Abstract
Rheumatoid arthritis (RA) fibroblast-like synoviocytes (FLS) display unique aggressive behavior, invading the articular cartilage and promoting inflammation. Using an integrative analysis of RA risk alleles, the transcriptome and methylome in RA FLS, we recently identified the limb bud and heart development gene (LBH) as a key dysregulated gene in RA and other autoimmune diseases. While some evidence suggests that LBH could modulate the cell cycle, the precise mechanism is unknown and its impact on inflammation in vivo has not been defined. Our cell cycle analysis studies show that LBH deficiency in FLS leads to S phase arrest and failure to progress through the cell cycle. LBH-deficient FLS had increased DNA damage and reduced expression of the catalytic subunit of DNA polymerase α. Decreased DNA polymerase α was followed by checkpoint arrest due to phosphorylation of checkpoint kinase 1 (CHK1). Because DNA fragments can increase arthritis severity in pre-clinical models, we then explored the effect of LBH deficiency in the K/BxN serum transfer model. Lbh knockout exacerbated disease severity, which is associated with elevated levels of IL-1β and CHK1 phosphorylation. These studies indicate that LBH deficiency induces S phase arrest that, in turn, exacerbates inflammation. Because LBH gene variants are associated with type I diabetes mellitus, systemic lupus erythematosus, RA, and celiac disease, these results suggest a general mechanism that could contribute to immune-mediated diseases.
Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory disease characterized by synovial infiltration by lymphocytes and macrophages, hyperplasia of the synovial lining, and destruction of cartilage and bone [1, 2]. Current treatment strategies include the use of highly targeted biologics and, more recently, Janus kinase inhibitors. Despite improved outcomes, many patients do not respond to the available therapies and new approaches are needed [3, 4].
Fibroblast-like synoviocytes (FLS) are key players in rheumatoid synovial pathology and joint destruction through production of cytokines and extracellular matrix-degrading enzymes [5]. These synovial intimal lining cells contribute to cartilage damage and display a unique invasive phenotype. The aggressive behavior is maintained in tissue culture as well as in vivo transplantation into immune-deficient mice, indicating that RA FLS are imprinted [6-8]. New therapies that target FLS could be beneficial, but the molecular mechanisms responsible for abnormal phenotype are still poorly understood.
To address this need, we recently performed an integrative analysis of methylome, transcriptome, and sequence variations in RA FLS [9]. One of the genes identified through this analysis, namely limb bud and heart development (LBH), is a transcription cofactor that regulates cell proliferation and differentiation during development and is linked to cardiac, skeletal and mammary gland abnormalities in the partial trisomy 2p syndrome [10-16]. Several LBH polymorphisms have been associated with autoimmune diseases, including RA, celiac disease, and systemic lupus erythematosus (SLE), indicating a key role regulating immune function [17-20]. We then showed that the RA-associated single-nucleotide polymorphism (SNP) decreases LBH gene transcription, interacts with adjacent differentially methylated loci in regulatory regions, and that low levels of LBH alter the cell cycle [10, 21].
In the present study, we investigated the molecular mechanism of cell cycle regulation by LBH. We discovered that LBH deficiency causes S phase arrest by decreasing expression of the catalytic subunit of DNA polymerase α, increasing DNA damage, and activating the S phase checkpoint. Because SNPs that lead to low levels of LBH are associated with RA, we evaluated Lbh deficient mice and showed that lack of LBH leads to S phase arrest and increases inflammatory arthritis severity. These findings suggest that low LBH expression increases DNA damage, alters cell cycle kinetics, and enhances synovial inflammation.
Materials and Methods
Fibroblast-like synoviocytes and culture conditions
This study was approved by the Institutional Review Board of University of California, San Diego School of Medicine, and informed consent was obtained from all participants. Synovial tissue was obtained from patients with osteoarthritis (OA) and RA at the time of total joint replacement or synovectomy, as previously described. The diagnosis of RA conformed to American College of Rheumatology 1987 or 2010 criteria [22, 23]. Enzymatic disaggregation of synovium was performed as previously described [21]. Cells were allowed to adhere overnight and non-adherent cells were removed. Adherent FLS were split at 1:3 when 70–80% confluent and used from passages 4 through 7, when they are a homogeneous population of fibroblasts. Primary FLS were cultured (at 5% CO2, 37°C) in Dulbecco's modified Eagle's medium (DMEM) supplemented with L-glutamine, gentamicin, penicillin/streptomycin, and 10% heat-inactivated fetal bovine serum (FBS) [24]. For synchronization experiments, cells were serum starved for 2 days and transfected with siRNA in DMEM containing 1% FBS and supplements, followed by addition of 10% FBS.
Antibodies and reagents
The antibodies and reagents included: anti-phosphorylated CHK1 (S345), anti-CHK1, anti-phosphorylated CHK2 (T68), anti-CHK2, anti–phosphorylated CDK2 (T160), anti-POLA1, anti-Cyclin D1, anti-Cyclin E1 (Cell Signaling Technology), anti-CDK2, anti-Cyclin A, anti-GAPDH (Santa Cruz Biotechnology), anti-Rb (BD Pharmingen), anti-LBH or anti–β-actin antibodies (Sigma), and horseradish peroxidase (HRP)–conjugated goat anti-rabbit or mice IgG (Cell Signaling Technology) as secondary antibody. Hydroxyurea was purchased from Sigma.
Cell cycle analysis
RA FLS were washed and re-suspended in cold PBS, fixed in 70% ethanol, permeabilized, and stained with propidium iodide/0.1% Triton X-100 solution supplemented with RNase A. The cells were analyzed by flow cytometry using a LSR II Flow Cytometer (BD Biosciences). Data were analyzed using FlowJo software (Tree Star) and Watson Pragmatic algorithm, and percentages of cells in the S phases were calculated. EdU assay was performed using Click-iT plus EdU Alexa Fluor 488 Flow cytometry assay kit and FxCycle violet (Thermo Fisher) according to manufacturer's instructions.
