Abstract
Long interspersed element 1 (L1) is an autonomous non-LTR retroelement that is active in mammalian genomes. Although retrotranspositionally incompetent and functional L1 loci are present in the same genomes, it remains unknown whether non-functional L1s have any trans effect on mobilization of active elements. Using bioinformatic analysis, we identified over a thousand of human L1 loci containing at least one stop codon in their ORF1 sequence. RNAseq analysis confirmed that many of these loci are expressed. We demonstrate that introduction of equivalent stop codons in the full-length human L1 sequence leads to the expression of truncated ORF1 proteins. When supplied in trans some truncated human ORF1 proteins suppress human L1 retrotransposition. This effect requires the N-terminus and coiled-coil domain (C-C) as mutations within the ORF1p C-C domain abolish the suppressive effect of truncated proteins on L1 retrotransposition. We demonstrate that the expression levels and length of truncated ORF1 proteins influence their ability to suppress L1 retrotransposition. Taken together these findings suggest that L1 retrotransposition may be influenced by coexpression of defective L1 loci and that these L1 loci may reduce accumulation of de novo L1 integration events.
INTRODUCTION
Long interspersed element 1 (LINE-1, L1) is an autonomous, non-long terminal repeat retrotransposon that has contributed to the structural variability of mammalian genomes (1). L1 has a 5΄ untranslated region (UTR) followed by an open reading frame 1 (ORF1), an inter-ORF region, an open reading frame 2 (ORF2) and a 3΄ UTR with a polyA site and an associated polyA tail (2,3). L1 transcription generates full-length mRNAs that produce two proteins, ORF1p and ORF2p (4). The ORF1p and ORF2p interact with their parental L1 mRNA in cis to form a ribonucleoprotein particle (RNP) (5,6). All three RNP components are required for successful L1 retrotransposition (5–7). Strong cis preference of L1 proteins for their mRNA is beneficial for L1 retrotransposition and genome stability (8,9). This cis preference minimizes the possibility for functional L1s to rescue retrotranspositionally incompetent L1s (8,9), which are much more abundant than the active L1 loci (8,9). It also minimizes nonspecific mobilization of cellular mRNAs (8,9). However, L1-generated proteins do operate in trans when they mobilize non-autonomous human retroelements such as Alu and SINE/VNTR/Alu (SVA) (10–12). Although ORF2p alone is sufficient to mobilize Alu, ectopic L1 ORF1p expression enhances Alu mobilization through an unknown mechanism (12). Whether there is any trans effect of ORF1p expression on retrotransposition of human or mouse L1 elements remains unknown.
ORF1p has four recognized domains: an N-terminal domain, which contains two highly conserved phosphorylation sites critical for retrotransposition (13), a coiled-coil domain (C-C) containing a leucine zipper motif, an RNA recognition motif (RRM) and a C-terminal domain (14). These domains are present in both mouse and human ORF1 proteins (13,15–17). Mouse and human ORF1 proteins function as homotrimers, which are formed through the C-C domain (15–20). The ORF1p also binds to RNA and has nucleic acid chaperone activity (6,8,9,15,21). ORF1p is generated in excess of the ORF2p (16–18,22). Although ORF1p expression has been used as a correlative measure of L1 activity (23–26), it remains unknown whether the amount of endogenous ORF1p adequately reflects L1 retrotransposition in vivo. Existing experimental data demonstrated that codon optimization of mouse and human L1 elements resulted in a substantial boost in L1 mRNA and protein expression of both L1 elements. This increase led to a dramatic increase in retrotransposition of the mouse L1 (27), but only a modest increase in the human L1 mobilization (28–30). Recently published data support that the relative ratio of L1 proteins may influence retrotransposition (22), with L1 RNPs containing few ORF1p potentially being more efficient at integration relative to those containing abundant ORF1p trimers. It is also not known whether all generated ORF1p molecules retain strict cis preference for their parental L1 mRNA. The trans effect of the ORF1p on Alu retrotransposition (10,12) and its requirement for SVA and host mRNA mobilization (11) demonstrates that some ORF1p is available to act in trans of their parental L1 mRNA.
L1 ORF1 protein homotrimerization has been observed in vitro (15–19). An interaction between mouse L1Tf and L1A ORF1 proteins has been observed using a yeast two hybrid system (20). It has been proposed that ORF1p molecules generated from the same parental mRNA associate to form ORF1p homotrimers that are involved in retrotransposition in mammalian cells (8,9). We have previously reported that ORF1p generated from different expression plasmids can form heterocomplexes in mammalian cells (31). This ability to heterotrimerize is species-specific, i.e. human L1 ORF1 proteins can heterodimerize with human (but not mouse) ORF1p in either mouse or human cells (31). This finding suggested the possibility that in mammalian cells, ORF1p produced from different L1 loci may have a trans effect on L1 retrotransposition in a manner similar to the effect of ORF1p on Alu mobilization (10,12). The ability of defective L1 loci to trans-complement each other to restore efficient L1 retrotransposition was previously tested and was ruled out (9,32). However, the possibility of the full-length ORF1p or truncated ORF1 proteins to affect retrotransposition of active L1s has not been investigated. The human genome contains thousands of full-length L1 loci (33), many of which have acquired premature stop codons within their ORF2 sequence (33–40). L1 mRNAs uniquely mapping to some of these retrotranspositionally-incompetent L1 loci were recovered from human cell lines (35,40) supporting the potential for expression of truncated ORF1p.
RNAseq analysis of authentic endogenous L1 mRNAs from three human cell lines confirmed that L1 loci containing stop codons in their ORF1 sequence are expressed (41). Our data demonstrate that transient or stable expression of the full-length or truncated human ORF1 proteins suppresses human L1 retrotransposition in human and mouse cells. This suppressive effect is species-specific, as expression of the full-length or truncated mouse ORF1p has no effect on human L1 mobilization. The dominant negative effect of the truncated human ORF1p on human L1 mobilization requires the N-terminus and an intact C-C domain of the ORF1p. We demonstrate that mutagenesis of key leucine residues in the C-C domain of the truncated human ORF1 protein abolishes its ability to form heterocomplexes and suppress L1 retrotransposition in trans. Confirmation of endogenous mRNA expression from L1 loci containing stop codons in their ORF1 sequence combined with our observations that stably expressed defective L1 elements suppress retrotransposition of transiently transfected active L1 in HeLa cells suggest that L1 loci expressing truncated ORF1 proteins may have a suppressive effect on retrotransposition of endogenously expressed functional L1 loci. These findings suggest that the unique spectrum of expressed non-functional L1 loci may differentially influence the efficiency of retrotransposition of functional L1s.
MATERIALS AND METHODS
Cells
HeLa (ATCC CCL2) and NIH 3T3 (ATCC CRL 1658) cells were maintained as previously described (42).
Plasmids
hL1wt is a plasmid expressing a full-length human wild-type (wt) L1.3 element (L1.3 plasmid as reported in (9)). L1Neo (pJM101/L1.3) (5,43) is a plasmid designed to express a full-length human wt L1 element tagged with a neomycin resistance cassette, which enables detection of retrotransposition events upon transient transfection in mammalian cells. 119Stop (pJM108) (5) plasmid is designed to express a full-length untagged human L1.3 element containing a stop codon at the amino acid position 119 in ORF1. hORF1 (pBudORF1opt) (12) is a plasmid containing codon-optimized human L1 ORF1 sequence designed to express a full-length ORF1p. mORF1 (pBudORF1syn) (27) is a plasmid containing codon-optimized mouse L1 ORF1 sequence designed to express a full-length mORF1p. mL1wt is a plasmid designed to express a full-length mouse wt L1spa element tagged with a neomycin resistance cassette, which enables detection of retrotransposition events upon transient transfection in mammalian cells (44). pBud (Invitrogen) was used as the empty plasmid control. pCEP (Invitrogen) was used as the empty control for Figures 7B, C, 8B and C. ‘pIRES’ is pIRES2-GFP expression plasmid that was used in the toxicity assays (Supplementary Figure S2) and as a transfection efficiency control (Figure 8C and Supplementary Figure S16B) (45). L1 ‘H1 H2’ (L1PA1 Neo) (28), L1 ‘H1 M2’ (pBS-L1-1H,2Mmneo) (28), L1 ‘M1 H2’ (pBS-L1-1M,2Hmneo) (28) and L1 ‘M1 M2’ (pBS syn mL1mneo) (28) are plasmids containing human and/or mouse codon-optimized ORF1 and ORF2 sequences. GAL4-fused ORF1 plasmid is designed to express a full-length human ORF1p containing a N-terminal GAL4 tag (31).
Figure 7.
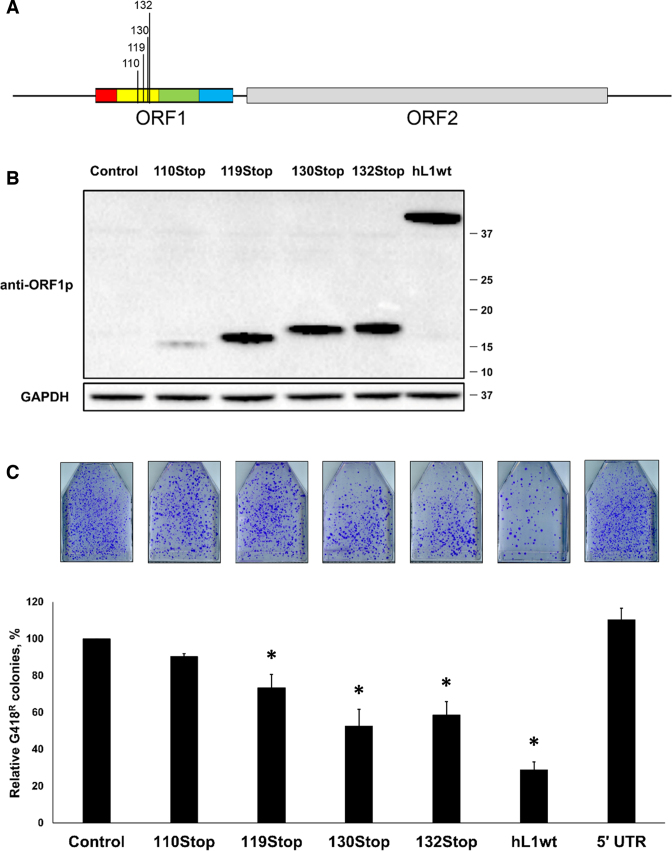
Human L1 retrotransposition is suppressed by full-length human L1s containing ORF1p stop codons. (A) A schematic of the L1 constructs generated to contain stop codons corresponding to amino acid positions 110, 119, 130 and 132 within the human L1 ORF1. (B) Western blot analysis (anti-ORF1p polyclonal antibodies) of ORF1 proteins generated from plasmids expressing full-length wt L1 (hL1wt) or full-length L1s containing stop codons (110Stop, 119Stop, 130Stop, 132Stop) in HeLa cells. A transfection with an empty plasmid was used as control (Control). Detection of GAPDH was used as a loading control. Molecular weight markers are shown on the right in kDa. (C) Result of L1 retrotransposition assay in HeLa cells using a Neo-tagged full-length human wt L1 expression plasmid co-transfected with an empty (control), hORF1, mORF1, 110Stop, 119Stop, 130Stop or 132Stop L1 expression plasmids. The number of G418 resistant colonies was normalized to the number of colonies determined for the Control flask for each independent experiment. Asterisk (*) denotes statistical significance between listed constructs and the control (n = 3, t-test, P < 0.05).
Figure 8.
