Summary
No disease-slowing treatments exist for Huntington’s disease (HD), but its monogenic inheritance makes it an appealing candidate for the development of therapeutics targeting pathogenic processes close to its root genetic cause. HD is caused by CAG repeat expansions in the HTT gene, which encodes huntingtin protein; targeting of HTT transcription and the translation of its mRNA are therefore an area of intense investigation. ‘Huntingtin-lowering’ strategies include antisense oligonucleotides (ASOs) and RNA interference compounds targeting mRNA; and zinc-finger transcriptional repressors (ZFTR) and newer CRISPR/Cas9 methods aiming to reduce transcription by targeting DNA. An intrathecally-delivered ASO targeting total huntingtin is now well into its first human clinical trial, with other ASO approaches expected to enter trials within the next 1-2 years, and virally-delivered RNAi and ZFTR in advanced testing in animal models. Meanwhile, recent advances to improve the design and delivery of targeted therapeutics are likely to improve their efficacy, safety, tolerability and duration of effect.
Introduction
Because all cases of Huntington’s disease (HD) are caused by the same basic mutation, it is an ideal candidate for the development of therapeutics targeting pathogenic processes close to its root genetic cause. Expansion of a CAG tract in the HTT gene results in ubiquitous expression of mutant huntingtin protein (mHTT), thought to be the predominant toxic species.1 Individuals destined to develop HD can be identified by genetic testing, so intervention in the long premanifest phase should postpone or prevent symptom onset.2
HD is autosomal dominantly inherited and thought to be substantially caused by toxic gains of function,3,4 so reducing mHTT production should alleviate its pathogenesis. There is some evidence that loss of function by mHTT (i.e. haploinsufficiency) contributes to HD,5 which could be exacerbated by reducing HTT production – an important consideration that human trials must be designed to detect.
It is a pivotal time for therapies targeting disease-associated genes and proteins. The first clinical success for such a therapy was recently reported: the US Food and Drug Administration (FDA) approved nusinersen, a lumbar intrathecally-administered antisense oligonucleotide (ASO) that extended survival in spinal muscular atrophy via targeted modulation of gene expression, increasing production of SMN2.6,7 We cannot assume general success from this single instance: this approach acted by restoring a missing protein function rather than reducing a toxic protein and SMA is typically more aggressive than HD, so potentially an easier disease in which to demonstrate a therapeutic effect in a relatively short efficacy trial. Nonetheless, the first proof that neurodegeneration can be slowed in humans by a targeted therapeutic modulating gene expression gives encouragement to similar programmes in HD.
The term ‘gene silencing’ is sometimes used for targeted reduction in gene expression. This could be interpreted as complete deactivation, which is probably neither desirable nor attainable. We therefore prefer ‘huntingtin lowering’. The term ‘gene therapy’ is reserved for approaches where genetic material of living cells is modified,8 resulting in host cells manufacturing non-native mRNA and sometimes protein; this has special safety and regulatory implications.
This review focuses on novel therapeutic strategies targeting the mHTT production pathway. Those designed to interact with HTT mRNA include antisense oligonucleotides (ASOs) and RNA interference (RNAi) compounds that accelerate degradation of the transcript; and orally bioavailable small molecules to reduce HTT through altering mRNA splicing. Agents that interact directly with HTT DNA include zinc finger transcriptional repressors (ZFTRs) and CRISPR/Cas9 ‘genome editing’ constructs.
Huntingtin and the molecular pathology of HD
The HTT gene is highly conserved,9 and its embryonic knockout is lethal,10 suggesting that the protein is essential for normal development. The function of wild-type huntingtin (wtHTT) is incompletely understood, but it is ubiquitously expressed, has many interaction partners and performs many likely roles including vesicular trafficking; the mediation of endocytosis, vesicular recycling and endosomal trafficking; coordination of cell division; transcriptional regulation; and metabolism.5 The toxic effects of mHTT are equally diverse, with numerous pathways deranged in its presence.11 Despite many attempts, no approach targeting these downstream pathways has succeeded,12 emphasising the difficulty of achieving clinical benefit in a disease with pleiotropic cellular derangements. This is perhaps one reason for the current emphasis on the therapies we describe here, which target the most proximal pathogenic events.
Some useful functions of huntingtin may already be impaired in HD mutation carriers because mHTT performs them less efficiently: for example, overexpressing wtHTT stimulates axonal vesicular transport of brain-derived neurotrophic factor (BDNF), but mHTT does not, suggesting loss of huntingtin function may contribute to BDNF deficiency in patient striatum.13
Lowering huntingtin could exacerbate these putative pathogenic contributions from haploinsufficiency. This concern needs to be balanced against the detrimental effects of the persistent presence of mHTT. Complete inactivation of HTT in the adult rodent brain may cause a progressive neurological phenotype14 but partial reduction to 50% or more in the adult is well-tolerated across model species, with the important caveat that follow-up times in these animal studies have generally been rather short.15–19 The longest primate study revealed no toxicity after 6 months’ partial suppression in the striatum.19 Importantly, no huntingtin-lowering strategy currently proposed for human use would produce complete knockdown. Nonetheless, it remains unknown whether long-term partial suppression of huntingtin in humans will prove safe, so human trials must be designed with long follow-up, and safety measures sensitive enough to detect toxicity from this on-target mechanism. For now, these concerns also favour the prioritisation of agents with shorter half-lives, that are reversible or that can be inactivated if problems emerge.
The desirability of suppressing only the mutant protein (‘allele-selective’ huntingtin-lowering), given the greater difficulty of achieving this, is an important focus of the field. Appropriately, efforts to pursue allele-selective and nonselective strategies are underway.
While a CAG-expanded HTT gene is certainly the proximal cause of HD, and mHTT is generally agreed to be harmful to neurons, recent years have seen growing debate about whether it is the sole pathogenic agent.