Western blot analysis
Western blot analysis was performed as previously described [25]. Protein was extracted from human FLS using lysis buffer (150 mM NaCl, 50 mM Tris, 1% Triton X-100, 100 mM NaF, 100 mM β-Glycerophosphate, protease inhibitor cocktail [Roche], and phosphatase inhibitor cocktail II and III [Sigma]). Blots were developed using an Immun-Star WesternC Chemiluminescence kit and analyzed using a VersaDoc imaging system and Quantity One software (Bio-Rad).
Gene knockdown
Gene knockdown was performed as previously described using the Amaxa system [10]. Although siRNA could have off-target effects, we used scrambled siRNA as a control. In addition, our previous transcriptome studies showed that the only gene to be consistently and significantly suppressed across all cell lines by LBH siRNA was LBH [10]. siRNA for LBH and control siRNA (ON-TARGET plus nontargeting pool) were purchased from Dharmacon. siRNAs (400 nM) were introduced into cultured cells using the Human Dermal Fibroblast Nucleofector Kit (Lonza).
Quantitative Real time PCR (qPCR)
Total RNA was isolated from FLS using RNASTAT-60 and reverse transcribed (Applied Biosystems, Foster City, CA). The cDNA served as template for amplification by qPCR using TaqMan Gene Expression assays (StepOnePlus Instruments, Applied Biosystems, Foster City, CA). The Ct values were normalized to ACTB or 18S as previously described [26].
Alkaline Comet Assay
Cells were prepared by mixing 4 × 105 cells with 1 mL of LMAgarose. The cell suspension was layered onto a glass microscope slide, and then the slide was placed in lysis buffer at 4 °C overnight. Electrophoresis was performed at 1 V/cm in electrophoresis buffer for 40 min. Thereafter, the slide was rinsed with water and 70% ethanol and stained with SYBR Gold for 30 min. The DNA damage levels were analyzed, and the slides were imaged using a fluorescence microscope. The comet assay is used to determine the “tail moment” measures of the smallest detectable size of migrating DNA (i.e., the “comet” tail length) and quantifies the number of fragments (i.e., the intensity of DNA in the tail). The Olive Tail Moment is calculated by measuring the [ (Tail mean) – (Head mean) × percentage of DNA in the Tail] / 100.
K/BxN Serum Transfer Model of Arthritis
Generation and phenotype of Rosa-26-Cre; Lbhloxp/loxp mice was previously described [27]. Rosa-26-Cre; Lbh+/+ and matched Rosa-26-Cre; Lbhloxp/loxp littermate mice were injected i.p. with 80 μl of pooled adult K/BxN mouse serum on days 0 and 2. Arthritis was evaluated for each paw as described in [28]. For tissue protein assays, snap-frozen ankle joints were homogenized in lysis buffer and assayed by western blotting. The other hind paws were fixed in 10% formalin and decalcified in EDTA. For tissue qPCR assays, RNA was isolated from snap-frozen ankle joints by RNeasy Lipid Tissue Mini kit (QIAGEN) and quantitative real-time PCR was performed.
Statistics
The 2-way ANOVA and 2-tailed paired or unpaired t test were performed using GraphPad Prism software. A comparison was considered significant if p was less than 0.05.
Results
LBH deficiency blocks S phase progression
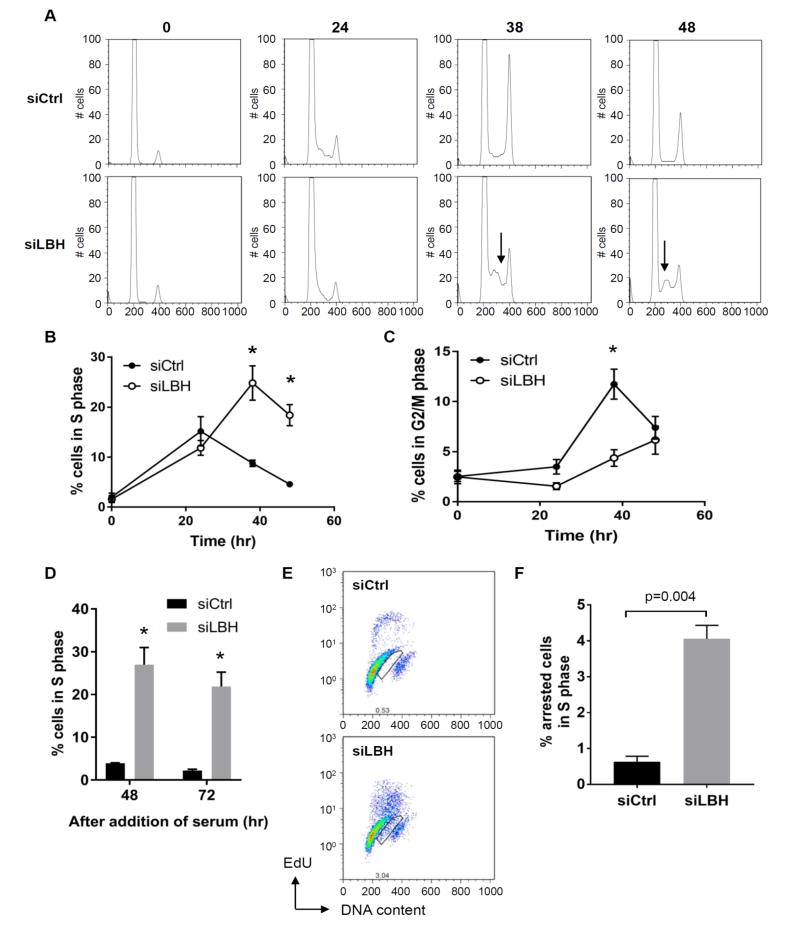
To define the role of LBH in the cell cycle, we evaluated the effect of LBH deficiency on cell cycle progression by flow cytometry. siRNA-mediated LBH deficiency (LBHlow) was induced and cell cycle analysis was performed on synchronized FLS using additional methods and more detailed kinetics than previously reported. Based on flow cytometry studies to evaluate DNA content, control FLS progressed past the G1 checkpoint into S phase and then rapidly entered G2 when cultured in serum (Figure 1, A, B and C). In contrast, LBH deficient cells remained in S phase, with delayed progression through the cell cycle (Figure 1, A and B). The effect of LBH deficiency on the cell cycle persisted with S phase arrest noted for at least 72 hr, although cells slowly entered G2/M towards the end of the culture period (Figure 1C and D). These data suggest that LBH is required for efficient progression through the S phase. To confirm the cell cycle data with a second technique, FLS were labeled with the thymidine analog EdU to quantify DNA synthesis in control and LBHlow cells. LBH deficiency increased population of non-replicating (EdU-negative) cells with DNA content corresponding to S phase after 48 hr (Figure 1, E and F).
