Abstract
A complex of two related mammalian proteins, SFPQ and NONO, promotes DNA double-strand break repair via the canonical nonhomologous end joining (c-NHEJ) pathway. However, its mechanism of action is not fully understood. Here we describe an improved SFPQ•NONO-dependent in vitro end joining assay. We use this system to demonstrate that the SFPQ•NONO complex substitutes in vitro for the core c-NHEJ factor, XLF. Results are consistent with a model where SFPQ•NONO promotes sequence-independent pairing of DNA substrates, albeit in a way that differs in detail from XLF. Although SFPQ•NONO and XLF function redundantly in vitro, shRNA-mediated knockdown experiments indicate that NONO and XLF are both required for efficient end joining and radioresistance in cell-based assays. In addition, knockdown of NONO sensitizes cells to the interstrand crosslinking agent, cisplatin, whereas knockdown of XLF does not, and indeed suppresses the effect of NONO deficiency. These findings suggest that each protein has one or more unique activities, in addition to the DNA pairing revealed in vitro, that contribute to DNA repair in the more complex cellular milieu. The SFPQ•NONO complex contains an RNA binding domain, and prior work has demonstrated diverse roles in RNA metabolism. It is thus plausible that the additional repair function of NONO, revealed in cell-based assays, could involve RNA interaction.
INTRODUCTION
Canonical nonhomologous end joining (c-NHEJ) is the default pathway for repair of DNA double-strand breaks (DSBs) in mammals (reviewed in (1,2)). Ku protein and the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) form an initial complex that promotes pairing of DNA ends and coordinates enzymatic processing (3–9). DNA-PKcs then undergoes autophosphorylation and dissociates, concomitant with recruitment of DNA ligase IV and its accessory factors, XRCC4 and XLF (10–12). These proteins form a nucleoprotein filament that promotes pairing and alignment of the DNA ends for ligation (9,13–19). Formation of this XLF-mediated filament is dispensable in some cell types, however, suggesting there could be a second, redundant mechanism that promotes substrate pairing in the ligation phase of the reaction (20).
In work that predated the discovery of XLF, we identified a heterodimer of two related proteins, SFPQ and NONO, as a candidate NHEJ factor. The purified SFPQ•NONO complex stimulates DNA ligase IV•XRCC4-mediated end joining by 10-fold or more in vitro (21,22). Although the mechanism of stimulation is incompletely understood, SFPQ has a DNA binding domain that mediates direct association of SFPQ•NONO dimers with the repair substrate. Structural studies, primarily of related complexes, indicate the potential for interaction between adjacent DNA-bound dimers to form a repeated, filamentous structure, which could serve as a platform for biomolecular interactions (23). Separately, biochemical assays using isolated SFPQ and SFPQ•NONO reveal properties that could be relevant to repair function. SFPQ, presumably as a homodimer, promotes homology-dependent DNA strand exchange in vitro (24–27). Other work shows that the SFPQ•NONO complex promotes capture of one DNA by another in a surface tethering assay and stimulates DNA-PKcs autophosphorylation in a protein kinase assay, both of which are consistent with an ability to promote association or aggregation of DNA substrates prior to ligation (28).
Cell and animal studies are consistent with a bona fide physiological role of SFPQ and NONO in DSB repair, although, again, the exact mechanism has not been established. Genetic deficiency in NONO, or mutations affecting DNA binding activity of SFPQ, are associated with a number of DNA damage sensitivity phenotypes. These include reduced NHEJ activity in a chromosomal reporter assay, delayed resolution of radiation-induced repair foci, increased frequency of radiogenic chromosome aberrations, and sensitivity to radiation-induced cell killing (29–33). An epistatic relationship between NONO deficiency and DNA-PKcs inhibition indicates that NONO is specifically involved in the c-NHEJ pathway of DSB repair (33). We recently extended these observations in vivo by demonstrating that Sertoli cells of NONO-deficient mice are compromised with respect to DSB repair, leading to hypersensitivity of male germ cells to radiation-induced cell death (Li, S., Shu, F.J., Li, Z., Jaafar, L., Zhao, S., Dynan, W.S. (2016) NONO (p54nrb), a multifunctional nuclear protein, protects male germ cells against radiation-induced cell death in vivo. Submitted). Consistent with a mechanism involving direct interaction with the repair substrate, the SFPQ•NONO complex mobilizes to sites of laser-induced DNA damage (30–32).
Interpretation of cell-based and in vivo studies is complicated, however, because SFPQ and NONO have a number of functions in addition to DSB repair. Together with a third protein, PSPC1, they make up the Drosophila Behavior, Human Splicing (DBHS) gene family, members of which form dimers in various combinations. All family members bind RNA via a shared globular domain that also makes up the dimerization interface, which is separable from the DNA binding domain unique to SFPQ. DBHS proteins bind regulatory noncoding RNAs, inosine-containing RNAs, pre-mRNA 5΄ splice sites, and other targets (reviewed in (23,34)). Regulation of genes involved in brain development and tumor progression are among many functions that have been reported (35–37). SFPQ and NONO do not appear to regulate DSB repair protein expression, based on a number of genes that have been analyzed (27,33). Nevertheless, it is difficult to exclude that some aspects of the DNA damage sensitivity phenotype associated with SFPQ or NONO deficiency arise indirectly, via effects on gene regulation.
In current work, we sought to clarify the mechanism by which SFPQ•NONO stimulates DSB repair in vitro and relate this to the currently well-established consensus model for c-NHEJ. Using an improved in vitro end joining system reconstituted from recombinant factors, we show that the SFPQ•NONO complex substitutes for the core c-NHEJ factor, XLF, on a nearly mole-for-mole basis, suggesting that SFPQ•NONO provides an alternative mechanism to facilitate DNA substrate pairing during the ligation phase of the reaction. We also performed gene knockdown studies to examine the relationship between the NONO subunit of the SFPQ•NONO complex and XLF in cells. In contrast to their redundant function in vitro, NONO and XLF have an epistatic relationship with respect to radiation survival and plasmid DNA end joining. This suggests that, in addition to a common function in promoting DNA pairing, each factor makes additional contributions to DSB repair in cells, which for NONO could involve RNA binding.
MATERIALS AND METHODS
Protein expression and purification
Repair factors were expressed Escherichia coli and purified by immobilized metal affinity chromatography, followed in most cases by Superdex S200 gel filtration. Procedures for expression and purification of factors other than SFPQ•NONO were based on modifications of existing protocols (38–41). Details are provided in Supplementary Data.
In vitro DNA end joining assays
Except as noted, reactions were performed using a 1503 bp BglII-linearized DNA fragment, which was propagated in an E. coli minicircle system (System Biosciences, Mountain View, CA, USA). Reactions were performed in a volume of 20 μl and contained 50 mM triethanolamine–HCl, 10 mM Tris–HCl (pH 7.9), 50 mM KOAc, 0.1 mM dithiothreitol, 5 mM Mg(OAc)2, 50 ng/μl bovine serum albumin, 1 nM ATP, 5 nM DNA, and repair proteins as indicated in the individual figure legends. Reactions were incubated for 1 h at room temperature and terminated by addition of 1 μl of 1% SDS and heating for 10 min at 70°C. Products (1 μl aliquot from each reaction) were analyzed using an Agilent DNA 12000 kit (Agilent Technologies, Santa Clara, CA, USA).