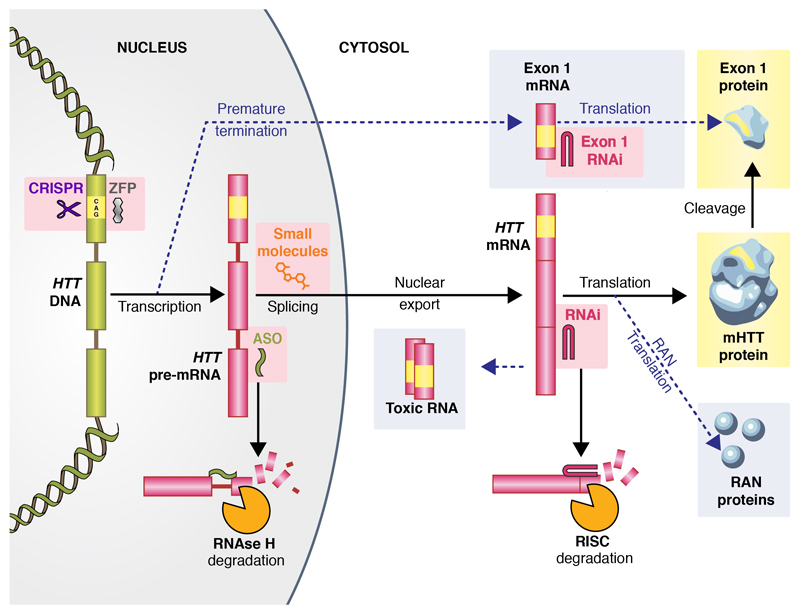
Huntingtin exon 1 contains the expanded polyglutamine tract and is sufficient to cause pathology.20 Fragments generated from cleavage of full-length mHTT are traditionally thought of as the source of this toxic species, but a variant of mHTT mRNA resulting from incomplete splicing was recently described that encodes a short exon 1 huntingtin protein. This transcript is formed more avidly when the HTT gene is CAG-expanded, and has been found in postmortem HD patient but not control brains.21 Others have shown that huntingtin mRNA is subject to repeat-associated non-ATG (‘RAN’) translation, which can generate many unusual protein species from both sense and antisense strands, some of which may be toxic.22 Finally, it has been proposed that CAG-expanded HTT RNA itself may be toxic.23 Figure 1 shows these proposed mechanisms, alongside the conventional pathway to mHTT and its toxic exon 1 fragment.
Figure 1.
The production of huntingtin protein, and targeted molecular therapies in development to reduce it. Yellow marks the pathogenic expanded CAG tract and its polyglutamine product. Therapeutic approaches are highlighted with pink boxes. Yellow boxes indicate the most widely accepted toxic species. Dotted arrows and grey boxes indicate proposed non-traditional mechanisms for the production of toxic species. The chief mechanisms of action of ASOs and RNAi compounds are shown at the bottom. The image of huntingtin protein is adapted from reference 24 under a creative commons licence (CC-BY-4.0).
The contribution of each putative alternative mechanism remains unclear, but their existence warrants special consideration, since the effect of each huntingtin-lowering approach on each pathogenic pathway will depend on precisely where the intervention targets the protein expression pathway. These, too, are highlighted in Figure 1. Targeting proximal events will be more likely to diminish these alternative mechanisms. This will need to be borne in mind, especially if an agent appears to have engaged with its target in a trial, but fails to ameliorate clinical features.
Targeting the RNA
Messenger RNA is accessible in the nucleus or cytosol and, in contrast to DNA, is unprotected by repair machinery. Thus, reducing translation of HTT mRNA ought to be easier to achieve than modulating transcription or altering the gene itself. Three main methods exist for attempting this: ASOs, RNA interference (RNAi) compounds and small-molecule splicing modulators (summarised in Table 1).
Table 1.
Current publicly-announced huntingtin-lowering programmes targeting mRNA.
| Class | Mechanism | Allele selectivity | Delivery | Vector | Sponsor | Key advantages | Key disadvantages | Ref |
|---|---|---|---|---|---|---|---|---|
| ASO | Pre-mRNA degradation | None | Intrathecal | None | Ionis | Single drug for all HD mutation carriers | Theoretical risk from reducing wtHTT | 25–28 |
| SNP-targeted | Intrathecal | None | Wave Life Sciences | Selective silencing of mutant allele |
|
29,30 | ||
| CAG repeat | Intrathecal | None | Biomarin | Selective silencing of mutant allele with a single drug for all mutation carriers | Will reduce expression of other important CAG-containing genes, risking off-target effects | 31 | ||
| RNAi | mRNA degradation | None | Intracranial | AAV2 | Spark | Single treatment provides sustained HTT reduction |
|
15–17 |
| Intracranial | AAV1 | Voyager | 32 | |||||
| Intracranial | AAV5 | UniQure N.V. | 33,34 | |||||
| Small molecule | Pre-mRNA splicing | Unknown | Oral | None | CHDI Foundation |
|
More difficult to achieve selectivity for HTT over other genes with non-nucleotide therapeutic | 35 |
ASOs and RNAi compounds are each nucleotide-based therapeutic molecules that bind to mRNA selectively through Watson-Crick complementarity, triggering the cell’s RNA degradation machinery to dispose of the transcript.
ASOs are synthetic single-stranded DNA molecules that principally bind pre-mRNA in the nucleus, targeting it for degradation by RNase H36. RNAi uses RNA-based therapeutic molecules – including short interfering RNA (siRNA), short hairpin RNA (shRNA) or microRNA (miRNA) depending on structure and sequence – that bind to mature, spliced cytosolic mRNA, targeting it for removal by argonaute 2, the RNAse enzyme within the RNA-induced silencing complex (RISC).37
Some differences between these approaches are apparent from their sites of action (Figure 1). Acting on pre-mRNA, ASOs can target exons and introns, permitting an increased choice of target sequences. They are expected to reduce the production of toxic mRNA and RAN proteins, but whether they can prevent formation of exon 1 truncated mRNA is unclear, since this might be generated and released before the addition of the downstream pre-mRNA sequence to which the ASO binds.
RNAi compounds act on spliced mRNA, so they could only reduce the formation of exon 1 mRNA fragments by directly targeting a sequence within that exon, and may not prevent the formation of toxic RNA or RAN proteins.
For reasons that remain obscure, single-stranded DNA diffuses well in the CNS and is taken up by neurons and other cells. Thus, injection of ASOs into the CSF (intraventricularly in mice or by lumbar intrathecal administration in larger mammals) results in fairly widespread delivery of drug to the brain and corresponding reductions in mRNA and protein.27 In contrast, double-stranded RNA has limited diffusion and cellular uptake in the CNS, so enhanced delivery methods or viral vectors such as adeno-associated virus (AAV) are needed to deliver the siRNA by injection into the brain parenchyma. Though more challenging, this mode of administration should permit lifelong treatment from a single dose.
Huntingtin lowering with ASOs and RNA-based compounds has been successfully achieved in numerous model systems (see Keiser et al. for a detailed inventory).38 Invariably, reduction in HTT mRNA produces a concomitant reduction in huntingtin protein, usually accompanied by amelioration of pathology and improvements in neurological deficits when given to symptomatic animals. When given to mutation-bearing animals before disease manifestations begin, onset is significantly postponed.
Despite these encouraging results, the limitations of animal models are substantial. No therapeutic HD animal model success has yet predicted benefit in a human trial.12 This is a particular concern for therapeutics administered directly into the CNS: a human brain is over 3,000 times larger than a mouse brain and is likely vastly different in respect of diffusion through the cerebrospinal fluid, into the brain parenchyma and within cells.
