Abstract
A major barrier to developing effective therapies for neurodegenerative diseases is our incomplete understanding of the underlying cellular mechanisms. Genetic screens in human induced pluripotent stem cell-derived neurons can elucidate such mechanisms. Genome-wide screens using CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) provide complementary biological insights and may reveal potential therapeutic targets.
Neurodegenerative diseases are devastating for patients and their families, and also a growing economic burden. Currently, there is no treatment that slows the progression of neurodegenerative diseases, let alone a cure. This lack of therapeutic options is due in large part, to our incomplete understanding of the molecular and cellular mechanisms driving neurodegeneration.
Intriguingly, specific neurons are selectively vulnerable to neurodegeneration, while other types of neurons are resilient. The pattern of selective vulnerability differs between neurodegenerative diseases. Knowledge of cellular factors that determine selective vulnerability would not only illuminate our understanding of cellular mechanisms of neurodegeneration, but may also point to potential therapeutic targets. Pharmacological targeting of the factors controlling selective vulnerability could turn susceptible neurons into resilient neurons, thus slowing, or perhaps blocking the progression of neurodegenerative diseases.
Efforts to understand determinants of selective vulnerability have mostly relied on the characterization of vulnerable and resilient cells using transcriptomics [1]. While transcriptomics and proteomics can catalog differences in gene expression between vulnerable and resilient neurons, the list of differentially expressed genes is long and only correlative. Moreover, it fails to establish causality. Therefore, a functional approach is needed that enables precise control of the neuronal expression of genes so as to establish their causal roles in selective vulnerability and to be able to consider them as putative therapeutic targets.
To address this need, in collaboration with Dr. Michael Ward, we are developing a platform for CRISPR-based genetic screens in human neurons to identify cellular factors controlling vulnerability and cellular processes underlying neurodegenerative diseases. The aim is to integrate the results from functional genetic screens in human neurons derived from induced pluripotent stem cells (iPSCs) using transcriptomics and proteomics analyses of vulnerable and resilient neurons (iPSC-derived or post-mortem brains of neurodegenerative disease patients). As such, we propose to identify relevant determinants of selective vulnerability that may lead to possible therapeutic candidates (Fig. 1A).
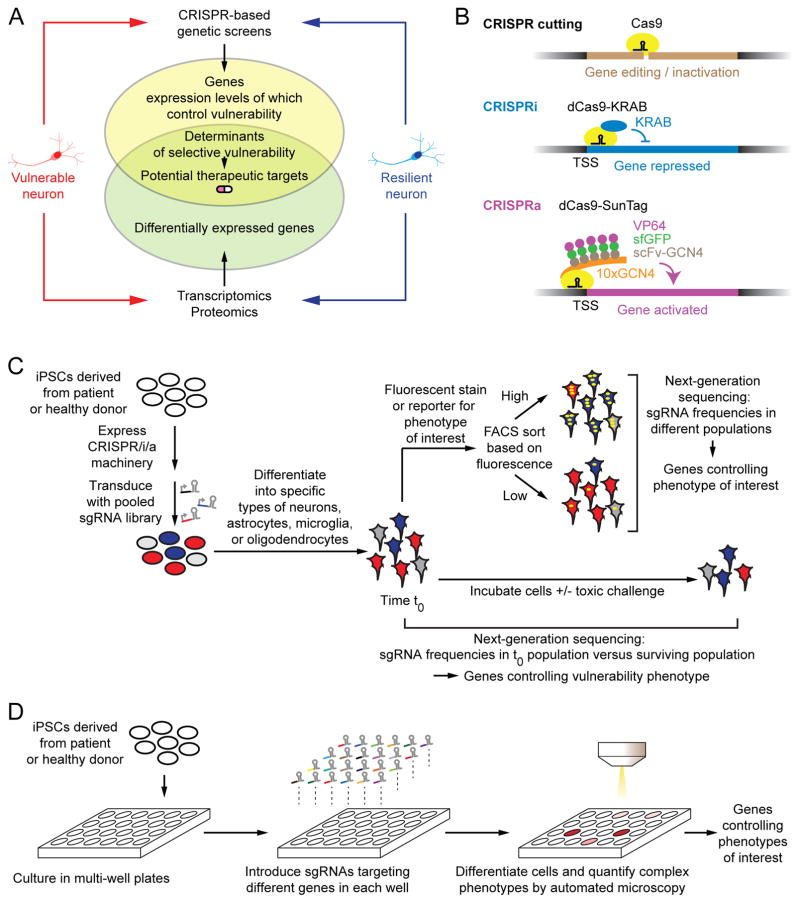
Figure 1. Strategy for CRISPR-Based Genetic Screens in Human iPSC-Derived Neurons.
