Abstract
Promoter methylation in a number of tumor-suppressor genes (TSGs) can play crucial roles in the development of thyroid carcinogenesis. The focus of the current meta-analysis was to determine the impact of promoter methylation of eight selected candidate TSGs on thyroid cancer and to identify the most important molecules in this carcinogenesis pathway. A comprehensive search was performed using Pub Med, Scopus, and ISI Web of Knowledge databases, and eligible studies were included. The methodological quality of the included studies was evaluated according to the Newcastle Ottawa scale table and pooled odds ratios (ORs); 95% confidence intervals (CIs) were used to estimate the strength of the associations with Stata 12.0 software. Egger’s and Begg’s tests were applied to detect publication bias, in addition to the “Metatrim” method. A total of 55 articles were selected, and 135 genes with altered promoter methylation were found. Finally, we included eight TSGs that were found in more than four studies (RASSF1, TSHR, PTEN, SLC5A, DAPK, P16, RARβ2, and CDH1). The order of the pooled ORs for these eight TSGs from more to less significant was CDH1 (OR = 6.73), SLC5 (OR = 6.15), RASSF1 (OR = 4.16), PTEN (OR = 3.61), DAPK (OR = 3.51), P16 (OR = 3.31), TSHR (OR = 2.93), and RARβ2 (OR = 1.50). Analyses of publication bias and sensitivity confirmed that there was very little bias. Thus, our findings showed that CDH1 and SCL5A8 genes were associated with the risk of thyroid tumor genesis.
Introduction
Thyroid cancer is the most common endocrine malignancy, and the incidence of thyroid cancer is increasing rapidly worldwide [1, 2]. Thyroid cancer includes several histological types and subtypes with various cellular origins and characteristics [3, 4] but is usually composed of two types of endocrine thyroid cells, i.e., follicular thyroid cells and para follicular C cells. The majority of thyroid malignancies, including papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), poorly differentiated thyroid cancer (PDTC), and anaplastic thyroid cancer (ATC), are derived from follicular thyroid cells[2, 4]. PTC and FTC are further classified into the differentiated thyroid cancer (DTC) group [5, 6].
Recently, scientists have demonstrated the involvement of genetic and epigenetic alterations in the development and progression of thyroid cancer. In addition to common gene mutations, such as mutations in BRAF [7–11], matrix metalloproteinase-2 [12], Ras family genes [13, 14], phosphatase and tensin homolog (PTEN) [15, 16], phosphatidylinositol 3-kinase (PIK3CA) [17, 18], anaplastic lymphoma kinase (ALK) [19], β-catenin 1 (CTNNB1) [20, 21], isocitrate dehydrogenase 1 (IDH1) [22], survivin [23], and epidermal growth factor receptor (EGFR) [24, 25], epigenetic alterations have also been shown to be important in thyroid cancer. Alterations in the genome by means of DNA methylation or histonemodification without altering the underlying DNA sequence, resulting in changes in the expression of target genes, are called epigenetic changes [26]. The most substantial epigenetic effects are observed by aberrant gene methylation, an epigenetic hallmark of human cancers, including thyroid cancer; methylation usually silences the gene when present in the promoter regions [27]. DNA methylation is a process in which methyl groups are added to the DNA molecule, altering the activity of the DNA segment without changing the nucleotide sequence. Several reports have shown that BRAF mutations are associated with hypermethylation of some TSGs, such as tissue inhibitor of metalloproteinases 3 (TIMP3), Ras association domain family member 1 (RASSF1), SLC5A, thyroid-stimulating hormone receptor (TSHR), death-associated proteinkinase1 (DAPK1), cyclin-dependent kinase inhibitor 2A (P16), and retinoic acid receptor-β (RARβ) [28–32]. Hypo methylation and over-expression of may genes are observed in various cancers [33].
In this study, we carried out a comprehensive meta-analysis of candidate genes associated with methylation in patients with thyroid cancer. Our results provide insights into the most effective TSGs, for which the promoter methylation has been considered a risk factor of progression towards thyroid carcinogenesis.
Materials and methods
The current meta-analysis was designed according to the latest version of the PRISMA checklist for meta-analysis guidelines S2 and S3 Figs.
Publication selection
This study (Prospero code: CRD42016033484) was conducted using PubMed, Scopus, and Web of Science search engines. Studies published between January 1, 2000 and October 1, 2016 were considered S1 Fig. The following key words were used: “methylation” or “hyper methylation” and “thyroid cancer” or “thyroid neoplasm” or “thyroid tumor” or “thyroid carcinoma” S4 Fig. Additionally, the references of the selected articles and related review articles were manually reviewed in order to identify any additional studies.
Inclusion and exclusion criteria
All nominated studies were reviewed by two authors independently. Studies that met our defined inclusion criteria were considered eligible for the meta-analysis. The purpose of our investigation was to identify definite gene promoter methylation in tumor tissue (fresh-frozen tissue, formalin-fixed paraffin-embedded [FFPE] samples, and plasma) from patients with thyroid cancer and normal sex- and age-matched controls. Normal controls were defined as normal adjacent tissue of thyroid cancer (NPTC), goiter, and nodular goiter (NG) samples. For normal controls, samples were the same or similar tissue type as those collected from patients with thyroid cancer. Methylation detection was based on methylation-specific polymerase chain reaction (MSP), quantitative MSP (QMSP), and combined bisulfite restriction analysis (COBRA) and methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) assays. All articles were published in English. Studies with insufficient data despite contacting the author were excluded.
Data collection
The following information was extracted from each study: first author, year of publication, country of research, type of sample, method for methylation determination, pathological stage, type of tissue (tumor and control), name of targeted gene(s), and frequency of promoter methylation in the target gene(s) in both tumor and normal thyroid tissues.
Quality assessment of individual studies
The quality of each study was assessed separately by two authors according to the Newcastle-Ottawa Scale (NOS) assessment tool [34]. Articles containing case control studies were scored according to the selection, comparability, and exposure. The assessment was made by scoring with stars ranging from zero to nine. Articles that scored six or more stars were qualified for inclusion into the meta-analysis.
Statistical analysis
Stata 12.0 (Stata Corporation, TX, and USA) was used in our meta-analysis. The odds ratios (ORs) and 95% confidence intervals (CIs) were applied to evaluate the association between promoter methylation of the eight TSGs and the risk of thyroid cancer [35]. Q-tests based on the χ2 and I2 statistics were used to investigate the heterogeneity among the studies [36–38]. If substantial heterogeneity existed (P<0.05 for the Q statistic or I2>50%), a random effect model was applied to pool the ORs; if not, a fixed effect model was applied [39, 40]. In addition, a meta-regression analysis was performed to discover the underlying reasons for statistical heterogeneity. Furthermore, subgroup analysis was conducted to determine the source of the heterogeneity. Sensitivity analysis was carried out to measure the effects of single studies on the overall estimate by ignoring one study at a time. A funnel plot, the trim-and-fill method, Begg’s test, and Egger’s test were applied to assess publication bias. All tests were two-sided, and results with P values of less than 0.05 were considered significant.
