Abstract
The full-length translation-regulating add adenine riboswitch (Asw) from Vibrio vulnificus has a more complex conformational space than its isolated aptamer domain. In addition to the predicted apo (apoA) and holo conformation that feature the conserved three-way junctional purine riboswitch aptamer, it adopts a second apo (apoB) conformation with a fundamentally different secondary structure. Here, we characterized the ligand-dependent conformational dynamics of the full-length add Asw by NMR and by single-molecule FRET (smFRET) spectroscopy. Both methods revealed an adenine-induced secondary structure switch from the apoB-form to the apoA-form that involves no tertiary structural interactions between aptamer and expression platform. This strongly suggests that the add Asw triggers translation by capturing the apoA-form secondary structure in the holo state. Intriguingly, NMR indicated a homogenous, docked aptamer kissing loop fold for apoA and holo, while smFRET showed persistent aptamer kissing loop docking dynamics between comparably stable, undocked and docked substates of the apoA and the holo conformation. Unraveling the folding of large junctional riboswitches thus requires the integration of complementary solution structural techniques such as NMR and smFRET.
INTRODUCTION
Riboswitches are cis-acting mRNA elements that regulate gene-expression in response to changes in the cellular concentration of a specific metabolite. Natural riboswitches occur predominantly in bacteria where they typically control mRNA transcription, mRNA translation or mRNA decay. The common mode of riboswitch action is that metabolite binding of a 5΄ aptamer domain induces an allosteric structural transition of a 3΄ effector domain termed expression platform to switch from a gene-OFF state to a gene-ON state or vice versa. To date, over 20 classes of riboswitches have been described that respond to a large variety of ligands involving nucleobases, amino acids and protein cofactors (1). For the majority of riboswitches, the isolated aptamer domain is well characterized in terms of structure and ligand-binding properties, but the regulatory refolding mechanism of the full-length mRNA sequence is poorly understood.
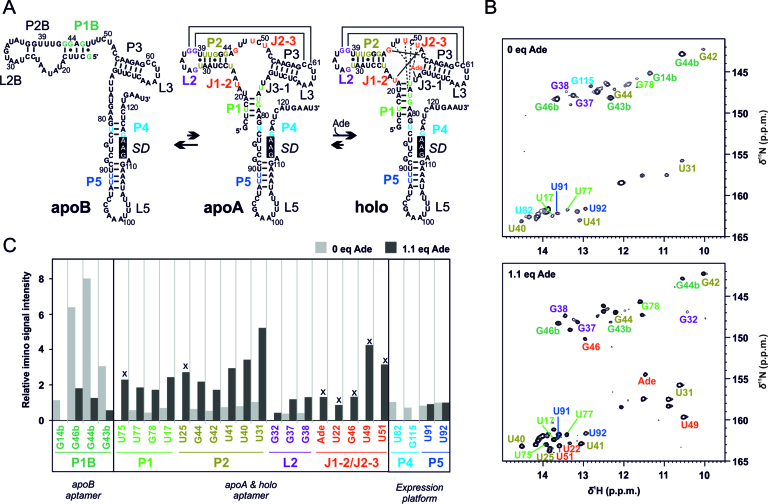
Our group has recently discovered a novel three-state translation-regulating mechanism in the full-length (112-nucleotide) add adenine riboswitch (Asw) from Vibrio vulnificus (Figure 1A) (2). The full-length add Asw has a bistable secondary structure in the apo state with an adenine-binding incompetent apoB conformation and an adenine-sensing apoA conformation. Adenine binding to apoA stabilizes a riboswitch fold with a solvent accessible Shine Dalgarno Sequence (SD) in the holo conformation and thus facilitates ribosome binding to the mRNA. The evolutionary advantage of a three-state over a two-state riboswitch is that the additional equilibrium can be modulated by an additional environmental stimulus to adjust the riboswitch response to the environment of its host. In case of the add Asw, the secondary structure pre-equilibrium between apoB and apoA is temperature-dependent and tunes the temperature response profile of the riboswitch (3). With its bistable secondary structure, the full-length add Asw has a more complex folding landscape than its previously characterized isolated aptamer domain, which exclusively adopted the apoA-form secondary structure of the apoA and the holo conformation.
Figure 1.
Adenine-induced switch in base pairing of the full-length add Asw analysed by NMR spectroscopy. (A) Model of the 3-state conformational equilibrium of the add Asw at 25°C. Guanosine and uridine imino reporter signals are color-coded according to structural elements. The Shine Dalgarno sequence (SD) is highlighted in black. The scheme was modified after Reining et al. (2). (B) Imino region of the 1H, 15N-BEST-TROSY spectra of the add Asw (0.3 mM) measured at 800 MHz in absence and presence of 1.1 eq adenine at 25°C and 5 mM Mg2+. The imino reporter signals are annotated. (C) Relative intensities of the imino reporter signals (normalized to U92) without adenine (light gray bars) and with 1.1 eq adenine (dark gray bars). Signals denoted with x report on the formation of the holo binding pocket.
In recent years, NMR and smFRET spectroscopies have unraveled valuable insight into the conformational dynamics of riboswitches that greatly improved our molecular-level understanding of these regulatory RNAs (4,5). The two spectroscopies are ideally suited for an integrated structural biology analysis of RNA folding, because they can reveal complementary structural information on similar timescales. By NMR of RNA imino protons, base pairing interactions can be characterized at nucleotide resolution for RNAs as large as 124 nucleotides (6). NZZ exchange and time-resolved NMR experiments give access to RNA refolding kinetics ranging from sub-seconds to hours (7). By smFRET of immobilized molecules, RNA tertiary structure and real-time tertiary structural dynamics can be monitored at the sub-second to minute timescale (8). However, NMR and smFRET operate at different solution conditions. NMR requires half-millimolar RNA concentration and isotope-labeled samples. smFRET uses dye-labeled surface-tethered RNA constructs at sub-nanomolar concentration. Given that RNA folding is highly sensitive to the ionic environment (9), the six-orders of magnitude difference between NMR and smFRET in the ratio of buffer cations over RNA might affect RNA folding. It is thus worthwhile also from a methodological point of view to compare NMR and smFRET data on the structural dynamics of RNA. To date, only one integrated NMR and smFRET study of RNA is available, a conformational dynamics study of the small pseudoknot SAM-II riboswitch (10). In case of the junctional add adenine riboswitch, the available NMR data (11,12) and smFRET data (13–15) cannot be directly compared, because they were all collected on aptamer constructs with differently modified P1 stems. Meanwhile, it is known that P1 stability has a strong impact on purine riboswitch folding. It modulates aptamer tertiary structure stability and regulatory activity (16,17).
