Abstract
Surgical site infection is a common cause of post-operative morbidity, often leading to implant loosening, ultimately requiring revision surgery, increased costs and worse surgical outcomes. Since implant failure starts at the implant surface, creating and controlling the bio-material interface will play a critical role in reducing infection while improving host cell-to-implant interaction. Here, we engineered a biomimetic interface based upon a chimeric peptide that incorporates a titanium binding peptide (TiBP) with an antimicrobial peptide (AMP) into a single molecule to direct binding to the implant surface and deliver an antimicrobial activity against S. mutans and S. epidermidis, two bacteria which are linked with clinical implant infections. To optimize antimicrobial activity, we investigated the design of the spacer domain separating the two functional domains of the chimeric peptide. Lengthening and changing the amino acid composition of the spacer resulted in an improvement of minimum inhibitory concentration by a three-fold against S. mutans. Surfaces coated with the chimeric peptide reduced dramatically the number of bacteria, with up to a nine-fold reduction for S. mutans and a 48-fold reduction for S. epidermidis. Ab initio predictions of antimicrobial activity based on structural features were confirmed. Host cell attachment and viability at the biomimetic interface were also improved compared to the untreated implant surface. Biomimetic interfaces formed with this chimeric peptide offer interminable potential by coupling antimicrobial and improved host cell responses to implantable titanium materials, and this peptide based approach can be extended to various biomaterials surfaces.
Keywords: Infections, bio-nanomaterial, interface, antimicrobial peptides, implants, biocoating, structural analysis, peptide design
1. Introduction
Bone and joint implants have revolutionized the healthcare of aging patients whose life expectancy has been increasing.1 Implants have been intensively used during the last 40 years in treating bone and joint degeneration, neoplasms, trauma and inflammation.1 Titanium and titanium alloys are used as implant biomaterials due to their biocompatibility, mechanical strength, and noncorrosive properties.2–5 However, nosocomial microbial attachment to the implant surface can result in infection and inflammation with implant loosening that requires surgical revision. In the first hours following surgery the implant surface is most vulnerable to bacterial colonization and the bacterial pathogens are also most susceptible to antimicrobial treatment.6,7 With time, bacteria populations multiply and co-operate to form biofilms that function as natural barriers against antibiotic effectiveness.8 Treatment for infection of this type is difficult and the revision surgery is more complex, adding to patient morbidity and overall health care costs. Despite improvements in implant technology including prophylactic therapy, most implant failures can be attributed to either infection or aseptic loosening resulting from poor integration with host tissue.9,10 Failure requiring revision surgery is caused by infection in 7.5% of total hip arthroplasty (THA) and 14.8% of total knee arthroplasty (TKA) and by aseptic loosening in 55.2% of THA and 29.8% of TKA.11 Immediate prevention of bacterial attachment on the implant surface is critical in prevention of infection related failure. However, host cell attachment and viability at the interface is also critical to host bone integration to prevent implant loosening. Therefore, an imperative clinical need exists to prevent bacterial colonization on the implant surface while not negatively affecting host cell response that could lead to poor integration of the implant material with the host. An implant surface with a fast-acting, broad-spectrum antimicrobial function prevents bacterial attachment to reduce biofilm formation while maintaining implant integration with the host tissue would prove to be a paradigm shift.
Multiple strategies have been developed with the aim of eliminating microbial attachment on the implant surface. Among them, the use of antibiotics have been commonly employed in daily practice as of today. For example, vancomycin powder is commonly used in posterior spinal wounds and has been shown to decrease surgical site infection. However, the rise of antibiotic resistance is lately becoming a major concern in dealing with bacteria, which also led to an increase in efforts to find alternative strategies.2–5 Silver, polyethylene glycol (PEG), or quaternary ammonia-based compounds (QACs) have been among the well-studied examples to bring the antimicrobial property by attaching them to the biomaterials using covalent chemical bonds.12–17 Another strategy is to improve the antibacterial properties of metals by doping them with elements such as bismuth and zinc.18,19 While promising, chemistry based immobilizations require complex steps, which may be not favorable within biological environment due to their harshness. Additionally, uniform coatings where bioactivity is both preserved and homogenously distributed throughout the biomaterial surface following their coupling onto the biomaterials are challenging to obtain.
Another infection prevention strategy is to coat the implant surface with antimicrobial peptides (AMPs). AMPs are abundant in nature and employed as natural innate immune system defense fighters. AMPs are fast-acting antimicrobial agents that are effective against a broad spectrum of gram-positive bacteria, gram-negative bacteria, viruses and fungi.20–22 AMPs offer an alternative to conventional antibiotics to which some pathogens develop resistance more readily.23 However, with the current technologies available, AMPs can be covalently immobilized onto the implant surfaces, but covalent immobilization of biomolecules has also proven to be less effective due to lack of control over the conformation of the biomolecules, which is critical to preserve their biofunctionality. In our previous studies, we demonstrated that AMPs can be immobilized on titanium implant surfaces through the engineering of chimeric peptides that use molecular recognition to attach and self-assemble on the implant surface as a novel biomolecular-coating.24–26 A chimeric peptide is a bifunctional single-chain relatively short peptide when compared to biological proteins, and it joins two functional domains through an engineered spacer. The functional domain joined to the AMP for immobilization on implant surface is a peptide that is identified using combinatorial biology based molecular libraries, i.e., phage and cell surface display libraries. These genome based screening process of the peptides allows to discovery of the potential candidates that can interact with the solid materials building upon molecular recognition, a feature found similar to Nature. Due to phenotype-genotype-based relations obtained for inorganic materials throughout the combinatorial biology-based selection process, these peptides are generally referred as genetically engineered peptide for inorganics (GEPIs). GEPIs offer the ability to use molecular recognition to self-assemble active peptide-based agents selectively on inorganic materials including titanium implants.27,28 Previous work has identified several titanium binding peptides (TiBP) that assemble onto the titanium surface with high affinity appropriate for the surface of titanium and titanium alloy-based implants.24,25 Peptide-based self-immobilization strategies therefore offer an opportunity to overcome the limitations and challenges associated with covalent immobilization of antibacterial agents on implant surfaces.
The current paper builds upon our studies suggesting that the function of an engineered chimeric peptide can be further improved through a spacer region that is placed in between the individual functional domains, i.e., TiBP and AMP. The novel design employed here allows the retention of AMP secondary structural features responsible for the antimicrobial activity without jeopardizing the implant self-assembling domain of the peptide. The changes offered in the spacer design induce enough structural alterations in the chimeric peptide to be more effectively displayed at the bio-materials interfaces. Herein, we demonstrate that engineering the length and composition of the spacer lead to improved antimicrobial function and favorable host cell response. Chimeric peptides offer a simple unifying strategy to immobilize AMPs as a uniform biocoating on titanium implant surfaces to combat implant failure due to infection.
