Abstract
Large-scale epigenetic changes take place when epithelial cells with cell-cell adhesion and apical-basal polarity transition into invasive, individual, mesenchymal cells through a process known as epithelial to mesenchymal transition (EMT). Importantly, cancers with stem cell properties disseminate and form distant metastases by reactivating the developmental EMT program. Recent studies have demonstrated that the epigenetic histone modification, H2BK5 acetylation (H2BK5Ac), is important in the regulation of EMT. For example, in trophoblast stem (TS) cells, H2BK5Ac promotes the expression of genes important to the maintenance of an epithelial phenotype. This finding led to the discovery that TS cells and stem-like claudin-low breast cancer cells share similar H2BK5Ac-regulated gene expression, linking developmental and cancer cell EMT. An improved understanding of the role of H2BK5Ac in developmental EMT and stemness will further our understanding of epigenetics in EMT-related pathologies. Here, we examine the binders and regulators of H2BK5Ac and discuss the roles of H2BK5Ac in stemness and EMT.
Keywords: H2BK5 acetylation, EMT, MAP3K4, CBP, HDAC6
Epithelial cells are nonmotile cells that maintain cell-cell adhesion and apical-basal polarity. The process of epithelial to mesenchymal transition (EMT) allows epithelial cells to acquire a mesenchymal phenotype with front-back polarity, diminished cell-cell adhesion, and invasiveness. The regulation of EMT is important during development, wound healing, organ fibrosis, and cancer metastasis. Several recent studies support the hypothesis that EMT is regulated by acetylation of histone H2B at lysine 5 (H2BK5Ac).
Contexts of EMT
EMT is a tightly regulated developmental process that generates cells with mesenchymal phenotypes in several distinct contexts [1, 2]. During development, trophoblast stem cells (TS cells) differentiate, undergo EMT, and establish implantation by invading the uterus and forming the placenta [3–5]. During tissue regeneration and wound healing, EMT generates new cells that migrate and promote reepithelialization [6–8]. Persistent inflammation can lead to aberrant EMT and organ fibrosis [9, 10]. In addition, epithelial carcinoma cells may undergo EMT in response to signaling in the tumor microenvironment, leading to metastatic disease [11]. Cancer cells with characteristics of EMT often show signs of stemness, including self-renewal and multipotency [11–14]. These invasive, stem-like cells digest through the basement membrane and into the vasculature, eventually undergoing mesenchymal to epithelial transition and initiating secondary tumors [13–15].
These distinct contexts of EMT activation share several molecular mechanisms. During EMT, epithelial cells lose cell-cell adhesion through the dissolution of cell-cell junctions [16, 17]. The loss of expression of cell-cell junction components like E-cadherin is accompanied by increased expression of mesenchymal markers, N-cadherin and Vimentin [17]. The transition from groups of cells to individual cells also involves a change from epithelial, apical-basal polarity to a more mesenchymal, spindle shaped front-back polarity [18]. In the nucleus, EMT progression is associated with the increased expression of Snail, Twist, and Zeb family EMT-inducing transcription factors (EMT-TFs). The most definitive and clinically relevant feature of EMT is the acquisition of invasiveness [18]. In many instances, these large-scale changes in cell phenotype have been associated with epigenetic changes in histone modification. In this review, we will briefly discuss histone modification and more specifically, examine the relationship between H2BK5Ac, stemness, and EMT.
Histone Modifications
The organization of the genome into nucleosomes containing ~147 base pairs wound around a core histone octamer allows cell types to access the genes necessary to specify their phenotype [19]. The core histones each have several transcript variants that are arranged in multi-copy clusters within the genome [20]. Once histones are transcribed, translated, and incorporated into nucleosomes, the modification of amino-terminal histone tails controls the availability of genes, allowing cells to adjust their phenotype [21, 22]. Further, histone modification allows for the specification of cell lineages at precise times during development. Many studies have detailed how covalent histone modifications either promote or repress specific transcriptional activity in developmental and pathological contexts. The most well studied histone modifications are methylation and acetylation of lysine residues. Other histone modifications include phosphorylation, sumoylation, ADP-ribosylation, deimination, proline isomerization, crotonylation, and ubiquitination [22–24].
