Abstract
This paper provides an overview of the discussion and presentations from the Workshop on the Management of Large CryoEM Facilities held at the New York Structural Biology Center, New York, NY on February 6–7, 2017. A major objective of the workshop was to discuss best practices for managing cryoEM facilities. The discussions were largely focused on supporting single-particle methods for cryoEM and topics included: user access, assessing projects, workflow, sample handling, microscopy, data management and processing, and user training.
Keywords: CryoTEM, automation, facility management, user training, single-particle, data management
Introduction
The cryoEM field has rapidly expanded over the past several years and there is an increasingly large demand for access to high resolution cryogenic transmission electron microscopes (cryoEM) equipped with direct detector cameras (Kuhlbrandt, 2014) (Elmlund et al., 2017). These instruments are expensive and complex to manage and maintain. In addition, producing cryoEM structures from these microscopes requires equipment for sample preparation and storage, and access to high performance computing capabilities. Currently, there are very few papers that discuss the management and operation of cryoEM facilities. The paper by Saibil et al. provides a good overview of the plans for the eBIC facility at the Diamond Light Source but does not go into the details on the management and the operation (Saibil et al., 2015). While there are several papers on managing and running core facilities for advanced light microscopes (Ferrando-May et al., 2016) (Trogadis, 2006) (DeMaggio, 2002) these are not generally applicable to setting up, managing and running a high resolution cryoEM facility.
A Workshop on the Management of Large CryoEM Facilities was organized by the National Resource for Automated Molecular Microscopy (NRAMM) and held at the Simons Electron Microscopy Center, New York Structural Biology Center, New York, NY on February 6–7, 2017. Twenty-four participants from 19 institutions attended the meeting and are co-authors of this paper. Most of the participants are actively engaged in the management and operation of multi-user group cryoEM facilities. Tables 1 and S1, show the wide range of institutions and services that were represented. These included large institutions, such as eBIC at Diamond Light Source and the New York Structural Biology Center, that support many users and multiple microscopes, as well as smaller facilities, such as EMBL Heidelberg, that primarily serve a more limited user community.
Table 1.
| Institution | # Active users per year | # Active projects | High End TEMS | # other TEMS | # Support Staff | Facility Website |
|---|---|---|---|---|---|---|
| Birckbeck College Institute of Structural and Molecular Biology | 30 | 30 | 1 Polara, 1 Krios (installed in 2018) | 3 | 2 EM specialists, 1 computer specialist | http://people.cryst.bbk.ac.uk/~ubcg16z/empage.html |
| California Institute of Technology | N/A | N/A | 1 Polara, 1 Krios | 2 | 2 | N/A |
| California NanoSystems Institute | 154 | 70 | 1 Krios and 1 low-base Titan S/TEM | 6 | 3 | http://www.EICN.ucla.edu |
| eBIC: Electron Bioimaging Center | 223 | N/A | 4 Krios, 1 Talos Artica | 0 | 5 microscopists | www.diamond.ac.uk/Science/Integrated-facilities/eBIC.html |
| EMBL Heidelberg | 22 | 2 iNext guests/month | 1 Polara, 2 Krios | 2 | 2 | N/A |
| Florida State University including the Southeastern Consortium for Microscopy of Macromolecula r Machines (SECM4) | 30 | Not tracked | 1 Krios | 2 | 3 | bsir.bio.fsu.edu |
| HMS: Harvard Medical School | 30 | Not tracked | 1 Polara | 3 | 2 | |
| JRC: HHMI Janelia Research Campus | 150 | Not tracked | 2 Krios | 1 | 4 | www.janelia.org/support-team/cryo-electron-microscopy |
| McGill Facility for Electron Microscopy Research | 160 (30 cryoTEM users) | 125 (30 cryo-EM projects) | 1 Krios | 5 | 1 staff member for the Krios lab, 5 staff members for other microscopes | www.mcgill.ca/femr |
| NeCEN: Netherlands Center for Electron Nanoscopy | 30 | 40 | 2 Krios | 3 EM specialists, 1 computing infrastructure, 1 program manager | www.necen.nl | |
| NYSBC: New York Structural Biology Center | 225 | 160 | 3 Krios | 3 | 5 microscopists | semc.nysbc.org |
| Northwestern University | 32 | Not tracked | 1 JEOL3200F S | 1 | 2 | www.facilities.research.northwestern.edu/browse-facilities/structural-biology-facility |
| OHSU Multiscale Microscopy Core | 15 | 20 | 1 Krios, 1 Talos Artica | 1 Tecnai | 3 | http://www.ohsu.edu/xd/research/research-cores/multi-scale-microscopy-core/ |
| Rockefeller University | 65 | Not tracked | 1 Krios | 1 | 2 | www.rockefeller.edu/cemrc |
| UCSF | 80 | Not tracked | 2 Krios,1 Polara | 4 | 3 EM specialists, 1 computer cluster specialist | msg.ucsf.edu/em/EMNEW2 |
| Yale University | 40 | N/A | 1 Krios | 4 + 1 (installation in progress) | 1 staff member at the Krios lab, 5 staff members for other microscopes | medicine.yale.edu/ccmi/em/cryoem.yale.edusciencehill-cryoem.yale.edu |
High End Microscopes: 300KeV TEMs equipped with direct detectors
Support staff: Scientists and technicians directly involved in supporting the instrumentation and users on the TEMS.
Over the course of two days, there were 18 presentations by the participants. The presentations were webcast to a larger audience, and the agenda and slides are accessible from the NRAMM website1. The presentations are also available on YouTube on the NRAMM/SEMC channel.2
A major objective of the workshop was to discuss best practices for managing cryoEM facilities and there was extensive discussion among the participants throughout the meeting. The discussions were largely focused on supporting single-particle methods for cryoEM and this paper provides a summary of the ideas and suggestions that arose from these discussions.
Multi-user cryoEM facilities
From the beginning, defining and clearly stating the purpose of a facility is critical for establishing the overall goals and to guide decision making during the operation and continued development of the facility. For example, NeCEN’s mission is “to be a center of excellence in high-resolution cryo-electron microscopy”. This entails providing access to cutting-edge cryoEM technology and expertise and implies that the instrumentation and technologies are regularly upgraded to avoid decline and loss of relevance. There are many different types of facilities ranging from those supporting a few groups that are proficient in cryoEM and are all from a single institution, to large national multi-user facilities with a wide range of users with varying levels of expertise. Some facilities are exclusively focused on data collection while others have a more extensive mandate including training and collaborations where facility staff are actively involved in the research project.
Two main models are presently emerging for cryoEM facilities. One type (eBIC and JRC) is modeled after the synchrotron facilities that X-ray crystallographers have been using for decades. These facilities typically provide high-end cryoEM for users and the focus is on providing large image collection sessions to many users who are typically geographically dispersed. The second type are facilities that are localized in an educational institution (UCLA, McGill, Caltech, OHSU) and have a broader mandate that includes not only providing high quality data to users but also user training across various levels. This second type of facility also frequently offers a much broader range of techniques and expertise. Most users in this type of facility are typically local and are affiliated with the institution hosting the core facility.
The Southeastern Consortium for Microscopy of Macromolecular Machines (SECM4), based at Florida State University, is an example of a regional facility that is hosted from the Biological sciences Imaging Resource (BSIR) at Florida State University (FSU). The BSIR is a core facility at FSU that provides access to most imaging modalities for biological research including light microscopes, SEM, and TEMs. The primary mission of the BSIR is to serve the imaging needs of the FSU community and surrounding universities. The Titan Krios is made available through the NIH U24 program to provide access to high-end instrumentation to experienced cryo electron microscopists. Through this mechanism, the SECM4 now collects high-end data for 21 investigators from 14 institutions.
