Abstract
Escherichia coli topoisomerase I (TopA), a regulator of global and local DNA supercoiling, is modified by Nε-Lysine acetylation. The NAD+-dependent protein deacetylase CobB can reverse both enzymatic and non-enzymatic lysine acetylation modification in E. coli. Here, we show that the absence of CobB in a ΔcobB mutant reduces intracellular TopA catalytic activity and increases negative DNA supercoiling. TopA expression level is elevated as topA transcription responds to the increased negative supercoiling. The slow growth phenotype of the ΔcobB mutant can be partially compensated by further increase of intracellular TopA level via overexpression of recombinant TopA. The relaxation activity of purified TopA is decreased by in vitro non-enzymatic acetyl phosphate mediated lysine acetylation, and the presence of purified CobB protects TopA from inactivation by such non-enzymatic acetylation. The specific activity of TopA expressed from His-tagged fusion construct in the chromosome is inversely proportional to the degree of in vivo lysine acetylation during growth transition and growth arrest. These findings demonstrate that E. coli TopA catalytic activity can be modulated by lysine acetylation–deacetylation, and prevention of TopA inactivation from excess lysine acetylation and consequent increase in negative DNA supercoiling is an important physiological function of the CobB protein deacetylase.
INTRODUCTION
Posttranslational modification is a reversible mechanism used by cells for rapid adaptation to changes in external environment without requiring the synthesis of new RNA and protein to fulfill the needs of adaptation to diverse growth conditions (1–3). Posttranslational modifications can alter the protein function, conformation, stability and localization. Nε-Lysine acetylation is one of the most abundant posttranslational modifications (4–6). Recently, proteomics studies have demonstrated the broad existence of lysine acetylation not only in eukaryotes, but also in diverse bacterial species including Escherichia coli (7–10). In addition, compelling evidence revealed the significant roles of Nε-lysine acetylation in bacterial physiology (6,11,12) for the regulation of central metabolism enzymes, motility and chemotaxis gene expressions and stress resistance systems (12–16). Studies of E. coli chemotaxis response regulator CheY, acetyl-CoA synthetase, RNA polymerase, regulators of capsule synthesis B (RcsB), RNase R, RNase II and N-hydroxyarylamine O-acetyltransferase (NhoA) explored the role of lysine acetylation in the regulation of these proteins (13,17–20). Two distinct mechanisms of lysine acetylation have been reported in E. coli (10). The first mechanism is enzymatic acetylation, which requires the Pka/YfiQ enzyme, a Gcn5-like acetyltransferase, or other acetyltransferases yet to be identified, to transfer the acetyl group from acetyl-coenzyme A (acCoA) to the ε-amino group of a lysine (10). The second mechanism is nonenzymatic acetylation, which is dependent on the direct acetyl group donation from acetyl-phosphate (acP) to the ε-amino group of a lysine (9,21). Both enzymatically acetylated-lysine and nonenzymatically acetylated-lysine modifications are reversed by the NAD+-dependent sirtuin deacetylase CobB, a predominant lysine deacetylase in E. coli that shows no preference for these two types of acetylated lysine sites (21).
Escherichia coli topoisomerase I, an enzyme encoded by the topA gene, regulates global and local DNA supercoiling. The level of supercoiling affects DNA-centered processes, including DNA replication, transcription, recombination and transposition (22–25). The level of DNA supercoiling itself is constantly affected by transcription (26), as well as changes in nutrient availability and growth environment (27–30). The homeostatic state of unconstrained DNA supercoiling is maintained mainly by the relaxation action of topoisomerase I (TopA) and the supercoiling action of DNA gyrase (25,31). Transcription of topA is increased when DNA is more negatively supercoiled while transcription of gyrA and gyrB is increased by DNA relaxation (32,33). In addition, the initiation of E. coli topA transcription at multiple promoters (34,35) is dependent on multiple sigma factors including σ70, σ32 and σs. The relaxation of transcription-driven negative supercoiling by TopA is important for the response to stress challenge including heat shock and oxidative stress (36–38). TopA function has also been shown to be important for survival following acid and antibiotics challenge (39–41). Posttranslational modification of TopA could be an additional mechanism for regulation of TopA activity and DNA supercoiling. In previous proteomics studies of protein acetylation in E. coli, multiple lysines of TopA were found to be acetylated (7–9,42–44). Lysine acetylation of TopA was found to be sensitive to acetyl phosphate (acP) levels (9,43). Therefore, it appears that TopA lysine acetylation can occur through the nonenzymatic acP-dependent mechanism. NAD+-dependent sirtuin CobB deacetylase, the predominant deacetylase in E. coli, is reported to strongly interact with TopA (45). Given the frequency of observation of TopA acetylation and its interaction with CobB, acetylation-deacetylation may have an impact on TopA function as well as other facets of cell physiology that can be influenced by DNA supercoiling.
In this study, we utilized a ΔcobB E. coli strain JW1106 from the Keio Collection and the parent E. coli strain BW25113 to evaluate the in vivo effects of TopA acetylation on cellular TopA catalytic activity, DNA supercoiling and topA transcription response. The direct effect of non-enzymatic acP-dependent lysine acetylation on TopA catalytic activity and the counter effect of purified CobB lysine deacetylase on TopA catalytic activity were demonstrated with in vitro biochemical experiments.
MATERIALS AND METHODS
Strains, plasmids and culture conditions
Escherichia coli strains BW25113 [Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514], JW1106-1 [Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− ΔcobB779::kan rph-1 Δ(rhaD-rhaB)568 hsdR514] (46) were provided by the Yale Coli Genetic Stock Center. The strain YN1434 containing a KanR cassette inserted immediately before the topA gene that encodes additional MRGSHHHHHHGS sequence at the N-terminus of the TopA protein was constructed using the phage lambda Red-mediated recombination system (47) with details provided in the Supplementary Information. pETOP plasmid is a derivative of pBAD/Thio (Invitrogen) expressing recombinant E. coli TopA (48). ASKA plasmid pCA24N that encodes CobB with an N-terminal His6 tag (49) was provided by NBRP (NIG, Japan): E. coli. The E. coli cobB gene was amplified using the primers: 5΄-TGACGATGACAAGCTCGCCCTTATGGAAAAACCAAGAGTACTCGTACTG-3΄, and 5΄-GGGATAGGCTTACCTTCAAGCTCGCCGGCAATGCTTCCCGCTTTTAATC-3΄ and ASKA plasmid pCA24NCobB as the template with Q5 High-fidelity DNA polymerase (New England Biolabs). The PCR product was cloned into pBAD/Thio to create plasmid pCobB using HiFi DNA assembly master mix (New England Biolabs). Cells were cultured in LB (Luria broth) with shaking at 200 rpm at 37°C. When required, antibiotics were added at the following final concentrations: carbenicillin 50 μg/ml, kanamycin 50 μg/ml.