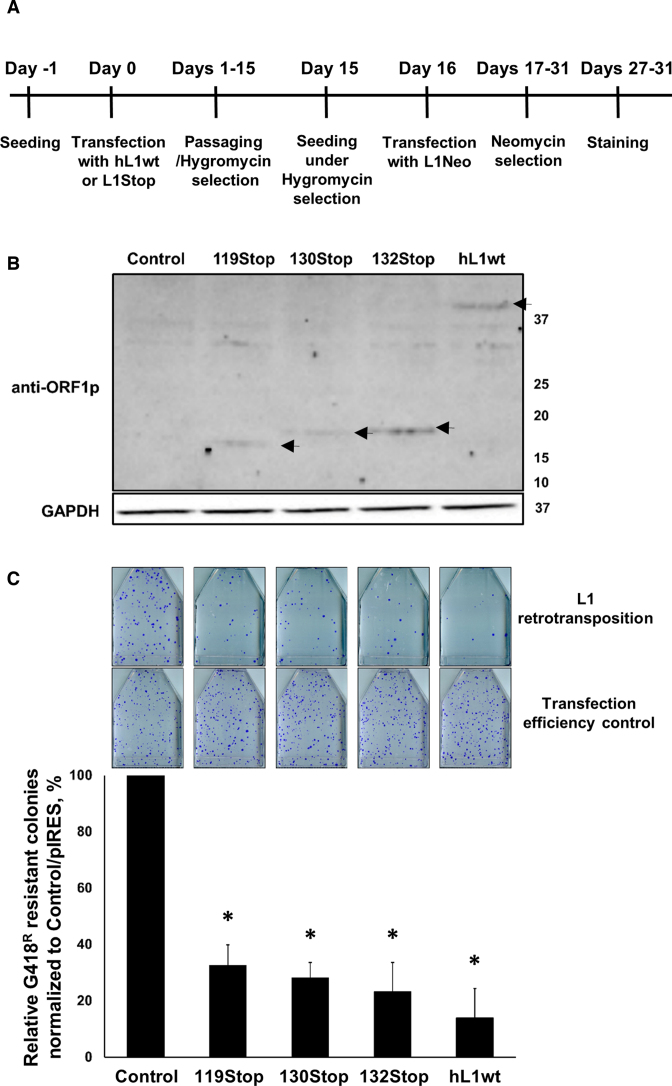
L1 retrotransposition is suppressed in HeLa cells constitutively expressing a full-length human L1 or full-length human L1s containing stop codons in their ORF1 sequence. (A) Schematic of experimental design. (B) Western blot analysis (anti-ORF1p polyclonal antibodies) of ORF1 proteins in engineered HeLa cells harboring full-length L1 (hL1wt), full-length L1s with stop codons (119Stop, 130Stop or 132Stop) or an empty (Control) expression plasmids. Molecular weight markers are shown on the right in kDa. Black arrows denote expected molecular weight of truncated or full-length ORF1 proteins generated from their respective expression plasmids. (C) Results of the wt human Neo-tagged L1 retrotransposition in HeLa cells constitutively expressing functional wt L1 or full-length wt L1s containing stop codons in their ORF1 sequence. Total number of G418 resistant colonies resulting from L1 retrotransposition was divided by the number of G418 resistant colonies resulting from the transfection efficiency control (transfection with the pIRES plasmid containing neomycin resistance) was normalized to the Control/pIRES ratio for each independent experiment. Asterisk (*) denotes statistical significance between the Control and L1 (n = 3, t-test, P < 0.05).
The DNA sequence of the 132M construct was synthesized (GenScript) to express a protein in which the leucine residues at positions 55, 73, 87, 90, 93, 100, 107 and 114 of human ORF1p were replaced with proline residues (CTG to CCC changes). The DNA sequence was subcloned into the pBud plasmid using HindIII and BamHI restriction endonucleases.
The DNA sequence of the mNhC-C construct was synthesized (GenScript) to contain codon-optimized sequences of the mouse ORF1 (corresponding to amino acids 1–46), human ORF1 (corresponding to amino acids 53–156) and a T7-tag. The DNA sequence of the hNmC-C construct was synthesized (GenScript) to contain codon-optimized sequence of the human ORF1 (corresponding to amino acids 1–52), mouse ORF1 (corresponding to amino acids 47–192), and a T7 tag. These sequences were subcloned into the pBud plasmid (Invitrogen) using HindIII and BamHI restriction endonucleases.
The previously reported consensus L1PA2 and L1PA3 ORF1 sequences (46) were used to synthesize (GenScript) full-length and truncated L1PA2 and L1PA3 ORF1 DNA sequence. The sequence AAG for lysine 14 was mutated to ACC to encode a threonine residue in order to match the epitope sequence of our hORF1p antibodies (custom rabbit polyclonal, epitope: TGNSKTQSASPPPK) (31). The truncated ORF1 sequences corresponding to endogenous (e) L1 ORF1 e259 (L1PA2 subfamily, chromosome 13, position 37724090–37730119 of UCSC genome browser hg19 build), e207 (L1PA2 subfamily, chromosome 1, position 174812365–174818381 of UCSC genome browser hg19 build) and e127 (L1PA2 subfamily, chromosome 12, position 96709723–96715749 of UCSC genome browser hg19 build) were codon-optimized using Primo Optimum 3.4 (http://www.changbioscience.com/primo/primoo.html) and commercially synthesized (GenScript). The numbers in the names of these constructs (259, 207 and 127) correspond to the amino acid position of the stop codons present in endogenous L1 loci. Full-length and truncated L1PA2, L1PA3 as well as endogenous L1 sequences were subcloned into the pBud plasmid using HindIII and BamHI restriction endonucleases.
Unless specifically mentioned otherwise, truncated constructs were generated by polymerase chain reaction (PCR) amplification of either wt (non-codon-optimized) or codon-optimized ORF1 DNA sequence. The resulting PCR products were subcloned into the pBud expression plasmid using HindIII and BamHI restriction endonucleases. See Supplementary Materials and Methods for primer sequences and generation of L1 expression plasmids containing stop codons.
Transfections
L1 retrotransposition assay
Adapted from (5), specific details for plasmid amounts, number of cells and transfection conditions for each figure are listed in the Supplementary Materials and Methods.
Total protein extraction
The cellular lysates were processed as previously described (31). See Supplementary Materials and Methods for details.
Toxicity assay
Adapted from (47). See Supplementary Materials and Methods for details.
Nuclear/cytoplasm fractionation
The processing of nuclear and cytoplasmic fractions was performed as previously described (31). See Supplementary Materials and Methods for details.
Western blot analysis
The western blot analysis was performed as previously described (31,48). See Supplementary Materials and Methods for details. The antibodies were diluted as listed below in a 3% milk (Bio Rad: 170–6404) in PBS-Tween: HRP-donkey anti-rabbit (Santa Cruz; sc-2317), HRP-donkey anti-goat (Santa Cruz; sc-2020) or HRP-goat anti-mouse (Santa Cruz; sc-2031) at 1:5000 dilution. GAPDH antibodies (Santa Cruz: sc-25778, 1:5000 dilution) and Lamin A/C (Santa Cruz 7293, 1:1000 dilution) were used as a fractionation and equal loading controls. anti-hORF1p is a custom rabbit polyclonal antibody (epitope: TGNSKTQSASPPPK, dilution 1:5000) (31), anti-mORF1p is a custom goat-polyclonal antibody (epitope: YRTPNRLDQKRNSS, dilution 1:1000) (31), T7-tag antibody (Cell Signaling; D9E1X, 1:10000 dilution) and 1:10000 dilution of Flag tag (Sigma Monoclonal Anti-Flag M2: F3165) antibody.
Sucrose cushion of the cytoplasmic extract
See Supplementary Materials and Methods for protocol details adapted from (28,32,49,50).
Co-immunoprecipitation
Adapted from (49). See Supplementary Materials and Methods for protocol details.
Bioinformatic analysis
L1Base (51) was used for analysis in Figure 6A and Supplementary Figure S10. L1Base (51) was utilized to identify full-length L1 loci containing stop codons in their ORF1 sequence. Human full-length >4500 nt LINE-1 elements Ens38.36 was used as the database to identify the L1 loci containing no gaps or frameshifts in ORF1 and containing one or more stop codons in ORF1, query start sites within 50 bp. This search identified 1474 L1 loci, which were then analyzed using the amino acid FASTA output for each of the ORF1 sequences to identify a methionine residue as the first amino acid of ORF1p. This approach identified 1244 L1 loci containing ORF1p with a start codon, which were further analyzed for the first stop codon position within the amino acid FASTA sequence. The frequency of stop codons at each ORF1 amino acid position was plotted (Figure 6A). The same approach was utilized for the mouse L1 loci using L1Base mouse full-length >5k nt LINE-1 elements Ens38.35 (51). The first 10 L1 loci entries identified on each mouse chromosome (a total of 198 loci) were subjected to the first stop codon position analysis. The frequency of stop positions at each amino acid of the mouse ORF1 protein was plotted (Supplementary Figure S10). Data from (Deininger et al.) (41) was used for Supplementary Figure S13A and B. See Supplementary Materials and Methods for additional details.
Figure 6.
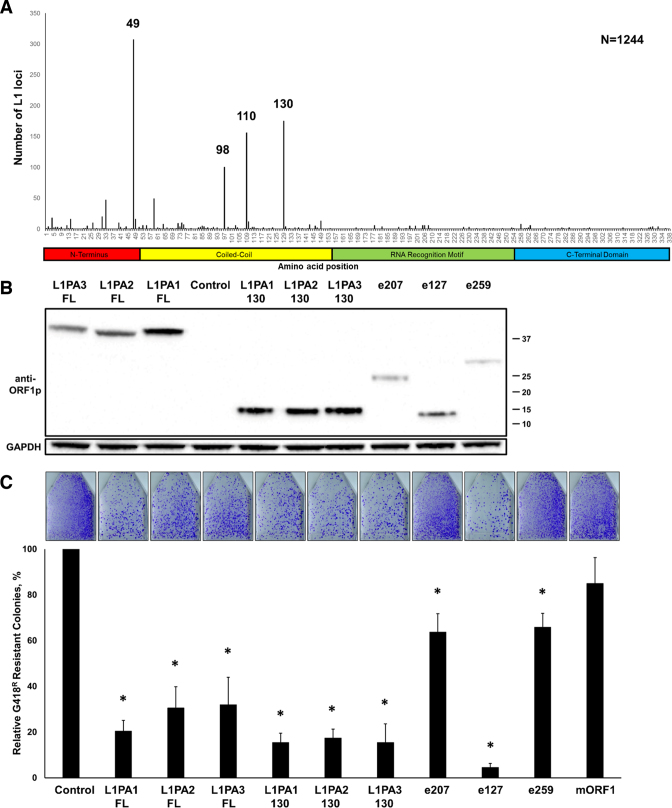
Bioinformatic analysis of stop codon positions in retrotranspositionally-incompetent human L1 loci. (A) Positions and frequencies of stop codons identified in the ORF1p sequence of 1244 full-length human L1 loci. The most frequent stop codon positions corresponding to amino acids 49, 98, 110 and 130 of the full-length human ORF1p are indicated. (B) Western blot analysis using anti-ORF1p Ab of full-length and truncated (130) ORF1 proteins generated from expression plasmids containing consensus sequences corresponding to L1PA1-3 subfamilies or sequences representing endogenously expressed L1s (e207, e127 or e259). Control lane indicates cells transfected with an empty plasmid. Positions of molecular markers are indicated on the right in kDa. (C) Results of L1 retrotransposition assay using a Neo-tagged, full-length human wt L1 expression plasmid cotransfected with an empty (Control), L1PA1-3 full-length or truncated (130, e207, e127 or e259) human ORF1 expression plasmids or a mORF1 expression plasmid. The number of G418 resistant colonies was normalized to the Control flask for each independent experiment. Asterisk (*) denotes statistical significance between listed constructs and the Control (n = 3, t-test, P < 0.05).