At the Oregon Health and Sciences University, the Multiscale Microscopy Core (MMC) was established in 2013 through the collaborative efforts of the OHSU Center for Spatial Systems Biomedicine (OCSSB) and Hillsboro-based FEI Co. This EM facility provides imaging services, technical support and training to both academic and corporate users. The MMC offers multiple levels of service tailored to the analysis and training that each user desires.
The HHMI CryoEM Shared Resource at Janelia Research Campus was one of the first shared cryoEM facilities open to a large group of users from diverse academic backgrounds and scattered at different geographic locations. It serves the entire Howard Hughes Medical Institute providing fair access to cutting-edge instrumentation and top-quality services. Depending on the users’ experiences and needs, the facility provides different types of services. For users with prior cryoEM experience, it offers high-throughput, high-quality data collection services for user-provided ready-to-image cryo grids. For users with limited cryoEM experience or no access to nearby equipment, it offers services to cover the entire cryoEM workflow from sample preparation to data collection to image processing and 3D reconstruction. Experienced facility staff take care of most of the technical tasks so users can focus on providing good samples and/or selecting targets for data collection. Two main experimental techniques, single particle analysis and electron cryotomography, are supported by the facility.
The Netherlands Centre for Electron Nanoscopy (NeCEN) is an open access cryoEM facility. It offers research institutes and companies, both Dutch and international, access to advanced cryo electron microscopy equipment and expertise specifically tailored to explore complex biological structures. NeCEN was initiated by cryoEM experts from eleven organizations in the Netherlands that foresaw that an individual university or institute could not sustainably support the costs for the emerging cryoEM equipment. As the mission of NeCEN is to be a center of excellence in high-resolution cryoEM, the NeCEN provides a variety of services, ranging from cryo specimen preparation and data collection to image processing and training. The overall access procedure is based upon pay-for-usage model. The primary goal is to provide the Dutch EM community access to the most advanced instrumentation at a reasonable cost. Secondary goals are to establish international collaborations and to be a showcase site for one of the major microscopy manufacturers. Starting in April 2015, after several changes in its organization, NeCEN is now embedded within the Leiden Institute of Biology (IBL) as part of the Faculty of Science. About 30% of its usage is for groups based in the Netherlands, 60% for groups outside the Netherlands, and 10% for industrial groups.
The Electron Bio-Imaging Centre (eBIC) at Diamond Light Source is a free-at-the-point of-access cryoEM facility funded by the MRC, BBSRC and Wellcome Trust. Of the total instrument uptime, 80% is available to the user program and 10% is dedicated to an in-house scientific research program. eBIC also offers training courses in sample preparation and microscope operation.
The Simons Electron Microscopy Center (SEMC) at the New York Structural Biology Center (NYSBC) offers access to high-end electron microscopes to nine local institutions. The center supports a wide range of users ranging from beginners in need of basic training to well established and experienced microscopy groups. Aside from the core mission of supporting the member institutions, the center has grant funded research for developing technology closely coupled to driving biological projects through the National Resource for Automated Molecular Microscopy.
Expectation Management
One issue that was universally agreed upon by all participants is that it is critical that users of the facility be provided with a detailed description of the expectations and requirements prior to their first use or visit to the facility. For example, users need specific details on the costs and payments associated with using the facility, how to prepare for a session and what supplies to provide, best practices for shipping samples and grids, what to expect during and after data collection, and how to retrieve and process data. A detailed up-to-date web site is crucial to provide this information to users and good examples of several such websites are listed in Table 1. Several facilities also request or require that the users and their PI’s sign an agreement that specifies expectations, costs, and limitations ahead of any activity. These agreements may outline authorship, confidentially and acknowledgment policies.
Project Assessment, User Access, and Allocation
National or regional facilities that primarily serve external users often assemble a review panel composed of respected scientists, preferably without any conflicts of interest, whose task is to prioritize projects based on both the sample readiness and the potential scientific impact. For university facilities that mainly serve internal users and thus also have an educational role, it can be a challenge to assess scientific impact when prioritizing the projects. Thus, it is often more realistic to focus on the technical aspects of the sample by reviewing preliminary data. Users who can provide evidence that their samples are of acceptable quality are often given priority for access to the instrument on a first-come, first-served basis.
In Europe, users can apply for data collection time at eBIC, NeCEN, EMBL, CSIS, and CEITEC through iNext (http://www.inext-eu.org/). The process normally proceeds as follows. A proposal is submitted, reviewed by an iNext panel, an appointed local site reviews data collection feasibility, and a time slot of one to three days is allocated depending on the site. The first day is typically spent with an experienced operator screening grids and searching for the most suitable areas on a sample for imaging, this is followed by one to several days of automated data collection.
In the United States at SEMC4, each user is allocated a specific number of microscope days per year that can be used at their discretion. Access priority is based on the order of requests and whether the user has recently used microscope time; users who haven’t had recent access are given higher priority. To maximize usage, any user can sign up for an unscheduled time slot regardless of whether they have already used their allocation.
At NYSBC, institutions are allocated a defined number of days per year according to membership level across all instrumentation. An advisory committee is composed of representatives from each member institution. All projects require a general user proposal and discussion with the staff to best match resources with the project. Krios time requires an additional request and is scheduled in consultation with institutional representatives. Institutions are generally given 2-day sessions, but longer sessions may be requested based on need. Time is specifically allocated in the schedule for routine maintenance, workflow development, and rapid access.
At eBIC, microscope access is available through three routes: block allocations of guaranteed microscope time to academic consortia, proprietary access, and a rapid proposal system that takes in proposals every 3 months whereby individual requests can be made for 48hr instrument allocations. All non-proprietary proposals are peer-reviewed by an external panel of cryoEM experts.
User access to the microscopes varies widely between facilities. At some facilities, users do not need to be present during data collection as facility staff performs all the sample loading, instrument setup, and data collection. At the other end of the scale are facilities that provide properly trained users full access to the microscopes and the user is completely responsible for the operation during their session. Most of facilities provide a mixed approach where users are present during the data collection, but facility staff manages the process; users help acquire data by selecting targets during data collection but have minimal access to the microscope controls and are not expected to perform microscope alignments and calibrations. In this way, users are engaged in the data collection process but are not required to be expert microscopists.
Microscope operation by users is also dependent on the type of instrument being used for the data collection. High-end instruments are typically operated by facility staff members that have extensive training and expertise on the specific instruments. At some facilities, users may operate middle range electron microscopes after they have received the applicable training. Often, appropriate access to these middle range instruments is a critical element within the training programs run by the facility, as time is too limited on the high-end instruments to fully train many users.
At SECM4, a staff cryoEM specialist typically collects data for the users. For routine single-particle projects, users are encouraged to ship samples and monitor the data collection online. Users typically get 2 days of microscope time but can receive up to 6 days depending on their needs and if they need to travel to be on-site for the data collection. Certain samples, like tomographic samples, require an experienced eye for sample targeting and it is helpful for the user to be on-site for those sessions. Grid screening usually takes between 3 and 5 hours, and throughput is typically ~1500 direct detector movies/day.
