Abstract
Besides translation, transfer RNAs (tRNAs) play many non-canonical roles in various biological pathways and exhibit highly variable expression profiles. To unravel the emerging complexities of tRNA biology and molecular mechanisms underlying them, an efficient tRNA sequencing method is required. However, the rigid structure of tRNA has been presenting a challenge to the development of such methods. We report the development of Y-shaped Adapter-ligated MAture TRNA sequencing (YAMAT-seq), an efficient and convenient method for high-throughput sequencing of mature tRNAs. YAMAT-seq circumvents the issue of inefficient adapter ligation, a characteristic of conventional RNA sequencing methods for mature tRNAs, by employing the efficient and specific ligation of Y-shaped adapter to mature tRNAs using T4 RNA Ligase 2. Subsequent cDNA amplification and next-generation sequencing successfully yield numerous mature tRNA sequences. YAMAT-seq has high specificity for mature tRNAs and high sensitivity to detect most isoacceptors from minute amount of total RNA. Moreover, YAMAT-seq shows quantitative capability to estimate expression levels of mature tRNAs, and has high reproducibility and broad applicability for various cell lines. YAMAT-seq thus provides high-throughput technique for identifying tRNA profiles and their regulations in various transcriptomes, which could play important regulatory roles in translation and other biological processes.
INTRODUCTION
Transfer RNAs (tRNAs) are universally expressed, non-coding RNAs measuring 70–90-nucleotides (nt) in length. tRNAs play a central role in translation machinery by converting information from mRNA codons to amino acids. The human nuclear genome encodes more than 600 tRNA genes for cytoplasmic tRNAs (cyto tRNAs) that function in the cytoplasmic translation system; in addition, the human mitochondrial genome encodes 22 mitochondrial tRNAs (mt tRNAs) that act in the mitochondrial translation system (1,2). Recently, the nuclear genome has been suggested to encode numerous cyto tRNA- and mt tRNA-lookalikes (3,4). The multitude of tRNA genes in the genome and the high stability of the expressed tRNAs have placed tRNAs among the most abundant RNA molecules in the cellular transcriptome.
Despite the abundance of tRNAs and their well-defined role in translation, demand for quantitative and qualitative analyses of the tRNA repertoire has been growing. Interestingly, the tRNA gene copy number varies among individuals (5), and tRNA abundance varies widely among different cells and tissues (6,7). This heterogeneity and variation have been implicated in the translational regulation of mRNA expression (8), animal development (9–11) and disease (12–15). Besides their role in translation, it has become increasingly apparent that tRNAs play a wide variety of non-canonical functions in many biological pathways such as nutrient sensing and apoptosis regulation (16,17). tRNAs further act as substrates for tRNA-derived short non-coding RNAs, many of which have been identified as functional molecules with significant roles in a wide variety of biological processes such as the small regulatory RNA pathway, stress responses and cell proliferation (17–25).
To unravel the emerging complexities of tRNA biology and molecular mechanisms underlying them, an efficient tRNA sequencing method is required. Although microarray-based methods have been proven to be useful for the quantification of many tRNA species (6,26), such methods require a custom-made array for each organism and do not completely evaluate all tRNA species including isodecoders that possess the same anticodon sequences but contain sequence variations in other parts. Because isodecoders can exhibit large functional variations (27), an efficient sequencing-based method capable of identifying the entire tRNA repertoire with all sequence variations is needed. However, the characteristic structure of tRNA presents a challenge to the development of such methods. Specifically, the 5΄-end of a mature tRNA binds tightly via base-pairs to the 3΄-end as an acceptor stem. This rigid terminal structure can prevent adapter access to the mature ends of tRNAs, thus hindering the adapter ligation step included in conventional RNA-seq methods and leading to an inefficient and biased representation of tRNAs in RNA-seq data. Instead of using conventional RNA-seq, recent studies have utilized the sequencing methods specialized for tRNAs. For example, Pang et al. employed a two-step ligation strategy to efficiently ligate adapters to tRNAs (28). A method described by Zhong et al. includes the addition of a poly-A tail to the deacylated 3΄-ends of mature tRNAs, thus facilitating the subsequent RT-PCR amplification of tRNAs (29). Furthermore, a DM-tRNA-seq devised by Zheng et al. included the successful reduction of sequence bias from tRNA post-transcriptional methylations by treating tRNAs with AlkB demethylase, followed by a template-switching reaction of thermostable group II intron reverse transcriptase for adapter attachment to tRNAs (30). However, these methods require relatively abundant starting RNA materials and include several purification steps and enzymatic reactions that are not necessary in conventional RNA-seq.
In this light, we report herein the development of Y-shaped Adapter-ligated MAture TRNA sequencing (YAMAT-seq), a convenient, efficient and specific mature tRNA sequencing method. The YAMAT-seq procedure includes a nick-ligation step catalyzed by bacteriophage T4 RNA Ligase 2 (Rnl2) (31), which catalyzes ligation at a 3΄-OH/5΄-P nick in double-stranded RNA or an RNA–DNA hybrid (32–34). This peculiar ligation activity toward double-stranded nucleotides makes Rnl2 an attractive tool for adapter ligation during cDNA preparation (35) and for the detection and quantification of SNPs (36), microRNAs (37), small RNA variants (38) and tRNAs (39). In our YAMAT-seq procedure, Rnl2 specifically ligates a Y-shaped adapter to mature tRNAs at a much greater level of efficiency than that achieved with conventional ligation reactions involving linear adapters and T4 RNA ligase 1 (Rnl1). Subsequent cDNA amplification and next-generation sequencing successfully identify the cyto tRNA and mt tRNA repertoire in the cellular transcriptome. YAMAT-seq requires only a small amount of total RNA extracted using a standard organic method, only a few experimental procedures and a time interval of approximately 2 days before submitting the samples for next-generation sequencing. YAMAT-seq is thus an efficient and convenient high-throughput technique for mature tRNA sequencing that should be applicable to the global identification of tRNA repertories in various cell types and conditions.
MATERIALS AND METHODS
Cell culture
Human breast cancer cell lines were used in this study. The BT-474 cell line was cultured in Dulbecco's modified Eagle's medium (Life Technologies) containing 10% fetal bovine serum (FBS). The SK-BR-3 cell line was cultured in RMPI-1640 medium (Life Technologies) containing 10% FBS. The BT-20 cell line was cultured in MEM medium (Life Technologies) containing 10% FBS. The MCF-7 cell line was cultured in MEM medium containing 10% FBS, 1 mM sodium pyruvate, 1× non-essential amino acids solution (Life Technologies) and 10 μg/ml insulin (Sigma).