2. Experimental Methods
2.1. Chimeric peptide design
TiBP and AMP domains previously demonstrated as viable in a chimeric peptide (TiBP-Spacer3-AMP) were selected for this work.24 Briefly, TiBP was selected by screening a bacterial surface display system, FliTrx (Invitrogen, Carlsbad, CA) against a titanium surface.27–29 After four rounds of bio-panning, 60 clones were selected and characterized based on their surface binding affinity using fluorescence microscopy techniques. The strongest binding sequence determined through these experiments was used in our chimeric peptide to bind to the titanium surface, anchoring the chimeric peptide. The AMP domain used in our chimeric peptide was computationally designed by data mining the available library of peptides.25,30,31 A novel spacer, Spacer5 was designed as an elongated link, joining TiBP with AMP to form the chimeric peptide, TiBP-Spacer5-AMP. TiBP-Spacer5-AMP was synthesized using solid phase peptide synthesis by KanPro (Lawrence, KS). Physical chemical data including, molecular weight, isoelectric point, charge and GRand AVerage of hydropathY (GRAVY) scores based on amino acid sequences for AMP, TiBP, TiBP-Spacer3-AMP, and TiBP-Spacer5-AMP were obtained using the ExPasy Proteonomics Server.32
2.2. Molecular structure modeling
To understand how the secondary structure of the chimeric peptides change in solution depending on the spacer sequence, we generated ensembles of 1,000 likely structures using the PyRosetta project software and identified secondary structures with the DSSP program.33,34 Structure generation is stochastic using a knowledge-based energy scoring function. An ensemble of structures was generated for each full chimeric peptide and each peptide domain to sample likely structural variations. Ramachandran plots were generated for the lowest energy structures for TiBP-Spacer3-AMP and TiBP-Spacer5-AMP structures. Chimera Software version 1.9 from University of California at San Francisco was used to visualize the structures.35
2.3. Antimicrobial “rule induction” method
A “rule induction” method was used to correlate the generated secondary structures with the probability of antimicrobial function. Rule induction is a data mining approach to learn associations between paired sets of data made of sets of cases. As previously published, our paired data is the computationally generated structure decoys for both chimeric AMPs and AMPs paired with the minimum inhibitory concentration (MIC) of the peptides in solution.1,24 Each structural decoy represents a single case in a set of cases. Given a list of cases where each case has a list of features and a selected outcome, rough-set theory approaches rule induction by looking for features which apply to the maximum number of cases and are selective for the selected outcome.36 For our project, the cases are structure decoys and the list of features are the secondary structure features found. The paired distinct outcome is the MIC result from the in-solution assay. The rough set theory implementation is based on MLEM2.37 Two secondary structure features, 4-amino-acid right-handed alpha helices and 5-amino-acid alpha helices were key features for rules inducted from our previous work.24 These rules associated with strong antimicrobial activity for the bacteria tested (S. epidermidis and S. mutans). The secondary structure feature frequencies of these two rules were compared against TiBP-Spacer3-AMP and TiBP-Spacer5-AMP. Higher frequencies of these secondary structure features associate with stronger antimicrobial activity.
2.4. Circular dichroism (CD) analysis
A solution containing 50 μM TiBP-Spacer5-AMP in phosphate buffered saline (PBS) at pH 7.4 was prepared for circular dichroism (CD) analysis. The spectrum is the average of four scans from 190–239 nm using a Jasco J-810 spectrometer (Easton, MD). Appropriate background buffer subtraction was performed and the instrument carefully calibrated. The averaged spectrum was subtracted from background and smoothed with the Savitzky–Golay algorithm. The spectrum was transformed for mean residue ellipticity in degrees · cm2/dmol. Two methods were used to estimate the secondary structure features from the CD spectra. The CAPITO method makes a comparison to reference spectra for helix (α-helix, 310-helix and π-helix), β-strands (β-sheets, β-bridge) and irregular secondary structures (bonded turns, bends and loops) using a liner regression method.38 The Raussens method is a concentration-independent estimation of α-helix, β-sheets and irregular secondary structure proportions.39
2.5. Bacterial maintenance and culturing
The antimicrobial activity of TiBP-Spacer5-AMP was evaluated against two bacterial strains, S. mutans (American Type Culture Collection (ATCC) 25175, Manassas, VA) and S. epidermidis (ATCC 29886). S. mutans cultures were prepared using Brain Heart Infusion Broth (BHI, BD Difco, Franklin Lakes, NJ) and S. epidermidis using Nutrient Broth (NB, BD Difco) according to ATCC protocols. Bacterial pellets obtained from ATCC were rehydrated in appropriate media of which several drops were used to streak either BHI or NB agar plates. Bacteria streaked agar plates were subsequently incubated for 24 h. Agar plates and cultures were incubated at 37°C in the presence of 5% CO2-supplemented atmosphere for S. mutans and in aerobic atmosphere and 200 rpm shaking for S. epidermidis. Overnight cultures were made by aseptically transferring a single-colony forming unit (CFU) into 10 mL of appropriate broth media followed by incubation in appropriate conditions for 16 h. Bacteria from overnight cultures were used to inoculate fresh media and grown to mid-log phase.
2.6. Antimicrobial activity in solution
The MIC of TiBP-Spacer5-AMP against S. mutans and S. epidermidis in solution was evaluated in 96 well plates (Corning Costar 3370, Corning, NY) spectrophotometrically over a period of 24 h by obtaining a measurement for the optical density at 600 nm (OD600) every two hours. Optical density at 600 nm was measured using a Cytation3 microplate reader (Bio Tek Instruments, Winooski, VT). Bacteria grown to mid-log phase at a density of 107 CFU/mL were cultured at appropriate growth conditions in appropriate broth media only as a control or in broth media containing a range from 5–70 μM of TiBP-Spacer5-AMP for S. mutans and 1-10 μM for S. epidermidis. The OD600 measurements obtained, relating optical density to bacteria CFUs/mL, were plotted versus time to generate standard growth curves. The minimum concentration of TiBP-Spacer5-AMP at which no increase in optical density measurement, corresponding to no bacterial growth occurring was designated as the MIC. AlamarBlue assay (Invitrogen, Carlsbad, CA) was used for determination of a minimum bactericidal concentration of TiBP-Spacer5-AMP. Bacteria in broth media only and with the TiBP-Spacer5-AMP concentrations described in the MIC experiments were prepared in 96 well plates. AlamarBlue reagent was added to experimental wells and incubated for two hours at 37°C. Experimental wells were observed and evaluated for color change. Wells corresponding to concentrations of TiBP-Spacer5-AMP where no color change occurred were determined to have bactericidal concentrations of the chimeric peptide.
2.7. Titanium surface preparation
Two surfaces, 99% pure titanium foil (Alfa Aesar 43677, Ward Hill, MA) and titanium implant discs cut from standard rods used in posterior lumbar surgery (University of Kansas Medical Center Department of Neurosurgery, Kansas City, KS) were used for evaluation of TiBP-Spacer5-AMP bio-coating antimicrobial activity. Titanium foils were cut into squares measuring 0.5 mm thick × 1 cm × 1 cm and 6 mm diameter implant rods were cut by the University of Kansas Medical Center Department of Neurosurgery with a standard orthopedic surgical rod cutter into 3 mm long disc segments. Surfaces were sterilized by soaking overnight in 70% bleach, followed by sonication for 15 min in each 1:1 acetone:methanol, isopropanol and filtered deionized water, dried under UV light in a biosafety cabinet and then autoclaved.
