Abstract
The human DEAD-box RNA-helicase DDX19 functions in mRNA export through the nuclear pore complex. The yeast homolog of this protein, Dbp5, has been reported to participate in translation termination. Using a reconstituted mammalian in vitro translation system, we show that the human protein DDX19 is also important for translation termination. It is associated with the fraction of translating ribosomes. We show that DDX19 interacts with pre-termination complexes (preTCs) in a nucleotide-dependent manner. Furthermore, DDX19 increases the efficiency of termination complex (TC) formation and the peptide release in the presence of eukaryotic release factors. Using the eRF1(AGQ) mutant protein or a non-hydrolysable analog of GTP to inhibit subsequent peptidyl-tRNA hydrolysis, we reveal that the activation of translation termination by DDX19 occurs during the stop codon recognition. This activation is a result of DDX19 binding to preTC and a concomitant stabilization of terminating ribosomes. Moreover, we show that DDX19 stabilizes ribosome complexes with translation elongation factors eEF1 and eEF2. Taken together, our findings reveal that the human RNA helicase DDX19 actively participates in protein biosynthesis.
INTRODUCTION
DEAD-box RNA helicases are found in all eukaryotes, as well as the majority of bacteria and some archaea (1,2). They play important roles in all aspects of RNA metabolism, from transcription to degradation of mRNA (1–3). These enzymes use the free energy of ATP hydrolysis to catalyze the separation of RNA duplexes into single stranded RNA and to remodel of RNA-protein complexes (4). DEAD-box RNA helicases are defined by the presence of the characteristic eponymous sequence Asp-Glu-Ala-Asp (DEAD) (5). One representative of this family is the human ATP-dependent RNA-helicase DDX19 (3). It is an essential protein that is conserved in eukaryotes (6,7). Like in all members of the family, the helicase core of DDX19 consists of two structurally similar domains called RecA-like domains, linked to each other by a flexible linker. Most conserved sequence motifs of this protein are located in the central cleft between two RecA domains, involved in binding and hydrolysis of ATP (3). The RNA is bound by both domains in a sequence-independent manner. The N-terminal part of DDX19 (residues 1–91) has auto-regulatory activity (8).
In the human genome two genes coding DDX19 (A and B) were identified. They are located close to each other on the chromosome 16. DDX19A consists of 478 amino acid residues; DDX19B comprises 479 amino acid residues. The amino acid sequences of these proteins are 96% identical; the two proteins differ only by 17 amino acid residues located in the N-terminal part between residues 17 and 44. The sequence in this region is variable in DEAD-box helicases, and therefore this part is probably not essential for activity. DDX19B is a homolog of well-studied yeast protein Dbp5, and DDX19A was annotated as DDX19-like protein. Importantly, the amino acid sequence of human DDX19B is only 46% identical to the yeast Dbp5 sequence (7). Consequently, they may have different functions.
DDX19 and Dbp5 have been shown to have a role in mRNA export through the nuclear pore complex (NPC) (7,9,10). They act in the final steps of mRNA export and were shown to interact with the NPC via the cytoplasmic nucleoporin Nup214 (Nup159 in yeasts) (7,11–13). The ATPase activity of DDX19 and Dbp5 in vitro is very low. At the nuclear rim, they are activated by Gle1 and its cofactor inositol 1,2,3,4,5,6-hexakiphosphate (IP6) (14,15). ADP-bound Dbp5 is able to displace recombinant Nab2 from mRNA–protein complex. In the presence of Gle1 and IP6, its ability increases (16). Mutations within the conserved motifs of the ATPase center inhibit mRNA export and are lethal in vivo. Furthermore, Dbp5 has been shown to be required for Mex67 dissociation (NXF1 in human) from exported mRNA (17), and the nuclear export factor RBM15 was shown to facilitate recognition of NXF1-mRNP complexes by DDX19 in humans (18).
Several studies provided evidence that Dbp5 and its homologous protein from Chironomus tentans participate in transcription (6). These studies were confirmed recently when DDX19 was shown to be required for nuclear import of MKL1, coactivator of the serum response factor (19), implicating human DDX19 in transcription regulation.
Using fluorescent fusion proteins, DDX19/Dbp5 was localized on the cytoplasmic side of the NPC and in the cytoplasm (6,7,9,10,12). Besides the most studied activity of this protein in mRNA transport through the nuclear pore, an additional cytoplasmic function was supposed. In 2007, Dbp5 was shown to participate in the translation termination (20), revealing its function in the cytoplasm. Termination of translation occurs when the release factors bind to the stop codon positioned in the decoding center of the ribosome. In eukaryotes, all three stop codons are recognized by release factor 1 (eRF1) (21). After stop codon recognition, release factor 3 (eRF3) hydrolyses GTP that induces a conformational change in eRF1 (22–26). As a result, the GGQ motif of eRF1 is accommodated in the peptidyl transferase center (PTC) of the ribosome and induces hydrolysis of peptidyl-tRNA. eRF3 is required to increase the efficiency of translation termination in eukaryotes; GTP hydrolysis by eRF3 stimulates peptidyl-tRNA hydrolysis in the PTC (27–29).
In yeast lysates, Dbp5 was detected in the cytoplasmic poly-ribosome fraction after sucrose density gradient (SDG) centrifugation (20). The addition of puromycin that disrupts polysomes leads to the displacement of Dbp5 into the fraction of mono-ribosomes. This confirmed that Dbp5 binds with the ribosomes. Subsequently, yeast experiments demonstrated that overexpression of DBP5 cannot suppress mutations in genes encoding initiation and elongation factors, but mutations in both genes encoding release factors SUP45 (eRF1 in human) and SUP35 (eRF3 in human) were suppressed by Dbp5. Additionally, different mutants of Dbp5 were synthetically lethal with the SUP45 or SUP35 mutants. Co-immunoprecipitation experiments indicated that Dbp5 interacts with the SUP45 in a RNA-independent manner. At the same time, no interaction between Dbp5 and SUP35 or Pab1 was identified in the absence of mRNA. Overexpression of DBP5 decreased stop codon read-through of the release factor mutants. The ATPase activity of Dbp5 was shown to be required for stop coon read-through suppression, and a mutation in the DEAD motif (E240Q) of Dbp5 eliminated this effect (20). Moreover, the cofactors of Dbp5 Gle1 and IP6 are implicated in translation termination as well (30,31). Gle1 mutants were identified which render yeast hypersensitive to antibiotics and increased stop codon read-through. Accordingly, Gle1 was detected in the polysome fractions after SDG centrifugation and found to interact with eRF1 (31). Strains lacking Ipk1 (IP6 producing kinase) are synthetically lethal with the mutant SUP45.
Rate control analyses of translation in yeast revealed that Dbp5 and eRF1 belong to a group of proteins, which are strongly associated with the important control points during protein synthesis (32). In fact, translation in yeast is very sensitive to the intracellular concentration of these proteins. A 20% decrease of the basal protein levels of Dbp5 or eRF1 in the cell has immediate negative effects on protein biosynthesis. It is important to note that the cellular concentration of eRF3 does not influence the rate of translation (a 60% decrease had no significant effect). These data confirm the important role of Dbp5 during translation in yeast.