The use of primates can lend support to a case that a compound is capable of attaining broad distribution and huntingtin-lowering, but these wild-type animals, lacking mHTT, are incomplete models for potential toxicity and cannot inform us at all about therapeutic effect. Large transgenic models such as sheep and pigs and are beginning to be used39 but the unparalleled complexity of the human brain, and the highly variable, slowly progressive course of HD in patients, mean that no portfolio of animal testing will ever be a guarantee of success. Combining multiple small and large animal models, with an awareness of the particular applicability and limitations of each model, must be paired with a cautious approach to clinical testing in order to maximise safety and the chance of successful translation.
Agents made from exogenous RNA and DNA have the potential for off-target effects. For instance, some synthetic oligonucleotides are recognised by toll-like receptors on immune cells, raising the risk of immunogenicity that may be difficult to predict from animal studies.40 Thrombocytopenia has been observed in some trials of systemically-administered ASOs, and worsening peripheral neuropathy with an siRNA drug. It is not clear whether these are class effects.41 The brain’s immunological privilege and smaller doses needed with direct CNS administration may mitigate these risks in neurodegeneration: this appears to have been the case with nusinersen,6 but this cannot be taken for granted with new therapeutics: vigilance is essential.
ASO approaches
Several ASOs are being investigated in HD model systems, but we begin by exploring in detail the first targeted huntingtin-lowering compound to enter a human trial.
IONIS-HTTRx
In September 2015, the first patients were dosed at University College London and the University of British Columbia with IONIS-HTTRx, an ASO targeting human huntingtin, in a phase 1b/2a clinical trial.25,42 Additional sites in Germany and the UK later joined the trial.28
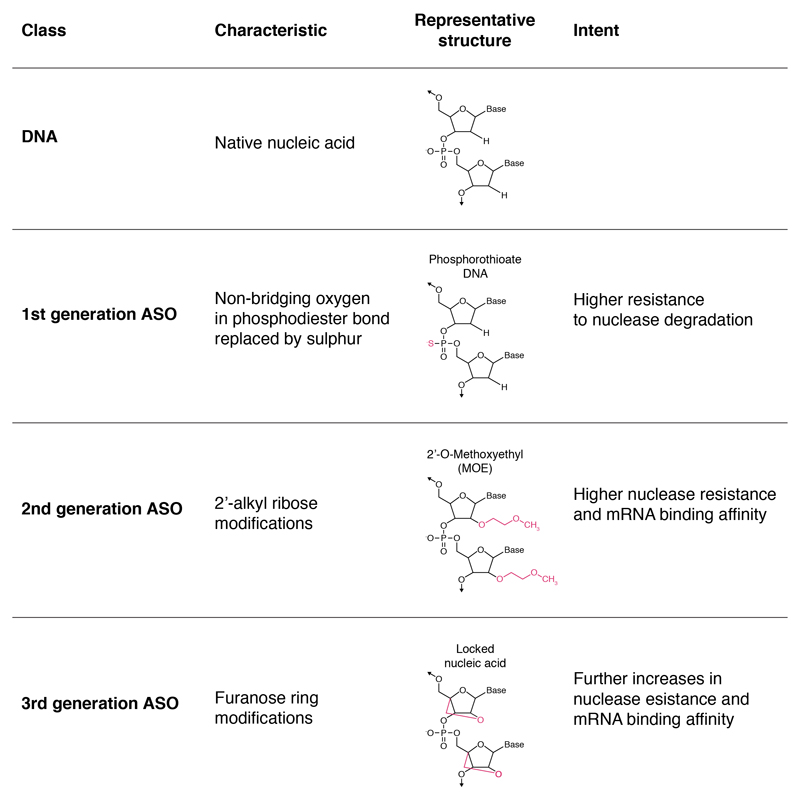
The investigational compound is a synthetic 20-nucleotides sequence in which many of the non-bridging oxygen atoms in the phosphate backbone have been substituted for sulfur to transform phosphodiester linkages to phosphorothioate. Additionally, the ASO has a DNA-like central region with 2’-O-methoxyethyl modifications at each end. This combination is intended to optimise CNS distribution, half-life, cellular uptake and RNAse activation (Figure 2).26
Figure 2.
First, second and third generation ASOs and native DNA. Each generation may contain numerous variations around the altered characteristic.43 HTTRx combines a phosphorothioate backbone (1st generation) with 2’-O-MOE modification (2nd generation) and is considered a 2nd generation ASO overall.26
After two weeks’ continuous intraventricular infusion into BACHD mice bearing a full-length human mutant HTT gene, huntingtin mRNA levels were reduced by up to 80% and protein levels by around two-thirds – reductions that persisted for 4 months. The ASO was widely distributed in brain across cell types including neurons and glia. Amelioration of disease phenotypes was seen in three different mouse models of HD, with near-complete restoration of motor deficits in young animals and partial improvement later in the disease in the slowly-progressive YAC128 and BACHD models; and increased survival and reduced brain atrophy in the rapidly-progressive R6/2 model.27
Curiously, the pathological and motor benefits of bolus ASO treatment outlasted the presence of the drug and reduction in huntingtin, suggesting that cells given a brief respite from mHTT can regain lost ground in the battle between damage and repair – a phenomenon dubbed the ‘huntingtin holiday’.44
Lumbar intrathecal infusion of a similar ASO, complementary to primate and human HTT mRNA, in wild-type rhesus monkeys, produced broad ASO distribution and huntingtin reduction in the cortex. This infusion was safe and well tolerated.27
The flow of CSF from lateral ventricles to spinal column to the brain’s external surface favours cortical more than striatal distribution, after intrathecal injection. However, ongoing primate pharmacokinetic/pharmacodynamics work has shown that lumbar intrathecal bolus ASO doses that produce ~50% cortical huntingtin reduction are associated with 15-20% reductions in the striatum, and were well-tolerated in these animals, providing support for human dose escalation.28 Wang and colleagues demonstrated that lowering mHTT expression in cortex or striatum each produce partial improvement in BACHD mice, but lowering in both regions produces a greater benefit than either alone.45 Lumbar intrathecal ASO injection in Yucatan pigs, whose spinal column is roughly the same length as a human’s, resulted in significant distribution to brain tissues.28 The striatum will probably receive lower doses than the cortex with intrathecal administration, but apparently attainable levels of lowering may be sufficient to produce clinical benefit without resorting to dual intrathecal/intracranial administration. Nonetheless this remains an option.