Results
Study selection and characteristics
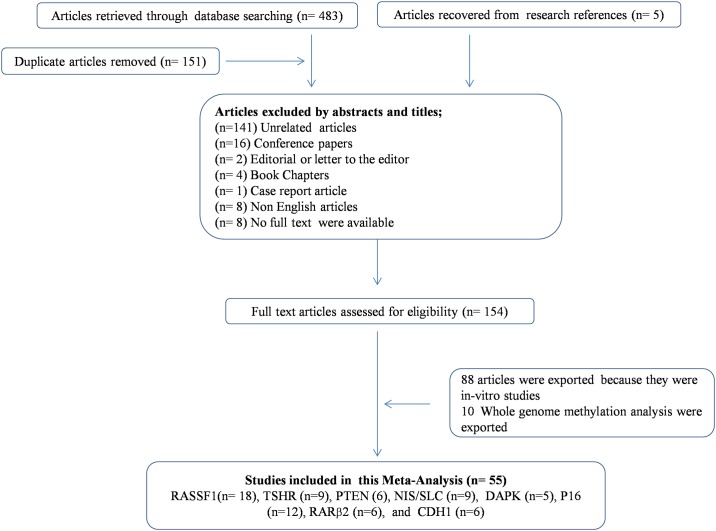
The strategies and results of study selection are presented in Fig 1. A total of 180 articles were excluded, and the remaining 154 related articles were analyzed further. Of these, 66 articles were case/control studies, and the remaining 88 articles were in- = vitro studies. Finally, 55 studies were chosen to evaluate promoter methylation and thyroid cancer because 10 studies were based on whole-genome methylation analysis. These 55 articles evaluated a total of 135 genes, including eight genes (RASSF1, TSHR, PTEN, electro genic sodium and chloride-dependent sodium-coupled solute transporter [SLC/NIS], DAPK, P16, RARβ2, and cadherin 1 [CDH1/E-cadherin] that have been shown to be associated with thyroid cancer in more than four studies; accordingly, we selected these eight TSGs for the meta-analysis. The frequency of promoter methylation in these eight TSGs was evaluated 55 studies; 18 studies evaluated RASSF1 (855 cases versus 379 controls), 12 studies evaluated P16 (626 cases versus 268 controls), nine studies evaluated TSHR (501 cases versus 258 controls), nine studies evaluated SLC/NIS (340 cases versus 201 controls), six studies evaluated PTEN (398 cases versus 189 controls), six studies evaluated RARβ2 (400 cases versus 132 controls), six studies evaluated CDH1 (355 cases versus 120 controls), and five studies evaluated DAPK (354 cases versus 117 controls) S1 Tables.
Fig 1. Flow diagram of study selection for the current meta-analysis.
Meta-analysis
Our results revealed that the frequency of promoter methylation of all eight TSGs increased in patients with thyroid cancer compared with that in controls under the random-effects model. Meta-analysis of the association between NIS hyper methylation and thyroid cancer, among 340 cases of thyroid cancer and 201 controls indicated a statistically significant difference (overall OR: 6.15, 95% CI: 2.62–14.40, p = 0.006). Subgroup analyses of NIS by the exact gene name indicated an OR of 7.33 for SLC5A8 and 4.31 for SLC5A5. Similar results were observed for the other seven genes, including RASSF1 (overall OR: 4.16, 95% CI: 2.57–6.73, p = 0.046), TSHR (overall OR: 2.93, 95% CI: 1.76–4.88, p = 0.133), P16 (overall OR: 3.31, 95% CI: 2.01–5.45, p = 0.345), PTEN (overall OR: 3.61, 95% CI: 0.70–18.76, p<0.001), DAPK (overall OR: 3.51, 95% CI: 0.41–30.16, p<0.001), CDH1(overall OR: 6.73, 95% CI: 3.29–13.75, p = 0.482), and RARβ2 (overall OR: 1.50, 95% CI: 0.94–2.41, p< 0.428), as shown in Fig 2. Subgroup analysis according to NOS quality grade for RASSF1 illustrated that studies with a quality of more than 7 yielded more specific thyroid cancer risk factors (OR: 2.67) in comparison with low-quality studies (OR: 7.39). Furthermore, subgroup meta-analyses of RASSF1 on the basis of the sampling method and tissue type showed that fresh-frozen tissues and paraffin-fixed embedded tissues were similar (OR: 4.32 and 3.37, respectively), in contrast to one study on blood samples (overall OR: 13.41, 95% CI: 4.00–44.97).
Fig 2. The result of meta-analysis (random-effects model.
Forest plot for evaluating the association between promoter methylation in the eight tumor-suppressor genes (RASSF1, P16, TSHR, PTEN, NIS/SLC, DAPK, RARβ2, and CDH1) and thyroid cancer risk. For RASSF1 and NIS/SLC, subgroup analyses are also presented. The random-effect model was used for all analyses.
Publication bias and sensitivity analysis
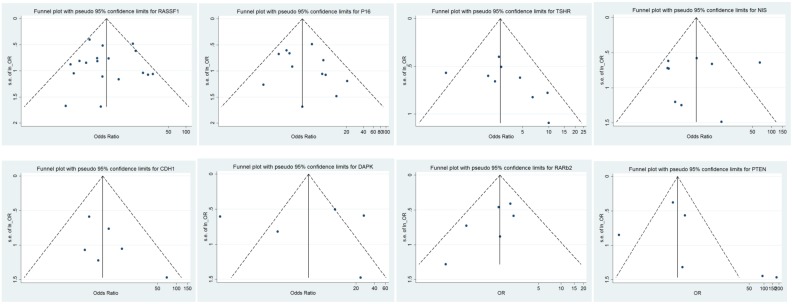
The publication bias of each individual TGS was evaluated separately using funnel plots, the trim-and-fill method, Begg’s linear regression test, and Egger’s linear regression. For RASSF1 and P16 genes (n > 10 reports each), funnel plots, Begg’s linear regression tests, and the trim-and-fill method were applied for analysis publication bias. However, for the other remaining six TSGs, funnel plots, Egger’s linear regression tests, and the trim-and-fill method were used for publication bias assessment because these TSGs were reported in less than 10 studies (Fig 3).
Fig 3. Funnel plot of publication bias.