Chloroquine gel analysis of DNA supercoiling
Plasmids were isolated from BW25113 or JW1106-1 transformants collected at the indicated growth stage using GeneJET Plasmid Mini prep Kit (Thermo Scientific). The superhelical density of the isolated plasmid DNA (1 μg) was compared using 2D electrophoresis analysis in 0.8% agarose gels with TAE buffer (40 mM Tris–acetate, pH 8.0, 2 mM EDTA). The first dimension gel and buffer contained 3 μg/ml chloroquine, and electrophoresis was carried out at 3 V/cm for 16 h. The gel was soaked and equilibrated in the running buffer containing 25 μg/ml chloroquine for at least 5 h. For the second dimension electrophoresis, the gel was rotated 90° in the gel tank and electrophoresed at 1.5 V/cm for 20 h. To remove chloroquine, the gel was washed by shaking in TAE buffer for 2 h with the buffer replaced every 30 min. The gel was then stained with ethidium bromide and visualized with UV light (AlphaImager Mini, ProteinSimple).
Assay of TopA relaxation activity in cell lysates
Wild type and ΔcobB mutant strains were grown in LB at 37°C with shaking until OD600 = 0.8. Cells were pelleted by centrifugation at 4°C and lysed by lysozyme treatment in lysis buffer (50 mM NaH2PO4, 0.3 M NaCl) on ice and three freeze-thaw cycles. The total soluble lysates were obtained as the supernatant fractions following centrifugation at 30 000 rpm at 4°C for 1 h. Protein concentrations were determined using the Bradford Protein Assay (Bio-Rad).
TopA activity in cell lysates was assayed in a standard reaction volume of 20 μl (10 mM Tris–HCl, pH 8.0, 50 mM NaCl, 0.1 mg/ml gelatin, 6 mM MgCl2, 150 ng of supercoiled pBAD/Thio plasmid DNA). Purified recombinant TopA was added to relax the DNA substrate in control reactions. After 20 min incubation at 37°C, the reaction was stopped by addition of 4 μl stop solution (50 mM EDTA, 50% glycerol, 0.5% (v/v) bromophenol blue). The DNA was electrophoresed in 0.8% agarose gel with TAE buffer containing 1 μg/ml chloroquine. After the removal of chloroquine, the gel was stained with ethidium bromide and visualized with UV light.
Western blot analysis of TopA protein in cell lysates
Twenty microgram of total cell lysates in SDS gel sample buffer (2% SDS, 62.5 mM Tris–HCl, pH 6.8) was heated at 100°C for 5 min. After separation by electrophoresis in 10% SDS-polyacrylamide gel, the total proteins in the gel was subjected to western blot analysis with mouse monoclonal antibody against E. coli TopA described previously (39). Signals were developed with ECL Plus reagents (Thermo Scientific), and detected with C-DiGit Blot Scanner (LI-COR). Bands were quantified using the Image Studio Digits Ver 4.0 software.
Quantitative PCR of topA mRNA
Wild type and ΔcobB cells were grown to A600 of 0.8. Total RNA was isolated using the Quick-RNA mini prep Kit (Zymo Research), and digested by DNase I (New England BioLabs). The cDNA template was synthesized in 20 μl reaction from 1 μg RNA using the iScript cDNA Synthesis Kit (Bio-Rad) and the following reaction conditions: 25°C for 5 min; 42°C for 30 min; 85°C for 5 min. RNA and cDNA concentrations were measured using the BioPhotometer Plus (Eppendorf). The qRT-PCR products were obtained using the following primers: topA forward: 5΄-ATCTGCCGGAAAGTCCGAATCAGT-3΄, reverse: 5΄-TCTGCGCATCTGCTTCCATATCCT-3΄; idnT forward: 5΄-CATCTGTTTAGCGAAGAGGAGATGC-3΄, reverse: 5΄ ACAAACGGCGGCGATAGC–3΄; hcaT forward: 5΄-GCTGCTCGGCTTTCTCATCC-3΄, reverse: 5΄-CCAACCACGCAGACCAACC-3΄ (50,51). The relative level of topA and internal reference idnT, hcaT transcripts was quantified in 10 μl reaction (10 ng cDNA, 0.25 μM primers, 1× SYBR Green supermix) with the RT-PCR detection system CFX96 (Bio-Rad). PCR amplification was carried out in triplicate using the following cycling conditions: 95°C for 30 s; followed by 40 cycles of 95°C for 20 s; 60°C for 20 s; 72°C for 30 s. The melting curve analysis was performed after qPCR reaction with the temperature set to rise from 65°C to 95°C. The topA expression level was normalized to idnT or hcaT expression level as the internal reference. Results obtained were analyzed with the CFX Manager Software (version 3.1, Bio-Rad).
Purification of recombinant E. coli TopA and CobB
Recombinant TopA was expressed and purified from LB cultures of E. coli BL21-AI (Invitrogen) transformant of TopA expression clone as described previously (52). Recombinant CobB with N-terminal His6 tag was expressed from the ASKA clone pCA24NCobB in BL21-AI by induction at 24°C with addition of 0.02% arabinose and 1 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG), followed by overnight growth at 24°C. The CobB protein in the soluble lysates of the induced culture was purified using a Ni Sepharose column (GE) according to published procedures (49).
In vitro non-enzymatic lysine acetylation and CobB mediated deacetylation reactions
In the in vitro nonenzymatic acetylation reaction, 1 μg of E. coli TopA was incubated with 2 mM and 5 mM acP at 37°C in 150 mM Tris–HCl (pH 8.0), 10% glycerol, and 10 mM MgCl2 for 4 h. The acetylation reaction was stopped by adding an equal volume of 2× SDS loading buffer and then boiling for 10 min before SDS PAGE and western blot analysis of lysine acetylation using mouse monoclonal anti-acetyllysine antibody (Cell Signaling Technology). The TopA protein on the membrane was then stained with Coomassie blue. For simultaneous in vitro acetylation–deacetylation, 1 μg of E. coli TopA was incubated with 0.2 μg CobB in 150 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 5mM acP, 0.25 mM NAD+ and 10% glycerol for 4 h at 37°C. To determine the effect of acetylation on TopA catalytic activity, 2 μl of the acetylation/deacetylation reaction was used for serial dilutions and assay of TopA relaxation activity.
Pull-down assays to study direct interaction between E. coli CobB and TopA
Purified recombinant TopA protein (5 nM) was mixed with purified N-terminal His-tagged CobB protein (10–50 nM) in pull-down buffer (10 mM HEPES, pH 7.4, 100 mM NaCl, 0.005% v/v Tween-20) overnight at 4°C. HisPur Cobalt Agarose resin (Thermofisher), pre-equilibrated in pull-down buffer, was then added to the CobB-TopA reaction and mixed at 4°C for 2 h. After centrifugation of the reactions, the resin was washed three times in pull-down buffer with 10 mM imidazole. The proteins bound to the resin were eluted in SDS sample buffer by boiling for 2 min. Eluates were electrophoresed in 10% polyacrylamide SDS gel, and TopA was detected by western blotting with monoclonal antibodies against E. coli TopA.