RESULTS
Full-length human ORF1p suppresses human L1 retrotransposition in trans
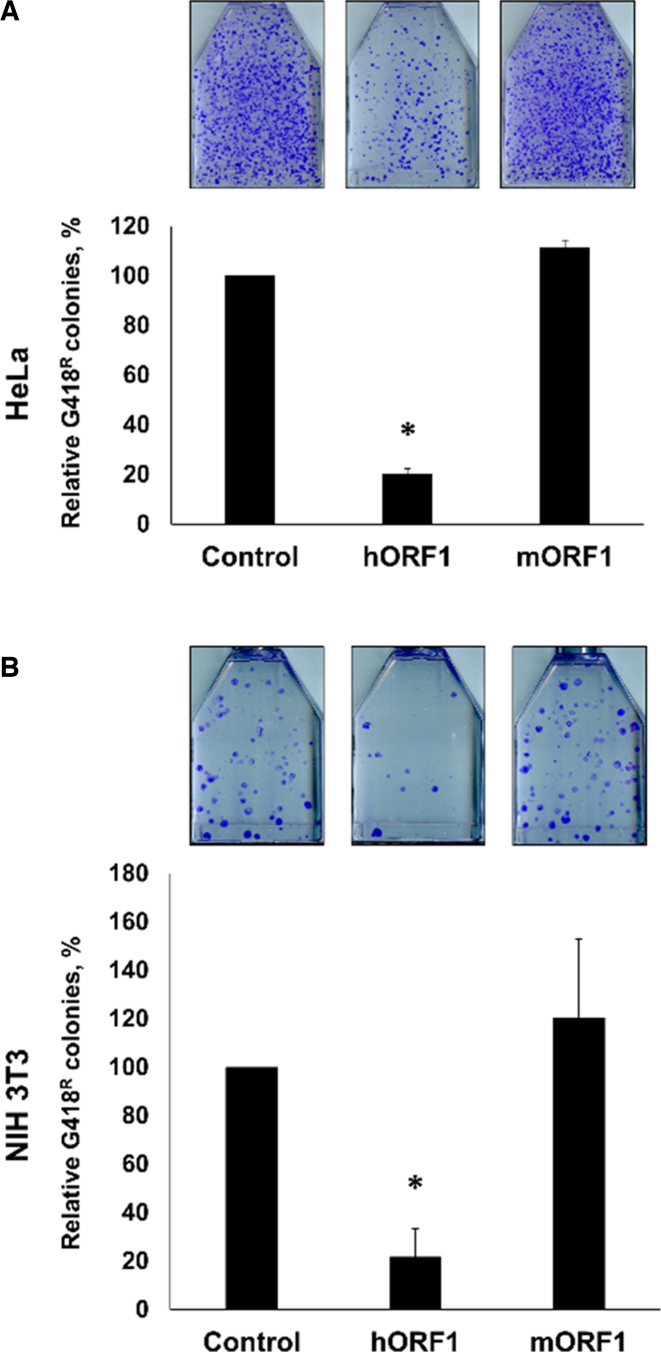
We have previously reported that ORF1 proteins generated from different mRNA molecules can trimerize in a species-specific manner in mammalian cells (31). Based on these observations (31) and the report that the levels of ORF1p may influence L1 retrotransposition (22), we hypothesized that transient expression of the full-length human ORF1p may have a trans effect on retrotransposition of human L1. This hypothesis was tested using HeLa cells transiently cotransfected with plasmids expressing human neomycin-tagged L1 (L1Neo) and full-length human or mouse ORF1 proteins (hORF1p or mORF1p). The L1Neo expression plasmid contained wt L1 sequence. The ORF1 expression plasmids contained codon-optimized human or mouse L1 sequence (28). This approach demonstrated that when expressed in trans human, but not mouse, ORF1p suppressed human L1 retrotransposition (Figure 1A and Supplementary Figure S1). The same experiment was carried out in NIH 3T3 cells to test the possibility of a human-specific host factor being involved in the effect. Similar to human cells, human L1 retrotransposition was suppressed when this element was coexpressed with hORF1p, but not mORF1p, in NIH 3T3 cells (Figure 1B).
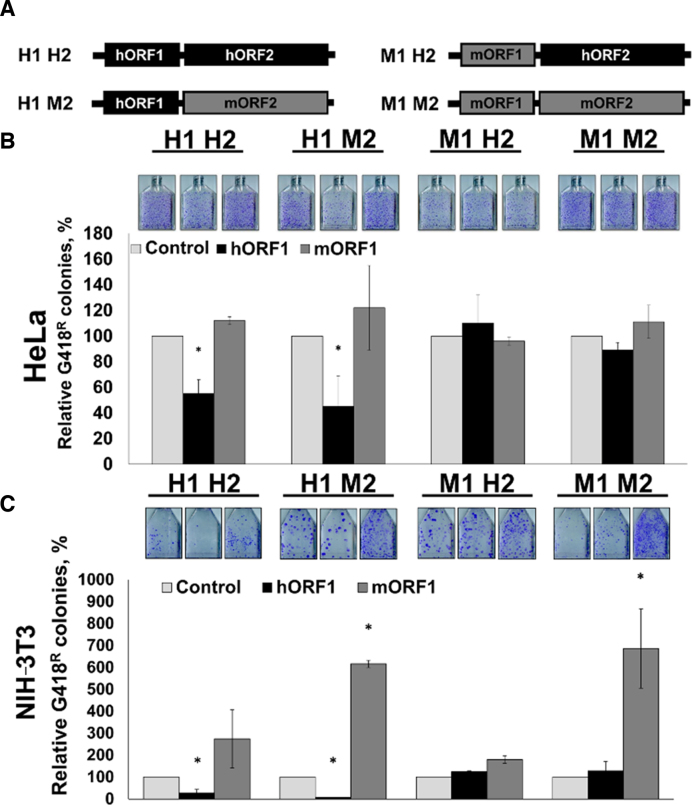
Figure 1.
Full-length human ORF1p suppresses human L1 retrotransposition in trans. Results of L1 retrotransposition assay in HeLa (A) or NIH 3T3 (B) cells using neomycin-tagged, full-length, wild-type (wt) human L1 cotransfected with an empty (control), hORF1 or mORF1 expression plasmids. The number of G418 resistant colonies was normalized to the Control flask (L1 construct cotransfected with empty plasmid) for each independent experiment. Asterisks (*) denote statistical significance between listed constructs and the control (n = 3, t-test, P < 0.05).
To assess which of the L1-encoded proteins may be involved in the response to ORF1p trans effect on L1 retrotransposition we used previously reported chimeric L1 constructs containing codon-optimized ORF1 and ORF2 sequences of either human or mouse origin (Figure 2A) (28). Specifically, we used the H1H2 and M1M2 L1s, which contain only human or mouse sequences, respectively, the H1M2 L1, which contains human ORF1 and mouse ORF2 sequences, and the M1H2 L1 that contains mouse ORF1 and human ORF2 sequences. Transient transfections of HeLa cells with these plasmids and with either an empty plasmid (control), or plasmids expressing hORF1 or mORF1 proteins demonstrated that hORF1p suppressed mobilization of L1 constructs containing human ORF1 sequence (Figure 2B, black bars: H1H2 and H1M2). mORF1p had no negative trans effect on retrotransposition of human, mouse, or chimeric L1s (Figure 2B, dark gray bars).
Figure 2.
A full-length human ORF1p suppresses retrotransposition of chimeric L1s containing human ORF1 sequence. (A) Schematic of neomycin-tagged, full-length chimeric L1s containing human and/or mouse codon-optimized ORF1 and ORF2 sequences. Human ORF1 and human ORF2 are designated as H1 and H2, mouse ORF1 and mouse ORF2 are designated as M1 and M2, respectively. (B) Results of L1 retrotransposition assay in HeLa cells using plasmids expressing H1 H2, H1 M2, M1 H2 or M1 M2 L1s cotransfected with an empty (control), hORF1 or mORF1 expression plasmids. (C) Results of L1 retrotransposition assay performed as in (B) using NIH 3T3 cells. The number of G418 resistant colonies was normalized to the control flask (L1 construct cotransfected with an empty plasmid) for each independent experiment. Asterisks (*) denote statistical significance between listed constructs and the control (n = 3, t-test, P < 0.05).
The same experiment was also carried out in NIH 3T3 cells. Consistent with the results obtained in HeLa cells, cotransfection of hORF1 expression plasmid with the H1H2 or H1M2 L1 constructs containing human ORF1 sequence suppressed their retrotransposition (Figure 2C, black bars: H1H2 and H1M2). Expression of hORF1p did not have any effect on retrotransposition of M1M2 or M1H2 L1 elements. Cotransfection of the mORF1p expression plasmid with the M1M2 or H1M2 L1 constructs containing mouse ORF2 (mORF2) sequence resulted in a significant (5–6-fold) increase in their retrotransposition in NIH 3T3 cells relative to the control plasmid (Figure 2C, dark gray bars versus light gray bars).
Toxicity associated with human ORF1p expression, an increase in L1 toxicity in the presence of trans ORF1p or ORF1p trans effect on retrotransposition through protein interactions with L1 or host proteins could explain the ORF1p-mediated decrease in L1 retrotransposition. ORF1p expression in trans did not have any adverse effect on cell viability when the full-length ORF1p was transiently expressed alone (Supplementary Figure S2A and B) or in combination with human L1 using a previously reported toxicity assay (29,45) (Supplementary Figure S2C). In combination with the results shown in Figure 2, these findings support the involvement of the L1-encoded ORF1 protein in the trans effect of ORF1p on human L1 retrotransposition.
The ORF1p fragment containing the N-terminus and C-C domain is sufficient to suppress human L1 retrotransposition
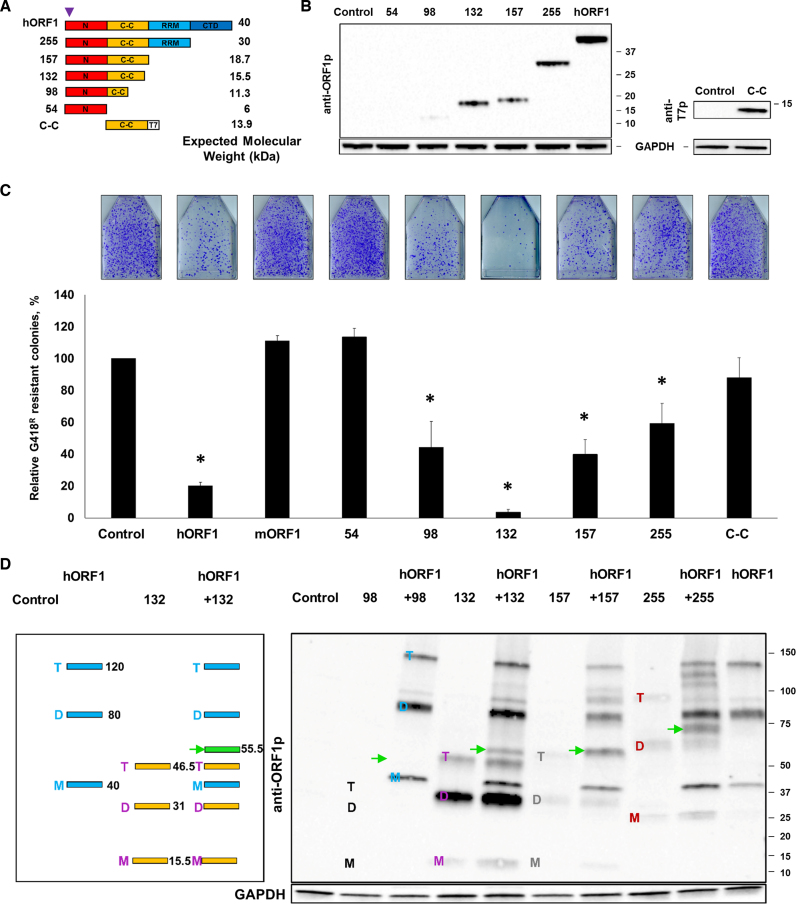
To identify which region(s) of the hORF1p is responsible for the suppression of L1 retrotransposition and to test the possibility that truncated ORF1 proteins may also suppress retrotransposition, we generated five plasmids designed to produce C-terminally truncated human ORF1p, with the construct name indicating the amino acid position at the site of truncation (Figure 3A). Plasmids 54, 157 and 255 are designed to produce truncated human ORF1 proteins containing the N-terminus, N-terminus/C-C, or N-terminus/C-C/RNA binding domains, respectively. Plasmids 98 and 132 are designed to produce truncated human ORF1 proteins containing the N-terminus and 6 or 11 out of the 14 heptads of the C-C domain, respectively. A plasmid termed C-C domain is designed to express a T7-tagged C-C domain of the human ORF1p because it is involved in the ORF1p trimerization (15–18). Western blot analysis using anti-ORF1p polyclonal antibodies recognizing an epitope in the N-terminus of the human ORF1p (31) or anti-T7 antibodies determined that other than the 54 construct, all plasmids expressed detectable levels of truncated ORF1 proteins in HeLa cells (Figure 3B). The 98 expression plasmid produced the lowest steady-state protein levels.