Funnel plot for evaluating the association of promoter methylation of eight tumor-suppressor genes with thyroid cancer risk.
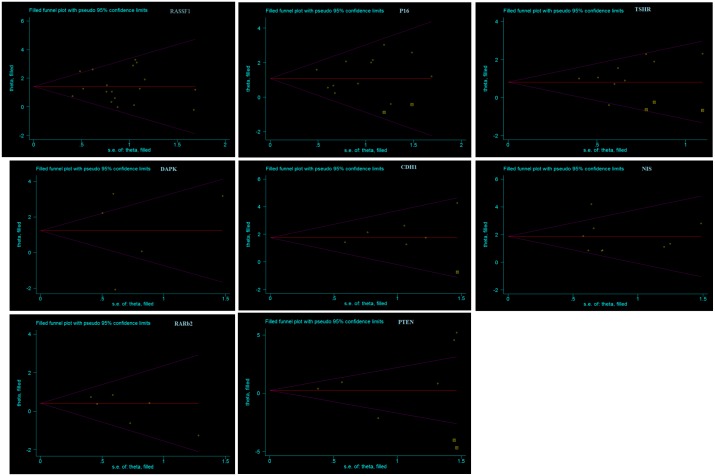
The results of Begg’s test and Egger’s test supported that no significant publication bias existed in our analysis of the association between promoter methylation in eight TSGs and thyroid cancer risk. Begg’s test results for RASSF1 (z = 0.08, p = 0.940) and P16 (z = 0.75, p = 0.451) and Egger’s graphs for TSHR, PTEN, CDH1, DAPK, SLC, and RARβ2 showed minimum bias. Sensitivity analysis by the trim-and-fill method specified that the results were stable in this meta-analysis, and removal of each study had no significant effect on the pooled ORs (Fig 4).
Fig 4. The result of Sensitivity analysis by the trim-and-fill method.
Funnel graph of the trim-and-fill method for evaluating publication bias and sensitivity for eight tumor-suppressor genes.
Discussion
In mammalian cells, epigenetic alternations play important role in regulation of gene expression. As the most common epigenetic alteration, DNA methylation usually occurs in CpG island regions of the gene promoter, causing activation or inactivation of gene function. The hypermethylation of TSG promoter suppresses genes expression by inhibiting transcription, thereby affecting cell signaling pathways. Two previous meta-analyses demonstrated the association between RASSF1A promoter methylation and PTC risk [31, 41]. In the current report, a comprehensive review was carried out to determine the relationship between thyroid cancer susceptibility and gene methylation. Among the eight TSGs identified in our study, hyper methylation in the promoter region of seven thyroid cancer-associated genes (RASSF1, TSHR, PTEN, SLC5A, DAPK, P16, and CDH1) was associated with an increased risk of thyroid cancer with an OR of 2 or more, and one gene (RARβ2) was associated with increased risk of thyroid cancer with an OR of 2 or less.
The CDH1 gene is a protein-coding gene that encodes a trans membrane glycoprotein localized in the adherents junctions of epithelial cells [42]. CDH1 gene inactivation by promoter methylation in cancer contributes to the increase in the proliferation, invasion, and metastasis of tumor cells [43–50]. For this gene, the pooled OR from six studies was 6.73, and only very minor publication bias and small study effects (p = 172) were observed, consistent with previous reports [44, 50, 51].
SLCs are a collection of membrane transport proteins comprising over 400 members [52, 53]. NIS member 5 (SLC5A5), also known as sodium/iodide co transporter or solute carrier family 5, is a protein encoded by the SLC5A5 gene in humans [54, 55]. This trans membrane glycoprotein (87 kDa and 13 trans membrane domains) transports two sodium cations (Na+) for each iodide anion (I–) into the cell [55]. In thyroid tissue, SLC5A5 plays a crucial role in thyroid hormone (TH) biosynthesis and uptake [56–58]. Our data showed that SLS5A5 was also identified as a risk factor (OR: 6.15). SLC5A is widely expressed in human tissues, including thyroid tissue [59–61] and is frequently methylated in human cancers [33–36], including thyroid cancer [61–64]. The SLC5A Meta bias test and resulting Egger graph showed that there was no the publication bias for association between the methylation of this gene and thyroid cancer. SLC5A8 as an another variant of SLC5 gene, transports iodide through a passive mechanism [65] and mono carboxylates short-chain fatty acids through a sodium-coupled mechanism [66]. SLC5A8 is a sodium-coupled mono carboxylate transporter; methylation of the SLC5A8 gene is essential in human cancers [67], and the protein encoded by this gene shows structural features of a sodium-iodide symporter with 48% homology to SLC5A5 (610 amino acids) [66]. We observed significant results for SLC5A5 and SLC5A8 in subgroup analysis using the exact SLC gene name. SLC5A8 showed the highest OR (7.33); therefore, SLC5A8 rather than CDH1 may be the most important gene showing hyper methylation in thyroid carcinogenesis.
Inactivation of the TSG RASSF1 has been reported in several studies [68]. RASSF1 protein contains a Ras-association domain and can control both the cell cycle and apoptosis pathways [69–71]. In some studies, RASSF1 promoter inactivation has been detected in more than 30% of thyroid tumors, representing the most and frequent event in thyroid cancers [72, 73]; however, in some reports, no significant correlation between RASSF1 methylation and thyroid cancer risk was detected [68, 74–78]. In two prior meta-analyses, researchers only examined the association between RASSF1 promoter methylation and PTC risk [31, 41]. Although the results of these prior meta-analyses demonstrated that the frequency of RASSF1 promoter methylation was significantly associated with increased risk of thyroid cancer, our study showed that the influence of RASSF1 promoter methylation was lower than those of CDH1 and SLC5A8 promoter methylation. Moreover, subgroup analysis demonstrated that RASSF1 was a risk factor with a higher OR in low-quality studies (OR: 7.39 versus 2.67). Additionally, subgroup analysis of the sampling source demonstrated major differences among blood samples, paraffin-fixed embedded tissues, and fresh-frozen tissues (OR: 5.65 versus 1.41 and 1.81). A significant association was also found for RASSF1 in both FTC and PTC [31]. The number of articles included in our meta-analysis of RASSF1 was 18 when conference presentations were excluded, whereas 12 studies, including two conference papers, were included in the previous study [31]. Thus, we could conclude that promoter methylation of RASSF1 was a risk factor for thyroid carcinogenesis, but that this gene was not as important as was previously thought. In contrast to our prediction, RASSF1 promoter hyper methylation was not the most important candidate for thyroid cancer association because higher ORs were observed for CDH1 and SLC5. The trim-and-fill method, which was not performed for RASSF1, implies that there was only very minor publication bias. Subgroup meta-analyses of RASSF1 according to the type of tissue showed that the fresh-frozen tissue and paraffin-fixed embedded tissue were similar, in contrast to blood samples. Due to the limited number of blood studies (only one), our investigation did not support the use of plasma or serum as the most reliable sample source for methylation analysis [79, 80].