Purification and characterization of chromosomally encoded His-tagged TopA during growth transition and growth arrest
TB7 broth (10 g/l tryptone, 100 mM potassium phosphate pH 7.0) supplemented with 0.4% glucose was inoculated with overnight culture of strain YN1434 at 1:750 dilution. Cells were cultured with shaking at 37°C for 4.5 h (1.5 l of culture at OD600 = 0.7), 5.5 h (750 ml of culture at OD600 = 1.7), 24 h (500 ml of culture at OD600 = 1.9). His-TopA expressed from the chromosome was purified from the cell pellets using GE Ni sepharose columns with the protease inhibitor PMSF (1 mM) and CobB inhibitor nicotinamide (10 mM) added to the lysis buffer. Based on Coomassie blue staining of purified His-TopA, equal amounts of His-TopA (1 μg) purified from the three growth conditions were electrophoresed in 10% SDS gel and transferred onto nitrocellulose membrane. The His-TopA on the membrane was stained with the MemCode (Thermo Scientific) reversible protein stain kit before western blot analysis with antibodies against acetylated lysine. The signals were quantified using the Image Studio Digits Ver 4.0 software.
RESULTS
Loss of CobB led to increase in DNA supercoiling
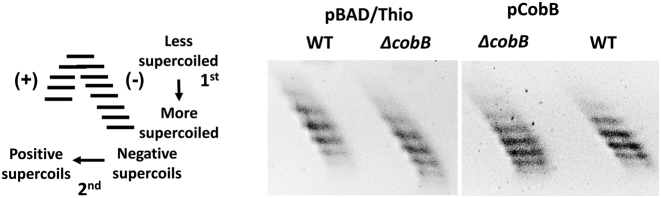
To assess the effect of lysine acetylation on DNA supercoiling in E. coli, wild-type BW25113 and ΔcobB mutant JW1106 transformed with plasmid pBAD/Thio were grown at 37°C until OD600 reached 0.8. The extracted DNA plasmids were electrophoresed in agarose gel in the presence of 3 μg/ml chloroquine in the first dimension and 25 μg/ml chloroquine in the second dimension to resolve the positively and negatively supercoiled DNA topoisomers. Plasmid pBAD/Thio from ΔcobB cells was more negatively supercoiled than DNA isolated from isogenic cobB+ cells (Figure 1). When the ΔcobB mutation was complemented in trans by cobB cloned into pCobB plasmid, the supercoiling level of the plasmid isolated from wild-type and ΔcobB mutant was identical (Figure 1). This result indicated that loss of CobB deacetylation function affects DNA supercoiling. Since TopA is the major activity for relaxation of negatively supercoils in E. coli and CobB has been shown by a previous study to associate with TopA (45), the increase in negative DNA supercoiling in ΔcobB cell may be due to reduction in TopA activity in the absence of CobB. Nonenzymatic lysine acetylation of TopA would require CobB for reversal of the post-translational modification. The influence of CobB on TopA activity and cell growth was investigated with further experiments.
Figure 1.
2-Dimensional chloroquine gel analysis comparing DNA supercoiling in BW25113 (wild-type) and JW1106 (ΔcobB) mutant strains. Plasmids pBAD/Thio and pCobB were extracted from LB cultures at OD600 = 0.8. The first dimension electrophoresis was carried out in 0.8% agarose gel with TAE buffer containing 3 μg/ml chloroquine at 3 V/cm for 16 h. The agarose gel was then rotated 90° for electrophoresis in the second dimension with 25 μg/ml chloroquine in the TAE buffer. The schematic diagram shows the expected distribution of positively and negatively supercoiled DNA topoisomers following electrophoresis.
Loss of CobB reduced cellular TopA activity while TopA expression was increased
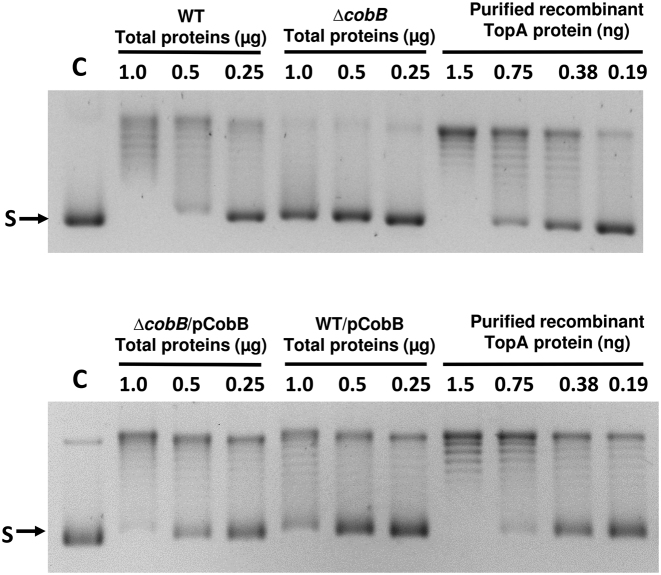
We hypothesized that increased lysine acetylation on TopA in the ΔcobB mutant led to reduction of TopA catalytic activity. The relaxation activities of TopA in total cell lysates of the wild type and ΔcobB strains were compared in assay with negatively supercoiled plasmid DNA as the substrate. The results showed that TopA from ΔcobB mutant cell lysate displayed reduced relaxation activity (Figure 2, top panel). The reaction buffer used in the Figure 2 experiments did not contain ATP so gyrase and topoisomerase IV present in the cell lysates would not be active on the plasmid DNA substrate under these experimental conditions. The reduction of TopA relaxation activity in the ΔcobB mutant may account for the increased negative DNA supercoiling observed in Figure 1. Complementation by plasmid pCobB restored the TopA relaxation activity in the ΔcobB mutant cell lysate (Figure 2, bottom panel). Overexpression of CobB in the wild-type cobB background did not result in higher relaxation activity. These results suggest that lack of normal physiological levels of CobB deacetylation activity could result in reduction of TopA catalytic activity, and increase of negative DNA supercoiling.
Figure 2.
Relaxation assay of TopA activity in total soluble cell lysates of BW25113 (wild-type) and JW1106 (ΔcobB) and their pCobB transformants. The indicated amount of total soluble proteins from BW25113 (wild-type) and JW1106 (ΔcobB) LB cultures at OD600 = 0.8 were incubated with 150 ng of negatively supercoiled plasmid DNA substrate (S) at 37°C for 20 min. DNA were electrophoresed in 0.8% agarose gel with TAE buffer containing 1 μg/ml chloroquine at 3 V/cm for 16 h. Reaction products from partial relaxation of the supercoiled plasmid DNA substrate by increasing amounts of purified recombinant TopA are included to show the effect of increasing TopA catalytic activity on plasmid electrophoretic mobility.
