Abstract
Background and Aims The root-knot nematode Meloidogyne graminicola is responsible for production losses in rice (Oryza sativa) in Asia and Latin America. The accession TOG5681 of African rice, O. glaberrima, presents improved resistance to several biotic and abiotic factors, including nematodes. The aim of this study was to assess the cytological and molecular mechanisms underlying nematode resistance in this accession.
Methods Penetration and development in M. graminicola in TOG5681 and the susceptible O. sativa genotype ‘Nipponbare’ were compared by microscopic observation of infected roots and histological analysis of galls. In parallel, host molecular responses to M. graminicola were assessed by root transcriptome profiling at 2, 4 and 8 d post-infection (dpi). Specific treatments with hormone inhibitors were conducted in TOG5681 to assess the impact of the jasmonic acid and salicylic acid pathways on nematode penetration and reproduction.
Key Results Penetration and development of M. graminicola juveniles were reduced in the resistant TOG5681 in comparison with the susceptible accession, with degeneration of giant cells observed in the resistant genotype from 15 dpi onwards. Transcriptome changes were observed as early as 2 dpi, with genes predicted to be involved in defence responses, phenylpropanoid and hormone pathways strongly induced in TOG5681, in contrast to ‘Nipponbare’. No specific hormonal pathway could be identified as the major determinant of resistance in the rice-nematode incompatible interaction. Candidate genes proposed as involved in resistance to M. graminicola in TOG5681 were identified based on their expression pattern and quantitative trait locus (QTL) position, including chalcone synthase, isoflavone reductase, phenylalanine ammonia lyase, WRKY62 transcription factor, thionin, stripe rust resistance protein, thaumatins and ATPase3.
Conclusions This study provides a novel set of candidate genes for O. glaberrima resistance to nematodes and highlights the rice-M. graminicola pathosystem as a model to study plant-nematode incompatible interactions.
Keywords: Oryza sativa, Oryza glaberrima, Meloidogyne graminicola, root-knot nematode, resistance responses, RNA-Seq, transcriptomics, histology, hormones, QTL, differentially expressed genes, monocots
Introduction
Root-knot nematodes (Meloidogyne spp.) are obligate parasites that settle in roots and complete their life cycle by feeding from specially adapted host cells (Williamson and Gleason, 2003). These nematodes may significantly decrease rice production in Asia, West Africa and South America, where Meloidogyne graminicola (Golden and Birchfield, 1965), Meloidogyne incognita (Kofoid and White, 1919; Chitwood, 1949) and Meloidogyne javanica are prevalent (Fortuner and Merny, 1979; Soriano and Reversat, 2003). Asian rice (Oryza sativa) is an important staple cereal grain food for a large part of the world's human population (Seck et al., 2012). Meloidogyne graminicola is increasing in importance in rice cultivation wherever water conservation measures reduce the extent to which flooding controls this pest. Importantly, the grain nutritional quality and seed germination of Asian rice varieties may be drastically lowered by M. graminicola infection (Patil and Gaur, 2014).
Meloidogyne graminicola may infect rice roots and complete its life cycle in < 3 weeks under favourable temperatures (Fernandez et al., 2014; Nguyen et al., 2014). Meloidogyne spp. are highly adapted parasites able to escape from plant recognition, establish feeding sites and suppress host defence reactions. Nematode stage 2 juveniles (J2) enter roots close to the apex at the elongation zone, and then reach the root central cylinder, where they initiate a feeding site from a set of parenchyma cells. This process may be observed as soon as 2 d post-infection (dpi) (Nguyen et al., 2014). In response to nematode signals, the parenchyma cells differentiate into multinucleate and metabolically active giant cells (GCs), resembling transfer cells (Kyndt et al., 2013; Rodiuc et al., 2014), enabling M. graminicola to withdraw nutrients from the plant’s vascular system. The M. graminicola feeding site consists of several GCs enclosed in a macroscopically visible gall or root knot formed on the root system. A gall may contain several feeding sites and nematodes. The juvenile then goes through several developmental stages (J3, J4) to finally differentiate into a female (from 14 dpi), which will lay numerous eggs inside the root tissue (Nguyen et al., 2014). During root invasion, M. graminicola juveniles express a high number of genes encoding cell wall-degrading enzymes and virulence effectors to enter, migrate into roots and induce GC formation (Haegeman et al., 2013; Petitot et al., 2015). In addition, transcriptome data from this compatible interaction showed that plant defence pathways are suppressed in M. graminicola-infected rice (O. sativa ‘Nipponbare’) roots (Nahar et al., 2011; Kyndt et al., 2012a; Ji et al., 2013). In particular, the hormone-mediated resistance signalling pathways controlled by salicylic acid (SA), jasmonic acid (JA) and ethylene are repressed in rice roots soon after infection (Nahar et al., 2011; Nguyen et al., 2014).
Until now, the lack of effective resistance in Asian rice germplasm has hampered effective genetic breeding for M. graminicola resistance. Sources of specific resistance to Meloidogyne spp. were identified in the related species Oryza longistaminata and Oryza glaberrima (Soriano et al., 1999). Oryza glaberrima is a low-yielding species originating from Africa that has many interesting agricultural traits, such as resistance to biotic and abiotic stresses (Linares, 2002). In particular, the M. graminicola-resistant TOG5681 variety has been used as donor parent to develop some NERICA (New Rice for Africa) lines that provide good yield and are adapted to lowland ecosystems (Ndjiondjop et al., 2008; Agnoun et al., 2012). Genetic analyses of backcrossed TOG5681 introgression lines indicated that resistance to M. graminicola may be controlled by several genetic loci (Plowright et al., 1999; Bimpong et al., 2010); however, the mechanisms underlying resistance are poorly understood. For example, although high levels of resistance to M. graminicola have been found in many O. glaberrima accessions tested, including TOG5681, there is still a low level of nematode reproduction (Soriano et al., 1999; Bimpong et al., 2010; Cabasan et al., 2012), indicating that the nematode is able to penetrate and develop feeding sites in roots of resistant genotypes. It is unclear whether the low level of nematode infection in TOG5681 is mainly due to limited penetration and/or whether subsequent resistance reactions inhibit nematode development. In the incompatible plant-nematode interactions studied so far, three main resistance response types were described based on the timing: a rapid hypersensitive response that blocks GC initiation; a resistance that restricts GC expansion; and a third type, occurring later, impairing the function of GCs as active transfer cells (Goverse and Smant, 2014).
Transcriptome profiling of plant immune responses during the interaction with microorganisms allows monitoring of the activation or suppression of specific regulatory and metabolic pathways. A recent study based on an Arabidopsis thaliana gene co-expression network in response to diverse biotic stresses (not including nematodes) has identified meta-modules enriched in known pathogen-responsive genes, such as those coding for pathogenesis-related (PR) proteins, or those involved in the biosynthesis, modification and signalling of plant hormones (Amrine et al., 2015). A set of 66 hub genes, most of them upregulated in response to all types of pathogens tested, with a high degree of connectivity within and across meta-modules enriched for immune response functions, were proposed as potential key components of the core defence responses activated by the host plant (Amrine et al., 2015). However, little information is available on plant immune responses to nematodes. Recent transcriptomic studies identified a series of candidate genes involved in the resistance response to Meloidogyne spp. in a few host plant species, including soybean (Beneventi et al., 2013), peanut (Guimaraes et al., 2015) and alfalfa (Postnikova et al., 2015). In rice, no transcriptome data are yet available for the resistance response to M. graminicola. Recently, 11 quantitative trait loci (QTL) associated with resistance to M. graminicola were mapped based on a genome-wide association study (GWAS) in O. sativa (Dimkpa et al., 2015). A set of 493 positional candidate genes within 200 kb of these QTL were described, including stripe rust resistance, orthologues of the powdery mildew resistance of barley mla-1, and lectins known to be associated with plant disease resistance (Dimkpa et al., 2016). In O. glaberrima, resistance to the cyst nematode Heterodera sacchari is controlled by one major gene, Hsa-1Og, with codominance of susceptible and resistant alleles, which has been mapped on chromosome 11 (Lorieux et al., 2003).
The objective of this work was to compare the penetration of M. graminicola juveniles, their ability to establish feeding sites, and the molecular host responses to M. graminicola in resistant (O. glaberrima TOG5681) versus susceptible (O. sativa ‘Nipponbare’) rice. By using a combination of histology and in-depth transcriptomic (RNA-Seq) analyses, we show that TOG5681 is able to mount a series of resistance responses that act at several steps during the nematode life cycle to limit penetration and feeding site development, preventing efficient nematode reproduction. Additionally, we describe a small set of differentially expressed genes that may potentially be associated with rice resistance to M. graminicola.
Materials and Methods
Plants
O ryza sativa ‘Nipponbare’ and Oryzaglaberrima TOG5681 genotypes of rice were used in this study. ‘Nipponbare’ is susceptible and TOG5681 is resistant to Meloidogynegraminicola (Soriano et al., 1999; Bimpong et al., 2010). Rice plants were maintained in a growth chamber under controlled conditions at 26 °C/24 °C day/night temperature, under a 12-h day/night light regime and 70 % relative humidity.
Nematode culture
The M. graminicola culture was originally isolated in Batangas, the Philippines (kindly provided by the International Rice Research Institute (IRRI). Meloidogynegraminicola pre-parasitic J2 were obtained as previously described (Nguyen et al., 2014).
Nematode infection experiments
Rice seeds were germinated on sand wetted with Hoagland solution for 7 d and then transferred to plastic tubes containing SAP (sand and absorbent polymer) substrate (Reversat et al., 1999) wetted with Hoagland solution. Three days after transplanting into SAP substrate, the ‘Nipponbare’ and TOG5681 plants were inoculated with 1 mL of water containing 100 or 400 J2 for penetration studies, and with 100 J2 (‘Nipponbare’) or 400 J2 (TOG5681) for RNA-Seq experiments. One day after inoculation, plants were transferred to a 50-mL hydroponic culture system containing Hoagland solution to synchronize the infection process.
For RNA-Seq experiments, root tips (1–2 mm) or visible galls were collected at 2, 4 and 8 dpi. Root tips from non-inoculated plants were collected at the same time as the 2-dpi plants, and were considered as the control. Samples were immediately frozen in liquid nitrogen and stored at − 80 °C until further use. A mean of 25 plants were pooled for each sample and collection time, and two independent biological replicates were performed.