More recently, it was shown that Dbp5 is also required for the nuclear export of both pre-ribosomal subunits. Dbp5 mutants were identified that lead to an accumulation of ribosomal subunits in the nucleus (33). During ribosome export, Dbp5 acts at the cytoplasmic side of the NPC as reported previously for mRNA export. This function of Dbp5 does not require ATPase activity. Most likely, Dbp5 binds with the ribosomal subunits at the cytoplasmic side of the NPC, escorts them to the cytoplasm and subsequently participates in translation.
Despite the fact that the cytoplasmic function of the yeast Dbp5 is described, the cytoplasmic activity of human DDX19 remained unclear. Using an in vitro reconstituted mammalian translation system, we found that DDX19 is involved in translation termination and elongation. DDX19 stabilizes termination complexes during stop codon recognition and increases the peptide release activity of eRFs. We demonstrated that DDX19 binds to pre-termination complexes in a nucleotide-dependent manner. Moreover, we revealed that DDX19 is able to stabilize elongation complexes in the presence of eEF1-aatRNA-GTP and eEF2-GTP/GMPPNP.
MATERIALS AND METHODS
Sucrose density gradient fractionation and Western blot analysis
HEK 293T cells were grown in a 15 cm dish to about 70% confluency. Prior to their lysis, cells were grown for another 15 min in the presence of 100 μg/ml cycloheximide. Then, the cells were lysed in 400 μl ice-cold polysome lysis buffer (20 mM Tris-HCl pH 7.5, 250 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 100 μg/ml cycloheximide) and centrifuged for 20 min at 16 000 g at 4°C. The supernatant was loaded onto a linear 12 ml 10–60% (w/v) sucrose gradient (20 mM Tris-HCl pH 7.5, 250 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 10% and 60% of sucrose). The samples were centrifuged at 40 000 rpm using a SW40 rotor (Beckman) at 4°C for 90 min and subsequently fractionated into 600 μl fractions with a continuous monitoring of the absorbance at 260 nm. Each fraction was precipitated with 10% (w/v) trichloracetic acid. The resulting pellet was dried and finally resuspended in 50 μl of SDS sample buffer. Samples were separated on a 10% SDS-PAGE and analyzed by Western blotting using different antibodies. For Western blots, anti-RPS15, anti-RPL15, anti-eRF1, anti-DDX19B antibodies (Abcam) were used for detection.
Ribosomal subunits and translation factors
The 40S and 60S ribosomal subunits as well as eukaryotic translation factors eIF2, eIF3, eEF1H and eEF2, were purified from rabbit reticulocyte lysate as described (22). Human translation factors eIF1, eIF1A, eIF4A, eIF4B, ΔeIF4G, ΔeIF5B, eIF5, DDX19A, DDX19B, DDX19B mutants, wt eRF1, eRF1 mutants (eRF1(AGQ), eRF1(K83N)) and eRF3c lacking the N-terminal domain (138 amino acid residues) were produced as recombinant proteins in E. coli strain BL21 and subsequently purified via Ni-NTA agarose and ion-exchange chromatography (22). Human full-length eRF3a (GSPT1) was kindly provided by Dr Christiane Schaffitzel. It was cloned into the pFastBac-Htb vector (Life Technolgies) and expressed in insect cells Sf21 using the EMBacY baculovirus from the MultiBac expression system (34). Hydroxylated form of human eRF1 (eRF1-OH), expressed in E. coli (35), was kindly provided by Dr Mathew Coleman.
In vitro transcription of mRNA
Using T7 RNA polymerase mRNA was transcribed in vitro from the MVHL-stop plasmid that contains a T7 promoter, four CAA repeats, the β-globin 5΄-untranslated region, an open reading frame (encoding for the peptide MVHL), followed by the UAA stop codon and a 3΄-untranslated region comprising the rest of the natural β-globin coding sequence (36). For run-off transcription, the MVHL-stop plasmid was linearized by restriction digest with XhoI.
Assembly of ribosomal complexes in vitro
Initiation complexes were assembled in 30 μl at 0°C and contained 2 pmol mRNA, 6 pmol Met-tRNAiMet, 4.5 pmol 40S and 60S ribosomal subunits, 7.5 pmol eIF2, eIF3, eIF4A, ΔeIF4G, eIF4B, eIF1, eIF1A, eIF5, ΔeIF5B each, supplemented with buffer composed of 20 mM Tris acetate, pH 7.5, 100 mM KAc, 2.5 mM MgCl2, 2 mM DTT, 0.3 U/μl RNAse inhibitor, 1 mM ATP, 0.25 mM spermidine, 0.2 mM GTP. The reaction mixture was kept at 37°C for 15 min to allow ribosomal-mRNA complex formation, and 10 μl sample subsequently was applied in the toe-print assay.
Peptide elongation was performed in 20 μl by addition of 3 pmol total tRNA (acylated with all or individual amino acids), 8 pmol eEF1H and 2 pmol eEF2 to the initiation complex and was incubated for 15 min at 37°C. A 10 μl sample, containing preTC, subsequently was applied in the toe-print assay.
The last 10 μl sample, containing preTC, was supplemented with 5 pmol eRF1 and 5 pmol eRF3a/c. The reaction mixture was incubated at 37°C for 15 min and subsequently was applied in the toe-print assay.
Purified preTCs assembly and termination analyses
The preparative amount of preTC was assembled in vitro as described (37) and used in peptide release assay and conformational rearrangement analysis (38). Briefly, 37 pmol of mRNA were incubated for 30 min in buffer A (20 mM Tris acetate, pH 7.5, 100 mM KAc, 2.5 mM MgCl2, 2 mM DTT) supplemented with 400 U RNAse inhibitor, 1 mM ATP, 0.25 mM spermidine, 0.2 mM GTP, 75 μg total tRNA (acylated with all or individual amino acids and [35S]Met), 75 pmol 40S and 60S purified ribosomal subunits, 125 pmol eIF2, eIF3, eIF4A, ΔeIF4G, eIF4B, eIF1, eIF1A, eIF5, ΔeIF5B each, 200 pmol eEF1H and 50 pmol eEF2 and then centrifuged in a Beckman SW55 rotor for 95 min at 4°C, 50 000 rpm in a 10–30% (w/w) linear sucrose density gradient prepared in buffer A with 5 mM MgCl2. Fractions corresponding to preTC complexes according to optical density and the presence of [35S]Met were combined, diluted 3-fold with buffer A containing 1.25 mM MgCl2 (to a final concentration of 2.5 mM Mg2+) and used in peptide release assay or conformational rearrangement analysis.
For peptide release assay 30 μl aliquots containing 0.09 pmol preTCs were incubated at 37°C with 0.5 pmol of eRF1 and 10 pmol of DDX19B for 15 min, or with 0.1 pmol of eRF1, eRF3c with 0.2 mM GTP and 10 pmol of DDX19B for 3 min. Ribosomes and tRNA were pelleted with ice-cold 5% TCA and centrifuged at 14 000 g at 4°C. The amount of released [35S]-containing peptide was determined by scintillation counting of supernatants using an Intertechnique SL-30 liquid scintillation spectrometer.