In the current IONIS-HTTRx trial, patients with early HD receive four lumbar intrathecal bolus doses of drug or placebo (randomized 3:1) at monthly intervals. Several dose escalations have taken place within the multiple ascending dose design.25 As well as safety and tolerability, the trial seeks to study the pharmacokinetics of IONIS-HTTRx in the human CNS, and will examine target engagement biomarkers46 including the CSF mHTT concentration, which can be reliably quantified using a novel immunoassay.47
Progress in other neurodegenerative diseases provides encouragement for intrathecally-delivered ASOs. A safety trial of an ASO in superoxide-dismutase 1 (SOD1) mutation-associated amyotrophic lateral sclerosis revealed no concerns48 and another is underway with a more potent ASO.49 The striking efficacy of nusinersen in SMA demonstrates disease-modification by an ASO in a previously unalterable human neurodegenerative condition. Notably, post-mortem brain tissue from the trial showed ASO in neuronal and non-neuronal cells in cortex and brainstem, and increased SMN2 expression in multiple brain areas – the first evidence that lumbar intrathecal bolus ASO injection can achieve brain penetration and target engagement in humans.6 This result is encouraging for IONIS-HTTRx – with the important caveats already mentioned.
Allele-selective ASOs
IONIS-HTTRx is expected to reduce expression of both mutant and wild-type HTT alleles equally. It may be many years before we know whether lowering wtHTT is safe in humans, especially for long-term use in symptom-free gene carriers. Allele-selective methods for reducing mHTT by targeting the CAG tract or heterozygous single-nucleotide polymorphisms (SNPs) are under investigation.
A long CAG tract is not unique to HTT: numerous human genes contain polyCAG stretches.50 Thus, an ASO or RNAi drug targeting the causative mutation is likely to downregulate other genes, which may produce ‘off-target’ adverse effects. Since several polyCAG genes encode transcription factors, these effects could be profound. Nonetheless, this approach appears at least feasible. An ASO targeting the mRNA equivalent of 7 CAG repeats produced 83% reduction in mHTT mRNA in HD patient-derived fibroblasts, with a lesser 43% reduction in the wild-type transcript. It also reduced the transcripts of other polyCAG genes including ATXN3, ATXN1 and ATN1. On the plus side, this suggests a polyCAG-targeting ASO could be of value in other CAG-expansion diseases; however, the risk from long-term reduction in all these genes, as well as apparently non-trivial reductions in wtHTT, limits its potential as a candidate therapeutic.51 Nonetheless this strategy was advanced by Datson and colleagues, who administered the ASO to R6/2 and Q175 knock-in mice, producing potent mHTT-lowering accompanied by clinical benefit.31
HTT has many common single-nucleotide polymorphisms (SNPs).52 Specialist sequencing techniques can establish which allele a SNPs is located upon – known as ‘phasing’.53 Thanks to haplotypes, certain SNPs frequently accompany CAG mutation; it is estimated that drugs targeting the commonest three SNPs would be sufficient to treat about 80% of the population – with the caveat that these studies were largely based on European-descended populations54 (HD is more prevalent in such populations than others but is present in all ethnicities and regions studied to date).55 Each SNP-targeting candidate must be developed and trialled separately – a considerable undertaking. The success of this approach depends on several incompletely understood factors. First, the true frequency of the SNPs in the population seeking treatment may differ from that in the prevalence studies. Second, the procedure for phasing the SNP to the mutant allele is critically important: accidental selective silencing of wtHTT, leaving mHTT untouched, may cause considerable harm. Third, “allele-specificity” is relative, not absolute: the real-world selectivity of a compound for mutant over wild-type may be less than expected, given the close overall sequence similarity between the two alleles. Fourth, the need to target SNP-bearing regions dramatically reduces the repertoire of candidate sequences, limiting the ability to select a potent drug candidate with minimal off-target effects. Nonetheless, patients bearing a suitable heterozygous SNP may benefit from the availability of such compounds, especially if liabilities emerge from nonselective lowering.
Plans to initiate parallel trials of two ASOs targeting SNP-containing regions of HTT mRNA have been announced.29 Potential participants will be screened for the presence of these SNPs within the mutant but not wild-type allele before allocation to one or other compound. The ASOs were selected on the basis not only of nucleotide sequence but also on the stereochemistry of the backbone, which may enable greater selectivity between alleles, as well as optimizing other aspects of pharmacokinetics.30
Future directions
A potentially important recent advance is the development of peptide-conjugated ASOs that, after intravenous injection, produced broad CNS distribution and dramatically extended survival in SMA mice.56 This technology warrants investigation for use in HD.
RNA interference approaches
No approach using RNA-based nucleotide agents has reached human trials, but many efforts are underway. This reflects the added delivery and distribution challenges. RNAi does not distribute well into brain tissue even after intraventricular administration, so stereotactic surgery is needed to deliver the agent directly to the brain parenchyma, probably using a viral vector to encourage spread and longevity.
Virally-delivered RNA therapeutics permanently transduce CNS cells, effectively turning them into factories making a drug that suppresses manufacture of mHTT. Thus, these drugs are gene therapy. Virally-delivered gene therapy has been attempted in neurodegeneration, for instance a phase 2 trial in Alzheimer’s disease patients of AAV2-encapsulated nerve neurotrophic factor RNA. The technique was safe and well-tolerated, albeit without apparent clinical benefit.57
siRNA in HD animal models
These approaches have been successful in multiple animal systems.38 The most advanced published work comes from Davidson and colleagues who, having established safety, potency of huntingtin reduction and phenotypic benefits from AAV-delivered siRNA in several HD rodent models,15,16 went on to deliver a microRNA targeting the human and primate HTT transcript, encapsulated in an AAV2 vector, by injection into the putamen of wild-type rhesus macaques. A fluorescent reporter indicated successful transfection of a variety of cell types in a small volume of tissue (approximately 100mm3 or 100 µL) around the injection site, accompanied by reduction of HTT transcripts by about half. No significant adverse effects were detected after comprehensive histological, biochemical and clinical assessment. The development of dystonia in one sham-injected animal highlights the risk with any neurosurgical delivery technique.17
Enhanced delivery methods
Stiles and colleagues used convection-enhanced delivery to infuse radiolabeled ‘naked’ siRNA (not packaged in a viral vector) under constant pressure into the primate putamen continuously for several days or weeks, producing siRNA distribution and HTT suppression over the entire striatum.18 Other non-viral approaches have been proposed to improve CNS delivery and distribution of RNAi compounds. Single-stranded RNA molecules (ssRNA) distribute well through brain parenchyma and into cells;58 and exosomes or other ‘Trojan horse’ methods may eventually permit intravenous dosing.59,60
Even with enhanced delivery methods, limited tissue distribution remains a significant challenge for RNA-based compounds, necessitating consideration of either multiple injection sites or prioritizing one region over others. Though the striatum is disproportionately affected in prodromal and early HD,61 whether that is because of intrinsic mHTT production or loss of trophic support from the cortex is debated.4 Either way, HD is undoubtedly a whole-brain condition. As discussed above, the cortex and striatum are each desirable targets,45 but whether mHTT-lowering in either is necessary or sufficient to produce clinical benefit in patients remains unknown. It would be preferable to act upon both – and indeed beyond, if possible. Any neurosurgical procedure carries risk, so early work will likely err on the side of caution, perhaps expanding to more ambitious approaches in time.