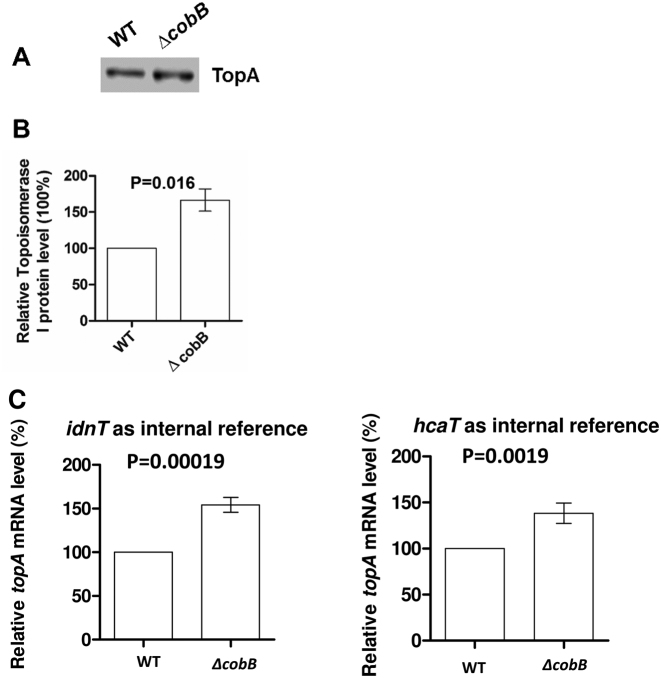
However, an alternate possibility to explain the reduction of TopA relaxation activity in the ΔcobB mutant is that the intracellular level of TopA protein in this strain is lower, resulting in decreased relaxation activity in the total cell lysate. To examine this possibility, TopA protein and topA mRNA levels were analyzed. We first determined the TopA protein level in wild-type and ΔcobB cells by western blotting analysis. Total soluble lysates obtained from wild type and ΔcobB cells were processed in parallel and the same amount of total proteins was loaded onto an SDS-PAGE gel. Equal loading of total cellular proteins based on Bradford protein assay results was confirmed by Coomassie blue staining (Supplementary Figure S1). This allows us to compare the relative abundance of TopA from the same amount of total protein. TopA protein level was significantly increased in the ΔcobB mutant, approximately 1.6-fold higher than that in wild-type cells (Figure 3A and B). Measurement of topA mRNA level by qPCR analysis also showed that topA transcription is higher (1.4–1.5-fold with hcaT and idnT as internal references) in the ΔcobB mutant than in wild-type cells (Figure 3C). These results showed that the reduction in the TopA catalytic activity in the ΔcobB cell extract (Figure 2) was not due to decrease in the TopA protein level. The higher level of topA expression in the ΔcobB mutant is likely due to the stimulation of topA transcription by the increase in negative supercoiling in the homeostatic regulation of DNA supercoiling (32,53). However, even with the increased topA expression, there is a deficiency in the TopA catalytic activity affecting the level of DNA supercoiling in the ΔcobB mutant.
Figure 3.
TopA expression level in BW25113 (wild-type) and JW1106 (ΔcobB) mutant. (A) 20 μg total proteins were resolved by 10% SDS-PAGE, and TopA protein was detected by western blot analysis using monoclonal antibodies against TopA. (B) Quantification of four independent western blot experiments as shown in (A). The level of TopA in the wild-type strain in each experiment was set at 100%, and the corresponding level of TopA in the ΔcobB mutant strain is shown. (C) Quantitative PCR measurements of topA transcript levels from three independent experiments normalized against either idnT or hcaT transcript level as internal reference. The topA transcript level in the wild-type strain was set at 100% for each experiment. Error bars indicate standard deviation of relative topA transcript level in the ΔcobB mutant strain.
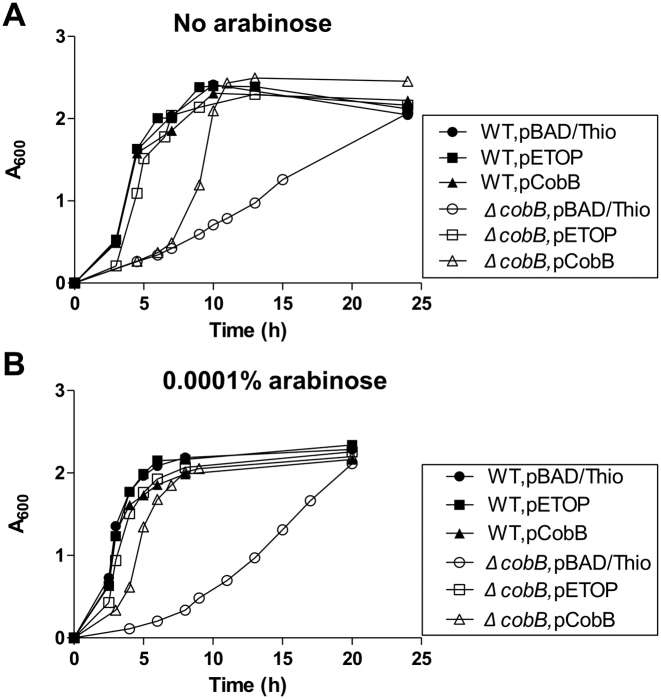
Overexpression of recombinant TopA partially compensated the delayed growth of the ΔcobB mutant
The growth kinetics of wild-type BW25113 and ΔcobB mutant JW1106 transformed with pBAD/Thio plasmid was monitored continuously for 24 h in LB culture grown with vigorous shaking at 37°C. The ΔcobB mutant strain grew much slower during the exponential phase when compared to the wild-type strain (Figure 4). The reduction of cellular TopA catalytic activity as well as increased negative supercoiling observed here for the ΔcobB mutant might account in part for its slow growth phenotype. A high copy number plasmid pETOP derived from the pBAD/Thio plasmid that overexpresses recombinant TopA was introduced into both strains for growth rate analysis under the same conditions. While wild-type cells with either pBAD/Thio or pETOP displayed similar growth rates, the delayed growth of the ΔcobB mutant was partially rescued by the presence of pETOP (Figure 4). This result indicates that reduction of TopA activity when acetylation-deacetylation regulation is perturbed is of physiological significance. Overexpression of recombinant CobB from the pCobB plasmid also partially rescued the slow growth of the ΔcobB mutant, with greater degree of complementation in the presence of 0.0001% arabinose to further induce the expression of recombinant CobB (Figure 4).
Figure 4.
Effect of recombinant TopA overexpression on the slow-growth phenotype of the ΔcobB mutant. (Filled circle) Wild-type BW25113 strain transformed with pBAD/Thio; (unfilled circle) ΔcobB JW1106 strain transformed with pBAD/Thio; (filled square) Wild-type strain transformed with pETOP for TopA overexpression; (unfilled square) ΔcobB mutant strain transformed with pETOP; (filled triangle) wild-type strain transformed with pCobB; (unfilled triangle) ΔcobB mutant strain transformed with pCobB. The LB medium contained no arabinose (A) or 0.0001% arabinose (B).
Acetylation by acP decreases TopA catalytic activity
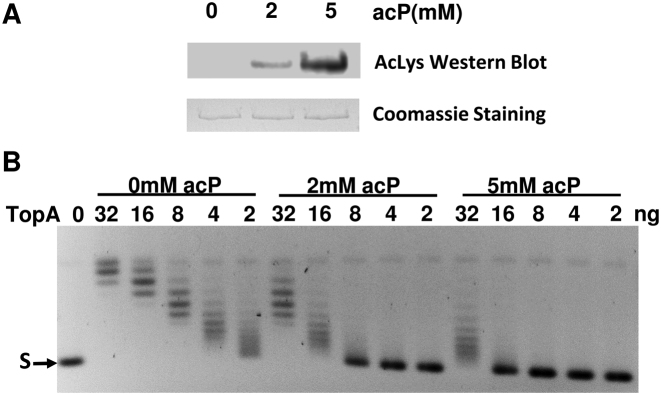
To examine the effect of lysine acetylation on TopA catalytic activity directly, we performed in vitro non-enzymatic lysine acetylation of TopA. Recombinant TopA expressed in a cobB+ genetic background, was purified and incubated with acetyl phosphate (acP). Increase in TopA lysine acetylation in the presence of 2 mM and 5 mM acP was observed by western blotting with antibodies against acetyllysine (Figure 5A). Assays with negatively supercoiled plasmid DNA as the substrate showed that the relaxation activity of TopA was reduced by ∼4-fold following acetylation by 2 mM acP and >8-fold following acetylation by 5 mM acP (Figure 5B).