Depending on the specific lysine, histone methylation can be associated with transcriptional activation, poised chromatin, or transcriptional repression. Lysine-methylation has been detected on all four core histones [25]. Tri-methylation of histone H3 lysines 9 or 27 (H3K9me3 or H3K27me3) is associated with transcriptional repression, while tri-methylation of histone H3 lysine 4 (H3K4me3) signals active transcription [23]. Regions of H3K27me3 containing smaller regions of H3K4me3 have been defined as bivalent chromatin using embryonic stem cells [26]. Bivalent or poised chromatin also controls ZEB1 gene expression in basal-like breast cancer, which allows interconversion of tumor cell and cancer stem cell phenotypes [27]. The methylation of H2A and H2B has been detected in several cell types, but the functions of these modifications have not been well characterized [25]. Although H2BK5 monomethylation and H2BK5Ac have both been associated with genes that are highly transcribed, data have shown that H2BK5Ac is a more accurate predictor of gene transcription [28].
Histone acetylation neutralizes negative charges on the DNA, promoting the formation of euchromatin and the recruitment of transcription factors [29]. Acetylation sites are found on each of the four histone tails, and most promote transcription when acetylated [30]. Deacetylation of histones results in the formation of heterochromatin, where the nucleosomes are condensed, occluding the access of transcriptional machinery [30, 31]. Recent data suggest that H2BK5Ac is a reliable predictor of gene expression and an important modification in the orchestration of the EMT program [4, 28, 32]. To better understand how H2BK5Ac contributes to cell phenotype, we will discuss the interacting partners and regulators of H2BK5Ac and their importance in EMT.
Interactors and Regulators of H2BK5Ac
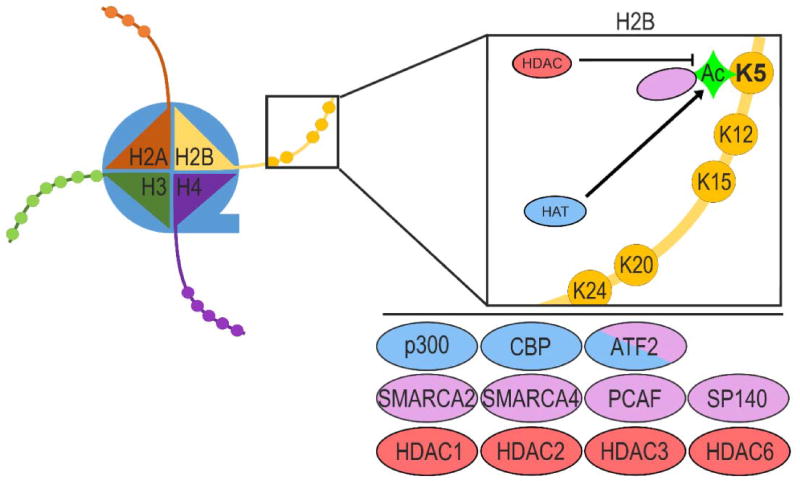
H2BK5Ac is a chromatin modification that recruits chromatin modifiers. For example, bromodomain containing proteins bind acetylated lysine residues to recruit the appropriate histone modifiers or modify the histone tails themselves. Importantly, several bromodomain containing proteins, including the SWI/SNF catalytic subunits, SMARCA4 and SMARCA2, bind H2BK5Ac (Figure 1). Using NMR, Shen et al. showed SMARCA4 binding to H2BK5Ac [33]. SMARCA2 binding of H2BK5Ac was detected by Filippakopoulos et al. using solid phase oligosaccharide tagging (SPOT) peptide arrays [34]. We hypothesize that SMARCA4 or SMARCA2 and the SWI/SNF complex may bind H2BK5Ac and shift nucleosomes to optimize binding of epigenetic modules and transcription factors during EMT. SPOT analysis also detected strong H2BK5Ac binding by the p300/CBP-associated histone acetyl-transferase (HAT), PCAF, as well as the nuclear body protein, SP140. These findings are less surprising, as these proteins each bind most acetyl-histone marks (Figure 1) [34]. Our understanding of H2BK5Ac as a binding target for other chromatin modifying complexes indicates that multiple layers of epigenetic changes regulate the expression of target genes. Importantly, enzymes that acetylate or deacetylate H2BK5 often modify other histone lysine residues as well, further demonstrating the multiple layers of epigenetic regulation controlling gene expression.