In this work, we performed a combined NMR and smFRET spectroscopic analysis of the native full-length 112-nucleotide add adenine riboswitch from Vibrio vulnificus to probe the response of its three-state conformational equilibrium to varying conditions of adenine and Mg2+ at the level of both secondary and tertiary structure. To map the tertiary structural changes of the full-length add Asw, we compared smFRET data of the wild-type and two variants, a mutant with stabilized apoB-form secondary structure and a mutant with stabilized apoA-form secondary structure. Finally, we discuss similarities and differences in the folding revealed by NMR and smFRET of this junctional purine riboswitch.
MATERIALS AND METHODS
Riboswitch preparation for NMR spectroscopy
15N-labeled 112-nucleotide add Asw was synthesized by in vitro transcription with T7 RNA polymerase from a PCR template with a 5΄-hammerhead ribozyme (Supplementary Table S1) as described (18) and purified as described (19). The RNA was folded by 5 min denaturation at 95°C at a concentration of 0.3 mM followed by immediate dilution to 0.03 mM with ice cold water and then exchanged into NMR buffer (25 mM K2HPO4/KH2PO4, 50 mM KCl, pH 6.2) using Vivaspin centrifugal concentrators (Sartorius AG). NMR samples were prepared with 10% D2O and 100 μM DSS as chemical shift standard.
NMR spectroscopy
NMR experiments were performed on Bruker spectrometers (AV600, AV800, AV900) equipped with a 5-mm z-axis gradient TXI-HCN cryogenic probe. 1H,15N-BEST-TROSY experiments were recorded using a pulse program implementing the modifications proposed by Brutscher et al. (20,21) 1H,1H-NOESY (150 ms mixing time) spectra were recorded with jump-return echo water suppression technique (22), while 15N-HSQC spectra were acquired using the binomial WATERGATE water suppression scheme (23). Data processing was performed in Topspin 3.1 (Bruker Biospin). The signal intensities of 1H,15N-BEST-TROSY spectra were parsed out as peak heights using the software Sparky 3.114 (24).
Riboswitch preparation for smFRET spectroscopy
Fluorescent dye labeled 112-nucleotide add Asw was synthesized by DNA-splinted enzymatic ligation of three RNA fragments (Supplementary Table S2) with the FRET pair Cy3/Cy5 at two of three 5-amino-allyl modified uridine residues (U36, U62 and U92) and a 3΄-terminal biotin modification. Modified RNA oligonucleotides were purchased from Dharmacon in 2΄-ACE protected form, coupled with the amine reactive dye (Cy3 or Cy5 mono-Reactive Dye Pack, respectively; GE Healthcare) and deprotected in deprotection buffer (Dharmacon) according to the manufacturers’ protocols. Dye-labeled RNA oligonucleotides were purified by HPLC on a C8 column (Kromasil 100) equilibrated with 100 mM triethylamine acetic acid, pH 7.0, employing an acetonitrile gradient. For ligation, the three RNA fragments and the DNA splint were annealed at 2 μM concentration in T4 RNA ligase 2 buffer (New England Biolabs) by 3 min heating at 75°C and 10 min cooling at room temperature, and subsequently incubated with 0.2 U/μl T4 RNA ligase 2 (New England Biolabs) at 37°C for 1 h, followed by splint digestion with 70 U/ml Turbo DNase (Ambion) at 37°C for 0.5 h. After extraction of the mixture with Roti®-Aqua-P/C/I (Carl Roth) and chloroform, the full-length RNA was purified by denaturing urea PAGE (10%). Target bands were identified by eye and eluted by shaking in 0.5 M ammonium acetate at room temperature overnight, followed by ethanol precipitation. For sample preparation, an aliquot of the FRET construct was folded by 3 min denaturation at 85°C at a concentration of 1 nM in immobilization buffer (25 mM K2HPO4/KH2PO4, 50 mM KCl, pH 7.0) followed by 15 min cooling at room temperature. 100 pM RNA samples were prepared in immobilization buffer, supplemented with the indicated concentrations of MgCl2 and adenine and chilled on ice until measurement within the same day.
smFRET spectroscopy
Glass slides (Carl Roth) were washed in 1 M KOH and in deionized MilliQ water for 15 min, glass cover slips (Carl Roth) were plasma cleaned in N2 plasma for 10 min (Diener Electronic). The slides and cover slips were assembled to measurement slides with ∼10 μl channels that were coated with biotinylated-BSA and streptavidin as described (25). The channels were flushed with 200 μl immobilization buffer with adjusted concentrations of MgCl2 and adenine, repeatedly flushed with 30 μl of a 100 pM RNA sample until a sufficient surface density of single molecules was achieved (∼0.1 molecules/μm2) and flowed with 125 μl imaging buffer (25 mM K2HPO4/KH2PO4 pH 7.0, 50 mM KCl, 9% (w/w) glucose, 14 U/ml glucose oxidase, 1000 U/ml catalase, sat. trolox and MgCl2 and adenine as specified) for measurement. All measurements were carried out at ambient temperature (22°C) with 532 nm laser excitation on an objective-type total internal reflection microscopy setup with an EMCCD camera (iXon, Andor Technology) at 100 ms integration time.
smFRET analysis
Background corrected single-molecule fluorescence intensities of Cy3 and Cy5 were extracted from the raw movie files using custom scripts in IDL (Exelis) as described (25) and imported into MATLAB (MathWorks) to calculate the corresponding FRET efficiencies E = IA/(IA + ID). The acceptor intensities were corrected for about 10% leakage of the donor fluorescence into the acceptor channel. For FRET histograms, the FRET efficiencies of single molecules were averaged over the first three frames (300 ms), binned in intervals of 0.025 and exported for Gaussian distribution fitting in Origin 9.0G (OriginLab). The donor-only population at E = 0 was removed from the histograms by subtraction of the corresponding Gaussian. For L2/L3-labeled add Asw, the fraction of docked molecules was determined from the Gaussian peak areas of the undocked and docked state and the error calculated from the fitting errors by Gaussian error propagation. For analysis of smFRET time traces of L2/L3-labeled add Asw, traces with a trace time ≥5 s until single-step photobleaching were pre-processed for Hidden Markov modeling (HMM) of the normalized donor and acceptor trajectories alongside the corresponding FRET trajectories as described by Blanco et al. (26). HMM analysis was performed in HaMMy (27) with an initial estimate of four states at the default values. The HaMMy output was further analysed in MATLAB with a custom script that identified transitions, created TODPs and exported dwell-times. Transitions were identified based on the following criteria: (i) change in FRET efficiency ΔE > 0.1 (ii) Anti-correlated change in donor and acceptor fluorescence. Dwell-time histograms of the undocked state (E < 0.6) and the docked state (E > 0.6) were created in Origin 9.0G with a bin width of 0.5 s and fitted with single-exponential decay functions to extract the docking and undocking rate constants.