For data collection at JRC, a user can decide whether to visit onsite or to work remotely. In either case, facility staff will load the sample, operate the microscope, optimize imaging conditions/parameters and set up automated data collection while the user can focus on evaluating and selecting targets for data collection. Facility staff will monitor the progress of data collection and periodically check data quality by performing motion correction and CTF estimation. For an HHMI lab or its collaborators to access the JRC cryoEM facility for data collection service, they need to make a request to the facility and provide proof that they have sample ready for high quality, high throughput data collection. They then will be put into a waiting queue and data collection time is allocated based on the order of requests received. A 3-day data collection session is typically given to each user.
At NYSBC, most instrument access is staff assisted. There are several training programs to bring users to varying levels of competence, ranging from ability to target and collect data once the instrument is set up, all the way to complete independence. Bringing users to a fully independent level generally requires them to be “embedded” with the group for at least six months, so this can only be provided to a very limited number of individuals. Training many semi-independent users frees staff from having to manage all aspects of every project.
At eBIC, microscopes are operated entirely by facility personnel. However, advanced users designated as their block allocation group “superuser” are offered training to enable autonomous use.
The California NanoSystems Institute at UCLA has a large user base and certified users are allowed, and indeed are highly encouraged, to operate all EM and support instruments on their own. Once trained for a certain instrument, the user can directly reserve time on the instrument once he or she has passed the training requirements unless the instrument is under high demand. Occasional users may request staff assisted use at an extra cost. The staff’s tasks include training of users, technical consultation to new users, instrument alignment, coordination of service and repair, and occasional assisted usage. This model gives users a large degree of flexibility and allows the facility to support a large user base with a limited number of staff members.
At UCSF, users are allowed direct access to the scheduling system and they can book any available slots within a defined period of time (typically 30 days) and with respect to their quota (usually 2 days). This model allows users to plan their experiments in advance. This method may not be practically scalable as the user base grows in size, and an alternative model based on a queue of access requests may be needed. An unavoidable challenge to scheduling is cancellation, either by the user, or the facility. In the former, there should be a clear policy as to the number of days in advance the user may cancel and the penalties that may arise. The latter often happens when instrument maintenance is required and may be on very short notice. In this case, the user has priority to book the next available time slot to compensate for their missed session. If the user cancels a session, every effort should be made to maximize the use of the microscope. This can be accomplished by moving up the next scheduled user, allowing facility staff to collect data for their own projects or collecting data to evaluate the performance of the microscope and data processing work.
Workflow
A typical workflow for a single-particle cryoEM project is shown in Figure 1. Initially, a sample is usually screened using negative stain EM methods. The objective of this screening is to qualitatively assess particle heterogeneity and, if the sample is promising, sufficient data are acquired to perform a 2D class average analysis and simultaneously obtain initial low-resolution map(s) using ab initio methods. Some groups acquire tilt pairs at the same time to generate low resolution maps and assess 3D heterogeneity using random conical tilt methods (Campbell et al., 2014a) (Radermacher et al., 1987) (Voss et al., 2010). This initial screening can be performed using a few hours of data collection on a 120KeV transmission electron microscope (TEM) equipped with a standard CCD camera. Some groups have integrated data collection with a processing pipeline (particle selection, CTF estimation, 2D class average analysis) that allows data to be simultaneously processed during acquisition; examples include using Appion (Lander et al., 2009), Scipion (de la Rosa-Trevin et al., 2016), and many customized scripts and other ad hoc solutions.
Figure 1.
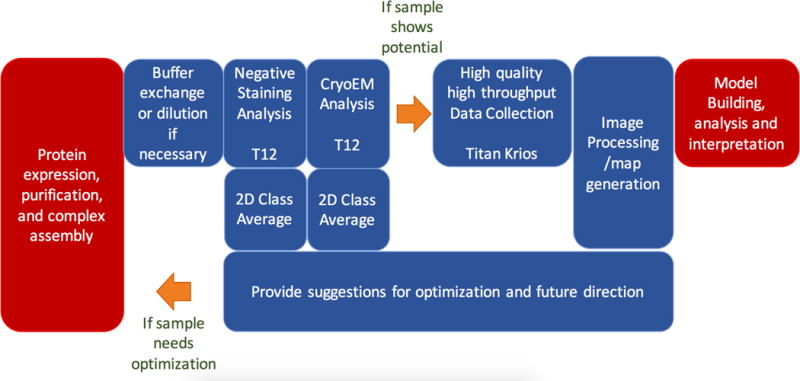
Typical workflow for single-particle CryoEM. The work modules shown in red are generally performed in the user’s own laboratory while work modules shown in blue are services that are offered by many facilities.
If the initial negative stain screening indicates that the sample is likely to be amenable to a structural analysis, the sample is then prepared using cryoEM vitrification methods and usually screened on a midrange TEM (200 KeV FEG) to assess vitreous ice thickness, particle density distribution, particle quality, particle homogeneity, and preferred orientation. If the sample is promising, a larger set of images may be acquired on the midrange instrument to facilitate further 2D and 3D analyses. In the best case, a moderate resolution 3D map may be obtained at this stage using these midrange TEMs (200KeV, FEG), equipped with direct detector cameras, which are capable of acquiring moderate to high-resolution (< 4Å) data (Campbell et al., 2014b) (Liang et al., 2015) (Ahmed et al., 2016) (Ripstein et al., 2017) (Li et al., 2017).
Once suitable cryoEM grids have been identified by these screening methods, a high-end TEM (300KeV, FEG) is used for collection of a large dataset with the goal of providing the highest possible resolution map. Today, the prevailing microscope for high-resolution cryoEM is an FEI Titan Krios equipped with a direct detector. At this writing, 180 maps at a resolution of better than 3.5Å have been deposited in the Electron Microscopy Data Bank3 and more than 80% of these maps used the Titan Krios microscope at 300KeV.
Once data has been acquired, it is further analyzed and refined to produce 3D maps. While the steps shown in Figure 1 may appear straightforward, the process is typically iterative and even after obtaining high quality data, the entire project may need to go back to the beginning to revise biochemical experiments and improve the quality of sample.
At the end of the data collection and initial processing at any cryoEM facility, the user should ideally be supplied with the documentation of the workflow and the metadata associated with the data collection and processing. Standardized templates for reporting are recommended and clear procedures for documenting the project should be implemented.
Sample and Grid Handling and Tracking
Facilities modeled based on the X-ray crystallography synchrotrons typically do not provide staff services for cryoEM grid preparation, and users are expected to provide grids that have been previously screened for proper ice quality, thickness, and particle distribution. Other types of facilities, e.g. NYSBC, frequently provide access to equipment and training in EM grid preparation and screening.
When users are required to ship cryoEM samples or grids to a facility, the facility should provide guidelines for users to transfer material to the facility in the most efficient and safe manner. Experience has shown that users need to be educated on the best way to transfer cryoEM grids to a facility. Typically, grids are sent in dry shippers and users are requested to include the facility manager’s email on the shipping form so the facility can be made aware of the shipment status, this is especially critical if shipment is delayed during transport. Communication between the user and facility staff is critical so that the facility is aware of incoming shipments and receipt of the sample can be acknowledged. Samples should be shipped early in a working week so that they are not delayed over a weekend or holiday period. Users should be requested to send a prepaid shipping label so that dry shippers can be easily returned. One consideration for preparing shipments is that grids will usually have to be clipped into cartridges after shipping. Since most grid clipping stations can only accommodate circular grid boxes, users should be requested to provide their grids in this format.
