Abstract
Water soluble matrix metalloproteinases (MMPs) have been regarded as diffusing freely in the extracellular matrix. Yet multiple MMPs are also observed at cell surfaces. Their membrane-proximal activities include sheddase activities, collagenolysis, bacterial killing, and intracellular trafficking reaching as far as the nucleus. The catalytic domains of MMP-7 and MMP-12 bind bilayers peripherally, each in two different orientations, by presenting positive charges and a few hydrophobic groups to the surface. Related peripheral membrane associations are predicted for other soluble MMPs. The peripheral membrane associations may support pericellular proteolysis and endocytosis. The isolated soluble domains of MT1-MMP can also associate with membranes. NMR assays suggest transient association of the hemopexin-like domains of MT1-MMP and MMP-12 with lipid bilayers. Peripheral association of soluble MMP domains with bilayers or heparin sulfate proteoglycans probably concentrates them near the membrane. This could increase the probability of forming complexes with membrane-associated proteins, such as those targeted for proteolysis.
Keywords: peripheral membrane protein, membrane recognition, membrane trafficking, sheddase, compartmentalization, protease
1. Introduction
The earliest clues that soluble MMPs might bind near cell surfaces came from their affinities for heparin, which is derived from the heparin sulfate chains radiating from proteoglycans (HSPGs) embedded in plasma membranes. Woessner and colleagues proposed that several MMPs bind HSPGs based on heparin extraction of MMP-2, 7, 9 and 13 from the uterus [1], affinities of MMP-1, 7 and 13 for heparin-Sepharose [2–4], heparin binding localized to the hemopexin-like (HPX) domains of MMP-1, 2 and 9 [5–7], and potential binding sites at basic residues of the MMP-1, 2, 7 and 13 catalytic domains [1]. Interactions of glycosaminoglycans (GAGs) and HSPGs with metalloproteinases such as MMP-1, 2, 7, 9, 13, and 16 were reviewed [8–10]. Effects of GAG interactions with selected MMP, ADAM, and ADAMTS enzymes in compartmentalization, activation, and linkage to substrates from cell surfaces continue to receive attention [9–19], but are topics outside the emphasis of this article.
MMP-9 is found expressed on the surface of activated polymorphonuclear neutrophils (PMNs) where it is highly active on protein substrates including elastin, collagen IV, gelatin, and α1-proteinase inhibitor [20]. Similarly, most MMP-8 secreted by activated PMNs is present on the plasma membrane, with about 40% of the bound protease in higher molecular weight forms and 60% in processed forms [21]. There the active fraction digests collagens I and II and α1-proteinase inhibitor [21]. PMN-bound MMP-8 and -9 are readily inhibited by small inhibitors but are less sensitive to TIMP protein inhibitors by up to two orders of magnitude [20, 21]. When bound to colon cancer cells, MMP-7 is also active and susceptible to inhibition by small inhibitors, but less so by TIMP-2 [22, 23]. These results imply that MMP-7, 8 and 9 bound to cells have open active sites, but with partial steric obstructions nearby that are absent from the enzymes free in solution. MMP-12 exhibits activity at the surface of activated macrophages, but not in the surrounding aqueous milieu [24]. In contrast, it is the latent zymogens of MMP-7, 8 and 12 that are found free in solution [21, 23, 24].
The plasma membrane, visited by these and other soluble MMPs, is characterized by a phospholipid to cholesterol ratio of 1:1, which promotes a mixture of liquid ordered and disordered phases of the bilayer [25]. The predominance of glycospingholipids (GSL), sphingomyelin (SM), and phosphatidylcholine (PC) on the external leaflet of the plasma membrane and on the luminal leaflet of the Golgi is maintained by lack of transport of these lipids to the cytosolic leaflet [25]. MMPs may in principle encounter the GSL, SM, and PC in the Golgi during secretion and on cell surfaces once secreted. Phosphatidylserine (PS) and phosphatidylethanolamine (PE), however, are transported to the cytosolic face of the plasma membrane and Golgi by P4 ATPases [25]. The plasma membrane is divided into compartments by the molecular skeleton of actin filaments (on the cytosolic face) to which some transmembrane proteins are anchored like picket fences rising out of the extracellular face [26]. MT1-MMP may contribute to such picket fences, as it is anchored to F-actin in invadopodia and podosomes [27, 28]. Within the compartments defined by actin and the attached pickets lie lipid rafts and domains of dynamic protein complexes [26]. This article considers some simplified, dynamic protein complexes.
The best understood plasma membrane regulation of the activation of an MMP regards the maturation of proMMP-2. At tumor cell surfaces, membrane type 1 (MT1) MMP activates proMMP-2 to active MMP-2 [29], via a ternary complex with TIMP-2 [30, 31]. Similar ternary complexes containing either TIMP-2 or TIMP-3 enable MT3-MMP to activate proMMP-2 on the surface of cultured cells [32]. Chondroitin sulfate proteoglycans also serve to connect MT3-MMP with proMMP-2 in order to activate the latter [14]. Perhaps analogously, the tetraspanin CD151 activates proMMP-7 on carcinoma cells [33]. Specific interactions of MMPs with many components of cell surfaces extend beyond HSPGs [11, 12, 14, 34–36], tetraspanins [33, 37, 38] and TIMP bridging to MT-MMPs [30, 32] to include integrins [39–43], low-density lipoprotein receptor-related proteins (LRPs) [44, 45], other proteins, and cholesterol sulfate (CS), as reviewed [46, 47]. MMP-7 associates with lipid rafts and their sulfated lipid components CS and the sulfoglycolipid SM3 when binding cancer cell lines and inducing the cells’ aggregation and aggressiveness [48]. Binding of CS was confirmed, but so was HSPG binding, in an independent study analyzing high affinity binding of MMP-7 (matrilysin) to cancer cells [23]. Anionic lipids are required for the high affinity of MMP-7 for vesicles [49, 50]. This article considers a few functional implications of peripheral interactions of MMPs with cell membrane bilayers, the interfaces of MMP catalytic and hemopexin-like (HPX) domains for lipid bilayers, and possibilities for lipid bilayer interactions with additional soluble MMP domains.
2. Influences of peripheral membrane interactions on MMP activities
2.1. Sulfated lipids and proteolysis by MMP-7
Numerous proteins are shed from the surface of colon cancer cells when MMP-7 binds the cells and CS is present in their plasma membranes [48]. MMP-7 bound to cancer cells via CS or the sulfoglycolipid SM4 (sulfatide) in their plasma membranes hydrolyzes markedly increased amounts of laminin-332 (laminin-5) and fibronectin because these substrates also bind the sulfated lipids CS and SM4 [51, 52]. This promotes detachment of the cancer cells from surfaces coated by laminin-332 or fibronectin, and ensuing homotypic aggregation of the cells. Since both the cell detachment and aggregation are relevant to metastasis, the authors proposed CS interactions with MMP-7 as a target for therapeutic development [51].
2.2. Pericellular collagenolysis
MMP-8 is also known as collagenase-2 and neutrophil collagenase. 92% of the type I collagenase activity of MMP-8 in mice is present at the plasma membranes of activated neutrophils [21]. The collagen-invasive activity of MT1-MMP requires tethering to the cell surface by a transmembrane anchor, which is a C-terminal transmembrane helix [53–55]. An alternative view has been that dimerization of MT1-MMP is critical to its pericellular collagenase activity [56]. The dependence of the pericellular collagenase activity of MT1-MMP on membrane anchoring and dimerization might be related, in that transmembrane helices routinely dimerize [57–59]. In the absence of the transmembrane helix, the collagen triple-helical peptidase activity of the ectodomain of MT1-MMP is enhanced by 50% by the addition of bilayered micelles called bicelles [60], i.e., bicelles with a lipid composition conferring the morphology of sheets with holes like “Swiss cheese” [61]. The study localized bicelle binding to the HPX domain [60].
MT1-MMP sheds CD44 and syndecan-1, which promotes cell migration [62, 63]. The transmembrane anchor of MT1-MMP could help in positioning the catalytic domain strategically to digest these key HSPGs. Analogous to the anionic membrane-associated MMP-7 digestion of laminin-332 / laminin-5 [51, 52], MT1-MMP also digests laminin-5 to stimulate cell motility [64–66].
2.3. Maturation of prothrombin by MMP-12 at membranes
The catalytic cleft of MMP-12 being open and active when bound to bilayers and cell membranes [24, 67] suggests the hypothesis of MMP-12 digesting membrane-associated physiological targets. At inflamed sites in vivo, i.e., murine peritonitis, MMP-12 inactivates antithrombin and activates prothrombin to thrombin, a key factor in blood coagulation, and thereby enhances the rate of coagulation [68]. Prothrombin and the coagulation cascade associate with membranes, where they are active. The associations require physiological [Ca2+] and are fostered by PS in the membranes [69–72]. To test if MMP-12 activation of prothrombin is aided by membranes, the emergence of the proteolytic activity of thrombin was monitored using a thrombin-specific colorimetric substrate. Small unilamellar vesicles (SUVs) of DMPC increase, in a concentration-dependent fashion, the amount of thrombin activity released by MMP-12-dependent maturation of human prothrombin (Fig. 1). Saturating additions of SUVs of 100 to 300 μM DMPC monomers increase the initial velocity of thrombin activity by 2.5-fold, at the enzyme concentrations and incubation times used. This suggests more efficient maturation of prothrombin by MMP-12 on the liposomes, as well as the pertinence of the association of these enzymes with plasma membranes.
