Abstract
Cross-regulation of Toll-like receptor responses by cytokines is essential for effective host defense, avoidance of toxicity, and homeostasis, but the underlying mechanisms are not well understood. A comprehensive epigenomic approach in human macrophages showed that the proinflammatory cytokines TNF and type I IFNs induce transcriptional cascades that alter chromatin states to broadly reprogram TLR4-induced responses. TNF tolerized inflammatory genes to prevent toxicity, while preserving antiviral and metabolic gene induction. Type I IFNs potentiated TNF inflammatory function by priming chromatin to prevent silencing of inflammatory NF-κB target genes. Priming of chromatin enabled robust transcriptional responses to weak upstream signals. Similar chromatin regulation occurred in human diseases. Our findings reveal that signaling crosstalk between IFNs and TNF is integrated at the level of chromatin to reprogram inflammatory responses, and identify new functions and mechanisms of action of these cytokines.
Introduction
Tumor necrosis factor (TNF) is important in innate immunity, inflammation, and host defense against microbial pathogens1. TNF is also a key pathogenic cytokine and driver of chronic inflammation in multiple autoimmune and inflammatory diseases2. Classical inflammatory activation of cells by TNF is mediated by canonical NF-κB and MAPK signaling that activates well known inflammatory genes such as IL1 and IL6. TNF also has potent paradoxical anti-inflammatory functions (discussed in ref3) that limit inflammation-associated toxicity4. Perhaps the most potent suppressive mechanism induced by TNF is ‘cross-tolerance’3, which resembles classical endotoxin tolerance5 in that various TLR ligands and inflammatory stimuli are unable to induce transcription of select inflammatory genes. Much less is known about cross-tolerance induction by TNF than endotoxin tolerance, and about how the tolerizing functions of TNF are over-ridden such that TNF is able to drive chronic inflammation.
Type I interferons (IFNs) activate the Jak-STAT signaling pathway to induce expression of interferon-stimulated genes (ISGs) that are activated by STAT proteins via binding to conserved ISRE and GAS DNA elements6. ISGs include antiviral proteins, chemokines, and antigen-presenting molecules, and thus type I IFNs can promote antiviral and immune responses, and have been implicated in autoimmune diseases. However, type I IFNs can also play a suppressive role in certain chronic infections and in multiple sclerosis6. Distinct IFN functions may be related to context dependent effects on inflammatory NF-κB-driven genes, as IFNs can either suppress cytokines or contribute to increased cytokine production in diseases such as SLE, increased inflammation when bacterial infections follow viral infections, and to microbiota-mediated priming of cytokine responses6–9. Mechanisms by which type I IFNs regulate inflammatory NF-κB target genes, which are not targets of the Jak-STAT signaling pathway, are not known10,11.
Potent inflammatory activators of macrophages such as TLR ligands activate signaling via NF-κB, MAPK-AP-1 and IRF3 pathways to induce expression of inflammatory cytokine genes12. The ability of signal-responsive transcription factors (TFs) to induce transcription is modulated by chromatin states at gene regulatory elements (promoters and enhancers)13–17. Activation of many TLR-inducible genes, including Il6, Il12b and Ifnb, requires increasing chromatin accessibility by deposition of positive histone marks and remodeling of nucleosomes to create nucleosome-free regions at promoters and enhancers13–15. More recent work has shown that environmental cues can fine tune the macrophage enhancer repertoire18–23. Induction of new enhancers can explain tissue-specific macrophage gene expression, and raises the possibility that enhancer remodeling can alter cellular responses to environmental signals. However, analysis of the effects of epigenomic remodeling on cellular responses to secondary inflammatory challenges has been limited. Furthermore, little is known about whether signaling crosstalk occurs in the nucleus at the level of chromatin during an inflammatory TLR-driven response.
It was previously reported that TNF induces a state of ‘cross-tolerance’ in which various TLR ligands are unable to induce transcription of the canonical inflammatory NF-κB-dependent cytokines IL-1β, IL-6 and TNF, and which protects mice from endotoxin lethality in vivo3. In the current study, we wished to understand TNF-induced cross-tolerance in greater depth, and to test whether macrophages could escape from cross-tolerance; abrogation of this feedback mechanism could provide an explanation for how TNF can drive sustained unremitting inflammation in a chronic setting. We took a comprehensive and integrated genome-wide approach using RNA-seq, ChIP-seq, and ATAC-seq24 with digital footprinting25 to investigate the regulation of TLR4 responses by TNF. We discovered that type I IFNs effectively abrogate TNF-induced cross-tolerance by priming chromatin to enable robust transcriptional responses to weak signals. These findings reveal mechanisms by which cytokine signaling crosstalk is integrated at the epigenomic level, and identify a new function and mechanism of action for type I IFNs.
Results
TNF Reprograms the LPS response in human macrophages
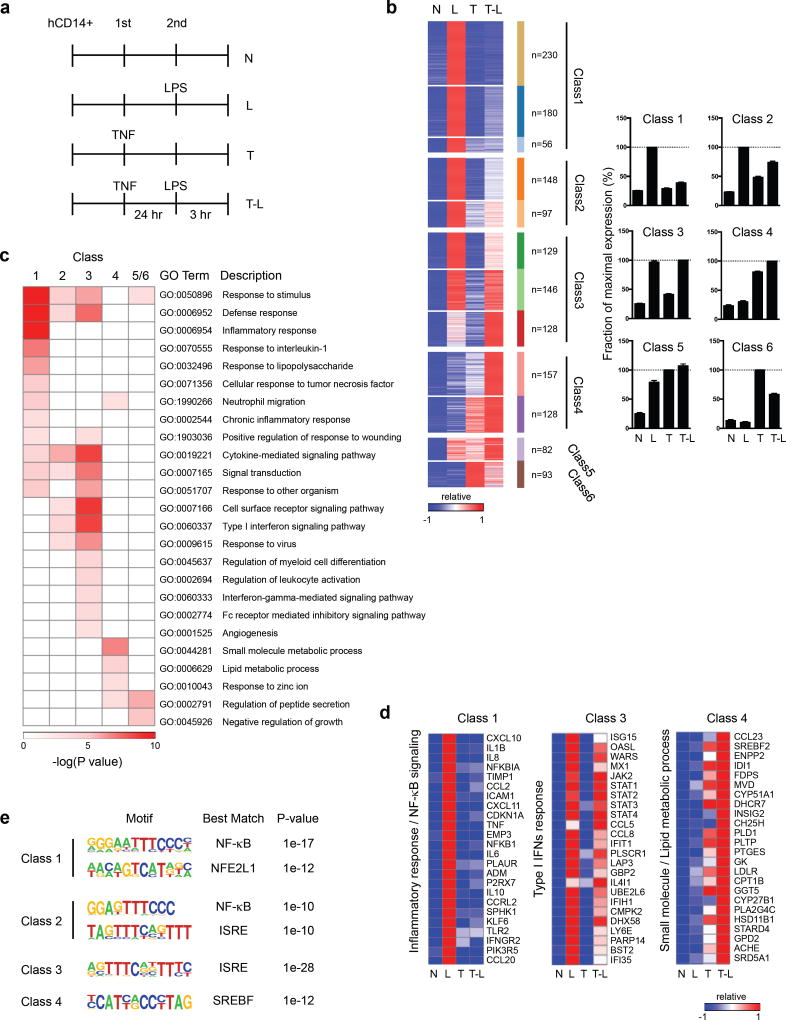
Previous work showed that TNF pretreatment strongly attenuates LPS-induced signaling and chromatin remodeling upon secondary challenge3,5. To gain insight into gene regulation in TNF-induced tolerance, we performed transcriptomic analysis using RNA sequencing (RNA-seq). We used our previously established system in which primary human macrophages are treated with TNF for 24 hours prior to LPS challenge3 (Fig. 1a) and focused our analysis on the 1,574 genes that were strongly induced (>3-fold) in response to LPS. Clustering of LPS-inducible genes by patterns of gene expression revealed 12 clusters that could be assembled into 6 major classes (Fig. 1b, Supplementary Fig. 1a and Methods). Two classes of robustly LPS-inducible genes were minimally (Class 1, n = 466) or weakly (Class 2, n = 245) induced by LPS in macrophages pretreated with TNF (Fig. 1b). Following previous nomenclature, we termed these ‘tolerized’ or ‘T’ genes5,26. Class 1 was enriched for gene ontology (GO) terms related to ‘defense response’ and ‘inflammatory response’ (Fig. 1c), and included many pro-inflammatory cytokines (Fig. 1d) and NF-κB target genes (Fig. 1e). Thus, TNF-induced cross-tolerance is broadly similar to endotoxin tolerance26,27 in transcriptional silencing of inflammatory NF-κB target genes.
Figure 1.
Pretreatment with TNF reprograms subsequent TLR4 response in human macrophages. (a) Experimental design: N, no treatment; L, no pretreatment, followed by LPS challenge; T, pretreatment with TNF; T-L, pretreatment with TNF and challenge with LPS; hCD14+, human CD14 positive monocyte-derived macrophages. (b) K-means clustering (K = 12) of 1,574 LPS-induced genes (>3-fold) in the indicated conditions; heat map shows gene expression relative to maximum, set at 1. 12 clusters were assembled into 6 major Classes (see Methods). Bar graphs represent pooled data from three biological replicates (% of maximum value) for a given class. (c) Functionally enriched Gene Ontology (GO) categories of the gene classes in Fig. 1b. (d) Heatmaps of representative genes from Classes 1, 3, and 4 that correspond to distinct biological functions. (e) Motifs enriched in the promoters (−300bp < TSS < +50bp) of given Class genes using GC-corrected background set of all other promoters using HOMER. Data (c–e) are representative of three biological replicates with similar results.