Once samples are received at the facility, they need to be carefully stored and tracked. If facility staff are responsible for clipping grids into cartridges, the grids must first be visually screened to identify and discard bent or severely damaged grids. This is especially important for the Krios autoloaders as a bent or damaged grid can cause jamming of the autoloader leading to subsequent weeks of downtime in a worst-case scenario. The state of grids should then be clearly communicated to the user and discarded grids can be shipped back to the user at their request. It is thus important that grid boxes and positions are clearly labeled and that the user provide an identification key for each grid.
At most facilities, cryoEM grid storage is still rather primitive. Vitrified grids are stored in small boxes, placed in 50 mL conical tubes, and suspended into a cryogenic dewar using strings threaded through holes in the conical tube to which simple labels are affixed. Though it may seem trivial, there are some important considerations for the preparation of the storage tube. The string attached to the tube is required to remove it from the long-term liquid nitrogen dewar. It is best if the string is attached to the tube body instead of the cap and that the string also is threaded through a hole in the cap. That way if the cap accidentally falls off, the cap remains in place and the tube can still be removed from the liquid nitrogen. Holes for threading the string should be drilled in the tube ~1/4 of the length from the top. This allows nitrogen to flow into the tube during filling and storage. Finally, it may be a good idea to weight the bottom of the conical tube so that it doesn’t float out of the liquid nitrogen during filling. These issues may be eliminated in the future as the community may adopt a more robust puck storage system developed by the X-ray crystallography community and recently adapted to EM storage (Scapin et al., 2017). This system also has the potential to provide high-density storage within a dewar (576–720 grid boxes) well suited to large cryoEM facilities.
The length of time that grids are retained and stored varies between facilities. Some facilities do not store grids at all while others will store grids for up to one year. At JRC, unused grids are stored for up to 6 months. Longer-term grid storage means that grids must be regularly inventoried and discarded once the storage time is exceeded. Again, a clear policy must be communicated so that users do not expect that they can retrieve their valuable grids indefinitely. For large facilities that store many grids, a bulk storage system that automatically performs LN2 fills may be a wise investment.
Sample Preparation
The cryoEM grid preparation method is not straightforward. Obtaining a vitreous ice film of the desired thickness with particles distributed evenly across the holes remains a challenge and is highly dependent on the sample (Glaeser et al., 2016) (Dobro et al., 2010). Most of the current vitrification methods are not particularly reproducible and small changes in the settings can dramatically affect the results. Screening of these grids is highly inefficient, as the entire grid must be searched to identify potentially only a few areas that may be suitable for data collection. Grid exchange on a microscope with a side entry holder is slow and throughput is low (3–10 grids/a day depending on ambient conditions and user skill) so only a limited number of conditions can be screened during a typical working day.
Microscopes with autoloaders make grid exchange easier, but grids must all be clipped and loaded into cartridges, which are costly and not reusable. The cartridge loading step also requires skill and must be done correctly to avoid jamming the autoloader possibly leading to extended instrument downtime. It is thus usually recommended that grids should be clipped and loaded by facility staff or a limited number of trusted users. Grids that arrive already clipped into cartridges should be closely inspected before loading them into the microscope. It should be noted that clipping grids into cartridges has the potential to damage the grids and thus some centers require users to clip their own grids to minimize liability. Autoloaders are also not generally compatible with the ability to iteratively try one condition after another, making a grid, loading it into the microscope for a quick check, and using this feedback to adjust the vitrification conditions. However, autoloaders do have the advantage that, if a suitable grid is obtained during the screening process, it is straightforward to transfer the same clipped grid to a high-end instrument for high quality data collection. Unclipped grids can also be “rescued” from a side entry holder but this usually requires a high level of skill and has the risk of accumulating ice contamination onto the grid during the transfer.
Microscope operation
Usually, facility staff are responsible for aligning and setting up the microscopes for each session and some facility managers are adamant that users should “not touch the lenses” as this could necessitate realignment of the microscopes.
Setting up microscopes for the highest quality data collection can be a complex process requiring many iterative steps. Once the microscope is correctly aligned and ready to go, there is often a series of steps that must be followed to ensure ideal data collection conditions. Some facilities have adopted the use of checklists (Gawande, 2009) and an example is shown in Figure 2. The checklists, though simple in concept, are used to ensure that a step is not missed resulting in a sub-optimal or a completely wasted data collection session. Though users may be present, they may not be knowledgeable about the data quality or collection parameters that are needed. The checklists can be used for setting up a single session or used for daily, weekly, and monthly scheduling of tasks.
Figure 2.
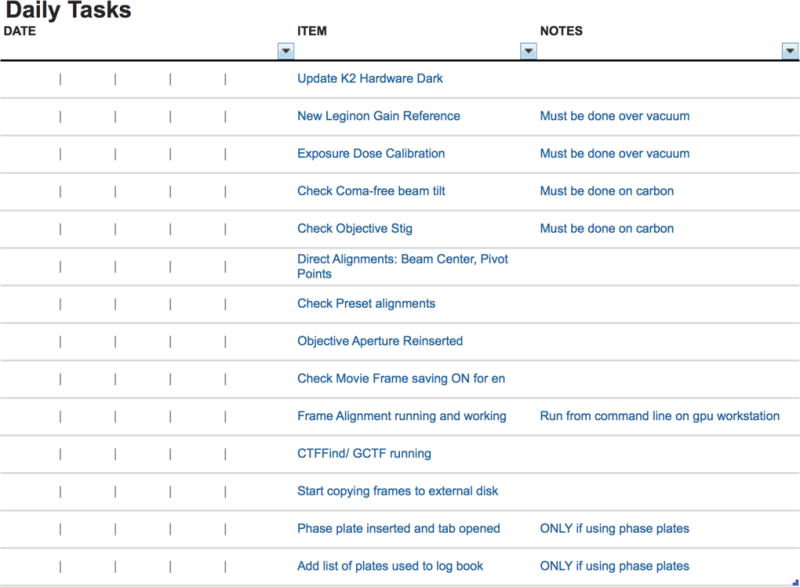
Example of daily checklist for operation of the Krios Microscope at NYSBC. These tasks take approximately 30–60 min to perform and the checklist, though not a how to guide or detailed protocol, aids in memory recall of important tasks. The tasks outlined range from acquiring images for gain normalization to microscope alignments and setting up the data processing pipeline.
For high-end TEM sessions, typically 3–6 grids will be loaded into the microscope and these grids will be screened to find the one most suitable for high-resolution data collection. Most facilities report that this initial screening process takes ~4–8 hours of microscope time, and the goal is to try to find a grid and select targets so that data is automatically collected during the subsequent ~16–36 hours. Most facilities have sessions that last either 24 or 48 hours, so a significant fraction of the high-end microscope time is spent on the screening process. Once data collection has started, a throughput of 750–2500 high magnification movies in 24 hours is reasonable if automated data collection software is used.