2.8. Chimeric peptide coating on surfaces
Sterilized titanium surfaces were transferred to sterile 24 well plates (Costar 3738) with the bactericidal concentrations (60 μM for S. mutans and 10 μM for S. epidermidis) of TiBP-Spacer5-AMP dissolved in PBS at pH 7.4 and incubated at 37°C, constant agitation (200 rpm) for 4 h.24 Following incubation substrates were washed twice by pipetting with PBS to remove unbound peptide and transferred to sterile 24 well plates to be used in experiments.
2.9. Antimicrobial activity on substrates
Antimicrobial activity of TiBP-Spacer5-AMP bio-coated titanium surfaces against each bacterial strain was evaluated by culturing bacteria in 24 well plates containing bio-coated surfaces or bare, untreated control surfaces. Bacteria grown to mid-log phase at a concentration of 107 CFU/mL were harvested by centrifugation at 2000 × g for 5 min followed by resuspension in 500 μL of appropriate media, transferred to sterile 2 mL centrifuge tubes, and then centrifuged at 2000 × g for three minutes.24 The supernatant was carefully removed from the pellet and the pellet resuspended in PBS at final concentration of 108 CFU/mL and 500 μL of suspension was added to wells containing foil surfaces and 1000 μL to wells with implants. Well plates with TiBP-Spacer5-AMP bio-coated surfaces were incubated for two hours at 37°C in the presence of 5% CO2-supplemented atmosphere for S. mutans and in aerobic atmosphere and 200 rpm shaking for S. epidermidis. Following incubation all surfaces were washed with PBS to remove unbound bacteria. Bacteria were fixed with 1 mL of 2% glutaraldehyde solution for 30 min and then dehydrated in 50%, 70%, 90% and 100% ethanol baths, 10 min for each ethanol concentration. Bacteria were stained with SYTO 9 green fluorescent dye (Life Technologies L7012, Carlsbad, CA), incubated for 15 min at room temperature protected from light and excess dye was removed by washing twice with PBS. Stained bacteria were imaged with a fluorescence microscope (Olympus Spin Disk Epifluorescent microscope, Richmond Hill, Ontario, Canada) at an excitation/emission wave number provided by the manufacturer. Five representative fluorescence images were taken for each sample (n = 3) and the bacteria were quantified using ImageJ Software and then subjected to statistical analysis.
2.10. Host cell response
Host cell response was evaluated with a fibroblast cell line (NIH/3T3 ATCC CRL-1658). The fibroblast cells were cultured following the ATCC protocol. Briefly, cells were grown in DMEM media (Gibco 11995073, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco 10437036) and 1% penicillin-streptomycin (Gibco 15070063) and incubated at 37°C in a 5% CO2 atmosphere. Fibroblasts were passaged using 0.25% Trypsin-EDTA (Gibco 25200072) and cells were counted to ensure correct seeding concentrations.
Fibroblast cell response to 60 μM TiBP-Spacer5-AMP bio-coating, 200 μg/mL collagen (Sigma C7661, St. Louis, MO) coating (positive control) and bare, untreated (negative control) titanium foil and implant surfaces was studied. Fibroblast cells at a concentration of 8 × 105 cells/mL were added to sterile 24 well plates containing TiBP-Spacer5-AMP coated, collagen coated, or bare, untreated foils or implants and incubated for 24 h at 37°C in a 5% CO2 atmosphere. Fibroblast attachment and spreading were evaluated by fixing fibroblasts on titanium surfaces with 2% glutaraldehyde solution, followed by dehydration in 10%, 30%, 60%, 90% and 100% ethanol. Fixed fibroblasts on titanium surfaces were washed twice with PBS, permeabilized with TritonX (Sigma T8787), sealed with BSA (Fisher BioReagents BP671-10, Carlsbad, CA), and stained with Alexa Fluor488-Phalloidin dye (Invitrogen). Unbound dye was removed by washing with PBS and substrates were imaged with a fluorescent microscope at 4, 10 and 20 times magnification. Five representative images of each surface (n = 3) were obtained and analyzed with ImageJ Software and then subjected to statistical analysis. Cell attachment was determined as number of cells per square millimeter and the percentage of the image surface covered by attached cells. Another measure of viability, metabolic activity was determined using a MTT (3-(4,5-dimethylthiazol-2-yl)-2-5-diphenyltetrazolium bromide) assay (Sigma M5655). Following incubation of fibroblasts with TiBP-Spacer5-AMP, collagen, or bare, untreated titanium surfaces for 24 h at 37°C in a 5% CO2 atmosphere, one tenth of the well liquid volume was removed and replaced by the same volume of 5 mg/mL MTT reagent. The substrates with MTT reagent were incubated for 3 h then transferred to a sterile 24 well plate. The formazan crystals were dissolved in the detergent reagent according to the manufacture's protocol. Absorbance was measured at 570 nm.
3. Results and Discussion
Here, we engineered and evaluated a chimeric peptide composed of a titanium binding and an antimicrobial domain linked by a novel spacer design (TiBP-Spacer5-AMP). Our objective was to preserve the secondary structural features of both the TiBP and the AMP so as to impart an effective antimicrobial activity against two bacteria commonly associated with nosocomial implant infections, S. mutans and S. epidermidis.40,41 Data from a similar chimeric peptide with identical functional domains, but a shorter spacer sequence (TiBP-Spacer3-AMP) and the AMP peptide alone were used to evaluate the effect of the new engineered spacer design.24–26 Table 1 contains the sequences and physical chemical properties for each chimeric peptide and their functional domains. Despite the physical chemical similarity to one another, we observed improved antimicrobial activity with the altered amino acid composition designed into the longer spacer called Spacer5. The interfacial activity model suggests that antimicrobial activity depends on amino acid composition and physical chemical properties.42 Interfacial activity is the electrostatic and hydrophobic interactions between peptides and the lipid surface of the bacterial cell wall. Literature suggests several mechanisms leading to cell death following interaction between the peptide and the lipid surface including a compromised bacterial cell wall which initiates a cascade of effects including cellular respiration, DNA damage and altered gene expression. Recent publications indicate the production of reactive oxygen species (ROS) when AMPs attack bacteria.43–45 Much of the literature characterize AMP activity based on either structure-function relationships or physical chemical properties.42 The effect of the engineered spacer design was evaluated through independent, but corroborating approaches, including: computational and direct structural analysis coupled with measurement of antimicrobial activity of the chimeric peptide in solution, as well as when bound to titanium substrates against common nosocomial microorganisms allowing us to suggest that the restored antimicrobial activity is due to the preserved structure associated with the Spacer5 design.
Table 1.