Figure 1. S phase arrest induced by LBH deficiency.
RA FLS were serum starved for 2 days and transfected with siRNA, followed by addition of 10% FBS to release cells into S phase, and at time intervals afterwards, the cells were analyzed. (A) Representative histograms of DNA content in RA FLS lines transfected with siCtrl and siLBH. Arrows show the increased cells in S phase. (B, C) The percentage of cells in S and G2/M phase. Values are averages of 4 different RA FLS lines. (D) The percentage of cells in S phase. Values are averages of 3 different RA FLS lines (E) Representative flow cytometry profiles of control and LBH-knockdown cells. Box regions indicate S-phase arrested cells. (F) Quantification of S phase arrested cells. The average of 4 different RA FLS lines is shown. Values are the mean ± SEM. Data were analyzed using the 2-tailed paired t test.
LBH regulation of cell cycle proteins
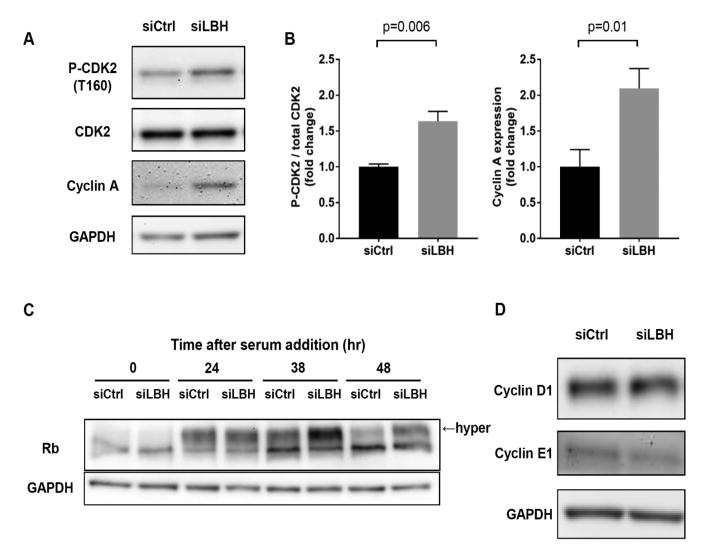
Western blot analysis was then performed to determine the mechanism of LBH-mediated cell cycle regulation. The initial experiments evaluated the effect of LBH deficiency on CDK2 and demonstrated increased phosphorylation at the T160 site (Figure 2, A and B). Cyclin A, which activates CDK2 during S phase, was increased in LBHlow cells compared with control (Figure 2, A and B). Finally, we evaluated the effect of LBH deficiency on Retinoblastoma protein (Rb), which is a substrate for CDK2-cyclin A or cyclin E complex. As shown in Figure 2, Rb in LBHlow cells was hyper-phosphorylated at 38 and 48 hr compared with control cells that had progressed through S, G2 and M phases (Figure 2C; p<0.004 comparing LBHlow to control cells). We did not find a significant effect on cyclin D1 and E1 expression in LBHlow cells (Figure 2D).
Figure 2. Effects of LBH deficiency on cell cycle proteins.
(A, B) Western blots for phosphorylated CDK2 (T160) and total levels of cyclin A 48 hr after serum addition. Data are presented as the fold increase in the level of phosphorylated CDK2 and cyclin A expression, normalized to siCtrl treated cells. Values are the mean ± SEM from 5 different cell lines. Data were analyzed using the 2-tailed paired t test. (C) Western blot for Rb at the indicated time after serum addition. Arrow indicates hyper-phosphorylated form of Rb (n=4; p<0.004). (D) Expression of cyclin D1 and E1 in RA FLS transfected with siCtrl and siLBH, assessed by Western blots, normalized to β–actin. Values are the mean ± SEM from 3 different cell lines.