Figure 1. Interactors and Regulators of H2BK5Ac.
Several proteins bind and/or modify H2BK5Ac. The expression and activity levels of these proteins control cell phenotype by regulating the availability of DNA encoding phenotype-specific genes. Histone acetyl-transferases (HATs) that promote H2BK5Ac are shown in blue. Histone deacetylaces (HDACs) associated with loss of H2BK5Ac are shown in red. Proteins known to directly bind H2BK5Ac are shown in pink.
Although direct binding has not been demonstrated, several proteins have been shown to increase H2BK5Ac levels. Researchers have demonstrated the effects of CBP/p300 HAT activity on H2BK5Ac in several contexts. CBP and p300 have many overlapping functions with regard to histone acetylation and transcriptional activation due to their homologous bromodomains and HAT domains [35]. In humans, mutations in CBP or p300 lead to Rubinstein-Taybi syndrome, resulting in mental and developmental retardation, abnormal facial patterning, and tumors of the brain and neural crest stem cell-derived tissues [36]. Another study showed that lymphoblastoid cell cultures from patients with heterozygous mutations of CBP showed decreased acetylation of H2A and H2B [37]. In mice, a 50% reduction in CBP expression resulted in skeletal malformation and defects in axial skeleton patterning [38]. The same CBP-heterozygous mice also showed selective loss of H2BAc in the hippocampus [39]. In mouse epithelial TS cells expressing CBP shRNAs, there was selective loss of H2A/H2BK5Ac that correlated with the gain of mesenchymal morphology and invasiveness [4]. In contrast, TS cells expressing p300 shRNAs showed selective loss of H3K9/H4K8Ac, indicating that CBP specifically promoted H2A/H2BK5Ac [4]. These data suggest that CBP HAT activity is important in the maintenance of H2BK5Ac and an epithelial morphology (Figure 1). Further, CBP mediated H2BK5Ac promotes proper timing of developmental EMT events, leading to skeletal formation and neural tube closure.
The transcription factor ATF2 also acetylates H2B (Figure 1). Using purified proteins, Kawasaki et al. demonstrated that ATF2 directly acetylates H2BK5 [40]. Another study found that when bound to amino acid response elements, ATF2 acetylates H2B [41]. Unlike CBP, neither ATF2 deletion nor mutation of ATF2 phosphorylation sites resulted in EMT-related developmental defects [42, 43]. These data suggest that compensation by other HATs may maintain H2BK5Ac in the absence of ATF2.
Histone deacetylases (HDACs) antagonize the activity of HATs by removing acetylation from lysine residues of histone tails. Loss of CBP is associated with loss of H2BK5Ac in several model systems, and inhibition of HDAC activity using broad-spectrum inhibitors (HDACi) partially restores H2BK5Ac. For example, HDACi partially restored H2A/H2BAc in lymphoblastoid cell lines from patients carrying heterozygous CBP or p300 mutations, demonstrating the dual-control of H2BAc by HATs and HDACs [37] (Figure 1). Overexpression of class I, IIa, and IIb HDACs has been implicated in several human cancers, making HDACi an attractive therapeutic strategy [44]. Selective inhibition of HDAC1 and HDAC2 has been shown to increase tumor H2BK5Ac in an epithelial human colorectal carcinoma (HCT116) xenograft model [45]. In multiple myeloma cell lines, treatment with entinostat, an inhibitor selective for HDAC1 and HDAC3, resulted in greater increases in H2BK5Ac than treatment with Merck60, an inhibitor selective for HDAC1 and HDAC2 [46]. Together, these studies demonstrate that H2BK5Ac can be controlled by HATs and HDACs, and that an overall shift towards hypoacetylation of histones is associated with several EMT related pathologies (Figure 1).