Total RNA isolation and deacylation treatment
Total RNA was extracted from cultured cells using TRIsure (Bioline) according to the manufacturer's protocol. Total RNAs were incubated at 37°C for 40 min in 20 mM Tris-HCl (pH 9.0) to remove amino acids from mature tRNAs (deacylation treatment), followed by ethanol precipitation.
Y-shaped adapter design
A DNA adapter containing Illumina 3΄-adapter sequences (Y-3΄-AD) and four different DNA/RNA hybrid adapters containing Illumina 5΄-adapter sequences and a 3΄-terminal A (Y-5΄-AD-A), G (Y-5΄-AD-G), C (Y-5΄-AD-C) or U (Y-5΄-AD-U) were synthesized by Integrated DNA Technologies. The adapter sequences were as follows (capital and small letters designate DNA and RNA, respectively): Y-3΄-AD, 5΄-5phos/GTATCCAGTTGGAATTCTCGGGTGCCAAGG/3ddC-3΄; Y-5΄-AD-A, 5΄-GTTCAGAGTTCTACAGTCCGACGATCACTGGATACTGga-3΄; Y-5΄-AD-G, 5΄-GTTCAGAGTTCTACAGTCCGACGATCACTGGATACTGgg-3΄; Y-5΄-AD-C, 5΄-GTTCAGAGTTCTACAGTCCGACGATCACTGGATACTGgc-3΄; and Y-5΄-AD-U, 5΄-GTTCAGAGTTCTACAGTCCGACGATCACTGGATACTGgu-3΄. The Y-shaped adapters comprising Y-3΄-AD/Y-5΄-AD-A, Y-3΄-AD/Y-5΄-AD-G, Y-3΄-AD/Y-5΄-AD-C and Y-3΄-AD/Y-5΄-AD-U are expected to hybridize mature tRNAs with discriminator bases of U, C, G and A, respectively.
Annealing and ligation of Y-shaped adapters to mature tRNAs
Four different Y-5΄-AD adapters (10 pmol each) and 40 pmol of the Y-3΄-AD adapter were incubated with 1 μg of deacylated total RNA in a 9-μl reaction volume at 90°C for 2 min. After adding 1 μl of 10× annealing buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM ethylenediaminetetraacetic acid and 100 mM MgCl2, the 10-μl mixture was subjected to annealing at 37°C for 15 min. To ligate the annealed adapter to mature tRNAs, 10 μl of 1× reaction buffer containing 1 unit of Rnl2 (New England Biolabs) was added to the mixture. The entire mixture (20 μl) was incubated at 37°C for 1 h, followed by overnight incubation at 4°C.
cDNA amplification and Illumina sequencing
cDNA amplification was performed using the TruSeq Small RNA Sample Preparation Kit (Illumina) according to the manufacturer's protocol. Briefly, 6 μl of ligated RNA and 1 μl of RTP primer were incubated at 70°C for 2 min and then placed on ice. Reverse transcription was subsequently performed by adding 5.5 μl of a RT reaction mixture comprising 2 μl of 5× First Strand Buffer, 0.5 μl of 12.5 mM each dNTP, 1 μl of 100 mM dithiothreitol, 1 μl of RNase inhibitor and 1 μl of SuperScript III Reverse Transcriptase (Life Technologies), followed by incubation at 55°C for 60 min. The resultant cDNAs were amplified by polymerase chain reaction (PCR) (11 cycles) using a PCR enzyme mix (PML) and primers included in the Illumina kit. PCR products were developed using 8% native polyacrylamide gel electrophoresis (PAGE), and mature tRNA amplified regions were gel-purified. To confirm the amplified cDNA sequences, purified cDNAs were cloned using StrataClone Blunt PCR Cloning Kit (Agilent Technologies). The cDNAs were further sequenced (100 nt single-read) on an Illumina HiSeq 2500 system after PhiX library clustering by the Next-Generation Sequencing Core at University of Pennsylvania.
Examination of ligation efficiency to synthetic tRNA
To determine ligation efficiency, a synthetic human cyto tRNAAspGUC (5΄-/5Phos/UCCUCGUUAGUAUAGUGGUGAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCGAUUCCCCGACGGGGAGCCA-3΄) was synthesized by Integrated DNA Technologies. Synthetic tRNA was subjected to either Y-adapter ligation or Illumina adapter ligation (conventional method). For Y-adapter ligation, 2 pmol of synthetic tRNA was incubated with 10 pmol each of Y-5΄-AD-C and Y-3΄-AD and subjected to annealing and Rnl2 ligation as described above. For conventional ligation, 2 pmol of synthetic tRNA was subjected to 5΄- and 3΄- adapter ligation using the TruSeq Small RNA Sample Preparation Kit (Illumina). The respective ligation products (1/100 volume, 20 fmol of tRNA) were subjected to reverse transcription using SuperScript III Reverse Transcriptase (Life Technologies) in a total reaction volume of 20 μl; 0.4 μl of each reaction mixture was further subjected to Real-time PCR using SsoFast™ EvaGreen® Supermix (BioRad) and a StepOne PlusTM Real-time PCR machine (Applied Biosystems). To determine the efficiency of the ligation between the 5΄-adapter and tRNA, the amounts of ligated products were quantified by RT-PCR, in which the region from the 5΄-adapter to nucleotide position (np) 35 of the tRNA (np is according to the nucleotide numbering system of tRNAs (40)) was amplified using the following primers: forward, 5΄-GTTCAGAGTTCTACAGTCCGACGATC-3΄; and RT/reverse, 5΄- ACAGGCGGGGATACTCACCACTA-3΄. To determine the efficiency of the ligation between the 3΄-adapter and tRNA, the following primers were used to amplify the region from np 40 of the tRNA to the 3΄-adapter: forward, 5΄-CGGGAGACCGGGGTTCGATT-3΄; and RT/reverse, 5΄-GCCTTGGCACCCGAGAATTCCA-3΄. PCR products were cloned using a StrataClone cloning kit (Agilent Technologies) to generate standard curves; these were subsequently used to calculate and determine the amounts of ligated products from the respective ligation procedures.