Novel viral vectors
Recent years have seen progress in viral vectors for CNS delivery. Many AAV serotypes exist, each of which may be combined with different cargos, enhancers and promoters. In 2015, Stanek and colleagues reported impressive success using an RNAi agent delivered by AAV1 under control of a hybrid cytomegalovirus enhancer / chicken beta actin promoter, which when injected into primate striatum produced widespread transduction of cells in the cortex, thalamus and hippocampus.32 Miniarikova and colleagues demonstrated efficacy of an optimized AAV5-encapsulated miRNA in rodents – now being readied for clinical use.33,34
A striking recent development was work by Deverman and colleagues, who used Cre recombination-based AAV targeted evolution (‘CREATE’) to develop AAV strains with novel capsids, selecting them for their ability to transduce neuronal tissue. One variant, AAV-PHP.B, demonstrated 40-fold improvement efficiency over AAV9, producing widespread distribution and transduction in the rodent brain (Figure 2) even after intravenous administration. However, there was also transfection of systemic organs (liver, heart and muscle), raising the possibility of peripherally-mediated side-effects; and whether the virus transfects a sufficient diversity of CNS cell-types to produce meaningful huntingtin reduction remains to be seen.62 Nonetheless, this novel AAV, and the methods used to develop it, are of interest to the CNS gene therapy field.
Such progress is a two-edged sword: researchers must decide whether to proceed to clinical trials now, using familiar but less widely-distributed vectors, or await trial-readiness of improved viruses and surgical techniques. Again, an incremental process will likely be adopted. The immunogenicity of AAVs in the human brain, especially with repeated administration, is also unclear. Although repeated dosing with any particular agent will not be needed, there is a chance that patients who volunteer for early trials may later find themselves unable to receive more advanced treatments because of the risk of immunogenicity from receiving multiple different AAV constructs.63
‘Small molecule’ approaches
A brain-penetrant, orally bioavailable small molecule acting on the protein manufacture pipeline to lower huntingtin expression is desirable. Naryshkin and colleagues used a screening process to identify small molecules that alter the splicing of SMN2 pre-mRNA, favouring the production of a functional SMN protein (a mechanism proven effective by nusinersen). In patient fibroblasts, these increased SMN concentration in a dose-dependent manner, and improved outcomes in an SMA mouse model – albeit through uncertain mechanisms.64 A phase 1b/2a clinical trial in SMA of the lead compound, RG7800, was terminated after ocular complications emerged in ongoing animal studies,65 perhaps highlighting an increased risk of off-target effects with protein-modulating compounds that lack the specificity conferred by nucleotide base-pairing. Nonetheless, a phase 1 study of a second compound, RG7916, was recently completed.66 Similar work is now underway to identify small molecule agents to lower mHTT levels.35
Targeting the DNA
Designing and implementing a therapeutic construct that interacts directly with DNA to reduce transcription of the mutant HTT gene brings fresh challenges but the potential for greater rewards. DNA-targeting therapeutics would be expected to ameliorate all aspects of HD, including those mediated by alternative splicing, non-RAN translation or any other mechanism we might postulate. Moreover, ‘editing’ the DNA raises the future prospect of germ-line treatment in living patient tissue, that could benefit future offspring as well as the mutation-carrier. This of course brings its own ethical issues.67
The two DNA-targeting gene therapies currently under investigation are zinc finger proteins (ZFPs) and CRISPR/Cas9 (Table 2). Each uses a protein-coding sequence encapsulated in a viral vector, injected intracranially, that transduces cells, causing them to produce a functional, non-native therapeutic protein.
Table 2.
Current publicly-announced huntingtin-lowering programmes targeting DNA.
| Class | Mechanism | Allele selectivity | Delivery | Vector | Sponsor | Key advantages | Key disadvantages | Ref |
|---|---|---|---|---|---|---|---|---|
| ZFTR | Transcriptional repression | CAG repeat | Intracranial | AAV | Shire |
|
|
68 |
| Imperial College London | As above, plus use of host-species proteins reduces inflammatory effects | Use of human proteins in clinical candidate compound may limit utility of animal work | 69 | |||||
| CRISPR/Cas9 | Genome editing | SNP-targeted | Intracranial | AAV | Harvard University |
|
|
70 |
| Nonselective HTT depletion by polyQ domain deletion | Emory University | 75 |
Zinc finger proteins
Zinc fingers are naturally-occurring structural motifs that can bind specific DNA sequences; they can be generated synthetically and used as DNA-targeting therapeutic compounds. ZFPs tooled for therapeutic use typically contain a zinc finger array specific to the DNA sequence of interest – one ‘finger’ per three bases – fused to a functional domain intended to act upon the DNA. Examples include zinc finger nucleases (ZFNs) to cleave DNA, and zinc finger transcription factors (ZFTRs) to modulate gene expression.71
Though ZFNs are theoretically capable of targeted ‘genome editing’, the process is not currently sufficiently precise or predictable for therapeutic application in postmitotic patient brain cells. Unfortunately, polyCAG stretch is particularly undesirable as a target for ZFN-mediated cleavage.69 However, zinc fingers are sufficiently reliable to contemplate clinical development of a therapeutic ZFTR. In order to bring the transcriptional repressor into proximity with the HTT promoter, the zinc finger array must bind a sequence near the 3’ end of the DNA sense strand. This limits the selection of target sequences and makes it difficult to avoid unwanted binding to other genes.
Fortuitously, while numerous genes contain polyCAG stretches,50 this tract in HTT is closer to the 3’ end than in other genes, so a CAG-targeting ZFTR will have selectivity both for HTT over other genes, and for the mutant over the wild-type allele. Such constructs have been developed by two groups. Both have shown early promise in animal models, effectively reducing mHTT protein expression in neurons without adversely affecting the expression of other genes or wtHTT.68,69 To our knowledge this is the only allele-selective huntingtin-lowering approach currently being readied for clinical use in which a single agent should selectively suppress mHTT long-term in all mutation carriers.
One limitation of zinc finger therapeutics is the production of non-native proteins that can trigger inflammatory and immune reactions, resulting in neuronal death and limited duration of effect. Agustín-Pavón and colleagues combined a polyCAG-targeting ZFTR with a non-viral promoter and a novel repressor element redesigned to be homologous to the host (mouse) protein. This construct produced more sustained silencing than others using non-native protein sequences, though the mHTT level gradually rebounded over six months.72 Such optimisations could make a difference across the lifespan of a human HD mutation-carrier, especially as the ultimate goal is to treat once, well before symptoms begin, to prevent the onset of the disease.