For penetration studies, plants were collected at 1 and 7 dpi and roots were stained with acid fuchsin as described in Bybd et al. (1983) with the following modification: after fuchsin coloration, roots were destained in a solution of lactic acid, acetic acid and water (1:1:1) for 24 h before placing in acidified glycerine. Nematodes inside the roots were visualized and counted using a Leica microscope (Leica Microsystems, France). Five plants of each genotype were used for each time point and the experiment was performed with three replications.
For the analysis of nematode development and for the chemical treatments, infections were performed with the mesocosm system (Reversat et al., 1999). Seeds were germinated on wet filter paper for 4 d at 30 °C and then transferred to SAP in polyvinyl chloride (PVC) tubes. Each plant was watered three times per week with 10 mL of Hoagland solution. When plants were 15 d old, each plant was inoculated with 250 J2. At 14 dpi, the root samples were stained with acid fuchsin and galls and nematodes inside the roots were visualized and counted using a Leica stereo microscope. Chemical treatment was conducted with salicyl hydroxamic acid (SHAM) to inhibit lipoxygenase-activity and l-2-aminooxy-3-phenylpropionic acid (AOPP) to inhibit phenylalanine ammonia lyase (PAL) activity. All chemicals were purchased from Sigma-Aldrich and dissolved in a few drops of ethanol before diluting in distilled water. Intact 14-d-old seedlings were sprayed with 200 µm SHAM or 100 µm AOPP 1 d before nematode inoculation. Eight plants of each genotype were used for each time point and the experiment was repeated three times.
Histology
Samples were prepared as described in Nguyen et al. (2014). Briefly, TOG5681 infected roots were harvested at 7, 11, 15, 19 and 22 dpi and galls were excised from each plant. Between 10 and 24 plants were used for each time point in order to recover at least five to ten galls. Galls were fixed in paraformaldehyde (Sigma-Aldrich), dehydrated in ethanol and embedded in epoxy resin (Technovit 7100, Kulzer Friedrichsdorf, Germany) according to the manufacturer’s protocol. Blocks containing root samples were sectioned (10 µm) and microscopically observed under UV light (UV filter set A2, Zeiss AXIO Imager). The same sections were subsequently stained (3 min at room temperature) with 0·05 % toluidine blue in 0·1 m sodium phosphate buffer, pH 5·5. Images were taken with an Axiocam digital camera (Leica microscope) with standard bright-field optics. The GC surface was measured on gall sections as indicated in Vieira et al. (2013).
Construction of RNA-Seq libraries and Illumina sequencing
Total RNA was extracted from rice root samples using an RNeasy Plant Mini Kit (Qiagen, France) with addition of an on-column DNase I digestion. RNA integrity was assessed based on RNA integrity number (RIN) using an Agilent RNA 6000 Nano chip. RNA was quantified using a Quant-iT™ RiboGreen® RNA Assay Kit, and 2 μg was used for library preparation. Individually indexed libraries were prepared using the TruSeq RNA Sample Preparation v2 kit according to the manufacturer’s instructions (Illumina Inc., USA). Each cDNA library was verified and quantified using a DNA 100 chip on a Bioanalyzer 2100. The libraries were sequenced at the Centre National de Séquençage (Genoscope, Evry, France) on a HiSeq2000 system (Illumina Inc.) generating 100-bp paired-end reads. Raw data have been submitted to ENA/SRA (accession number ERS715982).
Differential expression analyses
Reads were processed as previously described in Petitot et al. (2015) and only read pairs were kept for subsequent analyses. Then, the read pairs were aligned to the O. sativa (‘Nipponbare’) reference genome (MSU7 release from the http://rice.plantbiology.msu.edu/ website) using TopHat2 (Trapnell et al., 2009). A gene transfer format (GTF) file was supplied using the -G option. Standard parameters were used except for the following options: -r 100, -mate-std-dev 50, -i 30, -I 20000, -a 8, -m 1, --no-coverage-search, -g 10, --bowtie-n, -library-type fr-unstranded, --microexon-search, -N 2 (for ‘Nipponbare’ libraries) or 5 (for TOG5681 libraries). The Binary Alignment/Map (BAM) files generated by TopHat were converted to count files using htseq-count (Anders et al., 2015). Reads with more than one alignment on the rice genome were discarded during this step. A list of 875 candidate genes was compiled based on MAPMAN pathways (Thimm et al., 2004), Oryza orthologues of Arabidopsis core biotic stress responsive genes (Amrine et al., 2015), genes annotated as resistance proteins in MSU7 and QTL of Dimkpa et al. (2015). Differential gene expression was determined on count files using edgeR, a Bioconductor software package (Robinson et al., 2010). Differential expression of any transcript under consideration was analysed only if the sum of its counts in all the compared samples was >20. Analyses were performed with the generalized linear models method and the common dispersion as a dispersion estimate method. Adjustment of the false discovery rate was based on the Benjamini-Hochberg algorithm. Genes were considered as induced or repressed only when their log2 fold change (FC) was >2 or <−2, respectively, and their false discovery rate was <0·05.
RNA extraction, cDNA synthesis and real-time quantitative PCR analyses
Extraction of RNA and quantitative reverse transcription PCR (RT-qPCR) analyses were performed according to Nahar et al. (2011). Specific primers were designed from the O. sativa ‘Nipponbare’ sequence using Beacon Designer 5.0 software (Premier Biosoft International, Palo Alto, CA, USA). Primer sequences were checked on the O. glaberrima TOG5681 sequence, and new primers were designed when sequences were divergent (Supplementary Data Table S1). Primers were designed from the 3′ region of the gene and were synthesized by Eurogentec (Belgium). For each primer pair, a preliminary real-time assay was performed on non-infected O. sativa or O. glaberrima and mixed rice-M. graminicola cDNA samples to evaluate the amplification of non-specific products or primer-dimer artefacts. Rice gene expression was normalized with reference genes OsEIF5C, OsREF3, osREF4, OsREF5 or OsEXPNAR (Table S1). Based on the comparative Cq method, data are expressed as log (base 2) FC compared with the respective control samples.
Results
Nematode penetration and development
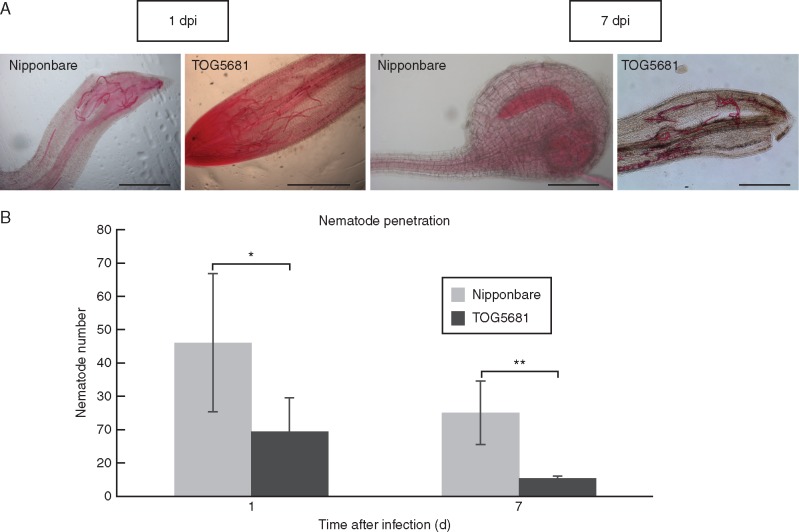
Penetration and development differed between the two rice genotypes examined. Observation of fuchsin-stained roots showed that in the resistant roots 40 % fewer (P<0·05) nematodes had penetrated at 1 dpi compared with susceptible roots (Fig. 1). At this time point, the J2 were found either clustered within the root apex or migrating upwards into the developing vascular cylinder. Root phenotypic difference between the two genotypes could only partially explain the differential J2 invasion, with 20 % fewer (P < 0·001) root tips in TOG5681 compared with ‘Nipponbare’ at the time of inoculation (mean root tip number 5·3 ± 0·7 and 3·9 ± 0·3, n = 30, respectively). At 7 dpi, galls had formed in both genotypes (mean number 7 ± 1·8 in susceptible, and 1·9 ± 0·4 in resistant, n = 15), although galling was sporadic in TOG5681, the majority of plants being without galls. A 75 % reduction in nematode numbers was observed in the galls of TOG5681 at 7 dpi when compared with 1 dpi. This contrasted with a 50 % reduction in numbers in ‘Nipponbare’ over the same time period. Overall, 80 % (P < 0·01) fewer nematodes were found in the resistant plants at 7 dpi than in the susceptible genotype. Additionally, all nematodes were swollen and larger at 7 dpi in ‘Nipponbare’ than at 1 dpi, indicating nematode feeding (Fig. 1A). In contrast, in the resistant genotype only a few nematodes appeared to have been feeding, with most J2 vermiform and the same size as observed at 1 dpi.
Fig. 1. Meloidogyne graminicola penetration in rice roots (Oryza sativa ‘Nipponbare’ and Oryza glaberrima TOG5681) at 1 or 7 dpi. (A) Light microscope photographs of nematode juveniles after fuchsin staining of infested rice roots. Bar indicates 1 mm. (B) Mean nematode number in rice roots after plant inoculation with 400 second-stage juveniles (J2). Bars are the mean numbers (± s.d.) of nematodes per plant in three independent biological replicates, with 20 plants per condition (n = 5;3). Stars indicate statistical differences determined by one-way ANOVA (**P < 0·01; *P < 0·5, n = 5).
At 14 dpi, the number of galls on the resistant and susceptible cultivars was evaluated using the mesocosm growth system. In TOG5681, 84 % fewer galls were formed compared with the susceptible cultivar ‘Nipponbare’ (5·0 ± 2·8 versus 31·6 ± 7·0, P < 0·001, Student’s t-test). The number of nematodes inside these galls was 86 % less in TOG5681 (6·7 ± 3·3) in comparison with ‘Nipponbare’ (48·0 ± 13·1, P < 0·001, Student’s t-test), confirming our earlier observation of reduced nematode penetration in the resistant line.
Histological analysis of TOG5681-M. graminicola interaction
Analysis of toluidine blue-stained gall sections (Fig. 2 and Supplementary Data Fig. S1) showed that nematodes infected both lines similarly, but abortion of gall development was distinctively observed in the resistant genotype at a later time point.