Fig. 1.
Maturation of prothrombin by MMP-12 is increased by lipid bilayers, exemplifying the hypothesis of membranes drawing soluble MMPs together with protein substrates. The emergence of thrombin activity is evident as increased A405 upon digestion of thrombin-specific substrate S-2238 (100 μM). Human prothrombin (400 nM) was inactive until proteolytically activated by 16 h of incubation with human MMP-12 catalytic domain (5 nM) at 37 °C in 20 mM Tris·HCl (pH 7.3), 100 mM NaCl, 5 mM CaCl2, and 100 μM ZnCl2. The increases of proteolytic activation by MMP-12 by increasing additions of DMPC liposomes (SUVs) are plotted in blue, pink, green, and navy blue.
2.4. How might membranes aid proteolysis by MMPs?
The increases of MMP-7 digestion of matrix proteins co-localized with it at sulfolipids [51, 52], as well as liposome-enhanced MMP-12 maturation of prothrombin (Fig. 1), appear to be cases of membrane bilayers bringing peripherally associated MMPs together with substrate proteins for more productive collisions. Localization on the plasma membrane could fit one of two general hypotheses regarding the mechanistic value. The early and dominant general hypothesis in molecular biophysics has been the acceleration of diffusion by reducing the dimensionality, thereby expediting the search for specific targets [73–75]. This hypothesis may apply to the nonspecific 1D diffusion of proteins sliding on DNA to specific sites [76, 77]. Reduced dimensionality has been invoked to account for peripheral proteins gliding on a 2D membrane to meet specific partners [78, 79]. However, the hypothesis of molecular encounters hastened by diffusion in two dimensions rather than three was criticized as resulting in only small to no advantage, and even retardation of encounters [80]. A strong alternative hypothesis argues that membrane localization increases the interactions of proteins and signal transduction by concentrating dilute proteins tremendously in a smaller volume near the membrane [81]. The latter hypothesis generalizes to both sides of the plasma membrane and probably to other membrane organelles. This concept may apply to the formation of multiple MMP complexes with substrates or receptors at membranes. In cases of co-localization on a membrane patch such as a lipid raft, acceleration of encounters by 2D diffusion within the patch is considered likely to enhance rates of association [81]. Since the plasma membrane is partitioned into many patches < 300 nm in breadth [26], the hypothesis of accelerated diffusion potentially applies widely to cells.
2.5. MMP internalization, trafficking, and peripheral membrane interactions
Macrophage-secreted MMP-12 enters cells infected by respiratory viruses and then appears in the nucleus. There it acts as a transcription factor upon the NFKBIA promoter to drive the transcription of many gene products [82]. Live cell imaging using MMP-12 catalytic domain tagged with a membrane-responsive fluor placed at membrane-binding sites offers Insight into this internalization and trafficking [67]. Labeled MMP-12 fluoresces brightly not only on the plasma membrane, but also accumulates especially in a perinuclear membrane network and on the nuclear envelope where it accumulates within 5 min after addition at 37 °C. MMP-12 is distributed similarly in HeLa cells incubated on ice to impede internalization, with the bulk of the fluorescence emitted from vesicles in the cytoplasm and a smaller portion emitted from features (invaginations or vesicles) at the plasma membrane (Fig. 2). The cold impeding the ingress of MMP-12 suggests endocytosis [67]. Similarly, proMMP-7 binds the plasma membrane of colon cancer cells, is internalized, and accumulates in cytoplasmic vesicles within 5 min after addition [50]. Since GSL, SM, and PC are enriched on the luminal face of endosomes [25], MMPs are likely to meet these lipids during endocytosis. Live cell imaging of the development of membrane-responsive fluorescence suggests the participation of peripheral membrane interactions of MMP-7 and 12 in endocytosis.
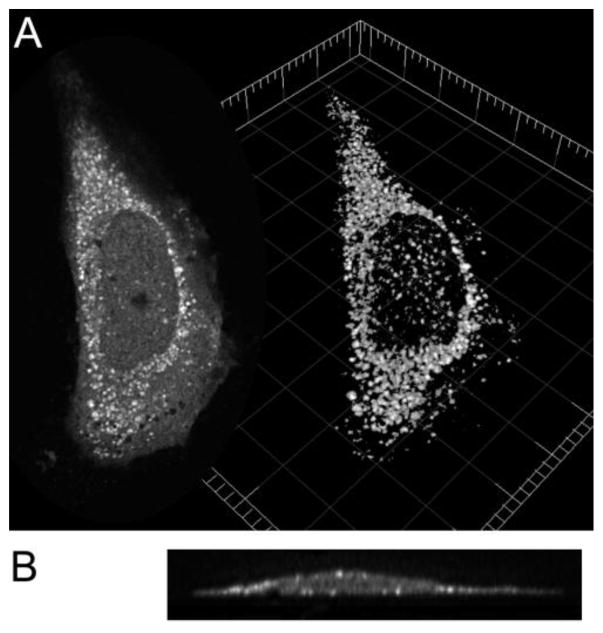
Fig. 2.
MMP-12 bound to vesicles of a HeLa cell. Human MMP-12(G155C) catalytic domain (100 nM) labeled with a membrane-sensitive fluor was incubated with HeLa cells in DMEM without phenol red for 2 h on ice to slow membrane trafficking. After extensive washing with PBS on ice and fixing with 4% paraformaldehyde, cells were imaged with a Zeiss LSM 510 Meta confocal microscope. (A) At left is shown an optical slice through a HeLa cell. At right is shown a cell volume rendering (using Imaris Scientific software) through a series of slices each separated by 0.35 μm in the Z-direction. (B) A YZ section through the Z-stack of slices is pictured and shows bright vesicles with labeled MMP-12 at the periphery of the cell profile.
The internalization of soluble MMP-7 and -12 within 5 min [50, 67] is faster than the internalization of transmembrane-anchored MT1-MMP in 30 to 45 min [83], GPI-anchored MT4-MMP in 45 to 60 min [84], or GPI-anchored MT6-MMP which is also slow [85]. Deletion of the transmembrane helical anchor of MT1-MMP is disruptive to trafficking and recycling [83]. In contrast, MMP-12 and MMP-7 lack such an anchor to depend on. MT1-MMP undergoes endocytosis by both clathrin-dependent and caveolin-dependent pathways [86]. Endocytosis of MT4-MMP proceeds by neither pathway, but rather through early endosomes via the pathway of clathrin-independent carriers/GPI-enriched early endosomal compartments [84]. Soluble MMP-13 is rapidly endocytosed by association of its HPX domain with LRP-1 [87]. How other soluble MMPs undergo endocytosis will require investigation.
MMP-12 accumulates within the nucleus of a live cell within 3 to 12 min after addition [67]. This is consistent with MMP-12 acting as a transcription factor [82]. Likewise, proMMP-7 binds HeLa cell surfaces, is internalized, accumulates with membranes in the cytoplasm, and continues into the nucleus. Endosomal membranes fusing with other membranes can be hypothesized for delivery of soluble MMPs to other membrane compartments. MMP-1*, 2*, 3*, 9, 13*, 14 and 26 are also observed in nuclei, as reviewed [88]. They and MMP-7, 8*, and 11 digest nuclear proteins [88]. Those marked with an asterisk have a putative nuclear localization signal (NLS), as do seven other MMPs not listed [89]. MMP-3 traffics to the nucleus, requiring its NLS [89]. Nuclear MMP-3 boosts transcription at the CTGF promoter [90]. MT1-MMP (MMP-14) also traffics inward and reaches the nucleus where it binds the p110δ promoter and dampens the macrophage immune response via wide effects on transcription and translation [91]. The presence of MMPs in the nucleus, their NLS sequences, and predictions that other soluble MMPs bind membranes (see below) raise the question of mechanisms of trafficking to the nucleus.
3. Interfaces with lipid bilayers
MMP-12 catalytic domain and proMMP-7 each visit two distinct orientations in binding the head group region of bilayers [50, 67], according to NMR measurements using spin-labeled bicelles [92].