The PTEN gene, encoding a phosphatase enzyme found in virtually all tissues, is located on chromosome 10q23.3 and has a major impact on the phosphatidylinositol 3-kinase (PI3K)/AKT pathway. In fact, PTEN functions to block this signaling pathway by converting PIP3 into PIP2, thereby regulating cell proliferation and differentiation [81, 82]. PTEN point mutations, deletions, and promoter methylation have been reported in thyroid carcinoma [83–87]. Our results showed that the pooled OR was 3.61, confirming the findings of previous studies. Funnel plots with the pseudo value of the 95% CI for PTEN, which covered the zero point in the Egger graph, detected no publication bias. Moreover, visual inspection of the funnel plot indicated no asymmetry, suggested that there was no publication bias in the evaluation of PTEN methylation and thyroid cancer risk. Moreover, Egger’s test did not provide statistical evidence of the asymmetry. However, application of the trim-and-fill method and addition of two studies caused the estimated OR to be similar to the original estimate, indicating the reliability and strength of our analyses.
DAPK is a large 160-kDa protein composed of various functional domains that interacts with cytoskeleton-associated serine/threonine kinase [88–90]. The tumor-suppressor function of the DAPK gene was distinguished, and methylation-mediated silencing was verified in many human cancers [91]. Our findings of the pooled OR of DAPK methylation and its association with thyroid cancer risk supported the involvement of this gene in thyroid tumor genesis, consistent with previous reports [29, 44, 46, 49, 85, 92]. A Meta bias study with Egger’s test showed that there was no small study effect (p = 976) and no publication bias.
The P16 (CDKN2a/INK4a) gene is an important TSG that is involved in the P16/cyclin-dependent kinase/retinoblastoma pathway and acts as a negative regulator of the cell cycle [93, 94]. The results of our pooled OR for P16, with the exclusion of Meta bias (which showed no publication bias), demonstrated that P16 was a thyroid cancer risk factor, consistent with previous studies [87, 95, 96], with the exception of one study showing opposite results [74].
The TSHR gene, located on chromosome 14q31, is a G protein-coupled receptor that regulates signaling across the cell membrane of follicular cells in the thyroid tissue. After activation by TSH, TSHR generates corresponding effects in the cell using second messengers. Deregulation of TSHR plays a crucial role in thyroid carcinogenesis [97–99]. Hyper methylation of the TSHR promoter is frequently found in thyroid carcinoma, although the promoter is un-methylated in normal and benign thyroid tumors [28, 85, 98–100]. The pooled OR in our study indicated that TSHR had an important role in thyroid cancer. Using Egger’s test results and the trim-and-fill method, we found that there was no publication bias.
In PTC, several methylation studies have shown that RARβ promoter methylation is significantly altered [28, 46, 101]. Quantitative assessments of RARβ and its association with BRAF mutations have revealed promoter methylation of this gene in thyroid tumor genesis [28, 29, 85, 92, 102]. RARβ has been shown to be associated with thyroid cancer recurrence; however, in our meta-analysis, it was not possible to examine this concept by subgroup analyses due to the low sample size and lack of data related to recurrence in the identified studies. Our results for pooled ORs highlighted the accuracy of the previous studies. Funnel plots with pseudo 95% CIs for RARβ2 and covering the zero point in Egger graphs detected no publication bias. Moreover, visual inspection of the funnel plots showed no asymmetry; therefore, there was no publication bias in our evaluation of PTEN methylation and thyroid cancer risk. Egger’s test also did not display statistical evidence of asymmetry (P = 0.115). The trim-and-fill method was applied to check the precision of our results.
In this meta-analysis, minimum publication bias was detected in the qualified studies for all eight TSGs. Begg’s test, Egger’s test, and funnel plots indicated that the data did not show a significant discrepancy among all studies. Furthermore, consistent results were found in the sensitivity analysis. However, there were several limitations to this analysis, including the influence of the small sample size in cases and controls and the non achievable cut-off point of methylation.
Conclusion
Taken together, our findings showed that promoter methylation of the CDH1 gene played an important role in thyroid cancer initiation and progression. Promoter methylation in this gene was found to be a promising biomarker for the early diagnosis of thyroid cancer. Other TSGs were also important, including SLC5A8.
Supporting information
(DOC)
The PRISMA (Preferred Reporting Items for Systematic Reviews and Meta_Analyses) Checklist that is a 27 checklist items pertain to the content of a systematic review and meta-analysis. Discussion.
(DOC)
A 19 checklist items pertain to the content of a systematic review and meta-analysis on genetic association studies.
(DOC)
The syntax and mesh terms that was used in this Meta-analysis.
(DOC)
The final candidate studies which were chosen for Meta-analysis.