Figure 5.
Decrease in TopA activity from acetyl phosphate mediated nonenzymatic lysine acetylation. (A) Western blot analysis of nonenzymatically acetylated TopA. Purified E. coli TopA (1 μg) was incubated with 2 and 5 mM acP at 37°C for 4 h. Acetylated TopA was visualized by western blot analysis with anti-acetyllysine antibody. TopA on the membrane was stained with Coomassie blue. (B) Following incubation with and without acetyl phosphate, serial dilutions of TopA (32, 16, 8, 4, 2 ng) were incubated with 150 ng negatively supercoiled plasmid DNA (S) at 37°C for 30 min to compare the relaxation activity.
Lysine deacetylation by CobB can counter the effect of non-enzymatic lysine acetylation on TopA catalytic activity
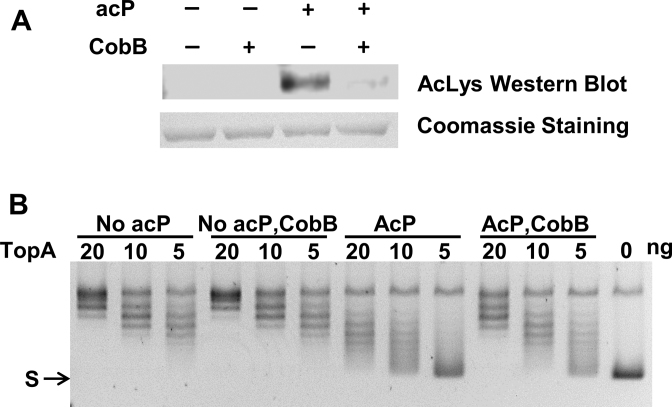
Lysine acetylation is a reversible post-translational modification, and acetylation–deacetylation can take place at the same lysine residues. We showed evidence that TopA relaxation activity in ΔcobB cell lysate is reduced, and nonenzymatic acetylation of purified TopA resulted in a decrease of its catalytic activity. Hence, the effect of CobB lysine deacetylase on TopA inactivation by AcP-mediated acetylation was examined next. TopA was incubated with both 2 mM acP and purified CobB, so that acetylation and deacetylation can both take place. Notably, the acetylation level of TopA was reduced by CobB (Figure 6A). Accordingly, as shown in Figure 6B, the presence of CobB partially protected TopA catalytic activity from the inhibitory effect of acP-dependent lysine acetylation. These results indicate that CobB is important for maintaining TopA deacetylation and TopA catalytic activity.
Figure 6.
CobB deacetylation counters the effect of acetyl phosphate mediated lysine acetylation on TopA relaxation activity. Purified E. coli TopA (1 μg) and 2 mM acP was incubated with or without CobB (0.65:1 molar ratio to TopA) at 37°C for 4 h. (A) Acetylation level of TopA was compared by western blot analysis with anti-acetyllysine antibody. TopA protein was visualized by Coomassie blue staining. (B) Following incubation with acetyl phosphate in the absence or presence of CobB, serial dilutions of TopA (20, 10, 5 ng) were incubated with 150 ng negatively supercoiled plasmid DNA (S) at 37°C for 30 min to assay the TopA relaxation activity.
TopA interacts directly with CobB in the absence of DNA
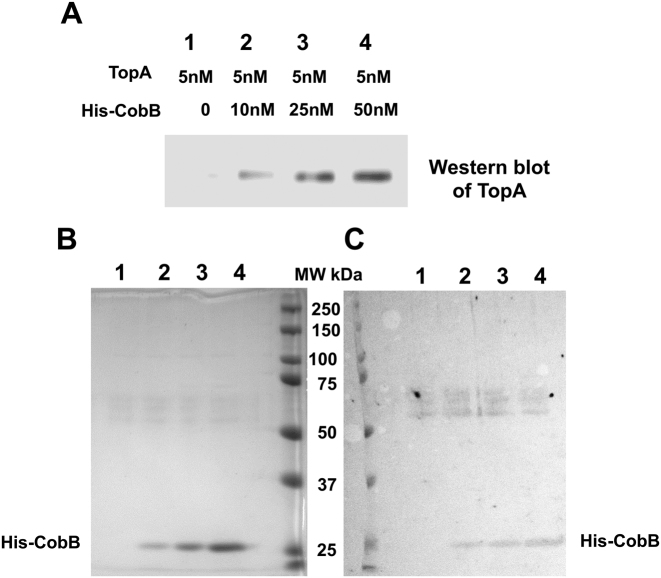
TopA has been demonstrated in a previous study (45) to be a CobB binding protein using a proteome microarray as well as bio-layer interferometry analysis. To confirm that TopA interacts directly with CobB in the absence of DNA, purified recombinant TopA and CobB proteins were mixed in binding buffer. HisPur Cobalt resin was used to pulldown TopA in complex with the N-terminal His-tagged CobB. Western blot analysis of the proteins bound to the cobalt resin with antibodies against TopA (Figure 7A) showed that the presence of His-tagged CobB allows binding of TopA to the Cobalt resin and co-elution with the His-CobB (Figure 7B and C).
Figure 7.
Direct interaction between TopA and CobB. Purified recombinant TopA and His-tagged CobB were allowed to interact before pull-down by HisPur Cobalt Agarose Resin. The presence of TopA in the bound proteins eluted from the cobalt resin was visualized by western blot analysis with anti-TopA antibodies (A). The co-eluted His-CobB was stained by Coomassie blue following SDS PAGE (B) or silver on the transfer membrane (C).