Examination of ligation efficiency to native tRNAs in total RNA fraction
Deacylated BT-474 total RNA (1 μg) was subjected to either Y-adapter ligation or Illumina adapter ligation (conventional method), followed by cDNA synthesis as described in the above cDNA amplification section. Resultant reverse transcription reaction mixture (0.4 μl from total volume of 20 μl) was further subjected to Real-time PCR as described above. To determine the ligation efficiency between the 5΄-adapter and tRNA, the following primers were used to amplify the region from the 5΄-adapter to anticodon arm: universal forward, 5΄-AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA-3΄; RT/reverse for tRNAAspGUC, 5΄-CGGGGATACTCACCACTATACTAACGAGGA-3΄; RT/reverse for tRNAGluCUC, 5΄-CGAATCCTAACCACTAGACCAC-3΄; RT/reverse for tRNALysCUU, 5΄-ACTGAGCTAGCCGGGC-3΄; and RT/reverse for tRNAPheGAA, 5΄-CTAACGCTCTCCCAACTGAGCTAT-3΄. To determine the efficiency of the ligation between the 3΄-adapter and tRNA, the following primers were used to amplify the region from anticodon arm to the 3΄-adapter: forward for tRNAAspGUC, 5΄-CGGGAGACCGGGGTTCGATT-3΄; forward for tRNAGluCUC, 5΄-GGGTTCGATTCCCGGTCA-3΄; forward for tRNALysCUU, 5΄-GTTCGAGCCCCACGTT-3΄; forward for tRNAPheGAA, 5΄-AAGGTCCCTGGTTCGATCC-3΄ and universal RT/reverse, 5΄- CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3΄.
Bioinformatics analyses
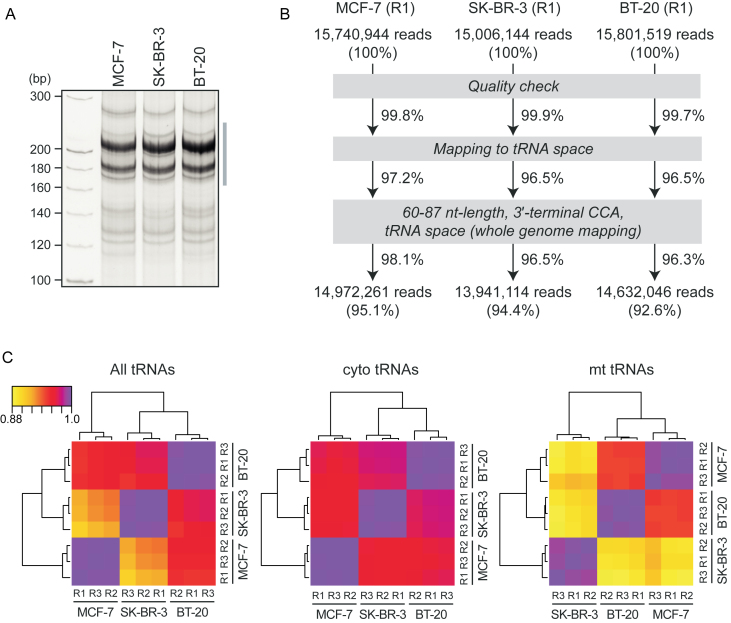
The numbers of YAMAT-seq raw reads obtained from total RNAs of MCF-7, SK-BR-3 and BT-20 with technical triplicates were shown in Figure 3B and Supplementary Figure S1; these are publicly available at NCBI's Sequence Read Archive (accession no. SRP096584). After quality check, SHRiMP2 (41) was used to map the reads to a set of 632 tRNA-reference genes (listed in gtRNAdb (1)) that comprised 610 nuclear-encoded cyto tRNAs from the GRCh37 assembly and 22 known mt tRNAs from tRNAdb (2). The 610 entries from gtRNAdb included 508 true tRNAs and 102 psuedo-tRNAs. We excluded tRNAs that mapped to contigs that are not part of the major chromosome assembly. We allowed non-unique mappings with a 10% mismatch rate, penalizing each mismatch and gap extension equally. We removed any introns from the reference set and CCA was added to the 3΄-ends of the tRNA-reference genes prior to mapping. To be conservative, we also mapped the reads (minus the CCA) to the full GRCh37 assembly using the same parameters and excluded the read if it mapped equally or better to non-tRNA space when compared with the tRNA-reference gene mapping. We confirmed that almost all reads that mapped to the tRNA reference also mapped to tRNAs during the full genome mapping. Lastly, we only kept reads that were 60–87 nt in length inclusive and ended in CCA. Because 100 nt reads, yielded by Illumina sequencing, contain 3΄-terminal sequences (13 nt) of Y-5΄-AD adapter and thereby 87 nt is the maximum read length for tRNAs, CCA sequences were not found in the reads of some long tRNAs (e.g. cyto tRNASeCUCA with 90 nt length). These long tRNA reads were retained as tRNA reads regardless of a lack of CCA sequences. Statistical analysis was performed using R (42) (https://www.R-project.org/). Heatmaps were built with the heatmap.2 function of the gplots package of R. Dendrograms were constructed and visualized with the amap and dendextend packages (43), respectively, with Euclidean distance as a metric for hierarchical clustering.
Figure 3.
Amplification and sequencing of mature tRNAs using YAMAT-seq. (A) Image of 8% native PAGE of amplified cDNAs resulting from YAMAT-seq of total RNAs from MCF-7, SK-BR-3 and BT-20 cells. The result of one of the triplicate, Replicate 1 (R1), from each cell line is shown. The region designated with a gray line was subjected to gel-purification and Illumina sequencing. (B) Flow diagram of the read numbers filtered by the indicated sequence analyses. The result of R1 from each cell line is shown, whereas the results of R2 and R3 are shown in Supplementary Figure S1. (C) Spearman's rank correlation analysis using tRNA reads of the technical triplicates (R1–R3) from each cell line. The results using the reads of all tRNAs (left), cyto tRNAs (middle) and mt tRNAs (right) are shown. The color bar indicates the strength of Spearman's correlation coefficient.
Northern blot analysis
Northern blot analysis was performed as described previously (44). Briefly, total RNA (500 ng) was resolved by 12% PAGE containing 7 M urea, transferred to Hybond N+ membranes (GE Healthcare) and hybridized to 5΄-end labeled antisense probes (cyto tRNALysCUU: 5΄-GTCTCATGCTCTACCGACT-3΄; cyto tRNAAlaAGC: 5΄-GCGCTCTACCACTGAGCTA-3΄; and mt tRNAValUAC: 5΄-GTGTTAAGCTACACTCTG-3΄). Typhoon 9400 and ImageQuant ver. 5.2 (GE Healthcare) were used for storage phosphor autoradiography visualization and quantification.