Gene silencing
Gene silencing was performed using lentiviral shRNA vectors (Sigma, St. Louis, MO, USA). Two shRNAs were tested for each gene and the more effective was used (for NONO, NM_007363, TRCN0000286693
(dCCGGGCCAGAATTCTACCCTGGAAACTCGAGTTTCCAGGGTAGAATTCTGGCTTTTTG); for XLF, NM_024782, TRCN0000275632
(dCCGGTACCATGGACTTTAGGTATATCTCGAGATATACCTAAAGTCCATGGTATTTTT)). The control shRNA was SHC002 (Sigma). Lentiviruses were generated by Lipofectamine 3000-mediated triple co-transfection of human HEK293FT cells with shRNA plasmid, psPAX2 (Addgene # 12260), and pMD2.G (Addgene #12259). Media was changed at 18 h, cells were incubated for an additional 30 h, and virus-containing supernatant was harvested and centrifuged to remove debris (1000g at 4°C for 15 min). Attenuation of protein expression was confirmed by immunoblotting using rabbit anti-NONO (GeneTex, GTX63618), rabbit anti-XLF (Bethyl Laboratories, A300-730A), rabbit anti-β-actin (Sigma, A5060), and horseradish peroxidase-conjugated anti-rabbit IgG secondary antibodies (GE Healthcare, NA 934V). Membranes were developed using Enhanced Chemiluminescence substrate (GE Healthcare) and were visualized using X-ray film.
Clonogenic survival assays.
Human (HeLa) cells were plated in 6-well plates and infected with lentiviral shRNA vectors (1 ml viral stock/well). After 60 h, cells were trypsinized, counted, and seeded in six-well plates at densities ranging from 150 to 10 000 cells per well. Cells were allowed to attach for 5–8 h. Cells were then exposed to 2–6 Gy of 137Cs γ-rays at a dose rate of 0.6 Gy min−1 or to 0.2–0.8 μM cisplatin. Incubation was continued at 37°C in a humidified 5% CO2 atmosphere for 10 days with one change of medium, fixed with 100% MeOH, and stained with 0.5% crystal violet in 25% methanol. Colonies of ≥50 cells were counted.
In vivo plasmid rejoining assay
Plasmid rejoining assays were performed as described (42). HeLa cells were infected with lentiviral shRNA vectors as described in the preceding section. After 48 h, cells were trypsinized and 2 μg of EcoRV- and AfeI-digested plasmid pDVG94 was introduced by electroporation (Nucleofector II, Amaxa). Cells were plated, and after a further 48 h, episomal DNA was recovered by Hirt extraction and junctional sequences were amplified as described (42). Amplified DNA was digested with BstXI to distinguish c-NHEJ products (resistant) and alternative, microhomology mediated end joining products (sensitive). We noted that the resistant product consistently migrates as a closely-spaced doublet in the Bioanalyzer system, which would not have been resolved in earlier studies using conventional electrophoresis and 32P labeling (42). The doublet was assumed to represent conformational isomers of the junctional fragment, as the bands are present in variable stoichiometry, it was present only in reporter-transfected cells, and it was seen with two different primer sets (data not shown).
53BP1 focus assay
HeLa cells were plated on eight-well chamber slides and infected with lentiviral shRNA vectors. At 48 h post-infection, cells received 1 Gy of 137Cs gamma radiation at a dose rate of 0.6 Gy min−1. They were allowed to recover for 30 min or 4 h, fixed with 4% paraformaldehyde, permeabilized with PBS containing 0.5% Triton X-100, and blocked with PBS containing 10% goat serum. Samples were stained using rabbit anti-53BP1 (Novus Biologicals, NB100-904, 1:1000) and Alexa Fluor 594-conjugated secondary antibody and counterstained with 4΄,6-diamidino-2-phenylindole. Images were collected using a DeltaVision microscope, deconvolved, and projected, and the number of foci per cell was scored.
Reproducibility and statistical analysis
Cell survival assays were performed in triplicate, and differences between treatment groups were analyzed by Student's t-test. In vitro end joining assays were performed at least twice with similar results, and results of a single representative experiment are shown. Repair factors were prepared at least twice independently and the purity and activity were demonstrated to be similar.
RESULTS
SFPQ•NONO and XLF independently stimulate end joining in vitro
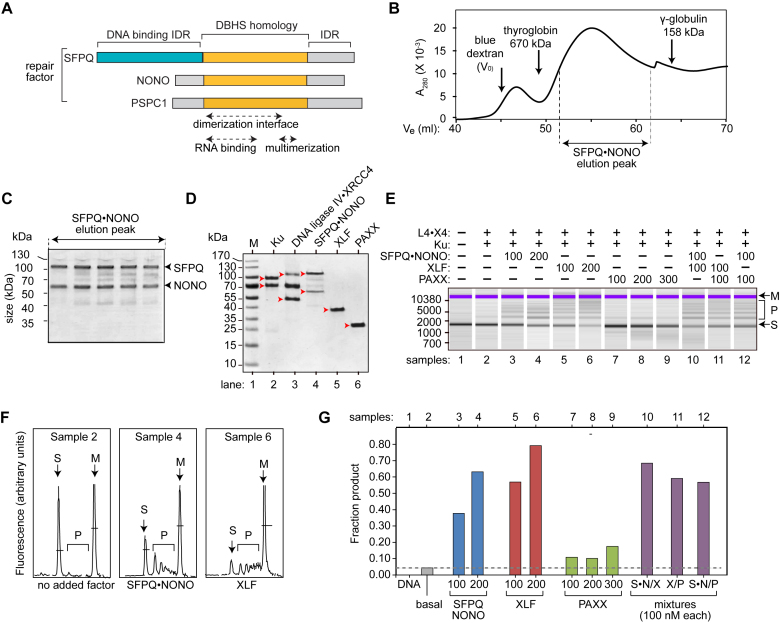
Figure 1A shows the relevant structural features of SFPQ, NONO and the related protein, PSPC1, which together make up the DBHS family. All three proteins contain a region of homology that mediates RNA binding, dimerization, and higher-order multimerization, flanked by divergent N- and C-terminal intrinsically disordered regions (IDRs) (23). Although family members form mixed dimers in all possible combinations, only SFPQ-containing complexes have DNA binding activity, which is mediated by unique sequences in the N-terminal IDR (31,43).
Figure 1.
SFPQ•NONO and XLF independently stimulate end joining in vitro. (A) Alignment of human DBHS proteins. DBHS homology region (orange) mediates RNA binding, dimerization, and higher-order multimerization. N-terminal IDR of SFPQ (teal) mediates DNA binding. Other IDRs (grey) are shown. (B) SFPQ•NONO Superdex 200 gel filtration profile, showing A280 as a function of elution volume. Positions of SFPQ•NONO elution peak and molecular weight standards (run independently) are indicated. Refer to Supplementary Data for selectivity curve. (C) SDS-PAGE analysis of indicated region of gel filtration profile. Positions of SFPQ and NONO are indicated at right, and molecular weight markers at left (kDa). (D) SDS-PAGE analysis of NHEJ factor preparations. Red arrowheads indicate relevant polypeptides. The 70 kDa species in the DNA ligase IV•XRCC4 preparation is believed to be an unrelated contaminant. (E) Results of end joining reactions. Assays contained 100 nM of 1503 bp BglII-linearized DNA substrate, 100 nM DNA ligase IV•XRCC4 (L4•X4), 100 nM Ku, and other factors as indicated (nM). Size markers are indicated at left (bp). Substrate (S), product (P), and upper dye marker (M, 17 000 bp) are indicated at right. (F) Selected samples from Panel E, displayed as electropherogram traces. Peaks are labeled as in panel E. All panels are presented at the same scale. (G) Fraction of substrate converted to product. Mixtures are as follows S•N/X, SFPQ•NONO/XLF; X/P, XLF/PAXX; S•N/P, SFPQ•NONO/PAXX. Dotted line indicates level of activity in basal reactions without stimulatory factors.