Fluorescence anisotropy measurements on Cy3 and Cy5
Bulk fluorescence anisotropy measurements were performed at 20°C on a FluoroMax-4 spectrophotometer (Horiba Scientific) in L-format geometry. Samples containing 8 nM free cyanine dye or single-fluorophore-labeled 112-nucleotide add Asw in immobilization buffer (25 mM K2HPO4/KH2PO4, 50 mM KCl, pH 7.0) were heated at 85°C for 3 min, subsequently cooled at room temperature for 10 min, diluted with an equal volume of 4 mM MgCl2 in immobilization buffer and then kept on ice until measurement. After a temperature equilibration time of 2 min, the fluorescence anisotropies were measured in accumulations of 20 scans at the excitation / emission wavelengths 525 nm / 565 nm for Cy3 and 625 nm / 665 nm for Cy5. The integration time was 100 ms and the excitation and emission bandwidth were 7 nm. The fluorescence anisotropy r was calculated as r = (IVV – G*IVH)/(IVV + 2G*IVH), where IVV denotes the intensity of vertically polarized emission and IVH the intensity of horizontally polarized emission detected at excitation with vertically polarized light. The correction factor G = IHV/IHH (0.71 for Cy3 and 0.51 for Cy5) was determined from the intensity IHV of vertically polarized emission and the intensity IHH of horizontally polarized emission measured at excitation with horizontally polarized light.
RESULTS
Adenine-dependent base pairing by NMR
We have previously reported the assignment of the imino NH resonances of the full-length 112-nucleotide add Asw (2). RNA imino resonances can be detected for guanosine and uridine residues in stable base pairs that protect the protons from solvent exchange. The intensity of these base pair reporter signals depends on the base pair occupancy and solvent accessibility. Here, we characterized the adenine-dependent base pairing of the full-length add Asw at 25°C and 5 mM Mg2+ by evaluating the adenine-dependence of imino signal intensities in different structural elements. We selected unique imino reporter signals for base pairs of the apoB aptamer (P1b), the apoA and holo aptamer (P1, P2, L2, J1-2/J2-3) and the expression platform (P4, P5; Figure 1A). The imino signal intensities were extracted from 1H,15N-BEST-TROSY spectra (Figure 1B). The signal pattern of these spectra at 25°C compared well with our previous data that were recorded at 10°C (Supplementary Figure S1). In absence of adenine, the imino signals were dominated by the apoB-specific helix P1b. The P1b signals were on average ∼7-fold higher than the signals of the apoA P1/P2/L2-module and ∼5-fold higher than those of the helices P4 and P5 in the expression platform (Figure 1C, light gray bars). The apoA aptamer exhibited a pre-folded L2/L3 kissing loop motif, as evidenced by the signals G37 and G38. P1 stem-terminal imino resonances and P4-specific signals of apoA were not detected suggesting that the apoA P1/P4 interface lacked persistent base pairs. The addition of 1.1 eq adenine led to the appearance of the imino signals for J1-2/J2-3, U75 (P1) and U25 (P2), which report on the formation of the holo conformation. Additionally, we observed that adenine induced a ∼5-fold decrease in the average imino signal intensity of P1b, a ∼4-fold increase for the P1/P2/L2-module and the disappearing of the P4 signals (Figure 1C, dark gray bars). The imino signal intensity ratio of L2 over P2 was not significantly affected by adenine. This finding indicates a similar stability of the L2/L3 kissing loop motif in the apoA and the holo aptamer. In summary, the effect of adenine on the imino signal intensities revealed three coupled structural changes. Adenine binding induced folding of the aptamer core. This folding stabilized the apoA secondary structure over apoB and melted the P4 helix in the expression platform.
Adenine-dependent long-range tertiary structural interactions by smFRET
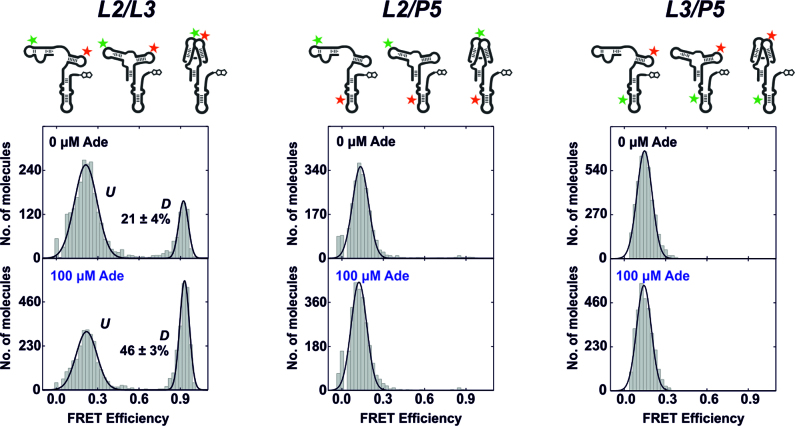
To elucidate global tertiary structural changes that accompany the adenine-induced switch in base pairing of the full-length 112-nucleotide add Asw, we performed smFRET measurements on three different FRET constructs that were each labeled with the fluorescent dye pair Cy3 and Cy5 (Figure 2). Based on existing literature, we chose two labeling sites in loop L2 (U36) and L3 (U62) of the aptamer domain. It was shown in previous studies that the L2/L3-labeled construct reliably reports on the ligand-modulated folding of the kissing loop motif of the isolated add Asw aptamer (13,14). In addition, we placed a third dye label in the expression platform helix P5 (U92) to explore possible interactions between the expression platform loops and the aptamer domain with an L2/P5- and an L3/P5-labeled construct. The P5-internal labeling site was chosen to avoid interference with the dynamics of P4. To investigate the degree of interaction between the FRET dyes and surrounding RNA nucleotides in the three FRET constructs, bulk fluorescence anisotropy measurements were performed. For these measurements, single-fluorophore labeled full-length riboswitches were prepared with Cy3 at L2, Cy5 at L3, Cy3 at P5 and Cy5 at P5. The fluorescence anisotropies of both cyanine dyes were significantly increased in the RNA-coupled form compared to the free form (Supplementary Table S3). The anisotropy of Cy3 was 0.285 at L2 and 0.296 at P5, and the anisotropy of Cy5 was 0.238 at L3 and 0.281 at P5. These values were comparable to the anisotropies of Cy3 (0.26) and Cy5 (0.25) in a FRET construct that was recently used to study the folding of the diels alderase ribozyme (28). This suggests that Cy3 and Cy5 did not exhibit specific interactions with their distinct environment at L2, L3 or P5 in our FRET constructs of the full-length add Asw.
Figure 2.
Adenine-dependent global tertiary structural changes of the full-length add Asw analyzed by smFRET. FRET histograms of the L2/L3-labeled, the L2/P5-labeled and the L3/P5-labeled add Asw in absence and presence of 100 μM adenine at 2 mM Mg2+. A schematic representation of each FRET construct in the apo conformations is depicted above. For the FRET histograms of the L2/L3-labeled construct, the fractional population of the docked state D is indicated in percent.