For conformational rearrangement (or toe-printing) analysis 10 μl aliquots containing 0.03 pmol preTCs were incubated at 37°C for 15 min with 0.625 pmol of eRF1 or eRF1 mutants (eRF1(AGQ), eRF1(K83N), eRF1-OH), eRF3c/a with 0.2 mM GTP/GMPPNP and 6.25 pmol of DDX19B or DDX19B mutants. Samples were analyzed using a primer extension protocol. Toe-printing analysis was performed with AMV reverse transcriptase and 5΄-FAM labeled primer 5΄-FAM-GCATTTGCAGAGGACAGG-3΄ complementary to β-globin mRNA nucleotides 197–214. cDNAs were separated by electrophoresis using standard GeneScan® conditions on an ABI Prism® Genetic Analyser 3100 (Applera).
Stop codon binding efficiency was calculated using the formula 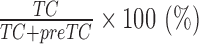 . All data were normalized to the stop codon binding efficiency of eRF1 alone.
. All data were normalized to the stop codon binding efficiency of eRF1 alone.
Elongation efficiency analyses
Purified preTCs were used to determine elongation efficiency in the presence of DDX19B or R372G mutant. For analysis of the eEF2-preTC complexes, 10 μl aliquots containing 0.03 pmol preTCs were incubated at 37°C for 15 min with 1 pmol eEF2 and 5 pmol DDX19B or mutant in the presence of 0.2 mM GTP/GMPPNP. Samples were analyzed using a primer extension protocol. The −1 shift efficiency was calculated using the formula 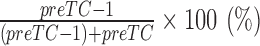 . All data were normalized to the eEF2-GMMPNP complex formation efficiency.
. All data were normalized to the eEF2-GMMPNP complex formation efficiency.
For analysis of translocation complexes, 10 μl aliquots containing 0.03 pmol preTCs were incubated at 37°C for 15 min with various amounts of eEF1 (0.5, 1, 1.5, 4 pmol), 8 pmol suptRNASer, 4 pmol eEF2 and 10 pmol DDX19B or with various amounts of eEF2 (0.1, 1, 4 pmol) 8 pmol suptRNASer, 4 pmol eEF1 and 10 pmol DDX19B. Samples were analyzed using a primer extension protocol. The +3 shift efficiency was calculated using the formula 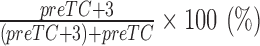 .
.
PreTC binding assay
Samples of 150 μl of purified preTCs (1.35 pmol) were incubated with either 10 pmol DDX19B or 10 pmol eRF1, 10 pmol eRF3c and 10 pmol DDX19B, in buffer A (20 mM Tris-HCl pH 7.5, 2.5 mМ MgCl2, 150 mM KAc, 2 mM DTT and 0.2 mM GTP with 0.2 mM MgCl2) at 37°C for 15 min in presence of different nucleotides. Complexes were centrifuged into a 10–30% (w/w) linear sucrose density gradient prepared with buffer A containing 5 mM MgCl2 for 95 min at 4°C and 50 000 rpm using a Beckman SW55 rotor. Each gradient was fractionated into 14 equal fractions followed by precipitation using 10% TCA. The protein pellets were dried and analyzed by Western blot.
ATPase assay
ATPase assays were carried out in the presence of 20 mM Tris-HCl pH 7.5, 15 mM NH4Cl, 5 mM MgCl2, 1% glycerol, 1 mM DTT, 0,1 mg/ml poly(A), 1 mg/ml bovine serum albumin, 1 mM ATP and 0.1 μl of [α-32P]ATP (20 μCi/μl, 3000 Ci/mmol). Reaction (10 μl) was initiated by the addition of 5 μg of purified DDX19 wild-type or mutants. After incubation for 30 min at 37°C, the reaction was stopped by adding 500 μl of a solution containing 5% charcoal in 25 mM NaH2PO4 to bind free ATP. Then the charcoal was precipitated by centrifugation, 375 μl of the reaction were subjected to liquid scintillation counting to quantify the released [32P] phosphate.
RESULTS
Human DDX19 is associated with polysomes
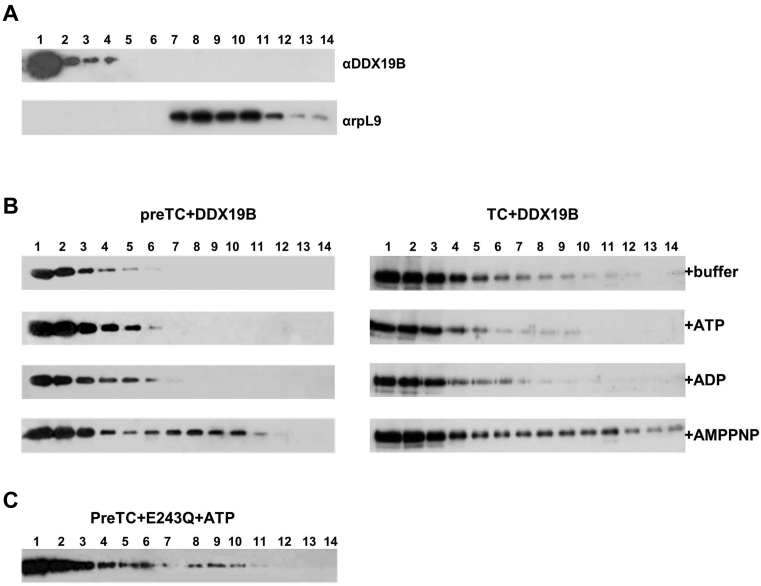
To investigate whether the cytoplasmic function of Dbp5 is conserved in higher eukaryotes, we have tested the association of DDX19 with polysomes in HEK293T cell lysate. Western blot analysis of fractions from sucrose density gradient centrifugation revealed that a significant amount of human DDX19 is found in polysome-containing fractions (Figure 1). Similarly, eRF1 is found in these polysome fractions. Based on this finding, we cloned the two annotated isoforms of human DDX19 (DDX19A and DDX19B), expressed them in Escherichia coli, purified the proteins and characterized their ATPase activities (Supplementary Data). We observed that enzymatic activities of the two isoforms were almost identical. Next, we analyzed both proteins by Western blotting using antibodies directed against DDX19B. We found that DDX19A also can be detected by these antibodies (data not shown). Therefore, we cannot discriminate based on Western blotting which of the two isoforms is found in polysome fraction. Since all published studies used DDX19B, and we observed no difference between the A and B forms in the ATPase assay, we decided to focus for all further experiments on DDX19B. However, in most assays we also tested the activity of DDX19A.
Figure 1.