For the current study, we developed an improved end joining assay system based entirely on E. coli-expressed repair factors. We co-expressed full-length SFPQ and NONO in E. coli and purified the soluble dimer by sequential affinity and gel filtration chromatography (Figure 1B and C). The physical properties of the recombinant complex were indistinguishable from the native HeLa cell factor used in previous studies (21,22,28). The complex eluted from the gel filtration column as a distinct peak with the two subunits present at 1:1 stoichiometry. Like the native complex (22), the recombinant factor eluted from the gel filtration column earlier than expected relative to globular protein standards (Figure 1B and Supplementary Data), perhaps reflecting an extended conformation adopted by the IDRs. Other core c-NHEJ factors were expressed in E. coli by modifications of existing methods (Figure 1D). These included Ku protein, DNA ligase IV•XRCC4, XLF, and PAXX, a recently described XRCC4/XLF paralog (41,44). DNA-PKcs was not used in the current experiments, as it is not essential and does not further stimulate SFPQ•NONO-dependent end joining (21).
End-joining reactions were performed using a 1503 bp linear DNA substrate, and products were resolved by a radiolabel-free capillary electrophoresis method. There was very little end joining in basal reactions containing Ku and DNA ligase IV•XRCC4 alone (Figure 1E, Sample 2), whereas addition of SFPQ•NONO increased the efficiency of the reaction by up to 15-fold in a concentration-dependent manner (samples 3–4). In separate reactions, addition of XLF had a very similar effect (samples 5–6). PAXX was much less active under the conditions used (samples 7–9). End joining was quantified by analysis of the Bioanalyzer peaks (Figure 1F). Notably, the stimulatory activities of SFPQ•NONO and XLF were comparable (within a factor of 2) on a mole-for-mole basis (Figure 1G).
There was no apparent synergy among SFPQ•NONO, XLF and PAXX. End-joining activity with mixtures of factors was equal to or less than the sum of their individual activities (Figure 1G). To exclude the possibility that the failure to observe synergy was because of saturation of the reaction, we also tested SFPQ•NONO and XLF at lower concentrations, and the activity of mixtures was again less than the sum of that in reactions containing the factors individually (Supplementary Data). The findings that SFPQ•NONO and XLF are interchangeable, and that the activity of mixtures is no more than additive, suggest that the two factors supply the same function. XLF is known to promote DNA substrate pairing, and by inference SFPQ•NONO does the same.
The in vitro end-joining products consisted primarily of linear concatemers, similar to those seen with native HeLa-cell derived SFPQ•NONO (21). At the DNA concentrations used, circular products should be favored (45), and they were seen in reactions containing T4 DNA ligase (Supplementary Data). One interpretation is that multi-copy binding of SFPQ•NONO or XLF enforces an extended DNA conformation, decreasing the efficiency of circularization. Multi-copy binding of Ku protein may have a similar effect, as concatemerization was also favored under basal conditions, in the absence of SFPQ•NONO and XLF.
Dependence of end joining efficiency on DNA end concentration
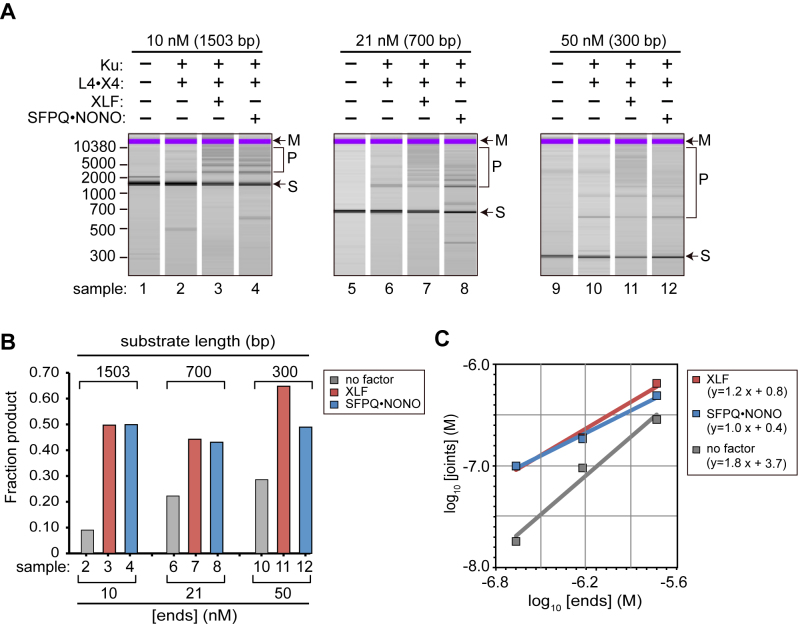
To gain further insight into the mechanism of SFPQ•NONO versus XLF-stimulated end joining, we compared the efficiency of end joining with various DNA substrates. We first tested the effect of DNA fragment length, under conditions where total DNA concentration was held constant. Substrate length varied five-fold, from 300 to 1503 bp. In effect, the experiment provided a way to vary the concentration of DNA ends while holding the overall protein:DNA ratio constant. That is, shorter substrates provide a higher molar concentration of DNA ends.
In basal reactions lacking SFPQ•NONO and XLF, increases in the DNA end concentration had quite a large effect on end joining, as expected for a bimolecular reaction requiring independent association of two substrate DNAs with the ligase (Figure 2A and B, samples 2, 6, 10). The dependence on substrate is even more evident when the raw data (fraction of substrate converted to product) are adjusted for differences in the initial numbers of DNA ends to calculate molar yield of DNA joints. A log-log plot of molar yield versus DNA end concentration has a slope of 1.8, indicating a nearly second-order dependency on DNA end concentration (Figure 2C).
Figure 2.
Dependence of end joining on DNA end concentration. (A) End-joining reactions were performed as in Figure 1 using equal masses of each of the indicated substrates. DNA end concentrations are indicated (nM). Reactions contained 100 nM of the indicated factors. Labeling as in Figure 1. (B) Quantification of Panel A, plotted as fraction substrate converted to product. (C) Quantification of Panel A, showing molar concentration of product (joints) versus initial concentration of substrate (ends) on a log–log scale.
In reactions containing SFPQ•NONO or XLF, the efficiency of end joining was less dependent on DNA end concentration. The fraction of substrate converted to product was approximately constant for the three substrates tested (Figure 2A and B). When adjusted for differences in initial numbers of DNA ends, log–log plots of molar yield versus DNA end concentration have slopes of 1.0–1.2, consistent with a first order dependency on DNA end concentration (Figure 2C).
Although the dataset is limited, there was a clear difference between the basal reactions and those containing SFPQ•NONO or XLF. Data here are consistent with the idea that SFPQ•NONO promote substrate pairing: because pairing increases the local concentration of one DNA end in the presence of another, reaction efficiency is less dependent on concentration of DNA ends in bulk solution.