We collected smFRET histograms of the three FRET constructs in absence and in presence of adenine at 2 mM Mg2+. The L2/L3-labeled construct showed a bimodal FRET distribution with a low-FRET state at E ∼ 0.2 and a high-FRET state at E ∼ 0.9 (Figure 2, left panel). The two FRET states matched those reported for the isolated aptamer domain (14), and were consequently assigned to molecules with undocked (U) and docked (D) aptamer loops. With 100 μM adenine, a ∼100-fold excess over the reported binding affinity of the add Asw (2), we observed a two-fold increase in the fractional population of the docked state. However, ∼50% of the ligand-saturated full-length add Asw remained in the undocked state, while the isolated aptamer domain was predominantly docked under comparable conditions (14,15). We confirmed the adenine binding competency of the L2/L3-labeled full-length add Asw with adenine titrations (Supplementary Figure S2). The half-saturating adenine concentration for aptamer kissing loop docking (5 ± 2 μM at 2 mM Mg2+) was in good agreement with the value determined for the isolated aptamer (3 ± 1 μM at 1 mM Mg2+) (15). The full-length L2/L3-labeled add Asw thus bound adenine with comparable affinity to the isolated aptamer, despite its persistent heterogeneity between states with undocked and docked aptamer loops. The FRET histograms of the L2/P5- and the L3/P5-labeled construct (Figure 2, middle and right panel) showed a single low-FRET state at E ∼ 0.15 independent of adenine. Based on these data, we can exclude that single stranded regions of the add Asw expression platform interacted with upstream sequence elements of the aptamer domain in the ligand-free and in the ligand-bound state. This confirms that the adenine-induced allosteric switch of the add Asw is spatially decoupled in that it involves no direct interaction between the two riboswitch domains.
smFRET analysis of the aptamer folding equilibrium in the full-length riboswitch
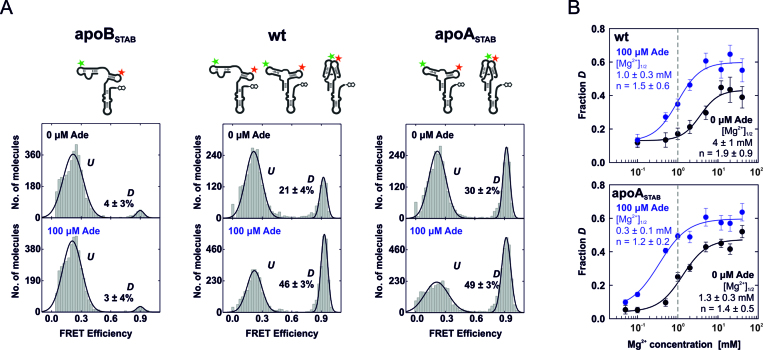
We next deciphered the heterogeneity between undocked and docked states in the ligand-free and the ligand-saturated L2/L3-labeled full-length add Asw by comparing the wild-type riboswitch (wt) with mutants that stabilized either the apoB conformation (apoBSTAB) or the apoA conformation (apoASTAB). We confirmed by NMR spectroscopy that apoBSTAB only forms the apoB-form secondary structure in absence and presence of adenine (Supplementary Figure S3). ApoASTAB was previously shown to exclusively adopt the apoA-form secondary structure and to bind adenine with a ∼5-fold higher affinity over wt (2). The FRET histograms of the L2/L3-labeled mutants apoBSTAB and apoASTAB at 2 mM Mg2+ showed identical FRET states to the wild-type (Figure 3A): the U state at E ∼ 0.2 and the D state at E ∼ 0.9. As expected, apoBSTAB almost exclusively populated the U state (∼95%) and was unresponsive to adenine. Wt and apoASTAB showed an adenine-dependent aptamer docking equilibrium. The fraction of docked molecules was decreased in adenine-free wt (∼20%) compared to adenine-free apoASTAB (∼30%) and equal in the two variants with 100 μM adenine (∼50%). These observations suggest that saturating ligand conditions suppressed apoB formation in wt by capturing the riboswitch in the holo state. Remarkably, the fractional population of the docked state in ligand-saturated wt was reproduced with apoASTAB. This indicates an equivalent tertiary structure heterogeneity in the holo state of the two variants. From apoASTAB, we derived that the adenine-induced stabilization of the docked apoA-form aptamer was ∼0.5 kcal/mol (Table 1). This value is strikingly similar to the related xpt guanine riboswitch aptamer, for which a ∼0.5 kcal/mol guanine-induced stabilization of the docked state was determined (29). The difference in the free docking enthalpy between ligand-free wt and apoASTAB was ∼0.3 kcal/mol (Table 1), which is of the same order of magnitude as the free enthalpy difference between the apoA and the apoB conformation previously determined by 1H,15N-HSQC NZZ-exchange NMR spectroscopy (∼0.6 kcal/mol) (2). This supports the hypothesis that the secondary structure bistability of wt accounts for it.
Figure 3.
Mutational smFRET analysis of aptamer docking of the full-length add Asw. (A) FRET histograms of L2/L3-labeled apoB-stabilized (apoBSTAB), wildtype (wt) and apoA-stabilized (apoASTAB) add Asw in absence and presence of 100 μM adenine at 2 mM Mg2+. A schematic representation of each variant in the apo conformations is depicted above. The fractional population of the docked state D is indicated in percent. (B) Fractional population of D in wt and apoASTAB as a function of the Mg2+ concentration without adenine (black) and with 100 μM adenine (blue). The data have been fitted using the Hill equation to obtain the Mg2+ concentration for half-maximal docking [Mg2+]1/2 and the Hill coefficient n. The dashed line indicates near-physiological free Mg2+ concentration (∼1 mM).
Table 1. Folding parameters for aptamer docking of wt and apoASTAB obtained by smFRET.
| Variant | Mg [mM] | Ade [μM] | ΔG(UD) Hista [kcal/mol] | kdockb [s−1] | kundockb [s−1] | ΔGdockKinc [kcal/mol] |
|---|---|---|---|---|---|---|
| Wt | 2 | 0 | 0.8 ± 0.1 | 0.7 ± 0.1 | 0.98 ± 0.03 | 0.2 ± 0.1 |
| 20 | 0 | 0.2 ± 0.1 | 0.7 ± 0.1 | 1.18 ± 0.02 | 0.3 ± 0.1 | |
| 2 | 100 | 0.1 ± 0.1 | 0.7 ± 0.1 | 1.5 ± 0.2 | 0.4 ± 0.1 | |
| apoASTAB | 2 | 0 | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.9 ± 0.1 | 0.3 ± 0.1 |
| 20 | 0 | 0.2 ± 0.1 | 0.5 ± 0.1 | 1.1 ± 0.1 | 0.5 ± 0.1 | |
| 2 | 100 | 0.0 ± 0.1 | 0.49 ± 0.04 | 1.2 ± 0.3 | 0.5 ± 0.1 |
aFree enthalpy of the docked state relative to the undocked state determined from the FRET distribution observed in the FRET histogram as ΔG(UD) = –RT*ln(D/U). The reported error was calculated from the fitting errors of the FRET populations by Gaussian error propagation.
bThe reported rate constants are mean values with standard deviations from single-exponentially fitted dwell-time histograms of two independent experiments.
cFree docking enthalpy determined from the observable kinetics as ΔGdock = –RT*ln(kdock/kundock). The reported error was calculated from the standard deviations of the rate constants from two independent experiments by Gaussian error propagation.