Physical chemical properties and amino acid sequences for TiBP, AMP, and two chimeric peptides TiBP-Spacer3-AMP and TiBP-Spacer5-AMP.
| Name | Sequence | Spacer length | MW (kDa) | PI | Charge | GRAVY | |
|---|---|---|---|---|---|---|---|
| TiBP | RPRENRGRERGL | N/A | 1.4956 | 11.82 | +3 | −2.633 | |
| AMP | LKLLKKLLKLLKKL | N/A | 1.6923 | 10.70 | +6 | 0.500 | |
| TiBP-Spacer 3-AMP | RPRENRGRERGL | −GGG LKLLKKLLKLLKKL | 3 | 3.3411 | 11.85 | +9 | −0.890 |
| TiBP-Spacer 5-AMP | RPRENRGRERGL | GSGGG LKLLKKLLKLLKKL | 5 | 3.4852 | 11.85 | +9 | −0.871 |
Notes: MW, molecular weight; pI, isoelectric point; GRAVY, GRand Average Value of hydropathicitY; and “−”, gap inserted for sequence alignment. Despite chimeric peptide similarity to one another, we observed improved antimicrobial activity with the altered amino acid composition of the longer peptide spacer, Spacer5.
3.1. Computational structure and function predictions
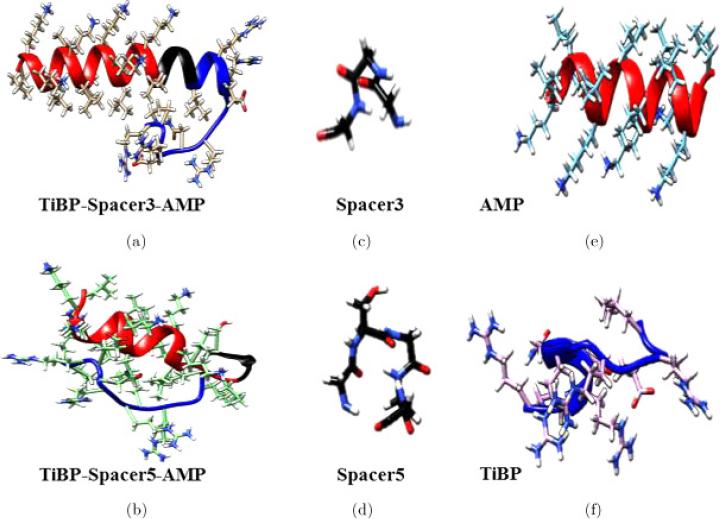
Computational molecular structures were generated using the PyRosetta structural ensemble generation method.33 One thousand likely energy minimized structures were generated for each chimeric peptide, for each spacer sequence, and for each functional domain. The lowest energy structure for each is depicted in Fig. 1 with TiBP, spacer domain, and AMP designated with blue-, black-, and red-shading, respectively. The images shown in Fig. 1 represent likely structures that are modeled in solution. Ramachandran plots were generated for the lowest energy chimeric peptide structures and are shown in the supporting information as Figs. S1 and S2 for TiBP-Spacer3-AMP and TiBP-Spacer5-AMP, respectively. The Ramachandran plots simulates the contribution of hydrogen bonding among backbone atoms and can be interpreted to correlate the contribution of the α-helix or β-sheet structural features depicted in the energy minimized structures.
Fig. 1.
Lowest energy structures modeled in solution for (a) TiBP-Spacer3-AMP chimeric peptide; (b) TiBP-Spacer5-AMP chimeric peptide; (c) Spacer3 (GGG); (d) Spacer5 (GSGGG); (e) AMP; (f) TiBP. The peptide backbone is represented as a ribbon to show secondary structure for peptides with side chains represented by full atoms. TiBP domains, spacer domains, and AMP domains are designated with blue-, black-, and red-shading, respectively. The. TiBP-Spacer3-AMP (a) has an α-helix feature beginning with the AMP domain and preserved through Spacer3, whereas TiBP-Spacer5-AMP (b) has a shorter α-helix ends at Spacer5. Both functional domains, AMP (e) and TiBP (f) have α-helix secondary structure, with a stronger prominence in the AMP domain (color online).
Previous published analysis using the “rule induction method” suggest that increasing the number of short α-helices is associated with antimicrobial activity, therefore we first examined the structure of the chimeric peptides.24 The computational structures in Fig. 1 show the secondary structure features for the chimeric peptides and their component parts. The structure of both TiBP and AMP peptides show features of alpha helicity with a stronger helicity prominence in the AMP domain [Fig. 1(e)]. TiBP-Spacer3-AMP [Fig. 1(a)], has an α-helix feature beginning within the AMP domain and preserved through Spacer3. From the Ramachandran plot we conclude the α-helix feature is approximately 26 amino acids long and confirm that backbone angles consistent with α-helix features are present though AMP, Spacer3, and almost the entire TiBP. All but three amino acids correspond to psi/phi angles (−90°, −60°) consistent with α-helix. Spacer3 consists of but three glycine amino acid residues; therefore the minimal side chain size of glycine in Spacer3 could allow the alpha helix feature to be preserved across the spacer domain and into the TiBP, producing longer alpha helices. The alpha helix feature in TiBP-Spacer5-AMP is comparatively much shorter. The Ramachandran plot for TiBP-Spacer5-AMP shows the psi/phi angles (−90°, −60°) corresponding to alpha helicity are assigned to the AMP domain, while psi/phi angles (−90°, +120°) corresponding to β-sheet/random coil secondary structures are observed in the rest of the molecule. We interpret these finding to suggest that the Spacer5 segregates the AMP domain from the rest of the chimeric peptide, allowing its antimicrobial activity to be preserved.
The Spacer5 (GSGGG) is composed of four glycine and a single serine amino acid residues, and the presence of a polar serine residue could produce a slight “ST staple” feature in the spacer region producing a backbone bend that prevents the continuity of the alpha helix feature observed in TiBP-Spacer3-AMP. The α-helix property that most accurately predicts antimicrobial activity by the “rule induction method” against S. mutans and S. epidermidis is the number of five amino acid- and four amino acid-right-handed-helices. The “rule induction method” also predicts antimicrobial function based on the percentage of these features present in the energy minimized PyRosetta generated structures. The “rule induction method” was used to predict the antimicrobial activity of TiBP-Spacer5-AMP and TiBP-Spacer3-AMP with the results shown in Table 2. Of the 1000 structures generated in the ensembles for each chimeric peptide, TiBP-Spacer5-AMP had a larger percentage of structural topologies represented with four or five amino acid residue alpha helix features. This is consistent with what we observed with the detailed structure analysis conducted for the lowest energy structure of each chimeric peptide. Based on previously published data validating the “rule induction method”, we predict that TiBP-Spacer5-AMP should have greater antimicrobial activity in solution against S. mutans and S. epidermidis.24 To further corroborate our analysis, we next turned to CD analysis which can directly measure secondary structure of TiBP-Spacer5-AMP.
Table 2.