Remote operation of the microscopes enables 24/7 access of high-end instruments and is desirable to users both local and afar. After a cryoEM grid is loaded into the instrument by staff, two major user tasks remain for a typical high-resolution cryoEM data collection session: targeting of appropriate sample areas to build up an imaging queue and periodic monitoring of imaging progress after the queue is submitted. Both tasks are performed by data acquisition software packages, such as Leginon (Suloway et al., 2005), SerialEM (Mastronarde, 2005) and FEI Co. EPU, and can thus be performed remotely over the internet. At UCLA’s Electron Imaging Center for Nanomachines (EICN), most users take advantage of the remote monitoring and operation capabilities implemented in Leginon. Remote monitoring can be conveniently done through the web viewing interface automatically installed with the Leginon server. Both local and remote users take advantage of the Leginon web portal to monitor imaging progress and to assess data quality while away from the instrument. To remotely control the instrument and perform sample area targeting, EICN users coordinate with a staff member to enable a Remote Desktop session as a VNC client and take control of the Leginon computer. This Leginon computer also provides a tunnel to control both the Titan Krios main computer and electron detector computer remotely to perform occasionally required alignment and calibration operations.
Users at the HHMI Janelia cryoEM facility can either visit the facility or access the microscopes remotely. If they choose to travel on site, they can stay at a hotel conveniently located on campus and get temporary badges to enter the facility. If they choose a remote session, they need to ship cryo grids to the facility 1–5 business days before their scheduled session and communicate with the facility beforehand on a priority list of grids to load and preferred imaging conditions/parameters. CryoEM facility staff will take care of most of the set up work such as loading grids, optimizing microscope/camera performance, setting up imaging conditions/parameters, setting up automated data collection, monitoring the data collection and transferring data to users on the fly. Users only need to help evaluate the quality of a cryo grid and choose areas for data collection. The software Teamviewer4 is used for users to remotely log into the workstation hosting data collection software. With this software, users can see live images when facility staff screen the loaded grids and help choose a grid for data collection. During the process of setting up automated data collection, users can remotely choose or add areas of interest, typically holes with ice within a certain thickness range, for data collection with help from facility staff when needed. While automated data collection is under way, facility staff will periodically monitor the process and check data quality by performing motion correction and CTF estimation. Users are welcome to remotely monitor the data collection process as well as to identify possible issues or interruptions and notify cryoEM staff. For users that can provide access to their network drive or cloud storage, cryoEM staff will upload data on the fly. Users can process the data immediately and notify the facility staff if they believe changes are needed to the imaging parameters or the grid needs to be changed. Currently, about 50% of the JRC users choose remote data collection sessions as they can obtain high quality data without having to travel to the facility.
After a data collection session, some facilities provide a report that summarizes collection parameters and data quality as shown in Figure 3. Ideally, the generation of these reports should be automated; e.g. for facilities that use Leginon for their data collection a session summary report is automatically generated from the database.
Figure 3.
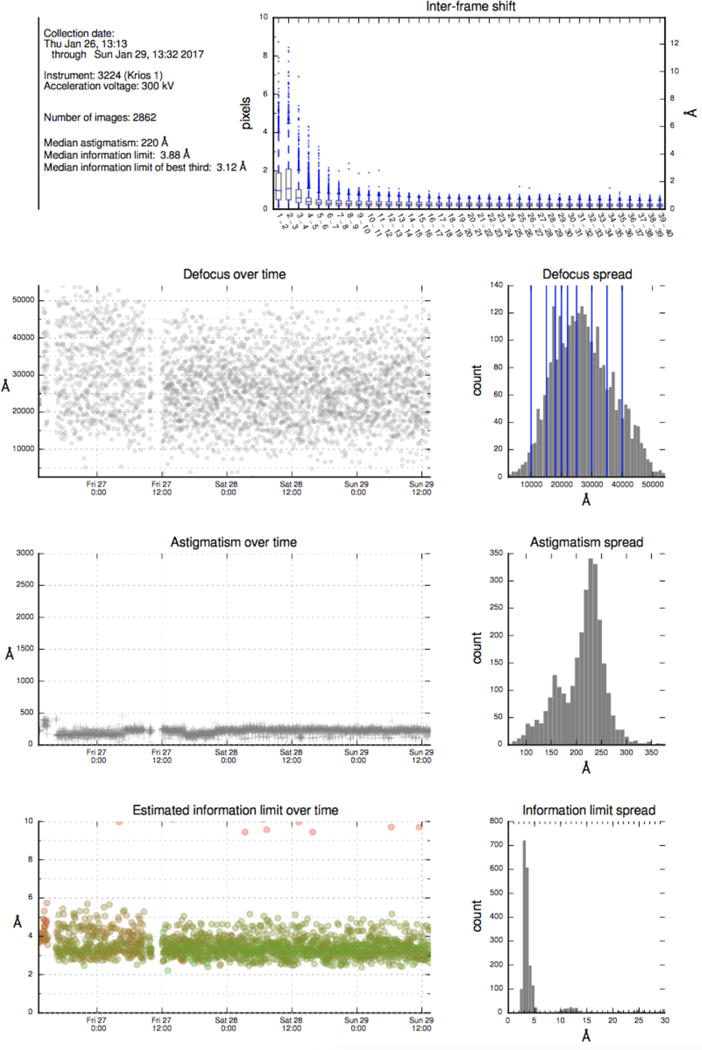
Example of report from NeCEN that provides an overview of data collection parameters and data quality. The top plot shows the inter-frame shift for alignment of frames in each movie acquired. The bottom three frames show time series plots and histograms of defocus, astigmatism and information limit of the images acquired during a data collection session.
Microscope Performance and Quality Checks
Regular performance evaluation and quality checks on the microscope are critical. This may be as simple as evaluating the Thon rings from an amorphous carbon film. For example, Leginon will automatically evaluate the CTF from the Thon rings as in Figure 4. Evaluating the DQE of the camera should also be performed regularly as sensor performance may change over time as shown in Figure 5 (Ruskin et al., 2013). A contamination rate test may reveal problems with the vacuum system in the microscope. These additional tests do not have to be performed every day but should be performed frequently enough to identify potential problems over time.
Figure 4.
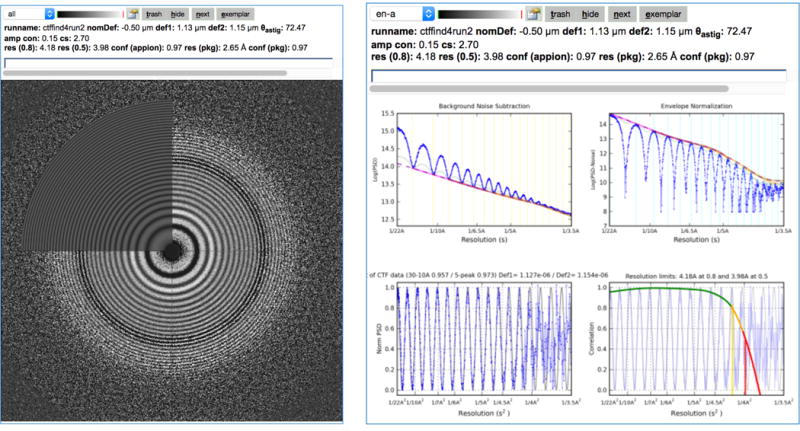
Image quality can be evaluated by analysis of the image power spectrum. This figure shows the Thon ring evaluation and contrast transfer function (CTF) analysis as displayed in the Leginon image viewer. The CTF is analyzed as images are being acquired using a choice of several CTF estimation packages (ACE, ACE2 (Yoshioka et al., 2007); CTFFIND4, (Rohou and Grigorieff, 2015); gCTF, (Zhang, 2016)).
Figure 5.