LPS effectively and paradoxically induced expression of various genes in TNF-treated cells (Fig. 1b, Classes 3–6, and Supplementary Fig. 1a and 1b), despite minimal LPS-induced signaling3 (see also below). In line with previous reports of endotoxin tolerance, we termed these ‘nontolerized’ or ‘NT’ genes26,28. Class 3 (n = 403) is comprised of genes strongly induced by LPS in both naïve and TNF-treated cells; Class 3 NT genes are most clearly regulated in a directly opposite manner from T genes. In contrast, genes in Classes 4 (n = 285) and 5 (n = 82) were substantially expressed in TNF-treated cells and superinduced by secondary LPS challenge (Fig. 1b and Supplementary Fig. 1a), thus revealing cooperation and even synergy (Class 4A, Supplementary Fig. 1a) between the ‘tolerizing’ factor TNF and LPS.
In addition to different expression patterns, the NT gene classes had distinct functions as revealed by GO analysis. Class 3 genes were enriched for cytokine and IFN signaling via Jak-STAT pathway, Class 4 genes were enriched for metabolic processes, and Class 5 and 6 contain additional genes important in lipid metabolic processes (Fig. 1c and 1d; complete list of genes in all six classes is provided in Supplementary Table 1).
The gene classes also differed in transcription factor binding motifs that were enriched in their promoters (Fig. 1e). Class 1 T gene promoters were most significantly enriched in NF-κB and NFE2L1 motifs, Class 3 NT in ISREs (bind type I IFN-activated ISGF3), and Class 4 NT genes in binding sites for SREBP, which drives expression of cholesterol pathway and lipid metabolism genes. Thus, in addition to different patterns of expression and different functions, the gene classes have distinct mechanisms of regulation. As predicted, various Class 3 NT genes were dependent on type I IFN signaling, whereas Class 4 NT genes were not (Supplementary Fig. 1c).
The gene classes could also be distinguished based on kinetics of induction by TNF (Supplementary Fig. 2a). Class 1 (T genes) is largely composed of early TNF-induced genes, which peak at 1–3 hr and decrease in expression by 24 hr. In contrast, Class 3 and Class 4 genes exhibited delayed induction kinetics; unlike the gradual increase of expression of Class 3 genes, Class 4 exhibited induction only at the late (24 hr) time point. We then compared the behavior TNF-induced (n=433, 1 or 3 hr) and LPS-induced (n=1,219, 3 hr) genes to test the possibility that TNF tolerance only affects TNF-induced genes. Strikingly, TNF tolerized a large fraction of LPS-inducible genes that were not induced by TNF (Supplementary Fig. 2b), supporting the idea of crosstolerance.
Collectively, the results reveal that TNF extensively reprograms the LPS response, with TNF-induced ‘cross-tolerance’ representing one component of ‘reprogramming’. The TNF-reprogrammed state appears to differ from classical endotoxin tolerance by the expression of IFN/cytokine-driven genes (Class 3) and lipid metabolic genes (Class 4–6) upon LPS challenge.
TNF regulates chromatin and TFs to reprogram LPS response
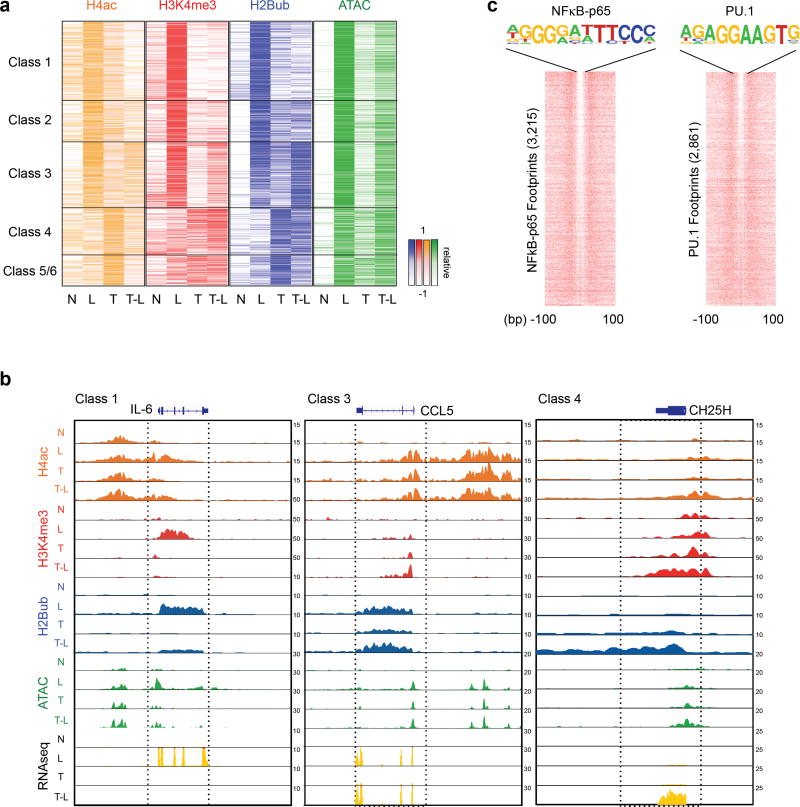
TLR4 signaling is almost completely abrogated in macrophages pretreated with TNF or endotoxin, and epigenetic mechanisms have been implicated in tolerance. Consistent with an epigenetic mechanism, TNF-induced tolerance was sustained for at least 48 h after the washout of TNF (Supplementary Fig. 2c). As epigenetic regulation has been studied for only a small number of T genes, and not for NT genes, we performed genome wide analysis of 9 histone marks using ChIP-seq, and of chromatin accessibility using ATAC-seq to gain greater insight into the role of chromatin regulation in TNF-induced reprogramming of the TLR4 response.
We found that tolerization with TNF attenuated LPS-induced increases of the positive histone marks H4-Ac and H3K4me3 (associated with open chromatin and transcription), and of increased chromatin accessibility (ATAC-seq reads) (Fig. 2a, quantitation shown in Supplementary Fig. 3a; representative gene tracks are shown in Fig. 2b and Supplementary Fig. 3b). These results are in accord with a model that in ‘tolerized’ cells LPS is unable to generate a sufficiently strong signal to induce chromatin remodeling that is required for effective induction of transcription3,26,29,30. We also found that tolerization attenuated LPS-induced increases in H2BK120 ubiquitination (H2Bub) (Fig. 2a), a positive mark that serves to increase H3K4me3 and open chromatin and is a prerequisite for H3K4me3 in other systems. This implies that inability of LPS to generate signals that lead to H2Bub contributes to the tolerization of these genes.
Figure 2.
Distinct epigenetic landscape at different TLR4-induced gene classes. (a) Heatmaps of H4ac, H3K4me3, H2Bub ChIP-seq and ATAC-seq normalized tag densities at the promoters (−2kb < TSS < +2kb) of a given gene class based on Fig. 1b. The order of genes in each column is the same for all heatmaps and Fig. 1b. (see Supplementary Fig. 3a for quantitation). (b) Representative UCSC Genome Browser tracks displaying normalized tag density profiles for H4ac, H3K4me3, H2Bub ChIP-seq, ATAC-seq and RNA-seq signals at IL6 (Class 1), CCL5 (Class 3), and CH25H (Class 4) genes in the indicated conditions. Boxes enclose genomic regions showing differential regulation. Data (a and b) show results from one representative donor; results from biological replicates (ChIP-seq) and pooled data from 3–5 replicates (ATAC-seq) are shown in Supplementary Fig. 4a. (c) Heatmaps showing per nucleotide ATAC-seq cleavage sites for NFκB-p65 and PU.1 motifs in LPS-stimulated human primary macrophages ranked by tag density. The number of ATAC-seq footprints for each TF is shown on y-axis. 200 bp windows are shown centered at the midpoints of the ATAC-seq footprint. Footprinting was performed with two independent ATAC-seq replicates.
Genes in nontolerized Classes 3–6 exhibited distinct chromatin regulation profiles. Notably, Class 3 NT genes, functionally related to IFN/cytokine Jak-STAT signaling, were ‘marked’ by H2Bub and H4-Ac after TNF treatment, which was associated with substantial inducibility of H3K4me3, opening of chromatin, and robust induction of gene expression in response to weak signaling upon LPS challenge (Fig. 2a and Supplementary Fig. 3a; representative gene tracks are shown in Fig. 2b and Supplementary Fig. 3b). This suggests that marking or ‘priming’ of Class 3 NT genes by H2Bub and H4-Ac enables chromatin remodeling and transcriptional responses even to weak signals.
Class 4–6 genes differed from Class 1–3 genes in that H2Bub, H3K4me3, and open chromatin (ATAC-seq read density) were lower at baseline in naive macrophages and weakly inducible by LPS (Fig. 2a and Supplementary Fig. 3a, c). Instead, these positive marks and opening of chromatin were induced during TNF stimulation (Fig. 2a, b and Supplementary Fig. 3a, b), suggesting that their epigenetic profile is ‘primed’ by TNF. Patterns of regulation were confirmed in additional replicates (Supplementary Fig. 4a), and for select genes by ChIP-qPCR and FAIRE in additional donors (Supplementary Fig. 4b and data not shown). Exceptions where histone marks did not correlate with transcriptomic changes are discussed in Supplementary Note 1; ChIP-seq data on other less informative or repressive histone marks are shown in Supplementary Fig. 3c–e. Regulation of histone marks and chromatin accessibility at enhancers in TNF-treated cells paralleled that of promoters but was quantitatively less dynamic (Supplementary Fig. 5a). In addition, CpG islands11, super-enhancers31, and latent enhancers20 did not correlate with patterns of regulation of the different gene classes (Supplementary Fig. 5b–d). Overall, the data support a model whereby TNF alters chromatin states at TLR4-inducible gene promoters to reprogram the TLR4-induced gene response to silence expression of inflammatory NF-κB-dependent genes, while augmenting the expression of cytokine-induced, antiviral, and metabolic genes.