Specific activity of His-TopA expressed from the chromosome is inversely proportional to degree of lysine acetylation during growth transition and growth arrest
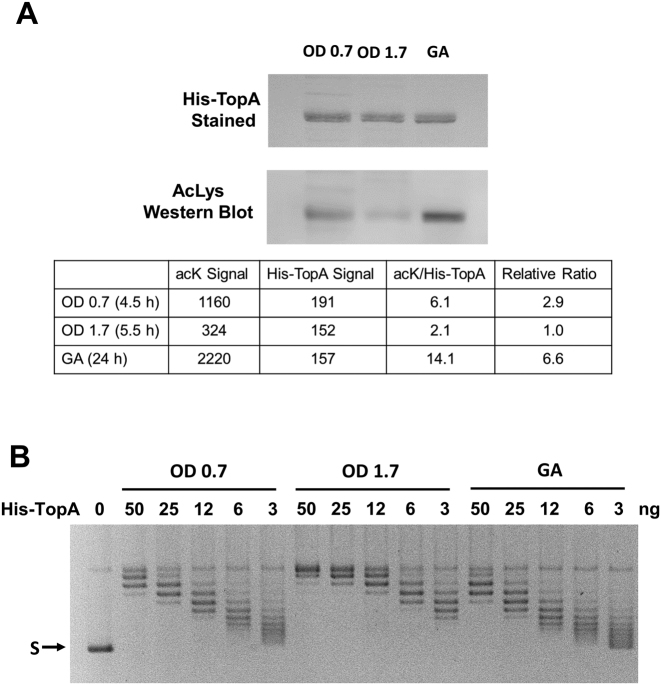
To demonstrate directly the effect of in vivo lysine acetylation on the specific activity of TopA, we utilize E. coli strain YN1434 that expresses TopA with N-terminal His-tag from the chromosome. Cells were collected from cultures in buffered TB7 broth supplemented with 0.4% glucose following growth for 4.5 h (OD600 = 0.7), 5.5 h (OD600 = 1.7) and growth arrest at 24 h (OD600 = 1.9). Acetyl phosphate-dependent acetylation has been shown to occur mostly following entry into stationary phase under these experimental conditions (12,43). The His-TopA was purified with Ni-affinity chromatography and analyzed for level of lysine acetylation (Figure 8A) and specific activity using supercoiled plasmid DNA as substrate (Figure 8B). The results showed that the level of lysine acetylation found in the purified His-TopA first decreased as cells entered stationary phase at 5.5 h. and then increased in growth arrest at 24 h (Figure 8A). The specific activity of the purified His-TopA was found to be inversely proportional to the degree of lysine acetylation (Figure 8B). The His-TopA purified from the growth arrest (GA) culture had the lowest specific activity (∼2-fold lower than OD600 = 0.7 culture, and 4–8-fold lower than OD600 = 1.7 culture). This further confirmed that TopA catalytic activity can be modulated in vivo based on the degree of lysine acetylation modification of the enzyme.
Figure 8.
Topoisomerase I specific activity is inversely proportional to degree of lysine acetylation during growth transition and growth arrest. (A) His-TopA expressed from the chromosome in strain YN1434 was purified from buffered and glucose supplemented TB7 cultures at exponential phase (OD600 = 0.7), entry into stationary phase (OD600 = 1.7) or growth arrest by Ni-affinity column. Following transfer of equal amounts of His-TopA (1 μg) from SDS gel onto nitrocellulose membrane, the His-TopA was stained with MemCode reversible stain and analyzed for level of lysine acetylation with antibodies against acetylated lysine (acK). The signals of His-TopA staining was used to normalize the acK signal for comparison of degree of lysine acetylation. (B) Serial dilutions of the purified His-TopA was assayed for relaxation activity using supercoiled plasmid DNA (S) as substrate.
DISCUSSION
Bacterial DNA topoisomerase I (TopA) is involved in various essential DNA-centered processes. TopA is highly efficient for catalyzing the removal of negative DNA supercoiling generated during rapid transcription. The utilization of different sigma factors for transcription of the topA gene and the effect of topA mutation on survival following stress challenge in E. coli (34,35,41,54,55) indicate the importance of maintaining cellular TopA activity level under different growth conditions for adaptation and survival (56). Transcription of topA is increased when DNA is more negatively supercoiled (32,53). The results we present here suggest that in addition to regulation at the transcription level, cellular TopA activity can be significantly affected by the post-translational modification of lysine acetylation-deacetylation, with global impact on DNA topology. These results expand our understanding about TopA regulation as well as the control of DNA topology by DNA topoisomerases. In a previous study of global response to loss of DNA supercoiling (57), cobB transcription was found to be repressed by inhibition of DNA gyrase or gyrB mutation. Based on the effect of the ΔcobB mutation on DNA supercoiling observed in Figure 1, decreased CobB activity in the presence of gyrase mutation or inhibitor may contribute to countering the effect of reduced gyrase supercoiling activity via reduction of TopA relaxation activity for the homeostatic regulation of DNA supercoiling.
Evidence has been provided in this report that in vitro acP-mediated nonenzymatic lysine acetylation of TopA decreases its catalytic activity. TopA activity in the total cell lysate is significantly reduced as a result of the deletion of the cobB deacetylase gene, leading to an increase in negative supercoiling in vivo, even though topA mRNA transcription level detected by qPCR analysis is increased, and TopA protein expression level detected by western blot analysis is also increased. On the basis of these results, we propose that the reduction of TopA relaxation activity due to the missing deacetylase function in the ΔcobB mutant increases DNA negative supercoiling, which in turn upregulates topA gene transcription and TopA expression.
Lysine acetylation affects multiple cellular functions, including cell motility, carbon source utilization central metabolism, and stress responses (6,13–16). Previous proteomics studies identified a large number of acetylated proteins in E. coli (7,9,44). In this report, we observe a slow growth phenotype in entering the exponential phase associated with the ΔcobB mutation. Nutrient should not be limiting in the LB medium under the growth condition of the experiment. The slow growth phenotype could potentially be due to the global effect of acetylation-deacetylation on a wide range of E. coli proteins. Interestingly, when the ΔcobB mutant is complemented with a high copy number plasmid expressing recombinant TopA, the slow growth phenotype is partially reversed. This could be due to the global effect of TopA via its influence on DNA supercoiling. TopA activity has been reported to be important for the removal of localized transcription-driven negative supercoiling at the ribosomal operon rrnB (58). Reduction in TopA catalytic activity as a result of the cobB deletion may lead to hypernegative supercoiling at the highly transcribed rRNA loci as the cells exit the growth arrest to enter the exponential phase under the culture conditions used in this study.
According to a previous study, the intracellular concentration of acP in E. coli reaches at least 3 mM, and may be as high as 4.5 mM in wild-type cells (59). Nonenzymatic acP-dependent acetylation is the predominant mechanism of lysine acetylation in E. coli cells (9,43). Our results showed that following in vitro acP-dependent acetylation, the activity of acP-acetylated TopA is reduced. Meanwhile, the NAD+-dependent sirtuin lysine deacetylase CobB can partially remove the lysine modification on TopA and maintain the TopA activity at a higher level. These results confirm our hypothesis that acetylation on TopA reduces its activity, and the deacetylation of TopA by CobB is of physiological significance.
We further established the correlation between degree of lysine acetylation and specific activity of topoisomerase I by purifying His-tagged TopA expressed from the chromosome following growth transition and growth arrest. Non-enzymatic acetylation of TopA by acetyl phosphate is expected to be highest at growth arrest following 24 h incubation in buffered TB7 media supplemented with glucose (12,43). A relatively low level of relaxation specific activity was observed along with the high level of acetylated lysine for His-TopA purified from growth arrest cells. Further studies are needed to determine the basis for the decrease in TopA acetylation that was observed during the initial transition from exponential phase to stationary phase.