Figure 3.
Analysis of truncated ORF1 proteins expression and their potential to suppress L1 retrotransposition. (A) ORF1p domains are indicated as an N-terminal domain (N), a coiled-coil domain (C-C), an RNA recognition motif (RRM) and a C-terminal domain (CTD). The numbers listed on the left indicate the amino acid position of truncation. Expected molecular weights corresponding to each truncated protein are listed on the right. The approximate position of the epitope for the anti-ORF1p polyclonal antibodies is shown (purple triangle). The C-C construct has a T7 tag (T7). (B) (Left) Western blot analysis of truncated ORF1 proteins transiently expressed in HeLa cells. Control lane indicates cells transfected with an empty plasmid. Numbers listed on the right correspond to molecular weight markers in kDa. Detection of GAPDH was used as a loading control. (Right) Western blot analysis of C-C domain protein expression using anti-T7 tag polyclonal antibodies (anti-T7p). (C) Results of L1 retrotransposition assay in HeLa cells using a plasmid expressing a Neo-tagged, full-length human wt L1 cotransfected with an empty (control), hORF1, mORF1, 54, 98, 132, 157, 255 or C-C domain expression plasmids. The number of G418 resistant colonies was normalized to the control flask (L1 construct cotransfected with an empty plasmid) for each independent experiment. Asterisk (*) denotes statistical significance between listed constructs and the control (n = 3, t-test, P < 0.05). (D) (Left) A schematic of the expected banding pattern resulting from the coexpression of the full-length (hORF1, blue) and 132 (132, yellow) ORF1 proteins when analyzed using non-reducing conditions. Blue and purple M, D, T letters and numbers (black) on the right correspond to monomers, dimers, trimers and their expected molecular weights. ORF1p 132 lane illustrates an appearance of a novel band with a unique molecular weight, if the coexpressed proteins form heterodimers (green band and arrow (55.5 kDa)). (Right) Western blot analysis of proteins generated from codon-optimized 98, 132, 157, 255 and hORF1 constructs transfected individually or cotransfected in HeLa cells (non-reducing conditions, anti-ORF1p polyclonal antibodies (anti-ORF1p)). Green arrows indicate expected heterodimers between hORF1p and 98 (51.3 kDa), 132 (55.5 kDa), 157 (58.7 kDa) and 255 (70 kDa). M, D, T on the left of each lane indicate monomers, dimers and trimers (black is 98, blue is ORF1, purple is 132, gray is 157 and 255 is red). Control lane indicates cells transfected with an empty plasmid. Numbers on the right of the image are molecular weight markers in kDa. GAPDH was used as a loading control.
To assess the trans effect of these truncated human ORF1 proteins on human L1 retrotransposition, HeLa cells were transiently cotransfected with L1Neo and each of the above plasmids as described in Figure 1. This approach demonstrated that when expressed in trans, the 54 and C-C domain proteins did not suppress L1 retrotransposition (Figure 3C). Cotransfection of the 98, 132, 157 or 255 expression constructs with the human L1Neo significantly reduced L1 mobilization. The 132 construct reduced L1 retrotransposition to the lowest levels at 4% of the control, which surpassed the extent of inhibition observed with the full-length ORF1p expression plasmid (Figure 3C).
A potential mechanism that could explain this negative trans effect of the truncated ORF1 proteins on L1 retrotransposition is through a direct interaction between the truncated ORF1p and the full-length ORF1p, which would produce ORF1p trimers containing one or two defective monomers. To test whether trans interactions between the truncated and full-length ORF1 proteins exist, HeLa cells were transiently cotransfected with plasmids containing truncated ORF1 sequences and an expression plasmid containing full-length ORF1 sequence. Cotransfection of these plasmids allows for an assessment of the ability of the proteins they produce to heterodimerize using a previously reported western blot analysis carried out under non-reducing conditions (31). The premise is that the cysteine residues present in the C-C domain of the human ORF1 protein cross link interacting ORF1 proteins upon oxidation during protein harvest resulting in bands with unique molecular weights corresponding to these heterodimers (Figure 3D, a green band). Consistent with their ability to suppress L1 mobilization in trans, the 132, 157 and 255 truncated ORF1 proteins formed heterodimers of expected molecular weights with the full-length ORF1p (Figure 3D, green arrows). Because the 98 construct expressed significantly less protein than the 132, 157 or 255 constructs (Figure 3B), our anti-ORF1p antibodies were not sensitive enough to detect monomers, dimers, trimers and heterodimers formed by this truncated protein under non-reducing conditions (Figure 3D). The C-C construct did not form heterodimers with the full-length ORF1p when coexpressed in HeLa cells, consistent with its inability to suppress L1 retrotransposition (Supplementary Figure S3).
Combined, these data demonstrate that the ORF1p fragment containing the N-terminus and the heptads of the C-C domains of the human ORF1 protein is sufficient to suppress human L1 retrotransposition in trans. Our results also demonstrate that the inclusion of the RRM domain reduces the suppressive effect of truncated ORF1p on L1 mobilization. Also, our finding that the C-C domain alone does not suppress L1 retrotransposition in trans, suggests that the N-terminus may be important for this effect (Figure 3C).
Truncated chimeric ORF1 proteins composed of human and mouse sequences do not suppress human L1 retrotransposition
The fact that a truncated ORF1p containing only the N-terminal sequence of the ORF1 protein is not stable in HeLa cells prevented us from testing its sole effect on the suppression of L1 retrotransposition. To test the contribution of the N-terminal domain to the ORF1p-mediated trans effect on retrotransposition by other approaches, we generated constructs designed to express chimeric proteins hNmC-C and mNhC-C. The hNmC-C protein contains the N-terminal sequence of the human ORF1p and the C-C domain of the mouse ORF1p. The mNhC-C protein contains the N-terminus of the mouse ORF1p and the C-C domain of the human ORF1p (Supplementary Figure S4A). Both chimeric constructs contain a T7 tag to allow for their detection by western blot analysis. Western blot analysis of total cellular lysates collected from HeLa cells transiently transfected with the chimeric plasmids detected chimeric proteins of expected molecular weights (Supplementary Figure S4A). Western blot analysis using non-reducing conditions demonstrated that the mNhC-C protein did not heterodimerize with the full-length hORF1p when coexpressed in HeLa cells (Supplementary Figure S4B). These chimeric NC-C constructs had no effect on L1 retrotransposition when cotransfected with the Neo-tagged human L1 expression plasmid in HeLa cells (Supplementary Figure S4C). These findings, combined with the lack of the suppressive effect of the truncated C-C domain protein on L1 retrotransposition, support that the N-terminus as well as the C-C domain of the human ORF1p are needed to reduce L1 retrotransposition in trans.
Mutations within the C-C domain of the truncated human ORF1p abolish its ability to suppress human L1 retrotransposition
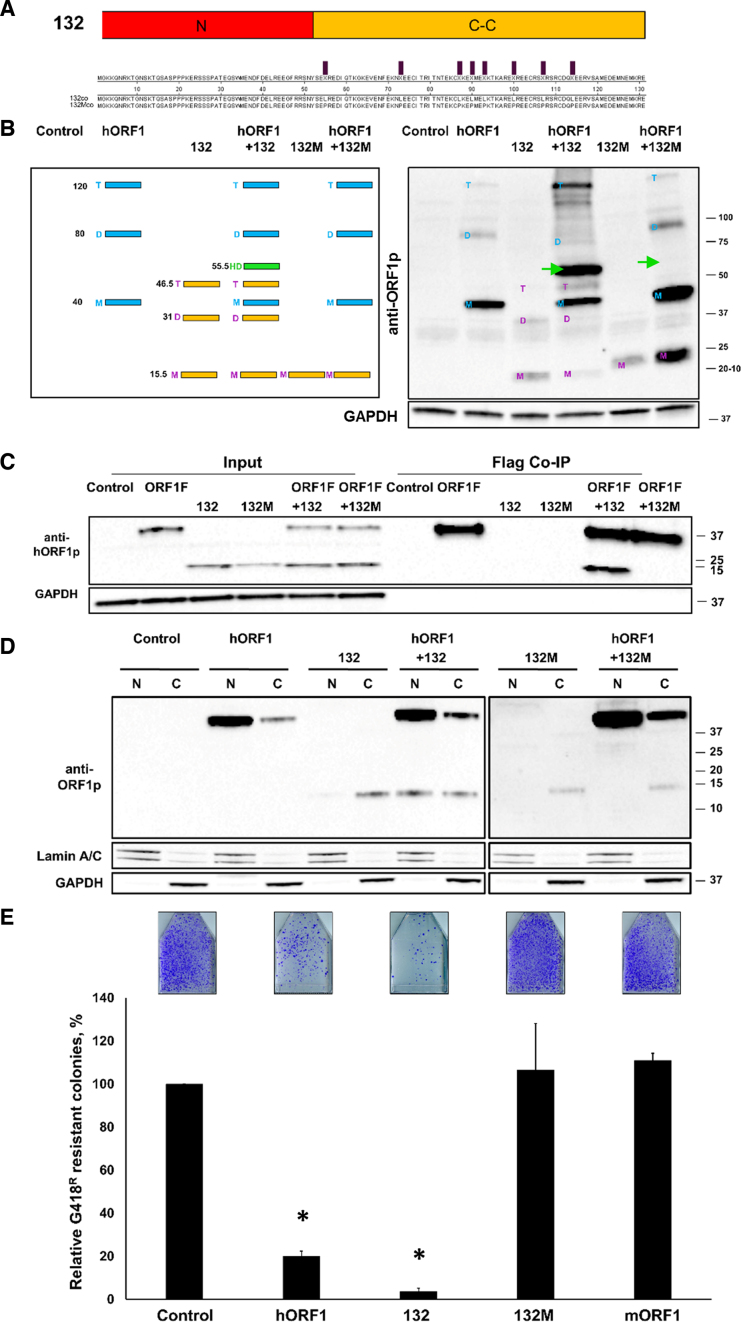
The confirmation that the N-terminal half of the human L1 ORF1p protein is sufficient to suppress L1 retrotransposition in trans prompted genetic manipulations to identify the requirements for this effect. To test the hypothesis that efficient heterodimerization through the C-C domain is required for the ORF1p suppressive effect on L1 retrotransposition, 8 leucine (L) amino acids present in the C-C domain of the human ORF1p were mutated into proline (P) amino acids to generate the 132 mutant construct (132M) (Figure 4A). This approach was chosen based on the previously published work demonstrating that the L to P mutations within C-C domains or leucine zipper motifs of other proteins abolished their interactions (52–54). Western blot analysis using anti-ORF1p polyclonal antibodies confirmed that the 132M protein was expressed in HeLa cells (Figure 4B, 132M lane in the right panel). Western blot analysis of cellular lysates harvested using non-reducing conditions demonstrated that the 132M protein was unable to form heterodimers with the hORF1p when both proteins were coexpressed in HeLa cells (Figure 4B, hORF1/132M lane in the right panel). The heterodimeric complexes formed by the wt 132 and full-length ORF1 proteins was readily detected using the same experimental conditions (Figure 4B, green arrows). To further confirm the loss of interaction between the 132M and full-length ORF1 proteins, we utilized a coimmunoprecipitation (co-IP) approach. We generated a C-terminally fused Flag-tagged ORF1 construct and confirmed that the protein generated from this construct heterodimerizes with the protein generated from a construct containing GAL4-fused ORF1 sequence in HeLa cells using non-reducing western blot approach (Supplementary Figure S5). HeLa cells were cotransfected with this Flag-tagged ORF1p expression plasmid and either the wt or mutant 132 expression plasmids. The resulting protein lysates were subjected to a pull down using anti-Flag antibody. The input and pull-down protein fractions were analyzed by western blot analysis using ORF1-specific polyclonal antibodies (31). This approach demonstrated that that the wt 132 protein co-IPed with the full-length human ORF1p, while the 132M protein did not (Figure 4C).