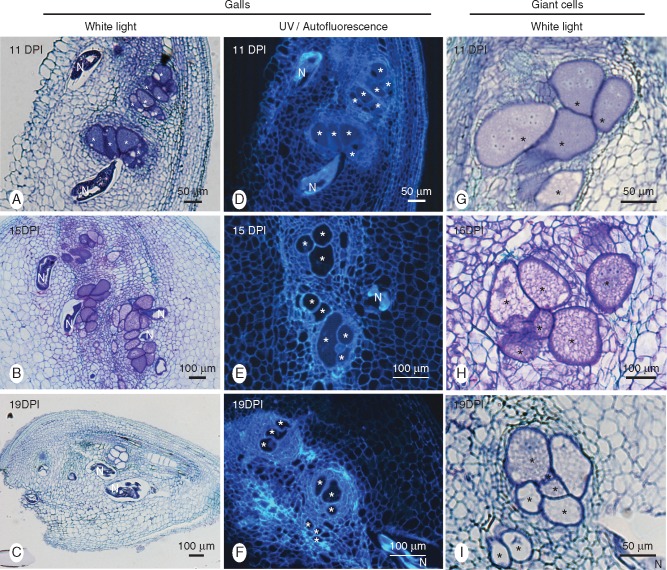
Fig. 2.
Histological analysis of galls induced by Meloidogyne graminicola in resistant rice (Oryza glaberrima TOG5681) roots. Gall cross-sections (10 µm) obtained at 11, 15 and 19 dpi were observed under UV light (middle panel) or were stained with toluidine blue and observed with white light (left and right panels). (A, B, C) Multiple nematode feeding on giant cells within the same gall. Note degraded nematode bodies and empty giant cells at 19 dpi (C). (D, E, F) Autofluorescence of sectioned galls. Note the absence of fluorescence accumulation around giant cells and nematodes. (G, H, I) Time course of giant cell degradation. (G) 11 dpi: dense cytoplasm and a large number of nuclei encircled by numerous neighbouring parenchymatic cells. (H) 15 dpi: vacuolated cytoplasm. (I) 19 dpi: degraded, devoid of cytoplasm and nuclei. Asterisk, giant cell; N, nematode.
At 7 and 11 dpi, several nematodes were found in galls formed on TOG5681 roots, and feeding sites near each nematode had developed. Vermiform nematodes were swollen, indicating they were actively feeding. At 7 dpi all nematodes visualized were at the J2 parasitic stage. At 11 dpi nematodes at the late J2/J3 stage were observed Fig. 2 A, D, G). On average, feeding sites were composed of six GCs surrounded by a series of neighbouring cells forming a tight crown around them (Fig. 2). At these time points, the GCs that had developed in TOG5681 were similar to those formed in ‘Nipponbare’, containing densely stained cytoplasm and multiple nuclei with large nucleoli stained dark blue.
At 15 dpi, the examined galls contained several feeding sites with nematodes still at the J2/J3/J4 stage. The GCs in the resistant genotype looked phenotypically similar to those in the susceptible genotype. However, in TOG5671 some GCs presented a higher number of vacuoles compared with giant cells in ‘Nipponbare’ (Fig. 2H). At 19 dpi, the giant cells contained significantly less cytoplasm in the resistant compared with the susceptible genotype. Nuclear shape was also modified, appearing less rounded. Some GCs were already devoid of cytoplasm and the nematodes had degenerated in most sections examined. Around 15 galls were examined at 19 and 22 dpi in TOG5671, where we observed only one egg-laying female and one male (Fig. S1). Female nematodes were small and for the egg-laying female fewer eggs were visible compared with those observed in ‘Nipponbare’. However, GCs still appeared similar to ‘Nipponbare’ GCs at 19 dpi. These observations are in accordance with the low nematode reproduction observed in TOG5681. In ‘Nipponbare’, eggs may be observed as soon as 14 dpi, and all observed M. graminicola females had laid eggs at 21 dpi. At 22 dpi, all galls examined in TOG5681 contained only dead nematodes and collapsed GCs. Nematode bodies were shrunken, and the empty space around them was indicative of the size of the nematode when alive. The female nematodes that had successfully reached the adult stage (rounded bodies) in the roots of TOG5681 were smaller in size compared with those generally found in the susceptible ‘Nipponbare’ (Fig. 2 and Fig. S1). Overall, the GC surface was smaller in the resistant genotype compared with the susceptible genotype (Kruskal-Wallis test, P < 0·05, n = 83).
To examine whether GC degradation and nematode collapse could be associated with the production of phenolic compounds by the plant against the nematode, 7- to 22-dpi galls and uninfected root sections were analysed by optical microscopy under UV light in order to detect the autofluorescence related to phenolic compounds. The same amount of fluorescence was observed next to nematode bodies or GCs in resistant roots (Fig. 2D, E, F) and in susceptible roots (Supplementary Data Fig. S2) at 7–22 dpi, indicating that the accumulation of phenolic compounds in TOG5681 roots is not the main reason for GC and nematode degradation.
Transcriptome analysis of resistant and susceptible genotypes of rice
To focus on early responses, ‘Nipponbare’ and TOG5681 infected roots were collected at 2, 4 and 8 dpi in two independent biological replicates. Total RNA samples were used to prepare Illumina RNA-Seq libraries and between 30 and 47 M read pairs were obtained for each library (Supplementary Data Table S2). Paired reads were aligned to the reference genome sequence of ‘Nipponbare’ (MSU7 annotation) and count files were generated to determine the expression levels of rice genes in each library. Differential gene expression analyses were performed between nematode-infected tissues and control root tips at each time point. Among the 29 984 genes for which expression was detected, the numbers of up- and downregulated genes were determined (Table 1).
Table 1.
Numbers of differentially expressed genes (DEG) in Meloidogyne graminicola-infected and control root tips in susceptible rice (Oryza sativa ‘Nipponbare’; Nip) and resistant rice (Oryza glaberrima TOG5681; Tog)
| Category | Expressed genes | Rice species | Induced genes |
Repressed genes |
Total DEGs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 dpi | 4 dpi | 8 dpi | Total | 2 dpi | 4 dpi | 8 dpi | Total | ||||
| All MSU7 genes | 29 984 | Nip | 362 (1·2 %) | 1972 (6·6 %) | 2823 (9·4 %) | 3494 (11·6 %) | 46 (0·2 %) | 937 (3·1 %) | 1221 (4·1 %) | 1503 (5·0 %) | 4997 (16·7 %) |
| Tog | 1228 (4·1 %) | 2641 (8·8 %) | 2801 (9·3 %) | 3584 (11·9 %) | 49 (0·2 %) | 347 (1·2 %) | 339 (1·1 %) | 548 (1·8 %) | 4132 (13·8 %) | ||
| Common | 199 | 982 | 1331 | 1828 | 1 | 162 | 160 | 270 | |||
| Signalling | 16 | Nip | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Tog | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| SA | 14 | Nip | 1 | 2 | 2 | 3 | 0 | 0 | 0 | 0 | 3 |
| Tog | 4 | 4 | 2 | 4 | 0 | 0 | 0 | 0 | 4 | ||
| Lignin | 21 | Nip | 0 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | 3 |
| Tog | 3 | 2 | 2 | 4 | 0 | 0 | 0 | 0 | 4 | ||
| Flavonoids and terpenoids | 64 | Nip | 3 | 12 | 7 | 16 | 0 | 8 | 8 | 11 | 27 |
| Tog | 9 | 11 | 12 | 18 | 4 | 4 | 10 | 12 | 30 | ||
| JA | 26 | Nip | 1 | 4 | 4 | 7 | 0 | 0 | 0 | 0 | 7 |
| Tog | 3 | 5 | 4 | 5 | 0 | 0 | 0 | 0 | 5 | ||
| Ethylene | 19 | Nip | 0 | 2 | 2 | 3 | 0 | 2 | 3 | 3 | 6 |
| Tog | 0 | 2 | 3 | 3 | 0 | 0 | 0 | 0 | 3 | ||
| Callose | 4 | Nip | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tog | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| PR proteins | 4 | Nip | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 | 2 |
| Tog | 4 | 4 | 3 | 4 | 0 | 0 | 0 | 0 | 4 | ||
| PR13: thionins | 25 | Nip | 1 | 3 | 5 | 5 | 2 | 11 | 8 | 11 | 16 |
| Tog | 0 | 1 | 0 | 1 | 0 | 6 | 8 | 9 | 10 | ||
| QTL | 323 | Nip | 2 | 28 | 44 | 49 | 1 | 15 | 17 | 24 | 73 |
| Tog | 14 | 34 | 39 | 45 | 1 | 6 | 3 | 7 | 52 | ||
| Resistance | 359 | Nip | 2 | 26 | 35 | 43 | 1 | 11 | 11 | 12 | 55 |
| Tog | 16 | 47 | 45 | 56 | 2 | 3 | 2 | 4 | 60 | ||
| Candidate genes | 875 | Nip | 11 (1·1 %) | 82 (8·6 %) | 102 (10·6 %) | 131 (13·7 %) | 4 (0·4 %) | 48 (5 %) | 48 (5 %) | 62 (6·5 %) | 193 (22·1 %) |
| Tog | 53 (5·5 %) | 110 (11·5 %) | 110 (11·5 %) | 140 (14·6 %) | 7 (0·7 %) | 19 (2·0 %) | 23 (2·4 %) | 32 (3·3 %) | 172 (19·7 %) | ||
Upon nematode infection, a total of 3494 genes (11·6 %) and 3584 genes (11·9 %) were induced in ‘Nipponbare’ and TOG5681 infected plants, respectively, among which 1828 genes showed a similar upregulated expression profile in both genotypes. A stronger response to the infection was observed for TOG5681 at 2 dpi: 1228 genes were induced in TOG5681 whereas only 362 genes were induced in ‘Nipponbare’. In addition, 1028 (84 %) of the 1228 TOG5681 induced genes were not altered in ‘Nipponbare’, suggesting that the nematode was able to inhibit plant responses in the compatible interaction. A high number of genes differentially expressed in TOG5681 were involved in immune or defence-associated pathways, such as genes encoding pathogen receptors and signalling proteins (R proteins, receptor-like protein kinases) or transcriptional regulators (AP2, MYB and WRKY transcription factors), and genes of the phenylpropanoid pathway or encoding Pathogen related (PR) proteins and secondary metabolism enzymes (CYP450).