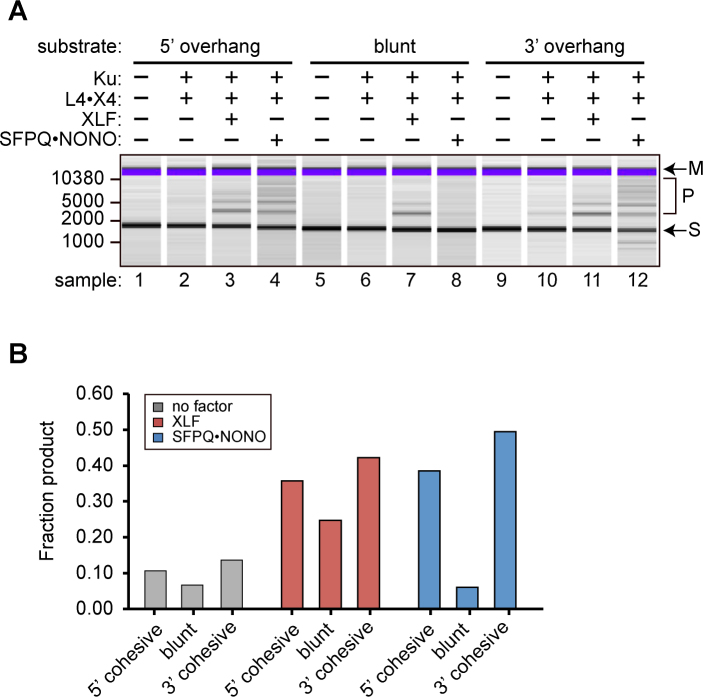
Joining of cohesive versus blunt ends
We next performed an experiment using substrates of the same length, but with different DNA end structures. Complementary single-strand overhangs promote transient base pairing, which helps position the DNA ends in the ligase active site. XLF, which cooperates with DNA ligase IV and XRCC4 to enforce precise end-to-end alignment in paired complexes (9,13–19), might obviate a requirement for cohesive ends. Whether SFPQ•NONO would do the same is unclear, given that it does not appear to contact the extreme termini (22,28).
Consistent with the structural models for XLF-directed substrate pairing, reactions containing XLF showed only a modest preference for cohesive ends (Figure 3A and B). Reactions containing SFPQ•NONO, by contrast, showed a very marked preference of cohesive versus blunt ends, suggesting that the geometry of the paired complex is different and does not force the ends into precise alignment. As shown in a subsequent experiment, NONO (and by implication the SFPQ•NONO complex) is able to promote joining of blunt-end DNA fragments in a cell-based assay. Its inability to do so in vitro could reflect the minimal nature of the biochemical assay system, which lacks additional factors that might hold the ends into alignment in the cellular environment.
Figure 3.
Dependence of end joining on DNA end structure. Substrates were of uniform 1503 bp length and had cohesive 5΄ overhang (BglII), blunt (SmaI), or cohesive 3΄ overhang (SacI) ends. (A) Results of end joining assay. Reactions contained 100 nM of indicated factors. Labeling as in Figure 1. (B) Quantification of results from panel A.
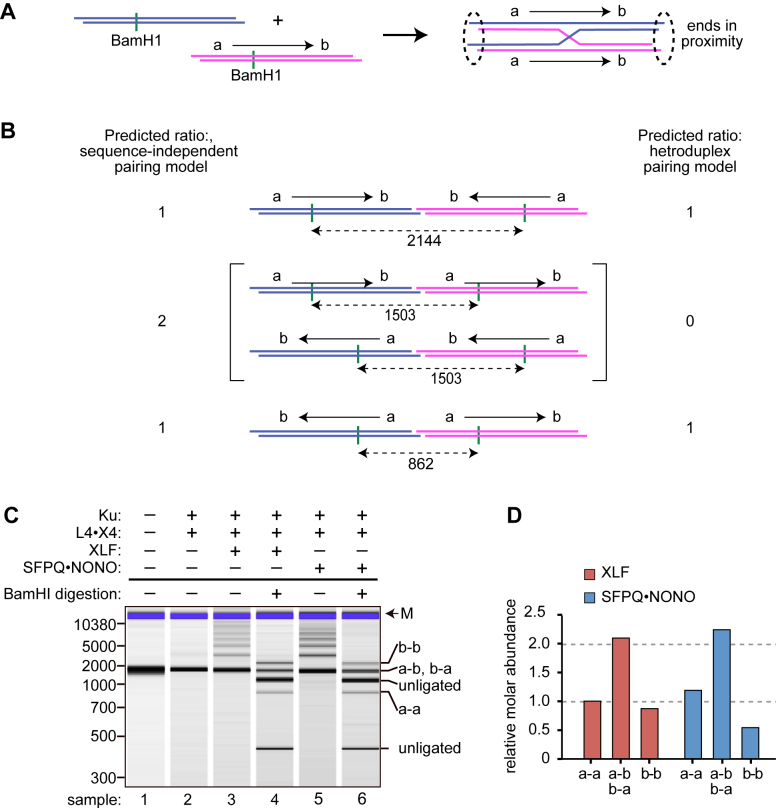
Sequence-independent pairing mechanism
Isolated SFPQ has a demonstrated potential to promote sequence-dependent DNA strand exchange and D-loop formation (24–27), and one hypothesis is that SFPQ•NONO- mediated substrate pairing involves formation of a transient heteroduplex intermediate between internally homologous sequences in the two substrate molecules. This model is attractive for end joining in vitro, where all substrate molecules are identical, and could also be relevant in a physiological setting (see Discussion). Heteroduplex formation constrains pairing to a parallel sequence orientation, which introduces a preference for joining to create inverted repeats (ab-ba and ba-ab joints in the nomenclature of Figure 4). By contrast, a protein-mediated, sequence-independent pairing model predicts a random half-and-half mixture of inverted and direct repeats (ab-ba, ab-ab, ba-ba, and ba-ab in Figure 4).
Figure 4.
Polarity of end joining. (A) Heteroduplex pairing model. Ends are designated as ‘a’ or ‘b’ with arrow denoting a to b polarity. Substrates have an off-center BamHI site to facilitate analysis of product distribution. In this model, pairing is constrained to a side-by-side, parallel arrangement, favoring a-a and b-b junctions. (B) End joining in different relative orientations creates diagnostic BamHI fragments as indicated. Distribution of reaction products discriminates between a heteroduplex pairing model and a sequence-independent, protein-mediated pairing model. (C) Results of end joining assay. Reactions contained 100 nM of indicated factors. Indicated samples were subjected to limit digestion with BamHI. Positions of size markers are indicated at left (bp), and diagnostic BamHI fragments arising from unligated substrate and different types of joints at right. (D) Quantification of results from panel C.
To distinguish these models, we performed standard end joining reactions in the presence of SFPQ•NONO or XLF. Products were cleaved with BamHI at an off-center site to produce restriction fragments that are diagnostic of each type of junction (Figure 4B). Digestion products were resolved by capillary electrophoresis (Figure 4C) and relative molar yields were determined (Figure 4D). Data show an almost exactly equal mixture of inverted and direct repeats, consistent with a sequence-independent, random joining model for both SFPQ•NONO and XLF. Results strongly disfavor any model where DNA fragments are constrained in a particular sequence orientation prior to ligation.