Figure 4.
Mutations in the C-C domain of truncated ORF1 protein abolishes its suppressive effect on L1 retrotransposition. (A) A schematic of the 132 construct (N-terminus and C-C) and alignment of functional (132) and mutant (132M) protein sequences. The positions of eight leucine residues mutated to proline residues are indicated by purple bars. (B) (Left) A schematic of the expected band pattern resulting from heterodimerization of the full-length ORF1p and truncated 132 ORF1p (green band). Numbers indicate molecular weights (kDa) for monomers (M), dimers (D) and trimers (T) and heterodimers (green band). (Right) Western blot analysis using anti-ORF1p polyclonal antibodies (anti-ORF1p) of proteins generated from codon-optimized 132, hORF1 and 132M constructs transiently transfected in HeLa cells (non-reducing conditions). Green arrows indicate the expected position of the heterodimer (55.5 kDa). Numbers on the right of the image are molecular weight markers. GAPDH is used as a loading control. (C) Co-immunoprecipitation of the Flag-tagged full-length ORF1p (ORF1F) with the wt (132) or mutant (132M) truncated ORF1p. Western blot analysis was performed using anti-ORF1p polyclonal antibodies (anti-ORF1p). Control lane indicates cells transfected with an empty plasmid. GAPDH is used as a loading control. Positions of molecular markers are indicated on the right in kDa. (D) Western blot analysis (reducing conditions, human-specific ORF1p polyclonal antibodies) of nuclear (N) and cytoplasmic (C) fractions collected from HeLa cells expressing hORF1, 132co or 132Mco proteins. GAPDH and Lamin A/C were used as loading controls. (E) Result of L1 retrotransposition assay using a Neo-tagged, full-length human wt L1 expression plasmid cotransfected with an empty (control), hORF1, 132, 132M or mORF1 expression plasmids. The number of G418 resistant colonies was normalized to the Control flask for each independent experiment. Asterisk (*) denotes statistical significance between listed constructs and the control (n = 3, t-test, P < 0.05).
To further characterize the 132p/ORF1p interactions we determined their subcellular localization. Western blot analysis using anti-ORF1p polyclonal antibodies demonstrated that the full-length ORF1p was observed in the nuclear fraction of HeLa cells transfected with the human ORF1 expression plasmid, consistent with our previous observations (31). The truncated, functional and mutant 132 proteins were detected predominantly in the cytoplasmic fraction of HeLa cells transfected with their respective expression plasmids (Figure 4D, lanes 132 and 132M). Cotransfection of the wilf-type 132 and full-length ORF1 plasmids in HeLa cells resulted in the detection of the wt 132 protein in the nuclear fraction (Figure 4D, compare lanes 132 and ORF1+132). In contrast, the 132M protein remained cytoplasmic in the presence of the full-length ORF1p (Figure 4D, compare lanes ORF1+132 and ORF1+132M). Consistent with its inability to heterodimerize with the full-length ORF1p as detected by western blot analysis, co-IP and subcellular localization, the 132M protein had no effect on L1 mobilization when transiently coexpressed with the L1Neo in HeLa cells (Figure 4E).
Expression levels and combination of truncated ORF1 proteins influence their trans effect on human L1 retrotransposition
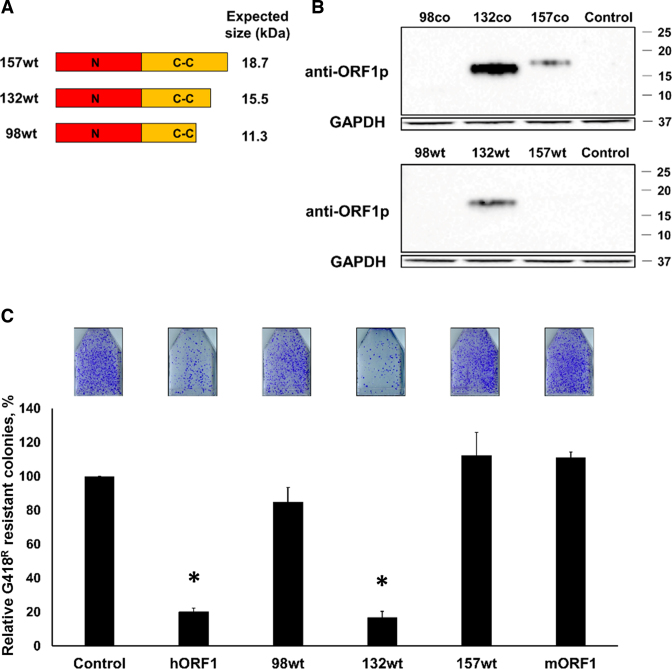
To determine whether expression levels alter the efficiency with which the truncated ORF1 proteins suppress L1 retrotransposition in trans, we generated C-terminally truncated 157, 132 and 98 ORF1p expression constructs shown in Figure 3A utilizing human non-codon-optimized wt ORF1 sequence (Figure 5A). Western blot analysis with anti-ORF1p polyclonal antibodies detected reduced steady-state levels of all proteins produced by the wt constructs compared to the corresponding constructs containing codon optimized L1 ORF1 sequences (Figure 5B and Supplementary Figure S6). Very low 98 and 157 wt protein expression resulted in the corresponding loss of their suppressive trans effect on L1 retrotransposition using 1:1 ratios of the transfected plasmids (Figure 5C). Despite its reduced expression, the 132 wt construct still significantly suppressed L1 retrotransposition (Figure 5C, 132 wt).
Figure 5.
Expression and effect of truncated ORF1 proteins generated from plasmids containing wt L1 sequence on L1 retrotransposition. (A) A schematic of the C-terminally truncated ORF1 expression constructs containing wt L1 sequence (98wt, 132wt and 157wt). (B) Western blot analysis using ORF1-specific antibodies of truncated ORF1 proteins generated from codon-optimized (co) or wt 98, 132, 157 constructs transiently transfected in HeLa cells. Control lane indicates cells transfected with an empty plasmid. GAPDH is used as a loading control. Positions of molecular weight markers are indicated on the right in kDa. (C) Result of L1 retrotransposition assay using a Neo-tagged, full-length, human wt L1 expression plasmid cotransfected with an empty (control), hORF1, 98wt, 132wt, 157wt or mORF1 constructs. The number of G418 resistant colonies was normalized to the control flask for each independent experiment. Asterisk (*) denotes statistical significance between listed constructs and the control (n = 3, t-test, P < 0.05).
It has been previously reported that both functional and retrotranspositionally-incompetent L1 loci are coexpressed (35,40). These published results suggest that the relative ratio of the functional and non-functional L1 loci as well as the make-up of truncated ORF1p expressed from non-functional L1 loci may vary among cells. To test the effect of coexpression of different amounts and forms of truncated ORF1 proteins on L1 retrotransposition, 0.2 or 0.4 μg of the 98 and 157 wt plasmids were cotransfected individually or together with the Neo-tagged human L1 in HeLa cells (Supplementary Figure S7A). The coexpression of 98 and 157 wt truncated ORF1 proteins with the Neo-tagged human L1 suppressed L1 retrotransposition when compared to the individual effects of each truncated ORF1 protein under the same transfection conditions (98 or 157 wt) (Supplementary Figure S7B). The same result was observed when truncated ORF1 proteins produced from the plasmids containing codon-optimized sequences were coexpressed (98co and 157co). As with the wt proteins, the coexpression of 98co and 157co proteins resulted in a greater suppression of L1 retrotransposition than their individual ability to limit L1 mobilization under the same transfection conditions (Supplementary Figure S7B). These results support that a specific combination of truncated ORF1 proteins expressed from multiple L1 loci as well as the levels of their expression may differentially influence L1 retrotransposition.
Bioinformatic analysis of genomic L1 loci identifies species-specific distribution of stop codon positions
It has been previously reported that retrotranspositionally incompetent L1 loci are expressed (35,40). However, it has not been determined how many full-length L1 loci containing stop-codons in their ORF1 sequence are present in the human genome. We analyzed the human genome to identify full-length L1 loci harboring stop codons within their ORF1 sequence that would have the potential to generate truncated ORF1 protein. Using L1Base (51), we identified 1244 human L1 loci containing ORF1 with one or more stop codons. These 1244 L1 loci were further analyzed to identify the position of their first stop codon in the ORF1 sequence. This analysis determined that the majority of identified human L1 loci (59%) contained their first stop codon at amino acid positions 49, 98, 110 or 130 (Figure 6A). The distribution of codons that can give rise to stop codons within the human ORF1 sequence via a single point mutation is shown in the Supplementary Figure S8. Out of the 1244 total L1 loci containing at least one stop codon in their ORF1 sequence 375 L1 loci harbored a stop codon within the sequence corresponding to the 99-132 amino acid region of the full-length ORF1p. Based on our experimental data, ORF1 proteins truncated within this region could be the most efficient in suppressing L1 retrotransposition in HeLa cells (Figures 3D and 5C). Bioinformatic analysis of these 375 loci determined that over 75% of them belonged to the three youngest L1 subfamilies (L1PA1-3). Specifically, 8.5% of loci belonged to the L1 HS subfamily, 35.7% belonged to the L1PA2 subfamily and 32.8% belonged to the L1PA3 subfamily (Figure 6A). Using the UCSC browser, we also identified that in four out of randomly chosen 50 full-length L1 loci, the presence of the stop codon in the ORF1 sequence was polymorphic (Supplementary Figure S9). The estimated allele frequencies of the L1 variants containing the stop codon ranged from 0.1 to 0.58 (Supplementary Figure S9).
The above described approach was also utilized to identify mouse L1 loci fitting the same criteria. We chose the first 10 L1 loci entries identified on each mouse chromosome, which resulted in the total of 198 loci. These L1 loci were used for further analysis of the position of the first stop codon in their ORF1 sequence. Among mouse L1 loci, the most common position for the first stop codon was at amino acid 251 (Supplementary Figure S10). Collectively, our bioinformatic analysis of the mouse and human L1 loci demonstrates that many full-length human and mouse L1 loci present in their respective genomes contain premature stop codons at specific dominant positions within their ORF1 sequence and that these prevalent positions differ between the mouse and human elements. All of these full-length L1 loci are retrotranspositionally incompetent, yet they have the potential to be transcribed and translated to generate truncated ORF1 proteins with a possible suppressive effect on retrotransposition of functional L1 loci.
