-regulatory changes act in opposite directions in the same lineage, resulting in trans/trans compensation, the net result would likely be a reduced trans-regulatory difference compared with either trans-regulatory change alone. As a consequence, trans-regulatory divergence would be relatively small, even though several substitutions had occurred. If such compensation becomes increasingly common as sequence divergence increases, as is expected given the continuous nature of the mutational process, then trans-regulatory divergence will eventually become saturated, resulting in a plateau in trans-regulatory divergence relative to sequence divergence. This process can also occur between cis-regulatory changes in opposite directions, resulting in cis/cis compensation. However, because trans-regulatory mutations have a greater mutational target size than cis-regulatory mutations (Gruber et al. 2012; Metzger et al. 2016), the possibility for trans/trans compensation should be higher, and happen more quickly during evolution, than for either cis/trans compensation or cis/cis compensation. Thus, the high frequency of cis/trans compensation observed between these species suggests that trans/trans compensation is an important but underappreciated mechanism governing the evolution of gene regulation. Consistent with this idea, measurements of cis- and trans-regulatory divergence using introgression lines, which contain only small portions of the genome integrated into an otherwise constant genetic background and are therefore less likely than F1 hybrids to contain compensatory trans/trans pairs, often find a greater role for trans-regulatory differences than is observed using F1 hybrids (Takahasi et al. 2011; Gordon and Ruvinsky 2012; Meiklejohn et al. 2014; Guerrero et al. 2016). Future work will be needed to directly estimate the frequencies of both trans/trans and cis/cis compensation and determine the extent to which they contribute to the evolution of gene expression and regulation.
The Evolution of Gene Expression and Regulation May Follow a Consistent Pattern across Taxa
A plateau in the rate of gene expression divergence with increasing evolutionary divergence has previously been observed in Drosophila, mammals, and yeast (Bedford and Hartl 2009; Coolon et al. 2014). Using the expected number of substitutions per site as an estimate of sequence divergence suggests that the relationship between sequence divergence and expression divergence may be consistent across these systems (supplementary additional file 1, fig. S3, Supplementary Material online). This pattern may reflect stabilizing selection acting on gene expression levels (Lemos et al. 2005; Bedford and Hartl 2009; Schraiber et al. 2013; Hodgins-Davis et al. 2015) and is consistent with recent work showing a common evolutionary rate among metazoan transcriptional networks (Carvunis et al. 2015). These results are consistent with the observation that the relationship between regulatory divergence and mode of inheritance is stable over long evolutionary timescales. Together, these observations suggest that there may be mechanisms responsible for the evolution of gene expression that are consistent between distantly related taxa.
We propose that the observations to date suggest a simple model for the evolution of gene expression that relies on weak, but widespread, stabilizing selection on expression levels, differences in the target size of cis- and trans-regulatory mutations, and opposing differences in the fixation probability of cis- and trans-regulatory changes due to differences in additivity (and/or pleiotropy). The combination of weak stabilizing selection on expression levels and large target size for trans-regulatory changes is consistent with trans-regulatory differences being the most common form of regulatory change observed within species. Differences in the fixation probability of cis- and trans-regulatory mutations due to differences in additivity and pleiotropy are then expected to cause a disproportionate increase in cis-regulatory changes over time. In addition, the combination of stabilizing selection on expression levels and the large target size for trans-regulatory mutations is expected to cause trans–trans compensation for expression at relatively low sequence divergence levels. As a consequence, net trans-regulatory divergence should stop increasing prior to net cis-regulatory divergence, further contributing to an increase in the proportion of regulatory divergence due to cis-regulatory changes between species. Finally, selection against changes in expression should cause the preferential fixation of compensatory cis/trans changes relative to reinforcing changes and a corresponding reduction in the rate of gene expression divergence with time. Work is now needed to test these hypotheses and determine the extent to which these patterns and mechanisms are present across a range of taxa.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
B.P.H.M., P.J.W., and J.D.C. conceived of study. B.P.H.M. performed all analyses. B.P.H.M. and J.D.C. wrote the manuscript with input from P.J.W. All authors read and approved the final manuscript.