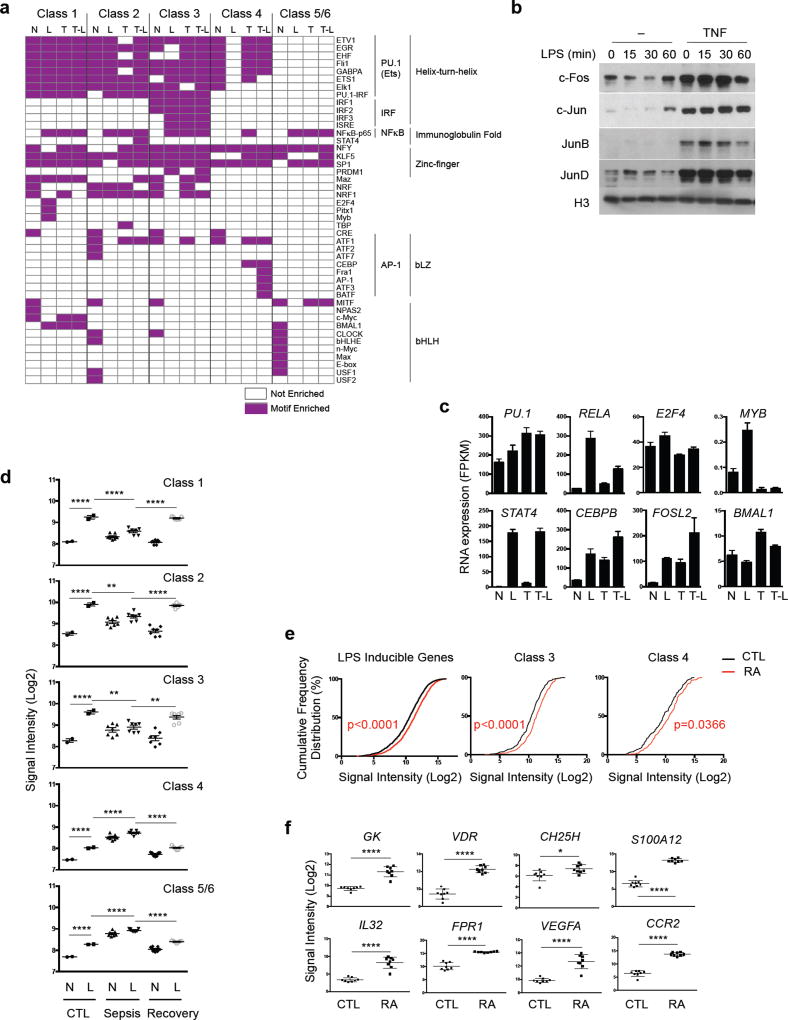
To identify candidate transcription factors that could explain the differential expression and regulation of the distinct gene classes, we identified digital footprints (p < 10−10) ‘underneath’ ATAC-seq promoter peaks followed by matching to all known transcription factor motifs. This approach has the advantage compared to motif enrichment in that it identifies sites that are actually bound by TFs, rather than just motifs that have the potential to bind TFs. Examples of well-delineated TF footprints are shown in Fig. 2c and Supplementary Fig. 4c. Footprinting analysis recovered binding of PU.1/Ets elements, of NF-Y and SP-1 core promoter elements32, and inducible NF-κB element binding, thus supporting the validity of the approach (Fig. 3a). Strikingly, footprinting analysis clearly identified distinguishing features among the promoters in the different gene classes (Fig. 3a). The most salient class-specific characteristics were inducible occupancy of IRF and ISRE sites at Class 3 NT gene promoters, which is consistent with regulation by IFNs and cytokines, and of AP-1 sites under T and TL conditions in Class 4 gene promoters. Accordingly, TNF induced sustained expression of AP-1 proteins (Fig. 3b) and induction of these genes was sensitive to MAPK inhibitors (ref.26 and data not shown). Patterns of expression of members of relevant transcription factor families are shown in Fig. 3c and Supplementary Table 1. Additional detailed description of footprinting results is provided in Supplementary Note 2.
Figure 3.
(a–c) Distinct transcription factor binding at different gene classes. (a) Heatmap of significantly enriched motifs (p < 10−5) within ATAC-seq footprints (p < 10−10) in gene class promoter regions (−2kb < TSS < +2kb) in indicated conditions. Motifs are grouped according to transcription factor families. (b) Immunoblot of nuclear lysates from macrophages stimulated overnight with TNF. (c) Bar graphs show cumulative values for representative TFs that match motifs in (a) from three replicates. (d–e) Expression of inflammatory gene classes in sepsis monocytes and RA synovial macrophages. (d) Expression of genes belonging to each gene class in healthy donor monocytes (CTL, n = 2), or monocytes from patients during sepsis (Sepsis, n = 7) and after recovery from sepsis (Recovery, n = 7) stimulated ex vivo with or without LPS. Each dot represents the average (log2) of a gene class in an individual donor. Data are presented as mean ± SEM. The gene expression data are from GSE46955. ****p < 0.0001, **p<0.01, analysis of variance and Dunnett’s multiple comparison post hoc test. (e) Cumulative distribution plot of normalized gene expression (log2) of LPS-inducible, Class 3 and Class 4 genes in RA synovial macrophages (RA, n=8) and control macrophages (CTL, n=8). p-value: CTL vs. RA, Kolmogorov-Smirnov test. (f) Expression of representative Class 4 genes in control or RA macrophages. Data are presented as mean ± SEM. **** p < 0.0001, *** p < 0.001, ** p < 0.01, * p < 0.05, unpaired Student’s t-test.
Footprinting analysis of enhancers showed inducible binding of NF-κB, IRF, AP-1 and C/EBP-AP-1 sites (Supplementary Fig. 5e) similar to inducible binding that was detected at promoters, suggesting that enhancers are also responsive to TNF and LPS. However, the occupancy of TF binding sites at enhancers was more similar among the six gene classes than was occupancy of promoters. In accord with the footprinting analysis, de novo motif enrichment analysis under enhancer ATAC-seq peaks using HOMER showed similarity among the gene classes (Supplementary Fig. 5f). Overall, the ChIP-seq and footprinting results provide insight into the distinct regulatory logic of the different gene classes, and suggest a role for IRF/ISGF3 (Class 3 NT) or AP-1 (Class 4 NT) for opening chromatin and allowing these genes to escape tolerance.
Expression of inflammatory gene classes in human diseases
To address whether our findings reflect inflammatory gene regulation in vivo, we examined expression of the six classes of LPS-inducible genes in human disease states. First, we analyzed gene expression in monocytes from sepsis patients collected during sepsis (where they are exposed to both endotoxin and TNF) and after recovery, and stimulated ex vivo with LPS28. Strikingly, genes in Class 1 and 2 exhibited tolerization in vivo that was reversed when patients recovered (Fig. 3d), thus recapitulating our in vitro model. Genes in classes 4–6 also exhibited similar expression patterns in sepsis patients as in our system, but Class 3 genes exhibited limited inducibility by LPS ex vivo. This reinforces the notion that inducibility of IFN target genes is a feature that distinguishes TNF-induced reprogramming from endotoxin tolerance. We also found that expression of genes in two non-tolerized gene sets, Class 3 and Class 4, was significantly elevated in synovial macrophages from patients with RA, a condition where inflammation is driven by TNF (Fig. 3e and 3f). Overall, these results support that the patterns of gene regulation and classes of inflammatory genes we identified in our model system reflect aspects of inflammatory gene regulation in vivo in infectious/inflammatory disease states.
Type I IFNs abrogate TNF-mediated tolerance
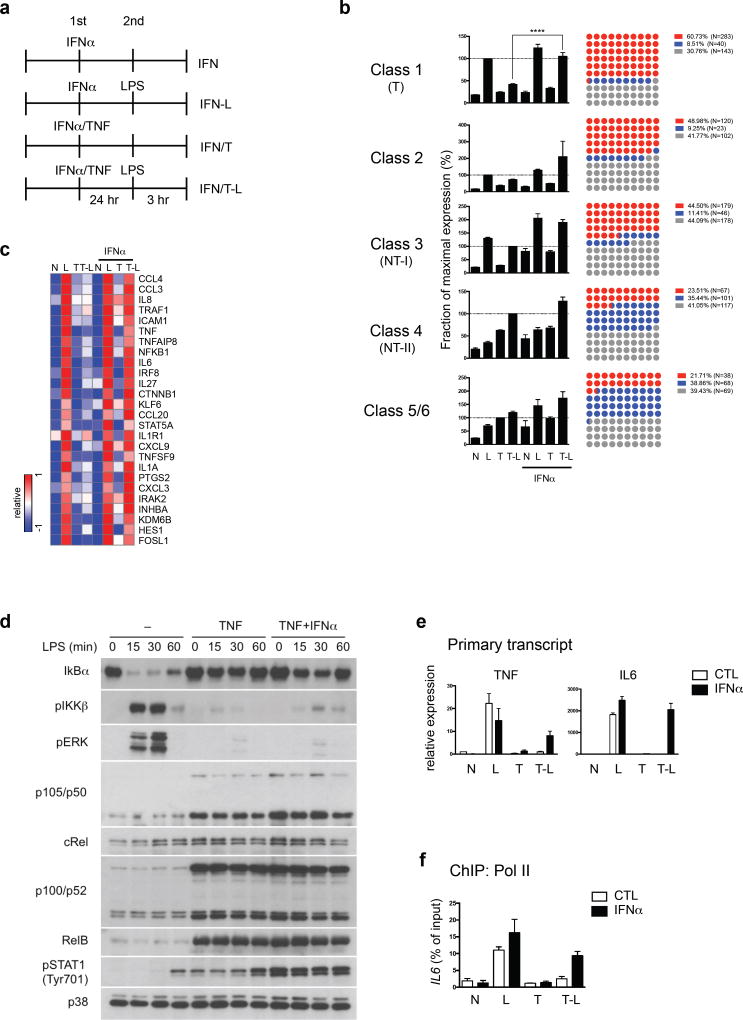
Building on previous work that inhibition of glycogen synthase kinase 3 (GSK3) reverses TNF-induced tolerance and GSK3 regulates IFN production3,5,33, we found that reversal of tolerization of IL6 by GSK3 inhibition was mediated by increased type I IFNs (Supplementary Fig. 4d–f). We tested the effects of type I IFNs on TNF-induced tolerance genome-wide using RNA-seq. Addition of exogenous IFN-α together with TNF (Fig. 4a) significantly restored LPS-inducibility to the majority of Class 1 T genes (60.7%, 283/466) (Fig. 4b and 4c), indicating a broad but gene-specific reversal of tolerance. IFN-α differentially affected expression of the 6 TLR4-induced gene classes and few genes in the NT Classes 4–6 (21–23%) were upregulated by IFN-α (Fig. 4b). Notably, type I IFN did not reverse tolerization of TNF and IL6 by LPS in the classic endotoxin model (Supplementary Fig. 4g). IFN-α increased LPS-induced Class 1 gene expression only in TNF-treated cells (Fig. 4b), in accord with extensive literature that Class 1 T genes are not canonical ISGs. Thus, crosstalk between IFN and TNF couples IFN signaling with Class 1 NF-κB target genes and prevents their tolerization.