To further characterize the different aptamer docking equilibria of all variants, we performed Mg2+ titrations (0–40 mM) without and with 100 μM adenine. For apoBSTAB, Mg2+ induced only a statistically insignificant increase in the fractional population of the docked state, regardless of the presence of adenine (Supplementary Figure S4). Wt and apoASTAB both exhibited strongly Mg2+-dependent aptamer docking (Figure 3B, and Supplementary Figure S5). These variants showed a two-state docking transition in absence and presence of adenine. We used a Hill-equation fit to determine the Mg2+ concentration [Mg2+]1/2 required for half-maximal docking. The [Mg2+]1/2 for wt (4 ± 1 mM) and apoASTAB (1.3 ± 0.3 mM) were decreased ∼4-fold with adenine in line with an increased Mg2+-affinity of the adenine-bound state. The Hill coefficients showed a minor decrease with adenine and indicated binding of 1-2 Mg2+ ions by the aptamer domain upon docking. Strikingly, it is at near-physiological free Mg2+ concentration (∼1 mM) (30) where the aptamer docking equilibrium of wt and apoASTAB showed greatest adenine sensitivity, and where the difference between wt and apoASTAB was most pronounced. At saturating Mg2+ concentration, wt and apoASTAB were indistinguishable in the fraction of docked molecules with ∼40% docked in absence and 60% docked in presence of adenine. This indicates that the conformational dynamics of the add Asw are adapted to physiological levels of Mg2+.
smFRET analysis of the aptamer folding dynamics
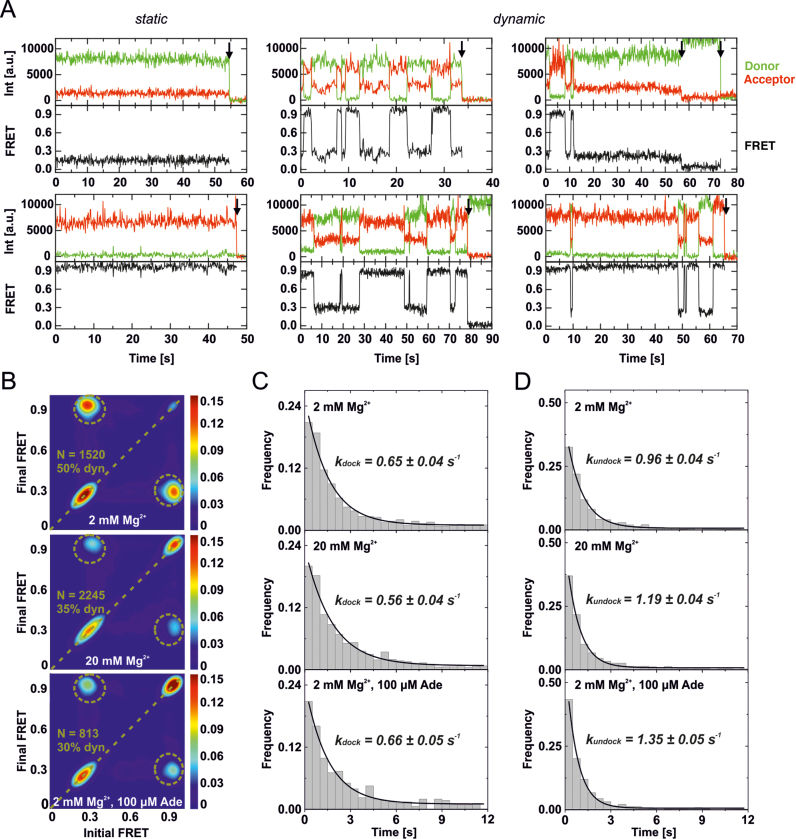
We also collected time-resolved smFRET data of L2/L3-labeled wt and apoASTAB to investigate the aptamer docking dynamics. We compared sub-saturating, near-physiological Mg2+ concentration (2 mM) without and with adenine (100 μM), with saturating Mg2+ concentration (20 mM). Because the dynamic properties of wt and apoASTAB were highly similar, only the data for wt are shown in the main text (Figure 4). Data for apoASTAB can be found in the Supplementary Information (Supplementary Figure S6). Under all conditions investigated, the smFRET traces of wt (Figure 4A) and apoASTAB (Supplementary Figure S6A) were heterogeneous. The traces showed static or dynamic behavior. Static traces (Figure 4A and Supplementary Figure S6A, left panel) exhibited a long-lived undocked or a long-lived docked state. Dynamic traces showed docking and undocking transitions between short-lived states (Figure 4A and Supplementary Figure S6A, middle panel) and also exhibited long-lived states (Figure 4A and Supplementary Figure S6A, right panel). A long-lived undocked or docked state with a comparable dwell-time to our average observation window until photobleaching (∼30s) was observed in ∼5% of all traces. These excursions of dynamic traces into the static regime strongly suggest that the heterogeneity between static and dynamic traces reflected a dynamic heterogeneity of the folding equilibrium of the add Asw.
Figure 4.
Mg2+- and adenine-dependent aptamer docking dynamics of the full-length wt add Asw analysed by smFRET. (A) Representative smFRET time traces of L2/L3-labeled wt collected at 2 mM Mg2+ without adenine. The aptamer domain of single riboswitches either statically remained in a long-lived undocked or docked state (left panel), or exhibited dynamics between short-lived and long-lived undocked and docked states (middle and right panel). Photobleaching events are indicated by a black arrow. (B) Transition occupancy density plots (TODPs) for smFRET traces collected at 2 mM Mg2+, at 20 mM Mg2+ and at 2 mM Mg2+ with 100 μM adenine. The fraction of molecules that exhibited dynamics (dyn) is indicated in percent. N indicates the number of traces included in each TODP. (C) Dwell-time histograms of the undocked state created from the dynamic smFRET traces designated in the corresponding TODP in (B). The data were fitted using single-exponential decay functions to extract the indicated docking rate constants. (D) Dwell-time histograms of the docked state created from the dynamic smFRET traces designated in the corresponding TODP in (B). The data were fitted using single-exponential decay functions to extract the indicated undocking rate constants.