H2BK5Ac and Maintenance of Epithelial Stem Cell Phenotype
The dual-control of H2BK5Ac by HATs and HDACs has been demonstrated in detail in mouse TS cells. Mice with a targeted, inactivating mutation of MAP3K4 (K1361R) (KI4 mice) show EMT-related defects [3, 47]. In addition to failed neural tube closure, KI4 mice are born with craniofacial defects similar to Rubenstein-Taybi patients and abnormal axial skeleton patterning like CBP-deficient mice [47]. As stem cells, mouse TS cells can be cultured indefinitely and undergo EMT upon differentiation by withdrawal of FGF4 and mouse embryonic fibroblast conditioned medium. TS cells from KI4 mice (TSKI4 cells) retain stemness properties, but also display characteristics of EMT including mesenchymal morphology, loss of epithelial keratins and tight junction proteins, increased expression of EMT-TFs, and increased invasiveness [4, 32]. In vivo, TSKI4 cells exhibit hyperinvasiveness, resulting in defective decidualization [4]. TSKI4 cells show a loss of gene expression that overlaps significantly with that of mesenchymal claudin-low breast cancer lines, highlighting the commonality of EMT in development and disease [4].
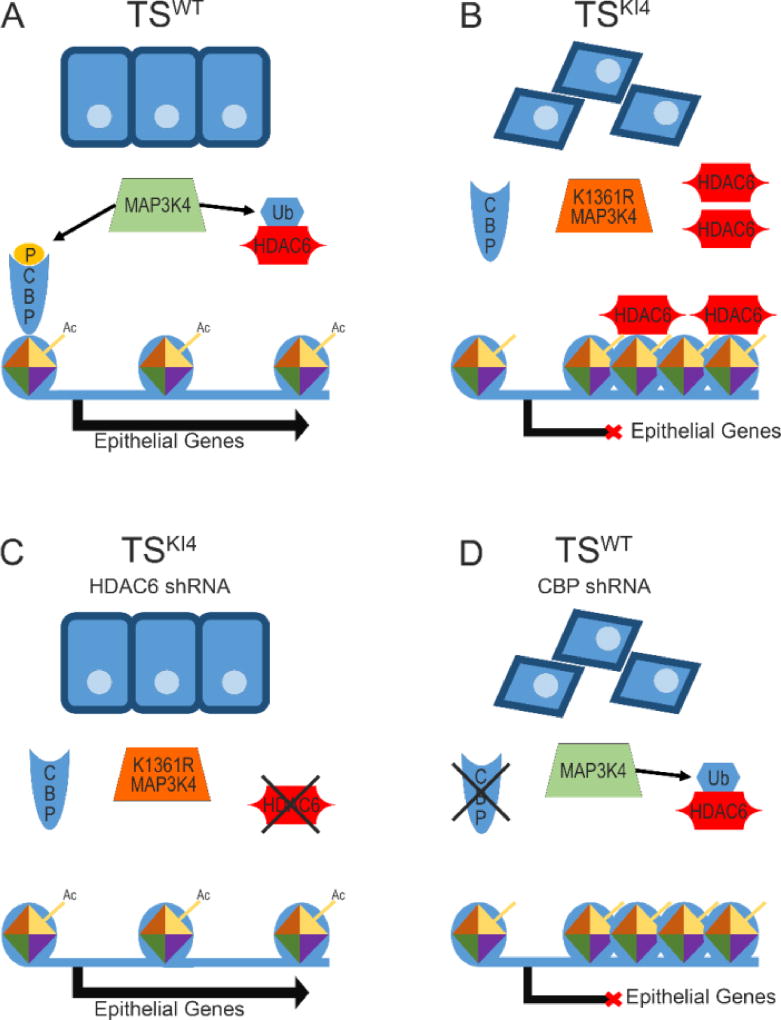
In TS cells CBP (HAT) and HDAC6 (HDAC) control H2BK5Ac [4, 32]. MAP3K4 phosphorylates and activates a signaling cascade that leads to the phosphorylation and activation of CBP. This pathway allows the epithelial TS cell phenotype to be maintained through CBP-mediated H2A/H2BK5Ac (Figure 2A). Additionally, MAP3K4 activity is required for the ubiquitination and destruction of HDAC6 [32]. Importantly, during normal TS cell differentiation-induced EMT HDAC6 message, protein, and activity levels are elevated [32]. Loss of MAP3K4 signaling in TSKI4 cells results in the loss of CBP-mediated acetylation of H2BK5 and an increase in HDAC6 protein and activity levels in the cytoplasm and nucleus [4, 32]. Increased cytoplasmic HDAC6 leads to decreased acetylation of tubulin [32]. In the nucleus, increased HDAC6 activity, coupled with decreased CBP HAT activity, leads to a global reduction in promoter H2BK5Ac and ultimately loss of epithelial gene expression. (Figure 2B) [4, 32]. shRNA knockdown of HDAC6 in TSKI4 cells restores H2BK5Ac at specific gene promotors, allowing the expression of genes important to the epithelial phenotype (Figure 2C) [32]. In TSKI4 cells with HDAC6 knockdown, acetylation is restored at the promoters of many genes important to the epithelial phenotype, including several tight junction components, Cldn6, Ocln, and Tjp1 [32]. HDAC6 knockdown in TSKI4 cells increases epithelial barrier formation that is lost in TSKI4 