Acknowledgments
We thank Fabien Duveau, Jennifer Lachowiec, and Mo Sidiq for providing comments on the manuscript. We thank J.J. Emerson for help accessing data. This work was supported by the National Science Foundation (MCB-1021398) and National Institutes of Health (1 R01 GM108826) for P.J.W., the University of Michigan Rackham Graduate School and National Institutes of Health Genome Sciences training grant (T32 HG000040) to B.P.H.M., and Wesleyan University startup funds for J.D.C. No funding body was involved in the design, collection, analysis, interpretation of data, or writing of the manuscript.
Literature Cited
- Bedford T, Hartl DL. 2009. Optimization of gene expression by natural selection. Proc. Natl. Acad. Sci. U. S. A. 106:1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GDM, Kane NC, Rieseberg LH, Adams KL. 2013. RNA-seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol Evol. 5:1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. [Google Scholar]
- Brawand D, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478:343–348. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. [DOI] [PubMed] [Google Scholar]
- Carvunis A-R, et al. 2015. Evidence for a common evolutionary rate in metazoan transcriptional networks. Elife 4:e11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nolte V, Schlötterer C. 2015. Temperature stress mediates decanalization and dominance of gene expression in Drosophila melanogaster. PLoS Genet. 11:e1004883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J, McManus CJ, Stevenson KR, Graveley BR, Wittkopp PJ. 2014. Tempo and mode of regulatory evolution in Drosophila. Genome Res. 24:797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon JD, et al. 2015. Molecular mechanisms and evolutionary processes contributing to accelerated divergence of gene expression on the Drosophila X chromosome. Mol Biol Evol. 32:2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. 2002. Detection of regulatory variation in mouse genes. Nat Genet. 32:432–437. [DOI] [PubMed] [Google Scholar]
- Davidson JH, Balakrishnan CN. 2016. Gene regulatory evolution during speciation in a songbird. G3 6:1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, et al. 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 37:544–548. [DOI] [PubMed] [Google Scholar]
- Dujon B. 2006. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 22:375–387. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, et al. 2010. Natural selection on cis and trans regulation in yeasts. Genome Res. 20:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Li W-H. 2010. The genetic basis of evolutionary change in gene expression levels. Philos Trans R Soc Lond B Biol Sci. 365:2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fog A. 2015. BiasedUrn: Biased Urn Model Distributions. R package version 1.07. https://CRAN.R-project.org/package=BiasedUrn.
- Goncalves A, et al. 2012. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 22:2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KL, Ruvinsky I. 2012. Tempo and mode in evolution of transcriptional regulation. PLoS Genet. 8:e1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze RM, McIntyre LM, Main BJ, Wayne ML, Nuzhdin SV. 2009. Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics 183:547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber JD, Vogel K, Kalay G, Wittkopp PJ. 2012. Contrasting properties of gene-specific regulatory, coding, and copy number mutations in Saccharomyces cerevisiae: frequency, effects and dominance. PLoS Genet. 8:e1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Posto AL, Moyle LC, Hahn MW. 2016. Genome-wide patterns of regulatory divergence revealed by introgression lines. Evolution 70:696–706. [DOI] [PubMed] [Google Scholar]
- Hittinger CT. 2013. Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 29:309–317. [DOI] [PubMed] [Google Scholar]
- Hodgins-Davis a, Rice DP, Townsend JP. 2015. Gene expression evolves under a house-of-cards model of stabilizing selection. Mol Biol Evol. 32:2130–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. 2007. Genetic properties influencing the evolvability of gene expression. Science 317:118–121. [DOI] [PubMed] [Google Scholar]
- Landry CR, et al. 2005. Compensatory cis–trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171:1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon ZH, Bukowski R, Sun Q, Doebley JF. 2014. The role of cis regulatory evolution in maize domestication. PLoS Genet. 10:e1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL. 2008. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc Natl Acad Sci U S A. 105:14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Meiklejohn CD, Cáceres M, Hartl DL. 2005. Rates of divergence in gene expression profiles of primates, mice, and flies: stabilizing selection and variability among functional categories. Evolution 59:126–137. [PubMed] [Google Scholar]