In the above described experiments (Figures 3–5 and Supplementary Figure S7), we determined that various truncated human ORF1 proteins suppress human L1 retrotransposition. However, whether truncated mouse ORF1 proteins behave in a similar manner is not known. Based on the L1Base analysis of selected mouse L1 loci shown in the Supplementary Figure S10, we generated a construct designed to express a 250 amino acid long mouse ORF1 protein tagged with a T7 sequence (m251) (Supplementary Figure S11A). Western blot analysis using polyclonal anti-mORF1p (31) or anti-T7 antibodies detected truncated m251 protein expression in HeLa cells (Supplementary Figure S11B). To test its ability to trans-interact with the full-length mORF1p, the m251 expression plasmid was cotransfected with the full-length mORF1 plasmid in HeLa cells. Western blot analysis using non-reducing conditions and either anti-mORF1 (31) or anti-T7 tag polyclonal antibodies detected a unique band that was consistent with the expected molecular weight of a m251/ORF1 protein heterodimer (Supplementary Figure S11C, green arrows). Transient cotransfection of HeLa cells with the neomycin-tagged mouse L1 and the m251 expression constructs demonstrated that when expressed in trans, the m251 protein significantly suppressed mouse L1 retrotransposition (Supplementary Figure S11D, m251). As expected, coexpression of the m251 protein with the Neo-tagged human L1 did not affect human L1 retrotransposition (Supplementary Figure S12). Combined these data demonstrate that, when coexpressed in trans, specific truncated mouse and human ORF1 proteins have a similar suppressive ‘dominant-negative’ effect on the retrotransposition of their respective full-length L1 elements.
L1 loci containing one or more stop codons within the ORF1 sequence are expressed in human cell lines
In order to determine whether any of the L1 loci containing stop codons in their ORF1 sequence identified in the human genome are expressed in human cells, we utilized an RNAseq approach that identifies individual endogenous L1 loci expressed in HeLa and HEK 293 cells (41). We determined the subfamily of the top 50 endogenous L1 loci identified to be expressed in HEK 293 or HeLa cells (41), and analyzed them for the presence of stop codons in their ORF1 sequence. This analysis determined that the majority (49/50) of the expressed L1 loci identified in HEK 293 cells belonged to the three youngest L1 subfamilies (the L1 HS 16/50, L1PA2 23/50 and L1PA3 10/50) (Supplementary Figure S13A). The endogenous L1 loci expressed in HeLa cells belonged to the PA1-PA6 subfamilies (L1HS 1/50, L1PA2 4/50, L1PA3 9/50, L1PA4 16/50, L1PA5 15/50 and L1PA6 5/50) (Supplementary Figure S13B). We determined that the majority of endogenously expressed L1 loci contain stop codons in the 99 through 132 aa region of the ORF1p belonged to the L1PA2 or L1PA3 subfamilies (Supplementary Tables S1 and S2). In agreement with RNAseq results identifying endogenous L1 mRNA expression, western blot analysis using anti-ORF1p polyclonal antibodies detected a band consistent with the expected size of the full-length ORF1 protein in HEK 293 cells (Supplementary Figure S14). No bands consistent with truncated ORF1 proteins were detected. Analysis of protein sequence corresponding to the top 50 L1 loci identified by RNAseq (41) determined that 66% of these loci code for proteins that have one or more subsititutions within the epitope recognized by our Abs. Most of the L1 loci containing no stop codons within the ORF1 sequence had a perfect match with the epitope. These findings demonstrate that retrotranspositionally competent L1 loci and L1 loci containing stop codons in their ORF1 sequence are coexpressed in mammalian cells.
ORF1p generated from L1PA2 and L1PA3 suppress L1 retrotransposition in trans
The RNAseq analysis determined that many endogenously expressed L1 loci harboring stop codons in their ORF1 sequence belong to the L1PA2 and L1PA3 subfamilies (Supplementary Table S1). To determine whether truncated ORF1 proteins generated by the L1PA2 and L1PA3 subfamilies can suppress L1 retrotransposition as effectively as the modern L1 ORF1, we generated plasmids containing commercially synthesized full-length or truncated ORF1 consensus sequences representing the L1PA2 and L1PA3 elements (46). We also generated three additional plasmids containing ORF1 sequences corresponding to specific endogenous L1PA2 loci identified in our analysis of the RNAseq expression data from HEK 293 cells (41). These plasmids referred to as endogenous (e) e207, e259, and e127 constructs are expected to produce truncated ORF1 proteins of 206, 258 or 126 amino acid long, respectively (Supplementary Table S1). Western blot analysis using anti-ORF1p polyclonal antibodies determined that all plasmids expressed detectable levels of ORF1 proteins of the expected molecular weights when transiently transfected in HeLa cells (Figure 6B). Western blot analysis of cellular lysates harvested using non-reducing conditions demonstrated that the full-length L1PA2 and L1PA3 ORF1p readily formed heterodimers with the GAL4-fused L1PA1 ORF1p when coexpressed in HeLa cells (Supplementary Figure S15A, green arrows). The truncated L1PA1, L1PA2, L1PA3 ORF1p also formed heterodimers with the full-length L1PA1 ORF1p when coexpressed in HeLa cells (Supplementary Figure S15B, green arrows). Consistent with these findings, both the full-length and truncated (130 aa) PA1, 2 and 3 proteins efficiently suppressed modern L1 mobilization when transiently coexpressed with the L1Neo in HeLa cells (Figure 6C).
The e259, e207, and e127 proteins produced by their respective plasmids were expressed at lower levels than the L1PA1-3 130 proteins (Figure 6B). They formed heterodimers with the full-length modern ORF1p when coexpressed in HeLa cells (Supplementary Figure S15C, green arrows) and suppressed L1 mobilization with various efficiencies (Figure 6C). The very efficient suppression of the L1.3 retrotransposition by the e127 protein compared to the statistically significant, but inefficient, suppression of L1 retrotransposition by the e207 and e259 proteins is consistent with our data shown in Figures 3B, C, 5B and C. These results show that truncated ORF1 proteins with lower expression levels and longer truncated ORF1 proteins are less efficient at suppressing L1 retrotransposition than the ORF1p truncated around the amino acid position 130. These data demonstrate that the full-length and truncated ORF1 proteins representing L1PA2 and L1PA3 subfamilies are able to efficiently interact with the modern L1 ORF1p and suppress L1 retrotransposition in trans.
Full-length wt human L1 elements containing stop codons in their ORF1 sequence suppress human L1 retrotransposition in trans
Based on the bioinformatic analysis of human L1 loci, we generated human L1.3 expression plasmids containing mutations that introduce stop codons at positions corresponding to amino acid 110 (110Stop), 130 (130Stop) or 132 (132Stop) of the human ORF1p (Figure 7A). A previously reported L1.3 plasmid containing a stop codon at position 119 of the ORF1p (119Stop) (5) was also included in the study (Figure 7A). Western blot analysis with anti-ORF1p polyclonal antibodies demonstrated that all L1 stop constructs expressed detectable levels of truncated ORF1p with expected molecular weights (Figure 7B). The 110Stop L1 produced the lowest steady-state levels of truncate ORF1 protein (Figure 7B). We next tested the effect of these L1 elements on L1 retrotransposition using transient cotransfections of HeLa cells. Consistent with the expression result, the 110 L1Stop construct had no significant impact on L1 retrotransposition (Figure 7C). Consistent with the previously published data (5) the 119Stop construct reduced L1 retrotransposition to about 80% of the control. The 130Stop and 132Stop containing L1 elements had the most suppressive effect on retrotransposition of the cotransfected human L1Neo in HeLa cells (Figure 7C). L1 was previously shown to have an antisense promoter (55,56) and to express ORF0 protein (56). A plasmid supporting luciferase expression driven by the L1 5΄UTR was used to control for a potential effect of the antisense mRNA generated by the 5΄ UTR (57,58). Cotransfection of the L1Neo plasmid with this L1 5΄UTR construct demonstrated that the trans L1 5΄ UTR had no negative effect on L1 mobilization (Figure 7C).
wt L1 retrotransposition is suppressed in HeLa cells constitutively expressing full-length wt L1 elements containing premature stop codons in their ORF1 sequence
To assess the effect of truncated ORF1 proteins on L1 retrotransposition in a more biologically relevant manner, we generated HeLa cell lines each harboring either untagged functional human L1 (hL1wt) or one of the above-described L1 Stop elements (119Stop, 130Stop or 132 Stop) (Figure 8A). All constitutively expressed truncated ORF1 proteins were detected at low levels in these engineered HeLa cells using western blot analysis with anti-hORF1p antibodies (Figure 8B). To assess L1 retrotransposition, human L1Neo plasmid was transiently transfected into each HeLa cell line (harboring a control plasmid, the L1 Stop or wt L1 elements) (Figure 8C). Equal transfection efficiency for each cell line was confirmed using pIRES, a neomycin expressing plasmid, that was transiently transfected into each engineered HeLa cell line under the same experimental conditions (Figure 8C, transfection efficiency control row). All engineered HeLa cell lines harboring the L1 Stop constructs supported less L1 retrotransposition relative to the control HeLa line (Figure 8C). This effect depended on the expression of truncated ORF1 proteins because the suppressive effect was reversed when the expression of truncated ORF1 proteins was lost in these engineered cell lines (Supplementary Figure S16A and B).
DISCUSSION
In human genomes, retrotranspositionally incompetent full-length L1 loci outnumber functional L1 loci by about 50-fold (33,39,59). Our bioinformatic analysis of the human genome identified 1244 full-length human L1 loci containing a stop codon in their ORF1 sequence (Figure 6A and Supplementary Figure S10). Many of these L1 loci (59%) contain stop codons corresponding to amino acid positions 49, 98, 110 or 130 of the ORF1p. In contrast, the majority of mouse L1s contain a stop codon corresponding to the amino acid position 251 of the mouse ORF1p. This bias is most likely due to the presence of CpGs which results in C to T mutations caused by spontaneous deamination. Regardless of the mechanism underlying the origin of these stop codons, these results demonstrate that the human and mouse genomes contain L1 loci that, if expressed, have the potential to generate truncated ORF1 proteins.