Considering down-regulated genes, 1503 genes (5 %) were repressed upon nematode infection in ‘Nipponbare’, while only 548 genes (1·8 %) were repressed in TOG5681. A total of 270 genes showed a similar downregulated expression profile in both genotypes. Major differences were evident at 4 and 8 dpi, when the number of repressed genes in ‘Nipponbare’ was around 3-fold higher than in TOG5681. Because no specific pathway was altered in this unbiased analysis, we focused our study on genes involved in JA, SA and ethylene hormonal pathways, phenylpropanoids and other antimicrobial molecules in the rice-root-knot nematode interaction. In addition, we examined genes annotated as homologues of R genes, genes associated with QTL for nematode resistance and genes described as responsive to pathogens [so-called core biotic stress-responsive (CBRS) genes, as identified by Amrine et al., 2015]. For all the selected genes, we present RNA-Seq expression data generated by edgeR analyses (Supplementary Data Table S3). For the genes showing the highest degree of differential expression, we verified their expression by RT-qPCR assays (Table 2, Supplementary Data Table S4).
Table 2.
List of differentially expressed candidate genes potentially associated with Meloidogyne graminicola resistance in Oryza glaberrima TOG5681. Means of normalized reads counts obtained by edgeR are indicated for susceptible plants (Oryza sativa ‘Nipponbare’) and TOG5681 resistant plants in the RNA-Seq columns. Differential gene expression (log fold change, Padj<0·05) between nematode-infected and control root tips and between Oryza species control root tips is shown by the colour code on the right. Results of quantitative RT-PCR assays are indicated in the Q-PCR column (OK, validation; nt, not tested). See Table S4 for Q-PCR details
| Locus | Pathway | Function | RNA-seq |
Q-PCR |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ‘Nipponbare’ |
TOG5681 |
|||||||||||||
| C | 2 dpi | 4 dpi | 8 dpi | C | 2 dpi | 4 dpi | 8 dpi | |||||||
| LOC_Os04g15920 | Lignin biosynthesis | Dehydrogenase, CAD6 | 419 | 475 | 293 | 130 | 172 | 823 | 586 | 379 | OK | |||
| LOC_Os08g38910 | Lignin biosynthesis | Caffeoyl-CoA O-methyltransferase, CCoAOMT | 12 | 13 | 67 | 309 | 26 | 112 | 297 | 788 | nt | |||
| LOC_Os08g38920 | Lignin biosynthesis | Caffeoyl-CoA O-methyltransferase, CCoAOMT | 17 | 8 | 5 | 6 | 192 | 339 | 381 | 196 | nt | |||
| LOC_Os04g01354 | Chalcones | Chalcone synthase | 5 | 6 | 15 | 2 | 31 | 544 | 288 | 21 | nt | Log FC | ||
| LOC_Os10g40880 | Flavonols | Flavonol synthase/flavanone 3-hydroxylase | 2 | 3 | 0 | 0 | 746 | 485 | 613 | 747 | OK | < −2 | ||
| LOC_Os04g09900 | Terpenoids | Ent-kaurene synthase, chloroplast precursor | 16 | 134 | 1158 | 62 | 1063 | 8902 | 3501 | 503 | OK | < −1 | ||
| LOC_Os04g10060 | Terpenoids | Ent-kaurene synthase, chloroplast precursor | 3 | 11 | 186 | 10 | 3 | 465 | 221 | 43 | OK | >1 | ||
| LOC_Os11g28530 | Terpenoids | Terpene synthase | 265 | 635 | 1289 | 276 | 554 | 2604 | 1106 | 308 | nt | >2 | ||
| LOC_Os02g41670 | SA biosynthesis | Phenylalanine ammonia lyase, OsPAL3 | 83 | 195 | 748 | 84 | 148 | 1009 | 819 | 217 | nt | |||
| LOC_Os02g41680 | SA biosynthesis | Phenylalanine ammonia lyase, OsPAL4 | 242 | 368 | 976 | 491 | 251 | 1411 | 1474 | 988 | OK | Tog/Nip | ||
| LOC_Os04g43800 | SA biosynthesis | Phenylalanine ammonia lyase, OsPAL6 | 12 | 97 | 170 | 58 | 119 | 1001 | 1117 | 394 | nt | >2 | ||
| LOC_Os09g25070 | SA response | WRKY62 | 604 | 425 | 647 | 180 | 206 | 1805 | 2504 | 1377 | OK | |||
| LOC_Os03g12500 | JA biosynthesis | Allene oxide synthase 2 | 6 | 16 | 98 | 26 | 12 | 406 | 629 | 297 | OK | |||
| LOC_Os08g39840 | JA biosynthesis | Lipoxygenase 7, chloroplast precursor | 12 | 32 | 195 | 21 | 9 | 243 | 140 | 48 | nt | |||
| LOC_Os11g08380 | Ethylene biosynthesis | 1-Aminocyclopropane-1-carboxylate oxidase | 296 | 229 | 138 | 144 | 1819 | 870 | 1151 | 955 | OK | |||
| LOC_Os06g31280 | PR | THION1, plant thionin family protein precursor | 7 | 20 | 1 | 24 | 6339 | 2410 | 2528 | 783 | OK | |||
| LOC_Os06g31800 | PR | THION2, plant thionin family protein precursor | 83 | 15 | 5 | 105 | 278829 | 104220 | 66448 | 50602 | OK | |||
| LOC_Os06g31890 | PR | THION3, plant thionin family protein precursor | 60 | 26 | 0 | 9 | 761 | 312 | 173 | 106 | nt | |||
| LOC_Os06g32240 | PR | THION9, plant thionin family protein precursor | 0 | 1 | 0 | 4 | 733 | 288 | 195 | 118 | nt | |||
| LOC_Os07g33780 | Resistance | Pleiotropic drug resistance protein 5 | 677 | 587 | 1364 | 1031 | 752 | 3914 | 4156 | 1338 | OK | |||
| LOC_Os09g34160 | Resistance | Resistance protein | 21 | 26 | 77 | 30 | 53 | 543 | 575 | 199 | OK | |||
| LOC_Os11g37860 | Resistance and QTL | Stripe rust-resistance protein Yr10 | 395 | 439 | 1067 | 1997 | 318 | 1435 | 6272 | 5256 | OK | |||
| LOC_Os01g73604 | QTL | LSM domain containing protein | 3 | 4 | 3 | 7 | 310 | 252 | 324 | 335 | nt | |||
| LOC_Os03g45740 | QTL | Transferase family protein | 62 | 20 | 1 | 5 | 551 | 339 | 179 | 233 | OK | |||
| LOC_Os03g45960 | QTL | Thaumatin | 35 | 57 | 53 | 42 | 146 | 954 | 1050 | 1312 | OK | |||
| LOC_Os03g46040 | QTL | Expressed protein | 0 | 0 | 0 | 1 | 488 | 471 | 573 | 441 | OK | |||
| LOC_Os03g46060 | QTL | Thaumatin family domain containing protein | 91 | 121 | 549 | 732 | 55 | 479 | 1278 | 612 | OK | |||
| LOC_Os03g46070 | QTL | Thaumatin | 111 | 158 | 474 | 128 | 34 | 1280 | 746 | 168 | OK | |||
| LOC_Os04g01470 | QTL | O-methyltransferase | 1 | 2 | 91 | 12 | 14 | 753 | 1248 | 147 | OK | |||
| LOC_Os04g04240 | QTL | Sterol 3-β-glucosyltransferase | 0 | 0 | 0 | 0 | 177 | 161 | 168 | 143 | nt | |||
| LOC_Os05g03920 | QTL | DUF26 | 9 | 14 | 119 | 132 | 38 | 124 | 314 | 393 | OK | |||
| LOC_Os12g28710 | QTL | ATPase 3 | 3 | 5 | 5 | 5 | 70 | 239 | 437 | 194 | OK | |||
Signalling
When a plant is attacked by a pathogen, specific pathogen-associated molecular patterns are detected by pattern recognition receptors at the plasma membrane, provoking pathogen-triggered immunity (PTI) responses in the cytoplasm, including signalling through mitogen-associated protein kinases (MaPK), and reactive oxygen species (mainly H2O2). This leads to physiological changes such as cell wall lignification, callose deposition, flavonoid production, hormonal changes and PR protein expression. Here, we investigated the transcriptional changes in these pathways (Table S3). No specific alteration of known PTI receptors occurred in either rice genotype during nematode infection. Also, MaPK expression was not influenced by nematode infection in either genotype. Seven rice genes known to be involved in H2O2 production were not altered, except for a slight induction of LOC_Os11g33120.
Callose
The expression pattern of genes involved in callose biosynthesis was similar when comparing infected ‘Nipponbare’ with infected TOG5681. Basal transcript levels of a callose degradation enzyme (LOC_Os01g71340) were higher in ‘Nipponbare’. However, upon infection this gene was strongly activated in TOG5681.
Phenylpropanoid pathway
Phenylalanine ammonia lyase is the first enzyme in the phenylpropanoid pathway that is responsible for the biosynthesis of different plant defence-related metabolites, like lignin precursors, flavonoids and hydroxycinnamic acid esters and SA (Vogt, 2010). In rice, nine PAL genes are known. The expression of seven PAL genes was detected in TOG5681 (Table S3). Three PAL genes were activated upon nematode infection in both ‘Nipponbare’ and TOG5681, although TOG5681 displayed an earlier (2 dpi) and stronger induction. In particular, LOC_Os02g41670 (OsPAL3), LOC_Os02g41680 (OsPAL4) and LOC_Os04g43800 (OsPAL6) were 6- to 8-fold upregulated upon nematode infection in TOG5681 at 2 dpi.
Lignin
Several enzyme families participate in the biosynthesis of lignin, including cinnamyl alcohol dehydrogenase (CAD) and caffeoyl-CoA O-methyltransferase (CCoAOMT). One CAD gene was slightly but significantly activated in TOG5681 (LOC_Os04g15920), mainly at 2 dpi. For the six genes encoding CCoAOMT, one was significantly induced in ‘Nipponbare’ and TOG5681 (LOC_Os08g38910), but this increased transcript level attained higher levels and appeared earlier in the resistant accession. In addition, another CCoAOMT (LOC_Os08g38920) presented higher expression in TOG5681 than in ‘Nipponbare’, both in uninfected tissues and throughout the infection process. Among the other genes examined in the lignin pathway, very few were significantly altered by nematode infection (Table S3).