Figure 4.
Type I IFNs block TNF-mediated tolerization of inflammatory genes without affecting LPS signaling. (a) Experimental design: IFN, treatment with IFN-α (25 ng/ml); IFN-L, treatment with IFN-α, followed by LPS challenge (10 ng/ml); IFN/T, treatment with IFN-α and TNF (10 ng/ml); IFN/T-L, treatment with IFN-α and TNF, followed by LPS. (b) Bar graphs represent cumulative values for a given gene class in RNA-seq analysis (left) from three replicates (% of maximum value). Error bars indicate SEM. The dot plots (right) show percent of genes in each class that were up-regulated, down-regulated or not changed by IFN-α (>1.5-fold, T-L vs. IFN/T-L). Each dot represents 1% of genes; red = upregulated, blue = downregulated, grey = not changed. ****p < 0.0001, *p<0.05, analysis of variance and Dunnett’s multiple comparison post hoc test. (c) Heatmap showing inflammatory Class 1 genes whose tolerization is reversed by IFN-α treatment. (d) Immunoblot analysis of IκBα, p105/p50, cRel, p100/p52, RelB and phosphorylated IKKβ, ERK and STAT1 in primary macrophages cultured for 24 h with TNF (10 ng/ml) with or without IFN-α (25 ng/ml), and challenged for the indicated times with LPS (10 ng/ml). Data are representative of four experiments. (e) RT-qPCR analysis of TNF and IL6 primary transcripts normalized relative to HPRT. Data are representative of five independent donors and error bars indicate SEM. (f) ChIP assays for recruitment of Pol II to IL6 promoter in the indicated conditions. Data are representative of 4 different donors.
We tested whether IFN-α could augment LPS-induced signaling in tolerized macrophages. As expected3, robust LPS-induced I-κBα degradation and activation of IKK and ERK were observed in naïve macrophages but strongly blunted in TNF-tolerized macrophages (Fig. 4d). In contrast to its effects on gene expression, IFN-α did not reverse these defects in proximal LPS-induced TLR signaling (Fig. 4d, lanes 5–8 versus 9–12). IFN-α also did not affect TNF-induced expression of noncanonical NF-κB proteins suggested to play a role in tolerance27,34. These results indicate that IFN-α strongly affects transcriptional responses of Class 1 and 2 genes without substantially altering proximal TLR signaling defects.
The gene-specific effects of IFN-α (Fig. 4b, right panels), together with its ability to enable robust induction of T genes in response to very weak TLR4-induced proximal signals in TNF-pretreated cells, suggested that IFN-α exerts its effects in the nucleus at the level of gene regulation. To test this idea, we first confirmed that IFN-α-mediated abrogation of gene tolerization occurs at the level of transcription by measuring primary transcripts (Fig. 4e). Furthermore, ChIP-qPCR experiments showed that IFN-α overcame the TNF-induced block in RNA polymerase II (pol II) recruitment to the tolerized gene IL6 (Fig. 4f). These results support the notion that IFN-α signaling acts in the nucleus to regulate gene expression.
Crosstalk between IFN and TNF primes chromatin at T genes
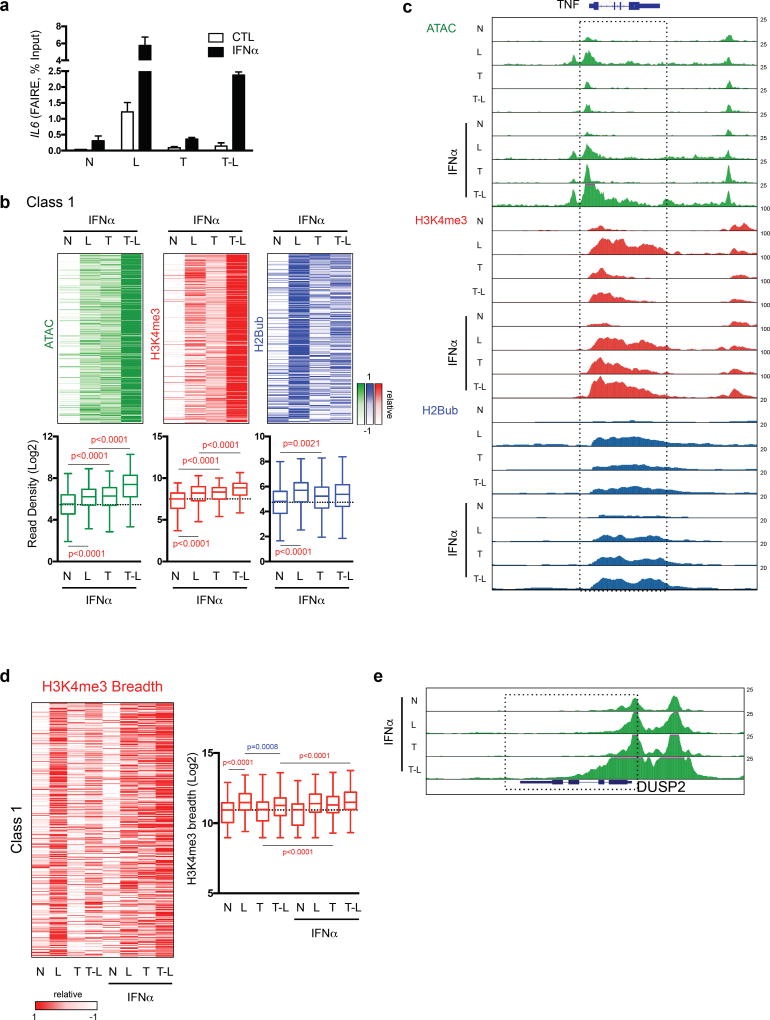
We reasoned that IFN-α could amplify transcriptional responses of Class 1/2 genes to weak LPS signals in TNF-treated macrophages by remodeling chromatin. This notion was supported by FAIRE assays showing that IFN-α promoted opening of chromatin at the IL6 promoter in TNF-treated macrophages (Fig. 5a). We then used ATAC-seq and ChIP-seq to analyze chromatin accessibility and the positive H2Bub and H3K4me3 marks genome-wide. IFN-α did not significantly increase ATAC-seq read counts at the majority of Class 1 genes in resting or LPS-stimulated naïve cells (Supplementary Fig. 6b). However, the combination of IFN-α and TNF resulted in increased chromatin accessibility at Class 1 gene promoters, with a further increase upon LPS stimulation (Fig. 5b and Supplementary Fig. 6b). A similar signal-responsive but quantitatively less dynamic pattern was observed at Class 1 gene enhancers (Supplementary Fig. 6c). Representative gene tracks are shown in Fig. 5c and Supplementary Fig. 6d.
Figure 5.
Integration of signaling crosstalk between IFN and TNF at the chromatin level. (a) Formaldehyde-assisted isolation of regulatory elements (FAIRE) assay at IL6 gene under indicated conditions. Data are representative of 4 experiments; error bars show SEM. (b) Heatmaps of ATAC-seq and H3K4me3, H2Bub ChIP-seq normalized tag densities at the promoters (−2kb < TSS < +2kb) of Class 1 genes ordered as in Fig. 1b (upper). Box graphs represent quantitation of the normalized tag densities (log2) for the indicated conditions. p value, Kolmogorov-Smirnov test. (c) Representative UCSC Genome Browser tracks displaying normalized profiles for ATAC-seq, H3K4me3 and H2Bub ChIP-seq signals at TNF gene under indicated conditions. ATAC-seq data represents pooled data from three to five biological replicates. (d) Heatmap of breadth of H3K4me3 ChIP-seq peaks at Class 1 gene promoters under indicated conditions (upper). Box graphs represent quantification of the H3K4me3 breadth (log2) for Class 1 genes (lower). p-value, Kolmogorov-Smirnov test. (e) Representative UCSC Genome Browser tracks displaying normalized profiles for ATAC-seq signals at IL1B, DUSP2 and NFKBIA (Class 1) genes under indicated conditions. Boxes enclose ATAC-seq peaks that extended into gene bodies in IFN/T-L condition.