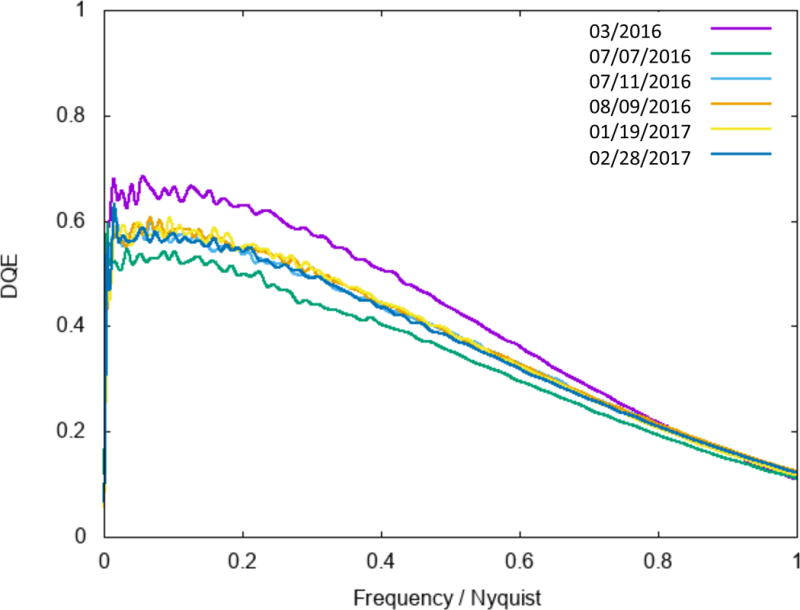
DQE camera analysis for a Gatan K2 detector over a period of 12 months. Regular checks of the camera can help to monitor the sensor performance over time. NYSBC performs this check every 3–4 months using the procedure outlined in Ruskin et al. 2013.
It is a good practice to acquire an information limit image at least once every month using a standard gold/carbon cross-grating. Some labs always keep a cross-grating grid available in the cassette and, while it occupies one slot in the multi-specimen holder, it facilitates microscope alignment, and frequent quality checks. A quick check of the information limit using a cross-grating helps verify proper operation of the microscope and avoids potential missteps, sometimes as simply as forgetting to turn off the autoloader turbo pump. This 2D visualization of the 1D contrast transfer function is a sensitive probe for microscope functionality. Any compromise in resolution, regardless of the source, will manifest itself in the information limit acquisition, and this test proves to be a good indicator of optimal microscope function.
Ideally, the results of these tests should be tracked over time using quality control charts and correlated with modifications or repairs to the instrumentation. FEI Co. has recently introduced system health monitoring that remotely monitors system parameters on the microscope and checks to make sure these are within control limits. The goal of this monitoring is the early identification of failure situations.
Some parameters can easily be monitored during the data collection session. For example, simple monitoring of the mean pixel value of the high magnification images during data collection can indicate when the FEG tip is terracing (Bronsgeest and Kruit, 2010). This event may last several hours and will likely degrade data quality during the intensity dip as shown in Figure 6.
Figure 6.
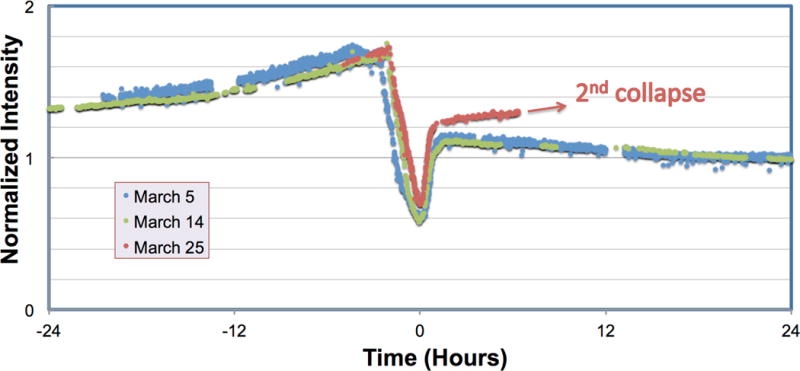
Monitoring of the average image intensity during data collection can predict FEG tip ring collapse. Three separate ring collapse events are shown for the same FEG tip showing the characteristic increase in average image intensity prior to the ring collapse. The intensity change pattern and duration of ring collapse depends on emitter tip temperature on the same tip but is fairly consistent.
Less routinely, a complete system benchmark test may be run through the data collection and processing pipeline for a standard test sample. For example, at NYSBC a 20S proteasome or Apoferritin test sample is used and a data collection session of ~8 hours followed by standardized processing steps typically yields a sub 3Å map. Performing these tests ensures that microscope, camera, and processing pipelines are optimal. These complete system benchmark tests can also help to evaluate performance differences between microscopes and to assess the value of peripherals such as energy filters, phase plates and correctors. The benchmark experiment using a well-characterized sample can also give us a more accurate pixel size by comparing the reconstruction with its known atomic model. It is not uncommon to see a ~5% error in the pixel size determined at the factory.
Routine maintenance tasks for the microscopes should be regularly scheduled and documented. With the advent of autoloaders, grids are being stored for a significant period of time in that system. Checks of ice contamination buildup over time on the grid should be monitored and may be detected via complete system benchmarks on the same grid after loading and after storage in the autoloader for some time. Cryo-cycling of the microscopes is critical to avoid buildup of contamination. The length and frequency of cryo-cycles varies widely between facilities ranging from one 48 hour cryo-cycle every 3–4 weeks to a 24 hour cryo-cycle once a week. It is generally believed that less frequent long duration cryo-cycles are preferable to shorter more frequent cycles. Camera annealing cycles also vary but are typically performed for 8 hours every 1–2 weeks. The Gatan K2 detector requires regular annealing cycles because the working temperature of K2 detector is −20°C, and it thus acts as a cold trap. Annealing at 50°C helps release the contamination and extensive annealing sometimes can fade burn marks caused by accidental exposure to an intense electron beam.
After any servicing of the microscope column, the information limit should be tested using a standard cross-grating test sample. If the microscope column is serviced, a helium leak test can also be done to confirm there are no small vacuum leaks compromising the long-term quality of the vitrified samples in the microscope.
Processing
The processing capabilities after data collection vary widely between facilities. Some facilities provide no additional data processing; users are given a disk with raw movie frames, and the user is responsible for all subsequent processing. Some facilities have integrated processing pipelines that simultaneously perform the processing during data collection. These steps may include gain normalization of the images, motion correction of the frames and CTF estimation. The user receives raw movie frame data and aligned average frames (weighted and unweighted) along with CTF estimates and quality assessments of each image. At NYSBC and SECM4, additional processing is also available using the web-based Appion pipeline for particle selection, stack creation and 2D class average analysis (Lander et al., 2009). Similarly, NeCEN and eBIC use Scipion to provide a similar processing pipeline (de la Rosa-Trevin et al., 2016). The user should be well informed of the processing deliverables and this should be part of the initial expectation management agreement.