“Rule Induction” method predictions of antimicrobial activity based on secondary structure features of four and five amino acid alpha helicity present in computationally generated structures.24 Increasing antimicrobial activity is associated with increasing percent of helix frequency over either a four or five amino acid average. The “rule induction method” predicts that TiBP-Spacer5-AMP possesses a secondary structure associated with antimicrobial activity to a greater extent than the secondary structure of the TiBP-Spacer3-AMP.
| Peptide | 4 aa α-helix frequency (%) | 5 aa α-helix frequency (%) |
|---|---|---|
| TiBP-Spacer3-AMP | 10.4 | 5.6 |
| TiBP-Spacer5-AMP | 17.6 | 8.0 |
3.2. Structure determination with CD
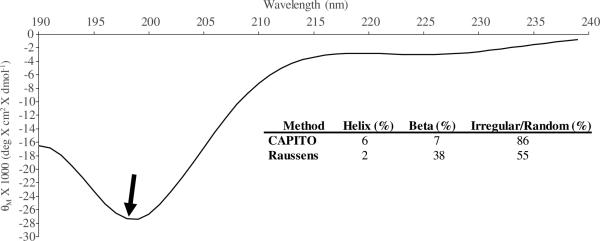
The chimeric peptide was prepared at a concentration of 50 μM in PBS at pH of 7.4 for secondary structure analysis by CD. Two complementary methods, the CAPITO and the Raussens method were used to quantify the results obtained from the CD spectra.38,39 We applied both the concentration dependent CAPITO method and the concentration independent Raussens method for these predictions to corroborate outcomes. Both approaches are regression methods used to transform CD spectral data in order to identify corresponding structural information from a protein database. The CD spectrum for TiBP-Spacer5-AMP is depicted in Fig. 2 with inset table containing results from analysis with the regression methods. The spectrum for the chimeric peptide with Spacer5 indicates a greater preference for right-circularly polarized light absorbance compared to the previously published spectrum for Spacer3, indicating that the predominance of α-helix secondary structure present in TiBP-Spacer3-AMP is not preserved through the newly designed Spacer5.26 The CD structural prediction results are consistent with the computationally predicted secondary structure analysis, indicating that a majority of the secondary structure of TiBP-Spacer5-AMP is β-sheet or random coil. Moreover, both the CAPITO and Raussens method assigns 86% and 55% secondary structure to irregular or random coil features, for TiBP-Spacer5-AMP, respectively. In addition to random coil features, the Raussens method assigns 38% of TiBP-Spacer5-AMP secondary structure to beta sheet features. The Raussens method also corroborates the Ramachandran plot prediction for analysis computationally generated structures. These secondary structure features are predicted by the “rule induction method” to also produce a greater antimicrobial activity for TiBP-Spacer5-AMP. These structural analyses are related only to the in solution secondary structure of the chimeric peptides not their structures when bound to titanium surfaces. Currently, a computational model for proteins bound to a titanium surface does not exist. While this limits our ability to describe the exact structural contributions to antimicrobial activity on the titanium implant surface, we can however measure the antimicrobial activity of the chimeric peptides in solution and empirically apply those findings to titanium surfaces.
Fig. 2.
CD spectrum for TiBP-Spacer5-AMP chimeric peptide at a concentration of 50 μM in PBs, pH 7.4. The feature designated by the arrow indicates a greater preference for right-circularly polarized light absorbance compared to the previously published spectrum for chimeric peptide with Spacer3.26 The CAPITO and Raussens methods indicates a predominance of irregular and random coil features in the spectrum consistent with what is observed in the computationally generated secondary structure for TiBP-Spacer5-AMP shown in Fig. 1(b).38,39
3.3. Chimeric peptide function
3.3.1. Antimicrobial effect in solution
Antimicrobial activity in solution was elucidated by determining the MIC of TiBP-Spacer5-AMP required to inhibit growth for two bacterial strains commonly recovered from infected implants, S. mutans and S. epidermidis.40,41 Previously published MIC values for TiBP-Spacer3-AMP and AMP alone were used for comparison.24 MIC data for AMP, TiBP-Spacer3-AMP, and TiBP-Spacer5-AMP are depicted in Table 3. The MIC value of TiBP-Spacer5-AMP against S. mutans and S. epidermidis are 50 μM and 8 μM, respectively. We observed a remarkable three fold improvement of MIC antimicrobial activity for the TiBP-Spacer5-AMP against S. mutans. This can be attributed to the increased frequency of secondary structural features corresponding to antimicrobial activity as predicted by the “rule induction method”, corroborating the importance of secondary structure features in AMP design. The design of the spacer offers an opportunity to fine-tune the structural properties of the chimeric peptide so as to improve its antimicrobial potential. The use of the Spacer5 results in a chimeric peptide displaying shorter α-helix structural features compared to Spacer3 and yields improved antimicrobial activity. In contrast however, the antimicrobial activity of TiBP-Spacer5-AMP against S. epidermidis appears to be slightly diminished compared to TiBP-Spacer3-AMP. We cannot yet account for why TiBP-Spacer5-AMP was less effective against S. epidermidis, than S. mutans and we are conducting further experiments to investigate this observation.
Table 3.
MIC of TiBP-Spacer5-AMP, TiBP-Spacer3-AMP, and AMP alone in solution against S. mutans and S. epidermidis. There is a three-fold decrease in TiBP-Spacer5-AMP MIC against S. mutans.
| Peptide | S. mutans (μM) | S. epidermidis (μM) |
|---|---|---|
| AMP | 38 | 4 |
| TiBP-Spacer3-AMP | 153 | 5 |
| TiBP-Spacer5-AMP | 50 | 8 |
The bactericidal concentration for TiBP-Spacer5-AMP against each bacteria was also determined using the AlamarBlue assay.46 The bactericidal concentration for TiBP-Spacer5-AMP was found to be 60 μM for S. mutans and 10 μM for S. epidermidis. These concentrations are only slightly higher than the observed MIC values indicating that TiBP-Spacer5-AMP corroborating these complementary methods of killing bacteria. Next, we used the bactericidal concentrations determined from the AlamarBlue assay to assess the antimicrobial activity of medical implants coated with TiBP-Spacer5-AMP by assessing bacterial growth on their surfaces.