RESULTS AND DISCUSSION
Design scheme of YAMAT-seq for high-throughput mature tRNA sequencing
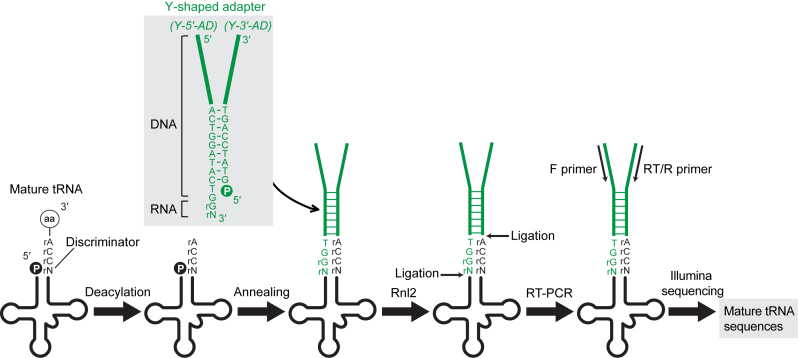
We designed YAMAT-seq to selectively and efficiently sequence mature tRNAs bearing conserved protruding 3΄-ends comprising a 4-nt sequence of the trinucleotides 5΄-CCA-3΄ and a preceding discriminator base (5΄-NCCA-3΄). This method includes the following four steps (Figure 1). First, amino acids at the 3΄-ends of mature aminoacylated tRNAs are removed by incubating total RNA in a high pH buffer (deacylation treatment). Second, a DNA/RNA hybrid Y-shaped adapter (Y-AD) is specifically hybridized and ligated to the deacylated tRNAs via Rnl2 nick ligation. Third, Y-AD-tRNA ligation products are amplified by RT-PCR. Fourth, the resultant cDNAs are gel-purified and subjected to Illumina sequencing.
Figure 1.
Schematic representation of mature tRNA sequencing by YAMAT-seq. Initially, amino acids at the 3΄-ends of mature aminoacylated tRNAs are removed by deacylation treatment. A DNA/RNA hybrid Y-shaped adapter is then specifically hybridized to these deacylated mature tRNAs. The bold line regions of the adapter contain identical sequences to those used for Illumina sequencing. Following hybridization, Rnl2 ligates nicks between the adapter and mature tRNA, and the resultant ligation product is amplified, gel-purified and subjected to Illumina sequencing.
Y-AD comprises two strands containing either Illumina 5΄-adapter sequences (Y-5΄-AD) or 3΄-adapter sequences (Y-3΄-AD) (Figure 1). Y-5΄-AD contains 5΄-OH and 3΄-OH ends and is composed of DNA, except for the two 3΄-terminal RNAs. Y-3΄-AD contains 5΄-P and 3΄-OH ends and its entire nucleotides are DNA. The two united strands form a 4-nt protruding end, 5΄-TGGN-3΄, which recognizes and hybridizes the common protruding 3΄-end of mature tRNAs (5΄-NCCA-3΄). Because the tRNA discriminator base can comprise any four nucleotides, four different Y-5΄-ADs, containing 3΄-terminal A, G, C or U, are mixed and used to capture all mature tRNA species. Hybridization unites the tRNA acceptor stem and Y-AD stem to form long double-stranded nucleotides containing two nicks: ‘Y-5΄-AD-3΄/5΄-tRNA’ and ‘tRNA-3΄/5΄-Y-3΄-AD’. The respective nick structures are ‘RNA-OH-3΄/5΄-P-RNA’ and ‘RNA-OH-3΄/5΄-P-DNA’, and both are efficient substrates for Rnl2 ligation (32–34). No significant ligation efficiency bias is expected between different nucleotides (45). Because the Rnl2 ligation products contain Illumina adapter sequences, they can be amplified by RT-PCR using an Illumina kit. The dependency of RT-PCR amplification on Rnl2 nick ligation allows highly specific amplification and sequencing of mature tRNAs among total RNAs.
Efficient one-step adapter ligation in YAMAT-seq drastically improves mature tRNA amplification efficiency
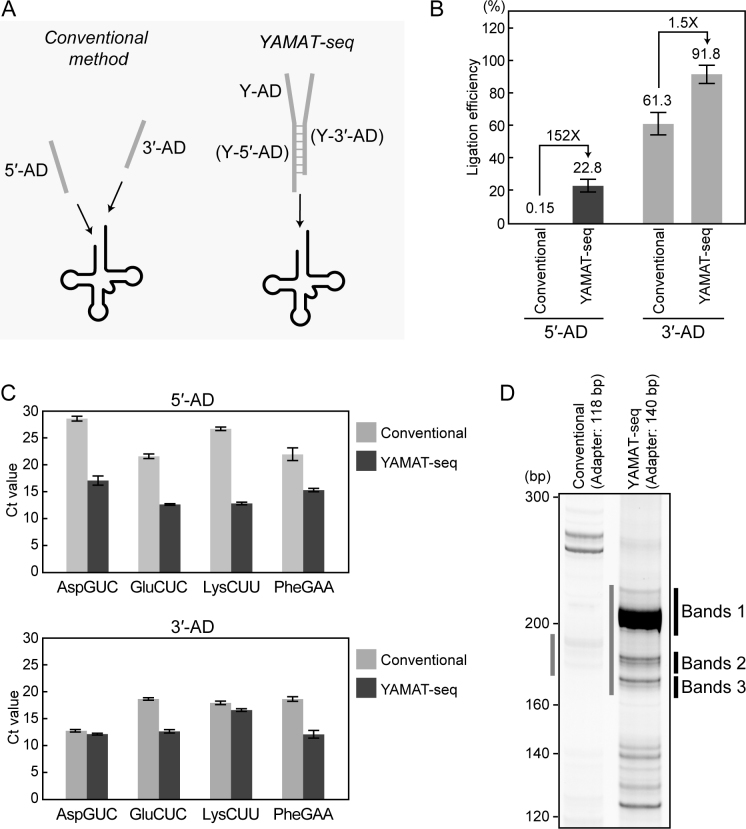
The conventional RNA-seq method utilizes two-step ligation reactions in which single-stranded 5΄- and 3΄-adapters (5΄- and 3΄-AD) are ligated individually to the respective 5΄- and 3΄-ends of tRNAs (Figure 2A). To examine the efficiency of these ligation reactions toward mature tRNAs, synthetic human cyto tRNAAspGUC was subjected to 3΄-AD ligation followed by 5΄-AD ligation, using the Illumina TruSeq Small RNA Sample Preparation Kit. Real-Time qRT-PCR quantification of the ligated products revealed 5΄- and 3΄-AD ligation efficiencies of 0.15% and 61.3%, respectively (Figure 2B). The extremely inefficient ligation between 5΄-AD and 5΄-end of tRNA might be attributable to the rigid terminal structure of the mature tRNA, in which 5΄-end forms base-pairs with 3΄-end to form an acceptor stem. The relatively high 3΄-AD ligation efficiency was likely attributable to the protruding 3΄-ends of tRNAs that would not be much affected by rigid tRNA structure.