(DOC)
Acknowledgments
Special thanks to Endocrinology and Metabolism Research Center, Endocrinology and Metabolism Clinical Sciences Institute, Tehran University of Medical Sciences, Tehran, Iran.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Shirazi HA, Hedayati M, Daneshpour MS, Shafiee A, Azizi F. Analysis of loss of heterozygsity effect on thyroid tumor with oxyphilia cell locus in familial non medullary thyroid carcinoma in Iranian families. Indian journal of human genetics. 2012;18(3):340–3. doi: 10.4103/0971-6866.107989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larijani B, Shirzad M, Mohagheghi M, Haghpanah V, Mosavi-Jarrahi A, Tavangar S, et al. Epidemiologic analysis of the Tehran cancer institute data system registry (TCIDSR). Asian Pac J Cancer Prev. 2004;5(1):36–9. [PubMed] [Google Scholar]
- 3.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13(3):184–99. doi: 10.1038/nrc3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haghpanah V, Soliemanpour B, Heshmat R, Mosavi-Jarrahi A, Tavangar S, Malekzadeh R, et al. Endocrine cancer in Iran: based on cancer registry system. Indian journal of cancer. 2006;43(2):80 [DOI] [PubMed] [Google Scholar]
- 5.Howlader N, Noone A, Krapcho M, Neyman N, Aminou R, Altekruse S, et al. SEER cancer statistics review, 1975–2009 (vintage 2009 populations). Bethesda, MD: National Cancer Institute; 2012:1975–2009. [Google Scholar]
- 6.Larijani B, Shirzad M, Mohagheghi M, Haghpanah V, Jarahi AM, Tavangar S, et al. Epidemiologic feature of thyroid cancer based on cancer registry data system. Iranian Journal of Public Health. 2005;34(4):1–7. [Google Scholar]
- 7.Trovisco V, Soares P, Preto A, de Castro IV, Lima J, Castro P, et al. Type and prevalence of BRAF mutations are closely associated with papillary thyroid carcinoma histotype and patients’ age but not with tumour aggressiveness. Virchows Archiv. 2005;446(6):589–95. doi: 10.1007/s00428-005-1236-0 [DOI] [PubMed] [Google Scholar]
- 8.Xing M, Westra WH, Tufano RP, Cohen Y, Rosenbaum E, Rhoden KJ, et al. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. The Journal of Clinical Endocrinology & Metabolism. 2005;90(12):6373–9. [DOI] [PubMed] [Google Scholar]
- 9.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocrine reviews. 2007;28(7):742–62. doi: 10.1210/er.2007-0007 [DOI] [PubMed] [Google Scholar]
- 10.Khatami F, Larijani B, Tavangar SM. Circulating Tumor BRAF Mutation and Personalized Thyroid Cancer Treatment. Asian Pac J Cancer Prev. 2017;18(2):293–4. doi: 10.22034/APJCP.2017.18.2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohammadi-Asl J, Larijani B, Khorgami Z, Tavangar S, Haghpanah V, Mehdipour P. Prevalence of BRAFV600E mutation in Iranian patients with papillary thyroid carcinoma: a single-center study. J Appl Sci. 2009;9(19):3593–7. [Google Scholar]
- 12.Sanii S, Saffar H, Tabriz HM, Qorbani M, Haghpanah V, Tavangar SM. Expression of matrix metalloproteinase-2, but not caspase-3, facilitates distinction between benign and malignant thyroid follicular neoplasms. Asian Pacific Journal of Cancer Prevention. 2012;13(5):2175–8. [DOI] [PubMed] [Google Scholar]
- 13.Howell GM, Hodak SP, Yip L. RAS mutations in thyroid cancer. The oncologist. 2013;18(8):926–32. doi: 10.1634/theoncologist.2013-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciampi R, Romei C, Pieruzzi L, Tacito A, Molinaro E, Agate L, et al. Classical point mutations of RET, BRAF and RAS oncogenes are not shared in papillary and medullary thyroid cancer occurring simultaneously in the same gland. Journal of Endocrinological Investigation. 2017;40(1):55–62. doi: 10.1007/s40618-016-0526-5 [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Ni Y, Saji M, Ringel MD, Jaini R, Eng C. Cowden syndrome-associated germline succinate dehydrogenase complex subunit D (SDHD) variants cause PTEN-mediated down-regulation of autophagy in thyroid cancer cells. Human molecular genetics. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alsina J, Alsina R, Gulec S. A Concise Atlas of Thyroid Cancer Next-Generation Sequencing Panel ThyroSeq v. 2. Molecular Imaging and Radionuclide Therapy. 2017;26(Suppl 1):102 doi: 10.4274/2017.26.suppl.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abubaker J, Jehan Z, Bavi P, Sultana M, Al-Harbi S, Ibrahim M, et al. Clinicopathological analysis of papillary thyroid cancer with PIK3CA alterations in a Middle Eastern population. The Journal of Clinical Endocrinology & Metabolism. 2008;93(2):611–8. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Hou P, Ji M, Guan H, Studeman K, Jensen K, et al. Highly prevalent genetic alterations in receptor tyrosine kinases and phosphatidylinositol 3-kinase/akt and mitogen-activated protein kinase pathways in anaplastic and follicular thyroid cancers. The Journal of Clinical Endocrinology & Metabolism. 2008;93(8):3106–16. [DOI] [PubMed] [Google Scholar]
- 19.Murugan AK, Xing M. Anaplastic thyroid cancers harbor novel oncogenic mutations of the ALK gene. Cancer research. 2011;71(13):4403–11. doi: 10.1158/0008-5472.CAN-10-4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Rostan G, Tallini G, Herrero A, Thomas G, Carcangiu ML, Rimm DL. Frequent mutation and nuclear localization of β-catenin in anaplastic thyroid carcinoma. Cancer research. 1999;59(8):1811–5. [PubMed] [Google Scholar]
- 21.Garcia-Rostan G, Camp RL, Herrero A, Carcangiu ML, Rimm DL, Tallini G. β-catenin dysregulation in thyroid neoplasms: down-regulation, aberrant nuclear expression, and CTNNB1 exon 3 mutations are markers for aggressive tumor phenotypes and poor prognosis. The American journal of pathology. 2001;158(3):987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murugan AK, Bojdani E, Xing M. Identification and functional characterization of isocitrate dehydrogenase 1 (IDH1) mutations in thyroid cancer. Biochemical and biophysical research communications. 2010;393(3):555–9. doi: 10.1016/j.bbrc.2010.02.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haghpanah V, Shooshtarizadeh P, Heshmat R, Larijani B, Tavangar SM. Immunohistochemical analysis of survivin expression in thyroid follicular adenoma and carcinoma. Applied Immunohistochemistry & Molecular Morphology. 2006;14(4):422–5. [DOI] [PubMed] [Google Scholar]