3.1. Dual modes of MMP-12 binding to bilayers
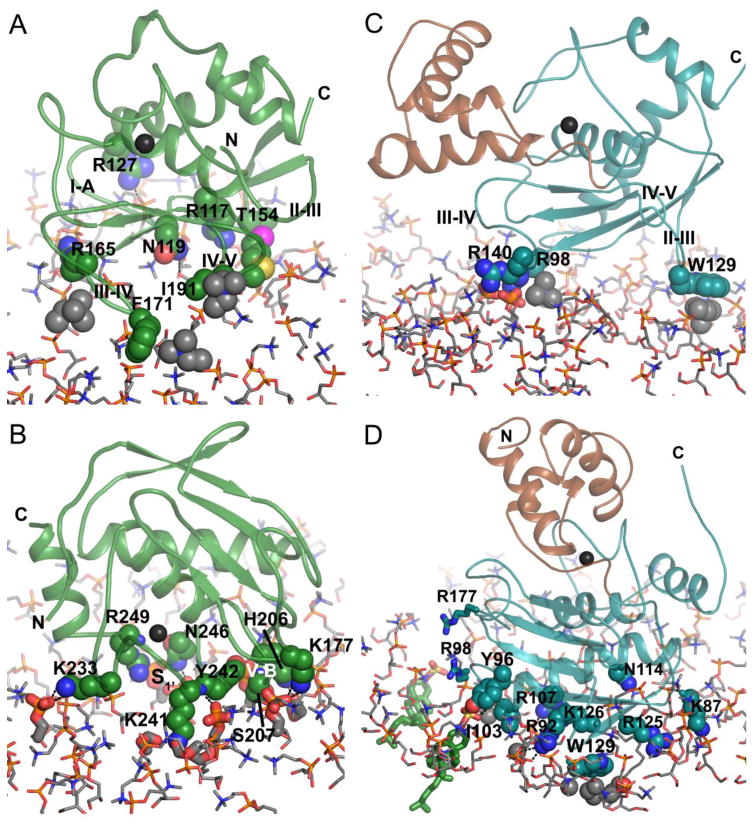
The β-sheet of MMP12 catalytic domain faces the bilayer such that the loops connecting β-strands make contact with the DMPC head groups [67]. Choline head groups make contact with the hydrophobic moieties in the side chains of the S-shaped III–IV loop and the IV–V loop connecting β-strands IV and V (Fig. 3A). Arginine residues 117, 127, and 165 appear to form transient salt bridges with phosphate groups in the head groups (Fig. 3A). Asn119 (I-A loop) and Thr154 (II–III loop) may donate fleeting hydrogen bonds. Overall, these polar contacts with the bilayer are distant from the active site, while the hydrophobic contacts with choline head groups are nearer to the active site (Fig. 3A). In contrast, the mode of MMP-12 binding near the α-helices is entirely polar and closer to the active site (Fig. 3B). Lysine side chains at opposite extremes of this so-called α-interface each form an apparent salt bridge with the phosphate of a DMPC head group [67], at left and right in Fig. 3B. Transitory hydrogen bonds appeared to be donated by side chains from the V-B loop (His206 and Ser207) and side chains from the S1’ loop (Lys241, Tyr242, Asn246, and Arg249) to the phosphoesters of the head groups (Fig. 3B).
Fig. 3.
Dual modes of binding of the catalytic domains of MMP-12 and MMP-7 to bicelle mimics of lipid bilayers. The complexes of DMPC bilayers with MMP-12 catalytic domain at the (A) β-interface (PDB ID: 2MLS) or (B) α-interface (PDB: 2MLR) are pictured. The proMMP-7 complexes with (C) zwitterionic DMPC bilayer (PDB: 2MZH) or (D) DMPC doped with anionic CS (green sticks; PDB: 2MZI) are depicted. In (C,D), the pro-domain is brown and the catalytic domain cyan. The color code by atom type is: nitrogen blue, oxygen red, phosphorus gold, sulfur yellow, lipid carbon gray, MMP-12 carbon green, and MMP-7 carbon cyan or brown. A black sphere marks the zinc in the active site. Side chains of amino acids with lipid contacts in the experiment models are plotted with spheres. Phospholipid carbons with apparent hydrophobic protein contacts are plotted with gray spheres. Phosphoesters with apparent polar contacts with the enzyme are plotted with heavier sticks. The fatty acyl chains of DMPC are omitted for clarity, apart from acyl carbons touching Trp129 and Ile103 in (D).
3.2. Charge-switched partial insertion of proMMP-7
The binding of proMMP-7 to zwitterionic bicelles is analogous to that of MMP-12 via its β-interface, but is more superficial in the case of proMMP-7. Three of the four corresponding loops of proMMP-7 superficially touch the bilayers without net charge: the I-A loop (Arg98), the II–III loop (Trp129), and the III–IV loop (Arg140) (Fig. 3C). The arginine residues appear to form salt bridges with DMPC phosphates while Trp129 contacts hydrophobic choline head groups (Fig. 3C). The lack of contact with the IV–V loop is related to the angle of approach of the β-face of MMP-7 differing by > 30° from that of MMP-12 (c.f. Fig. 3A). ProMMP-7 is slowed more modestly in its tumbling by small, zwitterionic bicelles [50] than is the MMP-12 catalytic domain [67]. The smaller, more superficial interface of proMMP-7 with zwitterionic bilayers correlates with greater transiency.
When proMMP-7 encounters anionic lipids in bilayers, particularly those with greater negative charge from sulfate or phosphate groups, the enzyme associates more tightly with the bilayer [50], with reports of IC50 and apparent KD values for CS ranging from low μM to low nM [23, 49, 52]. Upon binding bicelles doped with CS, the zymogen slows in rotational diffusion in proportion to the [CS] and draws roughly 11 Å closer to the bilayer, buries 3.6-fold more of its surface area with the bilayer, and rotates nearly 80° [50]. This rotation of the β-α-β at the back of the catalytic domain presents to the lipid head groups five basic side chains (Lys87, Arg92, Arg107, Arg125, and Lys126), polar Tyr96 and Asn114, and hydrophobic Ile103 (Fig. 3D). With CS present, the side chains of Trp129 and Ile103 reach into the hydrophobic interior of the membrane [50]. The side chains of Ile103, Arg107, Arg125, and Trp129 are required for association with the anionic lipids CS, sulfatide (SM4) or cardiolipin [52, 93], in excellent agreement with NMR and fluorescence results [50]. The wide and tight interface with CS present features the remarkable ordering and shifting of lipid head groups out around the inserted Trp129 and Ile103 side chains [50]. What favors this partial insertion? The CS molecules exert an electrostatic pull on seven or eight basic residues on the back of the MMP-7 catalytic domain, e.g., the sulfate group of one CS molecule and five basic residues draw as close as possible (Fig. 3D). This CS molecule appears to be drawn into a hydrogen bond with Tyr96 and a polar contact with the partial positive charge at the N-terminal end of the long helix nearby. Lys87, Arg92, Arg107, Asn114, Arg125, and Lys126 seem to engage in favorable but transient hydrogen bonds and/or salt bridges with phosphoester portions of the DMPC head groups [50]. The hydrophobic contacts of Trp129 and Ile103 with the fatty acyl chains are also energetically favorable. However, the electrostatic attraction between anionic bilayers and highly basic MMP-7 dominates this interface. MMP-7 is attracted to other anionic lipids as well, including the PS and sphingosine 1-phosphate signaling molecules [50]. This electropositive region of MMP-7 must also be attracted to the polyanionic GAG chains of glycosphingolipids, which are confined to the apical surfaces of epithelial cells [25] where MMP-7 also localizes [1].
3.3. Allostery between bilayer interfaces and the active site of MMP-7
Binding of lipids triggers long-range communication with the MMP-7 active site. A zinc-chelating, competitive inhibitor decreases MMP-7 affinity for CS and cardiolipin [52]. The inhibitor and anionic lipids bind on opposite sides of the domain. Binding of proMMP-7 to small bicelles, with or without CS embedded in them, results in auto-activation of the wild-type enzyme and alteration of the structure of the E195A-inactivated enzyme; the cysteine coordination of the catalytic zinc that maintains latency is lost [50]. NMR spectra suggest a path of long-range communication from Trp129 in the bilayer interface to the active site and affected cysteine [50].
3.4. Shared characteristics of the bilayer interfaces
Basic residues making polar contacts with head group phosphoesters accounts for the largest share of attraction between bilayers and MMP-7 or MMP-12 (Fig. 3). Basic residues for recognizing phosphate groups also stand out in the structural characterizations of intracellular signaling proteins called conditional peripheral membrane proteins, which have basic pockets to bind specific phosphoinositides, or flat basic surfaces to bind a variety of phosphoinositides promiscuously [79]. Other types of lipid-binding proteins nonspecifically recognize general properties of membranes such as the negative charge of the inner leaflet of the plasma membrane [79]. MMP-7 and MMP-12 likewise prefer anionic lipids, apparently nonspecifically [50, 67], but present on the outer leaflet of plasma membranes accessible by MMPs. For example, CS is present in skin [94] and cancerous lesions [95, 96]. PS exposure is a signature of apoptosis [97]. The importance of charge complementarity to MMP-12 association is evident from its sensitivity to salt [67]. The breadth of the membrane interfaces of MMP-7 and -12 of about 2400 and 2500 Å, respectively [50, 67], may support their charging-sensing properties.
3.5. Switching and adjustability of bilayer binding orientations
The dramatic charged-based switching in the binding of MMP-7 to lipid bilayers (Fig. 3) may trigger its attack upon proteins of the cell surface (exemplified by fibronectin and laminin-332) by the anionic lipids CS, sulfatide, and cardiolipin [48, 51, 52, 93]. The association with CS (and other anionic lipids) appears to rotate the pro-enzyme around 75°, to point the active site away from the bilayer surface for widest accessibility (Figs. 3D, 4A). CS recruits a partial insertion of the pro-enzyme into the bilayer, drawing it in toward the bilayer by at least 10 Å (Fig. 4A). This accounts for the slowing of its tumbling by CS and implies increased residence time of the enzyme on the bilayer [50]. The CS-induced changes in orientation [79] and increases in activity upon protein substrates at membrane surfaces [52, 93] support the hypothesis that CS increases productive collisions of MMP-7 with CS-recruited substrate proteins.
Fig. 4.
Adaptability of orientations of MMP-7 and 12 on DMPC bilayers. (A) The complexes of zwitterionic bilayers with proMMP-7 (cyan) and anionic bilayers with proMMP-7 (magenta) are superimposed in order to show the rotation and partial insertion induced by adding cholesterol sulfate (CS, pink) to the bilayers. The catalytic domain of MMP-12 is colored with its contacts with TIMP-2 inhibitor protein in purple and its contacts with small disk-like bicelles containing DMPC at the α-face (B) and β-face (C). Arrows symbolize minimal rotations of the catalytic domain in order to relieve the steric conflict with the TIMP-2 bound. The density of DMPC molecules is higher in panel A because it plots two bilayers superimposed.