A similar pattern of increased H3K4me3, which marks open chromatin, was observed when IFN-α was added in the T and TL conditions (Fig. 5b, c and Supplementary Fig. 6b, d). The breadth of H3K4me3 peaks and extension of this ‘promoter mark’ into gene bodies, which is associated with increased transcription35, was increased by IFN-α in a similar manner (Fig. 5d). Interestingly, IFN-α and TNF cooperated to increase H3K4me3 peak breadth. Finally, H2Bub, a prerequisite for H3K4me3 in other systems, was also increased when IFN-α and TNF were added together. These results are strikingly different from results obtained in the absence of IFN-α, where chromatin remained closed (Fig. 2a and Supplementary Fig. 6b). The results show that addition of IFN-α together with TNF conditions or ‘primes’ chromatin at Class 1 gene promoters, which can facilitate transcriptional responses to weak LPS signals. In accord with H3K4me3 marking open chromatin, in IFN-α- plus TNF-treated macrophages stimulated with LPS (IFN/T-L condition) we observed broad ATAC-seq peaks that extended into gene bodies (Fig. 5c, e). Thus, opening of chromatin at Class 1 genes is a salient feature of IFN-α action in TNF-treated macrophages, and IFN-α and TNF cooperate to prevent gene silencing.
IFN and TNF cooperate to recruit TFs to Class 1 promoters
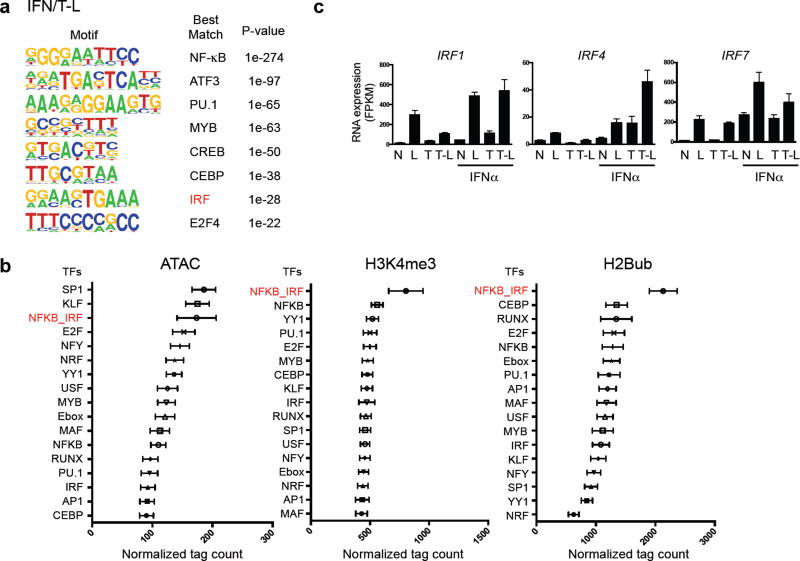
We addressed the possibility that opening of chromatin at Class 1 gene promoters under tolerizing conditions requires cooperation between TFs induced by IFN-α and TNF. De novo motif analysis of TF footprints under ATAC-seq peaks in IFN/T-L conditions revealed that under IFN-stimulated conditions these promoters newly gain occupancy of IRF sites (Fig. 6a). A similar ISRE site was enriched in Class I gene enhancers (Supplementary Fig. 7a). Motif analysis of TF footprints in the most ‘primed’ chromatin, as defined by peaks with high read density of ATAC-seq, H2Bub or H3K4me3, revealed enrichment of NF-κB and IRF motifs (Fig. 6b). Most strikingly, peaks with the highest chromatin accessibility were associated with a composite NF-κB/IRF motif (Fig. 6b, red font), and several IRFs were induced in IFN-α-treated tolerized macrophages (Fig. 6c and Supplementary Fig. 7b). This suggested that coordinate binding by TNF-induced NF-κB and IFN-induced IRFs to Class I promoters contributes to increased chromatin accessibility.
Figure 6.
IFN and TNF prime chromatin by cooperatively recruiting transcription factors to Class 1 promoters. (a) De novo motif enrichment analysis of promoter regions (−2kb < TSS < +2kb) of Class 1 genes using ATAC-seq footprints of IFN/T-L condition. Random background regions were selected as a control. (b) Graphs represent association of occupied transcription factor binding sites (footprints) with chromatin accessibility and histone modifications. Data are presented as mean ± SEM. Relative chromatin accessibility or histone modification (x-axis, normalized ATAC-seq, H2Bub or H3K4me3 tag counts) is measured as the mean intensity of ATAC-seq, H2Bub or H3K4me3 peaks containing the indicated motif (y-axis) across all experimental conditions. (c) Bar graphs represent cumulative values for IRF1, 4 and 7 in RNA-seq analysis from three replicates.
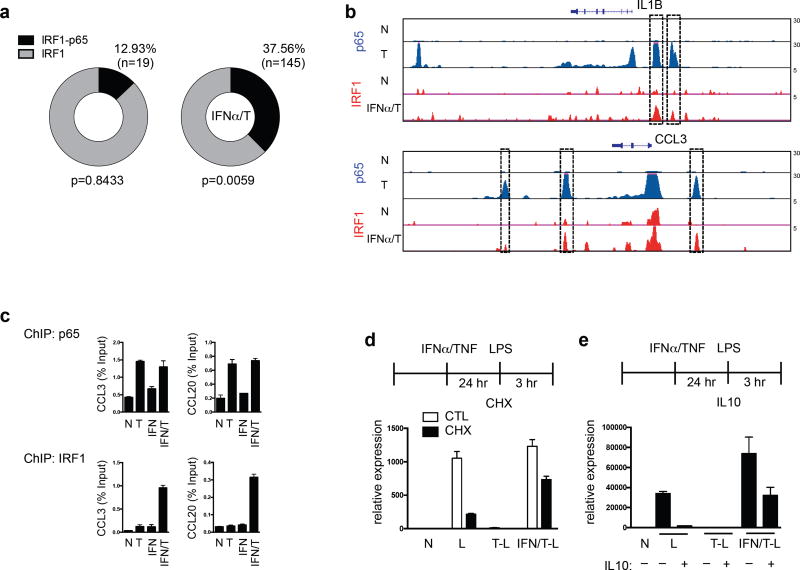
This notion was supported by ChIP-seq data that upon TNF + IFNα treatment a large fraction of IRF1 binding peaks at Class 1 gene promoter elements (ATAC-seq peaks) colocalized with NF-κB p65 peaks (Fig. 7a, p = 0.0059 relative to random genes). Representative gene tracks showing alignment of p65 and IRF1 binding peaks are depicted in Fig. 7b and Supplementary Fig. 7c). Increased co-recruitment of IRF1 and p65 after TNF + IFNα treatment was confirmed for a subset of genes by ChIPq-PCR (Fig. 7c and Supplementary Fig. 7d). Significantly increased colocalization of IRF1 and p65 binding was also observed when an independent data set (GSE43036) was used to define IRF1 peaks (Supplementary Fig. 7e, f). Overall, these results support a model (Supplementary Fig. 8) whereby TNF-induced NF-κB and IFN-α-induced IRFs bind coordinately to promoters of Class 1 tolerized genes and cooperate to maintain an open and ‘primed’ chromatin state, which enables strong transcriptional responses to weak LPS-induced signals.
Figure 7.
(a–c) Colocalization of IRF1 and NF-kB p65 in IFN-α and TNF-treated macrophages. (a) Circle represents 100% of Class 1 gene promoter IRF1 Chip-seq peaks in unstimulated (left, n = 147) or IFNα/T conditions (right, n = 386). The peak fraction that overlaps with p65 ChIP-seq peaks is shaded in black. P value was calculated relative to random genes. (b) Representative UCSC Genome Browser tracks displaying normalized profiles for p65 and IRF1 ChIP-seq signals at IL1B and CCL3 genes in indicated conditions. Boxes enclose co-localization of p65 and IRF1 binding peaks in the same genomic regions. (c) ChIP-qPCR analysis of recruitment of p65 and IRF1 to CCL3 and CCL20 promoters. Data are representative of three independent experiments. (d–e) Altered transcriptional requirements in IFN-α + TNF-treated macrophages. Naïve, TNF-, or IFN-α + TNF-treated macrophages were stimulated with LPS (10 ng/ml) with or without CHX (d, 20 µg/mL) or IL-10 (e, 10 ng/ml) and RT–qPCR was performed. Data are representative of three independent donors, and error bars indicate SEM.
Escape of IL6 transcription from suppression by IL-10
One potential consequence of ‘priming’ of chromatin in IFN-α + TNF-treated macrophages is a change in transcriptional requirements for LPS-induced gene activation. We tested this idea for IL6, a Class 1 gene whose induction by LPS in naïve macrophages is dependent on de novo protein synthesis (which is required for increased chromatin accessibility)11,30, and is suppressed by the potent anti-inflammatory cytokine IL-10. In contrast to naïve LPS-stimulated macrophages (Fig. 7d, bars 3, 4), in IFN-α + TNF-treated macrophages IL6 expression was strongly induced by LPS in the presence of the protein synthesis inhibitor cycloheximide (Fig. 7d, bars 7, 8). IL-10 essentially completely suppressed LPS-induced IL6 expression in naïve macrophages, but only partially in IFN-α-primed tolerized macrophages (Fig. 7e). These results indicate that ‘priming’ by crosstalk between IFN-α and TNF changes the regulatory logic of gene expression and can make genes resistant to suppression by anti-inflammatory stimuli, thereby promoting sustained TNF-driven inflammation.