cells, indicating that epithelial function is also restored [32]. Conversely, TS cells with CBP knockdown show loss of epithelial features and loss of H2BK5Ac (Figure 2D) [4]. Relative to the embryonic stem cells of the inner cell mass, TS cells show low levels of H3K27me3, suggesting that TS cells rely on the deacetylation of histones to repress gene expression at stages before implantation [48]. These data illustrate a dual-control network of epigenetic regulation in a developmental model of EMT that relies on histone deacetylation to repress gene expression.
Figure 2. Modifiers of H2BK5Ac control the expression of epithelial genes in TS cells.
(A) In epithelial TSWT cells, MAP3K4 activity promotes the phosphorylation of the HAT, CBP, which acetylates histones at the promoters of epithelial genes. HDAC6 is targeted for destruction by MAP3K4-dependent ubiquitination, which prevents the deacetylation of the promoters of epithelial genes. The resulting euchromatin allows transcription of epithelial genes. (B) In mesenchymal TSKI4 cells, the loss of MAP3K4-activity leads to a loss of H2BK5Ac at the promoters of epithelial genes, condensing the chromatin. MAP3K4-dependent CBP phosphorylation is lost, and HDAC6 expression and activity increase due to decreased ubiquitination and destruction. (C) shRNA-mediated knockdown of HDAC6 in TSKI4 cells partially restores H2BK5Ac. The increase in H2BK5Ac promotes the formation of euchromatin, which favors the expression of genes maintaining the epithelial phenotype. Although CBP activity is still diminished, decreased HDAC6 expression is sufficient to reduce invasiveness and restore epithelial features like tight junction formation and epithelial barrier function. (D) shRNA-mediated knockdown of CBP in TSWT cells decreases H2BK5Ac and induces a mesenchymal phenotype. Decreased H2BK5Ac at the promoters of genes important to the epithelial phenotype condenses the chromatin and prevents transcription.
Stemness and EMT: Future Directions
The investigation of TS cells allows for the isolation and manipulation of stemness and EMT features in cells with a normal karyotype. TS cells regulate stemness and EMT using different sets of histone acetylation marks. Differentiated TS cells show loss of acetylation of all four core histones (H2A, H2B, H3, and H4) [4, 32]. However, the mesenchymal TSKI4 cells show selective loss of H2A/H2BAc and retain stemness features. Together, these data indicate that H2A/H2BK5Ac may be less important in maintaining the stem cell state and more important in promoting an epithelial phenotype, suggesting a role for H2BK5Ac in protection against EMT-related pathologies.