3.3.2. Antimicrobial effect on surfaces
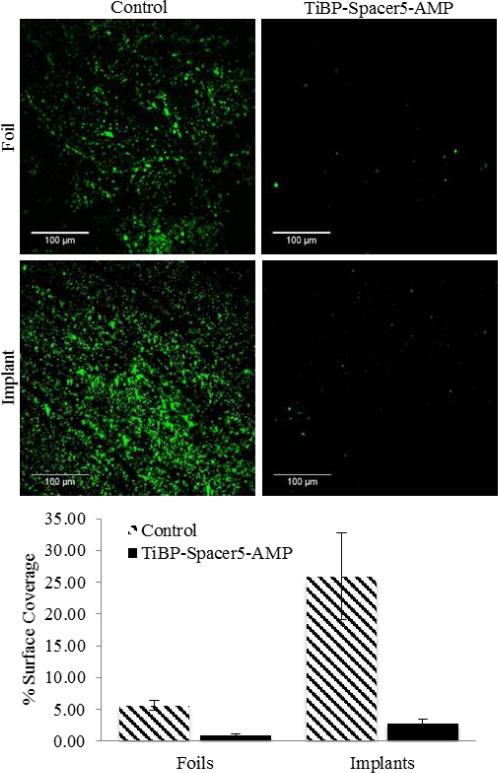
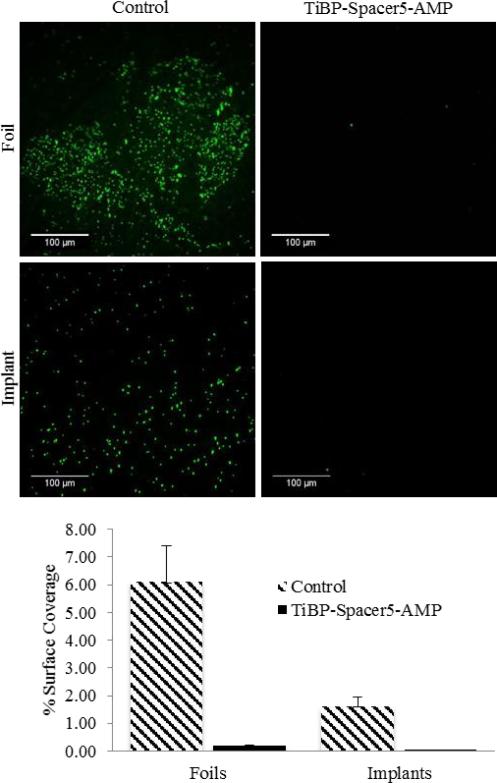
TiBP-Spacer5-AMP at 60 μM for S. mutans and 10 μM for S. epidermidis were permitted to self-assemble on selected titanium surfaces and evaluated for their antimicrobial activity. Titanium foils were selected for their ease of use, while discs cut from stock titanium orthopedic bar material were used to ascertain their effectiveness directly on a clinically relevant surface. For both surfaces, infectious organisms common to clinical infections, S. mutans and S. epidermids, were used to evaluate the antimicrobial activity of the bio-coating. Previous studies had established the binding characteristics and affinity for the TiBP as part of a chimeric molecule.24 Following incubation, the unbound peptide was removed by repeated washing, suggesting the antimicrobial activity observed for either titanium surface was the result of the chimeric peptide bound to the surface representing antimicrobial activity. The observed effectiveness of TiBP-Spacer5-AMP antimicrobial effect against S. mutans is shown in Fig. 3 and against S. epidermidis in Fig. 4. The images are representative areas, and the percent of the total surface area covered by bound bacteria was identified by bacterial staining and quantified by analysis with ImageJ. In all cases, TiBP-Spacer5-AMP bio-coating reduced the number of bacteria attached to the surface compared to uncoated control surfaces. The fold reduction for the number of bacteria on titanium surfaces with TiBP-Spacer5-AMP bio-coating is depicted in Table 4. There is a six–nine-fold reduction for S. mutans, with a 33–48-fold improvement noted for S. epidermidis on foil or implant surfaces, respectively, due to the presence of the TiBP-Spacer5-AMP bio-coating. These data suggest that the TiBP-Spacer5-AMP bio-coating is an effective strategy to combat infections and consequential implant failure by reducing bacterial colonization which ultimately transform to a complex biofilm that can resist systemic administration of antibiotics and lead to implant failure.47 Alternatively, the coating formed by the TiBP-Spacer5-AMP may interfere with bacterial attachment by forming a biomimetic surface that is less fouling than the bare titanium or titanium alloy surface.31 The increasing frequency of antibiotic resistant bacteria in hospital settings contributing to nosocomial infections and the increasing number of patients with co-morbidities can both contribute to a diminished ability of the host to resist and clear bacteria at surgical sites which lead to implant failure. Whether by antimicrobial activity or reduced attachment, the reduction in the number of pathogenic bacterial by the TiBP-Spacer5-AMP would result in improved patient outcomes. Lastly, we evaluated host cell response on titanium surfaces coated with the TiBP-Spacer5-AMP chimeric peptide.
Fig. 3.
Fluorescent microscope images (Scale Bar is 100 μm) of S. mutans bacteria on 99% pure titanium foils and orthopedic implant discs with TiBP-Spacer5-AMP bio-coating and bare, bare untreated controls. Chart depicts the percent surface coverage quantified by ImageJ of bacteria on the titanium surfaces.
Fig. 4.
Fluorescent microscope images (Scale Bar is 100 μm) of S. epidermidis bacteria on 99% pure titanium foils and orthopedic implant discs with TiBP-Spacer5-AMP bio-coating and bare, bare untreated controls. Chart depicts the percent surface coverage quantified by ImageJ of bacteria on the titanium surfaces.
Table 4.
Fold improvement calculated from fluorescent microscopy image analysis of S. mutans and S. epidermidis bacteria on titanium foil and implant surfaces with TiBP-Spacer5-AMP bio-coating, compared to bare, uncoated control surfaces. There is in resistance to bacteria as a result of the TiBP-Spacer5-AMP bio-coating on foil and implant surfaces.
| Fold improvement compared to uncoated Ti surfaces | ||
|---|---|---|
| Foils | Implants | |
| TiBP-Spacer5-AMP against S. mutans | 6 | 9 |
| TiBP-Spacer5-AMP against S. epidermidis | 33 | 48 |
3.3.3. Host cell attachment and viability
Host cell attachment and viability was evaluated in vitro using a fibroblast cell line (NIH/3T3) by measuring cell attachment, morphology/spreading, and viability response to TiBP-Spacer5-AMP coated substrates. The results are shown in Fig. 5 for titanium foils and those for orthopedic implants are shown in Fig. 6. The number of fibroblasts that attached to the TiBP-Spacer5-AMP bio-coated foils was not statistically different compared to an untreated control surface. However, the cells attached on the chimeric peptide bio-coated foil surface did demonstrate greater coverage, suggesting they spread more effectively compared to cells grown on untreated control surfaces. As expected, collagen-coated surfaces, the gold-standard used as a positive control, did outperform the TiBP-Spacer5-AMP chimeric peptide bio-coating. Interestingly, for studies with fibroblasts seeded onto titanium implant surfaces, the chimeric peptide bio-coated surfaces showed statistically greater cell attachment and spreading properties than observed for the unmodified implant substrates. Additionally, the TiBP-Spacer5-AMP bio-coated implant surfaces showed adhesion and spreading results that were statistically comparable to the positive collagen controls. These results suggest that bio-coating orthopedic medical implants with TiBP-Spacer5-AMP would result in an improved host cell response at the implant-tissue interface.
Fig. 5.