- 24.Murugan AK, Dong J, Xie J, Xing M. Uncommon GNAQ, MMP8, AKT3, EGFR, and PIK3R1 mutations in thyroid cancers. Endocrine pathology. 2011;22(2):97–102. doi: 10.1007/s12022-011-9155-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabriz HM, Adabi K, Lashkari A, Heshmat R, Haghpanah V, Larijani B, et al. Immunohistochemical analysis of nm23 protein expression in thyroid papillary carcinoma and follicular neoplasm. Pathology-Research and Practice. 2009;205(2):83–7. [DOI] [PubMed] [Google Scholar]
- 26.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes & Development. 2009;23(7):781–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing M. Gene methylation in thyroid tumorigenesis. Endocrinology. 2007;148(3):948–53. doi: 10.1210/en.2006-0927 [DOI] [PubMed] [Google Scholar]
- 28.Mohammadi-asl J, Larijani B, Khorgami Z, Tavangar SM, Haghpanah V, Kheirollahi M, et al. Qualitative and quantitative promoter hypermethylation patterns of the P16, TSHR, RASSF1A and RARβ2 genes in papillary thyroid carcinoma. Medical Oncology. 2011;28(4):1123–8. doi: 10.1007/s12032-010-9587-z [DOI] [PubMed] [Google Scholar]
- 29.Hu S, Liu D, Tufano RP, Carson KA, Rosenbaum E, Cohen Y, et al. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. International journal of cancer. 2006;119(10):2322–9. doi: 10.1002/ijc.22110 [DOI] [PubMed] [Google Scholar]
- 30.Hou P, Liu D, Xing M. Genome-wide alterations in gene methylation by the BRAF V600E mutation in papillary thyroid cancer cells. Endocrine-related cancer. 2011;18(6):687–97. doi: 10.1530/ERC-11-0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shou F, Xu F, Li G, Zhao Z, Mao Y, Yang F, et al. rassF1a promoter methylation is associated with increased risk of thyroid cancer: a meta-analysis. OncoTargets and therapy. 2017;10:247 doi: 10.2147/OTT.S124417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Rodero S, Delgado-Álvarez E, Díaz-Naya L, Nieto AM, Torre EM. Epigenetic modulators of thyroid cancer. Endocrinología, Diabetes y Nutrición. 2017. [DOI] [PubMed] [Google Scholar]
- 33.White MG, Nagar S, Aschebrook-Kilfoy B, Jasmine F, Kibriya MG, Ahsan H, et al. Epigenetic Alterations and Canonical Pathway Disruption in Papillary Thyroid Cancer: A Genome-wide Methylation Analysis. Annals of surgical oncology. 2016;23(7):2302–9. doi: 10.1245/s10434-016-5185-4 [DOI] [PubMed] [Google Scholar]
- 34.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25(9):603–5. doi: 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 35.Woolf B. On estimating the relation between blood group and disease. Annals of human genetics. 1955;19(4):251–3. [DOI] [PubMed] [Google Scholar]
- 36.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed). 2003;327(7414):557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled clinical trials. 1986;7(3):177–88. [DOI] [PubMed] [Google Scholar]
- 38.DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Statistics in medicine. 1996;15(12):1237–48; discussion 49–52. doi: 10.1002/(SICI)1097-0258(19960630)15:12<1237::AID-SIM301>3.0.CO;2-N [DOI] [PubMed] [Google Scholar]
- 39.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21(11):1539–58. doi: 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 40.Thakkinstian A, McElduff P, D'Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Statistics in medicine. 2005;24(9):1291–306. doi: 10.1002/sim.2010 [DOI] [PubMed] [Google Scholar]
- 41.Jiang JL, Tian GL, Chen SJ, Xu L, Wang HQ. Promoter methylation of p16 and RASSF1A genes may contribute to the risk of papillary thyroid cancer: A meta-analysis. Experimental and therapeutic medicine. 2015;10(4):1549–55. doi: 10.3892/etm.2015.2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends in biochemical sciences. 1999;24(2):73–6. [DOI] [PubMed] [Google Scholar]
- 43.Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. Journal of cell science. 2013;126(Pt 2):393–401. doi: 10.1242/jcs.100115 [DOI] [PubMed] [Google Scholar]
- 44.Wang D, Cui W, Wu X, Qu Y, Wang N, Shi B, et al. RUNX3 site-specific hypermethylation predicts papillary thyroid cancer recurrence. American journal of cancer research. 2014;4(6):725 [PMC free article] [PubMed] [Google Scholar]
- 45.Czarnecka K, Pastuszak-Lewandoska D, Migdalska-Sek M, Nawrot E, Brzezinski J, Dedecjus M, et al. Aberrant methylation as a main mechanism of TSGs silencing in PTC. Front Biosci (Elite Ed). 2011;3:137–57. [DOI] [PubMed] [Google Scholar]
- 46.Hoque M, Rosenbaum E, Westra W, Xing M, Ladenson P, Zeiger M, et al. Quantitative assessment of promoter methylation profiles in thyroid neoplasms. The Journal of Clinical Endocrinology & Metabolism. 2005;90(7):4011–8. [DOI] [PubMed] [Google Scholar]
- 47.Migdalska-Sek M, Pastuszak-Lewandoska D, Czarnecka K, Nawrot E, Domanska D, Brzezinski J, et al. Methylation profile of selected TSGs in non-cancerous thyroid tissue adjacent to primary PTC. Wspolczesna Onkol. 2011;15:191–7. [Google Scholar]
- 48.Rocha AS, Soares P, Seruca R, Máximo V, Matias-Guiu X, Cameselle-Teijeiro J, et al. Abnormalities of the E-cadherin/catenin adhesion complex in classical papillary thyroid carcinoma and in its diffuse sclerosing variant. The Journal of pathology. 2001;194(3):358–66. doi: 10.1002/path.905 [DOI] [PubMed] [Google Scholar]
- 49.Smith JA, Fan C-Y, Zou C, Bodenner D, Kokoska MS. Methylation status of genes in papillary thyroid carcinoma. Archives of Otolaryngology–Head & Neck Surgery. 2007;133(10):1006–11. [DOI] [PubMed] [Google Scholar]
- 50.Jensen K, Patel A, Hoperia V, Larin A, Bauer A, Vasko V. Dynamic changes in E-cadherin gene promoter methylation during metastatic progression in papillary thyroid cancer. Experimental and therapeutic medicine. 2010;1(3):457–62. doi: 10.3892/etm_00000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun W, Yang S, Ma Q, Zheng S, Wang J, Chen D, et al. SLUG promotes invasion and metastasis of anaplastic thyroid cancer cells through repression of E-cadherin. INTERNATIONAL JOURNAL OF CLINICAL AND EXPERIMENTAL PATHOLOGY. 2016;9(8):8373–9. [Google Scholar]
- 52.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Archiv: European journal of physiology. 2004;447(5):465–8. doi: 10.1007/s00424-003-1192-y [DOI] [PubMed] [Google Scholar]
- 53.Perland E, Fredriksson R. Classification Systems of Secondary Active Transporters. Trends in pharmacological sciences. 2017;38(3):305–15. doi: 10.1016/j.tips.2016.11.008 [DOI] [PubMed] [Google Scholar]