We created transition occupancy density plots (TODPs) to visualize the proportion of static and dynamic smFRET traces. In TODPs, static traces appear at their respective FRET efficiency on the diagonal and dynamic traces at every off-diagonal FRET pair (Initial FRET, Final FRET) for which at least one transition was observed (26). The TODPs of wt (Figure 4B) as well as apoASTAB (Supplementary Figure S6B) demonstrate that adenine and Mg2+ shifted the FRET distribution of the static traces towards the docked state while decreasing the fraction of dynamic traces from ∼50% to ∼30%. The fact that ∼30% of all traces were dynamic for adenine-saturated wt and apoASTAB corroborates the hypothesis of a holo conformation in dynamic equilibrium between undocked and docked aptamer folds. To analyze the docking and undocking kinetics of wt and apoASTAB, we created dwell-time histograms of the undocked and the docked state (Figure 4C, D and Supplementary Figure S6C, D). These data were fitted with single exponential decay functions to extract the rate constants for docking and undocking, which are summarized in Table 1. Remarkably, the adenine- and Mg2+-dependent differences in the docking equilibrium did not manifest themselves in the observed kinetics. The observable docking rate constant of wt (kdock = 0.7 ± 0.1 s−1) and apoASTAB (kdock = 0.5 ± 0.1 s−1) was independent of the concentrations of adenine and Mg2+. The observable undocking rate constant of wt (kundock = 0.98 ± 0.03 s−1) and apoASTAB (kundock = 0.9 ± 0.1 s−1) was slightly increased by adenine and Mg2+. For both of the riboswitch variants, the ligand-induced increase in kundock translated to a minute ∼0.2 kcal/mol increase of the free docking enthalpy calculated from the ratio of kdock over kundock. This is in contrast to the ligand-induced stabilization of the docked state deduced from the FRET histograms (Table 1). However, the TODPs revealed a major proportion of static traces (≥50%) in a long-lived undocked or docked state at all investigated conditions (Figure 4B and Supplementary Figure S6B). The long-lived states were not susceptible to dwell-time analysis, as their dwell-times were in the same range as our average observation window until photobleaching (∼30 s). Apparently, the kinetics that accounted for the observed differences in the fractional population of D remained hidden in the long-lived states. The here characterized largely ligand-independent aptamer kissing loop docking kinetics of the full-length add Asw must correspond to similar substates of apoA and holo, as discussed below in greater detail.
DISCUSSION
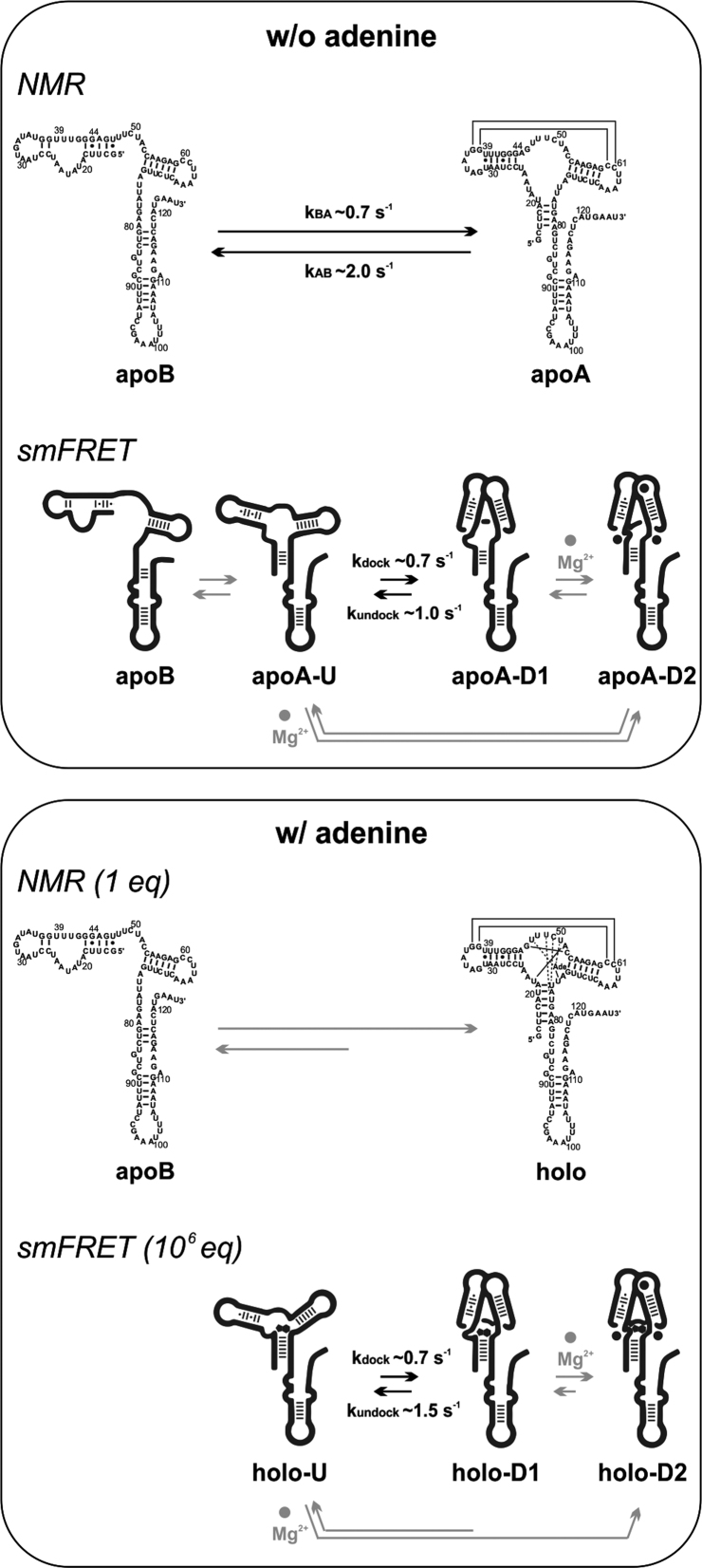
We have investigated the adenine-dependent folding of the native full-length translation-regulating add adenine riboswitch from Vibrio vulnificus at near-physiological Mg2+ concentration and ambient temperature by NMR and by smFRET spectroscopy. The add Asw exhibited persistent equilibrium dynamics between different conformational states both in the NMR and in the smFRET experiments. Figure 5 illustrates the conformational dynamics of this riboswitch as observed by the two solution-structural spectroscopies. NMR of the add Asw imino protons revealed an adenine-dependent equilibrium between two mutually exclusive secondary structures, the apoB-form and the apoA-form. In presence of adenine, the apoA-form was stabilized in the holo conformation by encapsulation of the adenine ligand in the aptamer core. The apoA and the holo aptamer had a docked aptamer kissing loop motif, as indicated by the single set of imino resonances observed for the P2/L2-hairpin. By smFRET in three different labeling schemes, we confirmed our NMR-derived hypothesis that adenine-dependent tertiary structural interactions of the full-length add Asw are confined to the aptamer domain. Further, the smFRET analysis of the wt along with an apoB-stabilized mutant and an apoA-stabilized mutant in the L2/L3-labeling scheme demonstrated that saturating adenine concentrations eliminated folding of the apoB conformation in wt. This proved that the mechanistic model of an adenine-induced secondary structure switch from the apoB-form to the apoA-form is valid at vastly different concentration ratios of Mg2+ over RNA, ranging from ∼17 equivalents in the case of NMR to ∼107 equivalents in the case of smFRET.