LBH deficiency increases accumulation of DNA damage
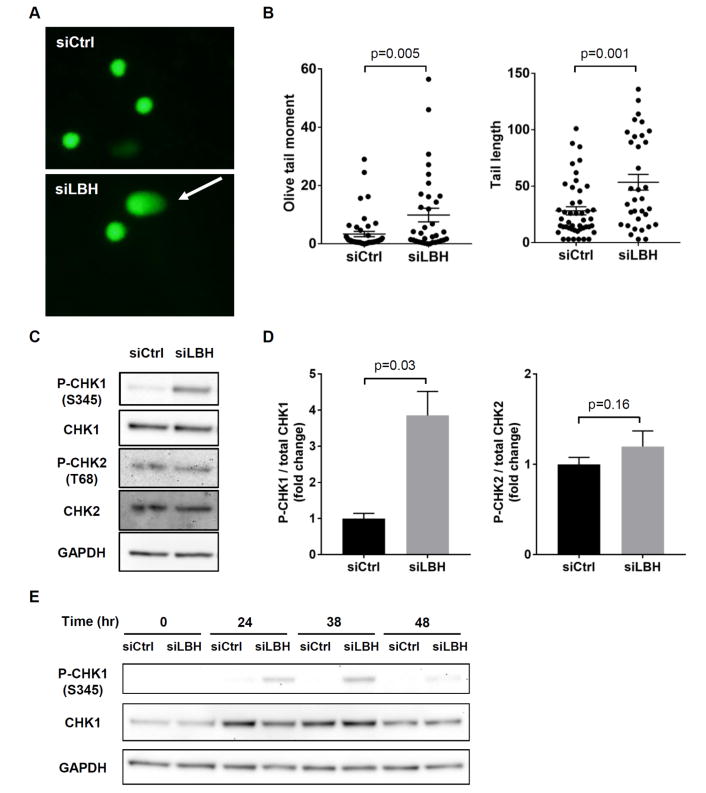
S phase arrest is often due to DNA replication stress and damage. To address this possibility, we explored whether LBH deficiency allows DNA damage to accumulate. We used a comet assay that quantifies broken DNA fragments or damaged DNA migration away from the cell nucleus (see Methods). As shown in Figure 3A, LBHlow FLS had comets with longer tails, while control cells had relatively shorter tails. There was a significant increase in the olive tail moment and tail length in LBHlow cells, indicating a significant increase in the amount of DNA damage in LBHlow cells compared to control (Figure 3B).
Figure 3. Activation of S phase checkpoint in LBHlow cells.
(A) Representative images of the alkaline comet assay at 48hr after serum addition. The cell looks like a comet with a distinct head consisting of intact DNA, and a tail that contains damaged or broken pieces of DNA shown by arrow. (B) A significant increase was observed in olive tail moment and tail length in LBHlow cells compared with control. Each symbol represents a single subject, and the mean ± SEM are also shown. (C, D) Western blots for CHK1 (S345) and CHK2 (T68) phosphorylation. Bars represent the average of 5 different cell lines. Values are the mean ± SEM. Data were analyzed using the 2-tailed paired t test. (E) Western blots for CHK1 (S345) at the indicated time after serum addition.
LBH deficiency activates S phase checkpoint
Evidence of S phase arrest and DNA damage in LBHlow cells led us to assess the activation status of the intra-S phase checkpoint that allows DNA repair and prevents propagation of mutated or fragmented DNA. The checkpoint kinases, CHK1 and CHK2, are activated in response to DNA damage or stalled replication forks during S phase. Figure 3C shows that CHK1 phosphorylation increased four-fold greater in LBH deficient cells than control cells within 24 hr. Phospho-CHK1 accumulation occurred before the increase in the percentage of cells in S phase was detected in LBHlow cells (Figure 3, C-E). However, CHK2 was not activated (Figure 3, C and D).
LBH deficiency delays replication by decreasing DNA polymerase α
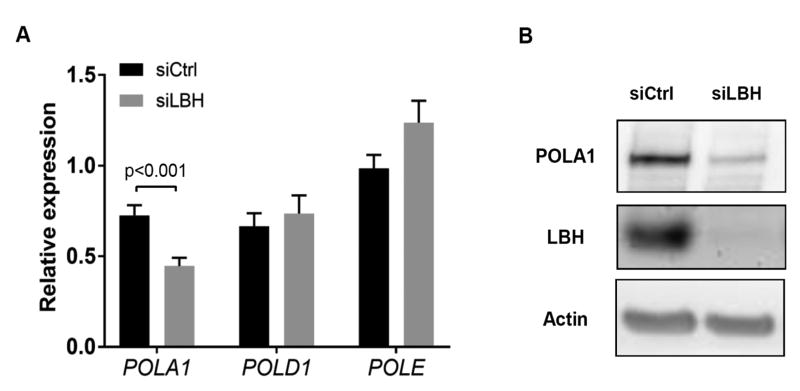
Three main polymerases are responsible for repair and propagation in S phase and were then assayed in control and LBHlow cells. LBH deficiency reduced POLA1 mRNA by nearly 40% (Figure 4A) and POLA1 protein by 86% (p<0.01) compared with control (Figure 4B). However, POLD1 and POLE were not affected by LBH silencing (Figure 4A). We then determined whether POLA1 regulation is specific for DNA damage associated with LBH deficiency or whether general S phase checkpoint activation can participate. FLS were exposed to hydroxyurea (HU), which is a DNA replication inhibitor that decreases the elongation and initiation phases of replication and triggers the intra-S phase checkpoint. The percentage of cells in S phase decreased from 16% to 4% in control cells, while the percentage of HU-treated FLS in S phase increased from 2% to 22% from 24 hr to 38 hr (suppl Fig. 1A). HU-treatment significantly increased CHK1 phosphorylation (suppl Fig. 1B). However, HU-treatment did not reduce POLA1 (suppl Fig. 1C).
Figure 4. Effects of LBH deficiency on DNA polymerases expression.
(A) Expression of POLA1, POLD1 and POLE mRNA. Ct values were normalized to actin expression. Values are the mean ± SEM of 7 different FLS lines, assessed by qPCR. Data were analyzed using the 2-tailed paired t test. (B) Expression of POLA1 in RA FLS transfected with siCtrl and LBH siRNA, assessed by Western blot analysis.