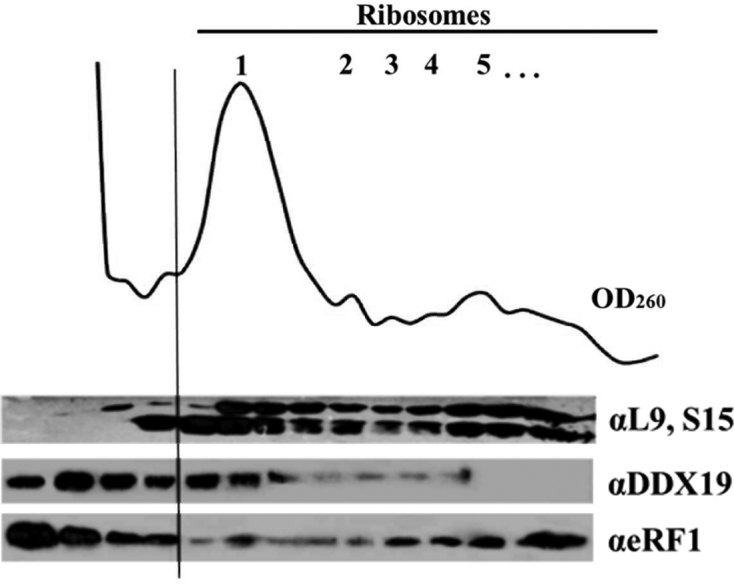
DDX19 is associated with polysomes during translation. Supernatant of HEK 293 cell lysate were loaded onto a 10–60% sucrose gradient, centrifuged, fractionated and subjected to Western blot analysis. Antibodies against ribosomal proteins L9 and S15 were used to detect ribosomal subunits. The absorbance at 260 nm (OD, optical density) shows the distribution of ribosomes. DDX19 and eRF1 are detected by specific antibodies.
Because DDX19 was found in polysome fractions, we decided to study whether and how DDX19 affects eukaryotic translation in vitro. DDX19B was added to all translational complexes (initiation, elongation and termination) that were assembled in vitro from individual components on the MVHL-UAA mRNA. To verify translation complex formation toe-print analyses were performed. Surprisingly, we observed no difference in the efficiency of the assembly of ribosomal complexes in the presence or absence of DDX19B (Supplementary Data).
Previously it was shown that the yeast analog Dbp5 binds eRF1 in a RNA-independent manner (20). Therefore, we tested whether DDX19 interacts directly with human eRF1. After pre-incubation of DDX19B with eRF1, the reaction was applied onto a gel filtration column. We could not detected complex formation in the absence of the ribosomes indicating that there is no direct interaction or a low affinity interaction of eRF1 and DDX19 (Supplementary Data).
DDX19 binds preTCs in a nucleotide-dependent manner
Since DDX19 was detected in the polysomal fractions of HEK293T cell lysates, we next asked whether this protein could bind to purified preTCs and TCs in the presence or absence of ATP, ADP or a non-hydrolysable ATP analog (AMPPNP). Ribosomal complexes purified by SDG centrifugation were incubated with DDX19B, release factors and nucleotides. The complexes were centrifuged into a SDG again. DDX19B was detected in the fractions by Western blotting.
To exclude that the resulting profile is caused by aggregation of DDX19B, we tested the distribution of this protein in the SDG in the absence of ribosomal complexes. DDX19B alone can be detected only in the top fractions of the gradient (fractions 1–4) whereas ribosomal complexes migrate in the fractions 8–14 (Figure 2A).
Figure 2.
preTC binding experiments with DDX19 and release factors. (A) Western blot analyses of DDX19B and purified preTCs separated via SDG centrifugation using antibodies raised against DDX19B and ribosomal protein L9, respectively. (B) Western blot analysis of purified preTCs or TCs, with the complex of release factors eRF1-eRF3a-GTP, bound with DDX19B in the presence of ATP, ADP and AMPPNP and separated via SDG centrifugation. Antibodies raised against DDX19B were used for detection. (C) Western blot analysis of purified preTCs bound with DDX19B E243Q in the presence of ATP and separated via SDG centrifugation. Antibodies raised against DDX19B were used for detection.
In the absence of nucleotides as well as in the presence of ATP and ADP DDX19B did not form stable complexes with preTCs and TCs. The addition of AMPPNP to the reaction mix increased complex formation and/or stabilized DDX19B binding to ribosomal complexes (Figure 2B). It is noteworthy that release factors eRF1 and eRF3a (full-length eRF3, isoform a) virtually did not affect DDX19B-ribosome complexes formation.
We speculated that the presence of non-hydrolysable analog of ATP (AMPPNP) prevents dissociation of DDX19B from the ribosomal complexes. To verify this hypothesis we generated a DDX19B mutant (E243Q) which is defective in ATP hydrolysis (7). As expected the absence of ATPase activity stabilized the DDX19B(E243Q)-preTC complexes in the presence of ATP during SDG centrifugation (Figure 2C).
DDX19 stimulates the efficiency of TC formation
Since DDX19B binds purified preTCs and TCs in the presence of AMPPNP we decided to study influence of this protein on the translation termination using purified complexes. We assembled preTCs in vitro from individual components on MVHL-UAA mRNA and subsequently purified by SDG centrifugation. Then, we used them to test the TCs formation in the presence of DDX19B. For this purpose, toe-print analysis of formed complexes was performed (Figure 3). The essence of the method is to detect positions of stable ribosomal complexes on the mRNA via synthesis of cDNA products by reverse transcriptase (Figure 3A). cDNA molecules are synthesized with fluorescently labeled primers. In Figure 3B, we show examples of raw data of the toe-print analyses, obtained by capillary electrophoresis of cDNA.
Figure 3.
DDX19B increases TC formation. (A) A scheme of toe-print analysis of a ribosome–mRNA complex formation. Stable ribosomal complex on the mRNA stops reverse transcriptase (AMV) at a certain position generating cDNA products of specific lengths. cDNA molecules synthesized with fluorescently labeled primers are separated and detected using fragment analysis. (B) Examples of raw data from capillary electrophoresis of cDNA products obtained using fluorescently labeled primers for toe-print analysis. PreTCs are shown after SDG purification, TC formation is induced by addition of eRF1 and eRF3c to the preTCs. Positions of preTCs and TCs are labeled by white and black triangles, respectively. (C) Toe-print analysis of termination complexes formed by addition to the preTCs of eRF1, eRF1+eRF3c+GTP, eRF1+eRF3a+GTP and DDX19B+ATP. Rfu – relative fluorescence unit. Positions of preTCs and TCs are labeled by white and black triangles, respectively. (D) Relative quantitative analysis of the stop codon binding efficiency of eRFs in the presence of DDX19B. Stop codon binding efficiency of eRF1 alone was set as 100%. The error bars represent the standard deviation, stars (**) mark a significant difference from the respective control P < 0.01 (n = 3).
During stop codon recognition of eRF1, the ribosome protects additional nucleotides on the mRNA, which can be detected in toe-print assays as a two-nucleotide shift of the ribosomal complex (Figure 3C) (22,39,40). In preTCs the stop codon is found in the ribosomal A-site. The addition of eRF1 leads to the appearance of a peak, shifted for two nucleotides forward, corresponding to TCs formation during stop codon recognition by eRF1. For our experiments, we applied limiting concentrations of release factors in order to simulate of physiological conditions. This allowed us to detect any change of release factor activity in the presence of DDX19.
Toe-print analysis revealed that addition of DDX19B to preTCs with eRF1 increases the amount of TC formed (Figure 3C and D). Using different quantities of DDX19B, we confirmed the specificity of the stimulatory effect of DDX19B on TC formation (Supplementary Data). Interestingly, DDX19A showed the same activity as DDX19B (Supplementary Data).