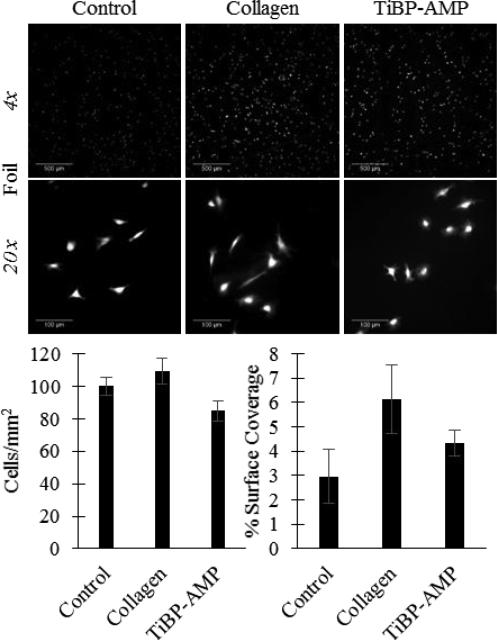
Fluorescent images of NIH/3T3 fibroblast attachment on titanium foils: Control (no treatment), Collagen (200 μg/mL collagen coating positive control), or TiBP-AMP (60 μM TiBP-Spacer5-AMP bio-coating). Scale bar represents 500 μm for 4× images and 100 μm for 20× images. TiBP-Spacer5-AMP bio-coated foils had fewer fibroblasts attach compared to untreated control, however the fibroblast surface coverage for TiBP-Spacer5-AMP was greater indicating the cells spread more.
Fig. 6.
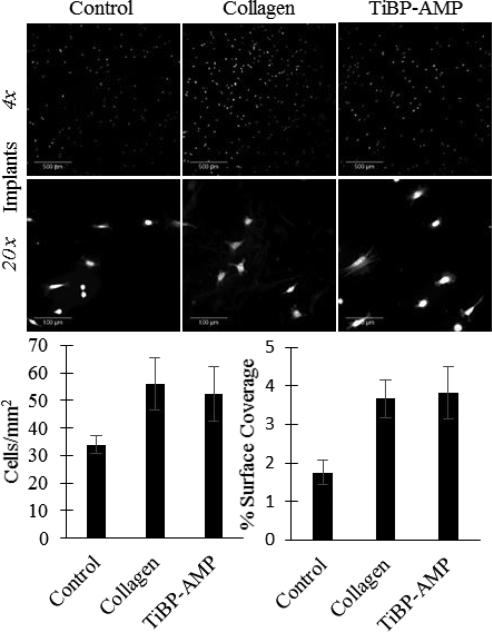
Fluorescent images of NIH/3T3 fibroblast attachment on titanium orthopedic implants: Control (no treatment), Collagen (200 μg/mL collagen coating positive control), or TiBP-AMP (60 μM TiBP-Spacer5-AMP bio-coating). Scale bar represents 500 μm for 4× images and 100 μm for 20× images. TiBP-Spacer5-AMP bio-coated implants showed greater cell attachment and spreading compared to untreated controls and attachment and spreading were comparable to collagen positive controls.
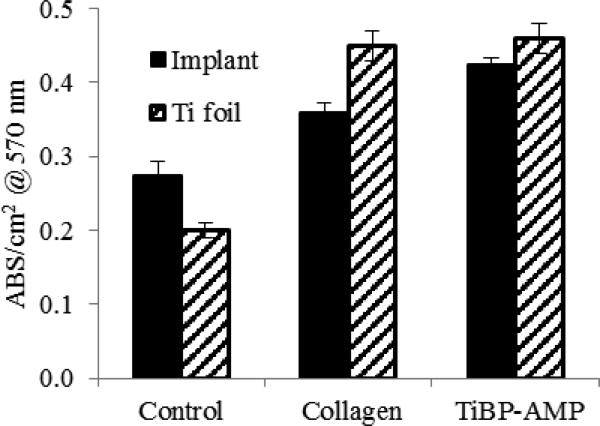
The MTT assay was used as a live-dead discrimination assay for fibroblasts grown on various surfaces. We found that cell viability on either titanium foils or implant surfaces treated with chimeric peptide were similar to values observed for the positive control collagen coated surfaces (Fig. 7), with approximately 50% greater cell viability observed compared to untreated surfaces.
Fig. 7.
NIH/3T3 fibroblast metabolism on titanium foils and implants measured by MTT assay. Control (no treatment), collagen (coated with 200 μg/mL collagen), TiBP-AMP (coated with TiBP-Spacer5-AMP at 60 μM).
4. Conclusion
A titanium binding, antimicrobial chimeric peptide with novel spacer design (TiBP-Spacer5-AMP) was rationally engineered. Computational structure analysis revealed secondary structural features that were dependent on the length and composition of the spacer. These features were confirmed through direct evaluation with CD. Specifically, TiBP-Spacer5-AMP has multiple short α-helix features with predominately irregular or random coil secondary structure corroborated by Ramachandran plot analysis of energy minimized structures and CD. The previously developed “rule induction method” was applied and predicted the beneficial effect of structural features induced by the spacer that resulted in greater antimicrobial activity. In fact, a three-fold decrease in MIC that indicates increased antimicrobial activity was observed against bacteria common to nosocomial implant infection. TiBP-Spacer5-AMP was assembled on titanium foils and orthopedic implant surfaces as a biomimetic coating which reduced bacterial numbers nine-fold against S. mutans, a bacteria common to dental implant infections, and 48-fold against S. epidermidis bacteria common to orthopedic implant infections. The potential of the chimeric peptide bio-coating to promote host cell attachment was evaluated using a fibroblast cell line. On chimeric peptide bio-coated surfaces, the cells attached, spread and exhibited 50% greater viability measured by a metabolic assay compared to identical cells on bare, untreated titanium surfaces. Data from the TiBP-Spacer5-AMP point to the importance of optimal design of the spacer between two functional domains within the chimeric peptide in order to optimize the function of each domain, namely binding and self-assembling onto titanium surfaces and the displayed antimicrobial activity on the biomaterial surface. The ability to create an antimicrobial bio-coating on titanium medical implants that serve to overcome complications associated with implant failure due to nascent infection and their eventual loss by infection that contributes to increasing medical costs and patient morbidity has interminable value.
Supplementary Material
Acknowledgments
This investigation was supported by Research Grants: R21 AR062249 support from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) and R01 DE025476 from the National Institute of Dental and Craniofacial Research (NIDCR), National Institutes of Health (NIH)–United State, Bethesda, MD 20892. We are also grateful to Madison & Lila Self Graduate Fellowship (CW) at the University of Kansas. We thank Mr. Dwight Deay for his assistance with CD experiments.
Contributor Information
Cate Wisdom, Bioengineering Program, University of Kansas, 3135A Learned Hall, 1530 W 15th Street Lawrence, Kansas 66045, USA.
Sarah Kay VanOosten, Bioengineering Program, University of Kansas, 3135A Learned Hall, 1530 W 15th Street Lawrence, Kansas 66045, USA.
Kyle W. Boone, Bioengineering Program, University of Kansas, 3135A Learned Hall, 1530 W 15th Street Lawrence, Kansas 66045, USA.
Dmytro Khvostenko, Bioengineering Research Center (BERC), University of Kansas, 3138 Learned Hall, 1530 W 15th Street Lawrence, Kansas 66045, USA, dmytro.khvostenko@ku.edu.