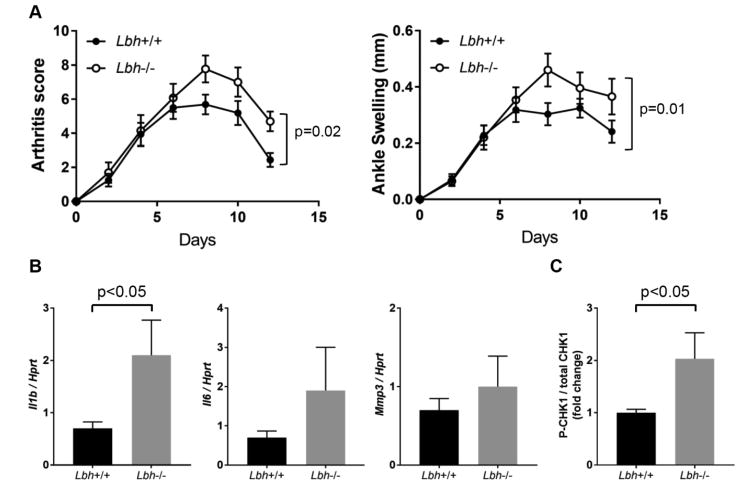
LBH deficiency increases arthritis severity
Low LBH expression and functional SNPs decrease LBH expression are associated with RA [17-21]. Based on those studies and our data indicating that LBH regulates the cell cycle, we hypothesized that LBH deficiency would exacerbate inflammatory arthritis. To examine the contribution of LBH in the pathogenesis of RA, passive K/BxN arthritis was studied in wild type (WT) and LBH deficient mice [27]. Arthritis severity was significantly increased in the Lbh-/- mice, primarily in the later stages of the model (Figure 5A). In addition, articular IL-1β gene expression was significantly higher in the joints of the Lbh-/- mice with arthritis compared to WT (Lbh+/+). (Figure 5B). There was a trend toward increased expression of IL-6 and MMP-3 in Lbh-/- mice compared with WT that did not reach statistical significance. To determine whether S phase arrest occurred in the arthritic mice, western blot analysis was performed to quantify phospho-CHK1 levels. There was a significant increase in ankle CHK1 phosphorylation in Lbh-/- mice compared with WT (Figure 5C). These results confirm that LBH deficiency led to exacerbation of joint inflammation and that DNA damage and S phase checkpoint activation occur in vivo.
Figure 5. Effect of LBH deficiency on passive K/BxN arthritis in vivo.
(A) At 7 to 16 weeks of age, male Rosa26Cre-Lbh+/+ (Lbh+/+, n = 16) and Rosa26Cre-Lbhloxp/loxp (Lbh-/-, n = 13) mice were injected i.p. with K/BxN sera to induce arthritis on days 0 and 2. Clinical score and ankle swelling was measured every other day after serum transfer until day 12. Mean ± SEM is shown. Data were analyzed using 2-way ANOVA. (B) Relative expression of mRNA for Il1β, Il6, Mmp3 genes. Lbh+/+ (n = 7) and Lbh-/- (n = 6) mice were administered K/BxN serum, and after 12 days, the ankle joints were harvested and homogenized, and mRNA expression was analyzed by quantitative polymerase chain reaction. Values are mean ± SEM. Data were analyzed using the 2-tailed unpaired t test. (C) Western blots for phospho-CHK1 (S345) and total CHK1 in ankle joints of Lbh+/+ (n = 7) and Lbh-/- (n = 6) mice. Bars represent the average. Values are the mean ± SEM. Data were analyzed using the 2-tailed unpaired t test.
Discussion
RA FLS display an aggressive phenotype and invade into the articular extracellular matrix. However, no current therapies target this cell lineage in inflammatory arthritis. To address this unmet need, we previously integrated RA risk alleles, differentially methylated genes and differentially expressed genes to identify key pathogenic pathways in RA [10]. LBH, a highly conserved transcription co-regulator emerged from this analysis and particularly interesting because it is associated with several autoimmune diseases, including SLE, type I diabetes, RA and celiac disease. Subsequent studies showed that a causative mutation in RA is located in an upstream enhancer and decreases LBH gene transcription. Furthermore, this SNP occurs adjacent to a nearby differentially methylated locus in RA. We hypothesized that the disease-associated polymorphism in regulatory regions decrease LBH expression and contribute to RA pathogenesis.
The precise function of LBH, especially in immune-mediated diseases, has been elusive. Forced expression of LBH in embryos increases cardiomyocyte growth but blocks cell differentiation and alters bone formation [11, 14]. The phenotype of LBH knockout mice is relatively mild, with reduced breast epithelial progenitor cell proliferation [16]. LBH has been implicated in Wnt signaling [29] and possibly MAP kinase signaling [13, 30]. However, we did not observe these effects in RA FLS [10]. Our recent functional studies showed that LBH expression in FLS is induced by platelet-derived growth factor (PDGF) and that low levels of LBH expression alters cell cycle dynamics. However, the precise impact on cell proliferation was not defined. In the present study, we explored the mechanisms of cell cycle regulation by LBH and its impact on genomic integrity.
Our initial interpretation of cell cycle data suggested that LBH can potentially rapid progression through the G1 checkpoint. More detailed kinetic analysis confirmed normal progression through G1, but also showed that cells accumulate in S phase in LBH-deficient cells. Delay in the cell cycle was associated with Rb hyper-phosphorylation, CDK2 activation and elevated cyclin A expression. Recent evidence suggests that cyclin A-CDK2 complex activation is important for drug-induced S phase arrest and that hyper-phosphorylated Rb is associated with an accelerated S phase entry [31, 32]. However, we observed that the cyclin A-CDK2 complex is activated in LBHlow cells after S phase arrest occurs, which implicates alternative mechanisms.