In the presence of both release factors (eRF1 and eRF3) and GTP, DDX19B stimulated the two-nucleotide shift as well (Figure 3C). A quantitative analysis indicated that the stimulatory effect of DDX19B is more pronounced in the presence of eRF1 alone (3-fold) compared to eRF1+eRF3a/c and GTP (approximately 2-fold) (Figure 3D). This is likely due to a higher efficiency of TC formation. Moreover, DDX19B stimulated TC formation to a similar extend in the presence of different eRF3 variants, full-size eRF3a and an N-terminally truncated version eRF3c lacking the first 138 amino acids (Figure 3D). This indicates that the stimulatory effect of DDX19B is independent of the N-terminal domain of eRF3a. In the absence of release factors, no effect of DDX19B on preTCs was observed in toe-print assays (Figure 3C).
DDX19 increases peptidyl-tRNA hydrolysis by the release factors
Toe-print assays demonstrated that DDX19 stimulates TC formation. Thus, in order to investigate whether DDX19 affects peptidyl-tRNA hydrolysis we assembled preTCs on the MVHL-UAA mRNA using S35-labeled initiator-tRNA. The efficiency of peptidyl-tRNA hydrolysis was determined by quantification of radioactive MVHL peptide released from ribosomal complexes that were incubated with saturating amounts of eRF1, eRF3c-GTP and DDX19B in the presence of ATP or AMPPNP. We found that the efficiency of peptide release is increased after addition of DDX19B to the termination reaction (Figure 4). Incubation of DDX19B with preTC and eRF1 increases the efficiency of peptidyl-tRNA hydrolysis about 5-fold (Figure 4A). In contrast, addition of DDX19B to the termination reaction, induced by eRF1+eRF3c+GTP, stimulates peptidyl-tRNA hydrolysis by approximately 2-fold (Figure 4B).
Figure 4.
DDX19B increases the efficiency of peptidyl-tRNA hydrolysis induced by release factors. (A) Hydrolysis of peptidyl-tRNA induced by the addition of eRF1 in the presence/absence of DDX19B with ATP or AMPPNP (n = 3). (B) Hydrolysis of peptidyl-tRNA induced by the addition of eRF1 and eRF3c in the presence/absence of DDX19B with ATP or AMPPNP (n = 3). The error bars represent the standard deviation, stars (**) mark a significant difference from the respective control P < 0.01.
We tested the requirement of DDX19 dissociation from the ribosomal complexes for activation of peptide release. In preTC binding experiments, we showed that AMPPNP prevents dissociation of DDX19B from the ribosomal complexes (Figure 2B). Therefore, we compared activities of DDX19B in the presence of ATP and AMPPNP in peptide release assay (Figure 4). DDX19B increases hydrolysis of peptidyl-tRNA equally in the presence of ATP or AMPPNP with both eRF1 and eRF1+eRF3c+GTP. Thus, peptide release stimulation by DDX19 occurs before its dissociation from the ribosome.
Activation of translation termination occurs during stop codon recognition
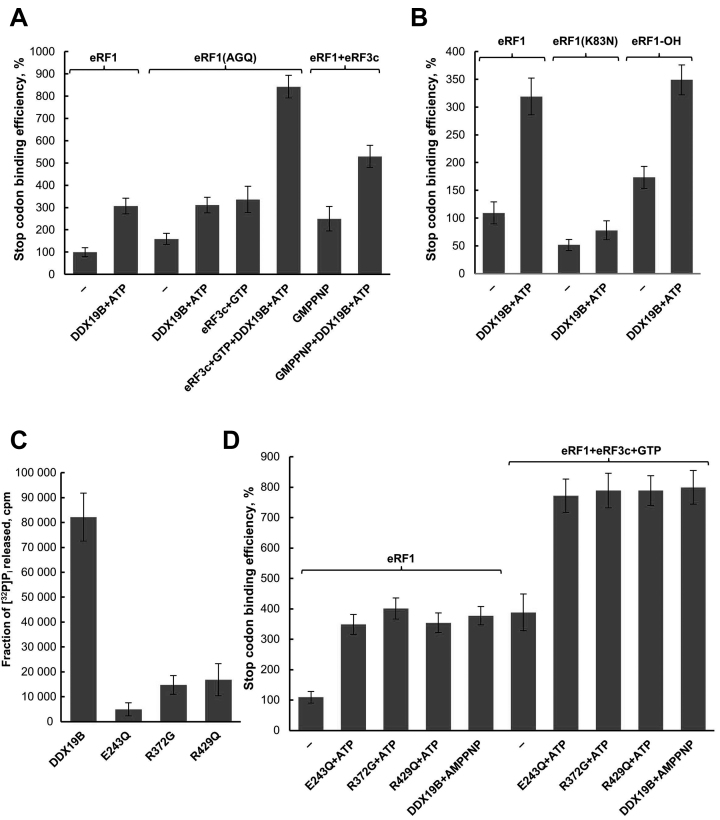
To determine the stage when activation of translation termination by DDX19B occurs, before or during peptidyl-tRNA hydrolysis, we used eRF1(AGQ) mutant which is unable to induce peptide release (27) in toe-print analysis. We found that DDX19B activates the stop codon recognition of eRF1(AGQ) to a similar extend as observed for wt eRF1 (Figure 5A, Supplementary Data).
Figure 5.
DDX19B activity in translation termination in the presence of eRFs variants. Relative quantitative analysis of the stop codon binding efficiency of (A) eRF1(AGQ), eRF1(AGQ)+eRF3c+GTP, eRF1+eRF3c+GMPPNP and (B) eRF1(K83N), hydroxylated eRF1 (eRF1-OH) in the presence of DDX19B. Stop codon binding efficiency of eRF1 alone was set as 100% (n = 3). (C) ATPase activity of DDX19B mutants E243Q, R372G and R429Q (n = 3). (D) Relative quantitative analysis of the stop codon binding efficiency of eRF1, eRF1+eRF3c+GTP and DDX19B mutants in the presence of ATP or AMPPNP. Stop codon binding efficiency of eRF1 alone was set as 100% (n = 3). The error bars represent the standard deviation.
To test whether the activation of termination by DDX19B is coupled to GTP hydrolysis by eRF3, we added a non-hydrolysable analog of GTP (GMPPNP) to the toe-print assays. We observed that, in the presence of GMPPNP, DDX19B also activates TC formation (Figure 5A, Supplementary Data). Therefore, stimulation of translation termination occurs before GTP hydrolysis by eRF3. Consequently, it precedes the accommodation of the M domain of eRF1 into the PTC of the ribosome and peptidyl-tRNA hydrolysis.
Dbp5, the yeast analog of DDX19, was shown to decrease stop codon readthrough in strains with a mutant eRF1 (SUP45-2 K80N) (20). We therefore decided to test how DDX19B influences on the activity of human eRF1 mutant with the same substitution (K83N). Similarly to wt eRF1, stop codon recognition by eRF1(K83N) is stimulated by addition of DDX19B (Figure 5B, Supplementary Data).