Figure 2.
Adapter-tRNA ligation efficiencies with a conventional method and YAMAT-seq. (A) Schematic representation of the adapter ligation reactions used in the conventional and YAMAT-seq methods. In the conventional method, tRNA was subjected to 3΄-adapter (3΄-AD) and 5΄-adapter (5΄-AD) ligations catalyzed by truncated Rnl2 and Rnl1, respectively, using the Illumina TruSeq Small RNA Sample Preparation Kit. In contrast, in the YAMAT-seq method, a Y-shaped adapter (Y-AD) is ligated to a tRNA by Rnl2. (B) Ligation products of synthetic human cyto tRNAAspGUC and 5΄-AD or 3΄-AD were quantified by real-time qRT-PCR. The amounts of ligation products were determined based on standard curves, and ligation efficiencies were calculated as percentages of ligated tRNA versus input tRNA (set as 100%). Each data set represents the average of three independent experiments with bars showing the SD. (C) Ligation products of the indicated native cyto tRNAs in BT-474 total RNA and 5΄-AD or 3΄-AD were quantified by real-time qRT-PCR. Each data set represents the average Ct values of three independent experiments with bars showing the SD. (D) Image of 8% native polyacrylamide gel electrophoresis (PAGE) of amplified cDNAs resulting from BT-474 total RNA sequencing by conventional and YAMAT-seq procedures. The total adapter lengths of the conventional and YAMAT-seq procedures are 118 and 140 bp, respectively. The YAMAT-seq-derived cDNAs in the designated Bands 1–3 regions were gel-purified and subjected to cloning; the identified sequences are shown in Supplementary Table S1. The cDNAs in regions designated with gray lines are expected to contain tRNA fraction.
The ability of YAMAT-seq to capture the mature tRNA repertoire relies on the efficient and specific ligation of Y-AD to mature tRNAs. Hybridization and Rnl2 ligation of Y-AD to synthetic human cyto tRNAAspGUC yielded ligation efficiencies of 22.8% and 91.8% for Y-5΄-AD and Y-3΄-AD, respectively (Figure 2A and B). The difference in efficiency is attributable to the substrate preference of Rnl2, as demonstrated previously by the finding that Rnl2-catalyzed ligation efficiency is higher at a nick of 5΄-RNA-OH/P-DNA-3΄ with a complementary DNA strand than at a nick of double-stranded RNA (31). Compared with the two-step linear adapter ligation reaction of the conventional method, the one-step Y-AD ligation reaction in YAMAT-seq improved the efficiency of 5΄-end ligation by ∼150 fold. Together with the 1.5-fold enhancement in 3΄-end ligation efficiency, YAMAT-seq increased the efficiency of adapter ligation to mature tRNA by more than 200-fold from that of the conventional method.
To further compare ligation efficiencies toward native mature tRNAs, total RNA extracted from BT-474 breast cancer cells was subjected to the conventional and YAMAT-seq methods. Real-time qRT-PCR reactions of the adapter-ligated cyto tRNAAspGUC, tRNAGluCUC, tRNALysCUU and tRNAPheGAA commonly yielded much lower Ct values in YAMAT-seq method than in conventional method, and the differences in Ct values were especially apparent for 5΄-end ligation (Figure 2C). These results confirmed that YAMAT-seq procedure greatly increased the adapter ligation efficiency to native mature tRNAs, especially regarding their recessed 5΄-end, compared to the conventional method.
In the conventional RNA-seq method, poor adapter ligation efficiency would be expected to cause the inefficient amplification of cDNAs derived from mature tRNAs. Indeed, despite the abundant existence of mature tRNAs as observed by PAGE (data not shown), Illumina kit-based cDNA amplification from the BT-474 total RNA yielded only faint bands with the length of ∼190 bp (adapter's length: 118 bp) in the region corresponding to the lengths of mature tRNAs (Figure 2D). In contrast, consistent with the improvement in adapter ligation efficiency, YAMAT-seq specifically amplified abundant cDNA bands with lengths of 170–230 bp (via comparison with marker DNAs in native PAGE; Figure 2D) from the same BT-474 total RNA. Given the length of Y-AD, the inserted RNA lengths were estimated to be 30–90 nt, whose range was wider than expected for the specific amplification of mature tRNAs. However, as shown in Supplementary Table S1, several clones obtained from cDNA bands in three major regions (Bands 1–3; Figure 2D) were unanimously derived from mature tRNAs, suggesting that YAMAT-seq efficiently and specifically amplified mature tRNAs; in addition, the band sizes of cDNAs of Y-AD-ligated mature tRNAs may be shifted relative to marker cDNAs, a characteristic that is likely due to the highly-structured sequences of these molecules. Taken together, the rigid mature tRNA structure prevents adapter ligation in conventional RNA-seq methods, thereby causing mature tRNAs to be inefficiently amplified. YAMAT-seq drastically improves mature tRNA amplification efficiency by employing efficient one-step Y-AD ligation scheme.
High specificity, sensitivity and reproducibility of YAMAT-seq
Given the success of mature tRNA amplification from BT-474 cells, we further applied the YAMAT-seq method to total RNAs isolated from three additional breast cancer cell lines, MCF-7, SK-BR-3 and BT-20, and each performed in triplicate. As shown in Figure 4A, amplified cDNAs from the total RNAs of different cell lines exhibited similar band patterns on native PAGE, indicating the feasibility and general versatility of YAMAT-seq for the amplification of mature tRNAs among total RNAs from various cells. Illumina next-generation sequencing of the amplified cDNA bands yielded ∼15–16 million raw reads, high percentage (>96%) of which were actually mapped to tRNA space (Figure 4B and Supplementary Figure S1). This is a significant improvement compared to the conventional RNA-seq method in which the Illumina sequencing reads of BT-474 cDNAs, derived from tRNA region (Figure 2D), showed only 25% mapping rate to tRNA space (data not shown). We further focused on mature tRNA-derived reads defined by the presence of 3΄-terminal CCA sequences and lengths of 60–87 nt, from which approximately 92–95% of total reads were eventually extracted. Whole genome mapping ensured that almost all extracted sequences were actually derived from tRNA space. The high rate of CCA-containing mature tRNA-derived sequences demonstrated the successful execution of selective mature tRNA amplification and sequencing with this YAMAT-seq scheme.