MMP-12 association with its bilayers by its α-face imposes some steric conflict upon the accessibility of the unprimed subsites of the active site (Figs. 3B, 4B), i.e. near His206, Ser207, Lys241, and Tyr242. Bilayer association with MMP-12 by its β-face leaves the active site more accessible (Figs. 3A, 4C) to membrane-associated protein substrates. The active site is better separated from the membrane surfaces and nearly parallel to them (Fig. 4C), and can face a protein neighbor on the membrane. This mode of binding might support membrane-proximal activities such as the activation of prothrombin (Fig. 1). The membrane association should also concentrate the MMP-12 at the membrane, thereby increasing the number of productive collisions with membrane-associated substrates, following the “piggyback” mechanism postulated for many signaling complexes [81].
Striking adaptability in the orientations of binding membranes is highlighted by steric competition of TIMP-2 and bilayers for both the α and β membrane interfaces of MMP-12 [67] (Figs. 4B,C). MMP-12 catalytic domain seems to respond by rolling away on the membrane to accommodate TIMP-2 occupying part of each membrane interface [67] (Figs. 4B,C). To relieve the steric clash between bilayer and TIMP-2, the MMP-12/TIMP-2 complex may rotate on the membrane (i) at the α-face more than 30° away from the conflict with the C-terminal domain of TIMP-2 (Fig. 4B) and (ii) at the β-face more than 10° away from the conflict with the AB loop of TIMP-2 (Fig. 4C). It is unclear whether the enzyme inherently rocks and rolls on fluid bilayers or hops and rebinds in orientations altered by a bound TIMP inhibitor. ProMMP-7 appears to rotate, slide, and bob on the bilayer surface [50]. The reorientations of MMP-12 promoted by TIMP-2 are larger than the smaller simulated fluctuations of proMMP-7.
4. Hypothesis that soluble domains from multiple MMPs interact with bilayers
4.1. Catalytic domains
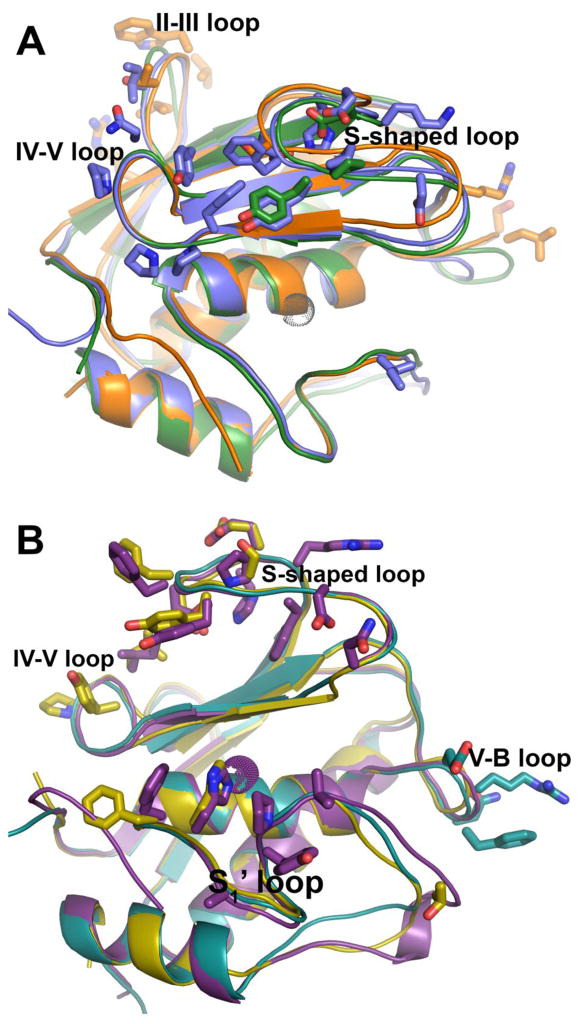
The practice and benefit of including the lipid-mimicking detergent Brij-35 in proteolytic activity assays of MMPs in vitro [98] might hint at the possibility that several soluble MMPs tend to interact with lipids. Nonspecific interactions of proteins with membrane bilayers can be predicted from atomic-resolution structural coordinates using web servers named Positioning of Proteins in Membranes (PPM) [99–102] or Membrane Optimal Docking Area (MODA) [103, 104]. The interfacial residues predicted by MODA are marked in Fig. 5 and agree with the interfaces predicted by PPM, except where noted. MMP-7 is predicted to bind bilayers using its II–III loop and V-B loop (orange in Fig. 5A), which are two of the three loops making superficial contacts with zwitterionic head groups in the NMR structure (Fig. 3C; PDB: 4MZH). Two other catalytic domains with forecasts of binding bilayers via the β-face, i.e. the loops around the β-sheet, are those of MMP-2 and 13 (Fig. 5A). However, PPM predicts that MMP-13 binds instead by the α-face, similarly to MMP-12 bound by its α-face to the bilayer (Fig. 3B). MMP-1 is predicted to bind using the α-face as well (Fig. 5B). Analogous to bilayer-MMP-12 complexes [67] (Fig. 3A,B), stromelysins 1 and 2, i.e. MMP-3 and 10, are predicted to bind bilayers by both the α-face and β-face (Fig. 5B). Judged against MMP-12 and proMMP-7, the predictions constitute only a subset of the interactions measured experimentally.
Fig. 5.
Predictions that catalytic domains of several soluble MMPs bind membrane bilayers via similar interfaces. The bilayer binding sites proposed by the MODA server [103] (http://molsoft.com/~eugene/moda/modamain.cgi) from structural coordinates are plotted with side chains. (a) Catalytic domains predicted to interact with bilayers via the same loops emanating from the β-sheet (the β-face) are represented by MMP-2 (PDB ID: 1QIB), MMP-7 (2Y6D), MMP-13 (1XUC) colored green, orange and blue, respectively. (b) Catalytic domains also predicted to interact with bilayers using loops of the α-face include MMP-1 (2TCL) in cyan, MMP-3 (1CIZ) in violet, and MMP-10 (3V96) in cyan, violet and yellow, respectively.
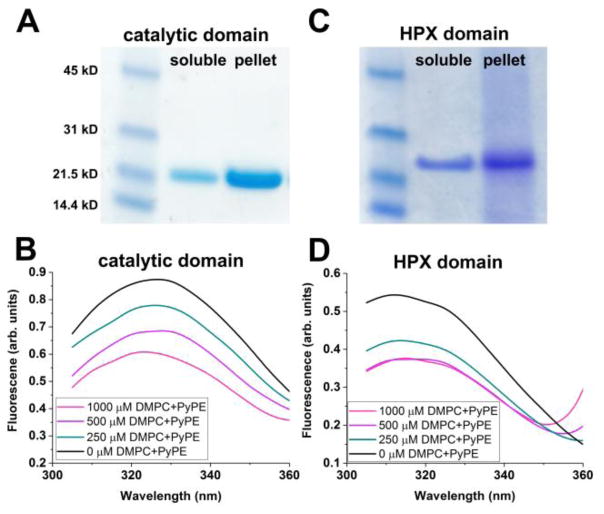
The catalytic domains of MT1-MMP and MT3-MMP are also predicted to bind bilayers, but with less agreement between the servers’ predictions regarding their orientation. MODA predicts that both enzymes should bind by the β-face, and MT1-MMP also by the α-face. PPM predicts that both enzymes bind by the α-face. Coordinates of GPI-anchored MT-MMPs are not yet available for structure-based predictions. Treatment of GPI-anchored MT6-MMP with phosphatidylinositol-specific phospholipase C liberates the enzyme from cell surfaces [85], casting doubt on the sufficiency of its domains to associate peripherally with membranes. Membrane interactions of the catalytic domain of MT1-MMP are supported by assays using SUVs composed of DMPC. The catalytic domain sediments together with SUVs (Fig. 6A). FRET assays indicate that tryptophan side chains of MT1-MMP catalytic domain draw near to the pyrene quencher attached to the head group of phosphatidylethanolamine (PyPE) embedded in SUVs of DMPC, increasingly as the concentration of these labeled SUVs is raised (Fig. 6B).
Fig. 6.
Evidence for association of the soluble domains of MT1-MMP with liposomes. The catalytic (A) and HPX (C) domains (10 μM and without linker sequences) were incubated with SUVs composed of DMPC (1 mM in monomers) for 1 h at 25 °C in 20 mM Tris·HCl (pH 7.2), 150 mM NaCl and 10 mM CaCl2. The mixtures were sedimented by ultracentrifugation at 180,000 × g for 20 min at 4°C. The pellets were suspended in 1/8th of the original sample volumes for SDS-PAGE. Quenching of intrinsic Trp fluorescence emission by PyPE-containing SUVs detects proximity [112] to the catalytic (B) and HPX (D) domains, held constant at 10 and 5 μM, respectively. Incubations were 30 min at 25 °C in the buffer listed above. The PyPE was present at 2 mol% and DMPC at 98 mol%. Excitation was at 275 nm with emission measured with a BioTek Synergy MX plate reader.