Chromatin accessibility in SLE monocytes
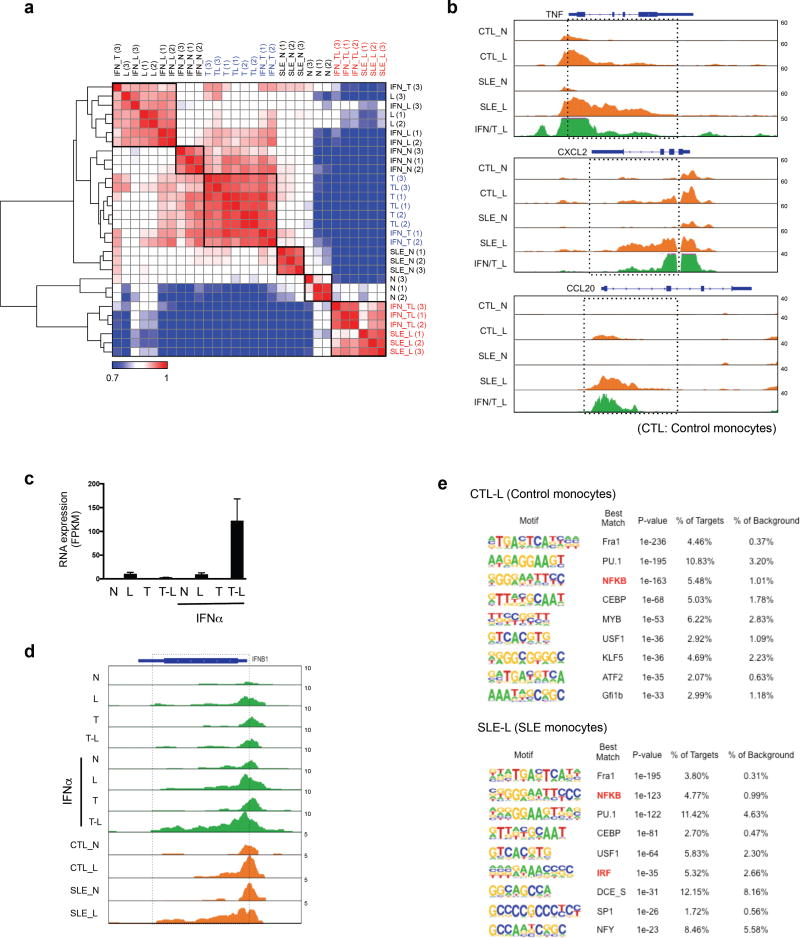
Type I IFNs have been proposed to contribute to SLE pathogenesis by opposing tolerance induction36. We reasoned that SLE monocytes, which have been exposed to IFNs, TNF and TLR ligands in vivo, might exhibit an altered epigenomic/chromatin landscape that reflects some of the IFN-mediated regulatory mechanisms we have described in this study. Pearson correlation analysis showed that the chromatin accessibility profile of SLE monocytes stimulated with LPS ex vivo (labelled SLE-L) was closely correlated with the profile of IFN-α-treated tolerized cells stimulated with LPS in our system (labelled IFN/T-L) (Fig. 8a, shown in red font), but did not correlate with LPS-stimulated naïve (Fig. 8a, rows 2,4,5) or tolerized (rows 13–16) cells. Examination of gene tracks of individual genes (Fig. 8b) revealed that in SLE monocytes LPS induced a broad region of chromatin accessibility that extended into the gene bodies, reminiscent of results obtained with IFN-α-treated tolerized cells (IFN/T-L condition, Fig. 8b and 5c, e). This is in accord with the broad H3K4me3 peaks in our system (Fig. 5d, e) and reported in SLE monocytes37.
Figure 8.
Chromatin accessibility in SLE monocytes. (a) Correlation matrix heatmap based on unsupervised Pearson correlation coefficients comparing normalized ATAC-seq tag densities at promoters (−2kb < TSS < +2kb) of Class 1 genes across all indicated conditions and replicates. (SLE-N: untreated SLE monocytes, SLE-L: LPS (10 ng/ml)-treated SLE monocytes). (b) Representative UCSC Genome Browser tracks displaying normalized profiles for ATAC-seq signals at TNF, CXCL2 and CCL20 genes in indicated conditions. (CTL: control monocytes, SLE: SLE monocytes). Boxes enclose a broad region of chromatin accessibility that extends into the gene bodies in SLE monocytes. (c) Bar graph represents cumulative values for IFNB1 in RNA-seq analysis under indicated conditions from three replicates. (d) UCSC Genome Browser tracks displaying normalized profiles for ATAC-seq signals at IFNB1 under indicated conditions. (CTL: control monocytes, SLE: SLE monocytes). Boxes enclose a broad region of chromatin accessibility that extends into the gene bodies in IFN/T-L and SLE-L conditions. (e) De novo motif enrichment analysis of Class 1 gene promoters using ATAC-seq footprints of LPS-stimulated control monocytes (CTL-L) and SLE monocytes (SLE-L).
Given the importance of type I IFNs in SLE, we next turned our attention to the IFNB1 gene. Strikingly, IFN-α led to a massive superinduction of IFNB1 by LPS, but only in cells that were also treated with TNF (Fig. 8c, bar 8). Thus, IFNB1 resembles Class 1 genes in that induction requires preconditioning by IFN-α and TNF followed by LPS challenge. LPS challenge of IFN-α + TNF-treated cells induced a broad region of chromatin accessibility at IFNB1 that extended from the promoter into the gene body, and similar results were obtained using LPS-stimulated SLE, but not control, monocytes (Fig. 8d). To further compare SLE monocytes with IFN-α-primed tolerized monocytes, we performed de novo motif analysis underneath ATAC seq peaks in LPS-challenged cells. Similar to IFN/T-L cells (Fig. 6a), SLE monocytes stimulated with LPS showed enrichment of IRF motifs, which likely is related to in vivo exposure to type I IFNs, and was not observed in control monocytes stimulated with LPS (Fig. 8e). Overall, the results show similarities in LPS-induced chromatin accessibility in Class 1 genes between SLE monocytes and IFN-α-treated tolerized monocytes, suggesting that our model system mimics aspects of chromatin regulation in an IFN-mediated disease in vivo.
Discussion
Epigenomic reprogramming has been linked to tissue-specific macrophage phenotypes18–23,38,39, but how reprogramming affects inflammatory responses is not well understood15,40–42. In this study we found that TNF and IFN-α reprogram the human macrophage epigenome to alter inflammatory responses to TLR4 stimulation. TNF induces a balanced response that limits potentially toxic induction of inflammatory NF-κB target genes, while enabling expression of antiviral, metabolic, and Jak-STAT target genes. Type I IFNs potentiated TNF inflammatory function by preventing the silencing of inflammatory genes. Mechanistically, type I IFNs and TNF cooperate to induce signals and transcription factors that prime chromatin at inflammatory gene promoters to make them responsive to weak signals, and also resistant to suppression by IL-10. Overall, our findings identify a new function of type I IFNs, and reveal that signaling crosstalk between IFN-α and TNF is integrated at the level of chromatin to crossregulate transcriptional responses to LPS.
There are important differences between TNF-induced reprogramming and previously described ‘endotoxin tolerance’. These include the abrogation of TNF-induced silencing of T genes by type I IFNs and the nature of the ‘NT’ response, which determines the macrophage functional phenotype. The ability of an endogenous cytokine like IFN-α to prevent silencing of T genes suggests that such ‘tolerization’ is a physiological process that is regulated by cytokines to fine tune the magnitude, duration, and qualitative nature of inflammatory responses, rather than a ‘last ditch effort’ to prevent endotoxin toxicity that can lead to profound immunosuppression and death. On the other hand, increased expression of type I IFNs, as occurs in several autoimmune diseases, will inactivate an important TNF-induced homeostatic mechanism that places a ‘brake’ on inflammatory gene expression and can contribute to inflammatory pathogenesis.
Our results provide substantial new insights about the functions and mechanisms regulating expression of genes that are effectively or synergistically activated by LPS in TNF-pretreated macrophages (the ‘ NT response’). One notable finding is discovery of Class 3, which is comprised of NT genes important for cytokine-Jak-STAT and IFN-antiviral responses. Induction of these genes is functionally important, as it allows cells exposed to TNF-driven inflammation to preserve antiviral host defense. Interestingly, Class 3 genes are tolerized (silenced) in the classical endotoxin tolerance model, as endotoxin induces additional and stronger tolerance mechanisms than does TNF3. Accordingly, compromised antiviral responses, superinfection, and reactivation of latent viruses are major complications in sepsis patients with endotoxin-tolerized cells. We have also identified new classes of ‘NT genes’, new functions, potential roles for transcription factors including SREBP2, AP-1 and E box proteins, and chromatin-based mechanisms that enable robust NT gene induction. A primed chromatin state, especially high H3K4me3, can greatly reduce requirements for activation of gene transcription43 and facilitate robust transcriptional responses to weak signals. The results overall are consistent with a ‘sequential rheostat’ model where environmental cues can independently regulate the intensity of upstream signaling and the accessibility of downstream chromatin. In this model, closed chromatin can block a strong signal, whereas open/primed chromatin can amplify a weak signal, in a gene-specific manner.
Type I IFNs have pleiotropic immune stimulatory and suppressive effects6. Little is known about mechanisms that determine context-dependent type I IFN functions. Classically, type I IFNs promote inflammation/immunity by inducing transcription of ISGs that harbor binding sites for ISGF3/STATs/IRFs and encode chemokines and antigen-presenting molecules. Previous reports, in line with our results, showed that NF-κB-driven inflammatory cytokine genes, such as those that comprise Class 1, are not induced and if anything are suppressed by type I IFNs6,8,9,44. In sharp contrast, in the context of co-treatment with TNF, IFNAR signaling was coupled to NF-κB target genes, as it prevented tolerization of a large fraction of Class 1 and 2 genes. Thus, in addition to induction of canonical ISGs, type I IFNs regulate chromatin at distinct inflammatory and possibly other gene sets. This regulatory role likely extends beyond the system used in this study, and may contribute to phenomena such as innate immune training22,42 and maintenance of basal immune responsiveness by commensal microbiota, which has been linked to IFNs and increased H3K4me3 at Tnf and Il6 promoters7,45.
Genomic regulatory sequences show less conservation between human and mouse than do coding sequences. One advantage of using a human macrophage experimental system is that it enables direct comparisons with pathogenic macrophages obtained from clinical samples. It is encouraging that our results model aspects of gene expression or chromatin regulation patterns in monocytes/macrophages from patients with sepsis/recovery, RA and SLE37. SLE is characterized by IFN production and TLR activation in which TNF can be protective (consistent with tolerizing inflammatory genes). These similarities support the utility of our system to model and dissect mechanisms relevant for disease pathogenesis. It will be interesting to test in future work whether type I IFNs promote SLE pathogenesis in part by preventing tolerization of inflammatory ‘T’ genes, as has already been suggested36. Equally importantly, the mechanisms we have discovered and data sets we have developed provide molecular signatures linked to pathogenic cytokines and pathways that can motivate and help guide interpretation of studies using patient samples. Finally, our in vitro model can be exploited to develop and test the efficacy of therapeutic approaches that target epigenetic mechanisms that regulate cytokine production.