Improving our current understanding of EMT in pathologies like cancer requires an interdisciplinary effort to better understand the stable reactivation of developmental processes, the role of stemness, and how large-scale phenotypic changes are orchestrated by diseased cells in vivo. Some of the most malignant cancer cells exist in a stem-like state. These “cancer stem cells” produce stem-like tumor initiating cells and highly proliferative tumor-building cells, while displaying hallmarks of EMT [12]. Our work has led to the discovery of several genes with decreased promoter H2BK5Ac and expression in both mesenchymal TSKI4 cells and claudin low-breast cancer cells [4, 32]. There is great clinical value in understanding how cancer cells independently regulate stemness and EMT. However, epigenetic regulation of cancer cell stemness and EMT is undoubtedly more complex than that of the TS cells, and likely relies more heavily on gene repression by histone and DNA methylation. Cancer cells repress epithelial gene expression and activate mesenchymal gene expression through several epigenetic changes, including histone acetylation, histone methylation, DNA methylation, and nucleosome remodeling [49]. Further, a host of genetic mutations and chromosomal aberrations influence cancer cell gene expression. It is important to acknowledge that H2BK5Ac occurs in concert with a host of other histone modifications such as H2AK5Ac [4, 32]. Also, the specific nature of H2BK5Ac is difficult to define due to its multi-copy nature and multiple transcript variants. Specific roles have not been defined for each of the sixteen H2B isoforms, but it has been shown that the H2B isoforms have similar acetylation levels [50]. Further investigation in these fields of research will better define the significance of specific histone modifications in development and disease.
In an attempt to simplify a single aspect of pathological EMT, we have presented a basic, developmental model of EMT controlled by H2BK5Ac. Recent data demonstrate that important similarities between developmental and pathological EMT need to be investigated further. Our goal is that findings from the investigation of histone acetylation, stemness, and EMT in TS cells will translate into clinically relevant insight for EMT-related pathologies.
Acknowledgments
A.N.A. is supported by the Memphis Research Consortium and by the National Institutes of Health (grant GM116903).
Footnotes
Conflict of Interest: No conflicts declared.
References
- 1.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nieto MA, et al. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Abell AN, et al. Trophoblast stem cell maintenance by fibroblast growth factor 4 requires MEKK4 activation of Jun N-terminal kinase. Mol Cell Biol. 2009;29(10):2748–61. doi: 10.1128/MCB.01391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abell AN, et al. MAP3K4/CBP-regulated H2B acetylation controls epithelial-mesenchymal transition in trophoblast stem cells. Cell Stem Cell. 2011;8(5):525–37. doi: 10.1016/j.stem.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DaSilva-Arnold S, et al. Differentiation of first trimester cytotrophoblast to extravillous trophoblast involves an epithelial-mesenchymal transition. Placenta. 2015;36(12):1412–8. doi: 10.1016/j.placenta.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Krawczyk WS. A pattern of epidermal cell migration during wound healing. J Cell Biol. 1971;49(2):247–63. doi: 10.1083/jcb.49.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan C, et al. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor necrosis factor-alpha through bone morphogenic protein-2. Am J Pathol. 2010;176(5):2247–58. doi: 10.2353/ajpath.2010.090048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson LG, et al. Cutaneous wound reepithelialization is compromised in mice lacking functional Slug (Snai2) J Dermatol Sci. 2009;56(1):19–26. doi: 10.1016/j.jdermsci.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwano M, et al. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110(3):341–50. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grande MT, et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med. 2015;21(9):989–97. doi: 10.1038/nm.3901. [DOI] [PubMed] [Google Scholar]
- 11.Jolly MK, et al. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7(19):27067–84. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10(6):717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Abell AN, Johnson GL. Implications of Mesenchymal Cells in Cancer Stem Cell Populations: Relevance to EMT. Curr Pathobiol Rep. 2014;2(1):21–26. doi: 10.1007/s40139-013-0034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2014;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang RY, Guilford P, Thiery JP. Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J Cell Sci. 2012;125(Pt 19):4417–22. doi: 10.1242/jcs.099697. [DOI] [PubMed] [Google Scholar]