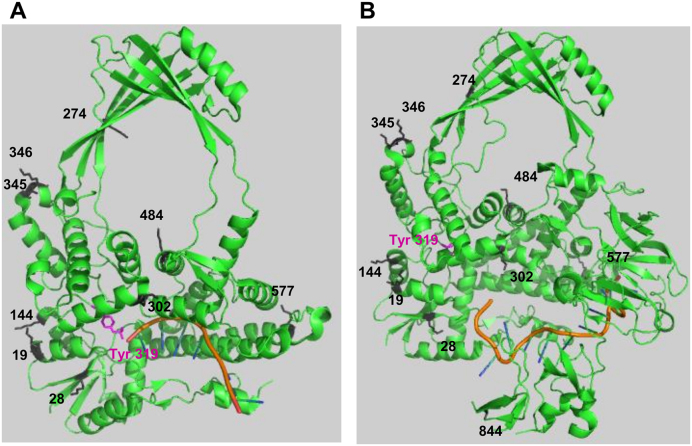
In previous proteomics studies, TopA was found to be acetylated at multiple lysine residues under various growth conditions (9,16). The locations of these acetylated lysines are shown (Figure 9) in the two available E. coli TopA crystal structures with ssDNA bound covalent to the active site, or non-covalently to the C-terminal domains (60,61). Most of the acetylated lysines are located in the solvent exposed positions. They would be accessible to the CobB deacetylase as substrate, and may influence initial association between TopA and chromosomal DNA for the formation of the catalytically competent TopA-DNA complex. The acetylation state of some of the TopA lysines could also potentially affect the enzyme conformational change required for the opening and closing of the gap between the 3΄-OH and 5΄-phosphotyrosine ends of the cleaved DNA single strand at the active site to allow passage of the complementary DNA single strand through the gap into the central hole of TopA. Lysine acetylation at the C-terminal domains of TopA may also influence its direct interaction with RNA polymerase during transcription elongation (41,62). The acetylation state of different TopA lysines could have differential effects on the protein-DNA or protein-protein interactions. The elucidation of the biochemical significance of individual TopA lysine acetylation sites would require future mechanistic and genetic studies.
Figure 9.
Location of lysines in E. coli TopA observed as acetylated lysines in proteomics studies. The lysine residues reported to be acetylated are shown in the crystal structures of the (A) TopA N-terminal domains D1–D4 in covalent complex with cleaved ssDNA substrate (PDB 3PX7); (B) full length TopA with ssDNA non-covalently bound to the C-terminal domains (PDB 4RUL).
The results presented here support modulation of TopA catalytic activity via lysine acetylation, and a physiological function of E. coli CobB in preventing TopA inactivation from excess lysine acetylation. There are questions that remain to be answered with regard to potential regulation of topoisomerase activity and DNA supercoiling as a global signal by acetylation-deacetylation in bacteria. Determination of the acetylation stoichiometry at specific TopA lysine residues under various growth conditions, including the growth conditions employed in this study, is currently incomplete. In addition to the confirmed lysine acetyltransferase Pka/YfiQ, there may be other E. coli acetyltransferases that remain to be identified and characterized. Lysine acetylation-deacetylation has been linked to stress response in bacteria (15), and may provide a more rapid mechanism for modification of TopA catalytic activity than the transcription response from the topA promoters.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge Yale CGSC and National BioResource Project (NIG, Japan): E. coli for providing strains and plasmids. We thank Shayna Sandhaus for assistance in editing the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (NIH) [R01 GM054226 to Y.T.]. Y.N.Z., D.J.J. were supported by Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Funding for open access charge: Florida International University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Fan J., Krautkramer K.A., Feldman J.L., Denu J.M.. Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 2015; 10:95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Su X., Wellen K.E., Rabinowitz J.D.. Metabolic control of methylation and acetylation. Curr. Opin. Chem. Biol. 2016; 30:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cain J.A., Solis N., Cordwell S.J.. Beyond gene expression: The impact of protein post-translational modifications in bacteria. J. Proteomics. 2014; 97:265–286. [DOI] [PubMed] [Google Scholar]
- 4. Thao S., Escalante-Semerena J.C.. Control of protein function by reversible Nε-lysine acetylation in bacteria. Curr. Opin. Microbiol. 2011; 14:200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 6. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y. et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010; 327:1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu B.J., Kim J.A., Moon J.H., Ryu S.E., Pan J.G.. The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 2008; 18:1529–1536. [PubMed] [Google Scholar]
- 8. Zhang K., Zheng S., Yang J.S., Chen Y., Cheng Z.. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res. 2013; 12:844–851. [DOI] [PubMed] [Google Scholar]
- 9. Weinert B.T., Iesmantavicius V., Wagner S.A., Scholz C., Gummesson B., Beli P., Nystrom T., Choudhary C.. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell. 2013; 51:265–272. [DOI] [PubMed] [Google Scholar]
- 10. Wolfe A.J. Bacterial protein acetylation: new discoveries unanswered questions. Curr. Genet. 2016; 62:335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernal V., Castano-Cerezo S., Gallego-Jara J., Ecija-Conesa A., de Diego T., Iborra J.L., Canovas M.. Regulation of bacterial physiology by lysine acetylation of proteins. N. Biotechnol. 2014; 31:586–595. [DOI] [PubMed] [Google Scholar]
- 12. Schilling B., Christensen D., Davis R., Sahu A.K., Hu L.I., Walker-Peddakotla A., Sorensen D.J., Zemaitaitis B., Gibson B.W., Wolfe A.J.. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 2015; 98:847–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li R., Gu J., Chen Y.Y., Xiao C.L., Wang L.W., Zhang Z.P., Bi L.J., Wei H.P., Wang X.D., Deng J.Y. et al. CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol. Microbiol. 2010; 76:1162–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liarzi O., Barak R., Bronner V., Dines M., Sagi Y., Shainskaya A., Eisenbach M.. Acetylation represses the binding of CheY to its target proteins. Mol. Microbiol. 2010; 76:932–943. [DOI] [PubMed] [Google Scholar]
- 15. Ma Q., Wood T.K.. Protein acetylation in prokaryotes increases stress resistance. Biochem. Biophys. Res. Commun. 2011; 410:846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Castano-Cerezo S., Bernal V., Post H., Fuhrer T., Cappadona S., Sanchez-Diaz N.C., Sauer U., Heck A.J., Altelaar A.F., Canovas M.. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol. Syst. Biol. 2014; 10:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thao S., Chen C.S., Zhu H., Escalante-Semerena J.C.. N ε-lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS One. 2010; 5:e15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lima B.P., Thanh Huyen T.T., Basell K., Becher D., Antelmann H., Wolfe A.J.. Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J. Biol. Chem. 2012; 287:32147–32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song L., Wang G., Malhotra A., Deutscher M.P., Liang W.. Reversible acetylation on Lys501 regulates the activity of RNase II. Nucleic Acids Res. 2016; 44:1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Q.F., Gu J., Gong P., Wang X.D., Tu S., Bi L.J., Yu Z.N., Zhang Z.P., Cui Z.Q., Wei H.P. et al. Reversibly acetylated lysine residues play important roles in the enzymatic activity of Escherichia coli N-hydroxyarylamine O-acetyltransferase. FEBS J. 2013; 280:1966–1979. [DOI] [PubMed] [Google Scholar]