Genetic interaction between NONO and XLF in cell survival assays
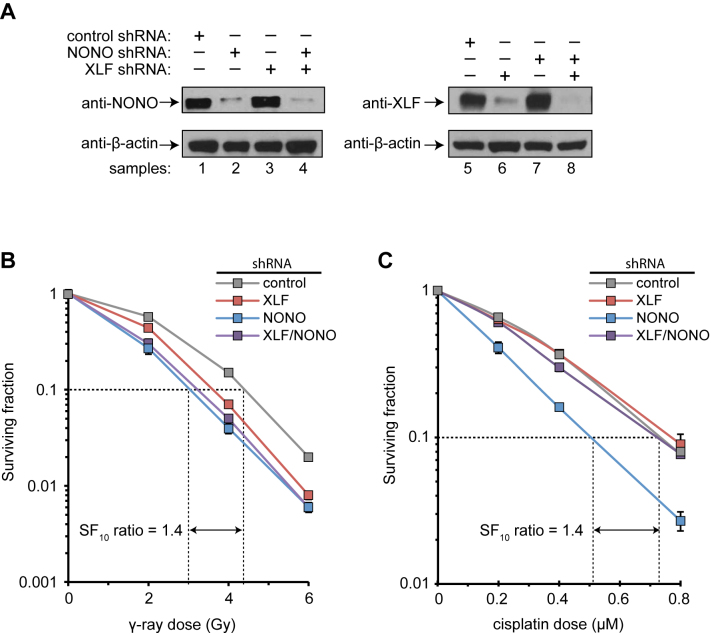
We next sought to determine the relationship between SFPQ•NONO and XLF-directed end joining in cells. If the two factors operate independently, dual deficiency should produce a stronger phenotype than deficiency in either one individually. If they cooperate in a single pathway, dual deficiency should produce the same phenotype as individual deficiency; that is, the factors will have an epistatic relationship. This and subsequent genetic analyses focused on the NONO subunit of the SFPQ•NONO complex because NONO deficiency is tolerated in human cells, whereas SFPQ deficiency is not (31,46). We used previously validated lentiviral shRNAs, which reduced NONO and XLF expression efficiently and selectively at the protein and mRNA levels (Figure 5A, Supplementary Data). Transduction with NONO shRNA did not affect expression of XLF and vice versa.
Figure 5.
Clonogenic survival assays. (A) Lentiviral shRNA-mediated knockdown of NONO and XLF. Proteins from total cell lysates were resolved by SDS-PAGE and immunoblotting was performed separately with anti-NONO, anti-XLF, and anti β-actin as a loading control. Panels are from same experiment. (B) Survival following γ-ray exposure at indicated doses. HeLa cells were transduced with indicated lentiviral shRNA vectors, exposed after 48 h to 137Cs γ-rays, and incubated to allow colony formation. Surviving fraction was normalized to reflect plating efficiency of non-irradiated control cells. Assays were performed in triplicate. Error bars (smaller than symbols in some groups) denote standard deviation. Dotted lines indicate dose reduction factor (NONO versus control shRNA) at 10% survival. (C) Survival following cisplatin exposure at indicated concentrations for 1 h. Results were pooled from two independent experiments, each performed in triplicate, for a total of six replicates for each condition. Surviving fraction was normalized to plating efficiency of control cells in each experiment. Error bars denote standard deviation.
We performed standard clonogenic survival assays using γ-rays, which kill cells primarily through induction of DSBs. Attenuation of NONO expression resulted in a modest but significant dose reduction factor of 1.4 at 10% survival (Figure 5B), an effect size that is consistent with prior studies (29,32). Attenuation of XLF expression had approximately the same effect. The strength of the XLF phenotype was less than with XLF knockout cells (47), which could reflect either residual expression or a cell line-specific difference. Dual deficiency in NONO and XLF had the same effect as deficiency in NONO alone, indicating an epistatic relationship where both factors cooperate in a single pathway in cells.
Prior work has shown that the genetic relationship between DSB repair factors can differ for ionizing radiation versus chemical agents that introduce more complex lesions (44). To investigate whether this was the case for XLF and NONO, we performed an additional survival experiment using cisplatin, a widely used chemotherapy agent that produces interstrand crosslinks and other lesions, which can lead indirectly to strand breaks. Attenuation of XLF expression had no effect on cisplatin survival, whereas attenuation of NONO resulted in a dose reduction factor of 1.4, similar to that for γ-rays (Figure 5C). Unexpectedly, attenuation of XLF and NONO together suppressed the cisplatin-sensitivity phenotype. The results suggest that XLF antagonizes a NONO-dependent mechanism that promotes cisplatin survival (see Discussion).
Genetic interaction between NONO and XLF in DSB repair assays
The epistatic relationship between NONO and XLF in γ-ray survival assays, and previous work showing an epistatic relationship between NONO and DNA-PKcs inhibition (33), indicate specific involvement of NONO in the c-NHEJ pathway. To confirm and extend this finding, we also performed plasmid rejoining assays that discriminate between c-NHEJ and a separate pathway of alternative, or microhomology-mediated end joining (MHMEJ). MHMEJ relies on a different ensemble of repair factors and serves as a slower backup pathway of DSB repair in c-NHEJ-deficient cells. Prior work has shown that XLF deficiency results in a shift from c-NHEJ to MHMEJ (47), and our hypothesis was that NONO deficiency would do the same.
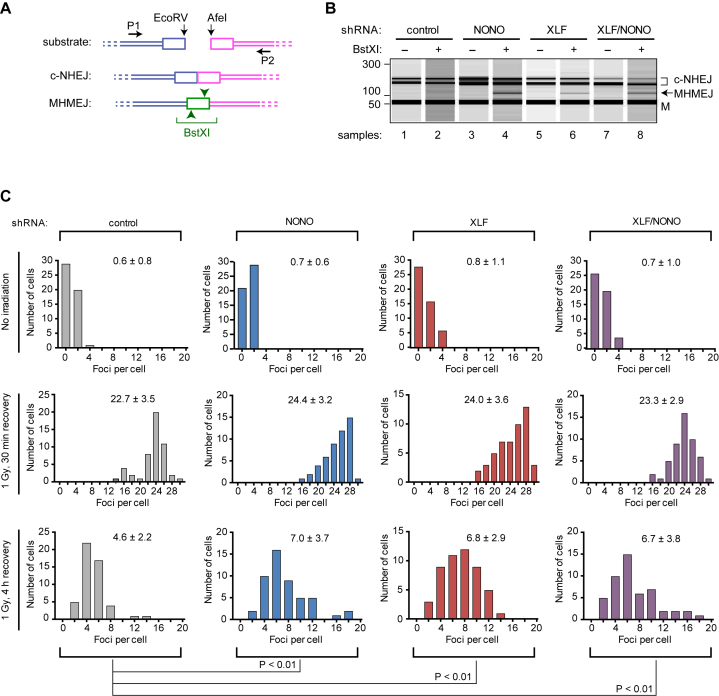
To measure pathway choice, cells were transfected with a linearized plasmid that has a six-nucleotide block of sequence identity at each end (42). Repair by c-NHEJ results in precise rejoining to create a tandem direct repeat (Figure 6A). Repair by MHMEJ involves limited 5΄ resection of DNA ends, annealing of 3΄ ends via the exposed region of microhomology, and ligation resulting in the loss of precisely one copy of the 6 bp repeat. Closed circular DNAs were recovered and the junctions were amplified by PCR. The MHMEJ-associated deletion creates a novel BstXI site, which can be used to discriminate between c-NHEJ and MHMEJ products.