- 54.Dai G, Levy O, Carrasco N. Cloning and characterization of the thyroid iodide transporter. Nature. 1996;379(6564):458–60. doi: 10.1038/379458a0 [DOI] [PubMed] [Google Scholar]
- 55.Dohan O, De la Vieja A, Paroder V, Riedel C, Artani M, Reed M, et al. The sodium/iodide Symporter (NIS): characterization, regulation, and medical significance. Endocr Rev. 2003;24(1):48–77. doi: 10.1210/er.2001-0029 [DOI] [PubMed] [Google Scholar]
- 56.Ravera S, Reyna-Neyra A, Ferrandino G, Amzel LM, Carrasco N. The Sodium/Iodide Symporter (NIS): Molecular Physiology and Preclinical and Clinical Applications. Annual Review of Physiology. 2017;79:261–89. doi: 10.1146/annurev-physiol-022516-034125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kogai T, Brent GA. The sodium iodide symporter (NIS): regulation and approaches to targeting for cancer therapeutics. Pharmacology & therapeutics. 2012;135(3):355–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kogai T, Taki K, Brent GA. Enhancement of sodium/iodide symporter expression in thyroid and breast cancer. Endocr Relat Cancer. 2006;13(3):797–826. doi: 10.1677/erc.1.01143 [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez A-M, Perron B, Lacroix L, Caillou B, Leblanc G, Schlumberger M, et al. Identification and characterization of a putative human iodide transporter located at the apical membrane of thyrocytes. The Journal of Clinical Endocrinology & Metabolism. 2002;87(7):3500–3. [DOI] [PubMed] [Google Scholar]
- 60.Lacroix L, Pourcher T, Magnon C, Bellon N, Talbot M, Intaraphairot T, et al. Expression of the apical iodide transporter in human thyroid tissues: a comparison study with other iodide transporters. The Journal of Clinical Endocrinology & Metabolism. 2004;89(3):1423–8. [DOI] [PubMed] [Google Scholar]
- 61.Porra V, Ferraro-Peyret C, Durand C, Selmi-Ruby S, Giroud H, Berger-Dutrieux N, et al. Silencing of the tumor suppressor gene SLC5A8 is associated with BRAF mutations in classical papillary thyroid carcinomas. The Journal of Clinical Endocrinology & Metabolism. 2005;90(5):3028–35. [DOI] [PubMed] [Google Scholar]
- 62.Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, et al. Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene. 2005;24(44):6646–56. doi: 10.1038/sj.onc.1208822 [DOI] [PubMed] [Google Scholar]
- 63.Zane M, Agostini M, Enzo MV, Ide EC, Del Bianco P, Torresan F, et al. Circulating cell-free DNA, SLC5A8 and SLC26A4 hypermethylation, BRAF V600E: A non-invasive tool panel for early detection of thyroid cancer. Biomedicine & Pharmacotherapy. 2013;67(8):723–30. [DOI] [PubMed] [Google Scholar]
- 64.Makhlouf A-M, Chitikova Z, Pusztaszeri M, Berczy M, Delucinge-Vivier C, Triponez F, et al. Identification of CHEK1, SLC26A4, c-KIT, TPO and TG as new biomarkers for human follicular thyroid carcinoma. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodriguez AM, Perron B, Lacroix L, Caillou B, Leblanc G, Schlumberger M, et al. Identification and characterization of a putative human iodide transporter located at the apical membrane of thyrocytes. The Journal of clinical endocrinology and metabolism. 2002;87(7):3500–3. doi: 10.1210/jcem.87.7.8797 [DOI] [PubMed] [Google Scholar]
- 66.Gopal E, Fei YJ, Sugawara M, Miyauchi S, Zhuang L, Martin P, et al. Expression of slc5a8 in kidney and its role in Na(+)-coupled transport of lactate. The Journal of biological chemistry. 2004;279(43):44522–32. doi: 10.1074/jbc.M405365200 [DOI] [PubMed] [Google Scholar]
- 67.Hong C, Maunakea A, Jun P, Bollen AW, Hodgson JG, Goldenberg DD, et al. Shared epigenetic mechanisms in human and mouse gliomas inactivate expression of the growth suppressor SLC5A8. Cancer Res. 2005;65(9):3617–23. doi: 10.1158/0008-5472.CAN-05-0048 [DOI] [PubMed] [Google Scholar]
- 68.Schagdarsurengin U, Gimm O, Hoang-Vu C, Dralle H, Pfeifer GP, Dammann R. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer research. 2002;62(13):3698–701. [PubMed] [Google Scholar]
- 69.Khaled H, Al Lahloubi N, Rashad N. A review on thyroid cancer during pregnancy: Multitasking is required. Journal of Advanced Research. 2016;7(4):565–70. doi: 10.1016/j.jare.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.St. Bernard R, Zheng L, Liu W, Winer D, Asa SL, Ezzat S. Fibroblast growth factor receptors as molecular targets in thyroid carcinoma. Endocrinology. 2005;146(3):1145–53. doi: 10.1210/en.2004-1134 [DOI] [PubMed] [Google Scholar]
- 71.Kondo T, Zheng L, Liu W, Kurebayashi J, Asa SL, Ezzat S. Epigenetically controlled fibroblast growth factor receptor 2 signaling imposes on the RAS/BRAF/mitogen-activated protein kinase pathway to modulate thyroid cancer progression. Cancer research. 2007;67(11):5461–70. doi: 10.1158/0008-5472.CAN-06-4477 [DOI] [PubMed] [Google Scholar]
- 72.Allen N, Donninger H, Vos M, Eckfeld K, Hesson L, Gordon L, et al. RASSF6 is a novel member of the RASSF family of tumor suppressors. Oncogene. 2007;26(42):6203–11. doi: 10.1038/sj.onc.1210440 [DOI] [PubMed] [Google Scholar]
- 73.Hesson LB, Cooper WN, Latif F. The role of RASSF1A methylation in cancer. Disease markers. 2007;23(1–2):73–87. doi: 10.1155/2007/291538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brait M, Loyo M, Rosenbaum E, Ostrow KL, Markova A, Papagerakis S, et al. Correlation between BRAF mutation and promoter methylation of TIMP3, RARβ2 and RASSF1A in thyroid cancer. Epigenetics. 2012;7(7):710–9. doi: 10.4161/epi.20524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xing M, Cohen Y, Mambo E, Tallini G, Udelsman R, Ladenson PW, et al. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer research. 2004;64(5):1664–8. [DOI] [PubMed] [Google Scholar]
- 76.Lee J-J, Geli J, Larsson C, Wallin G, Karimi M, Zedenius J, et al. Gene-specific promoter hypermethylation without global hypomethylation in follicular thyroid cancer. International journal of oncology. 2008;33(4):861 [PubMed] [Google Scholar]
- 77.Nakamura N, Carney JA, Jin L, Kajita S, Pallares J, Zhang H, et al. RASSF1A and NORE1A methylation and BRAFV600E mutations in thyroid tumors. Laboratory investigation. 2005;85(9):1065–75. doi: 10.1038/labinvest.3700306 [DOI] [PubMed] [Google Scholar]