Figure 4.
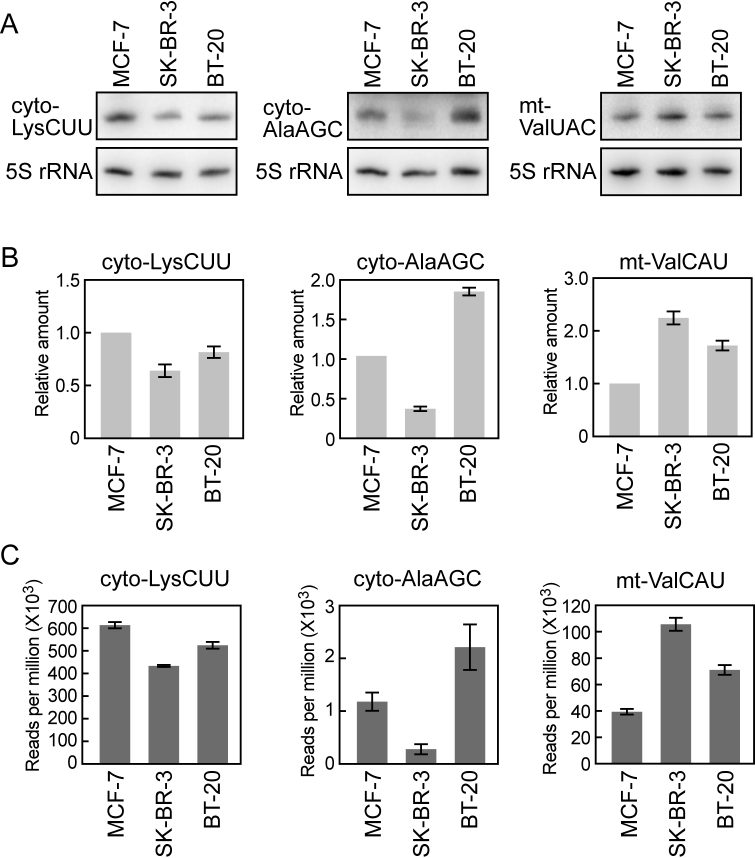
YAMAT-seq can quantitatively estimate identified mature tRNAs. (A) Cyto tRNALysCUU, cyto tRNAAlaAGC and mt tRNAValUAC were detected in the indicated cells by Northern blots. 5S rRNA was detected in the respective membranes as a loading control. (B) Quantification of Northern blot bands. The band intensities of tRNAs were normalized by those of 5S rRNA. Each data set represents the average amounts of three independent experiments, relative to those in the MCF-7 cells (set as 1), with bars showing the SD. (C) Reads per million of the three tRNAs present in YAMAT-seq libraries from the indicated cells. Each data set represents the average of technical triplicates with bars showing the SD.
Regarding the mapped isoacceptors with different anticodon sequences, all 22 mt tRNAs and 46 of 49 cyto tRNAs (93.9%) were successfully detected from all of the three cell lines (Supplementary Table S2 and S3). The high coverage of the identified tRNA species for all the examined cell lines demonstrated the sensitivity and general versatility of YAMAT-seq for mature tRNA sequencing analyses. Cyto tRNAAsnAUU and tRNATyrAUA were not detected in all of the libraries. In addition, cyto tRNAIleGAU and tRNAIleUAU were either not detected or detected as very low read numbers. These tRNAs may not be expressed or be expressed at extremely low levels. Alternatively, these tRNAs may contain several modifications that strongly interfere with reverse transcription.
To examine the reproducibility of YAMAT-seq, nine sequence libraries comprising technical triplicates from the respective three cell lines were subjected to statistical correlation analyses. Scatter plots of the sequence reads from any two libraries in the triplicate unexceptionally showed an excellent correlation with R2 of 0.99 or better (Figure S2). Furthermore, Spearman's rank correlation analysis using all the libraries showed strong correlations between libraries in each technical triplicate, while libraries from different cell lines showed lower correlations (Figure 3C). The consistency among the technical triplicates indicates the high reproducibility of YAMAT-seq for mature tRNA sequencing analyses.
High quantitative ability of YAMAT-seq
To analyze whether the obtained sequence read numbers reflected the actual quantities of expressed mature tRNAs, we selected three tRNAs, cyto tRNALysCUU, cyto tRNAAlaAGC and mt tRNAValUAC, whose read numbers have wide varieties (∼230–620,000 reads per million). The amounts of the three tRNAs among identical amounts of total RNAs from MCF-7, SK-BR-3 and BT-20 cells were examined by Northern blot. As shown in Figure 4A–C, their relative abundances via Northern blots were consistent with those estimated by YAMAT-seq read numbers, indicating the ability of the YAMAT-seq method to provide quantitative information regarding mature tRNA expression among different samples.