4.2. Hemopexin-like domains
The HPX domain of MT1-MMP also interacts with SUVs in assays, reaching near to saturation by 500 μM DMPC monomers (Fig. 6C,D). One formulation of bicelles also binds the ectodomain, HPX domain, and its blades III and IV, distal from the collagen binding site [60](Fig. 7A). Preliminary modes of binding to blades III and IV were proposed, but differ from preliminary modes of binding suggested by paramagnetic NMR using small disc-like bicelles (Marcink and Van Doren, unpublished). These results suggest that previous assumptions stating that the catalytic and HPX domains of MT1-MMP radiate away from the cell membrane may need to be reconsidered. The soluble domains of MT1-MMP may be located part-time on the membrane surface.
Fig. 7.
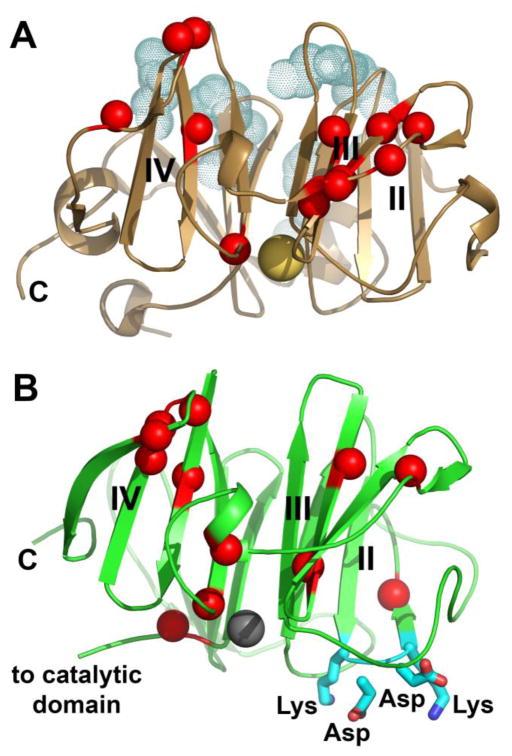
Functional sites and bilayer-affected sites of the HPX domains of (A) MT1-MMP and (B) MMP-12. Red spheres mark amide groups with NMR peaks severely broadened by addition of (A) sheet-like DMPC/CHAPS bicelles to 1% (w/v) [60] or (B) Nanodiscs. The Nanodiscs (100 μM) were prepared from DMPC and MSP1D1 to encircle the bilayer, resulting in discs with diameters near 10 nm [107, 111]. Full-length MMP-12(F171D/E219A) was present at 167 μM at 300 K in 25 mM Tris-HCl (pH 7.2), 10 mM CaCl2, 0.15 M NaCl, and 0.1 M acetohydroxamic acid. NMR peak assignments available [113] were used to interpret 15N-TROSY NMR spectra collected at 800 MHz. The blades of each β-propeller are labeled with Roman numerals. Side chains of functional binding site are plotted in cyan. (A) Shaded dots plot the side chains of the HPX domain of MT1-MMP that bind the collagen triple-helix [114]. The large sphere in gold marks a sodium ion. (B) The side chains of the hairpin loop critical to the bactericidal activity of the MMP-12 HPX domain [105] are plotted with sticks. The large sphere in gray marks a calcium ion.
The HPX domain of MMP-12 kills gram-positive and gram-negative bacteria and binds bacteria in phagolysozomes of macrophages [105]. (At pH 4.8 to 5.7 typical of phagolysozomes [106], MMP catalytic domains unfold while HPX domains remain stable). The bactericidal activity of the MMP-12 HPX domain suggests the distinct possibility that the domain may interact with bacterial membranes since antimicrobial peptides are often judged by their ability to destabilize membranes of their targets. To test for bilayer association, we prepared Nanodiscs, which are trusted and homogeneous bilayers confined to small discs [107–111]. The Nanodiscs broaden away amide NMR peaks of MMP-12(F171D/E219A) for at least 11 residues in blades III and IV, and the peak of another residue in blade II (Fig. 7B). The Nanodisc-induced NMR line broadening in blades III and IV of MMP-12 is similar to the line broadening of blades III and IV of MT1-MMP by DMPC/CHAPS bicelles (c.f. Fig. 7A). The bilayer-affected location in the MMP-12 HPX domain in blade II adjoins its β-hairpin loop critical for killing bacteria. This loop has the sequence of KDDK that is unique to the MMP-12 subfamily [105] and marked in Fig. 7B. The partial overlap of the KDDK loop with the bilayer-perturbed region raises the question of the nature and extent of the relationship between bactericidal activity and the interface for binding lipid bilayers.
The flexible linkers between the pairs of membrane-interacting catalytic and HPX domains of MMP-12 and MT1-MMP should enable their peripheral membrane interactions to adapt to membrane curvature that they might encounter in caveolae, clathrin-coated pits, and endosomes.
Conclusions
Peripheral membrane associations of soluble domains of MMPs appear to be coupled to pericellular proteolysis, endocytosis, and intracellular trafficking. The pericellular proteolysis includes cases of sheddase and maturase activities and collagenolysis. Binding of membrane components by soluble MMP-7, MMP-12, and MMP-13 is associated with the endocytosis of each. This appears analogous to the importance of membrane anchoring of MT-MMPs in endocytosis, but the mechanisms of endocytosis are diverse. The intracellular trafficking of MMP-7 and MMP-12 manifests association with membrane compartments as well. Bacterial killing by the HPX domain of MMP-12 can be hypothesized to be related to binding of membrane components of bacterial envelopes. Since the assembly of MT-MMPs upon membranes heavily influences their function, the possibility of transient associations of their soluble domains with bilayers complementing the transmembrane anchors should now be considered. Attraction of basic side chains to anionic lipids and phosphate moieties in lipid head groups are emerging themes in peripheral associations of soluble MMP domains with membranes. The modes of binding can adjust, especially in response to charge. Anionic lipids increase pericellular proteolysis by MMP-7, probably by increasing productive collisions. Peripheral membrane associations are predicted from the structural coordinates of the catalytic domains of several soluble MMPs. The HPX domains of MT1-MMP and MMP-12 bind bilayers as well. Peripheral association of soluble MMP domains with membrane components concentrates the MMPs and may accelerate their local diffusion, which may increase the effective affinity of their transient complexes with substrates and receptors formed at the membrane.
Highlights.
Peripheral membrane associations couple MMPs to pericellular proteolysis and trafficking
Multiple MMP catalytic domains and some hemopexin domains may bind lipid bilayers
Basic residues of some MMPs recruit them to anionic groups of lipids
The bilayer associations are dynamic and adaptable but concentrate MMPs with partners
Acknowledgments
This work received support from NIH grants R01GM057289 and R01CA098799, as well as S10RR022341 toward purchase of the NMR instrument. We thank Prof. Alexander Franz for comments on the manuscript and Prof. Marco Fragai for the expression vector for MMP-12.