The lack of effect on cyclins differs from the previous reports indicating that low LBH promotes cyclin D1 and E1 expression in nasopharyngeal cancer cells, leading to G1-S transition [33]. In contrast to transformed cells, LBH silencing did not affect these cyclins in FLS. The differences could be due to culture conditions, variations resulting from the biology of cell lineages, or, most likely, the role of LBH in primary non-transformed cells compared with prior studies using malignant cells.
Subsequent studies were designed to understand how LBH regulates S phase arrest in FLS. Previous studies in human cancer cell lines showed that DNA damage activates the S phase checkpoint, which enable DNA repair [34-37]. To test this hypothesis, we showed that LBH deficiency in FLS leads to accumulation of DNA damage followed by CHK1 phosphorylation and S phase checkpoint activation. These observations suggest that LBH could play a role in the genomic surveillance and that low levels prevent cells with damaged DNA from progressing through the cell cycle.
Accumulation of DNA damage in LBH deficient cells led us to evaluate whether DNA synthesis is impaired. LBH deficiency selectively reduced POLA1 expression, but not other DNA polymerases. The effect was selective for endogenous DNA damage because it was observed when S phase check point activation was initiated by hydroxyurea. POLA1 initiates de novo DNA synthesis at replication forks and discontinuously replicates DNA during lagging strand DNA synthesis [38, 39]. The other DNA polymerases appear to replicate lagging and leading DNA synthesis. Limiting lagging strand DNA synthesis due to LBH deficiency might cause the replication fork to stall, leading to fork collapse, which is consistent with activation of CHK1 but not CHK2 [34]. This result is also consistent with reports that POLA1 is a component of the signaling that activates S phase checkpoint through CHK1 activation [40-42]. Several studies have demonstrated that LBH may be a transcriptional cofactor, although the transcription factor partners are not known [11-13].
Because low LBH is associated with RA and altered cell cycle dynamics, we explored whether LBH deficiency promote inflammatory in a serum transfer arthritis model. This hypothesis was supported by previous studies showing that DNA fragment accumulation increases arthritis severity [43]. LBH deficiency in this model increased arthritis severity, especially in the later stages, and was associated with higher IL-1β expression. The effect is relatively modest, but it is reproducible and statistically significant. The degree of synovitis is similar to other target genes that increase arthritis severity, including p53, p38, and Bax/Bim [44-46]. Given the relatively modest effect of LBH SNPs on the odds ratio for autoimmune disease susceptibility, it is not surprising that the effect is subtle. Increased CHK1 phosphorylation was observed and suggests that S phase arrest occurs in vivo and exacerbates inflammation. It is also interesting to speculate that persistent DNA damage caused by low LBH might contribute to accumulation of somatic mutations in RA as described by several groups [47-50].
The K/BxN serum transfer arthritis model is dependent on FLS and innate immunity, especially IL-1β [51, 52]. The model also involves other cells that contribute to arthritis progression, including macrophages, neutrophils, and osteoclasts, Understanding the relative role FLS will involve future studies on conditional knockout mice when they are available.
In conclusion, we identified LBH as a critical gene that regulates immune-mediated diseases. The mechanism of action is related to altered cell cycle through accumulation of DNA damage accumulation and S phase arrest in the presence of continued cyclin A-CDK2 activity. The data suggest that the LBH pathway participates in RA and could be mined for potential new therapeutic targets.
Supplementary Material
Acknowledgments
Research supported, in part, by Rheumatology Research Foundation (Disease Targeted Research), NIH (NIAMS) R01AR065466 (GSF), NIH RO1 GM113256 (KB)
Abbreviations
- LBH
Limb Bud and Heart development gene
- RA
rheumatoid arthritis
- FLS
fibroblast-like synoviocytes
- CHK1
checkpoint 1
- siRNA
small interfering RNA
- CDK
cyclin-dependent kinase
- POL
DNA polymerase
Footnotes
Disclosures: The authors have declared that no conflict of interest exists.