In summary, our results reveal how signaling crosstalk between type I IFNs, TNF and TLR4 is integrated at the level of chromatin, and associate these chromatin changes with reprogramming of gene expression. They highlight the concept that chromatin is not just a target that propagates signaling cascades, but instead serves as an integration node that determines transcriptional output. These findings provide insights into regulation of inflammatory gene expression that can be used to develop approaches to modulate macrophage activation and cytokine production by targeting chromatin regulators.
METHODS
Cell culture, purification and stimulation
Primary human CD14+ monocytes were isolated from buffy coats purchased from the New York Blood Center using anti-CD14 magnetic beads (Miltenyi Biotec) as previously described, using a protocol approved by the Hospital for Special Surgery Institutional Review Board. Monocytes were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated defined FBS (HyClone Fisher), penicillin/streptomycin (Invitrogen), L-glutamine (Invitrogen), and 20 ng/ml human macrophage colony-stimulating factor (M-CSF; Peprotech). Recombinant human TNF and IFN-α were from Peprotech and PBL science respectively (endotoxin concentrations were below limit of detection (<0.1 pg/ g)). LPS (tlrl-3pelps) and SB216763 (S3442) were from Invivogen and Sigma respectively.
Analysis of mRNA and protein
Total RNA was extracted with an RNeasy Mini Kit (Qiagen) and was reverse-transcribed with a First Strand cDNA Synthesis kit (Fermertas). Real-time PCR was performed in triplicate with Fast SYBR Green Master Mix and 7500 Fast Real-time PCR system (Applied Biosystems). Whole-cell extracts were prepared as described and were fractionated by 7.5–10% SDS-PAGE, transferred to polyvinylide fluoride membranes (Millipore) and incubated with specific antibodies, then enhanced chemiluminescence was used for detection (Amersham). Antibody to phosphorylated IKKβ (2697), Erk (4377) and STAT1 (9171), and antibody to IκBα (4812), p105/p50 (3035), cRel (4727), p100/p52 (3017), and RelB (4922) were from Cell Signaling. Anti-p38 (sc-535) was from Santa Cruz Biotechnology.
RNA-sequencing
After RNA extraction, libraries for sequencing were prepared using the Illumina TruSeq Stranded Total RNA Library Prep Kit following the manufacturer’s instructions. High throughput sequencing (50 bp, paired end) was performed at the Weill Cornell Medicine Epigenomic Core Facility. More than 100 million reads were obtained for each sample. After quality filtering according to the Illumina pipeline, paired-end reads were mapped to reference human genome (hg19 assembly) using STAR aligner version 2.4.0 with default parameters. Transcript abundance was quantified using Cufflinks 2.2.1, and Cuffdiff version 2.2.1 was used to determine differentially expressed genes. The expression levels of genes in each sample were normalized by means of fragments per kilobase of exon per million fragments mapped (FPKM). Independently processed replicates from three different donors showed high similarity (the lowest r value across samples was 0.86 for all genes (Refseq) and 0.9316 for LPS-inducible genes (n = 1,574), Fig. 1 and Supplementary Fig. 1).
Chromatin Immunoprecipitation and ChIP-sequencing
Cells were crosslinked for 5 min at room temperature by the addition of one-tenth of the volume of 11% formaldehyde solution (11% formaldehyde, 50 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0) to the growth media followed by 5 min quenching with 100 mM glycine. Cells were pelleted at 4°C and washed with ice-cold PBS. The crosslinked cells were lysed with lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100) with protease inhibitors on ice for 10 min and washed with washing buffer (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) for 10 min. The lysis samples were resuspended and sonicated in sonication buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% Nlauroylsarcosine) using a Bioruptor (Diagenode) with 30 sec ON, 30 sec OFF on high power output for 18 cycles. After sonication, samples were centrifuged at 12,000 rpm for 10 minutes at 4°C and 1% of sonicated cell extracts was saved as input. The resulting whole-cell extract was incubated with Protein A Agarose for ChIP (EMD Millipore) for 1 hr at 4°C. Precleared extracts were then incubated with 50 µl (50% v/v) of Protein A Agarose beads for ChIP (EMD Millipore) with 5–10 µg of the appropriate antibody overnight at 4°C. ChIP grade antibodies against H3K 4me3 (ab8580), H3K27ac (ab4729), H3K36me3 (ab9050) and IRF1 (ab26109, ChIP-qPCR) were from Abcam. Antibody against H2Bub (5546), p65 (8242, ChIP-qPCR) was from Cell Signaling Technology. ChIP antibodies against H3K56ac (39281) and H3K79me2 (39143) were from Active Motif. Antibody against H4ac (06-866) was from EMD Millipore. Antibodies against Pol II (sc-899), IRF1 (sc-497, ChIPmentation) and p65 (sc-372) were from Santa Cruz Biotechnology. After overnight incubation, antibody-bound agarose beads were washed twice with sonication buffer, once with sonication buffer with 500 mM NaCl, once with LiCl wash buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 250 mM LiCl, 1% NP-40), and once with TE with 50 mM NaCl. ChIPmentation was perfomed as described46 using magnetic beads (Thermo Scientific, 26162). The beads were washed twice with 10 mM cold Tris-HCl, pH 8.0, to remove detergent, salts and EDTA. Subsequently, beads were resuspended in 30 µl of the tagmentation reaction buffer (10 mM Tris, pH 8.0, 5 mM MgCl2) containing 1 µl Tagment DNA Enzyme from the Nextera DNA Sample Prep Kit (Illumina) and incubated at 37 °C for 10 min. Following tagmentation, the beads were washed twice with TE with 50 mM NaCl. After washing, DNA was eluted in freshly prepared elution buffer (1% SDS, 0.1M NaHCO3). Cross-links were reversed by overnight incubation at 65°C. RNA and protein were digested using RNase A and Proteinase K, respectively and DNA was purified with ChIP DNA Clean & Concentrator™ (Zymo Research). For ChIP assays, immunoprecipitated DNA was analyzed by quantitative real-time PCR and normalized relative to input DNA amount. For ChIP-seq experiments, 10 ng of purified immunoprecipitated DNA per sample was ligated with adaptors, and 100–300 bp DNA fragments were purified to prepare DNA libraries using Illumina TruSeq ChIP Library Prep Kit following the manufacturer’s instructions. For ChIPmentation experiments, we amplified library fragments using 1× NEB next PCR master mix and 1.25 M of custom Nextera PCR primers as previously described46, using the following PCR conditions: 72 °C for 5 m in; 98 °C for 30 s; and thermocycling at 98°C for 10 s, 63°C for 30 s and 7 2°C for 1 min. The libraries were purified using a Qiagen PCR cleanup kit yielding a final library concentration of ~30 nM in 20 µL. Libraries were amplified for a total of 8–10 cycles. ChIP libraries were sequenced (50 bp single end reads) using an Illumina HiSeq 2500 Sequencer at the Weill Cornell Medicine Epigenomic Core Facility per manufacturer's recommended protocol. For input DNA to be used as control for background noise, we fragmented 1 ng of chromatin for each sample, which underwent all steps of the ChIP-seq protocol except for immunoprecipitation and washing. Then, sequenced reads were aligned to reference human genome (GRCh37/hg19 assembly) using Bowtie2 version 2.2.6 with default parameters, and clonal reads were removed from further analysis. A minimum of 10 million uniquely mapped reads were obtained for each condition. Data in figures are from one representative out of two independent experiments (H2Bub, H3K4me3 and H3K36me3) with different blood donors or one experiment (H4ac, H3K27ac, H3K56ac, H3K79me2, H3K27me3 and H3K9me3).
ATAC-seq
ATAC-seq was performed as previously described41. To prepare nuclei, we spun 50,000 cells at 500g for 5 min, which was followed by a wash using 50 mL of cold 1× PBS and centrifugation at 500g for 5 min. Cells were lysed using cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630). Immediately after lysis, nuclei were spun at 500g for 10 min using a refrigerated centrifuge. Immediately following the nuclei prep, the pellet was resuspended in the transposase reaction mix (25 µL 2× TD buffer, 2.5 µL transposase (Illumina) and 22.5 µL nuclease- free water). The transposition reaction was carried out for 30 min at 37 °C. Directly following transposition, the sample was purified using a Qiagen MinElute kit. Then, we amplified library fragments using 1× NEB next PCR master mix and 1.25 M of custom Nextera PCR primers as previously described, using the following PCR conditions: 72 °C for 5 min; 98 °C for 30 s; and thermocycling at 98°C for 10 s, 63°C for 30 s and 72°C for 1 min. The libraries were purified using a Qiagen PCR cleanup kit yielding a final library concentration of ~30 nM in 20 µL. Libraries were amplified for a total of 10–13 cycles and were subjected to high-throughput sequencing using the Illumina HiSeq 2500 Sequencer (single end). ATAC-seq data was aligned to the genome using the same pipeline as ChIP-seq data.
Peak calling and annotation
We used the makeTagDirectory followed by findPeaks command from HOMER version 4.7.2 (http://homer.salk.edu/homer/) to identify peaks of ChIP-seq and ATAC-seq. A false discovery rate (FDR) threshold of 0.001 was used for all data sets. The following HOMER command was used: cmd = findPeaks <sample tag directory> -style histone (for histone modifications and ATAC-seq) or factor (for transcription factors) -o <output file> -i <input tag directory for ChIP-seq>. Super-enhancers were identified using H3K27ac ChIP-seq data in HOMER using the findPeaks –style super, –L 0 and –i < input sample > options. The total number of mapped reads in each sample was normalized to ten million mapped reads. Peak-associated genes were defined based on the closest genes to these genomic regions using RefSeq coordinates of genes. We used the annotatePeaks command from HOMER to calculate ChIP-seq and ATAC-seq tag densities from different experiments and to create heatmaps of tag densities. Sequencing data were visualized by preparing custom tracks for the UCSC Genome browser.