- Lin MF, Deoras AN, Rasmussen MD, Kellis M. 2008. Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. PLoS Comput Biol. 4:e1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G. 2015. The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife 4:e05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A. 2012. The rapid evolution of X-linked male-biased gene expression and the large-X effect in Drosophila yakuba, D. santomea, and their hybrids. Mol Biol Evol. 29:3873–3886. [DOI] [PubMed] [Google Scholar]
- Mack KL, Campbell P, Nachman MW. 2016. Gene regulation and speciation in house mice. Genome Res. 26:451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean CJ, Metzger BPH, Yang J-R, Ho W-C, Moyers B, Zhang J., et al. 2017. Deciphering the genic basis of environmental adaptations by simultaneous forward and reverse genetics in Saccharomyces cerevisiae. In Review. [DOI] [PubMed] [Google Scholar]
- McManus CJ, et al. 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20:816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Coolon JD, Hartl DL, Wittkopp PJ. 2014. The roles of cis- and trans-regulation in the evolution of regulatory incompatibilities and sexually dimorphic gene expression. Genome Res. 24:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Malone JH, Clark AG. 2012. Faster-X evolution of gene expression in Drosophila. PLoS Genet. 8:e1003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger BPH, et al. 2016. Contrasting frequencies and effects of cis- and trans-regulatory mutations affecting gene expression. Mol Biol Evol. 33:1131–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad AB, Allard MW, Green ED. 2008. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 25:1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Carroll SB. 2007. Emerging principles of regulatory evolution. Proc Natl Acad Sci U S A. 104:8605–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: A Language and Environment for Statistical Computing. Available from: http://www.r-project.org/.
- Scannell DR, et al. 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 1:11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefke B, et al. 2013. Inheritance of gene expression level and selective constraints on trans- and cis- regulatory changes in yeast. Mol Biol Evol. 30:2121–2133. [DOI] [PubMed] [Google Scholar]
- Schraiber JG, Mostovoy Y, Hsu TY, Brem RB. 2013. Inferring evolutionary histories of pathway regulation from transcriptional profiling data. PLoS Comput Biol. 9:e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, et al. 2012. cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun. 3:950. [DOI] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution?. Evolution 62:2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H-M, et al. 2009. Roles of trans and cis variation in yeast intraspecies evolution of gene expression. Mol Biol Evol. 26:2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorov A, et al. 2013. Intra-specific regulatory variation in Drosophila pseudoobscura. PLoS One 8:e83547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi KR, Matsuo T, Takano-Shimizu-Kouno T. 2011. Two types of cis-trans compensation in the evolution of transcriptional regulation. Proc Natl Acad Sci U S A. 108:15276–15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy A. a, Barkai N. 2009. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324:659–662. [DOI] [PubMed] [Google Scholar]
- Voordeckers K, Pougach K, Verstrepen KJ. 2015. How do regulatory networks evolve and expand throughout evolution? Curr Opin Biotechnol. 34:180–188. [DOI] [PubMed] [Google Scholar]
- Wang D, et al. 2007. Expression evolution in yeast genes of single-input modules is mainly due to changes in trans-acting factors. Genome Res. 17:1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ. 2005. Genomic sources of regulatory variation in cis and in trans. Cell Mol Life Sci. 62:1779–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. 2004. Evolutionary changes in cis and trans gene regulation. Nature 430:85–88. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. 2008a. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 40:346–350. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. 2008b. Independent effects of cis- and trans-regulatory variation on gene expression in Drosophila melanogaster. Genetics 178:1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206–216. [DOI] [PubMed] [Google Scholar]
- Wray G. a, et al. 2003. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 20:1377–1419. [DOI] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO. 2009. Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics 182:943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Adams KL. 2007. Extensive allelic variation in gene expression in Populus F1 hybrids. Genetics 177:1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.