Figure 6.
DSB repair assays. (A) Plasmid rejoining assay design. Substrate is a linearized plasmid with EcoRV and AfeI ends. Boxes denote 6 bp of identical sequence at each end. Precise c-NHEJ-mediated joining results in a 6 bp direct repeat. Microhomology-mediated end joining (MHMEJ) results in a 6 bp deletion, leaving one copy of the repeat and creating a novel BstXI site. Positions of PCR primers are indicated. Drawing is not to scale. (B) Results of plasmid rejoining assay. HeLa cells were transduced with indicated lentiviral shRNA vectors and after 48 h transfected with EcoRV/AfeI-linearized reporter plasmid. After a further 48 h, episomal DNA was recovered by Hirt extraction, junctional sequences were amplified by PCR, and products were resolved using an Agilent Bioanalyzer. Indicated samples were subjected to limit digestion with BstXI. Positions of BstXI-resistant PCR product (180 bp, resolves as doublet, see text), larger BstXI cleavage product (120 bp), and lower size marker (M, 50 bp) are indicated. Lower size marker obscures the smaller BstXI cleavage product. (C) Results of repair foci assay. Assays were performed using HeLa cells as described in Materials and Methods. Cells were irradiated and allowed to recover as indicated, processed, and 50 cells per treatment group were scored for 53BP1 foci. Results are presented as histograms, with mean number of foci per cell and standard deviation indicated. Statistical analysis was performed by ANOVA.
Attenuation of NONO or XLF expression, individually or in combination, resulted in the appearance of a diagnostic 120 bp BstXI product, which indicates a shift toward MHMEJ (Figure 6B, samples 4 and 6). The shift was less complete than in a previous study using XLF-deficient HCT116 cells (47), but is nonetheless readily apparent. We noted that the 180 bp BstXI-resistant product runs as a doublet with variable stoichiometry. The lower band migrates as expected, and the upper band perhaps represents a conformational isomer (see Materials and Methods). Precise quantification was difficult because of low signal and variable background, but it appears that dual deficiency in NONO and XLF did not increase the severity of the phenotype, again indicating an epistatic relationship.
We also measured the effect of attenuation of NONO and XLF expression on repair of chromosomal DSBs, using 53BP1 foci as a marker of unrepaired DSBs. Prior work in mammalian cells has shown that the number of 53BP1 foci reaches a maximum at 30 min post-irradiation and declines thereafter (48–50). The number of residual foci at 4 h post-irradiation is significantly greater in c-NHEJ deficient cells than in their wild-type counterparts (51).
We compared groups transduced with control, NONO, XLF, or a combination of NONO and XLF shRNAs. There was no difference in the number of background foci in non-irradiated cells, or in the initial number of foci when cells were irradiated with 1 Gy of γ-rays and fixed after 30 min (Figure 6C). By contrast, when irradiated cells were fixed after 4 h, the XLF and NONO shRNA groups had 50% more residual foci than the control shRNA group. The effect was significant at the P < 0.01 level. There was no further increase in the number of residual foci in the cells that received a combination of XLF and NONO shRNAs. Together with the survival and plasmid rejoining assays, results provide a third line of evidence for an epistatic relationship between XLF and NONO in cell-based assays.
DISCUSSION
We report here two novel findings regarding the mechanistic role of SFPQ•NONO in DSB repair. First, SFPQ•NONO substitutes in vitro for the core c-NHEJ factor, XLF, on a nearly mole for mole basis, with no evident synergy when both factors are present simultaneously. We interpret this as an indication that both factors affect the same limiting step, which most likely involves DNA substrate pairing. Results are consistent with observations that XLF-dependent filament formation is dispensable in some cell types, suggesting the existence of an alternative mechanism for maintaining pairing of DNA ends during ligation (20). They are also consistent with previously reported results that the isolated SFPQ•NONO complex has a DNA capture activity (28). The second finding is that, in contrast to the redundant function of SFPQ•NONO and XLF in vitro, the NONO subunit of the SFPQ•NONO complex and XLF have an epistatic relationship with respect to repair of radiation- and enzymatically-induced DNA DSBs in cells. This implies that each factor has additional functions that, although dispensable in vitro, are important in the cellular environment.
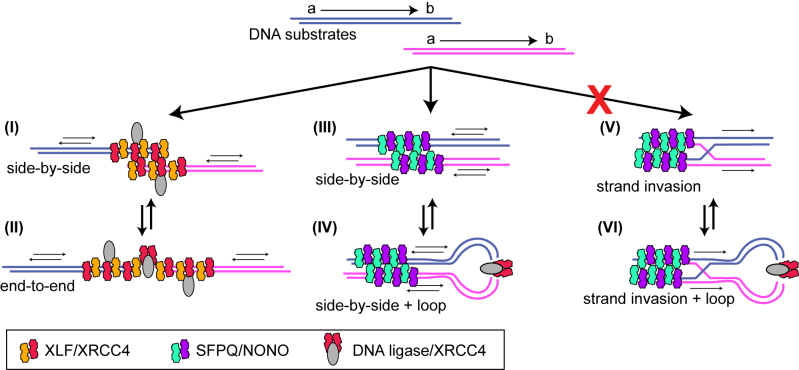
Although SFPQ•NONO and XLF function nearly interchangeably in vitro, evidently by promoting DNA pairing, it is likely that the mechanism of pairing differs in detail. XLF is recruited by Ku protein to free DNA ends, where it coassembles with XRCC4 and DNA ligase IV into a nucleoprotein filament (9,13–19,52). Evidence suggests that filaments formed on opposing DNA ends may form a transient, side-by-side complex (Figure 7, complex I) (18), which is followed by their coalescence into a continuous filament (II), in which ligase-compatible ends are aligned for covalent rejoining (9,13–19).
Figure 7.
Model. Substrate DNAs are shown at top, with ends designated as ‘a’ and ‘b’ and arrow showing a to b polarity. Complexes (I) and (II) depict side-by-side and end-to-end substrate pairing mediated by DNA ligase IV/XRCC4/XLF filament. Pairing is protein-mediated and DNA sequence-independent. Joining occurs with random polarity (equal probability of ab-ab, ba-ba, ab-ba, ba-ab joints). Complexes (III) and (IV) depict an alternative side-by side pairing geometry, which has not been observed directly but is consistent with known biochemical characteristics of SFPQ•NONO (see text). Loop structure allows joining by protomeric DNA ligase IV•XRCC4 complex (XLF not required). Joining occurs with random polarity. Complexes (V) and (VI) depict side-by-side pairing stabilized by sequence-dependent DNA strand invasion. Because substrates are constrained to a parallel geometry, inverted (ab-ba, ba-ab) joints are favored. This model is excluded by data in Figure 3. Complexes also contain Ku protein, although the stoichiometry and position relative to other factors is uncertain. For simplicity, Ku is not shown.
Based on analogy to related complexes, SFPQ•NONO is also likely to assemble into nucleoprotein filaments (53,54). Unlike XLF, recruitment of SFPQ•NONO to DNA requires neither free ends nor interaction with Ku protein. Rather, SFPQ•NONO appears to bind to internal sequences, away from the DNA ends. Biochemical data are consistent with a side-by-side pairing model (III), in which SFPQ•NONO filaments contact each other, but not other repair factors, and the DNA ends remain free to interact with DNA ligase IV•XRCC4 (III) (22,28). The side-by-side geometry is similar to that of the proposed initial XLF complex (I). Paired filaments have not been observed directly, perhaps because structural studies showing filament formation were performed using truncated proteins lacking the full IDRs. The IDRs of SFPQ, NONO, and a number of other proteins have been proposed to drive assembly of higher-order protein complexes via a liquid-liquid phase transition mechanism (reviewed in (55)).