Flavonoids
Chalcone synthase (CHS) is the first enzyme in the production of flavonoids. Remarkably, eight of the 22 detected CHS genes were expressed at significantly higher levels in uninfected TOG5681 versus ‘Nipponbare’. Upon infection, seven CHS genes were generally repressed in both TOG5681 and ‘Nipponbare’. In contrast, one gene (LOC_Os04g01354), encoding naringenin-chalcone synthase, was strongly activated in TOG5681 (log FC = 4) while not altered in ‘Nipponbare’. Another CHS gene (LOC_Os07g11440) was activated in upon infection in ‘Nipponbare’ from 2 dpi, but not differentially expressed in TOG5681.
When looking into other genes involved in flavonoid production (flavonols, dihydroflavonols, isoflavonoids), a similar transcriptional response upon nematode infection was observed in both ‘Nipponbare’ and TOG5681. However, here again the upregulated genes were induced earlier and to higher levels in TOG5681 than in ‘Nipponbare’. However, the flavonol synthase genes were negatively affected by nematode infection. One gene, encoding flavonol synthase/flavanone 3-hydroxylase (LOC_Os10g40880), had a higher basal expression level in TOG5681 than in ‘Nipponbare’. Among the eight genes detected in the isoflavonoid pathway, only an isoflavone reductase (LOC_Os01g13610) was markedly activated (80-fold) in TOG5681 from 2 dpi.
Terpenoids
When focusing on genes encoding enzymes of the terpenoid pathway, a high number of genes (17 out of 29) were found to be activated upon infection in both genotypes, whereas only three were repressed. In particular, one terpene synthase gene (LOC_Os11g28530) and two ent-kaurene synthase genes (LOC_Os04g09900 and LOC_Os04g10060) were activated in TOG5681 at 2 and 4 dpi, and their induction was stronger in TOG5681 than in ‘Nipponbare’. Remarkably, LOC_Os04g09900 (encoding an ent-copalyl diphosphate synthase) already had a very high basal expression in TOG5681 control root tips, and was strongly induced upon nematode infection in this genotype, at 2 and 4 dpi.
Salicylic acid pathway
In addition to its production via the phenylpropanoid pathway (Lee et al., 1995; pathway already described above), SA can also be synthesized via the isochorismate pathway (involving ICS1). Although only significant in TOG5681 at 4 and 8 dpi, a gradual reduction in ICS1 expression was observed in both ‘Nipponbare’ and TOG5681 upon nematode infection.
WRKY45, WRKY13 and WRKY62 are well-known SA-response genes. While the expression of WRKY62 and WRKY45 was significantly reduced in ‘Nipponbare’ at 8 dpi, both genes were induced upon infection in TOG5681 (only significant for WRKY62). For WRKY13, no significant differences were observed.
Jasmonic acid pathway
Nematode infection clearly influenced a series of genes coding for enzymes involved in JA biosynthesis [lipoxygenase (LOX) 7 and 8, allene oxide synthase (AOS) 2, 12-oxophytodienoate reductases] (Table S3). The highest induction was seen at 4 dpi in both genotypes, but this response seemed to occur earlier (2 dpi) and was sustained until 8 dpi in TOG5681. Notably, LOX7 and AOS2 were strongly induced, reaching a log FC of 5 as soon as 2 dpi. In contrast, two genes involved in the JA response, namely Coi1 (LOC_Os01g63420) and JAMyb (LOC_Os11g45740), were not significantly altered by nematode infection.
Ethylene pathway
In plants, ethylene is produced from S-adenosyl-l-methionine by two enzymes acting successively, 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS) and ACC oxidase (ACO). Interestingly, some differences in basal levels of expression were observed between ‘Nipponbare’ and TOG5681 for genes encoding these enzymes (Table S3). Notably, one ACO gene (LOC_Os11g08380) had a much higher basal expression level in TOG5681. In response to nematode infection, three ACO genes (of 13) were activated from 4 dpi in TOG5681. In contrast, a complex pattern of repression or activation was observed in ‘Nipponbare’. The two ACS as well as two ACO genes were suppressed from 4 dpi in ‘Nipponbare’. However, two other ACO genes were induced in both ‘Nipponbare’ and TOG5681. Regarding genes involved in the ethylene response pathway, like EIN2 (involved in ethylene perception) and ERF1 (ethylene response factor), no obvious differences in expression levels could be detected when comparing ‘Nipponbare’ and TOG5681 with or without infection.
PR genes
Among the PR genes detected, OsPR10/PBZ1, JiOsPR10 and OsPR2 were highly activated in response to infection in TOG5681 from 2 dpi until 8 dpi, while their induction was lower in ‘Nipponbare’ (Table S3). All genes were activated upon infection.
Interestingly, four thionin (PR13) genes presented a very high basal expression level in TOG5681 root tips, including LOC_Os06g31280 (THION1), LOC_Os06g31800 (THION2), LOC_Os06g31890 (THION3) and LOC_Os06g32240 (THION9). Notably, the THION2 gene displayed the highest read number (278 829) of all 55 987 loci analysed in the root tip transcriptome of the TOG5681 control plants (data not shown). These four genes were suppressed upon nematode infection, THION2 being reduced 4-fold.
Remarkably, upon infection, 11 thionin genes in ‘Nipponbare’ and nine thionin genes in TOG5681were repressed, especially those thionin genes located on chromosomes 3 and 6 (with highly similar sequences). Thionin genes were suppressed earlier and more strongly in ‘Nipponbare’ than in TOG5681, which may be related to the higher number of nematodes infecting the susceptible roots. Only one gene was slightly activated in TOG5681 (LOC_Os11g15250 THION31).
However, three thionin-like genes on chromosome 7 (LOC_Os07g25060, LOC_Os07g25050, LOC_Os07g24830), were strongly activated (log FC 5·2) from 4 dpi in ‘Nipponbare’, while expression was almost undetectable in TOG5681.
Transcriptome analysis of known resistance genes and QTL associated with resistance (positional candidate genes)
Next, the transcriptional activity of 404 rice genes annotated as ‘resistance (R) gene’ was evaluated. Among the 359 R genes detected in our study, 55 and 60 were altered by nematode infection in the susceptible and resistant genotype, respectively (Table S3). Almost all genes (56) were activated in TOG5681 between 2 and 4 dpi. In the susceptible genotype, only 35 genes were activated between 4 and 8 dpi and 12 were suppressed. The early and strongly differentially activated R genes in TOG5681 included three genes: LOC_Os07g33780, LOC_Os09g34160 and LOC_Os11g37860.
The last of these genes is an orthologue of the stripe rust resistance protein Yr10, which has previously been found to be associated with an M. graminicola resistance QTL in a GWAS study in O. sativa, by Dimkpa et al. (2016). This study reported 11 QTL and listed 493 positional candidate genes putatively associated with nematode resistance, including stripe rust resistance proteins, orthologues of the powdery mildew resistance gene of barley mla-1, and lectins. In order to investigate whether basal levels of expression of these positional candidate genes may be linked to the resistance response of TOG5681, we searched for differentially expressed genes in control root tips of both genotypes. In TOG5681 control plants, 25 genes had a higher basal expression level than in ‘Nipponbare’ control plants (Table S3). Among them, ten genes retained a higher expression level in TOG5681 than in ‘Nipponbare’ during all infection stages. In particular, LOC_Os03g46040 (expressed protein, QTL3,2) and LOC_Os04g04240 (sterol 3-β-glucosyltransferase, QTL4,3) were only expressed in TOG5681 (on average 470 and 160 reads, respectively), with no reads at all found in ‘Nipponbare’. Another gene, LOC_Os01g73604 (LSM domain containing protein, QTL1) was weakly expressed in ‘Nipponbare’ (three to seven reads), whereas it was on average 80-fold more expressed in TOG5681. Seven of these 25 genes with higher basal expression in TOG5681 were also differentially expressed during nematode infection in this line (six being activated, one repressed). Upon infection, two genes showed a much higher level of expression in TOG5681 compared with ‘Nipponbare’: LOC_Os04g01470 (O-methyltransferase, QTL4,1) and LOC_Os12g28710 (ATPase 3, QTL12) were highly activated at 2 and 4 dpi. LOC_Os04g01470 (O-methyltransferase) was also activated, but to a lesser extent, in susceptible plants, while LOC_Os12g28710 (ATPase 3) was not altered at all in ‘Nipponbare’. Finally, LOC_Os03g45740 (transferase family protein, QTL3,2), which has 9-fold higher basal expression in TOG5681 plants, was strongly repressed in ‘Nipponbare’ upon infection, but not in TOG5681. During nematode infection, 52 of the 323 positional candidate genes showed an altered expression level in TOG5681. We found 45 genes to be activated, including seven genes that were highly upregulated as soon as 2 dpi. These genes putatively encode thaumatins (LOC_Os03g45960, LOC_Os03g46070, LOC_Os03g46060), orthologues of cysteine-rich receptor kinases (LOC_Os05g03920) and stripe rust resistance protein Yr10 (LOC_Os11g37860), and two genes that have a higher basal expression in TOG5681, O-methyltransferase (LOC_Os04g01470) and ATPase 3 (LOC_Os12g28710). Interestingly, expression of thaumatin (LOC_Os03g45960) and ATPase 3 genes was not altered at all in ‘Nipponbare’ upon nematode infection. On the contrary, among the 73 genes altered in ‘Nipponbare’ during infection, a protein kinase (with lectin domain) (LOC_Os04g03579) was highly upregulated in ‘Nipponbare’ while not altered in TOG5681.
Rice candidate genes for nematode resistance
From Table S3, we selected a short list of candidate genes for M. graminicola resistance in rice (Table 2). We chose 32 genes displaying elevated levels of expression in TOG5681 either in control plants or upon infection and showing contrasting expression profiles in the resistant genotype versus the susceptible plant. We were able to design specific primers to measure and validate the expression of 21 genes by RT-qPCR assays (Table 2 and Table S4). For the other genes, primers were either not specific enough or efficiency was too low to get reliable data.