CRISPR/Cas9
Clustered regularly interspaced short palindromic repeats (CRISPR) and the accompanying CRISPR-associated system (Cas) together form the basis of a prokaryote immune system that recognises and destroys foreign DNA. Cas9 nuclease can be combined with synthetic guide RNA (gRNA) to produce a construct that can cut DNA with high precision at any chosen site. The use of such CRISPR/Cas9 complexes for targeted genome editing is a rapidly-evolving field with huge potential for the study and treatment of diseases including HD.73
The possibilities for HTT-directed CRISPR/Cas9 therapeutic strategies are legion, from returning alleles to harmless length by excising CAGs, to inactivating the mutant allele by inserting stop codons or missense mutations (perhaps, in tandem, up-regulating the wild-type allele to replace lost huntingtin function).74 CRISPR/Cas9 in HD is at an early stage of investigation. It was first used to inactivate the mutant HTT allele permanently and selectively in patient-derived fibroblasts, using two constructs to excise a large region of HTT DNA resulting in near-total reduction in both RNA and mHTT protein.70 Recently the method was successfully tested in an HD rodent model.75 This affirms the feasibility of this approach but much work is needed to bring these rapidly-evolving technologies to the clinic, especially given recent concerns about unexpected off-target mutations with CRISPR-Cas9 gene editing.76
Conclusions and future directions
Huntington’s disease is particularly suited to advanced experimental therapeutics that target DNA and RNA to modulate protein expression. The first human trial is underway of an antisense oligonucleotide that seeks to reduce the manufacture of mutant huntingtin protein by targeting HTT mRNA, and future ASO trials are planned, including agents that seek to lower mutant huntingtin selectively. RNA-based huntingtin-lowering agents require viral delivery, bringing the key advantage of potential lifelong treatment from a single dose, at the cost of increased invasiveness, the challenge of brain penetration and risks around long-term toxicity; nonetheless several programs are nearing clinical trials. Small molecule RNA-targeting compounds are appealing in terms of delivery, have been shown to be attainable in other conditions and are being investigated for HD. Zinc-finger transcriptional repressors may reduce huntingtin by targeting DNA without altering it, while CRISPR-Cas9 therapeutics bring the promise of permanently correcting the CAG expansion mutation that causes HD – but these approaches share the challenges of viral delivery and bring additional liabilities of their own, so are further from human trials.
This is a time of significant progress for molecular therapies targeting huntingtin expression. The coming years will certainly see more trials of novel agents, likely periodically boosted by technological improvements in CNS delivery and distribution. Early efficacy trials will focus on patients with early manifest disease, in whom both benefit and harm can be studied. Selecting suitable outcome measures and designing trials capable of identifying meaningful clinical benefit is an urgent focus of current work.
If efficacy can be demonstrated in manifest HD, the next hurdle will be the leap to ‘pre-manifest’ mutation carriers. This will need to be supported by a battery of clinical and imaging biomarkers that have been under concerted development for this purpose,61,77,78 plus relatively novel biochemical markers of target engagement and biological effect.46,47,79 Cost, availability and the infrastructural requirements for delivering invasive treatments are legitimate concerns, but first we must establish that disease modification is possible.
The HD community is distinguished by its trial-readiness, with global registries of well-characterised potential participants and strong networks of professionals and HD family members. While success is far from guaranteed, we are perhaps better placed than ever to make a meaningful impact to treat and ultimately prevent Huntington’s disease.
Search strategy
References for this Review were identified by searches of PubMed between 1969 and 6 July, 2017, and references from relevant articles. The search terms "Huntington’s disease”[MeSH term], “gene silencing”, “huntingtin lowering”, “antisense oligonucleotide”, “RNA interference”, “siRNA”, “miRNA”, “zinc finger”, “CRISPR”, “Cas9” and “AAV” were used. There were no language restrictions. The final reference list was generated on the basis of relevance to the topics covered in this Review.
Acknowledgements
EJW has research funding from the Medical Research Council (UK), CHDI Foundation Inc and European Huntington’s Disease Network. SJT has research funding from the Medical Research Council UK, the Wellcome Trust, the EU FP7 Health Call, European Huntington’s Disease Network, the Huntington’s Disease Association of the UK, the Rosetrees Trust, Takeda Pharmaceuticals, and CHDI Foundation. This work was supported in part by the National Institute for Health Research University College London Hospitals Biomedical Research Centre and the UCL Leonard Wolfson Experimental Neurology Centre.
Role of the funding source
No funding source had any role in the design, sourcing and writing of this review, nor in the decision to submit it for publication.
Footnotes
Contributors
EJW completed the literature search, created the figures and drafted the manuscript. SJT designed the Review, provided critical review and finalised the work with EJW. Both authors have approved the final version.
Declaration of interests
EJW has participated in scientific advisory boards with Hoffmann-La Roche Ltd, Ionis, Shire, Novartis and Wave Life Sciences and is an investigator on the HTTRx trial. SJT has participated in scientific advisory boards with Hoffmann-La Roche Ltd, Ionis, Shire, Teva Pharmaceuticals, GSK, Takeda Pharmaceuticals and Heptares Therapeutics, and is the global Principal Investigator on the HTTRx trial, for which she receives no personal salary or fees. All honoraria for these advisory boards were paid through UCL Consultants Ltd, a wholly owned subsidiary of UCL. Their host clinical institution, University College London Hospitals NHS Foundation Trust, receives funds as compensation for conducting clinical trials for Ionis Pharmaceuticals, Pfizer and Teva Pharmaceuticals.
References
- 1.Ross CA, Tabrizi SJ. Huntington's disease: from molecular pathogenesis to clinical treatment. The Lancet Neurology. 2011;10(1):83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 2.Wild EJ, Tabrizi SJ. In: Premanifest and Early Huntington’s Disease. Fourth ed Bates G, Jones L, Tabrizi SJ, editors. Huntington's Disease: Oxford: Oxford University Press; 2014. [Google Scholar]
- 3.Lee JM, Ramos EM, Lee JH, et al. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012;78(10):690–5. doi: 10.1212/WNL.0b013e318249f683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross CA, Aylward EH, Wild EJ, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10(4):204–16. [Google Scholar]
- 5.Saudou F, Humbert S. The Biology of Huntingtin. Neuron. 89(5):910–26. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Finkel RS, Chiriboga CA, Vajsar J, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. The Lancet. 2016;388(10063):3017–26. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 7.US Food and Drug Administration. FDA approves first drug for spinal muscular atrophy. [accessed 21 May 2017];2016 http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm534611.htm.