We suggest that an initial side-by-side complex evolves into a loop structure (IV), where the increase in local DNA end concentration facilitates ligation. In contrast to the XLF-mediated end-to-end pairing, the termini are not constrained in precise alignment, which explains why cohesive overhangs remain advantageous, at least under the conditions of the in vitro assays, where other repair or chromatin proteins, which might stabilize pairing in cells, are absent. Looping interactions control the efficiency of a wide range of processes in nucleic acid biochemistry, including transcriptional activation and DNA cleavage by type IIS restriction endonucleases (56,57), and the model is thus amenable to testing using structural and biophysical approaches analogous used to study these other systems.
Data in the present study argue against an alternative nucleic acid sequence-dependent pairing model (V and VI). This model was inspired by reports that SFPQ promotes strand invasion and D-loop formation in vitro (24–27). In theory, sequence-dependent pairing is possible in vitro where all substrate molecules have the same sequence, and perhaps in vivo for DSBs that occur in or near pairs of interspersed repetitive elements. However, heteroduplex formation requires that paired DNAs align in the same relative sequence orientation. This is not in agreement with the in vitro data, which show that ligation products exhibit random polarity. It should be noted that although a DNA strand exchange mechanism is excluded here, nucleic acid strand exchange activity of SFPQ•NONO could be important under other conditions or in respect to other repair processes in the dynamic cellular environment.
In contrast to redundant functions of SFPQ•NONO and XLF in vitro, the NONO subunit and XLF have an epistatic relationship in vivo with respect to radioresistance, plasmid end-joining, and repair foci resolution. The results of survival, plasmid rejoining and foci resolution assays are in good agreement: all show a modest effect of XLF and NONO depletion that does not increase when both proteins are depleted simultaneously. This indicates that, in addition to the common function revealed in vitro, each protein also has separate functions in a common pathway in vivo.
For XLF, additional functions in cells could include promoting DNA ligase IV re-adenylation (58). This function is perhaps dispensable in vitro where DNA ligase IV•XRCC4 complexes are present in large excess over DNA ends. For NONO, additional functions could involve RNA binding. It may be that NONO-containing complexes modulate expression of genes involved in the DNA damage response, or more speculatively, that they interact directly with DNA damage-inducible noncoding RNAs present at the sites of DNA damage or elsewhere. To further understand the role of RNA interaction, it will be of interest to create separation-of-function mutants that are deficient for RNA binding but where protein folding and dimerization are unperturbed. Recent reports identifying candidate RNA-binding surface residues will be helpful as a guide to mutant design (59,60).
In the course of the work, we made an incidental observation that NONO deficiency sensitizes cells to cisplatin, that XLF deficiency does not, and that attenuation of XLF expression suppresses the NONO-deficient phenotype. Results provide further evidence that repair functions of NONO and XLF are genetically separable. They also confirm and extend a finding that attenuation of Ku protein expression suppresses the Fanconi anemia C protein phenotype, which was interpreted in terms of a ‘corrupting’ influence of c-NHEJ on interstrand cross-link repair in a setting where the Fanconi anemia pathway was compromised (61). One interpretation is that Ku and XLF, which are known to directly interact directly (52), form a nonproductive complex with an inter-strand crosslink repair intermediate.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Murray Junop (McMaster University, Hamilton, ON, Canada), Dr Stephen Jackson (Cambridge University, UK), Dr Leslyn Hanakahi (University of Illinois-Rockford), Dr Dik van Gent (Erasmus University, Rotterdam, NL), and former members of the Dynan laboratory for plasmid expression vectors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health [2 R01 CA98239]. Funding for open access charge: Emory University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Williams G.J., Hammel M., Radhakrishnan S.K., Ramsden D., Lees-Miller S.P., Tainer J.A.. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst.). 2014; 17:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waters C.A., Strande N.T., Wyatt D.W., Pryor J.M., Ramsden D.A.. Nonhomologous end joining: a good solution for bad ends. DNA Repair (Amst.). 2014; 17:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pang D., Yoo S., Dynan W.S., Jung M., Dritschilo A.. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 1997; 57:1412–1415. [PubMed] [Google Scholar]
- 4. Cary R.B., Peterson S.R., Wang J., Bear D.G., Bradbury E.M., Chen D.J.. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ramsden D.A., Gellert M.. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998; 17:609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeFazio L.G., Stansel R.M., Griffith J.D., Chu G.. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002; 21:3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spagnolo L., Rivera-Calzada A., Pearl L.H., Llorca O.. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006; 22:511–519. [DOI] [PubMed] [Google Scholar]
- 8. Hammel M., Yu Y., Mahaney B.L., Cai B., Ye R., Phipps B.M., Rambo R.P., Hura G.L., Pelikan M., So S. et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010; 285:1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graham T.G., Walter J.C., Loparo J.J.. Two-stage synapsis of DNA ends during non-homologous end joining. Mol. Cell. 2016; 61:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cui X., Yu Y., Gupta S., Cho Y.M., Lees-Miller S.P., Meek K.. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 2005; 25:10842–10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reddy Y.V., Ding Q., Lees-Miller S.P., Meek K., Ramsden D.A.. Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J. Biol. Chem. 2004; 279:39408–39413. [DOI] [PubMed] [Google Scholar]
- 12. Cottarel J., Frit P., Bombarde O., Salles B., Negrel A., Bernard S., Jeggo P.A., Lieber M.R., Modesti M., Calsou P.. A noncatalytic function of the ligation complex during nonhomologous end joining. J. Cell Biol. 2013; 200:173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hammel M., Yu Y., Fang S., Lees-Miller S.P., Tainer J.A.. XLF regulates filament architecture of the XRCC4.ligase IV complex. Structure. 2010; 18:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammel M., Rey M., Yu Y., Mani R.S., Classen S., Liu M., Pique M.E., Fang S., Mahaney B.L., Weinfeld M. et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J. Biol. Chem. 2011; 286:32638–32650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ropars V., Drevet P., Legrand P., Baconnais S., Amram J., Faure G., Marquez J.A., Pietrement O., Guerois R., Callebaut I. et al. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andres S.N., Vergnes A., Ristic D., Wyman C., Modesti M., Junop M.. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012; 40:1868–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsai C.J., Chu G.. Cooperative assembly of a protein-DNA filament for nonhomologous end joining. J. Biol. Chem. 2013; 288:18110–18120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reid D.A., Keegan S., Leo-Macias A., Watanabe G., Strande N.T., Chang H.H., Oksuz B.A., Fenyo D., Lieber M.R., Ramsden D.A. et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2575–E2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ochi T., Wu Q., Blundell T.L.. The spatial organization of non-homologous end joining: from bridging to end joining. DNA Repair (Amst.). 2014; 17:98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roy S., de Melo A.J., Xu Y., Tadi S.K., Negrel A., Hendrickson E., Modesti M., Meek K.. XRCC4/XLF Interaction Is Variably Required for DNA Repair and Is Not Required for Ligase IV Stimulation. Mol. Cell. Biol. 2015; 35:3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bladen C.L., Udayakumar D., Takeda Y., Dynan W.S.. Identification of the polypyrimidine tract binding protein-associated splicing factor•p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005; 280:5205–5210. [DOI] [PubMed] [Google Scholar]