(A) Transcriptomics/proteomics of vulnerable and resilient neurons detects differentially expressed genes but does not reveal those functionally relevant for selective vulnerability. CRISPR-based screens can elucidate determinants of vulnerability, and hence, potential therapeutic targets.
(B) Uses of CRISPR/Cas9 technology: Cas9 nuclease cleaves DNA for gene editing or inactivation, as directed by a single guide RNA (sgRNA). Catalytically dead Cas9 (dCas9) can be used to recruit transcriptional repressors or activators to endogenous genes to enable inducible and reversible gene repression (CRISPRi) or activation (CRISPRa). Our screening platform [6] uses the KRAB repressor domain, and the SunTag, in which several copies of the activator VP64 are recruited to a dCas9-fused tandem repeat of the GCN4 epitope via a superfolder GFP (sfGFP)-stabilized nanobody targeting the GCN4 epitope (scFv-GCN4).
(C) Pooled genetic screens are carried out by stably transducing a population of induced pluripotent stem cells (iPSCs) expressing the CRISPRi or CRISPRa machinery shown in (B) with a lentiviral sgRNA library targeting a large number of genes. After differentiation into neurons or other relevant cell types, cells are either sorted according to phenotypes of interest by fluorescence-activated cell sorting (FACS), or samples are taken before and after culture, and in the presence or absence of a toxic challenge. Frequencies of cells expressing each sgRNAs are compared in different populations by next-generation sequencing, and genes controlling the phenotype of interest are detected.
(D) Arrayed screens are carried out in multi-well plates, where sgRNAs targeting different genes are introduced into iPSCs separately in each well. Cells are differentiated into neurons or other relevant cell types, and complex phenotypes can be evaluated by automated microscopy to identify candidate genes controlling these phenotypes.
Until recently, the method of choice for genetic screens in human cells consisted of RNA interference (RNAi). However, results from genome-wide RNAi-based screens were notoriously noisy [2], due in large part to the off-target effects of each RNAi reagent (short interfering RNA, siRNA, or short hairpin RNA, shRNA). We have been able to demonstrate that ultra-complex shRNA libraries, in combination with a rigorous statistical framework can overcome this limitation and produce robust screening results [3, 4]. However, the large size of such ultra-complex libraries is prohibitive for genome-wide screens in cell types such as neurons, culturing of which is less scalable.
Our ability to perturb gene function in a wide range of biological systems has been revolutionized by the use of CRISPR/Cas9 genome editing, which relies on a bacterial endonuclease that can be programmed by a single guide RNA (sgRNA) to cleave specific DNA sequences [5] (Fig. 1B). We recently co-developed a genetic screening platform that exploits catalytically dead Cas9 (dCas9) to recruit transcriptional repressors or activators to endogenous genes, to enable inducible and reversible repression (CRISPRi) or activation (CRISPRa) of genes in human cells (Fig. 1B). This approach has enabled genome-wide loss- and gain-of-function screens [6]. Importantly, CRISPRi overcomes the problem of off-target effects that has plagued RNAi-based approaches, resulting in high specificity of hit detection in genome-wide screens, while maintaining high sensitivity [6]. Its performance in genome-wide screens is comparable to CRISPR cutting-based platforms [7]. CRISPRi makes it possible to investigate the biological function of essential genes by enabling different levels of gene knockdown, essentially creating an allelic series of different gene expression levels [6]. Together, CRISPRi and CRISPRa screens yield rich, complementary insights [6].