Abbreviations
- CS
cholesterol 3-sulfate
- DMPC
dimyristoylphosphatidylcholine
- FRET
Förster resonance energy transfer
- HPX
hemopexin-like
- LRP
low-density lipoprotein receptor-related protein
- MMP
matrix metalloproteinase
- MT1-MMP
membrane type 1 MMP
- NLS
nuclear localization sequence
- NMR
nuclear magnetic resonance spectroscopy
- PyPE
1,2-dioleoyl-sn-glycero-3-phospho-[N-1-pyrenesulfonyl] ethanolamine
- SM3
lactosylceramide 3′-sulfate
- SM4
sulfatide or 3-O-sulfogalactosylceramide
- TIMP
tissue inhibitor of metalloproteinases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yu WH, Woessner JF., Jr Heparan sulfate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7) J Biol Chem. 2000;275:4183–4191. doi: 10.1074/jbc.275.6.4183. [DOI] [PubMed] [Google Scholar]
- 2.Gadher SJ, Eyre DR, Wotton SF, Schmid TM, Woolley DE. Degradation of cartilage collagens type II, IX, X and XI by enzymes derived from human articular chondrocytes. Matrix. 1990;10:154–163. doi: 10.1016/s0934-8832(11)80164-2. [DOI] [PubMed] [Google Scholar]
- 3.Cha J, Pedersen MV, Auld DS. Metal and pH dependence of heptapeptide catalysis by human matrilysin. Biochemistry. 1996;35:15831–15838. doi: 10.1021/bi962085f. [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto S, Sakamoto M, Goldhaber P, Glimcher MJ. Studies on the interaction between heparin and mouse bone collagenase. Biochimica et Biophysica Acta (BBA)-General Subjects. 1975;385:41–50. doi: 10.1016/0304-4165(75)90072-0. [DOI] [PubMed] [Google Scholar]
- 5.Crabbe T, O’Connell JP, Smith BJ, Docherty AJ. Reciprocated matrix metalloproteinase activation: a process performed by interstitial collagenase and progelatinase A. Biochemistry. 1994;33:14419–14425. doi: 10.1021/bi00252a007. [DOI] [PubMed] [Google Scholar]
- 6.Amour A, Slocombe PM, Webster A, Butler M, Knight CG, Smith BJ, Stephens PE, Shelley C, Hutton M, Knauper V, Docherty AJ, Murphy G. TNF-alpha converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 1998;435:39–44. doi: 10.1016/s0014-5793(98)01031-x. [DOI] [PubMed] [Google Scholar]
- 7.Wallon UM, Overall CM. The hemopexin-like domain (C domain) of human gelatinase A (matrix metalloproteinase-2) requires Ca2+ for fibronectin and heparin binding. Binding properties of recombinant gelatinase A C domain to extracellular matrix and basement membrane components. J Biol Chem. 1997;272:7473–7481. doi: 10.1074/jbc.272.11.7473. [DOI] [PubMed] [Google Scholar]
- 8.Theocharis AD, Gialeli C, Bouris P, Giannopoulou E, Skandalis SS, Aletras AJ, Iozzo RV, Karamanos NK. Cell-matrix interactions: focus on proteoglycan-proteinase interplay and pharmacological targeting in cancer. FEBS J. 2014;281:5023–5042. doi: 10.1111/febs.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadler-Olsen E, Fadnes B, Sylte I, Uhlin-Hansen L, Winberg J-O. Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 2011;278:28–45. doi: 10.1111/j.1742-4658.2010.07920.x. [DOI] [PubMed] [Google Scholar]
- 10.Tocchi A, Parks WC. Functional interactions between matrix metalloproteinases and glycosaminoglycans. FEBS J. 2013;280:2332–2341. doi: 10.1111/febs.12198. [DOI] [PubMed] [Google Scholar]
- 11.Yu WH, Woessner JF, Jr, McNeish JD, Stamenkovic I. CD44 anchors the assembly of matrilysin/MMP-7 with heparin-binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev. 2002;16:307–323. doi: 10.1101/gad.925702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Park PW, Wilson CL, Parks WC. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111:635–646. doi: 10.1016/s0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]
- 13.Gao G, Plaas A, Thompson VP, Jin S, Zuo F, Sandy JD. ADAMTS4 (aggrecanase-1) activation on the cell surface involves C-terminal cleavage by glycosylphosphatidyl inositol-anchored membrane type 4-matrix metalloproteinase and binding of the activated proteinase to chondroitin sulfate and heparan sulfate on syndecan-1. J Biol Chem. 2004;279:10042–10051. doi: 10.1074/jbc.M312100200. [DOI] [PubMed] [Google Scholar]
- 14.Iida J, Wilhelmson KL, Ng J, Lee P, Morrison C, Tam E, Overall CM, McCarthy JB. Cell surface chondroitin sulfate glycosaminoglycan in melanoma: role in the activation of pro-MMP-2 (pro-gelatinase A) Biochem J. 2007;403:553–563. doi: 10.1042/BJ20061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Echtermeyer F, Bertrand J, Dreier R, Meinecke I, Neugebauer K, Fuerst M, Lee YJ, Song YW, Herzog C, Theilmeier G, Pap T. Syndecan-4 regulates ADAMTS-5 activation and cartilage breakdown in osteoarthritis. Nat Med. 2009;15:1072–1076. doi: 10.1038/nm.1998. [DOI] [PubMed] [Google Scholar]
- 16.Ra HJ, Harju-Baker S, Zhang F, Linhardt RJ, Wilson CL, Parks WC. Control of promatrilysin (MMP7) activation and substrate-specific activity by sulfated glycosaminoglycans. J Biol Chem. 2009;284:27924–27932. doi: 10.1074/jbc.M109.035147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorensen HP, Vives RR, Manetopoulos C, Albrechtsen R, Lydolph MC, Jacobsen J, Couchman JR, Wewer UM. Heparan sulfate regulates ADAM12 through a molecular switch mechanism. J Biol Chem. 2008;283:31920–31932. doi: 10.1074/jbc.M804113200. [DOI] [PubMed] [Google Scholar]
- 18.Fulcher YG, Sanganna Gari RR, Frey NC, Zhang F, Linhardt RJ, King GM, Van Doren SR. Heparinoids activate a protease, secreted by mucosa and tumors, via tethering supplemented by allostery. ACS Chem Biol. 2014;9:957–966. doi: 10.1021/cb400898t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulcher YG, Prior SH, Zhang F, Linhardt RJ, Van Doren SR. Structure. 2017 Jul 5;25(7):1100–1110.e5. doi: 10.1016/j.str.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owen CA, Hu Z, Barrick B, Shapiro SD. Inducible expression of tissue inhibitor of metalloproteinases-resistant matrix metalloproteinase-9 on the cell surface of neutrophils. Am J Respir Cell Mol Biol. 2003;29:283–294. doi: 10.1165/rcmb.2003-0034OC. [DOI] [PubMed] [Google Scholar]
- 21.Owen CA, Hu Z, Lopez-Otin C, Shapiro SD. Membrane-Bound Matrix Metalloproteinase-8 on Activated Polymorphonuclear Cells Is a Potent, Tissue Inhibitor of Metalloproteinase-Resistant Collagenase and Serpinase. J Immunol. 2004;172:7791–7803. doi: 10.4049/jimmunol.172.12.7791. [DOI] [PubMed] [Google Scholar]
- 22.Kioi M, Yamamoto K, Higashi S, Koshikawa N, Fujita K, Miyazaki K. Matrilysin (MMP-7) induces homotypic adhesion of human colon cancer cells and enhances their metastatic potential in nude mouse model. Oncogene. 2003;22:8662–8670. doi: 10.1038/sj.onc.1207181. [DOI] [PubMed] [Google Scholar]
- 23.Berton A, Selvais C, Lemoine P, Henriet P, Courtoy PJ, Marbaix E, Emonard H. Binding of matrilysin-1 to human epithelial cells promotes its activity. Cell Mol Life Sci. 2007;64:610–620. doi: 10.1007/s00018-007-6415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cobos-Correa A, Trojanek JB, Diemer S, Mall MA, Schultz C. Membrane-bound FRET probe visualizes MMP12 activity in pulmonary inflammation. Nat Chem Biol. 2009;5:628–630. doi: 10.1038/nchembio.196. [DOI] [PubMed] [Google Scholar]
- 25.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 27.Yu X, Zech T, McDonald L, Gonzalez EG, Li A, Macpherson I, Schwarz JP, Spence H, Futo K, Timpson P, Nixon C, Ma Y, Anton IM, Visegrady B, Insall RH, Oien K, Blyth K, Norman JC, Machesky LM. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J Cell Biol. 2012;199:527–544. doi: 10.1083/jcb.201203025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Azzouzi K, Wiesner C, Linder S. Metalloproteinase MT1-MMP islets act as memory devices for podosome reemergence. J Cell Biol. 2016;213:109–125. doi: 10.1083/jcb.201510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 30.Imai K, Ohuchi E, Aoki T, Nomura H, Fujii Y, Sato H, Seiki M, Okada Y. Membrane-type matrix metalloproteinase 1 is a gelatinolytic enzyme and is secreted in a complex with tissue inhibitor of metalloproteinases 2. Cancer Res. 1996;56:2707–2710. [PubMed] [Google Scholar]
- 31.Itoh Y, Takamura A, Ito N, Maru Y, Sato H, Suenaga N, Aoki T, Seiki M. Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J. 2001;20:4782–4793. doi: 10.1093/emboj/20.17.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Bernardo MM, Osenkowski P, Sohail A, Pei D, Nagase H, Kashiwagi M, Soloway PD, DeClerck YA, Fridman R. Differential inhibition of membrane type 3 (MT3)-matrix metalloproteinase (MMP) and MT1-MMP by tissue inhibitor of metalloproteinase (TIMP)-2 and TIMP-3 rgulates pro-MMP-2 activation. J Biol Chem. 2004;279:8592–8601. doi: 10.1074/jbc.M308708200. [DOI] [PubMed] [Google Scholar]
- 33.Shiomi T, Inoki I, Kataoka F, Ohtsuka T, Hashimoto G, Nemori R, Okada Y. Pericellular activation of proMMP-7 (promatrilysin-1) through interaction with CD151. Lab Invest. 2005;85:1489–1506. doi: 10.1038/labinvest.3700351. [DOI] [PubMed] [Google Scholar]
- 34.Yu Q, Stamenkovic I. Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes Dev. 1999;13:35–48. doi: 10.1101/gad.13.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori H, Tomari T, Koshikawa N, Kajita M, Itoh Y, Sato H, Tojo H, Yana I, Seiki M. CD44 directs membrane-type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. EMBO J. 2002;21:3949–3959. doi: 10.1093/emboj/cdf411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suenaga N, Mori H, Itoh Y, Seiki M. CD44 binding through the hemopexin-like domain is critical for its shedding by membrane-type 1 matrix metalloproteinase. Oncogene. 2005;24:859–868. doi: 10.1038/sj.onc.1208258. [DOI] [PubMed] [Google Scholar]