- 22. Udayakumar D., Bladen C.L., Hudson F.Z., Dynan W.S.. Distinct pathways of nonhomologous end joining that are differentially regulated by DNA-dependent protein kinase mediated phosphorylation. J. Biol. Chem. 2003; 278:41631–41635. [DOI] [PubMed] [Google Scholar]
- 23. Knott G.J., Bond C.S., Fox A.H.. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 2016; 44:3989–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akhmedov A.T., Lopez B.S.. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 2000; 28:3022–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morozumi Y., Takizawa Y., Takaku M., Kurumizaka H.. Human PSF binds to RAD51 and modulates its homologous-pairing and strand-exchange activities. Nucleic Acids Res. 2009; 37:4296–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morozumi Y., Ino R., Takaku M., Hosokawa M., Chuma S., Kurumizaka H.. Human PSF concentrates DNA and stimulates duplex capture in DMC1-mediated homologous pairing. Nucleic Acids Res. 2012; 40:3031–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rajesh C., Baker D.K., Pierce A.J., Pittman D.L.. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011; 39:132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Udayakumar D., Dynan W.S.. Characterization of DNA binding and pairing activities associated with the native SFPQ.NONO DNA repair protein complex. Biochem. Biophys. Res. Commun. 2015; 463:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li S., Kuhne W.W., Kulharya A., Hudson F.Z., Ha K., Cao Z., Dynan W.S.. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009; 37:6746–6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salton M., Lerenthal Y., Wang S.Y., Chen D.J., Shiloh Y.. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010; 9:1568–1576. [DOI] [PubMed] [Google Scholar]
- 31. Ha K., Takeda Y., Dynan W.S.. Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells. DNA Repair (Amst.). 2011; 10:252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y.. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012; 40:10287–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li S., Li Z., Shu F.J., Xiong H., Phillips A.C., Dynan W.S.. Double-strand break repair deficiency in NONO knockout murine embryonic fibroblasts and compensation by spontaneous upregulation of the PSPC1 paralog. Nucleic Acids Res. 2014; 42:9771–9780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yarosh C.A., Iacona J.R., Lutz C.S., Lynch K.W.. PSF: nuclear busy-body or nuclear facilitator. Wiley Interdiscip. Rev. RNA. 2015; 6:351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schiffner S., Zimara N., Schmid R., Bosserhoff A.K.. p54nrb is a new regulator of progression of malignant melanoma. Carcinogenesis. 2011; 32:1176–1182. [DOI] [PubMed] [Google Scholar]
- 36. Mircsof D., Langouet M., Rio M., Moutton S., Siquier-Pernet K., Bole-Feysot C., Cagnard N., Nitschke P., Gaspar L., Znidaric M. et al. Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects. Nat. Neurosci. 2015; 18:1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu P.Y., Erriquez D., Marshall G.M., Tee A.E., Polly P., Wong M., Liu B., Bell J.L., Zhang X.D., Milazzo G. et al. Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. J. Natl. Cancer Inst. 2014; 106:dju113. [DOI] [PubMed] [Google Scholar]
- 38. Hanakahi L.A. 2-Step purification of the Ku DNA repair protein expressed in Escherichia coli. Protein Expr. Purif. 2007; 52:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Lamarche B.J., Tsai M.D.. Human DNA ligase IV and the ligase IV/XRCC4 complex: analysis of nick ligation fidelity. Biochemistry. 2007; 46:4962–4976. [DOI] [PubMed] [Google Scholar]
- 40. Andres S.N., Modesti M., Tsai C.J., Chu G., Junop M.S.. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell. 2007; 28:1093–1101. [DOI] [PubMed] [Google Scholar]
- 41. Ochi T., Blackford A.N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V.M., Robinson C.V. et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015; 347:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Verkaik N.S., Esveldt-van Lange R.E., van Heemst D., Bruggenwirth H.T., Hoeijmakers J.H., Zdzienicka M.Z., van Gent D.C.. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002; 32:701–709. [DOI] [PubMed] [Google Scholar]
- 43. Song X., Sun Y., Garen A.. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:12189–12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xing M., Yang M., Huo W., Feng F., Wei L., Jiang W., Ning S., Yan Z., Li W., Wang Q. et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 2015; 6:6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dugaiczyk A., Boyer H.W., Goodman H.M.. Ligation of EcoRI endonuclease-generated DNA fragments into linear and circular structures. J. Mol. Biol. 1975; 96:171–184. [DOI] [PubMed] [Google Scholar]
- 46. Kaneko S., Rozenblatt-Rosen O., Meyerson M., Manley J.L.. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3΄ processing and transcription termination. Genes Dev. 2007; 21:1779–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fattah F.J., Kweon J., Wang Y., Lee E.H., Kan Y., Lichter N., Weisensel N., Hendrickson E.A.. A role for XLF in DNA repair and recombination in human somatic cells. DNA Repair (Amst.). 2014; 15:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D.. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000; 151:1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Asaithamby A., Chen D.J.. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 2009; 37:3912–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cao Z., Kuhne W.W., Steeb J., Merkley M.A., Zhou Y., Janata J., Dynan W.S.. Use of a microscope stage-mounted Nickel-63 microirradiator for real-time observation of the DNA double-strand break response. Nucleic Acids Res. 2010; 38:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ahmed E.A., Velaz E., Rosemann M., Gilbertz K.P., Scherthan H.. DNA repair kinetics in SCID mice Sertoli cells and DNA-PKcs-deficient mouse embryonic fibroblasts. Chromosoma. 2016; doi:10.1007/s00412-016-0590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yano K., Morotomi-Yano K., Wang S.Y., Uematsu N., Lee K.J., Asaithamby A., Weterings E., Chen D.J.. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008; 9:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Passon D.M., Lee M., Rackham O., Stanley W.A., Sadowska A., Filipovska A., Fox A.H., Bond C.S.. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:4846–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee M., Sadowska A., Bekere I., Ho D., Gully B.S., Lu Y., Iyer K.S., Trewhella J., Fox A.H., Bond C.S.. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 2015; 43:3826–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Uversky V.N. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2016; 44:18–30. [DOI] [PubMed] [Google Scholar]
- 56. Levine M., Cattoglio C., Tjian R.. Looping back to leap forward: transcription enters a new era. Cell. 2014; 157:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pingoud A., Wilson G.G., Wende W.. Type II restriction endonucleases—a historical perspective and more. Nucleic Acids Res. 2014; 42:7489–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Riballo E., Woodbine L., Stiff T., Walker S.A., Goodarzi A.A., Jeggo P.A.. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 2009; 37:482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Duvignaud J.B., Bedard M., Nagata T., Muto Y., Yokoyama S., Gagne S.M., Vincent M.. Structure, dynamics and interaction of p54nrb/NonO RRM1 with 5΄ Splice Site RNA sequence. Biochemistry. 2016; 55:2553–2566. [DOI] [PubMed] [Google Scholar]
- 60. Knott G.J., Lee M., Passon D.M., Fox A.H., Bond C.S.. Caenorhabditis elegans NONO-1: insights into DBHS protein structure, architecture, and function. Protein Sci. 2015; 24:2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pace P., Mosedale G., Hodskinson M.R., Rosado I.V., Sivasubramaniam M., Patel K.J.. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010; 329:219–223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.