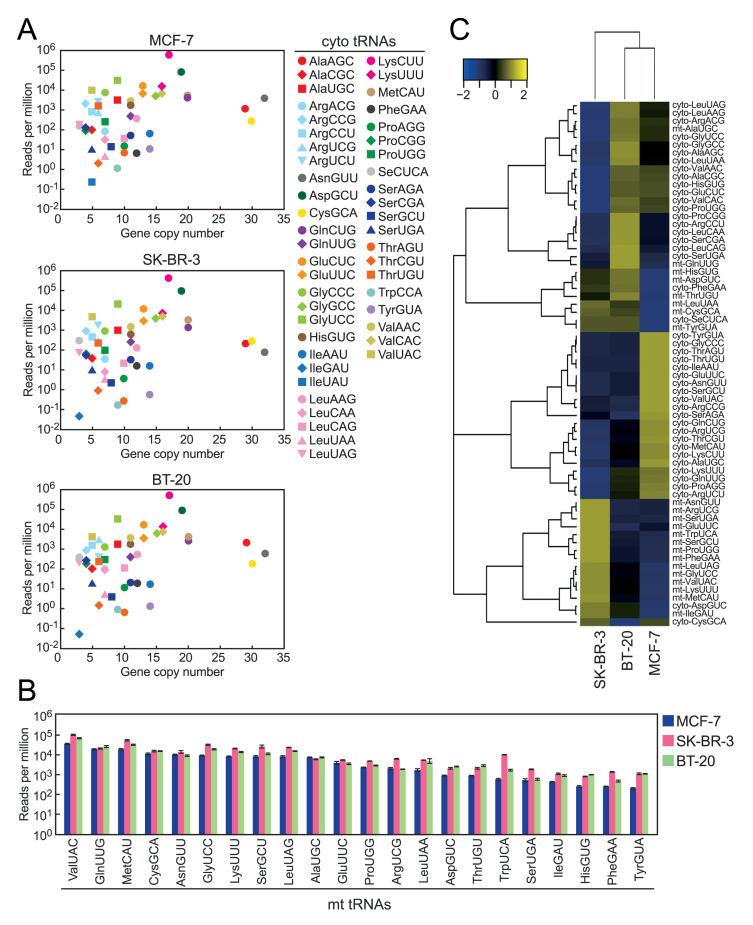
Because the tRNAs whose genes have high copy numbers in the genome tended to be abundant in YAMAT-seq data (Figure 5A), we reasoned that YAMAT-seq read numbers of each tRNA in the same library could reflect the actual abundance. To examine this, the amounts of cyto tRNALysCUU, cyto tRNAAspGUC, mt tRNAAlaUGC and mt tRNAGluUUC in MCF-7 total RNA were determined based on Northern blot analyses in which standard curves were obtained from corresponding synthetic tRNAs (Supplementary Figure S3A and S3B). Rough correlation between the determined amounts and YAMAT-seq read numbers (Supplementary Figure S3B) suggested that the read numbers contain quantitative information to some extent. However, the correlation did not show perfect linearity, probably because the efficiency of YAMAT-seq procedures, especially at the step of reverse transcription, is dependent on the structure and modification state of each tRNA.
Figure 5.
Mature tRNA repertoire and abundances identified using YAMAT-seq in three cell lines. (A) Scatter plots of cyto tRNAs for their YAMAT-seq reads per million from the indicated cells and copy number in human genome. The average reads per million of technical triplicate were used. (B) Reads per million of mt tRNAs in YAMAT-seq libraries from the indicated cells. Each data set represents the average of technical triplicates with bars showing the SD. tRNAs were aligned in order of the read numbers from MCF-7 cells. (C) Heatmap and dendrogram showing the Euclidean distance of tRNA expression pattern among YAMAT-seq libraries from the indicated cells. tRNAs with <1.0 reads per million (cyto tRNAIleGAU, tRNAIleUAU and tRNATrpCCA) were omitted, and the average reads per million of technical triplicate were used.
Each cell line exhibited a distinctive mature tRNA expression signature, and each mature tRNA species exhibited a wide variety of distinctive expression patterns in different cells (Figure 5A–C). The analyses of Spearman's correlation imply that the expression profiles of mt tRNAs could be more variable among cell lines than those of cyto tRNAs (Figure 3C). The wide variations in mature tRNA expression patterns have been suggested in previous reports (6–8), and the current findings suggest the usefulness of YAMAT-seq for identifying such variations.
Advantages of the YAMAT-seq method
YAMAT-seq circumvents the issue of inefficient adapter ligation, a characteristic of conventional RNA sequencing methods, by employing the efficient ligation of Y-AD to mature tRNAs via Rnl2. In addition to efficiency, YAMAT-seq features the advantages of convenience and specificity over previously-reported tRNA sequencing methods that require several steps involving the gel- or HPLC-purification of tRNA fractions or Poly-A-polymerase reaction (28–30). YAMAT-seq requires only 300 ng (3/10 of ligation mixture with 1 μg total RNA was subjected to reverse transcription) of total RNA extracted by standard organic extraction methods, with no purification/concentration steps to obtain the tRNA fraction. After total RNA is subjected to deacylation, Rnl2 ligation to Y-AD and Illumina RT-PCR are the only procedures required for specific mature tRNA amplification, which yields numerous mature tRNA sequences and a high level of coverage of isoacceptors. Given these advantages, we expect that the YAMAT-seq concept will be widely applicable to the identification of mature tRNA expression profiles in cells or tissues from any organism, as well as valuable clinical samples, provided that 300 ng of total RNA is available. Many post-transcriptional modifications existing in mature tRNAs are known to impair reverse transcription (46), leading to bias in sequencing results. However, recent studies have successfully removed some RT-impairing modifications, such as m1A and m1G, using AlkB demethylase and thus have improved the efficiency and quality of the amplification and sequencing of tRNAs and their fragments (30,47). Such demethylase pretreatment of total RNAs would be an option to reduce bias of the sequencing results in YAMAT-seq.