- 21. AbouElfetouh A., Kuhn M.L., Hu L.I., Scholle M.D., Sorensen D.J., Sahu A.K., Becher D., Antelmann H., Mrksich M., Anderson W.F. et al. The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites. Microbiologyopen. 2015; 4:66–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Z., Harshey R.M.. Crucial role for DNA supercoiling in mu transposition: A kinetic study. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sobetzko P. Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res. 2016; 44:1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Usongo V., Drolet M.. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet. 2014; 10:e1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drlica K. Control of bacterial DNA supercoiling. Mol. Microbiol. 1992; 6:425–433. [DOI] [PubMed] [Google Scholar]
- 26. Liu L.F., Wang J.C.. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldstein E., Drlica K.. Regulation of bacterial DNA supercoiling: Plasmid linking numbers vary with growth temperature. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:4046–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheung K.J., Badarinarayana V., Selinger D.W., Janse D., Church G.M.. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 2003; 13:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balke V.L., Gralla J.D.. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J. Bacteriol. 1987; 169:4499–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsieh L.S., Burger R.M., Drlica K.. Bacterial DNA supercoiling and [ATP]/[ADP]. changes associated with a transition to anaerobic growth. J. Mol. Biol. 1991; 219:443–450. [DOI] [PubMed] [Google Scholar]
- 31. Zechiedrich E.L., Khodursky A.B., Bachellier S., Schneider R., Chen D., Lilley D.M., Cozzarelli N.R.. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000; 275:8103–8113. [DOI] [PubMed] [Google Scholar]
- 32. Tse-Dinh Y.C. Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling. Nucleic Acids Res. 1985; 13:4751–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Menzel R., Gellert M.. Regulation of the genes for E. coli DNA gyrase: Homeostatic control of DNA supercoiling. Cell. 1983; 34:105–113. [DOI] [PubMed] [Google Scholar]
- 34. Lesley S.A., Jovanovich S.B., Tse-Dinh Y.C., Burgess R.R.. Identification of a heat shock promoter in the topA gene of Escherichia coli. J. Bacteriol. 1990; 172:6871–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qi H., Menzel R., Tse-Dinh Y.C.. Regulation of Escherichia coli topA gene transcription: Involvement of a sigmaS-dependent promoter. J. Mol. Biol. 1997; 267:481–489. [DOI] [PubMed] [Google Scholar]
- 36. Qi H., Menzel R., Tse-Dinh Y.C.. Effect of the deletion of the sigma 32-dependent promoter (P1) of the Escherichia coli topoisomerase I gene on thermotolerance. Mol. Microbiol. 1996; 21:703–711. [DOI] [PubMed] [Google Scholar]
- 37. Weinstein-Fischer D., Altuvia S.. Differential regulation of Escherichia coli topoisomerase I by fis. Mol. Microbiol. 2007; 63:1131–1144. [DOI] [PubMed] [Google Scholar]
- 38. Weinstein-Fischer D., Elgrably-Weiss M., Altuvia S.. Escherichia coli response to hydrogen peroxide: A role for DNA supercoiling, topoisomerase I and fis. Mol. Microbiol. 2000; 35:1413–1420. [DOI] [PubMed] [Google Scholar]
- 39. Stewart N., Feng J., Liu X., Chaudhuri D., Foster J.W., Drolet M., Tse-Dinh Y.C.. Loss of topoisomerase I function affects the RpoS-dependent and GAD systems of acid resistance in Escherichia coli. Microbiology. 2005; 151:2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu I.F., Aedo S., Tse-Dinh Y.C.. Resistance to topoisomerase cleavage complex induced lethality in Escherichia coli via titration of transcription regulators PurR and FNR. BMC Microbiol. 2011; 11:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang J., Annamalai T., Cheng B., Banda S., Tyagi R., Tse-Dinh Y.C.. Antimicrobial susceptibility and SOS-dependent increase in mutation frequency are impacted by E. coli topoisomerase I C-terminal point mutation. Antimicrob. Agents Chemother. 2015; 59:6195–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baeza J., Dowell J.A., Smallegan M.J., Fan J., Amador-Noguez D., Khan Z., Denu J.M.. Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem. 2014; 289:21326–21338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuhn M.L., Zemaitaitis B., Hu L.I., Sahu A., Sorensen D., Minasov G., Lima B.P., Scholle M., Mrksich M., Anderson W.F. et al. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One. 2014; 9:e94816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C.F., Grishin N.V., Zhao Y.. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics. 2009; 8:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu C.X., Wu F.L., Jiang H.W., He X., Guo S.J., Tao S.C.. Global identification of CobB interactors by an Escherichia coli proteome microarray. Acta Biochim. Biophys. Sin. (Shanghai). 2014; 46:548–555. [DOI] [PubMed] [Google Scholar]
- 46. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006; 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng B., Shukla S., Vasunilashorn S., Mukhopadhyay S., Tse-Dinh Y.C.. Bacterial cell killing mediated by topoisomerase I DNA cleavage activity. J. Biol. Chem. 2005; 280:38489–38495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H.. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005; 12:291–299. [DOI] [PubMed] [Google Scholar]
- 50. Zhou K., Zhou L., Lim Q., Zou R., Stephanopoulos G., Too H.P.. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011; 12:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peng S., Stephan R., Hummerjohann J., Tasara T.. Evaluation of three reference genes of Escherichia coli for mRNA expression level normalization in view of salt and organic acid stress exposure in food. FEMS Microbiol. Lett. 2014; 355:78–82. [DOI] [PubMed] [Google Scholar]
- 52. Narula G., Annamalai T., Aedo S., Cheng B., Sorokin E., Wong A., Tse-Dinh Y.C.. The strictly conserved Arg-321 residue in the active site of Escherichia coli topoisomerase I plays a critical role in DNA rejoining. J. Biol. Chem. 2011; 286:18673–18680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tse-Dinh Y.C., Beran R.K.. Multiple promoters for transcription of the Escherichia coli DNA topoisomerase I gene and their regulation by DNA supercoiling. J. Mol. Biol. 1988; 202:735–742. [DOI] [PubMed] [Google Scholar]
- 54. Qi H., Menzel R., Tse-Dinh Y.C.. Increased thermosensitivity associated with topoisomerase I deletion and promoter mutations in Escherichia coli. FEMS Microbiol. Lett. 1999; 178:141–146. [DOI] [PubMed] [Google Scholar]
- 55. Tse-Dinh Y.C. Increased sensitivity to oxidative challenges associated with topA deletion in Escherichia coli. J. Bacteriol. 2000; 182:829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006; 59:723–730. [DOI] [PubMed] [Google Scholar]
- 57. Peter B.J., Arsuaga J., Breier A.M., Khodursky A.B., Brown P.O., Cozzarelli N.R.. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004; 5:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Masse E., Phoenix P., Drolet M.. DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J. Biol. Chem. 1997; 272:12816–12823. [DOI] [PubMed] [Google Scholar]
- 59. Klein A.H., Shulla A., Reimann S.A., Keating D.H., Wolfe A.J.. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 2007; 189:5574–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tan K., Zhou Q., Cheng B., Zhang Z., Joachimiak A., Tse-Dinh Y.C.. Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I. Nucleic Acids Res. 2015; 43:11031–11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Z., Cheng B., Tse-Dinh Y.C.. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:6939–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cheng B., Zhu C.X., Ji C., Ahumada A., Tse-Dinh Y.C.. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J. Biol. Chem. 2003; 278:30705–30710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.