Earlier, we showed that C4 lysyl hydroxylation of the eRF1 K63 residue is required for optimal efficiency of translation termination (35). Recently published cryoEM structures of eukaryotic termination complexes confirmed that modified lysine 63 is directly involved in the recognition of the first nucleotide of the stop codon (39,40). The effect of DDX19B on the hydroxylated form of eRF1 (eRF1-OH) was tested. We found that the activation of eRF1 by DDX19B is independent from hydroxylation of eRF1 (Figure 5B, Supplementary Data).
Mutants of DDX19 activate translation termination
To confirm that the activity of DDX19B in termination does not depend on the hydrolysis of ATP, we tested in toe-print assay human DDX19 mutant that is defective in ATPase activity (DDX19(E243Q)) (7). The purified recombinant protein was characterized using ATPase assays (Figure 5C). As expected, the ATPase activity of DDX19(E243Q) was significantly decreased. This mutant stimulated TC formation to the same extent as wild-type DDX19B (Figure 5D, Supplementary Data). This confirms that the activation of translation termination occurs before ATP hydrolysis. Moreover, we performed toe-print analysis of termination complexes in the presence of AMPPNP (Figure 5D, Supplementary Data). As expected, DDX19B stimulated TC formation also in the presence of AMPPNP.
DDX19B also binds RNA (7), we therefore asked whether this ability changes its stimulatory effect on translation termination. We produced two mutant forms of DDX19B deficient in RNA-binding (DDX19(R372G) and DDX19(R429Q)) (7,41). As shown previously, these mutants were virtually inactive in ATPase assays (Figure 5C) (7). In toe-print assays, DDX19(R372G) and DDX19(R429Q) activated TC formations to a similar extent like DDX19 wild type (Figure 5D, Supplementary Data). Consequently, RNA binding activity of DDX19B is not required for its function in translation termination.
DDX19 stimulates activities of translation elongation factors in ribosome
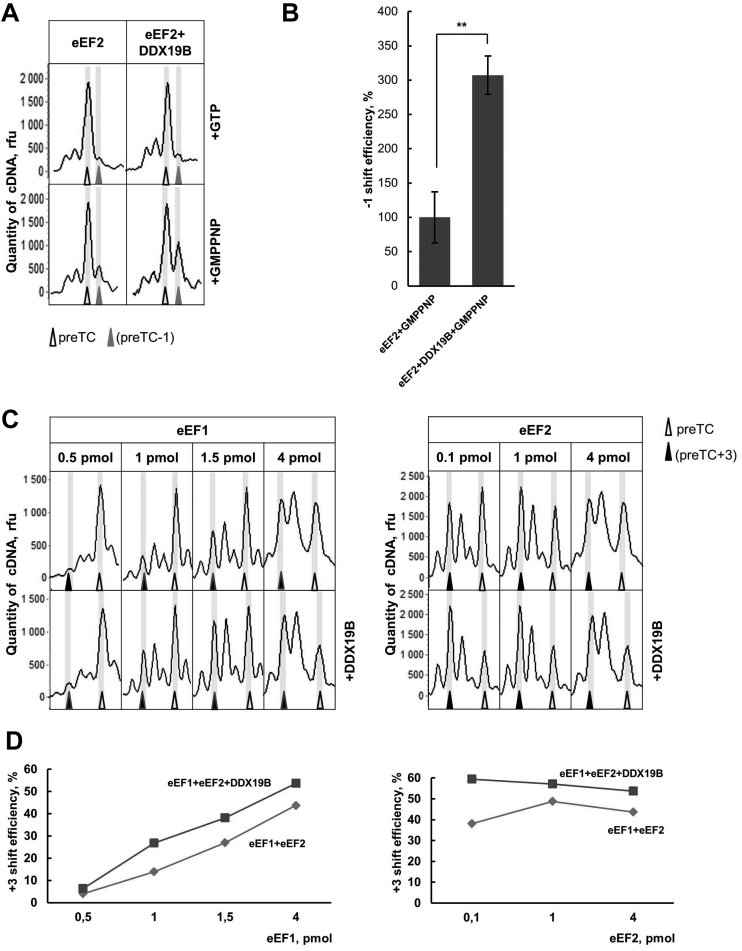
We showed that DDX19B efficiently binds preTCs (Figure 2B). However, besides eRFs, translation elongation factors eEF1 and eEF2 can interact with preTCs (22). eEF2 is able to bind to the vacant preTC in the presence of GTP/GMPPNP, which causes a −1 nt shift in toe-print assay (Figure 6A). This could be explained by a conformational change caused by interaction of eEF2-GMPPNP with the ribosome (42). Notably, in the presence of GTP any conformational change in the ribosome induced by eEF2 is not detected (22). Addition of DDX19B significantly increases the −1 shift of the ribosomal complex in the presence of eEF2 and GMPPNP (Figure 6A). We conclude that DDX19B stabilizes a ribosomal complex interacting with eEF2-GMPPNP. Our data were confirmed by a quantitative analysis of the −1 shift efficiency (Figure 6B), indicating a 3-fold stimulation of eEF2-GMPPNP complex formation by DDX19B.
Figure 6.
DDX19B stabilizes translation elongation complexes. (A) Toe-print analyses of eEF2-preTC complexes with GTP or GMPPNP in the presence of DDX19B. (C) Toe-print analyses of the ribosomal complexes obtained as a result of the decoding of the stop codon by eEF1-suptRNASer-GTP following the ribosome translocation induced by eEF2-GTP in the presence of DDX19B. Rfu – relative fluorescence unit. Positions of preTC are labelled by white triangles, preTC-1 – by grey, preTC+3 – by black. (B) Relative quantitative analysis of the −1 shift efficiency of eEF2-GMPPNP in the presence of DDX19B. −1 shift efficiency of eEF2-GMPPNP was set as 100%. The error bars represent the standard deviation, stars (**) mark a significant difference from the respective control P < 0.01 (n = 3). (D) A quantitative analysis of the translocation efficiency in the presence of DDX19B.
The elongation ternary complex (eEF1-aminoacyl-tRNA-GTP) corresponds most closely to the termination ternary complex (eRF1-eRF3-GTP). To reconstitute one step of elongation of translation in toe-print assay, we constructed ochre suppressor tRNA by site-specific mutagenesis of the anticodon of human tRNASer. The various amounts of eEF1-Ser-tRNAOchre-GTP complex and eEF2-GTP were added to the preTCs, and obtained translocated complexes were analyzed in toe-print assay (Figure 6C and D). We detected a +3 shift of the ribosomal complexes, which corresponds to the ribosome translocation by one codon forward. Addition of DDX19B to the translocation reaction, both in case of limiting amount of eEF1 and in case of limiting amount of eEF2, increased +3 shift efficiency (Figure 6C and D). Therefore, interaction of DDX19 with the ribosome stimulates activities of both translation elongation factors.
To study the influence of RNA binding activity of DDX19 on translation elongation, we added the DDX19(R372G) mutant, unable to bind RNA, to the elongation efficiency assays (Supplementary Data). We observed that the R372G mutant, as the wild-type DDX19, increases binding of both translation elongation factors to the ribosome. Therefore, RNA binding activity of DDX19 is not essential for stimulation of elongation.