Paul M. Arnold, Department of Neurosurgery, University of Kansas Medical Center, 3901 Rainbow Boulevard Kansas City, Kansas 66160, USA, parnold@kumc.edu
Malcolm L. Snead, Center for Craniofacial Molecular Biology, Herman Ostrow School of Dentistry of USC, The University of Southern California CSA 142, 2250 Alcazar Street Los Angeles, CA 90033, USA, mlsnead@usc.edu
Candan Tamerler, Mechanical Engineering Department and BERC, University of Kansas, 3138 Learned Hall, 1530 W 15th Street Lawrence, Kansas 66045, USA.
References
- 1.Carr AJ, et al. The Lancet. 2012;379:1331. doi: 10.1016/S0140-6736(11)60752-6. [DOI] [PubMed] [Google Scholar]
- 2.Al-Radha ASD, Dymock D, Younes C, O'Sullivan D. J. Dent. 2012;40:146. doi: 10.1016/j.jdent.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Gugwad R, Reddy GS, Reddy RG, Bhatt R, Nagaral SC. Clin. Dent. 2013;7:0974. [Google Scholar]
- 4.Gepreel MA-H, Niinomi M. J. Mech. Behav. Biomed. Mater. 2013;20:407. doi: 10.1016/j.jmbbm.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Verissimo NCC, et al. J. Biomed. Mater. Res. A. 2015;103:3757. doi: 10.1002/jbm.a.35517. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard EM, Valentin T, Panilaitis B, Omenetto F, Kaplan DL. Adv. Funct. Mater. 2013;23:854. doi: 10.1002/adfm.201201636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zilberman M, Elsner JJ. J. Control. Release. 2008;130:202. doi: 10.1016/j.jconrel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Bernthal NM, et al. J. Vis. Exp. e51612. 2014 doi: 10.3791/51612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridgeway S, et al. J. Bone Joint Surg. Bri. 2005;87:844. doi: 10.1302/0301-620X.87B6.15121. [DOI] [PubMed] [Google Scholar]
- 10.Olsen MA, et al. J. Bone Joint Surg. 2008;90:62. doi: 10.2106/JBJS.F.01515. [DOI] [PubMed] [Google Scholar]
- 11.Sadoghi P, et al. J. Arthroplasty. 2013;28:1329. doi: 10.1016/j.arth.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Cano A, Pacha-Olivenza M-Á, Babiano R, Cintas P, González-Martín M-L. Surf. Coat. Technol. 2014;245:66. [Google Scholar]
- 13.Peng S, Zhu Y. Chem. Lett. 2014;43:355. [Google Scholar]
- 14.Hickok NJ, Shapiro IM. Adv. Drug Deliv. Rev. 2012;64:1165. doi: 10.1016/j.addr.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ketonis C, Parvizi J, Jones LC. J. Am. Acad. Orthop. Surg. 2012;20:478. doi: 10.5435/JAAOS-20-07-478. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, et al. Surf. Coat. Technol. 2013;216:158. [Google Scholar]
- 17.Gottenbos B, van der Mei HC, Klatter F, Nieuwenhuis P, Busscher HJ. Biomaterials. 2002;23:1417. doi: 10.1016/s0142-9612(01)00263-0. [DOI] [PubMed] [Google Scholar]
- 18.He G, et al. J. Mater. Chem. B, Mater. Biol. Med. 2015;3:6676. [Google Scholar]
- 19.Lin D-J, et al. J. Biomater. Appl. 2013;27:553. doi: 10.1177/0885328211414942. [DOI] [PubMed] [Google Scholar]
- 20.Pasupuleti M, Schmidtchen A, Malmsten M. Crit. Rev. Biotechnol. 2012;32:143. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 21.Hilchie AL, Wuerth K, Hancock RE. Nat. Chem. Biol. 2013;9:761. doi: 10.1038/nchembio.1393. [DOI] [PubMed] [Google Scholar]
- 22.Cederlund A, Gudmundsson GH, Ager-berth B. Febs J. 2011;278:3942. doi: 10.1111/j.1742-4658.2011.08302.x. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Xiang Q, Zhang Q, Huang Y, Su Z. Peptides. 2012;37:207. doi: 10.1016/j.peptides.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Yucesoy D, et al. JOM. 2015;67:754. doi: 10.1007/s11837-015-1350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yazici H, et al. Acta Biomater. 2013;9:53415352. [Google Scholar]
- 26.Yazici H, et al. ACS Appl. Mater. Interfaces. 2016;8:5070. doi: 10.1021/acsami.5b03697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamerler C, Sarikaya M. Acta Biomater. 2007;3:289. doi: 10.1016/j.actbio.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Tamerler C, et al. Biopolymers. 2010;94:78. doi: 10.1002/bip.21368. [DOI] [PubMed] [Google Scholar]
- 29.Seker UO, et al. Langmuir. 2007;23:7895. doi: 10.1021/la700446g. [DOI] [PubMed] [Google Scholar]
- 30.Fjell CD, Jenssen H, Cheung WA, Hancock RE, Cherkasov A. Chem. Biol. Drug Des. 2011;77:48. doi: 10.1111/j.1747-0285.2010.01044.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Snead ML, Tamerler C. Nanomedicine. 2015;11:431. doi: 10.1016/j.nano.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkins MR, et al. Methods Mol. Biol. 1999;112:531. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 33.Chaudhury S, Lyskov S, Gray JJ. Bioinformatics. 2010;26:689. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leaver-Fay A, et al. Methods Enzymol. 2011;487:545. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, et al. J. Comput. Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Pawlak Z. Cybern. Sys. 1998;29:661. [Google Scholar]
- 37.Grzymala-Busse JW, Rzasa W. Fundam. Inform. 2010;100:99. [Google Scholar]
- 38.Wiedemann C, Bellstedt P, Görlach M. Bioinformatics. 2013;29:1750. doi: 10.1093/bioinformatics/btt278. [DOI] [PubMed] [Google Scholar]
- 39.Raussens V, Ruysschaert JM, Goormaghtigh E. Anal. Biochem. 2003;319:114. doi: 10.1016/s0003-2697(03)00285-9. [DOI] [PubMed] [Google Scholar]
- 40.Socransky S, Haffajee A, Lindhe J, Karring T, Lang N. Clinical Periodontology and Implant Dentistry. Wiley; NY: 2008. [Google Scholar]
- 41.Montanaro L, et al. Future Microbiol. 2011;6:1329. doi: 10.2217/fmb.11.117. [DOI] [PubMed] [Google Scholar]
- 42.Wimley WC. ACS Chem. Biol. 2010;5:905. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fjell CD, Hiss JA, Hancock RE, Schneider G. Nat. Rev. Drug Discov. 2012;11:37. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 44.Choi H, Rangarajan N, Weisshaar JC. Trends Microbiol. 2016;24:111. doi: 10.1016/j.tim.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, et al. Adv. Health. Mater. 2016;5:557. doi: 10.1002/adhm.201500712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rampersad SN. Sensors (Basel) 2012;12:12347. doi: 10.3390/s120912347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costerton JW, Stewart PS, Greenberg EP. Science. 1999;284:1318. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.