- 8.US Food and Drug Administration:Guidance for Industry. Guidance for Human Somatic Cell Therapy and Gene Therapy. Hum Gene Ther. 1998;9(10):1513–24. doi: 10.1089/hum.1998.9.10-1513. [DOI] [PubMed] [Google Scholar]
- 9.Tartari M, Gissi C, Lo Sardo V, et al. Phylogenetic Comparison of Huntingtin Homologues Reveals the Appearance of a Primitive polyQ in Sea Urchin. Mol Biol Evol. 2008;25(2):330–8. doi: 10.1093/molbev/msm258. [DOI] [PubMed] [Google Scholar]
- 10.Nasir J, Floresco SB, O'Kusky JR, et al. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81(5):811–23. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 11.Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nature Reviews Disease Primers. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 12.Mestre T, Ferreira J, Coelho MM, Rosa M, Sampaio C. Therapeutic interventions for disease progression in Huntington's disease. Cochrane database of systematic reviews (Online) 2009;(3):CD006455. doi: 10.1002/14651858.CD006455.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauthier LR, Charrin BC, Borrell-Pages M, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118(1):127–38. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26(3):300–6. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 15.Harper SQ, Staber PD, He X, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 2005;102(16):5820–5. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franich NR, Fitzsimons HL, Fong DM, Klugmann M, During MJ, Young D. AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington's disease. Mol Ther. 2008;16(5):947–56. doi: 10.1038/mt.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride JL, Pitzer MR, Boudreau RL, et al. Preclinical Safety of RNAi-Mediated HTT Suppression in the Rhesus Macaque as a Potential Therapy for Huntington's Disease. Mol Ther. 2011;19(12):2152–62. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stiles DK, Zhang Z, Ge P, et al. Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp Neurol. 2012;233(1):463–71. doi: 10.1016/j.expneurol.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 19.Grondin R, Kaytor MD, Ai Y, et al. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135(4):1197–209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangiarini L, Sathasivam K, Seller M, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 21.Sathasivam K, Neueder A, Gipson TA, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proceedings of the National Academy of Sciences. 2013;110(6):2366–70. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banez-Coronel M, Ayhan F, Tarabochia AD, et al. RAN Translation in Huntington Disease. Neuron. 2015;88(4):667–77. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rue L, Banez-Coronel M, Creus-Muncunill J, et al. Targeting CAG repeat RNAs reduces Huntington's disease phenotype independently of huntingtin levels. J Clin Invest. 2016;126(11):4319–30. doi: 10.1172/JCI83185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijayvargia R, Epand R, Leitner A, et al. Huntingtin's spherical solenoid structure enables polyglutamine tract-dependent modulation of its structure and function. eLife. 2016;5:e11184. doi: 10.7554/eLife.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ClinicalTrials.gov. Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of IONIS-HTTRx in Patients With Early Manifest Huntington's Disease. [accessed 21 May 2017];2015 https://clinicaltrials.gov/ct2/show/NCT02519036.
- 26.Bennett CF, Swayze EE. RNA Targeting Therapeutics: Molecular Mechanisms of Antisense Oligonucleotides as a Therapeutic Platform. Annu Rev Pharmacol Toxicol. 2010;50(1):259–93. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 27.Kordasiewicz Holly B, Stanek Lisa M, Wancewicz Edward V, et al. Sustained Therapeutic Reversal of Huntington's Disease by Transient Repression of Huntingtin Synthesis. Neuron. 2012;74(6):1031–44. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leavitt B, Tabrizi S, Kordasiewicz H, et al. Discovery and Early Clinical Development of ISIS-HTTRx, the First HTT-Lowering Drug to Be Tested in Patients with Huntington’s Disease. Neurology. 2016;86 (16 Supplement): (abstr) [Google Scholar]
- 29.Hersch S, Claassen D, Edmondson M, Wild E, Guerciolini R, Panzara M. Multicenter, Randomized, Double-blind, Placebo-controlled Phase 1b/2a Studies of WVE-120101 and WVE-120102 in Patients with Huntington’s Disease (P2.006) Neurology. 2017;88 (16 Supplement): (abstr) [Google Scholar]
- 30.Butler D, Iwamoto N, Meena M, Svrzikapa N, Verdine GL, Zlatev I. Chiral control. [accessed 21 May 2017];2015 https://www.google.com/patents/US20150211006. [Google Scholar]
- 31.Datson NA, Gonzalez-Barriga A, Kourkouta E, et al. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS One. 2017;12(2):e0171127. doi: 10.1371/journal.pone.0171127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanek L. Widespread gene delivery to the nonhuman primate brain for the treatment of Huntington’s disease. CHDI Foundation Annual Therapeutics Conference; Palm Springs, CA, USA: 2015. [Google Scholar]
- 33.Miniarikova J, Zanella I, Huseinovic A, et al. Design, Characterization, and Lead Selection of Therapeutic miRNAs Targeting Huntingtin for Development of Gene Therapy for Huntington's Disease. Mol Ther Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samaranch L, Blits B, San Sebastian W, et al. MR-guided parenchymal delivery of adeno-associated viral vector serotype 5 in non-human primate brain. Gene Ther. 2017;24(4):253–61. doi: 10.1038/gt.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doherty EM. Screening approaches to identify small-molecule modulators of huntingtin protein levels. CHDI Foundation Annual Therapeutics Conference; Malta. 2017. (abstr) [Google Scholar]
- 36.Larrouy B, Blonski C, Boiziau C, et al. RNase H-mediated inhibition of translation by antisense oligodeoxyribonucleotides: use of backbone modification to improve specificity. Gene. 1992;121(2):189–94. doi: 10.1016/0378-1119(92)90121-5. [DOI] [PubMed] [Google Scholar]
- 37.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 38.Keiser MS, Kordasiewicz HB, McBride JL. Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington's disease and spinocerebellar ataxia. Hum Mol Genet. 2016;25(R1):R53–R64. doi: 10.1093/hmg/ddv442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pouladi MA, Morton AJ, Hayden MR. Choosing an animal model for the study of Huntington's disease. Nat Rev Neurosci. 2013;14(10):708–21. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- 40.Agrawal S, Kandimalla ER. Role of Toll-like receptors in antisense and siRNA [corrected] Nat Biotechnol. 2004;22(12):1533–7. doi: 10.1038/nbt1042. [DOI] [PubMed] [Google Scholar]
- 41.Chi X, Gatti P, Papoian T. Safety of antisense oligonucleotide and siRNA-based therapeutics. Drug Discovery Today. 2017;22(5):823–33. doi: 10.1016/j.drudis.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 42.BBC News. Landmark Huntington's trial starts. [accessed 21 May 2017];2015 http://www.bbc.co.uk/news/health-34552041.