- 37.Takino T, Miyamori H, Kawaguchi N, Uekita T, Seiki M, Sato H. Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane-type 1 matrix metalloproteinase. Biochem Biophys Res Commun. 2003;304:160–166. doi: 10.1016/s0006-291x(03)00544-8. [DOI] [PubMed] [Google Scholar]
- 38.Yanez-Mo M, Barreiro O, Gonzalo P, Batista A, Megias D, Genis L, Sachs N, Sala-Valdes M, Alonso MA, Montoya MC, Sonnenberg A, Arroyo AG, Sanchez-Madrid F. MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood. 2008;112:3217–3226. doi: 10.1182/blood-2008-02-139394. [DOI] [PubMed] [Google Scholar]
- 39.Dumin JA, Dickeson SK, Stricker TP, Bhattacharyya-Pakrasi M, Roby JD, Santoro SA, Parks WC. Pro-collagenase-1 (matrix metalloproteinase-1) binds the alpha(2)beta(1) integrin upon release from keratinocytes migrating on type I collagen. J Biol Chem. 2001;276:29368–29374. doi: 10.1074/jbc.M104179200. [DOI] [PubMed] [Google Scholar]
- 40.Stricker TP, Dumin JA, Dickeson SK, Chung L, Nagase H, Parks WC, Santoro SA. Structural analysis of the alpha(2) integrin I domain/procollagenase-1 (matrix metalloproteinase-1) interaction. J Biol Chem. 2001;276:29375–29381. doi: 10.1074/jbc.M102217200. [DOI] [PubMed] [Google Scholar]
- 41.Stefanidakis M, Bjorklund M, Ihanus E, Gahmberg CG, Koivunen E. Identification of a negatively charged peptide motif within the catalytic domain of progelatinases that mediates binding to leukocyte beta 2 integrins. J Biol Chem. 2003;278:34674–34684. doi: 10.1074/jbc.M302288200. [DOI] [PubMed] [Google Scholar]
- 42.Bjorklund M, Heikkila P, Koivunen E. Peptide inhibition of catalytic and noncatalytic activities of matrix metalloproteinase-9 blocks tumor cell migration and invasion. J Biol Chem. 2004;279:29589–29597. doi: 10.1074/jbc.M401601200. [DOI] [PubMed] [Google Scholar]
- 43.Redondo-Munoz J, Ugarte-Berzal E, Terol MJ, Van den Steen PE, Hernandez del Cerro M, Roderfeld M, Roeb E, Opdenakker G, Garcia-Marco JA, Garcia-Pardo A. Matrix metalloproteinase-9 promotes chronic lymphocytic leukemia b cell survival through its hemopexin domain. Cancer Cell. 2010;17:160–172. doi: 10.1016/j.ccr.2009.12.044. [DOI] [PubMed] [Google Scholar]
- 44.Emonard H, Bellon G, Troeberg L, Berton A, Robinet A, Henriet P, Marbaix E, Kirkegaard K, Patthy L, Eeckhout Y, Nagase H, Hornebeck W, Courtoy PJ. Low density lipoprotein receptor-related protein mediates endocytic clearance of pro-MMP-2.TIMP-2 complex through a thrombospondin-independent mechanism. J Biol Chem. 2004;279:54944–54951. doi: 10.1074/jbc.M406792200. [DOI] [PubMed] [Google Scholar]
- 45.Van den Steen PE, Van Aelst I, Hvidberg V, Piccard H, Fiten P, Jacobsen C, Moestrup SK, Fry S, Royle L, Wormald MR, Wallis R, Rudd PM, Dwek RA, Opdenakker G. The hemopexin and O-glycosylated domains tune gelatinase B/MMP-9 bioavailability via inhibition and binding to cargo receptors. J Biol Chem. 2006;281:18626–18637. doi: 10.1074/jbc.M512308200. [DOI] [PubMed] [Google Scholar]
- 46.Murphy G, Nagase H. Localizing matrix metalloproteinase activities in the pericellular environment. FEBS J. 2011;278:2–15. doi: 10.1111/j.1742-4658.2010.07918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piccard H, Van den Steen PE, Opdenakker G. Hemopexin domains as multifunctional liganding modules in matrix metalloproteinases and other proteins. J Leukoc Biol. 2007;81:870–892. doi: 10.1189/jlb.1006629. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto K, Higashi S, Kioi M, Tsunezumi J, Honke K, Miyazaki K. Binding of active matrilysin to cell surface cholesterol sulfate is essential for its membrane-associated proteolytic action and induction of homotypic cell adhesion. J Biol Chem. 2006;281:9170–9180. doi: 10.1074/jbc.M510377200. [DOI] [PubMed] [Google Scholar]
- 49.Ganguly B, Banerjee J, Elegbede AI, Klocke DJ, Mallik S, Srivastava DK. Intrinsic selectivity in binding of matrix metalloproteinase-7 to differently charged lipid membranes. FEBS Lett. 2007;581:5723–5726. doi: 10.1016/j.febslet.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prior SH, Fulcher YG, Koppisetti RK, Jurkevich A, Van Doren SR. Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors. Structure. 2015;23:2099–2110. doi: 10.1016/j.str.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto K, Miyazaki K, Higashi S. Cholesterol sulfate alters substrate preference of matrix metalloproteinase-7 and promotes degradations of pericellular laminin-332 and fibronectin. J Biol Chem. 2010;285:28862–28873. doi: 10.1074/jbc.M110.136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto K, Miyazaki K, Higashi S. Pericellular proteolysis by matrix metalloproteinase-7 is differentially modulated by cholesterol sulfate, sulfatide, and cardiolipin. FEBS J. 2014;281:3346–3356. doi: 10.1111/febs.12865. [DOI] [PubMed] [Google Scholar]
- 53.Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J Cell Biol. 2000;149:1309–1323. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, Lopez-Otin C, Shapiro S, Inada M, Krane S, Allen E, Chung D, Weiss SJ. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li XY, Ota I, Yana I, Sabeh F, Weiss SJ. Molecular dissection of the structural machinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol Biol Cell. 2008;19:3221–3233. doi: 10.1091/mbc.E08-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itoh Y, Ito N, Nagase H, Evans RD, Bird SA, Seiki M. Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol Biol Cell. 2006;17:5390–5399. doi: 10.1091/mbc.E06-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 58.Bocharov EV, Volynsky PE, Pavlov KV, Efremov RG, Arseniev AS. Structure elucidation of dimeric transmembrane domains of bitopic proteins. Cell Adh Migr. 2010;4:284–298. doi: 10.4161/cam.4.2.11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martfeld AN, Rajagopalan V, Greathouse DV, Koeppe RE., 2nd Dynamic regulation of lipid-protein interactions. Biochim Biophys Acta. 2015;1848:1849–1859. doi: 10.1016/j.bbamem.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 60.Cerofolini L, Amar S, Lauer JL, Martelli T, Fragai M, Luchinat C, Fields GB. Bilayer Membrane Modulation of Membrane Type 1 Matrix Metalloproteinase (MT1-MMP) Structure and Proteolytic Activity. Sci Rep. 2016;6:29511. doi: 10.1038/srep29511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durr UH, Soong R, Ramamoorthy A. When detergent meets bilayer: birth and coming of age of lipid bicelles. Prog Nucl Magn Reson Spectrosc. 2013;69:1–22. doi: 10.1016/j.pnmrs.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kajita M, Itoh Y, Chiba T, Mori H, Okada A, Kinoh H, Seiki M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J Cell Biol. 2001;153:893–904. doi: 10.1083/jcb.153.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Endo K, Takino T, Miyamori H, Kinsen H, Yoshizaki T, Furukawa M, Sato H. Cleavage of syndecan-1 by membrane type matrix metalloproteinase-1 stimulates cell migration. J Biol Chem. 2003;278:40764–40770. doi: 10.1074/jbc.M306736200. [DOI] [PubMed] [Google Scholar]
- 64.Koshikawa N, Giannelli G, Cirulli V, Miyazaki K, Quaranta V. Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J Cell Biol. 2000;148:615–624. doi: 10.1083/jcb.148.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koshikawa N, Minegishi T, Sharabi A, Quaranta V, Seiki M. Membrane-type matrix metalloproteinase-1 (MT1-MMP) is a processing enzyme for human laminin gamma 2 chain. J Biol Chem. 2005;280:88–93. doi: 10.1074/jbc.M411824200. [DOI] [PubMed] [Google Scholar]
- 66.Udayakumar TS, Chen ML, Bair EL, von Bredow DC, Cress AE, Nagle RB, Bowden GT. Membrane Type-1-Matrix Metalloproteinase Expressed by Prostate Carcinoma Cells Cleaves Human Laminin-5 β3 Chain and Induces Cell Migration. Cancer Res. 2003;63:2292–2299. [PubMed] [Google Scholar]
- 67.Koppisetti RK, Fulcher YG, Jurkevich A, Prior SH, Xu J, Lenoir M, Overduin M, Van Doren SR. Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12. Nature communications. 2014;5:5552. doi: 10.1038/ncomms6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bellac CL, Dufour A, Krisinger MJ, Loonchanta A, Starr AE, Auf dem Keller U, Lange PF, Goebeler V, Kappelhoff R, Butler GS, Burtnick LD, Conway EM, Roberts CR, Overall CM. Macrophage matrix metalloproteinase-12 dampens inflammation and neutrophil influx in arthritis. Cell Rep. 2014;9:618–632. doi: 10.1016/j.celrep.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 69.Nelsestuen GL, Kisiel W, Di Scipio RG. Interaction of vitamin K dependent proteins with membranes. Biochemistry. 1978;17:2134–2138. doi: 10.1021/bi00604a017. [DOI] [PubMed] [Google Scholar]
- 70.Zwaal RF, Comfurius P, Bevers EM. Lipid-protein interactions in blood coagulation. Biochim Biophys Acta. 1998;1376:433–453. doi: 10.1016/s0304-4157(98)00018-5. [DOI] [PubMed] [Google Scholar]