- Designing research studies: SM, DH, DB, SD, WW, GSF
- Conducting experiments: SM, DH, KT
- Acquiring data: SM, DH, KT
- Analyzing data: SM, DH, KT
- Providing critical reagents: KB
- Writing the manuscript: SM, DH, KT, KB, DB, SD, WW, GSF
References
- 1.van Vollenhoven RF. Rheumatoid arthritis in 2012: Progress in RA genetics, pathology and therapy. Nature reviews Rheumatology. 2013;9(2):70–72. doi: 10.1038/nrrheum.2012.232. [DOI] [PubMed] [Google Scholar]
- 2.Catrina AI, Joshua V, Klareskog L, Malmstrom V. Mechanisms involved in triggering rheumatoid arthritis. Immunological reviews. 2016;269(1):162–174. doi: 10.1111/imr.12379. [DOI] [PubMed] [Google Scholar]
- 3.Stanczyk J, Ospelt C, Gay S. Is there a future for small molecule drugs in the treatment of rheumatic diseases? Current opinion in rheumatology. 2008;20(3):257–262. doi: 10.1097/BOR.0b013e3282fa13ee. [DOI] [PubMed] [Google Scholar]
- 4.Orme ME, Macgilchrist KS, Mitchell S, Spurden D, Bird A. Systematic review and network meta-analysis of combination and monotherapy treatments in disease-modifying antirheumatic drug-experienced patients with rheumatoid arthritis: analysis of American College of Rheumatology criteria scores 20, 50, and 70. Biologics : targets & therapy. 2012;6:429–464. doi: 10.2147/BTT.S36707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottini N, Firestein GS. Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nature reviews Rheumatology. 2013;9(1):24–33. doi: 10.1038/nrrheum.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller-Ladner U, Kriegsmann J, Franklin BN, Matsumoto S, Geiler T, Gay RE, Gay S. Synovial fibroblasts of patients with rheumatoid arthritis attach to and invade normal human cartilage when engrafted into SCID mice. The American journal of pathology. 1996;149(5):1607–1615. [PMC free article] [PubMed] [Google Scholar]
- 7.Steenvoorden MM, Tolboom TC, van der Pluijm G, Lowik C, Visser CP, DeGroot J, Gittenberger-DeGroot AC, DeRuiter MC, Wisse BJ, Huizinga TW, et al. Transition of healthy to diseased synovial tissue in rheumatoid arthritis is associated with gain of mesenchymal/fibrotic characteristics. Arthritis research & therapy. 2006;8(6):R165. doi: 10.1186/ar2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefevre S, Knedla A, Tennie C, Kampmann A, Wunrau C, Dinser R, Korb A, Schnaker EM, Tarner IH, Robbins PD, et al. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nature medicine. 2009;15(12):1414–1420. doi: 10.1038/nm.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitaker JW, Shoemaker R, Boyle DL, Hillman J, Anderson D, Wang W, Firestein GS. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome medicine. 2013;5(4):40. doi: 10.1186/gm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekwall AK, Whitaker JW, Hammaker D, Bugbee WD, Wang W, Firestein GS. The Rheumatoid Arthritis Risk Gene LBH Regulates Growth in Fibroblast-like Synoviocytes. Arthritis & rheumatology. 2015;67(5):1193–1202. doi: 10.1002/art.39060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briegel KJ, Baldwin HS, Epstein JA, Joyner AL. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor Lbh. Development. 2005;132(14):3305–3316. doi: 10.1242/dev.01887. [DOI] [PubMed] [Google Scholar]
- 12.Briegel KJ, Joyner AL. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Developmental biology. 2001;233(2):291–304. doi: 10.1006/dbio.2001.0225. [DOI] [PubMed] [Google Scholar]
- 13.Ai J, Wang Y, Tan K, Deng Y, Luo N, Yuan W, Wang Z, Li Y, Wang Y, Mo X, et al. A human homolog of mouse Lbh gene, hLBH, expresses in heart and activates SRE and AP-1 mediated MAPK signaling pathway. Molecular biology reports. 2008;35(2):179–187. doi: 10.1007/s11033-007-9068-4. [DOI] [PubMed] [Google Scholar]
- 14.Conen KL, Nishimori S, Provot S, Kronenberg HM. The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development. Developmental biology. 2009;333(2):348–358. doi: 10.1016/j.ydbio.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Ali H, Rieger ME, Seldeen KL, Harris TK, Farooq A, Briegel KJ. Biophysical characterization reveals structural disorder in the developmental transcriptional regulator LBH. Biochemical and biophysical research communications. 2010;391(1):1104–1109. doi: 10.1016/j.bbrc.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindley LE, Curtis KM, Sanchez-Mejias A, Rieger ME, Robbins DJ, Briegel KJ. The WNT-controlled transcriptional regulator LBH is required for mammary stem cell expansion and maintenance of the basal lineage. Development. 2015;142(5):893–904. doi: 10.1242/dev.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu ZY, Lu WS, Zuo XB, Hu J, Yao S, Li Y, Han JW, Sun LD, Cheng YL, Xu Q, et al. One novel susceptibility locus associate with systemic lupus erythematosus in Chinese Han population. Rheumatology international. 2013;33(8):2079–2083. doi: 10.1007/s00296-013-2697-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, Westra HJ, Fehrmann RS, Kurreeman FA, Thomson B, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS genetics. 2011;7(2):e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y, Sheng Y, Cheng Y, Lin Y, Zhu Z, Wen L, Yang C, Yang L, Liu L, Zhou F, et al. Downregulated expression of LBH mRNA in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. The Journal of dermatology. 2016;43(1):99–102. doi: 10.1111/1346-8138.13006. [DOI] [PubMed] [Google Scholar]
- 20.Kim K, Bang SY, Lee HS, Cho SK, Choi CB, Sung YK, Kim TH, Jun JB, Yoo DH, Kang YM, et al. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Annals of the rheumatic diseases. 2015;74(3):e13. doi: 10.1136/annrheumdis-2013-204749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammaker D, Whitaker JW, Maeshima K, Boyle DL, Ekwall AH, Wang W, Firestein GS. LBH Gene Transcription Regulation by the Interplay of an Enhancer Risk Allele and DNA Methylation in Rheumatoid Arthritis. Arthritis & rheumatology. 2016;68(11):2637–2645. doi: 10.1002/art.39746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis and rheumatism. 1988;31(3):315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 23.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, 3rd, Birnbaum NS, Burmester GR, Bykerk VP, Cohen MD, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis and rheumatism. 2010;62(9):2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 24.Rosengren S, Boyle DL, Firestein GS. Acquisition, culture, and phenotyping of synovial fibroblasts. Methods in molecular medicine. 2007;135:365–375. doi: 10.1007/978-1-59745-401-8_24. [DOI] [PubMed] [Google Scholar]