For single-particle EM, the computational resources are now becoming almost as important as the microscope. Most facilities do not have the computational resources to allow their users to run large-scale refinements that may occupy tens of thousands of hours of computer time. The computational landscape is rapidly evolving as users have a choice of software packages and hardware architectures. These range from single workstations with GPUs, to institutionally hosted large multi node clusters, to accessing Cloud computing resources (Cianfrocco and Leschziner, 2015). OHSU has excellent computational support where the microscope suite has 10 Gb/s internet connectivity using fiber and copper interconnects to the server room. Once the data reaches the cluster from the microscopes, it can be pre-processed in real-time. This pre-processing includes particle detection, frame alignment, and data-dependent compression. The preprocessed and compressed image data is then transferred to a larger ExaCloud cluster for in-depth processing and analysis. The preprocessing cluster has 9 compute nodes, each with 256 Gb of RAM and 24 cores giving a total of 216 cores for the cluster. The primary ExaCloud cluster includes over 6,600 Xeon cores with 35.5 TB of memory distributed across 250 compute nodes. A subset of the compute nodes are dedicated to image processing tasks, such as performing single-particle refinement.
eBIC benefits from being located at the Diamond Light Source, a high data rate and volume facility. Data is immediately moved from the microscopes onto a large GPFS parallel file system where it remains available to users for processing for 40 days. All data from each external group is securely stored and readable only by the group. From the secure individual area, users can copy the data onto local media or transfer it back to their own institutions. Microscope users also have shared access to clusters with an excess of 3000 cores of various architectures, and dedicated access to 10 × 20 core CPU clusters, each housing two GPU cards. eBIC users also benefit from the Diamond-wide experiment data policy5 which includes archiving a single copy of all data onto tape media.
NYSBC has a dedicated fiber backbone to connect direct detectors to dedicated buffer servers for on-the-fly computation and compression. Data is then moved to the main server for temporary storage of up to 2 months. Each buffer server is configured with 2 GPU cards and sufficient local storage for up to one week of data in case of system failure. External users are expected to have their own clusters for large-scale computations, however a 44-node cluster is available for collaborative use, each node with 256 GB RAM and a total of 1056 cores. Several GPU workstations are also on-site for use by programs optimized for GPU computation.
Keeping the potpourri of typical processing software packages updated and optimized is a non-trivial task and facilities need to have the appropriate support in place. For those facilities that do not have their own information technology staff, the SBGrid consortium6 provides the global structural biology community with support by deploying and maintaining software tools. Maintaining software may become more straightforward if containerization packages, such as Docker7, are widely adopted. The Biological Science Imaging Resource at Florida State uses this approach on their institutional cluster so that the application software is less dependent on the system software upgrades performed on the cluster.
Data Management
Since the advent of the high-frame rate direct detector, managing the large volumes of data from a high-end microscope can be a significant challenge. High-end facilities typically generate 1–3 TB of raw data per day per detector and this can quickly overwhelm a storage system. A week of data collection from a single microscope can consume up to 21 TB of disk space. Compounding this problem is that the data may need to be processed concurrently with data collection and simultaneously written off to external archive drives. These multiple simultaneous operations necessitate the use of a high-performance storage system and these can be quite expensive. For instance, at SECM4 the Lustre high-performance storage costs $1450/TB for five years of service compared to a commodity disk that costs ~$25 for a 1 TB disk. Given the expense of high performance storage, that space must be tightly managed. Another consideration is how to transfer the data to the end users. Small datasets can be transferred over the internet through rsync, but this is often prone to failure. Other services like Globus check the integrity and completeness of data transfer but require an additional layer of IT to implement. Currently, the simplest and most robust way of providing data to users is by shipping USB hard drives. SECM4 creates two copies of the USB drives to ensure data integrity. One copy is sent to the user and upon verification of the integrity of the data, the other copy can be shipped or recycled for another session. At the NYSBC and SEMC4, all steps of the data workflow are performed concurrently with data collection. As soon as images are taken, the frames data are transferred from the camera computer to the high-performance storage. Frames are aligned at approximately the same rate that they are collected, and raw data is automatically compressed and then transferred to external USB drives. Transfer is started soon after large-scale data collection is begun, to minimize the lag time between the end of a session and when users get their data. After all the data is transferred to a USB drive, that drive is cloned, at which point it is safe to delete the data from the high-performance storage. In practical terms, the raw compressed images are kept for two months, sufficient time for external users to be sure they have the data.
Training
The user training mission varies widely between facilities and typically consists of a combination of courses, workshops and one-on-one training covering a range of topics including:
-
-
Orientation to the facility: Includes overview of facility procedures and safety
-
-
Introduction to cryoEM: Overview of specimen preparation, data collection and processing for cryoEM
-
-
Introduction to sample preparation: Negative stain and cryo preparation
-
-
Basic operation of the microscope and software for data collection
-
-
Introduction to single-particle processing
-
-
Introduction to other data collection methods and processing
Many facilities do not expect their users to be expert microscopists, but they must have sufficient training to understand the overall workflow. If they use the microscope, they are expected to be able to perform the data collection once the microscope is setup and to understand minimal actions to take if something goes wrong during the process (i.e. close the column valves). At NYSBC, all users must undergo a basic one-day orientation session that includes an introduction to EM and a demonstration of sample preparation and the operation of the microscopes. After this orientation session, users are then allowed to book sessions on the microscopes concurrent with additional one-on-one training with a staff member until they become proficient. NYSBC also offers regular hands-on workshops for single-particle and tomographic data processing and these are recorded and made available on the NRAMM/SEMC YouTube channel.8
A more extensive model is NeCEN’s annual 9-week full time school for cryoEM. The goal is to provide intensive training to 6 students on all aspects of cryoEM from specimen preparation to processing. Students get hands-on experience on a Krios TEM and at the end of the course are expected to have sufficient expertise to independently practice the methods.
For theoretical training, Grant Jensen’s “Getting Started in CryoEM Course”9 is an excellent resource that consists of 47 well-produced lectures covering all aspects of cryoEM.10 The MRC Laboratory of Molecular Biology provides another very popular and professional lecture series11. Other online materials are available at the NRAMM/SEMC YouTube channel12, which includes lectures that supplement the Grant Jensen course as well as the proceedings of several specialized workshops. There are many other additional resources for EM including John Rodenberg’s tutorial in TEM13 and the Australian Microscopy and Microanalysis Research Facility site supports an online TEM simulator.14
How does a cryoEM facility measure success?
Success for a user-based facility is a balance between accomplishing the goals of the facility and meeting the needs of its users. This balance is inextricably tied to the broader scopes and missions of both. Success of facility performance can be quantified using a series of metrics, including those stemming from user productivity and satisfaction. Categories to consider in the measure for success include resource management, communications, impact, and planning (Turpen et al., 2016). Measurables include equipment, space, expertise, safety, compliance, publications, grants supported, and self-assessment (Turpen et al., 2016). The question “how to measure success?” invokes a host of answers that unveil the symbiotic relationship between users and the facility. Drivers of value will change over time, but the dependence between users and the facility will not.
The facility can use internal metrics to assess performance, for example, the hours of availability of instruments, quality of the data produced, performance of the instruments, etc. Users can also provide information such as the amount and quality of the data obtained, the number of structures solved, and other metrics. These are largely objective measures that can easily be obtained and that help identify potential problems or shortcomings that need to be solved. More difficult to quantitate are subjective metrics, such as user and staff satisfaction. Well-designed periodical surveys help to assess some of these more subjective metrics and can guide improvements. For example, at Northwestern University, the results of an annual user satisfaction survey of all users are analyzed to assess criteria such as timeliness of service, responsiveness of staff, quality of help, and support, etc. At eBIC all users are automatically requested to provide experimental feedback and reports through the Diamond Light Source User Administration System. All scores and comments are collated and discussed with the eBIC User Committee on a biannual basis. These data, together with metrics such as hours of operation, help produce a more global picture of the performance of the facility and thus provide other measures of success. Success in a facility usually means both excellence in internal performance, meaning that the facility is well-run and delivers the desired services, coupled with external productivity in terms of papers and grants, as well as overall user satisfaction.