CONCLUSIONS
We have described the development of YAMAT-seq, an efficient and convenient method for the selective amplification and sequencing of mature tRNAs. We have further demonstrated the high specificity of this method for mature tRNAs, high efficiency and sensitivity that cover most isoacceptors in a small amount of total RNA, high reproducibility that yields consistent results from technical triplicates, quantitative capability to estimate relative expression levels, and broad applicability for different cell lines. As our view of tRNA function is greatly being expanded, YAMAT-seq will provide a much-needed convenient and high-throughput method for the analysis of mature tRNA abundance and heterogeneity, which could play important regulatory roles in translation and other biological processes.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to their lab members for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM106047 to Y.K.]; W.M. Keck Foundation [to I.R.]; Institutional funds [Y.K. and I.R.]; JSPS Postdoctoral Fellowships for Research Abroad [M.S. and S.H.]. Funding for open access charge: National Institutes of Health [GM106047 to Y.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chan P.P., Lowe T.M.. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; 37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Juhling F., Morl M., Hartmann R.K., Sprinzl M., Stadler P.F., Putz J.. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009; 37:D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Telonis A.G., Loher P., Kirino Y., Rigoutsos I.. Nuclear and mitochondrial tRNA-lookalikes in the human genome. Front. Genet. 2014; 5:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Telonis A.G., Kirino Y., Rigoutsos I.. Mitochondrial tRNA-lookalikes in nuclear chromosomes: could they be functional?. RNA Biol. 2015; 12:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iben J.R., Maraia R.J.. tRNA gene copy number variation in humans. Gene. 2014; 536:376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dittmar K.A., Goodenbour J.M., Pan T.. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006; 2:e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pavon-Eternod M., Gomes S., Geslain R., Dai Q., Rosner M.R., Pan T.. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009; 37:7268–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gingold H., Tehler D., Christoffersen N.R., Nielsen M.M., Asmar F., Kooistra S.M., Christophersen N.S., Christensen L.L., Borre M., Sorensen K.D. et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014; 158:1281–1292. [DOI] [PubMed] [Google Scholar]
- 9. Rideout E.J., Marshall L., Grewal S.S.. Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marshall L., Rideout E.J., Grewal S.S.. Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. EMBO J. 2012; 31:1916–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmitt B.M., Rudolph K.L., Karagianni P., Fonseca N.A., White R.J., Talianidis I., Odom D.T., Marioni J.C., Kutter C.. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA-tRNA interface. Genome Res. 2014; 24:1797–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abbott J.A., Francklyn C.S., Robey-Bond S.M.. Transfer RNA and human disease. Front. Genet. 2014; 5:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daly N.L., Arvanitis D.A., Fairley J.A., Gomez-Roman N., Morton J.P., Graham S.V., Spandidos D.A., White R.J.. Deregulation of RNA polymerase III transcription in cervical epithelium in response to high-risk human papillomavirus. Oncogene. 2005; 24:880–888. [DOI] [PubMed] [Google Scholar]
- 14. Zhou D.H., Lee J., Frankenberger C., Geslain R., Rosner M., Pan T.. Anti-tumor effects of an engineered ‘killer’ transfer RNA. Biochem. Biophys. Res. Commun. 2012; 427:148–153. [DOI] [PubMed] [Google Scholar]
- 15. Grewal S.S. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim. Biophys. Acta. 2015; 1849:898–907. [DOI] [PubMed] [Google Scholar]
- 16. Raina M., Ibba M.. tRNAs as regulators of biological processes. Front. Genet. 2014; 5:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Phizicky E.M., Hopper A.K.. tRNA biology charges to the front. Genes Dev. 2010; 24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shigematsu M., Honda S., Kirino Y.. Transfer RNA as a source of small functional RNA. J. Mol. Biol. Mol. Imaging. 2014; 1:1–8. [PMC free article] [PubMed] [Google Scholar]
- 19. Anderson P., Ivanov P.. tRNA fragments in human health and disease. FEBS Lett. 2014; 588:4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gebetsberger J., Polacek N.. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013; 10:1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sobala A., Hutvagner G.. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip. Rev. RNA. 2011; 2:853–862. [DOI] [PubMed] [Google Scholar]
- 22. Garcia-Silva M.R., Cabrera-Cabrera F., Guida M.C., Cayota A.. Hints of tRNA-derived small RNAs role in RNA silencing mechanisms. Genes (Basel). 2012; 3:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Honda S., Loher P., Shigematsu M., Palazzo J.P., Suzuki R., Imoto I., Rigoutsos I., Kirino Y.. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Telonis A.G., Loher P., Honda S., Jing Y., Palazzo J., Kirino Y., Rigoutsos I.. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015; 6:24797–24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gapp K., Miska E.A.. tRNA fragments: novel players in intergenerational inheritance. Cell Res. 2016; 26:395–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dittmar K.A., Mobley E.M., Radek A.J., Pan T.. Exploring the regulation of tRNA distribution on the genomic scale. J. Mol. Biol. 2004; 337:31–47. [DOI] [PubMed] [Google Scholar]
- 27. Geslain R., Pan T.. Functional analysis of human tRNA isodecoders. J. Mol. Biol. 2010; 396:821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pang Y.L., Abo R., Levine S.S., Dedon P.C.. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 2014; 42:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhong J., Xiao C., Gu W., Du G., Sun X., He Q.Y., Zhang G.. Transfer RNAs mediate the rapid adaptation of escherichia coli to oxidative stress. PLoS Genet. 2015; 11:e1005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng G., Qin Y., Clark W.C., Dai Q., Yi C., He C., Lambowitz A.M., Pan T.. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods. 2015; 12:835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ho C.K., Shuman S.. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:12709–12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nandakumar J., Ho C.K., Lima C.D., Shuman S.. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 2004; 279:31337–31347. [DOI] [PubMed] [Google Scholar]
- 33. Nandakumar J., Shuman S.. Dual mechanisms whereby a broken RNA end assists the catalysis of its repair by T4 RNA ligase 2. J. Biol. Chem. 2005; 280:23484–23489. [DOI] [PubMed] [Google Scholar]
- 34. Bullard D.R., Bowater R.P.. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 2006; 398:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clepet C. RNA captor: a tool for RNA characterization. PLoS One. 2011; 6:e18445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park K., Choi B.R., Kim Y.S., Shin S., Hah S.S., Jung W., Oh S., Kim D.E.. Detection of single-base mutation in RNA using T4 RNA ligase-based nick-joining or DNAzyme-based nick-generation. Anal. Biochem. 2011; 414:303–305. [DOI] [PubMed] [Google Scholar]
- 37. Cheng Y., Zhang X., Li Z., Jiao X., Wang Y., Zhang Y.. Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew. Chem. Int. Ed. Engl. 2009; 48:3268–3272. [DOI] [PubMed] [Google Scholar]
- 38. Honda S., Kirino Y.. Dumbbell-PCR: a method to quantify specific small RNA variants with a single nucleotide resolution at terminal sequences. Nucleic Acids Res. 2015; 43:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Honda S., Shigematsu M., Morichika K., Telonis A.G., Kirino Y.. Four-leaf clover qRT-PCR: A convenient method for selective quantification of mature tRNA. RNA Biol. 2015; 12:501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S.. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998; 26:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. David M., Dzamba M., Lister D., Ilie L., Brudno M.. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics. 2011; 27:1011–1012. [DOI] [PubMed] [Google Scholar]
- 42. The-R-Core-Team R: a language and environment for statistical computing. R Found. Stat. Comput. 2015; http://www.R-project.org/. [Google Scholar]
- 43. Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015; 31:3718–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Honda S., Kirino Y., Maragkakis M., Alexiou P., Ohtaki A., Murali R., Mourelatos Z.. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA. 2013; 19:1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chauleau M., Shuman S.. Kinetic mechanism of nick sealing by T4 RNA ligase 2 and effects of 3΄-OH base mispairs and damaged base lesions. RNA. 2013; 19:1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kellner S., Burhenne J., Helm M.. Detection of RNA modifications. RNA Biol. 2010; 7:237–247. [DOI] [PubMed] [Google Scholar]
- 47. Cozen A.E., Quartley E., Holmes A.D., Hrabeta-Robinson E., Phizicky E.M., Lowe T.M.. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods. 2015; 12:879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.