- 25.Svensson CI, Inoue T, Hammaker D, Fukushima A, Papa S, Franzoso G, Schett G, Corr M, Boyle DL, Firestein GS. Gadd45beta deficiency in rheumatoid arthritis: enhanced synovitis through JNK signaling. Arthritis and rheumatism. 2009;60(11):3229–3240. doi: 10.1002/art.24887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle DL, Rosengren S, Bugbee W, Kavanaugh A, Firestein GS. Quantitative biomarker analysis of synovial gene expression by real-time PCR. Arthritis research & therapy. 2003;5(6):R352–360. doi: 10.1186/ar1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindley LE, Briegel KJ. Generation of mice with a conditional Lbh null allele. Genesis. 2013;51(7):491–497. doi: 10.1002/dvg.22390. [DOI] [PubMed] [Google Scholar]
- 28.Simelyte E, Rosengren S, Boyle DL, Corr M, Green DR, Firestein GS. Regulation of arthritis by p53: critical role of adaptive immunity. Arthritis and rheumatism. 2005;52(6):1876–1884. doi: 10.1002/art.21099. [DOI] [PubMed] [Google Scholar]
- 29.Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Molecular and cellular biology. 2010;30(17):4267–4279. doi: 10.1128/MCB.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tufegdzic Vidakovic A, Rueda OM, Vervoort SJ, Sati Batra A, Goldgraben MA, Uribe-Lewis S, Greenwood W, Coffer PJ, Bruna A, Caldas C. Context-Specific Effects of TGF-beta/SMAD3 in Cancer Are Modulated by the Epigenome. Cell reports. 2015;13(11):2480–2490. doi: 10.1016/j.celrep.2015.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding H, Han C, Guo D, Wang D, Chen CS, D’Ambrosio SM. OSU03012 activates Erk1/2 and Cdks leading to the accumulation of cells in the S-phase and apoptosis. International journal of cancer. 2008;123(12):2923–2930. doi: 10.1002/ijc.23896. [DOI] [PubMed] [Google Scholar]
- 32.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. eLife. 2014:3. doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q, Guan X, Lv J, Li X, Wang Y, Li L. Limb-bud and Heart (LBH) functions as a tumor suppressor of nasopharyngeal carcinoma by inducing G1/S cell cycle arrest. Scientific reports. 2015;5:7626. doi: 10.1038/srep07626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrett MD, Collins I. Anticancer therapy with checkpoint inhibitors: what, where and when? Trends in pharmacological sciences. 2011;32(5):308–316. doi: 10.1016/j.tips.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 35.Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Current opinion in cell biology. 2001;13(2):225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 37.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annual review of biochemistry. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 39.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annual review of biochemistry. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 40.Bhaumik D, Wang TS. Mutational effect of fission yeast polalpha on cell cycle events. Molecular biology of the cell. 1998;9(8):2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taricani L, Shanahan F, Parry D. Replication stress activates DNA polymerase alpha-associated Chk1. Cell cycle. 2009;8(3):482–489. doi: 10.4161/cc.8.3.7661. [DOI] [PubMed] [Google Scholar]
- 42.Chattopadhyay S, Bielinsky AK. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Molecular biology of the cell. 2007;18(10):4085–4095. doi: 10.1091/mbc.E06-12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, Nagata S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443(7114):998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 44.Guma M, Hammaker D, Topolewski K, Corr M, Boyle DL, Karin M, Firestein GS. Antiinflammatory functions of p38 in mouse models of rheumatoid arthritis: advantages of targeting upstream kinases MKK-3 or MKK-6. Arthritis and rheumatism. 2012;64(9):2887–2895. doi: 10.1002/art.34489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scatizzi JC, Bickel E, Hutcheson J, Haines GK, 3rd, Perlman H. Bim deficiency leads to exacerbation and prolongation of joint inflammation in experimental arthritis. Arthritis and rheumatism. 2006;54(10):3182–3193. doi: 10.1002/art.22133. [DOI] [PubMed] [Google Scholar]
- 46.Yamanishi Y, Boyle DL, Pinkoski MJ, Mahboubi A, Lin T, Han Z, Zvaifler NJ, Green DR, Firestein GS. Regulation of joint destruction and inflammation by p53 in collagen-induced arthritis. The American journal of pathology. 2002;160(1):123–130. doi: 10.1016/S0002-9440(10)64356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Firestein GS, Echeverri F, Yeo M, Zvaifler NJ, Green DR. Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(20):10895–10900. doi: 10.1073/pnas.94.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inazuka M, Tahira T, Horiuchi T, Harashima S, Sawabe T, Kondo M, Miyahara H, Hayashi K. Analysis of p53 tumour suppressor gene somatic mutations in rheumatoid arthritis synovium. Rheumatology (Oxford, England) 2000;39(3):262–266. doi: 10.1093/rheumatology/39.3.262. [DOI] [PubMed] [Google Scholar]
- 49.Roivainen A, Jalava J, Pirila L, Yli-Jama T, Tiusanen H, Toivanen P. H-ras oncogene point mutations in arthritic synovium. Arthritis and rheumatism. 1997;40(9):1636–1643. doi: 10.1002/art.1780400913. [DOI] [PubMed] [Google Scholar]
- 50.Cannons JL, Karsh J, Birnboim HC, Goldstein R. HPRT- mutant T cells in the peripheral blood and synovial tissue of patients with rheumatoid arthritis. Arthritis and rheumatism. 1998;41(10):1772–1782. doi: 10.1002/1529-0131(199810)41:10<1772::AID-ART9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 51.Ji H, Pettit A, Ohmura K, Ortiz-Lopez A, Duchatelle V, Degott C, Gravallese E, Mathis D, Benoist C. Critical Roles for Interleukin 1 and Tumor Necrosis Factor α in Antibody-induced Arthritis. The Journal of Experimental Medicine. 2002;196(1):77–85. doi: 10.1084/jem.20020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee DM, Kiener HP, Agarwal SK, Noss EH, Watts GF, Chisaka O, Takeichi M, Brenner MB. Cadherin-11 in synovial lining formation and pathology in arthritis. Science (New York, NY) 2007;315(5814):1006–1010. doi: 10.1126/science.1137306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.