Effect of inhibition of PAL activity and JA biosynthesis on TOG5681 resistance response
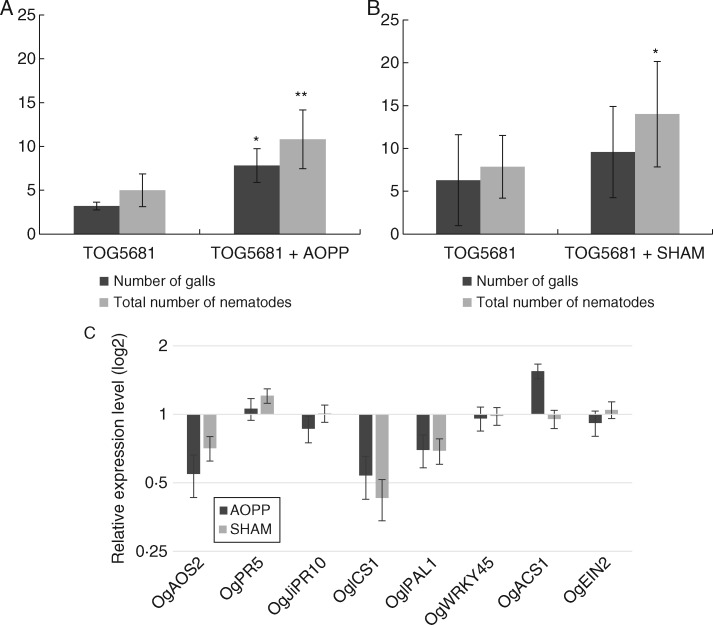
Transcriptome data showed that genes involved in phenylpropanoid and JA biosynthesis were activated earlier and more strongly in TOG5681 than in the susceptible rice genotype. To elucidate the role of the phenylpropanoid and jasmonate pathways in the early resistance response observed in TOG5681, chemical inhibitors were applied to the plants 24 h before nematode inoculation and the numbers of galls and nematodes on the root system were evaluated at 14 dpi. Samples for qPCR were also taken 24 h after chemical treatment, to investigate which pathways were suppressed inside the roots of the treated plants. In addition, we assayed the endogenous SA and JA levels and found that they were similar between roots of ‘Nipponbare’ and TOG5681 (data not shown). In plants treated with AOPP, an inhibitor of PAL, slightly more nematodes and significantly more galls were observed (Fig. 3A). In plants treated with SHAM, an inhibitor of LOX activity, the total number of nematodes inside the roots was significantly higher, but the number of galls was not significantly influenced (Fig. 3B). qRT-PCR on the roots of treated plants revealed that, for both inhibitor treatments, expression of genes involved in jasmonate biosynthesis (OgAOS2) as well as genes encoding PAL (OgPAL) and isochorismate synthase (OgICS1) was repressed (Fig. 3C). These observations suggest that both inhibitors repress the formation of phenylpropanoids, salicylate and jasmonate. Taken together, these results demonstrate that inhibiting these metabolites can only slightly impair the resistance response in TOG5681.
Fig. 3.
Role of PAL activity and JA biosynthesis in the resistance response of Oryza glaberrima TOG5681. (A, B) Number of galls and nematodes in rice roots 14 d after infection with 250 J2 Meloidogyne graminicola. Effects of (A) PAL inhibitor l-2-aminooxy-3-phenylpropionic acid (AOPP, 100 µm) and (B) lipoxygenase inhibitor salicyl hydroxamic acid (SHAM, 200 µm) on plant susceptibility to nematode infection. Leaves of eight plants were sprayed with inhibitor or water (as control) and plants were inoculated with nematodes 24 h later. The experiment was repeated three times. (C) Effect of treatments on gene expression levels in rice roots 24 h after foliar treatment with the inhibitors. Gene expression levels were measured by RT-qPCR assays and normalized using three internal reference genes: OsEIF5C, OsREF3 and OsEXPNAR. Data shown are relative transcript levels compared with control roots of water-sprayed plants (expression level set at 1). Bars represent the mean expression level ± s.e. from two independent biological replicates and three technical replicates, each containing a pool of six plants. Asterisks indicate significantly different expression levels (*P < 0·1; **P ≤ 0·05).
Core biotic stress-responsive genes
Until now, molecular plant immune responses to nematodes have been poorly documented. We examined in this study whether similar transcriptional changes occur in rice inoculated with M. graminicola and arabidopsis plants challenged with different pathogens. We searched for and compared the expression levels of a set of core biotic stress-responsive (CBSR) genes described in arabidopsis (Amrine et al., 2015). We found 78 rice orthologues corresponding to 46 of the 66 A. thaliana CBSR genes, for which expression was found for 73 of the rice orthologous genes in our data set (Supplementary Data Fig. S3). On average, half of the CBSR genes were altered by nematode infection in susceptible or resistant rice plants. However, we found only eight CBSR genes that have a similar response to M. graminicola as to other pathogens when we compared the response of the resistant or susceptible rice genotypes at 8 dpi with the A. thaliana CBSR gene responses (Amrine et al., 2015). Interestingly, three genes were annotated as receptor kinases, two are WRKY and MYB transcription factors and the third (PDCB3) is involved in callose deposition at plasmodesmata and thus in plant cell-to-cell communication. The two other genes had no annotation (Fig. S3).
Discussion
The African rice line TOG5681 presents high resistance to the root-knot nematode M. graminicola (Soriano et al., 1999). However, insights into the mechanisms operating in host resistance are lacking. To provide an extensive characterization of the rice resistance responses to M. graminicola we present sequence data generated by Illumina technology of the whole transcriptomes of O. glaberrima and O. sativa root tips, uninfected or after root-knot nematode infection. This is the first report presenting such data for O. glaberrima under biotic stress. Oryzaglaberrima is essentially cultivated in West Africa, where it is better adapted than O. sativa (Linares, 2002). A number of O. glaberrima genotypes present resistance to various biotic and abiotic stresses, including resistance or tolerance to viruses (Albar et al., 2003), root-knot nematodes (Soriano et al., 1999), bacteria, drought, iron toxicity and high salinity (Linares, 2002). The species is a rich genetic reservoir used for NERICA varieties, which combine the traits of resistance to abiotic and biotic stresses of the African rice species with the high yield and grain quality of the Asian rice species (Ndjiondjop et al., 2008; Agnoun et al., 2012). Improving our knowledge of this resource species, especially at the level of the molecular basis of resistance, is of great importance. Importantly, gene expression data could help in the annotation of the two recently published O. glaberrima genome sequences, for instance with novel genes – either novel transcribed regions or alternatively spliced forms (Wang et al., 2014; Zhang et al., 2014).
TOG5681 limits nematode penetration and development
When comparing nematode infection in TOG5681 and ‘Nipponbare’, we observed that significantly fewer M. graminicola juveniles penetrated and persisted in resistant roots (Fig. 1), but the few persisting nematodes were able to induce GCs (Fig. 2). However, as galls developed in TOG5681, GCs degenerated. Most parasitic nematodes did not reach the adult stage and were therefore not able to reproduce. In other resistant O. glaberrima lines tested, some HR-like necrosis of root cells surrounding the nematode or in the vicinity of GCs has been reported (Cabasan et al., 2013), but a low level of nematode penetration and late GC degradation was the most common plant defence response to M. graminicola. Transmission electron microscopy has proved to be a valuable tool to visualize ultrastructural changes in GCs or cells in close vicinity to the nematode, typical of an hypersensitive response (HR) cell death phenotype, including cytoplasmic shrinkage, chromatin condensation, mitochondrial swelling, and chloroplast disruption combined with vacuolization (Coll et al., 2011). Transmission electron microscopy could verify that resistance in African rice to M. graminicola is characterized by HR or HR-like responses, as occurs in other plant species, including tomato (Bleve-Zacheo et al., 1982), coffee (Anthony et al., 2005; Albuquerque et al., 2010), grapevine (Anwar and McKenry, 2002), pepper (Bleve-Zacheo et al., 1998), resistant Ma plums (Saucet et al., 2016) and wild grasses (Balhadère and Evans, 1995). In these resistant species, the HR is usually efficient in stopping nematode development (Albuquerque et al., 2010; Bleve-Zacheo et al., 1982, 1998; Khallouk et al., 2011) or is accompanied by poor development of nutrient feeding sites (Anthony et al., 2005; Fourie et al., 2013).
Early resistance of TOG5681 was associated with low levels of juvenile penetration and strong activation of defence genes. Whether activation of host defence is directly responsible for a lower penetration rate of juveniles is still an open question. Meloidogyne spp. J2s are attracted towards the plant by root exudates, but some compounds may have nematotoxic or nematostatic effects (Curtis et al., 2009; Dutta et al., 2012). Although it is known that rice roots produce small lipophilic molecules that can repel M. graminicola (Dutta et al., 2012), no data are currently available about nematode chemoattraction to O. glaberrima roots. Nevertheless, lower chemical attraction or higher production of repellents may occur in TOG5681 varieties.
TOG5681 transcriptional responses are expressed early
Transcriptome data of incompatible plant-Meloidogyne sp. interactions reported so far showed that there was no specific pathway induced in the resistant cultivar, but rather that several small changes in gene expression are associated with the resistance response. For instance, hormonal pathways are deeply altered in soybean—M. javanica interaction (Beneventi et al., 2013) and in peanut-Meloidogyne arenaria interaction (Guimaraes et al., 2015). General defence responses, including the phenylpropanoid pathway, are also strongly affected in soybean, peanut and alfalfa resistant genotypes (Tirumalaraju et al., 2011; Beneventi et al., 2013; Guimaraes et al., 2015; Postnikova et al., 2015). In addition, important changes in R gene expression were reported in the alfalfa-M. incognita interaction (Postnikova et al., 2015).
In this study we generated whole-transcriptome data for a susceptible and a resistant rice genotype at several time points (2, 4 and 8 dpi) after nematode challenge, making it possible to refine expression data chronologically. A higher number of genes was activated in TOG5681 versus ‘Nipponbare’ as soon as 2 d after M. graminicola infection (Table 1). In fact, we observed that 1028 (84 %) of the 1228 TOG5681-induced genes were not altered at all in ‘Nipponbare’ at 2 dpi. In addition, most of these genes were consistently expressed at high levels in the resistant infected roots at all time points analysed (2–8 dpi). These data are consistent with the resistance responses registered in other plant-pathogen interactions where a strong and rapid activation of plant defence usually differentiates the resistant genotype from the susceptible. A recent study in susceptible and resistant varieties of rice (O. sativa) showed similar contrasting data in the qPCR expression pattern for a limited set of genes (Kumari et al., 2016). However, only fragmented data have been reported so far in incompatible plant-nematode interactions. For instance, it was reported in the alfalfa-M. incognita interaction that the host response was significantly stronger in the susceptible compared with the resistant cultivar (Postnikova et al., 2015), but this analysis was conducted only at 7 dpi.