Here, we present our strategy for CRISPRi and CRISPRa genetic screening in human iPSC-derived neurons. CRISPRi in iPSCs has previously been demonstrated [8], and our own unpublished results have recently established its feasibility of pooled CRISPRi-based screens in iPSC-derived neurons. Such neurons are powerful tools to study cellular mechanisms of neurodegenerative diseases. iPSCs can be derived from skin biopsies of healthy donors as well as sporadic or familial neurodegenerative disease patients. Upon differentiation into neurons, iPSCs derived from patients can display pathological hallmarks of the neurodegenerative disease, such as increased levels of phosphorylated tau protein [9], and increased vulnerability to toxic insults such as oxidative stress [10].
To elucidate the molecular mechanisms underlying these disease-associated cellular phenotypes, CRISPRi and CRISPRa genetic screens can be conducted in either a pooled format (Fig. 1C) or in an arrayed format (Fig. 1D). For pooled screens, iPSCs are stably transduced with a lentiviral sgRNA expression library targeting the entire genome or a selected set of genes. The multiplicity of infection is chosen such that most cells express at most, one sgRNA. Cells are then differentiated into neurons or other cell types of interest, such as astrocytes, microglia or oligodendrocytes. Gene repression or activation is induced at a desired time point using inducible CRISPRi or CRISPRa machinery. Pooled screens are suitable for the investigation of two important classes of readouts: i) Phenotypes that can be monitored by a fluorescent signal (e.g. immunostaining of a protein of interest using a labeled antibody, or a genetically-encoded fluorescent reporter) (Fig. 1C, top); cell populations can be separated according to these phenotypes using fluorescence-activated cell sorting (FACS), and genes controlling the phenotype can be identified by comparing the frequencies of cells expressing each sgRNA in these populations as quantified by next-generation sequencing [11]. ii) Cells can be incubated in the absence or presence of a toxic challenge, such as toxic protein oligomers or high concentrations of glutamate (Fig. 1C, bottom); comparison of sgRNA frequencies in cells before and after the incubation period can reveal genes controlling cellular survival and vulnerability to toxicity [3, 6].
Arrayed screens (Fig. 1D) are more challenging to implement, since they require arrayed libraries of sgRNA reagents, and a high-throughput strategy for analyzing cellular phenotypes. However, arrayed screens enable the investigation of complex phenotypes that cannot be readily studied in pooled screens, which rely on physical coupling of the phenotype to the genotype. Such complex phenotypes – relevant in neurodegenerative diseases – include cellular morphology (e.g. dendrite structure), time-resolved phenotypes (e.g. electrophysiological properties), and non-cell autonomous phenotypes (e.g. processes in glial cells that affect neuronal function).
We expect that CRISPR-based genetic screens in iPSC-derived human neurons may reveal key factors controlling neurodegeneration and selective vulnerability of specific neurons, and thereby inform on possible therapeutic targets. Moreover, this methodology might also pave the way for future applications involving other iPSC-derived cell types and associated processes in other disease types.
Acknowledgments
We thank Michael Ward, Lea Grinberg and members of the Kampmann lab for discussions. Our research on neurodegeneration is supported by an NIH Director’s New Innovator Award (NIH/NIGMS DP2 GM119139), an Allen Distinguished Investigator Award (Paul G. Allen Family Foundation), the Tau Center Without Walls (NIH/NINDS U54 NS100717), the Chan-Zuckerberg Biohub and the Paul F. Glenn Center for Aging Research. We apologize to colleagues whose work we were not able to cite due to restrictions on the number of references.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang X, et al. Functional genomics of brain aging and Alzheimer’s disease: focus on selective neuronal vulnerability. Curr Genomics. 2010;11:618–633. doi: 10.2174/138920210793360943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science. 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kampmann M, et al. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2317–2326. doi: 10.1073/pnas.1307002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kampmann M, et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3384–E3391. doi: 10.1073/pnas.1508821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horlbeck MA, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 2016:5. doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandegar MA, et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Israel MA, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen HN, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sidrauski C, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife. 2015;4:e07314. doi: 10.7554/eLife.07314. [DOI] [PMC free article] [PubMed] [Google Scholar]