DISCUSSION
Here, we show that human DDX19 is associated with polysomes during translation in HEK293 cell lysates (Figure 1A), as described previously for the homolog Dbp5 in yeast (20). This indicates that DDX19 participates in protein biosynthesis or translational control. However, the efficiency of the assembly of ribosomal complexes in vitro is independent of the presence of DDX19 (Supplementary Data). Probably a large excess of translation factors in vitro with a small amount of mRNA masks the effect of DDX19. However, when preTCs are purified from all translational components, DDX19 stimulates elongation and termination of translation (Figures 3, 4 and 6). Using mutant eRF1(AGQ) we show that such stimulation occurs during stop codon recognition (Figure 5A). We did not find stable complexes formed by eRF1 and DDX19 in solution (Supplementary Data). This distinguishes human DDX19 from yeast Dbp5 which co-immunoprecipitates with eRF1 (20). Possibly, human DDX19 lost its ability to directly interact with eRF1 in solution and can form complexes only in the presence of ribosomes. Both forms of DDX19 (A and B) have similar activities in ATPase tests (Supplementary Data), ribosome binding experiments and toe-print assays (Supplementary Data). Both proteins are represented in most tissues. Probably they have the same function in translation.
In our experiments the N-terminal domain of eRF3a does not influence the activity of DDX19 in translation termination (Figure 3C and D). Moreover, we demonstrate that activity of DDX19 in termination independent from eRF3 (Figure 3C and D). This result is consistent with earlier data obtained for Dbp5, which also cannot bind eRF3a in yeast (20). The role of DDX19 in termination is further confirmed by the observed increased efficiency of stop codon recognition using the eRF1 mutant eRF1(K83N) (Figure 5B). Similarly, yeast Dbp5 has been shown to decrease stop codon readthrough in the presence of a mutant release factor SUP45-2 (K80N) (20). In this eRF1 mutant, the mutated amino acid residue is positioned in the N domain of eRF1, and in termination complexes lysine 83 is located very close to the tRNA in the ribosomal P site (39,40). Lysine 83 likely interacts with the tRNA in the P site via its positive charge and thereby stabilizes eRF1 binding in the A site of the ribosome (the decoding center). Mutation of K83 very likely disrupts the proper positioning of eRF1 in the A site of the ribosome and as a result suppresses stop codon recognition by eRF1.
Besides activation of the stop codon recognition by DDX19, we detected its stimulatory effect on peptidyl-tRNA hydrolysis (Figure 4A and B). However, using the conformational rearrangement analysis of TC obtained with eRF1(AGQ), we demonstrated that enhancement of stop codon recognition by DDX19 does not require peptidyl-tRNA hydrolytic activity of eRF1 (Figure 5A). Probably in the peptide release assay we observe a secondary effect of DDX19 activation of stop codon recognition by eRF1. Obviously, the definition of the precise mechanism of activation of translation termination by DDX19 requires further research.
The stable complexes between DDX19 and preTCs or TCs were detected only in the presence of AMPPNP (Figure 2B). We suggested that the DDX19 dissociates from the ribosome after ATP hydrolysis. This hypothesis was confirmed by a DDX19 mutant defective in ATP hydrolysis, which efficiently binds preTCs even in the presence of ATP (Figure 2C). We conclude that DDX19 interacts with preTCs in the presence of ATP and dissociates from the ribosome after ATP hydrolysis. It is known that AMPPNP and ADP stimulate the interaction of DDX19 with RNA to a similar extent (7). We demonstrated that DDX19 does not form stable complexes with preTCs or TCs in the presence of ADP. This indicates that the observed association of DDX19 with the ribosome is specific and independent of its RNA-binding activity. Further, we showed that the activation of stop codon recognition and peptide release by DDX19 is independent from ATP hydrolysis or DDX19's RNA binding ability (Figures 4 and 5D). In the presence of AMPPNP, DDX19 increases stop codon recognition and peptidyl-tRNA hydrolysis by eRF1. Consequently, DDX19 activates translation termination in its ATP-bound form.
It was shown that the overexpression of translation factors does not affect the rate of protein synthesis and cell growth (32). The only exception is eRF1 where a 2-fold increase in concentration results in enhanced protein synthesis and cell growth. Decrease of the cellular eRF1 concentration dramatically reduces the rate of translation. Possibly, the cellular concentration of eRF1 is strictly controlled and limited to allow an optimal balance between normal termination of translation, re-initiation and nonsense-mediated mRNA decay. In our experiments, we used 0.03 pmol preTCs (i.e. active ribosomes) and 0.625 pmol eRFs each. In our system, the efficiency of stop codon recognition is very low, and even in the presence of a 20-fold excess of eRFs relative to ribosomes DDX19 increases termination activity. In yeast, the copy number of eRF1 is estimated to be 22 000 molecules per cell, and about 200 000 ribosomes have been determined per cell (32). Thus, in cell the eRF1/ribosome ratio is about 0.1. This means that, additional supporting translation termination factors should present in the cytoplasm to maintain an appropriate level of peptide release in these conditions. Previously, we demonstrated that deacylated tRNA bound in the E site of the ribosome is one of the activating termination factors (38). Here, we show that DDX19 is a second stimulating translation termination factor. Therefore, one of the physiological roles of DDX19 in living cells is to stabilize ribosomal termination complexes.
We have shown that DDX19 binds with the preTCs, which consist of the ribosome and peptidyl-tRNA, independently of release factors (Figure 2B). We therefore propose that binding of DDX19 precedes termination of translation and could affect the elongation stage as well. Indeed, we demonstrate the stabilization of eEF1-aatRNA-GTP- and eEF2-GTP/GMPPNP-ribosome complexes by DDX19 (Figure 6). DDX19 stabilizes an optimal conformation of the ribosome to perform stop codon recognition by eRF1, which binds to the A site of the ribosome. Consequently, it is not surprising that DDX19 can also stabilize the optimal ribosomal complex for binding of the aminoacyl-tRNA-eEF1 or eEF2 to the same A site. Obviously, the activation of translation elongation should not affect the termination of translation, as they are independent and non-competitive stages of protein biosynthesis. We suppose that the actual substrate for DDX19 is the 80S ribosome in different states. Likely, binding of DDX19 to the ribosome slows down switches between various ribosome conformations, stabilizing both termination and different states of elongation. We conclude that DDX19 stabilizes 80S ribosomal complexes with different proteins bound to the A site.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Ludmila Frolova for providing plasmids encoding release factors, Mathew Coleman for providing the preparation of hydroxylated form of human eRF1 and to Tatyana Pestova and Christopher Hellen who provided us with plasmids encoding initiation factors. Sequencing of plasmids, coding mutant proteins and cDNA fragment analyses were performed by the center of the collective use ‘Genome’ of EIMB RAS. The authors would like to thank Christiane Schaffitzel for preparation of eRF3a and critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Science Foundation [14-14-00487]. Funding for open access charge: Russian Science Foundation [14-14-00487].