Most of the changes in plant gene expression critical to the initiation of defence pathways occurred at a time when the nematode is inducing GC formation (Nguyen et al., 2014). As described previously (Nahar et al., 2011; Kyndt et al., 2012b; Ji et al., 2013), our data confirm that the nematode is able to inhibit plant transcriptional responses in the compatible interaction. Meloidogyne graminicola candidate virulence effectors involved in the compatible interaction with ‘Nipponbare’ have been identified (Haegeman et al., 2013; Petitot et al., 2015). Their role in defence suppression is under investigation by our groups.
Interestingly, when focusing on the CBRS genes known to respond to a wide range of non-nematode pathogens (Amrine et al., 2015), we found a very limited number of similarities with the genes responding to M. graminicola in rice (Fig. S3). However, the study of Amrine et al. (2015) involved only one root pathogen, Plasmodiophora brassicae, which causes the disease cabbage club root. Using the NEMATIC database tool (Cabrera et al., 2014), we confirmed that almost no similarities in CBRS gene expression occur when comparing A. thaliana challenged with M. incognita (data not shown) versus other pathogens. This study thus highlighted that plant responses to nematodes involve distinct pathways, deserving further attention.
Role of hormonal pathways is complex
The role of jasmonic acid and related oxylipins in plant defence is well known (Blée, 2002). Previous research has demonstrated the role of the JA pathway in rice defence against M. graminicola (Nahar et al., 2011). The transcriptome data analysed here also suggested a potential role for JA biosynthesis in TOG5681 resistance to M. graminicola. OsLOX7 and OsAOS2 were strongly induced upon infection in both susceptible and resistant plants, but in TOG5681 they reached a log FC of 5 as soon as 2 dpi. The expression of these genes was consistently high, while in ‘Nipponbare’ they were only later and less strongly induced. Inhibition of JA through application of specific LOX inhibitors led to a slightly increased number of nematodes in the TOG5681 line, although it did not result in infection levels attaining those of the compatible interaction. This minor increase in penetration due to LOX inhibition probably reflects the role of JA or other oxylipins in basal defence against nematodes. Similarly, Bhattarai et al. (2008) showed that JA-dependent signalling does not play a role in Mi-1-mediated resistance to M. incognita in tomato.
The role of ethylene in plant-nematode interactions is complex (reviewed in Kyndt et al., 2013). Although ethylene biosynthesis genes responded slightly faster to nematode infection in TOG5681 in comparison with ‘Nipponbare’, a complex pattern of repression and activation was observed in both the incompatible and compatible interactions.
Salicylic acid can be produced via the phenylpropanoid or the isochorismate pathway. No indication of a role of the isochorismate pathway in TOG5681 resistance was found in our transcriptome analysis since it was repressed in both the compatible and the incompatible interactions. However, the phenylpropanoid pathway showed an earlier and stronger induction in TOG5681 upon infection. Specifically, an earlier (2 dpi) and stronger (6- to 8-fold) activation of OsPAL4 and OsPAL6 was observed. OsPAL4 and to a lesser extent OsPAL6 were shown to be key elements in resistance to several bacterial and fungal diseases governed by QTL (Tonnessen et al., 2015). Inhibition of PAL activity by AOPP application to TOG5681 led to increased gall formation, although this inhibition could not fully restore susceptibility. Next to SA biosynthesis, the phenylpropanoid pathway is involved in production of many other defence-related metabolites and hence further research is needed to decipher which one is specifically accumulating and potentially important in this incompatible interaction.
Rice candidate genes for nematode resistance
Although nematode infection triggered a broad range of biological responses, we selected a short list of candidate genes for M. graminicola resistance in rice (Table 2). They all displayed elevated levels of expression in TOG5681, either in control plants or upon infection, and showed contrasting expression profiles in the resistant genotype versus the susceptible plants. As expected, a subset of these genes encodes PR proteins (thaumatins, thionins) and components of the phenylpropanoid pathway that are known to be pathogen-responsive (Vogt, 2010). These genes participate in the general defence responses to a wide range of pathogens, but are not primary determinants of the outcome of an interaction between a plant and a pathogen. Remarkably, OsThion2 displayed the highest basal expression of the 55 987 expression profiles analysed in the TOG5681 control root tips. Thionins are small antimicrobial cysteine-rich peptides that are involved in plant defence and classified in the PR-13 family (Sels et al., 2008). In rice (O. sativa), a role for OsTHION7 in increased defence to M. graminicola infection has been recently demonstrated (Ji et al., 2015). Three other thionin genes (OsThion1, 3 and 9) presented a very high basal expression in TOG5681 root tips. However, one striking characteristic of the responses to M. graminicola infection was the repression of these genes in both the resistant and the susceptible rice genotypes. Other candidate genes highlighted in this study are four homologues of disease resistance genes characterized in other pathosystems, including the stripe rust resistance protein Yr10, a sterol 3-β-glucosyltransferase, an ATPase and an unknown protein. Interestingly, these last genes were previously identified as positional candidate genes for QTL involved in O. sativa resistance to M. graminicola (Dimkpa et al., 2015). Our study adds expression data to these 323 positional candidate genes, highlighting those genes for which regulation correlates with resistance to M. graminicola.
Concluding remarks
This investigation provides novel insights into the resistance responses of the African rice O. glaberrima to root-knot nematodes. We describe here for the first time RNA-Seq data for O. glaberrima under biotic stress. As observed by microscopy, resistance in African rice clearly involves the employment of different mechanisms during the rice-nematode interaction, initially with juvenile penetration restricted, and later with degeneration of GCs. Future research will focus on advancing understanding of the role of the upregulated rice genes in this resistance response. These data offer interesting perspectives for developing control strategies in rice towards Meloidogyne spp. based on rice genetic resources such as the African rice species O. glaberrima. Finally, this study again highlights the rice-M. graminicola pathosystem as a model to study plant-nematode incompatible interactions in monocotyledons (Fernandez et al., 2015).
Supplementary Data
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: histological analysis of galls induced by Meloidogyne graminicola in resistant rice (Oryza glaberrima TOG5681) and susceptible rice (Oryza sativa ‘Nipponbare’) roots. Figure S2: galls formed by M. graminicola in susceptible rice (O.sativa ‘Nipponbare’) roots at 7, 15 and 22 dpi. Sections (10 µm) were observed under UV light (×10 or ×20). Figure S3: common transcriptional regulation between Arabidopsis thaliana and rice core biotic stress-responsive (CBSR) genes after pathogen challenge. Table S1: list of genes, primer sequences and annealing temperatures used for quantitative reverse transcriptase PCR in this study. Table S2: description of the RNA-Seq libraries used in this study. Table S3: RNA-Seq expression data for selected sets of rice genes in O.sativa ‘Nipponbare’ and Oryza glaberrima TOG5681 roots. Table S4: real-time quantitative PCR expression data for candidate rice genes in O.sativa ‘Nipponbare’ and O.glaberrima TOG5681 roots.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr François Sabot (IRD) for TOG5681 nucleotide analyses. This work was supported by France Génomique (9-2012), Agropolis Fondation (1002-003), Ghent University Special Research Fund (BOF13/GOA/030), Fonds Wetenschappelijk Onderzoek - Vlaanderen (to T.K.) and the Syrian Ministry of Science (to R.H.).