Figure 5.
Adenine-dependent equilibrium folding of the full-length add Asw observed by NMR and smFRET spectroscopy at ambient temperature and near-physiological Mg2+ concentration. Without adenine, NMR revealed the equilibrium between the apoB and the apoA conformation with docked aptamer loops. The indicated rate constants were previously determined via NZZ exchange experiments (2). By smFRET we could infer the apoB conformation and three substates of the apoA conformation: ApoA-U with undocked aptamer, apoA-D1 with docked aptamer and apoA-D2 with the docked aptamer stabilized by Mg2+. For illustrative purposes, the three Mg2+ binding sites identified for the isolated aptamer domain are indicated in apoA-D2 (12). The kinetics observed by smFRET were specifically associated with the Mg2+-independent folding between apoA-U and apoA-D1. With adenine, NMR revealed an equilibrium shift from apoB to holo. By smFRET we observed equivalent substates to apoA in the holo conformation. The substate equilibrium was shifted towards holo-D2 while undocking of holo-D1 was slightly accelerated.
However, our smFRET data of L2/L3-labeled add Asw unambiguously showed tertiary structure heterogeneity of the aptamer kissing loop motif in the apoA and in the holo conformation. Both the wt and the apoA-stabilized mutant populated co-existing FRET states with undocked and docked aptamer kissing loops at ligand-free and at ligand-saturating conditions. The undocked and docked aptamer exhibited heterogeneous lifetimes on the second to minute timescale. Adenine and Mg2+ increased the fractional occupancy of docked aptamers without substantially affecting the measurable second timescale docking and undocking kinetics. The simplest possible kissing-loop docking mechanism that can describe these observations involves three distinct substates of the apoA-form secondary structure. We denoted these substates by U, D1 and D2. In absence of adenine, the undocked apoA-U state was in equilibrium with a weakly docked apoA-D1 state and with a firmly docked apoA-D2 state that was stabilized by the binding of Mg2+ ions. The Mg2+-induced folding into apoA-D2 could occur via the induced fit pathway from apoA-U to apoA-D2 or via the conformational capture pathway from apoA-U over apoA-D1 to apoA-D2. Both pathways were slower than the accessible timescale of our single-molecule dwell-time histograms (∼15s). The dwell-time histograms revealed the Mg2+-independent docking and undocking kinetics between apoA-U and apoA-D1 that were on the same timescale as the rates of interconversion between apoB and apoA previously determined by NZZ exchange NMR spectroscopy. Notably, the clear separation of the undocking kinetics of the two docked substates D1 and D2 is characteristic of the native full-length sequence and has not been observed by smFRET of the P1-stabilized add Asw aptamer domain in the identical labeling scheme (13,14). At saturating adenine conditions, we observed the equivalent three substates in the holo form. The conformational equilibrium between holo-U, holo-D1 and holo-D2 was shifted towards holo-D2 and the undocking of holo-D1 was slightly accelerated. Overall, the folding of the loop–loop interaction in apoA and in holo proceeded through strikingly similar folding pathways.
It is remarkable that smFRET revealed an adenine-directed equilibrium between comparably stable undocked and docked substates of the apoA-form aptamer, while NMR exclusively showed a docked aptamer kissing-loop fold. Since we used the same RNA sequence for NMR and smFRET, this difference must be associated (i) with the different sensitivity of the two spectroscopies toward tertiary structure heterogeneity and/or (ii) with the different in vitro conditions employed by the two methods. As for the first point, it is a known drawback of NMR of RNA that imino resonances can only be observed for structured parts of the RNA, while for transient base pairs or unpaired nucleotides, the imino resonances are broadened beyond detection due to solvent-exchange (31). As characteristic imino signals for the undocked apoA-form aptamer would correspond to the solvent-accessible, loop-terminal base pairs of the P2/L2 or the P3/L3 hairpin, it is plausible to assume that such signals were undetectable in our study as a result of solvent-exchange. This hypothesis is further supported by previously reported 1H,15N-correlation spectra of the isolated add Asw aptamer domain that were measured in the absence of both adenine and Mg2+ (11). These spectra lacked the imino reporter signals of the L2/L3 docked state but did not show additional signals that could be specifically assigned to an L2/L3 undocked state. Regarding the second point, one needs to consider potential effects of dye-labeling or surface-tethering on the stability of the docked state in the smFRET experiment. The here employed L2/L3 FRET-labeling scheme was carefully devised to avoid interference with the loop-loop interaction on the basis of the X-ray structure of the add Asw aptamer (13). Moreover, the fluorescence anisotropies of the RNA-coupled dyes suggested that there were no specific interactions between the dyes and their environment at the L2 or the L3 labeling site. It is thus unlikely that dye-labeling significantly affected the conformational equilibrium of the add Asw. Surface-tethering of the RNA was here achieved by 3΄-immobilization via a biotin-streptavidin tag. Although the 3΄-biotin (Dharmacon) was attached via a long (24-atom) chemical spacer that was adjacent to seven single stranded nucleotides of the add Asw, we cannot fully exclude that surface-tethering influenced the immobilized riboswitch molecules. Yet, we have considered this possibility in our construct design by choosing the 3΄-end of the add Asw for immobilization, so that the protein-coated surface mimics physiologically relevant conditions where the large 30S ribosome is expected to be bound downstream of the riboswitch at a standby site for translation initiation (32). Recent studies have demonstrated that the folding of the add Asw aptamer domain can respond to variations in environmental conditions such as different concentrations of low-molecular-weight solutes and macromolecular crowders (33,34). It is therefore possible that environmental effects associated with surface proximity, RNA concentration or method specific buffer additives could have changed the fractional population of folding states in smFRET compared to NMR.
Taken together, our NMR and smFRET data highlight the necessity to integrate complementary solution structural techniques to uncover the full conformational space of riboswitch RNAs. Yet, they raise the question which of the conformational transitions of the add Asw regulate translation initiation. Transcription-translation coupled luciferase in vitro expression assays with a luciferase reporter gene under riboswitch control have shown that the apoA-stabilized mutant is an ON-state mutant (2). This suggested that the secondary structure switch from the apoB-form to the apoA-form is the primary regulatory step in the molecular mechanism of the add Asw. That we here observed an adenine-dependent switch from the apoB-form to the apoA-form by both NMR and smFRET corroborates this hypothesis. However, in order to clarify to what extent tertiary structure folding dynamics between substates of apoA and holo contribute to translation initiation, the conformational dynamics of the add Asw must be characterized in complex with the 30S ribosome. Such studies are under way in our laboratory.
Supplementary Material
ACKNOWLEDGEMENTS
We kindly thank Robert Ernst and his group for access to their steady-state fluorescence spectrometer.