- 71.Tavoosi N, Davis-Harrison RL, Pogorelov TV, Ohkubo YZ, Arcario MJ, Clay MC, Rienstra CM, Tajkhorshid E, Morrissey JH. Molecular determinants of phospholipid synergy in blood clotting. J Biol Chem. 2011;286:23247–23253. doi: 10.1074/jbc.M111.251769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morrissey JH, Davis-Harrison RL, Tavoosi N, Ke K, Pureza V, Boettcher JM, Clay MC, Rienstra CM, Ohkubo YZ, Pogorelov TV, Tajkhorshid E. Protein-phospholipid interactions in blood clotting. Thromb Res. 2010;125(Suppl 1):S23–25. doi: 10.1016/j.thromres.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adam G, Delbruck M. Reduction of dimensionality in biological diffusion processes. In: Rich A, Davidson N, editors. Structural Chemistry and Molecular Biology. Freeman, Place Published; 1968. pp. 198–215. [Google Scholar]
- 74.Berg OG. Orientation constraints in diffusion-limited macromolecular association. The role of surface diffusion as a rate-enhancing mechanism. Biophys J. 1985;47:1–14. doi: 10.1016/S0006-3495(85)83870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 76.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 77.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 78.Gupte SS, Hackenbrock CR. Multidimensional diffusion modes and collision frequencies of cytochrome c with its redox partners. J Biol Chem. 1988;263:5241–5247. [PubMed] [Google Scholar]
- 79.Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCloskey MA, Poo MM. Rates of membrane-associated reactions: reduction of dimensionality revisited. The Journal of Cell Biology. 1986;102:88–96. doi: 10.1083/jcb.102.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kholodenko BN, Hoek JB, Westerhoff HV. Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol. 2000;10:173–178. doi: 10.1016/s0962-8924(00)01741-4. [DOI] [PubMed] [Google Scholar]
- 82.Marchant DJ, Bellac CL, Moraes TJ, Wadsworth SJ, Dufour A, Butler GS, Bilawchuk LM, Hendry RG, Robertson AG, Cheung CT, Ng J, Ang L, Luo Z, Heilbron K, Norris MJ, Duan W, Bucyk T, Karpov A, Devel L, Georgiadis D, Hegele RG, Luo H, Granville DJ, Dive V, McManus BM, Overall CM. A new transcriptional role for matrix metalloproteinase-12 in antiviral immunity. Nat Med. 2014;20:493–502. doi: 10.1038/nm.3508. [DOI] [PubMed] [Google Scholar]
- 83.Remacle AG, Rozanov DV, Baciu PC, Chekanov AV, Golubkov VS, Strongin AY. The transmembrane domain is essential for the microtubular trafficking of membrane type-1 matrix metalloproteinase (MT1-MMP) J Cell Sci. 2005;118:4975–4984. doi: 10.1242/jcs.02610. [DOI] [PubMed] [Google Scholar]
- 84.Truong A, Yip C, Paye A, Blacher S, Munaut C, Deroanne C, Noel A, Sounni NE. Dynamics of internalization and recycling of the prometastatic membrane type 4 matrix metalloproteinase (MT4-MMP) in breast cancer cells. FEBS J. 2016;283:704–722. doi: 10.1111/febs.13625. [DOI] [PubMed] [Google Scholar]
- 85.Radichev IA, Remacle AG, Shiryaev SA, Purves AN, Johnson SL, Pellecchia M, Strongin AY. Biochemical characterization of the cellular glycosylphosphatidylinositol-linked membrane type-6 matrix metalloproteinase. J Biol Chem. 2010;285:16076–16086. doi: 10.1074/jbc.M110.107094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Remacle A, Murphy G, Roghi C. Membrane type I-matrix metalloproteinase (MT1-MMP) is internalised by two different pathways and is recycled to the cell surface. J Cell Sci. 2003;116:3905–3916. doi: 10.1242/jcs.00710. [DOI] [PubMed] [Google Scholar]
- 87.Yamamoto K, Okano H, Miyagawa W, Visse R, Shitomi Y, Santamaria S, Dudhia J, Troeberg L, Strickland DK, Hirohata S, Nagase H. MMP-13 is constitutively produced in human chondrocytes and co-endocytosed with ADAMTS-5 and TIMP-3 by the endocytic receptor LRP1. Matrix Biol. 2016;56:57–73. doi: 10.1016/j.matbio.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cauwe B, Opdenakker G. Intracellular substrate cleavage: a novel dimension in the biochemistry, biology and pathology of matrix metalloproteinases. Crit Rev Biochem Mol Biol. 2010;45:351–423. doi: 10.3109/10409238.2010.501783. [DOI] [PubMed] [Google Scholar]
- 89.Si-Tayeb K, Monvoisin A, Mazzocco C, Lepreux S, Decossas M, Cubel G, Taras D, Blanc JF, Robinson DR, Rosenbaum J. Matrix metalloproteinase 3 is present in the cell nucleus and is involved in apoptosis. Am J Pathol. 2006;169:1390–1401. doi: 10.2353/ajpath.2006.060005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eguchi T, Kubota S, Kawata K, Mukudai Y, Uehara J, Ohgawara T, Ibaragi S, Sasaki A, Kuboki T, Takigawa M. Novel Transcription Factor-Like Function of Human Matrix Metalloproteinase 3 Regulating the CTGF/CCN2 Gene. Mol Cell Biol. 2008;28:2391–2413. doi: 10.1128/MCB.01288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shimizu-Hirota R, Xiong W, Baxter BT, Kunkel SL, Maillard I, Chen X-W, Sabeh F, Liu R, Li X-Y, Weiss SJ. MT1-MMP regulates the PI3Kδ·Mi-2/NuRD-dependent control of macrophage immune function. Genes Dev. 2012;26:395–413. doi: 10.1101/gad.178749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marcink TC, Koppisetti RK, Fulcher YG, Van Doren SR. Mapping Lipid Bilayer Recognition Sites of Metalloproteinases and Other Prospective Peripheral Membrane Proteins. In: Galea CA, editor. Matrix Metalloproteases: Methods and Protocols. Springer New York, Place Published; 2017. pp. 61–86. [DOI] [PubMed] [Google Scholar]
- 93.Higashi S, Oeda M, Yamamoto K, Miyazaki K. Identification of amino acid residues of matrix metalloproteinase-7 essential for binding to cholesterol sulfate. J Biol Chem. 2008;283:35735–35744. doi: 10.1074/jbc.M806285200. [DOI] [PubMed] [Google Scholar]
- 94.Elias PM, Williams ML, Choi E-H, Feingold KR. Role of cholesterol sulfate in epidermal structure and function: Lessons from X-linked ichthyosis. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2014;1841:353–361. doi: 10.1016/j.bbalip.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, Koch M, Ratliff TL, Cooks RG. Cholesterol Sulfate Imaging in Human Prostate Cancer Tissue by Desorption Electrospray Ionization Mass Spectrometry. Anal Chem. 2010;82:3430–3434. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kiguchi K, Iwamori M, Yamanouchi S, Ishiwata I, Saga M, Amemiya A. Coexpression of cholesterol sulfate and cytokeratin as tumor markers in well-differentiated squamous cell carcinoma of the human uterine cervix. Clin Cancer Res. 1998;4:2985–2990. [PubMed] [Google Scholar]
- 97.van den Eijnde SM, Boshart L, Baehrecke EH, De Zeeuw CI, Reutelingsperger CPM, Vermeij-Keers C. Cell surface exposure of phosphatidylserine during apoptosis is phylogenetically conserved. Apoptosis. 1998;3:9–16. doi: 10.1023/a:1009650917818. [DOI] [PubMed] [Google Scholar]
- 98.Knight CG, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 99.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 101.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct Biol. 2007;7:44. doi: 10.1186/1472-6807-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: a computational approach. Protein Sci. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kufareva I, Lenoir M, Dancea F, Sridhar P, Raush E, Bissig C, Gruenberg J, Abagyan R, Overduin M. Discovery of novel membrane binding structures and functions. Biochem Cell Biol. 2014;92:555–563. doi: 10.1139/bcb-2014-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marcink TC, Koppisetti RK, Fulcher YG, Van Doren SR. Mapping Lipid Bilayer Recognition Sites of Metalloproteinases and other Prospective Peripheral Membrane Proteins. In: Galea CA, editor. Matrix Metalloproteases: Methods and Protocols. Springer, Place Published; 2017. in press. [DOI] [PubMed] [Google Scholar]
- 105.Houghton AM, Hartzell WO, Robbins CS, Gomis-Ruth FX, Shapiro SD. Macrophage elastase kills bacteria within murine macrophages. Nature. 2009;460:637–641. doi: 10.1038/nature08181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Geisow MJ, D’Arcy Hart P, Young MR. Temporal changes of lysosome and phagosome pH during phagolysosome formation in macrophages: studies by fluorescence spectroscopy. The Journal of Cell Biology. 1981;89:645–652. doi: 10.1083/jcb.89.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 108.Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol. 2016;23:481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schuler MA, Denisov IG, Sligar SG. Nanodiscs as a new tool to examine lipid-protein interactions. Methods Mol Biol. 2013;974:415–433. doi: 10.1007/978-1-62703-275-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, Wagner G. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods. 2017;14:49–52. doi: 10.1038/nmeth.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 112.Tatulian SA. Interfacial Enzymes: Membrane Binding, Orientation, Membrane Insertion, and Activity, Methods Enzymol. Academic Press, Place Published; [DOI] [PubMed] [Google Scholar]
- 113.Bertini I, Calderone V, Fragai M, Jaiswal R, Luchinat C, Melikian M, Mylonas E, Svergun DI. Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12. J Am Chem Soc. 2008;130:7011–7021. doi: 10.1021/ja710491y. [DOI] [PubMed] [Google Scholar]
- 114.Zhao Y, Marcink TC, Sanganna Gari RR, Marsh BP, King GM, Stawikowska R, Fields GB, Van Doren SR. Transient collagen triple helix binding to a key metalloproteinase in invasion and development. Structure. 2015;23:257–269. doi: 10.1016/j.str.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]