Despite their inability to further propagation in the host genome, many inactive L1s may retain their ability to express because they contain a functional promoter present in the L1 5΄ UTR (Supplementary Figure S13 and Supplementary Tables S1 and 2). It has been reported that L1 mRNAs mapping to retrotranspositionally incompetent L1 loci were detected in human cells (35,40). However, the approaches utilized to identify these mRNAs are limited in their ability to discriminate between authentic L1 mRNA and L1 sequences included in cellular transcripts (60). Analysis of recently published data sets generated by strand-specific paired-end RNAseq and 5΄ RACE/Pacbio approaches that unambiguously identify individual expressed human L1 loci determined that multiple L1 loci containing stop codons at different positions of their ORF1 sequence are expressed in human cells (41). These L1 loci are not only expressed in HEK 293 and HeLa cells, but their mRNA is likely translated because it is detected in the polyribosomal fraction of analyzed cells (8,32,41). Western blot analysis of the polyribosomal fraction extracted from HEK 293 cells demonstrated that it contained a band consistent with the size of the full-length ORF1 protein (Supplementary Figure S14). This band was not detected in HeLa cells most likely because of the difference in the levels of endogenous L1Hs expression detected between the two cell lines with HEK 293 cells supporting higher expression of L1Hs L1s (41). The same approach did not detect any specific bands consistent with the presence of truncated ORF1p proteins in either HEK 293 or HeLa cells. Manual analysis of the protein sequence encoded by the expressed L1 loci containing stop codons in their ORF1 sequence demonstrated that the majority of them contain one or more amino acid substitutions in the epitope sequence (9-22 aa of L1PA1 ORF1) recognized by our anti-ORF1p antibodies. Additionally, because expressed L1 loci harbor stop codons at different positions within their ORF1 sequences, truncated proteins generated from different L1 mRNAs would not produce a cumulative signal like the one generated by the full-length ORF1 proteins translated from different L1 transcripts. Finally, some truncated ORF1 proteins exhibit lower steady state levels than others and the full-length ORF1p (Figures 3B, 5B, 6B and 7B). Combined, these data demonstrate that mRNA expression of functional L1 loci and those L1 loci containing stop codons in their ORF1 sequence occurs in mammalian cells. Further, the band detected with the anti-ORF1p antibodies in HEK 293 is consistent with endogenous L1 ORF1p expression, which varies between human cell lines analyzed here. This observation is consistent with the previous finding that various levels of ORF1p expression have been detected in cancerous and normal human tissues using immunohistochemistry (23,24,61–64) and human cell lines using western blot analysis (65).
The finding that non-functional L1 loci (including those containing stop codons in the ORF1 sequence) are expressed in mammalian cells along with functional L1 loci raises questions regarding the impact of this coexpression on L1 retrotransposition. It has been previously reported that retrotransposition of non-functional L1 loci is not efficiently rescued by functional L1 loci due to cis preference (8,9). However, both L1 proteins can function in trans of their parental mRNA as demonstrated by retrotransposition of SINEs and SVA elements that utilize L1 proteins (10–12). Although, ORF1p molecules produced from different mRNAs can assemble heterotrimers (31), it is not known whether non-functional L1 loci, specifically those that may produce truncated ORF1 proteins, have an impact on retrotransposition of active L1 elements. The conventional thinking assumes that L1 mRNA and/or ORF1 protein levels serve as a direct indicator of L1 retrotransposition (23–26). Consistent with this line of thinking, several examples of an increase in L1 mRNA and protein expression and corresponding upregulation in L1 retrotransposition upon elimination of suppressors of L1 expression have been reported (22,42,66–77). Further supporting a correlation between ORF1p expression and L1 retrotransposition, codon optimization of the mouse L1 dramatically (100-fold) increased its mRNA, protein expression and retrotransposition (27). Deviating from this straight forward relationship is the observation that codon optimization of the human L1s significantly increased its mRNA and protein expression, but this increase resulted in a modest 3–5-fold increase in the human L1 mobilization (27–30). Additionally, recently published data suggest that the ratio of ORF1/ORF2 proteins may influence L1 retrotransposition (22). Therefore, the expression levels of ORF1p may not always directly correlate with L1 retrotransposition. A relationship between the levels of endogenous L1 (mRNA or ORF1p) expression and L1 retrotransposition in vivo is further complicated by the fact that typically the total levels of these molecules are produced by multiple expressed L1 loci (41,65). Some of these L1 loci are functional and some are defective. Therefore, a specific spectrum of expressed functional and non-functional L1 loci contributing to the total mRNA and protein signal may be important for understanding the resulting impact on the host genome from active L1 loci. Our data suggest the possibility that the levels of the ORF1p detected in any given tissue may not always directly correspond to the amount of L1 retrotransposition, and that an inverse relationship between L1 retrotransposition and the relative amount of the total ORF1p may exist. This scenario is based on the observations that both the full-length and C-terminally truncated ORF1 proteins suppress retrotransposition of active human L1 in transient assays (Figures 3C, 5C, 6C, 7C and Supplementary Figure S7), as well as in engineered cells harboring the full-length wt L1 or L1s containing stop codons in their ORF1 sequence (Figure 8C). These data support that expression of non-functional L1 loci containing stop codons in their ORF1 sequence may be beneficial to the host genome as it may reduce accumulation of de novo L1 inserts.
While both the full-length and the truncated ORF1 proteins suppress human L1 retrotransposition when supplied in trans, the underlying mechanisms of their effects are likely different. One piece of experimental evidence that supports this likelihood is that the full-length human and mouse ORF1 proteins have contrasting effects on retrotransposition of their respective elements. While the full-length human ORF1p suppressed mobilization of the human L1 and chimeric L1s containing human ORF1 sequence, mouse full-length ORF1p increased retrotransposition of the codon-optimized mouse L1 and the chimeric L1s containing mouse ORF2p in NIH 3T3 cells (Figure 2). This differential effect of the mouse and human ORF1 proteins on L1 mobilization could be a factor contributing to the differences in retrotransposition rates of mouse and human L1 elements containing codon-optimized sequences (28–30). The suppressive trans effect of the human ORF1p on human L1 retrotransposition is also opposite from its stimulatory impact on Alu mobilization (12). Although understanding the underlying mechanism and species-specific differences of the full-length ORF1 effect on L1 retrotransposition is beyond the scope of this manuscript, our results are consistent with the recent observation that ORF1p/ORF2p ratio may influence human L1 integration (22). In conjunction with previously published results, our data regarding the trans effect of the full-length ORF1p on L1 retrotransposition are consistent with the proposed hypothesis that the ORF1p content within L1 RNPs may impact retrotransposition (22).
In contrast to the opposing effects of the full-length ORF1 proteins on human and mouse L1 retrotransposition, truncated ORF1 proteins from both species suppress mobilization of their respective full-length L1 elements (Figures 3C, 5C, 6C, 7C, Supplementary Figures S7B and S11D). Our results demonstrate that the minimal unit of the human ORF1p that is able to suppress L1 retrotransposition in trans consists of the N-terminal and C-C domains. The effect of truncated ORF1 proteins is consistent with a dominant negative mechanism of their action, as it depends on the ability of these proteins to form heterodimers with the full-length ORF1p (Figure 3D and Supplementary Figure S15). Genetically engineered disruption of the C-C domain function abolished the ability of truncated ORF1p to form heterodimeric complexes with the full-length ORF1p. These changes also eliminated the suppressive effect of the 132 protein on L1 retrotransposition (Figure 4B and E). The fact that longer truncated human ORF1 proteins are less efficient at suppressing L1 retrotransposition than those containing only the N-terminus and C-C domain is also consistent with the dominant negative mechanism of their interference. Although it is not known whether the truncated ORF1 proteins could be incorporated into L1 RNPs and interfere with L1 integration, our data demonstrate that the full-length ORF1p can ferry truncated wt 132 protein into the nucleus (Figure 4D, hORF1+132). In contrast, the C-C domain mutant 132 protein remains cytoplasmic in the presence of the full-length ORF1p (Figure 4D, hORF1+132M). This result also demonstrates that the full-length and truncated ORF1 proteins have different subcellular localization. This may explain some observed variability in localization of the ORF1p signal detected by immunohistochemistry in human samples (23,24,61,78), if the ORF1p-specific antibodies can recognize both the full-length and truncated ORF1 proteins generated by expressed L1 loci. This result also supports that the RRM/C-terminal portion of the protein is important for its nuclear localization and that having two (or potentially one) full-length ORF1 molecules within the trimer may be sufficient for gaining access to the nucleus.
The experimental evidence presented here supports that the efficiency of the dominant negative effect of the truncated L1 ORF1p proteins on retrotransposition is influenced by the levels of their expression and the extent of their truncation (Figures 3B, D, 5B, C, 6B, C, 7B, C and Supplementary Figure S7B). These findings have important biological implications, if the rules we have identified here apply in vivo. Analysis of endogenously expressed L1 loci harboring stop codons in their ORF1 sequence showed that their spectrum varies among different human cell lines (HeLa versus HEK 293) (Supplementary Tables S1 and 2). The expression levels, positions of stop codons and the composition of specific expressed L1 loci are therefore expected to influence the resulting amount of DNA damage generated from endogenous active L1s. For example, based on the results obtained using transient transfections of L1 loci with stop codons roughly corresponding to amino acid positions 110–130 of the human ORF1p may be optimal for guarding against accumulation of de novo L1 events. However, generation of engineered HeLa cells stably expressing different L1Stop elements supports that, even at low levels, truncated ORF1 proteins are able to suppress L1 retrotransposition when constitutively expressed in human cells (Figure 8). This outcome could be pointing to some biologically relevant differences related to the ORF1p life cycle during transient versus constitutive expression. It is worth noting that our findings demonstrate that the negative effect of L1 elements containing stop codons in their ORF1 sequence on L1 retrotransposition is reversible because the loss of constitutive expression of the full-length or truncated ORF1 proteins lead to the loss of suppression of L1 retrotransposition (Supplementary Figure S16A and B). This finding suggests that the protective effect from expression of L1s with ORF1 stop codons could be gained or lost following epigenetic changes due to aging, environmental exposures or cellular differentiation or transformation. Additionally, these results combined with the recently reported cell-type specific expression of L1 loci (41) suggest that the protective effect of L1 stop loci may be tissue specific. Furthermore, the impact of L1Stop loci on L1 retotransposition may be influenced by the fact that the presence of some of these stop codons within fixed L1 loci is polymorphic in the human population (Supplementary Figure S9).