Discussion
A common platform for sharing workflow and protocols among users would be greatly beneficial to the community. FEI Co. has introduced a commercial Apple iPad application-based project that provides detailed protocols and how-to videos for each step in the workflow. The application software is proprietary, but laboratories can document their own protocols within the application. The community would greatly benefit from a non-commercial, open source platform for sharing this information. Nature Publications supports the Nature Protocol Exchange15 that is a commercial enterprise but the protocols are under the Creative Commons License. This provides an on-line open resource where researchers may submit laboratory protocols without peer-review or editing, but one disadvantage is that it does not support uploading of videos. There are other data exchanges such as Zenodo16, an open science initiative supported by CERN that supports all forms of data types. Unfortunately, the cryoEM community has thus far not readily adopted any of these exchanges. A possible alternative would be for the Electron Microscopy Public Image Archive (EMPIAR)17 to support sharing of protocols as a supplement to the deposition of cryoEM images.
The “Management of Large CryoEM Facilities” workshop provided an opportunity to discuss many of the detailed operations required for running a high-end cryoTEM facility. These discussions do not often take place at scientific meetings and are not typically the subject of journal publications. Though there was a wide variation in the mission of the facilities represented at the workshop, there were common themes on how a facility should be run and managed. There was also open discussion of the challenges of running facilities for a technique that is in high demand. These challenges include: staff recruitment and retention, maximizing productivity of expensive instrumentation, and training a large, expanding, and demanding user base. There are many new high-end EM facilities being established and communication of best practices is critical for continuing to advance the field. These practices are also valuable for smaller facilities or individual labs interested in setting up and maintaining a functional and productive cryoEM suite.
Supplementary Material
Supplement
Table S1: Description of access and services offered at each facility
Acknowledgments
The workshop was part of the activities of the National Resource for Automated Molecular Microscopy at the Simons Electron Microscopy Facility that is supported by grants from NIH NIGMS GM103310 and the Simons Foundation (349247). Additional financial support for travel and other costs directly related to the workshop was provided by FEI Co./Thermo Fisher Scientific.
Footnotes
Literature References
- Ahmed T, Yin Z, Bhushan S. Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci Rep. 2016;6:35793. doi: 10.1038/srep35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronsgeest MS, Kruit P. ‘Collapsing rings’ on Schottky electron emitters. Ultramicroscopy. 2010;110:1243–1254. doi: 10.1016/j.ultramic.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Campbell MG, Smith BC, Potter CS, Carragher B, Marletta MA. Molecular architecture of mammalian nitric oxide synthases. Proc Natl Acad Sci U S A. 2014a;111:E3614–3623. doi: 10.1073/pnas.1413763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MG, Kearney BM, Cheng A, Potter CS, Johnson JE, Carragher B, Veesler D. Near-atomic resolution reconstructions using a mid-range electron microscope operated at 200 kV. J Struct Biol. 2014b;188:183–187. doi: 10.1016/j.jsb.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco MA, Leschziner AE. Low cost, high performance processing of single particle cryo-electron microscopy data in the cloud. Elife. 2015;4 doi: 10.7554/eLife.06664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rosa-Trevin JM, Quintana A, Del Cano L, Zaldivar A, Foche I, Gutierrez J, Gomez-Blanco J, Burguet-Castell J, Cuenca-Alba J, Abrishami V, Vargas J, Oton J, Sharov G, Vilas JL, Navas J, Conesa P, Kazemi M, Marabini R, Sorzano CO, Carazo JM. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J Struct Biol. 2016;195:93–99. doi: 10.1016/j.jsb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- DeMaggio S. Running and setting up a confocal microscope core facility. Methods Cell Biol. 2002;70:475–485. doi: 10.1016/s0091-679x(02)70015-0. [DOI] [PubMed] [Google Scholar]
- Dobro MJ, Melanson LA, Jensen GJ, McDowall AW. Plunge freezing for electron cryomicroscopy. Methods Enzymol. 2010;481:63–82. doi: 10.1016/S0076-6879(10)81003-1. [DOI] [PubMed] [Google Scholar]
- Elmlund D, Le SN, Elmlund H. High-resolution cryo-EM: the nuts and bolts. Curr Opin Struct Biol. 2017;46:1–6. doi: 10.1016/j.sbi.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Ferrando-May E, Hartmann H, Reymann J, Ansari N, Utz N, Fried HU, Kukat C, Peychl J, Liebig C, Terjung S, Laketa V, Sporbert A, Weidtkamp-Peters S, Schauss A, Zuschratter W, Avilov S, German BioImaging, n. Advanced light microscopy core facilities: Balancing service, science and career. Microsc Res Tech. 2016;79:463–479. doi: 10.1002/jemt.22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawande A. The Checklist Manifesto. Metropolitan Books; New York, NY: 2009. [Google Scholar]
- Glaeser RM, Han BG, Csencsits R, Killilea A, Pulk A, Cate JH. Factors that Influence the Formation and Stability of Thin, Cryo-EM Specimens. Biophys J. 2016;110:749–755. doi: 10.1016/j.bpj.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W. Cryo-EM enters a new era. Elife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou N, Chen W, Zhu B, Wang X, Xu B, Wang J, Liu H, Cheng L. Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV. J Mol Biol. 2017;429:79–87. doi: 10.1016/j.jmb.2016.11.025. [DOI] [PubMed] [Google Scholar]
- Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SP. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Ripstein ZA, Huang R, Augustyniak R, Kay LE, Rubinstein JL. Structure of a AAA+ unfoldase in the process of unfolding substrate. Elife. 2017;6 doi: 10.7554/eLife.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin RS, Yu Z, Grigorieff N. Quantitative characterization of electron detectors for transmission electron microscopy. J Struct Biol. 2013;184:385–393. doi: 10.1016/j.jsb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil HR, Grunewald K, Stuart DI. A national facility for biological cryo-electron microscopy. Acta Crystallogr D Biol Crystallogr. 2015;71:127–135. doi: 10.1107/S1399004714025280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapin G, Prosise W, Wismer MK, Strickland C. A novel storage system for cryoEM samples. J Struct Biol. 2017 doi: 10.1016/j.jsb.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Trogadis J. Issues in the management of a core imaging facility. Lab Manager Magazine. 2006;1:11–16. [Google Scholar]
- Turpen PB, Hockberger PE, Meyn SM, Nicklin C, Tabarini D, Auger JA. Metrics for Success: Strategies for Enabling Core Facility Performance and Assessing Outcomes. J Biomol Tech. 2016;27:25–39. doi: 10.7171/jbt.16-2701-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Lyumkis D, Cheng A, Lau PW, Mulder A, Lander GC, Brignole EJ, Fellmann D, Irving C, Jacovetty EL, Leung A, Pulokas J, Quispe JD, Winkler H, Yoshioka C, Carragher B, Potter CS. A toolbox for ab initio 3-D reconstructions in single-particle electron microscopy. J Struct Biol. 2010;169:389–398. doi: 10.1016/j.jsb.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka C, Pulokas J, Fellmann D, Potter CS, Milligan RA, Carragher B. Automation of random conical tilt and orthogonal tilt data collection using feature-based correlation. J Struct Biol. 2007;159:335–346. doi: 10.1016/j.jsb.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement
Table S1: Description of access and services offered at each facility