- 17.Wheelock MJ, et al. Cadherin switching. J Cell Sci. 2008;121(Pt 6):727–35. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 18.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154(1):8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 19.Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 20.Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum Genet. 1997;101(3):284–94. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- 21.Lin W, Dent SY. Functions of histone-modifying enzymes in development. Curr Opin Genet Dev. 2006;16(2):137–42. doi: 10.1016/j.gde.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H, et al. Quantitative proteomic analysis of histone modifications. Chem Rev. 2015;115(6):2376–418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 27.Chaffer CL, et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154(1):61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chitsazian F, Sadeghi M, Elahi E. Confident gene activity prediction based on single histone modification H2BK5ac in human cell lines. BMC Bioinformatics. 2017;18(1):67. doi: 10.1186/s12859-016-1418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marushige K. Activation of chromatin by acetylation of histone side chains. Proc Natl Acad Sci U S A. 1976;73(11):3937–41. doi: 10.1073/pnas.73.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DY, et al. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 31.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 32.Mobley RJ, et al. MAP3K4 Controls the Chromatin Modifier HDAC6 during Trophoblast Stem Cell Epithelial-to-Mesenchymal Transition. Cell Rep. 2017;18(10):2387–2400. doi: 10.1016/j.celrep.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen W, et al. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46(8):2100–10. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- 34.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–31. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68(6):1145–55. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 36.Roelfsema JH, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76(4):572–80. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Atalaya JP, et al. Histone acetylation deficits in lymphoblastoid cell lines from patients with Rubinstein-Taybi syndrome. J Med Genet. 2012;49(1):66–74. doi: 10.1136/jmedgenet-2011-100354. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka Y, et al. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci U S A. 1997;94(19):10215–20. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarcon JM, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–59. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki H, et al. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature. 2000;405(6783):195–200. doi: 10.1038/35012097. [DOI] [PubMed] [Google Scholar]
- 41.Bruhat A, et al. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Res. 2007;35(4):1312–21. doi: 10.1093/nar/gkm038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breitwieser W, et al. Feedback regulation of p38 activity via ATF2 is essential for survival of embryonic liver cells. Genes Dev. 2007;21(16):2069–82. doi: 10.1101/gad.430207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maekawa T, et al. Mouse ATF-2 null mutants display features of a severe type of meconium aspiration syndrome. J Biol Chem. 1999;274(25):17813–9. doi: 10.1074/jbc.274.25.17813. [DOI] [PubMed] [Google Scholar]
- 44.Spiegel S, Milstien S, Grant S. Endogenous modulators and pharmacological inhibitors of histone deacetylases in cancer therapy. Oncogene. 2012;31(5):537–51. doi: 10.1038/onc.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Methot JL, et al. Delayed and Prolonged Histone Hyperacetylation with a Selective HDAC1/HDAC2 Inhibitor. ACS Med Chem Lett. 2014;5(4):340–5. doi: 10.1021/ml4004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minami J, et al. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia. 2014;28(3):680–9. doi: 10.1038/leu.2013.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abell AN, et al. Ablation of MEKK4 kinase activity causes neurulation and skeletal patterning defects in the mouse embryo. Mol Cell Biol. 2005;25(20):8948–59. doi: 10.1128/MCB.25.20.8948-8959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rugg-Gunn PJ, et al. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci U S A. 2010;107(24):10783–90. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molden RC, et al. Multi-faceted quantitative proteomics analysis of histone H2B isoforms and their modifications. Epigenetics Chromatin. 2015;8:15. doi: 10.1186/s13072-015-0006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]