Footnotes
Present address: Philipp Höllthaler, Institute of Biochemistry, Biocenter, Johann Wolfgang Goethe-Universität, Frankfurt am Main, Hessen 60438, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [SFB 902 to B.F., Mi.He., Ma.He. and H.S., EXC115 to Mi.He., Ma.He. and H.S., SP1157 to H.S.]; Fonds of the Chemical Industry (Chemiefonds fellowship to S.W.); State of Hesse [BMRZ]. Funding for open access charge: Johann Wolfgang Goethe-Universität.
Conflict of interest statement. None declared.
REFERENCES
- 1. Serganov A., Nudler E.. A decade of riboswitches. Cell. 2013; 152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reining A., Nozinovic S., Schlepckow K., Buhr F., Fürtig B., Schwalbe H.. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013; 499:355–359. [DOI] [PubMed] [Google Scholar]
- 3. Fürtig B., Nozinovic S., Reining A., Schwalbe H.. Multiple conformational states of riboswitches fine-tune gene regulation. Curr. Opin. Struct. Biol. 2015; 30:112–124. [DOI] [PubMed] [Google Scholar]
- 4. Liberman J.A., Wedekind J.E.. Riboswitch structure in the ligand-free state. Wiley Interdiscip. Rev. RNA. 2012; 3:369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Savinov A., Perez C.F., Block S.M.. Single-molecule studies of riboswitch folding. Biochim. Biophys. Acta. 2014; 1839:1030–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ziegeler M., Cevec M., Richter C., Schwalbe H.. NMR studies of HAR1 RNA secondary structures reveal conformational dynamics in the human RNA. Chembiochem. 2012; 13:2100–2112. [DOI] [PubMed] [Google Scholar]
- 7. Rinnenthal J., Buck J., Ferner J., Wacker A., Fürtig B., Schwalbe H.. Mapping the landscape of RNA dynamics with NMR spectroscopy. Acc. Chem. Res. 2011; 44:1292–1301. [DOI] [PubMed] [Google Scholar]
- 8. Al-Hashimi H.M., Walter N.G.. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 2008; 18:321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Draper D.E., Grilley D., Soto A.M.. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005; 34:221–243. [DOI] [PubMed] [Google Scholar]
- 10. Haller A., Rieder U., Aigner M., Blanchard S.C., Micura R.. Conformational capture of the SAM-II riboswitch. Nat. Chem. Biol. 2011; 7:393–400. [DOI] [PubMed] [Google Scholar]
- 11. Lee M.-K., Gal M., Frydman L., Varani G.. Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:9192–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Noeske J., Schwalbe H., Wöhnert J.. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007; 35:5262–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemay J.-F., Penedo J.C., Tremblay R., Lilley D.M.J., Lafontaine D.A.. Folding of the adenine riboswitch. Chem. Biol. 2006; 13:857–868. [DOI] [PubMed] [Google Scholar]
- 14. Dalgarno P.A., Bordello J., Morris R., St-Pierre P., Dube A., Samuel I.D.W., Lafontaine D.A., Penedo J.C.. Single-molecule chemical denaturation of riboswitches. Nucleic Acids Res. 2013; 41:4253–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y., Holmstrom E., Zhang J., Yu P., Wang J., Dyba M.A., Chen D., Ying J., Lockett S., Nesbitt D.J. et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature. 2015; 522:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nozinovic S., Reining A., Kim Y.-B., Noeske J., Schlepckow K., Wöhnert J., Schwalbe H.. The importance of helix P1 stability for structural pre-organization and ligand binding affinity of the adenine riboswitch aptamer domain. RNA Biol. 2014; 11:655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marcano-Velázquez J.G., Batey R.T.. Structure-guided mutational analysis of gene regulation by the Bacillus subtilis pbuE adenine-responsive riboswitch in a cellular context. J. Biol. Chem. 2015; 290:4464–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Helmling C., Keyhani S., Sochor F., Fürtig B., Hengesbach M., Schwalbe H.. Rapid NMR screening of RNA secondary structure and binding. J. Biomol. NMR. 2015; 63:67–76. [DOI] [PubMed] [Google Scholar]
- 19. Stoldt M., Wöhnert J., Görlach M., Brown L.R., Allain F., Gubser C., Howe P., Nagai K., Neuhaus D., Varani G. et al. The NMR structure of Escherichia coli ribosomal protein L25 shows homology to general stress proteins and glutaminyl-tRNA synthetases. EMBO J. 1998; 17:6377–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Favier A., Brutscher B.. Recovering lost magnetization: polarization enhancement in biomolecular NMR. J. Biomol. NMR. 2011; 49:9–15. [DOI] [PubMed] [Google Scholar]
- 21. Solyom Z., Schwarten M., Geist L., Konrat R., Willbold D., Brutscher B.. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR. 2013; 55:311–321. [DOI] [PubMed] [Google Scholar]
- 22. Sklenář V., Bax A.. Spin-echo water suppression for the generation of pure-phase two-dimensional NMR spectra. J. Magn. Reson. 1987; 74:469–479. [Google Scholar]
- 23. Mori S., Abeygunawardana C., Johnson M.O., van Zijl P.C.. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. B. 1995; 108:94–98. [DOI] [PubMed] [Google Scholar]
- 24. Goddard T.D., Kneller D.G.. Sparky 3. 2008; Univ. California, San Fr
- 25. Hengesbach M., Kim N.-K., Feigon J., Stone M.D.. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew. Chem. Int. Ed. 2012; 51:5876–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blanco M. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 2010; 472:153–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKinney S.A., Joo C., Ha T.. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006; 91:1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kobitski A.Y., Nierth A., Helm M., Jaschke A., Nienhaus G.U.. Mg2+-dependent folding of a Diels-Alderase ribozyme probed by single-molecule FRET analysis. Nucleic Acids Res. 2007; 35:2047–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brenner M.D., Scanlan M.S., Nahas M.K., Ha T., Silverman S.K.. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010; 49:1596–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tyrrell J., McGinnis J.L., Weeks K.M., Pielak G.J.. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013; 52:8777–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fürtig B., Schnieders R., Richter C., Zetzsche H., Keyhani S., Helmling C., Kovacs H., Schwalbe H.. Direct 13C-detected NMR experiments for mapping and characterization of hydrogen bonds in RNA. J. Biomol. NMR. 2016; 64:207–221. [DOI] [PubMed] [Google Scholar]
- 32. de Smit M.H., van Duin J.. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 2003; 331:737–743. [DOI] [PubMed] [Google Scholar]
- 33. Trachman R.J., Draper D.E., Draper D.E.. Comparison of interactions of diamine and Mg2+ with RNA tertiary structures: similar versus differential effects on the stabilities of diverse RNA folds. Biochemistry. 2013; 52:5911–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tyrrell J., Weeks K.M., Pielak G.J.. Challenge of mimicking the influences of the cellular environment on RNA structure by PEG-induced macromolecular crowding. Biochemistry. 2015; 54:6447–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.