- 43.Chan JHP, Lim S, Wong WSF. Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol. 2006;33(5–6):533–40. doi: 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- 44.Lu X-H, Yang XW. “Huntingtin Holiday”: Progress toward an Antisense Therapy for Huntington's Disease. Neuron. 2012;74(6):964–6. doi: 10.1016/j.neuron.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang N, Gray M, Lu X-H, et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington's disease. Nat Med. 2014;20(5):536–41. doi: 10.1038/nm.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byrne LM, Wild EJ. Cerebrospinal Fluid Biomarkers for Huntington's Disease. Journal of Huntington's disease. 2016;5(1):1–13. doi: 10.3233/JHD-160196. [DOI] [PubMed] [Google Scholar]
- 47.Wild EJ, Boggio R, Langbehn D, et al. Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington’s disease patients. The Journal of Clinical Investigation. 2015;125(5):1979–86. doi: 10.1172/JCI80743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller TM, Pestronk A, David W, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12(5):435–42. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ClinicalTrials.gov. Single and Multiple Dose Study of BIIB067 (Isis-SOD1Rx) in Adults With Amyotrophic Lateral Sclerosis (ALS) [accessed 21 May 2017];2016 https://clinicaltrials.gov/ct2/show/NCT02623699.
- 50.Butland SL, Devon RS, Huang Y, et al. CAG-encoded polyglutamine length polymorphism in the human genome. BMC genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evers MM, Pepers BA, van Deutekom JC, et al. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PloS one. 2011;6(9):e24308. doi: 10.1371/journal.pone.0024308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JM, Gillis T, Mysore JS, et al. Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am J Hum Genet. 2012;90(3):434–44. doi: 10.1016/j.ajhg.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu W, Kennington LA, Rosas HD, et al. Linking SNPs to CAG repeat length in Huntington's disease patients. Nat Meth. 2008;5(11):951–3. doi: 10.1038/nmeth.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kay C, Collins JA, Skotte NH, et al. Huntingtin Haplotypes Provide Prioritized Target Panels for Allele-specific Silencing in Huntington Disease Patients of European Ancestry. Mol Ther. 2015;23(11):1759–71. doi: 10.1038/mt.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pringsheim T, Wiltshire K, Day L, Dykeman J, Steeves T, Jette N. The incidence and prevalence of Huntington's disease: a systematic review and meta-analysis. Mov Disord. 2012;27(9):1083–91. doi: 10.1002/mds.25075. [DOI] [PubMed] [Google Scholar]
- 56.Hammond SM, Hazell G, Shabanpoor F, et al. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proceedings of the National Academy of Sciences. 2016;113(39):10962–7. doi: 10.1073/pnas.1605731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rafii MS. A Phase II trial of AAV2-NGF in mild to moderate Alzheimer's disease. J Prev Alzheimers Dis. 2015;2015:274–5. [Google Scholar]
- 58.Yu D, Pendergraff H, Liu J, et al. Single-Stranded RNAs Use RNAi to Potently and Allele-Selectively Inhibit Mutant Huntingtin Expression. Cell. 2012;150(5):895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotech. 2011;29(4):341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 60.Niewoehner J, Bohrmann B, Collin L, et al. Increased Brain Penetration and Potency of a Therapeutic Antibody Using a Monovalent Molecular Shuttle. Neuron. 81(1):49–60. doi: 10.1016/j.neuron.2013.10.061. [DOI] [PubMed] [Google Scholar]
- 61.Tabrizi SJ, Scahill RI, Owen G, et al. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington's disease in the TRACK-HD study: analysis of 36-month observational data. The Lancet Neurology. 2013;12(7):637–49. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- 62.Deverman BE, Pravdo PL, Simpson BP, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotech. 2016;34(2):204–9. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mingozzi F, High KA. Immune responses to AAV in clinical trials. Curr Gene Ther. 2011;11(4):321–30. doi: 10.2174/156652311796150354. [DOI] [PubMed] [Google Scholar]
- 64.Naryshkin NA, Weetall M, Dakka A, et al. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345(6197):688–93. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 65.ClinicalTrials.gov. A Study of RO6885247 in Adult and Pediatric Patients With Spinal Muscular Atrophy (MOONFISH) [accessed 21 May 2017];2016 https://clinicaltrials.gov/ct2/show/NCT02240355.
- 66.ClinicalTrials.gov. A Study to Investigate the Safety, Tolerability, Pharmacokinetics and Pharmacodynamics of RO7034067 (RG7916) Given by Mouth in Healthy Volunteers. [accessed 21 May 2017];2016 https://clinicaltrials.gov/ct2/show/NCT02633709.
- 67.Vassena R, Heindryckx B, Peco R, et al. Genome engineering through CRISPR/Cas9 technology in the human germline and pluripotent stem cells. Hum Reprod Update. 2016;22(4):411–9. doi: 10.1093/humupd/dmw005. [DOI] [PubMed] [Google Scholar]
- 68.Zeitler B, Froelich S, Yu Q, et al. Allele-Specific Repression of Mutant Huntingtin Expression By Engineered Zinc Finger Transcriptional Repressors as a Potential Therapy for Huntington's Disease. Molecular Therapy. 2014;22(Supplement 1):S233. (abstr) [Google Scholar]
- 69.Garriga-Canut M, Agustín-Pavón C, Herrmann F, et al. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proceedings of the National Academy of Sciences. 2012;109(45):E3136–E45. doi: 10.1073/pnas.1206506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shin JW, Kim K-H, Chao MJ, et al. Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–31. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 72.Agustín-Pavón C, Mielcarek M, Garriga-Canut M, Isalan M. Deimmunization for gene therapy: host matching of synthetic zinc finger constructs enables long-term mutant Huntingtin repression in mice. Molecular Neurodegeneration. 2016;11(1):64. doi: 10.1186/s13024-016-0128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Translational Research. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 74.Cox DBT, Platt RJ, Zhang F. Therapeutic Genome Editing: Prospects and Challenges. Nat Med. 2015;21(2):121–31. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang S, Chang R, Yang H, et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J Clin Invest. 2017;127:2719–24. doi: 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schaefer KA, Wu W-H, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. Unexpected mutations after CRISPR–Cas9 editing in vivo. Nat Methods. 2017;14:547–48. doi: 10.1038/nmeth.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tabrizi SJ, Scahill RI, Durr A, et al. Biological and clinical changes in premanifest and early stage Huntington's disease in the TRACK-HD study: the 12-month longitudinal analysis. The Lancet Neurology. 2011;10(1):31–42. doi: 10.1016/S1474-4422(10)70276-3. [DOI] [PubMed] [Google Scholar]
- 78.Tabrizi SJ, Reilmann R, Roos RA, et al. Potential endpoints for clinical trials in premanifest and early Huntington's disease in the TRACK-HD study: analysis of 24 month observational data. Lancet Neurol. 2012;11(1):42–53. doi: 10.1016/S1474-4422(11)70263-0. [DOI] [PubMed] [Google Scholar]
- 79.Byrne LM, Rodrigues FB, Blennow K, et al. Neurofilament light protein in blood as a potential biomarker of neurodegeneration in Huntington’s disease: a retrospective cohort analysis. Lancet Neurology. 2017 doi: 10.1016/S1474-4422(17)30124-2. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]