Classification of chromatin regions
Gene promoters were assigned to the genomic region within ±2 kb of a TSS (hg19). To determine potential enhancers, peaks of H3K27ac and ATAC-seq within 2 kb of a gene TSS were filtered out, then only peaks of ATAC-seq which overlapped with H3K27ac peaks were selected for the further analysis. For latent enhancers, we used peaks that have (1) up-regulated tag density of H3K4me1 with ATAC-seq in T or TL conditions and (2) no H3K27ac in N or L conditions. All enhancer peaks between the different conditions in each comparison were merged into one peak set using mergePeaks –size given. Each enhancer was assigned to the nearest TSS. CpG database was retrieved from the UCSC Genome browser (hg19).
Clustering and correlation analysis
To generate the heatmap of K-mean clusters with RNA-seq, we used Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) K-means algorithm with the Euclidean distance metric. K was chosen at 12 because lower values failed to identify all meaningful clusters and higher values subdivided meaningful clusters. The clusters were grouped into gene classes based primarily upon pattern of gene expression in the 4 experimental conditions (Fig. 1b) also taking into account the quantitative pattern of expression of each cluster (Supplementary Fig. 1a). For clusters that were on the border between two classes and difficult to assign solely based on pattern of gene expression, class assignment was supported by gene ontogeny analysis and similarity in induction kinetics (Supplementary Fig. 2a). For the correlation heatmap in Figure 8a, we calculated the pairwise Pearson’s correlation between samples (and replicates) using the ATAC-seq read density of gene promoters. To generate heat maps, we used GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/) set to relative comparison. Representative genes in Fig. 1d and 4c were selected based on GSEA hallmark gene sets, which summarize and represent specific well-defined biological states and display coherent expression.
Gene ontology analysis
For expression data in Fig. 1c, GO annotation was determined using the Gorilla (http://cbl-gorilla.cs.technion.ac.il/) tool on each of the 6 classes in Fig. 1b (using a ranked list as input). One representative category based on p-value is depicted if more than one similar category was identified. For super enhancers, enriched pathways were compiled from the GREAT (http://bejerano.stanford.edu/great/public/html/) tool. In both cases, pathways were ranked using p-value or binomial p-value respectively.
Digital genomic footprinting
Our ATACseq datasets (number of samples: 28; mean of total aligned read counts: 1.425×108) and signal to noise ratios sufficient to undergo digital genomic footprinting. For this purpose, the wellington_footprints (http://pythonhosted.org/pyDNase/index.html) function of the Wellington suite was used with standard parameters (p-value score, threshold 10−10) on BED3-converted peaks. Then, the footprints associated with LPS-inducible genes (Fig. 1b) at the p value of 10−10 were selected for the further study. To visualize footprints as heatmap showing enrichment of known transcription factors (Fig. 2c), we used pyDNase command ‘dnase_to_javatreeview.py’ to generate a CSV file that can be used in JavaTreeView or GENE-E.
Motif enrichment analysis
We used findMotifs function of HOMER to analyze the promoters of genes for motifs that are enriched in target gene promoters relative to other promoters (−300bp < TSS < +50bp, Fig. 1e). For the motif analysis comparing different conditions in Fig. 3a and Supplementary Fig. 5e, we used the known motif results to find which motifs in the HOMER and JASPAR databases were enriched in our data sets according to HOMER2 (p ≤ 10−5). Motifs corresponding to 1) TFs not expressed in our experimental setup or 2) low % of targets sequences (<2%) were excluded from the analysis. De novo transcription factor motif analysis was performed with motif finder program findMotifsGenome from HOMER package, on given ATAC-seq footprints. Peak sequences were compared to random genomic fragments of the same size and normalized G+C content to identify motifs enriched in the targeted sequences.
Determining relationships between transcription factor motifs and chromatin regulation
To obtain the results shown in Fig. 6b, occurrences of motifs from the JASPAR database were identified by running HOMER on the hg19 reference sequence with a detection threshold of P < 10−5. Highly enriched motifs were manually chosen based on well-known functions of transcription factors for macrophage activation. For each of our experimental conditions we scored each motif’s association with chromatin accessibility and histone modifications. We then used the GraphPad Prism 6 to visualize the distribution of this motif across different experimental conditions.
Analysis of patient monocytes and macrophages
Peripheral blood was obtained from SLE patients and healthy donors using a protocol approved by the Institutional Review Board at the Hospital for Special Surgery. As described above, human CD14+ monocytes/macrophages were purified using anti-CD14 magnetic beads and were cultured in RPMI-1640 medium with 10% (vol/vol) FBS (Hyclone). For RA synovial macrophages, the microarray data described in Donlin et al., J Immunol, 2014, 193:2373 (GSE97779) were normalized by a quantile normalization method using the preprocessCore package in R. Normalized expression levels were averaged within the same condition (Fig. 3e, f). ATAC-seq with monocytes from healthy or patient donors (Fig. 8) was performed as described above with or without ex vivo LPS (10 ng/ml) stimulation. The datasets from sepsis patients (Fig. 3d) were retrieved from GSE46955.
Statistical analysis
Graphpad Prism 6 for Mac (GraphPad Software, Inc) was used for all statistical analysis. Detailed information about statistical analysis including tests and values used, and number of times experiments were repeated, is indicated in the figure legends. P values are provided in the text or the figure legends.
Supplementary Material
Acknowledgments
We thank A. Pernis and I. Rogatsky for helpful discussions and review of the manuscript and F. Barrat for providing SLE monocytes. This work was supported by grants from the N.I.H. and support for the David Z. Rosensweig Genomics Center from The Tow Foundation.
Footnotes
Author Contributions: Sung Ho P. conceptualized, designed and performed most of the experiments, performed bioinformatic analysis and wrote the manuscript. Kyuho K., Y.Q., G.K., and K. P.-M. contributed experiments and expertise. E.G. and Keunsoo K. performed bioinformatic analysis. L.B.I. conceptualized and oversaw the study and edited the manuscript. All authors reviewed and provided input on the manuscript.
Accession codes
The data in this paper have been deposited in the GEO data base with accession code GSE100383.
Data availability statement
The data sets that support the findings of this study and were generated by the authors as part of this study have been deposited in the Gene Expression Omnibus (GEO https://www.ncbi.nlm.nih.gov/geo/) with the accession code GSE100383. Additional data sets that support the findings in Figure 3 are publically available at GEO with accession codes GSE46955 and GSE 97779.
References
- 1.Brenner D, Blaser H, Mak TW. Regulation of tumour necrosis factor signalling: live or let die. Nature reviews. Immunology. 2015;15:362–374. doi: 10.1038/nri3834. [DOI] [PubMed] [Google Scholar]
- 2.Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nature reviews. Rheumatology. 2016;12:49–62. doi: 10.1038/nrrheum.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park SH, Park-Min KH, Chen J, Hu X, Ivashkiv LB. Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nature immunology. 2011;12:607–615. doi: 10.1038/ni.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends in immunology. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nature reviews. Immunology. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Ivashkiv LB. Type I interferon modulation of cellular responses to cytokines and infectious pathogens: potential role in SLE pathogenesis. Autoimmunity. 2003;36:473–479. doi: 10.1080/08916930310001605882. [DOI] [PubMed] [Google Scholar]
- 9.Prinz M, et al. Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity. 2008;28:675–686. doi: 10.1016/j.immuni.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annual review of immunology. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 14.Smale ST, Natoli G. Transcriptional control of inflammatory responses. Cold Spring Harbor perspectives in biology. 2014;6:a016261. doi: 10.1101/cshperspect.a016261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends in immunology. 2013;34:216–223. doi: 10.1016/j.it.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nature reviews. Molecular cell biology. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivashkiv LB, Park SH. Epigenetic Regulation of Myeloid Cells. Microbiology spectrum. 2016;4 doi: 10.1128/microbiolspec.MCHD-0010-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 21.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed S, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piper J, et al. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic acids research. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 27.Yan Q, et al. Nuclear factor-kappaB binding motifs specify Toll-like receptor-induced gene repression through an inducible repressosome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14140–14145. doi: 10.1073/pnas.1119842109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shalova IN, et al. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1alpha. Immunity. 2015;42:484–498. doi: 10.1016/j.immuni.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Ivashkiv LB. IFN-gamma abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19438–19443. doi: 10.1073/pnas.1007816107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez-Carrozzi VR, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes & development. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, et al. IFN-beta production by TLR4-stimulated innate immune cells is negatively regulated by GSK3-beta. Journal of immunology. 2008;181:6797–6802. doi: 10.4049/jimmunol.181.10.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, El Gazzar M, Yoza BK, McCall CE. The NF-kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. The Journal of biological chemistry. 2009;284:27857–27865. doi: 10.1074/jbc.M109.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benayoun BA, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi L, et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PloS one. 2014;9:e93846. doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, et al. H3K4 tri-methylation breadth at transcription start sites impacts the transcriptome of systemic lupus erythematosus. Clinical epigenetics. 2016;8:14. doi: 10.1186/s13148-016-0179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Qiao Y, et al. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nature immunology. 2016;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nature immunology. 2016;17:18–25. doi: 10.1038/ni.3325. [DOI] [PubMed] [Google Scholar]
- 42.Netea MG, et al. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lauberth SM, et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharif MN, et al. Twist mediates suppression of inflammation by type I IFNs and Axl. The Journal of experimental medicine. 2006;203:1891–1901. doi: 10.1084/jem.20051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nature methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.