LITERATURE CITED
- Agnoun Y, Sié M, Djedatin G. , et al . 2012. Molecular profiling of interspecific lowland rice progenies resulting from crosses between TOG5681 and TOG5674 (Oryza glaberrima) and IR64 (Oryza sativa). International Journal of Biology 4: 19–28. [Google Scholar]
- Albar L, Ndjiondjop MN, Esshak Z. , et al . 2003. . Fine genetic mapping of a gene required for rice yellow mottle virus cell-to-cell movement. Theoretical and Applied Genetics 107: 371–378. [DOI] [PubMed] [Google Scholar]
- Albuquerque E, Carneiro R, Costa P. , et al . 2010. . Resistance to Meloidogyne incognita expresses a hypersensitive-like response in Coffea arabica. European Journal of Plant Pathology 127: 365–373. [Google Scholar]
- Amrine KC, Blanco-Ulate B, Cantu D.. 2015. . Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co-expression network analysis. PLoS One 10: e0118731. doi:10.1371/journal.pone.0118731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony F, Topart P, Martinez A, Silva M, Nicole M.. 2005. . Hypersensitive-like reaction conferred by the Mex-1 resistance gene against Meloidogyne exigua in coffee. Plant Pathology 54: 476–482. [Google Scholar]
- Anwar SA, McKenry MV.. 2002. . Developmental response of a resistance-breaking population of Meloidogyne arenaria on Vitis spp. Journal of Nematology 34: 28–33. [PMC free article] [PubMed] [Google Scholar]
- Balhadère P, Evans AAF. . 1995. . Histopathogenesis of susceptible and resistant responses of wheat, barley and wild grasses to Meloidogyne naasi . Fundamental and Applied Nematology 18: 531–538. [Google Scholar]
- Bhattarai KK, Xie QG, Mantelin S. , et al . 2008. Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Molecular Plant Microbe Interactions 21: 1205–1214. [DOI] [PubMed] [Google Scholar]
- Beneventi MA, da Silva OB, Lisei de Sá ME. , et al . 2013. Transcription profile of soybean root-knot nematode interaction reveals a key role of phytohormones in the resistance reaction. BMC Genomics 14: 322. doi:10.1186/1471-2164-14-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimpong IK, Carpena AL, Mendioro MS. , et al. 2010. Evaluation of Oryza sativa x O. glaberrima derived progenies for resistance to root-knot nematode and identification of introgressed alien chromosome segments using SSR markers. African Journal of Biotechnology 9: 3988–3997. [Google Scholar]
- Blée E. 2002. Impact of phyto-oxylipins in plant defense. Trends in Plant Science s7: 315–322. [DOI] [PubMed] [Google Scholar]
- Bleve-Zacheo T, Zacheo G, Melillo MT, Lamberti F. . 1982. Ultrastructural aspects of the hypersensitive reaction in tomato root cells resistant to Meloidogyne incognita. Nematologia Mediterranea 10: 81–90. [Google Scholar]
- Bleve-Zacheo T, Bongiovanni M, Melillo MT, Castagnone-Sereno P.. 1998. . The pepper resistance genes Me1 and Me3 induce differential penetration rates and temporal sequences of root cell ultrastructural changes upon nematode infection. Plant Science 133: 79–90. [Google Scholar]
- Bybd DW, Kirkpatrick T, Barker KR.. 1983. An improved technique for clearing and staining plant tissues for detection of nematodes. Journal of Nematology 15: 142–143. [PMC free article] [PubMed] [Google Scholar]
- Cabasan MTN, De Waele D, Kumar A.. 2012. Comparison of migration, penetration, development and reproduction of Meloidogyne graminicola on susceptible and resistant rice genotypes. Nematology 14: 405–415. [Google Scholar]
- Cabasan MTN, Kumar A, Bellafiore S, de Waele D. . 2013. Histopathology of the rice root-knot nematode, Meloidogyne graminicola, on Oryza sativa and O. glaberrima. Nematology 16: 73–81. [Google Scholar]
- Cabrera J, Bustos R, Favery B, Fenoll C, Escobar C.. 2014. NEMATIC: a simple and versatile tool for the in silico analysis of plant-nematode interactions. Molecular Plant Pathology 15: 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL.. 2011. Programmed cell death in the plant immune system. Cell Death and Differentiation 18: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis RHC, Robinson AF, Perry RN.. 2009. Hatch and host location In: Perry R, Moens M, Starr J, eds. Root-knot nematodes. Wallingford, UK: CABI Publishing, 139–162. [Google Scholar]
- Dimkpa SO, Lahari Z, Shrestha R, Douglas A, Gheysen G, Price AH.. 2015. A genome-wide association study of a global rice panel reveals resistance in Oryza sativa to root-knot nematodes. Journal of Experimental Botany 67: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta TK, Powers SJ, Gaur HS, Birkett M, Curtis R.. 2012. Effect of small lipophilic molecules in tomato and rice root exudates on the behaviour of Meloidogyne incognita and M. graminicola. Nematology 14: 309–320. [Google Scholar]
- Fernandez D, Petitot AS, Grossi de Sá M, Nguyen VP, de Almeida-Engler J, Kyndt T. . 2015. . Recent advances in understanding plant-nematode interactions in monocots. Advances in Botanical Research 73: 189–219. [Google Scholar]
- Fernandez L, Cabasan MTN, De Waele D.. 2014. Life cycle of the rice root-knot nematode Meloidogyne graminicola at different temperatures under non-flooded and flooded conditions. Archives of Phytopathology and Plant Protection 47: 1042–1049. [Google Scholar]
- Fortuner R, Merny G. . 1979. Root-parasitic nematodes of rice. Revue de Nématologie 2: 79–102. [Google Scholar]
- Fourie H, Mc Donald AH, De Waele D, Jordaan A. . 2013. Comparative cellular responses in susceptible and resistant soybean cultivars infected by Meloidogyne incognita. Nematology 15: 695–708. [Google Scholar]
- Goverse A, Smant G.. 2014. . The activation and suppression of plant innate immunity by parasitic nematodes. Annual Review of Phytopathology 52: 243–265. [DOI] [PubMed] [Google Scholar]
- Guimaraes PM, Guimaraes LA, Morgante CV. , et al . 2015. Root transcriptome analysis of wild peanut reveals candidate genes for nematode resistance. PLoS One 10: e0140937. doi:10.1371/journal.pone.0140937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman A, Bauters L, Kyndt T, Rahman MM, Gheysen G.. 2013. Identification of candidate effector genes in the transcriptome of the rice root knot nematode Meloidogyne graminicola. Molecular Plant Pathology 14: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Gheysen G, Denil S. , et al . 2013. Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. Journal of Experimental Botany 64: 3885–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Gheysen G, Ullah C. , et al . 2015. The role of thionins in rice defence against root pathogens. Molecular Plant Pathology 16: 870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khallouk S, Voisin R, Van Ghelder C, Engler G, Amiri S, Esmenjaud D.. 2011. . Histological mechanisms of the resistance conferred by the Ma gene against Meloidogyne incognita in Prunus spp. Phytopathology 101: 945–951. [DOI] [PubMed] [Google Scholar]
- Kumari C, Dutta TK, Banakar P, Rao U.. 2016. Comparing the defence-related gene expression changes upon root-knot nematode attack in susceptible versus resistant cultivars of rice. Scientific Reports 6: 22846. doi: 10.1038/srep22846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt T, Denil S, Haegeman A. , et al . 2012a. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytologist 196: 887–900. [DOI] [PubMed] [Google Scholar]
- Kyndt T, Nahar K, Haegeman A, De Vleesschauwer D, Höfte M, Gheysen G.. 2012b. Comparing systemic defence-related gene expression changes upon migratory and sedentary nematode attack in rice. Plant Biology (Stuttgart, Germany) 14 Suppl 1: 73–82. doi:10.1111/j.1438-8677.2011.00524.x. [DOI] [PubMed] [Google Scholar]
- Kyndt T, Vieira P, Gheysen G, de Almeida-Engler J.. 2013. Nematode feeding sites: unique organs in plant roots. Planta 238: 807–818. [DOI] [PubMed] [Google Scholar]
- Lee HI, León J, Raskin I.. 1995. . Biosynthesis and metabolism of salicylic acid. Proceedings of the National Academy of Sciences of the USA 92: 4076–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares OF. 2002. African rice (Oryza glaberrima): history and future potential. Proceedings of the National Academy of Sciences of the USA 99: 16360–16365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux M, Reversat G, Diaz SXG. , et al . 2003. Linkage mapping of Hsa-1Og, a resistance gene of African rice to the cyst nematode, Heterodera sacchari. Theoretical and Applied Genetics 107: 691–696. [DOI] [PubMed] [Google Scholar]
- Nahar K, Kyndt T, De Vleesschauwer D, Höfte M, Gheysen G.. 2011. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiology 157: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndjiondjop MN, Semagn K, Sie M, Cissoko M, Fatondji B, Jones M. . 2008. . Molecular profiling of interspecific lowland rice populations derived from IR64 (Oryza sativa) and TOG5681 (Oryza glaberrima). African Journal of Biotechnology 7: 4219–4229. [Google Scholar]
- Nguyen VP, Bellafiore S, Petitot AS. , et al . 2014. Meloidogyne incognita–rice (Oryza sativa) interaction: a new model system to study plant root-knot nematode interactions in monocotyledons. Rice 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil J, Gaur HS.. 2014. The effect of root-knot nematode, Meloidogyne graminicola, on the quality and vigour of rice seed. Nematology 16: 555–564. [Google Scholar]
- Petitot AS, Dereeper A, Agbessi M. , et al. 2016. Dual RNA-seq reveals Meloidogyne graminicola transcriptome and candidate effectors during the interaction with rice plants. Molecular Plant Pathology 17: 860–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RA, Coyne DL, Nash P, Jones MP.. 1999. Resistance to the rice nematodes Heterodera sacchari, Meloidogyne graminicola and M. incognita in Oryza glaberrima and O. glaberrima x O. sativa interspecies hybrids. Nematology 1: 745–751. [Google Scholar]
- Postnikova O, Hult M, Shao J, Skantar A, Nemchinov LG.. 2015. Transcriptome analysis of resistant and susceptible alfalfa cultivars infected with root-knot nematode Meloidogyne incognita. PLoS One 10: e0118269. doi:10.1371/journal.pone.0118269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodiuc N, Vieira P, Banora MY, de Almeida Engler J.. 2014. On the track of transfer cell formation by specialized plant-parasitic nematodes. Frontiers in Plant Science 5: 160. doi:10.3389/fpls.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reversat G, Boyer J, Sannier C, Pando-Bahuon A.. 1999. A mixture of sand and water-absorbent synthetic polymer as substrate for the xenic culturing of plant-parasitic nematodes in the laboratory. Nematology 1: 209–212. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucet SB, Van Ghelder C, Abad P, Duval H, Esmenjaud D.. 2016. Resistance to root-knot nematodes Meloidogyne spp. in woody plants. New Phytologist 211: 41–56. doi:10.1111/nph.13933. [DOI] [PubMed] [Google Scholar]
- Seck PA, Diagne A, Mohanty S, Wopereis MCS.. 2012. Crops that feed the world 7: rice. Food Security 4: 7–24. [Google Scholar]
- Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF.. 2008. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiology and Biochemistry 46: 941–950. [DOI] [PubMed] [Google Scholar]
- Soriano IR, Reversat G.. 2003. Management of Meloidogyne graminicola and yield of upland rice in South-Luzon, Philippines. Nematology 5: 879–884. [Google Scholar]
- Soriano I, Schmit V, Brar D, Prot JC, Reversat G.. 1999. Resistance to rice root-knot nematode Meloidogyne graminicola identified in Oryza longistaminata and O. glaberrima. Nematology 1: 395–398. [Google Scholar]
- Thimm O, Bläsing O, Gibon Y. , et al . 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Tirumalaraju SV, Jain M, Gallo M.. 2011. Differential gene expression in roots of nematode-resistant and -susceptible peanut (Arachis hypogaea) cultivars in response to early stages of peanut root-knot nematode (Meloidogyne arenaria) parasitization. Journal of Plant Physiology 168: 481–492. [DOI] [PubMed] [Google Scholar]
- Tonnessen BW, Manosalva P, Lang JM. , et al . 2015. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Molecular Biology 87: 273–286. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. . 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P, Escudero C, Rodiuc N. , et al . 2013. . Ectopic expression of Kip-related proteins restrains root-knot nematode-feeding site expansion. New Phytologist 199: 505–519. [DOI] [PubMed] [Google Scholar]
- Vogt T. 2010. . Phenylpropanoid biosynthesis. Molecular Plant 3: 2–20. [DOI] [PubMed] [Google Scholar]
- Wang M, Yu Y, Haberer G. , et al . 2014. . The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nature Genetics 46: 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM, Gleason CA.. 2003. Plant-nematode interactions. Current Opinion in Plant Biology 6: 327–333. [DOI] [PubMed] [Google Scholar]
- Zhang QJ, Zhu T, Xia EH. , et al. 2014. Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proceedings of the National Academy of Sciences of the USA 111: E4954–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.