Conflict of interest statement. None declared.
REFERENCES
- 1. Linder P., Jankowsky E.. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. [DOI] [PubMed] [Google Scholar]
- 2. Linder P., Fuller-Pace F.V.. Looking back on the birth of DEAD-box RNA helicases. Biochim. Biophys. Acta. 2013; 1829:750–755. [DOI] [PubMed] [Google Scholar]
- 3. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011; 36:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cordin O., Banroques J., Tanner N.K., Linder P.. The DEAD-box protein family of RNA helicases. Gene. 2006; 367:17–37. [DOI] [PubMed] [Google Scholar]
- 6. Zhao J., Jin S.B., Bjorkroth B., Wieslander L., Daneholt B.. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 2002; 21:1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmitt C., von Kobbe C., Bachi A., Pante N., Rodrigues J.P., Boscheron C., Rigaut G., Wilm M., Seraphin B., Carmo-Fonseca M. et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999; 18:4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins R., Karlberg T., Lehtio L., Schutz P., van den Berg S., Dahlgren L.G., Hammarstrom M., Weigelt J., Schuler H.. The DEXD/H-box RNA helicase DDX19 is regulated by an {alpha}-helical switch. J. Biol. Chem. 2009; 284:10296–10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tseng S.S., Weaver P.L., Liu Y., Hitomi M., Tartakoff A.M., Chang T.H.. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998; 17:2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snay-Hodge C.A., Colot H.V., Goldstein A.L., Cole C.N.. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998; 17:2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. von Moeller H., Basquin C., Conti E.. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009; 16:247–254. [DOI] [PubMed] [Google Scholar]
- 12. Napetschnig J., Kassube S.A., Debler E.W., Wong R.W., Blobel G., Hoelz A.. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hodge C.A., Colot H.V., Stafford P., Cole C.N.. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 1999; 18:5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weirich C.S., Erzberger J.P., Flick J.S., Berger J.M., Thorner J., Weis K.. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006; 8:668–676. [DOI] [PubMed] [Google Scholar]
- 15. Alcazar-Roman A.R., Tran E.J., Guo S., Wente S.R.. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006; 8:711–716. [DOI] [PubMed] [Google Scholar]
- 16. Tran E.J., Zhou Y., Corbett A.H., Wente S.R.. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell. 2007; 28:850–859. [DOI] [PubMed] [Google Scholar]
- 17. Lund M.K., Guthrie C.. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell. 2005; 20:645–651. [DOI] [PubMed] [Google Scholar]
- 18. Zolotukhin A.S., Uranishi H., Lindtner S., Bear J., Pavlakis G.N., Felber B.K.. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 2009; 37:7151–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rajakyla E.K., Viita T., Kyheroinen S., Huet G., Treisman R., Vartiainen M.K.. RNA export factor Ddx19 is required for nuclear import of the SRF coactivator MKL1. Nat. Commun. 2015; 6:5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J.J., Seedorf M., Cole C.N., Krebber H.. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007; 315:646–649. [DOI] [PubMed] [Google Scholar]
- 21. Seit-Nebi A., Frolova L., Kisselev L.. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002; 3:881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V.. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006; 125:1125–1136. [DOI] [PubMed] [Google Scholar]
- 23. Saito K., Ito K.. Genetic analysis of L123 of the tRNA-mimicking eukaryote release factor eRF1, an amino acid residue critical for discrimination of stop codons. Nucleic Acids Res. 2015; 43:4591–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blanchet S., Rowe M., Von der Haar T., Fabret C., Demais S., Howard M.J., Namy O.. New insights into stop codon recognition by eRF1. Nucleic Acids Res. 2015; 43:3298–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wada M., Ito K.. A genetic approach for analyzing the co-operative function of the tRNA mimicry complex, eRF1/eRF3, in translation termination on the ribosome. Nucleic Acids Res. 2014; 42:7851–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conard S.E., Buckley J., Dang M., Bedwell G.J., Carter R.L., Khass M., Bedwell D.M.. Identification of eRF1 residues that play critical and complementary roles in stop codon recognition. RNA. 2012; 18:1210–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seit-Nebi A., Frolova L., Justesen J., Kisselev L.. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001; 29:3982–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng Z., Saito K., Pisarev A.V., Wada M., Pisareva V.P., Pestova T.V., Gajda M., Round A., Kong C., Lim M. et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009; 23:1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kong C., Ito K., Walsh M.A., Wada M., Liu Y., Kumar S., Barford D., Nakamura Y., Song H.. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. cell. 2004; 14:233–245. [DOI] [PubMed] [Google Scholar]
- 30. Alcazar-Roman A.R., Bolger T.A., Wente S.R.. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J. Biol. Chem. 2010; 285:16683–16692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bolger T.A., Folkmann A.W., Tran E.J., Wente S.R.. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008; 134:624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Firczuk H., Kannambath S., Pahle J., Claydon A., Beynon R., Duncan J., Westerhoff H., Mendes P., McCarthy J.E.. An in vivo control map for the eukaryotic mRNA translation machinery. Mol. Sys. Biol. 2013; 9:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neumann B., Wu H., Hackmann A., Krebber H.. Nuclear export of pre-ribosomal subunits requires Dbp5, but not as an RNA-Helicase as for mRNA export. PloS One. 2016; 11:e0149571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ivanov A., Mikhailova T., Eliseev B., Yeramala L., Sokolova E., Susorov D., Shuvalov A., Schaffitzel C., Alkalaeva E.. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 2016; 44:7766–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng T., Yamamoto A., Wilkins S.E., Sokolova E., Yates L.A., Munzel M., Singh P., Hopkinson R.J., Fischer R., Cockman M.E. et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol. Cell. 2014; 53:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alkalaeva E., Eliseev B., Ambrogelly A., Vlasov P., Kondrashov F.A., Gundllapalli S., Frolova L., Soll D., Kisselev L.. Translation termination in pyrrolysine-utilizing archaea. FEBS Lett. 2009; 583:3455–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kryuchkova P., Grishin A., Eliseev B., Karyagina A., Frolova L., Alkalaeva E.. Two-step model of stop codon recognition by eukaryotic release factor eRF1. Nucleic Acids Res. 2013; 41:4573–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Susorov D., Mikhailova T., Ivanov A., Sokolova E., Alkalaeva E.. Stabilization of eukaryotic ribosomal termination complexes by deacylated tRNA. Nucleic Acids Res. 2015; 43:3332–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown A., Shao S., Murray J., Hegde R.S., Ramakrishnan V.. Structural basis for stop codon recognition in eukaryotes. Nature. 2015; 524:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matheisl S., Berninghausen O., Becker T., Beckmann R.. Structure of a human translation termination complex. Nucleic Acids Res. 2015; 43:8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hodge C.A., Tran E.J., Noble K.N., Alcazar-Roman A.R., Ben-Yishay R., Scarcelli J.J., Folkmann A.W., Shav-Tal Y., Wente S.R., Cole C.N.. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011; 25:1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spahn C.M., Gomez-Lorenzo M.G., Grassucci R.A., Jorgensen R., Andersen G.R., Beckmann R., Penczek P.A., Ballesta J.P., Frank J.. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004; 23:1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.