- 78.QU F, XUE W. RASSF1A methylation and its clinical roles in papillary thyroid carcinoma. Journal of Nantong University (Medical Sciences). 2012;6:016. [Google Scholar]
- 79.Khatami F, Aghayan HR, Sanaei M, Heshmat R, Tavangar SM, Larijani B. The Potential of Circulating Tumor Cells in Personalized Management of Breast Cancer: A Systematic Review. Acta Med Iran. 2017;55(3):175–93. [PubMed] [Google Scholar]
- 80.Khatami F, Larijani B, Tavangar S. Circulating Tumor BRAF Mutation and Personalized Thyroid Cancer Treatment. Asian Pacific journal of cancer prevention: APJCP. 2017;18(2):293 doi: 10.22034/APJCP.2017.18.2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guerra A, Di Crescenzo V, Garzi A, Cinelli M, Carlomagno C, Tonacchera M, et al. Genetic mutations in the treatment of anaplastic thyroid cancer: a systematic review. BMC surgery. 2013;13(2):S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nozhat Z, Hedayati M. PI3K/AKT pathway and its mediators in thyroid carcinomas. Molecular diagnosis & therapy. 2016;20(1):13–26. [DOI] [PubMed] [Google Scholar]
- 83.Hou P, Ji M, Xing M. Association of PTEN gene methylation with genetic alterations in the phosphatidylinositol 3-kinase/AKT signaling pathway in thyroid tumors. Cancer. 2008;113(9):2440–7. doi: 10.1002/cncr.23869 [DOI] [PubMed] [Google Scholar]
- 84.Ng K, Shin V, Leung CP, Chan VW, Law FBF, Siu MT, et al. Elevation of methylated DNA in KILLIN/PTEN in the plasma of patients with thyroid and/or breast cancer. OncoTargets and therapy. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schagdarsurengin U, Gimm O, Dralle H, Hoang-Vu C, Dammann R. CpG island methylation of tumor-related promoters occurs preferentially in undifferentiated carcinoma. Thyroid. 2006;16(7):633–42. doi: 10.1089/thy.2006.16.633 [DOI] [PubMed] [Google Scholar]
- 86.Alvarez-Nuñez F, Bussaglia E, Mauricio D, Ybarra J, Vilar M, Lerma E, et al. PTEN promoter methylation in sporadic thyroid carcinomas. Thyroid. 2006;16(1):17–23. doi: 10.1089/thy.2006.16.17 [DOI] [PubMed] [Google Scholar]
- 87.Chang H, Shin B, Kim A, Kim H, Kim B. DNA methylation analysis for the diagnosis of thyroid nodules–a pilot study with reference to BRAFV600E mutation and cytopathology results. Cytopathology. 2015. [DOI] [PubMed] [Google Scholar]
- 88.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9(1):15–30. [DOI] [PubMed] [Google Scholar]
- 89.Bialik S, Kimchi A. The death-associated protein kinases: structure, function, and beyond. Annual review of biochemistry. 2006;75:189–210. doi: 10.1146/annurev.biochem.75.103004.142615 [DOI] [PubMed] [Google Scholar]
- 90.Ivanovska J, Mahadevan V, Schneider-Stock R. DAPK and cytoskeleton-associated functions. Apoptosis: an international journal on programmed cell death. 2014;19(2):329–38. [DOI] [PubMed] [Google Scholar]
- 91.Schneider-Stock R, Roessner A, Ullrich O. DAP-kinase—protector or enemy in apoptotic cell death. The international journal of biochemistry & cell biology. 2005;37(9):1763–7. [DOI] [PubMed] [Google Scholar]
- 92.Zhang B, Liu S, Zhang Z, Wei J, Qu Y, Wu K, et al. Analysis of BRAF V600E mutation and DNA methylation improves the diagnostics of thyroid fine needle aspiration biopsies. Diagnostic pathology. 2014;9(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ai L, Stephenson KK, Ling W, Zuo C, Mukunyadzi P, Suen JY, et al. The p16 (CDKN2a/INK4a) tumor-suppressor gene in head and neck squamous cell carcinoma: a promoter methylation and protein expression study in 100 cases. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2003;16(9):944–50. [DOI] [PubMed] [Google Scholar]
- 94.Boltze C, Zack S, Quednow C, Bettge S, Roessner A, Schneider-Stock R. Hypermethylation of the CDKN2/p16 INK4A promotor in thyroid carcinogenesis. Pathology-Research and Practice. 2003;199(6):399–404. [DOI] [PubMed] [Google Scholar]
- 95.Ishida E, Nakamura M, Shimada K, Higuchi T, Takatsu K, Yane K, et al. DNA hypermethylation status of multiple genes in papillary thyroid carcinomas. Pathobiology. 2007;74(6):344–52. doi: 10.1159/000110028 [DOI] [PubMed] [Google Scholar]
- 96.Lam AKY, Lo CY, Leung P, Lang BHH, Chan WF, Luk JM. Clinicopathological roles of alterations of tumor suppressor gene p16 in papillary thyroid carcinoma. Annals of surgical oncology. 2007;14(5):1772–9. doi: 10.1245/s10434-006-9280-9 [DOI] [PubMed] [Google Scholar]
- 97.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nature Reviews Cancer. 2013;13(3):184–99. doi: 10.1038/nrc3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kartal K, Onder S, Kosemehmetoglu K, Kilickap S, Tezel YG, Kaynaroglu V. Methylation status of TSHr in well-differentiated thyroid cancer by using cytologic material. BMC cancer. 2015;15(1):824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khan MS, Pandith AA, Masoodi SR, Wani KA, Hussain MU, Mudassar S. Epigenetic silencing of TSHR gene in thyroid cancer patients in relation to their BRAF V600E mutation status. Endocrine. 2014;47(2):449–55. doi: 10.1007/s12020-014-0319-6 [DOI] [PubMed] [Google Scholar]
- 100.Xing M, Usadel H, Cohen Y, Tokumaru Y, Guo Z, Westra WB, et al. Methylation of the thyroid-stimulating hormone receptor gene in epithelial thyroid tumors. Cancer Research. 2003;63(9):2316–21. [PubMed] [Google Scholar]
- 101.Kiseljak-Vassiliades K, Xing M. Association of cigarette smoking with aberrant methylation of the tumor suppressor gene RARβ2 in papillary thyroid cancer. Frontiers in endocrinology. 2011;2:99 doi: 10.3389/fendo.2011.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoque M, Rosenbaum E, Westra W, Xing M, Ladenson P, Zeiger M, et al. Quantitative assessment of promoter methylation profiles in thyroid neoplasms (vol 90, pg 4011, 2005). JOURNAL OF CLINICAL ENDOCRINOLOGY & METABOLISM. 2006;91(9):3278-. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
The PRISMA (Preferred Reporting Items for Systematic Reviews and Meta_Analyses) Checklist that is a 27 checklist items pertain to the content of a systematic review and meta-analysis. Discussion.
(DOC)
A 19 checklist items pertain to the content of a systematic review and meta-analysis on genetic association studies.
(DOC)
The syntax and mesh terms that was used in this Meta-analysis.
(DOC)
The final candidate studies which were chosen for Meta-analysis.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.