Numerous cellular factors and pathways have been identified to suppress L1 expression and integration, establishing the existence of significant redundancy in mechanisms downregulating L1 retrotransposition (22,42,66–77). The suppressive trans effect of hORF1p on human L1 retrotransposition is another potential mechanism that may influence L1-induced damage in vivo. Our findings support that coexpression of retrotranspositionally incompetent L1s containing stop codons in their ORF1 sequence with functional L1 loci may slow down accumulation of de novo L1 integration events. Our data, along with other recent reports (22,30,79), suggest that the relationship between detected ORF1p levels may not always be proportional to the extent of L1 retrotransposition, and that an inverse correlation between ORF1p expression levels and L1 retrotransposition may exist under certain circumstances. The factors influencing L1 retrotransposition in any given cell may include the relative ratios of the functional full-length ORF1p and truncated proteins, as well as the length of truncated ORF1 proteins expressed in a given cell.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Consortium of Mobile Elements at Tulane (COMET) for critical discussions of the data and Dr Kristine J Kines for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Louisiana State Board of Regents Graduate Research Fellowship (to M.S.) (in part); Life Extension Foundation (to V.P.B.); National Institutes of Health [P20GM103518 to V.P.B.]; Kay Yow Cancer Fund (to V.P.B.)]. Funding for open access charge: Tulane Cancer fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cordaux R., Batzer M.A.. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kazazian H.H., Wong C., Youssoufian H., Scott A.F., Phillips D.G., Antonarakis S.E.. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988; 332:164–166. [DOI] [PubMed] [Google Scholar]
- 3. Woods-Samuels P., Wong C., Mathias S.L., Scott A.F., Kazazian H.H. Jr, Antonarakis S.E.. Characterization of a nondeleterious L1 insertion in an intron of the human factor VIII gene and further evidence of open reading frames in functional L1 elements. Genomics. 1989; 4:290–296. [DOI] [PubMed] [Google Scholar]
- 4. Alisch R.S., Garcia-Perez J.L., Muotri A.R., Gage F.H., Moran J.V.. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006; 20:210–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H. Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996; 87:917–927. [DOI] [PubMed] [Google Scholar]
- 6. Kolosha V.O., Martin S.L.. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:10155–10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belancio V.P., Hedges D.J., Deininger P.. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008; 18:343–358. [DOI] [PubMed] [Google Scholar]
- 8. Kulpa D.A., Moran J.V.. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 2005; 14:3237–3248. [DOI] [PubMed] [Google Scholar]
- 9. Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V.. Human L1 retrotransposition: cisPreference versus trans complementation. Mol. Cell. Biol. 2001; 21:1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dewannieux M., Esnault C., Heidmann T.. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003; 35:41–48. [DOI] [PubMed] [Google Scholar]
- 11. Ostertag E.M., Goodier J.L., Zhang Y., Kazazian H.H. Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003; 73:1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wallace N., Wagstaff B.J., Deininger P.L., Roy-Engel A.M.. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008; 419:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cook P.R., Jones C.E., Furano A.V.. Phosphorylation of ORF1p is required for L1 retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:4298–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin S.L. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010; 7:706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin S.L., Branciforte D., Keller D., Bain D.L.. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl. Acad. Sci. 2003; 100:13815–13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Callahan K.E., Hickman A.B., Jones C.E., Ghirlando R., Furano A.V.. Polymerization and nucleic acid-binding properties of human L1 ORF1 protein. Nucleic Acids Res. 2012; 40:813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khazina E., Truffault V., Büttner R., Schmidt S., Coles M., Weichenrieder O.. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 2011; 18:1006–1014. [DOI] [PubMed] [Google Scholar]
- 18. Basame S., Wai-lun Li P., Howard G., Branciforte D., Keller D., Martin S.L.. Spatial assembly and RNA Binding stoichiometry of a LINE-1 protein essential for retrotransposition. J. Mol. Biol. 2006; 357:351–357. [DOI] [PubMed] [Google Scholar]
- 19. Naufer M.N., Callahan K.E., Cook P.R., Perez-Gonzalez C.E., Williams M.C., Furano A.V.. L1 retrotransposition requires rapid ORF1p oligomerization, a novel coiled coil-dependent property conserved despite extensive remodeling. Nucleic Acids Res. 2016; 44:281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin S.L., Li J., Weisz J.A.. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-11. J. Mol. Biol. 2000; 304:11–20. [DOI] [PubMed] [Google Scholar]
- 21. Martin S.L., Bushman F.D.. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001; 21:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor M.S., LaCava J., Mita P., Molloy K.R., Huang C.R.L., Li D., Adney E.M., Jiang H., Burns K.H., Chait B.T. et al. . Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell. 2013; 155:1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris C.R., Normart R., Yang Q., Stevenson E., Haffty B.G., Ganesan S., Cordon-Cardo C., Levine A.J., Tang L.H.. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer. 2010; 1:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen L., Dahlstrom J., Chandra A., Board P., Rangasamy D.. Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res. Treat. 2012; 136:129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malki S., van der Heijden G.W., O’Donnell K.A., Martin S.L., Bortvin A.. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev. Cell. 2014; 29:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soper S.F.C., van der Heijden G.W., Hardiman T.C., Goodheart M., Martin S.L., de Boer P., Bortvin A.. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev. Cell. 2008; 15:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Han J.S., Boeke J.D.. A highly active synthetic mammalian retrotransposon. Nature. 2004; 429:314–318. [DOI] [PubMed] [Google Scholar]
- 28. Wagstaff B.J., Barnerβoi M., Roy-Engel A.M.. Evolutionary conservation of the functional modularity of primate and murine LINE-1 elements. PLoS One. 2011; 6:e19672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wallace N.A., Belancio V.P., Deininger P.L.. L1 mobile element expression causes multiple types of toxicity. Gene. 2008; 419:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wagstaff B.J., Kroutter E.N., Derbes R.S., Belancio V.P., Roy-Engel A.M.. Molecular reconstruction of extinct LINE-1 elements and their interaction with nonautonomous elements. Mol. Biol. Evol. 2013; 30:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sokolowski M., deHaro D., Christian C.M., Kines K.J., Belancio V.P.. Characterization of L1 ORF1p self-interaction and cellular localization using a mammalian two-hybrid system. PLoS One. 2013; 8:e82021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kulpa D.A., Moran J.V.. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006; 13:655–660. [DOI] [PubMed] [Google Scholar]
- 33. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al. . Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 34. Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V.. LINE-1 retrotransposition activity in human genomes. Cell. 2010; 141:1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kines K.J., Sokolowski M., deHaro D.L., Christian C.M., Belancio V.P.. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. 2014; 42:10488–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Konkel M.K., Wang J., Liang P., Batzer M.A.. Identification and characterization of novel polymorphic LINE-1 insertions through comparison of two human genome sequence assemblies. Gene. 2007; 390:28–38. [DOI] [PubMed] [Google Scholar]
- 37. Huang C.R.L., Schneider A.M., Lu Y., Niranjan T., Shen P., Robinson M.A., Steranka J.P., Valle D., Civin C.I., Wang T. et al. . Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010; 141:1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ewing A.D., Kazazian H.H.. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011; 21:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H.. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rangwala S., Zhang L., Kazazian H.. Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009; 10:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deininger P., Morales M.E., White T.B., Baddoo M., Hedges D.J., Servant G., Srivastav S., Smither M.E., Concha M., DeHaro D.L. et al. . A comprehensive approach to expression of L1 loci. Nucleic Acids Res. 2016; e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perepelitsa-Belancio V., Deininger P.. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 2003; 35:363–366. [DOI] [PubMed] [Google Scholar]
- 43. Sassaman D.M., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H.. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997; 16:37–43. [DOI] [PubMed] [Google Scholar]
- 44. Naas T.P., DeBerardinis R.J., Moran J.V., Ostertag E.M., Kingsmore S.F., Seldin M.F., Hayashizaki Y., Martin S.L., Kazazian H.H.. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998; 17:590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gasior S.L., Wakeman T.P., Xu B., Deininger P.L.. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006; 357:1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khan H., Smit A., Boissinot S.. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006; 16:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kines K.J., Sokolowski M., deHaro D.L., Christian C.M., Baddoo M., Smither M.E., Belancio V.P.. The endonuclease domain of the LINE-1 ORF2 protein can tolerate multiple mutations. Mob. DNA. 2016; 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sokolowski M., DeFreece C., Servant G., Kines K., deHaro D., Belancio V.. Development of a monoclonal antibody specific to the endonuclease domain of the human LINE-1 ORF2 protein. Mob. DNA. 2014; 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Christian C.M., deHaro D., Kines K.J., Sokolowski M., Belancio V.P.. Identification of L1 ORF2p sequence important to retrotransposition using Bipartile Alu retrotransposition (BAR). Nucleic Acids Res. 2016; 44:4818–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Christian C.M., Sokolowski M., deHaro D., Kines K.J., Belancio V.P.. Involvement of conserved amino acids in the C-terminal region of LINE-1 ORF2p in retrotransposition. Genetics. 2017; 205:1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Penzkofer T., Dandekar T., Zemojtel T.. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Res. 2005; 33:D498–D500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Surks H.K., Mochizuki N., Kasai Y., Georgescu S.P., Tang K.M., Ito M., Lincoln T.M., Mendelsohn M.E.. Regulation of myosin phosphatase by a specific interaction with cGMP-dependent protein kinase Iα. Science. 1999; 286:1583–1587. [DOI] [PubMed] [Google Scholar]
- 53. Chen S.S., Lee C.N., Lee W.R., McIntosh K., Lee T.H.. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J. Virol. 1993; 67:3615–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheng H.Y., Schiavone A.P., Smithgall T.E.. A point mutation in the N-terminal C-C releases c-Fes tyrosine kinase activity and survival signaling in myeloid leukemia cells. Mol. Cell. Biol. 2001; 21:6170–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 2001; 21:1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Denli A.M., Narvaiza I., Kerman B.E., Pena M., Benner C., Marchetto M.C.N., Diedrich J.K., Aslanian A., Ma J., Moresco J.J. et al. . Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015; 163:583–593. [DOI] [PubMed] [Google Scholar]
- 57. Belancio V.P. Importance of RNA analysis in interpretation of reporter gene expression data. Anal. Biochem. 2011; 417:159–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Belancio V.P., Roy-Engel A.M., Deininger P.. The impact of multiple splice sites in human L1 elements. Gene. 2008; 411:38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beck C.R., Garcia-Perez J.L., Badge R.M., Moran J.V.. LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 2011; 12:187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deininger P., Belancio V.P.. Garcia-Pérez LJ. Detection of LINE-1 RNAs by Northern Blot. Transposons and Retrotransposons: Methods and Protocols. 2016; NY: Springer; 223–236. [DOI] [PubMed] [Google Scholar]
- 61. Doucet-O’Hare T.T., Rodić N., Sharma R., Darbari I., Abril G., Choi J.A., Young Ahn J., Cheng Y., Anders R.A., Burns K.H. et al. . LINE-1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4894–E4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rodic N., Steranka J.P., Makohon-Moore A., Moyer A., Shen P., Sharma R., Kohutek Z.A., Huang C.R., Ahn D., Mita P. et al. . Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 2015; 21:1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rodić N., Sharma R., Sharma R., Zampella J., Dai L., Taylor M.S., Hruban R.H., Iacobuzio-Donahue C.A., Maitra A., Torbenson M.S. et al. . Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 2014; 184:1280–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Doucet-O’Hare T.T., Sharma R., Rodić N., Anders R.A., Burns K.H., Kazazian H.H.. Somatically acquired LINE-1 insertions in normal esophagus undergo clonal expansion in esophageal squamous cell carcinoma. Hum. Mutat. 2016; 37:942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Philippe C., Vargas-Landin D.B., Doucet A.J., van Essen D., Vera-Otarola J., Kuciak M., Corbin A., Nigumann P., Cristofari G.. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife. 2016; 5:e13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. deHaro D., Kines K.J., Sokolowski M., Dauchy R.T., Streva V.A., Hill S.M., Hanifin J.P., Brainard G.C., Blask D.E., Belancio V.P.. Regulation of L1 expression and retrotransposition by melatonin and its receptor: implications for cancer risk associated with light exposure at night. Nucleic Acids Res. 2014; 42:7694–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Belancio V.P., Hedges D.J., Deininger P.. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006; 34:1512–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goodier J.L., Cheung L.E., Kazazian H.H.. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 2013; 41:7401–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang A., Dong B., Doucet A.J., Moldovan J.B., Moran J.V., Silverman R.H.. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 2014; 42:3803–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Goodier J.L., Pereira G.C., Cheung L.E., Rose R.J., Kazazian H.H. Jr. The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet. 2015; 11:e1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moldovan J.B., Moran J.V.. The zinc-finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet. 2015; 11:e1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang N., Kazazian H.H.. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 2006; 13:763–771. [DOI] [PubMed] [Google Scholar]
- 73. Goodier J.L., Cheung L.E., Kazazian H.H. Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012; 8:e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Horn A.V., Klawitter S., Held U., Berger A., Jaguva Vasudevan A.A., Hofmann H., Hanschmann K.-M.O., Trösemeier J.-H., Flory E. et al. . Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res. 2014; 42:396–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McLaughlin R.N., Gable J.T., Wittkopp C.J., Emerman M., Malik H.S.. Conservation and innovation of APOBEC3A restriction functions during primate evolution. Mol. Biol. Evol. 2016; 33:1889–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Servant G., Streva V.A., Derbes R.S., Wijetunge M.I., Neeland M., White T.B., Belancio V.P., Roy-Engel A.M., Deininger P.L.. The nucleotide excision repair pathway limits L1 retrotransposition. Genetics. 2017; 205:139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Belancio V.P. LINE-1 activity as molecular basis for genomic instability associated with light exposure at night. Mob. Genet. Elements. 2015; 5:46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Goodier J.L., Zhang L., Vetter M.R., Kazazian H.H.. LINE-1 ORF1 protein localizes in stress granules with other RNA-Binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007; 27:6469–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. An W., Dai L., Niewiadomska A., Yetil A., O’Donnell K., Han J., Boeke J.. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob. DNA. 2011; 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.