Abstract
Analysis of antigen-specific CD4+ T cells in mycobacterial infections at the transcriptome level is informative but technically challenging. While several methods exist for identifying antigen-specific T cells, including intracellular cytokine staining, cell surface cytokine-capture assays, and staining with peptide:MHC-II multimers, all of these have significant technical constraints that limit their usefulness. Measurement of activation-induced expression of CD154 has been reported to detect live, antigen-specific CD4+ T cells but this approach remains underexplored, and to our knowledge has not previously been applied in mycobacteria-infected animals. Here we show that CD154 expression identifies adoptively transferred or endogenous antigen-specific CD4+ T cells induced by Mycobacterium bovis Bacillus Calmette-Guérin (BCG) vaccination. We confirmed that antigen-specific cytokine production was positively correlated with CD154 expression by CD4+ T cells from BCG-vaccinated mice, and show that high quality microarrays can be performed from RNA isolated from CD154+ cells purified by cell sorting. Analysis of microarray data demonstrated that the transcriptome of the CD4+ CD154+ cells was distinct from that of CD154− cells, and showed major enrichment of transcripts encoding multiple cytokines and pathways of cellular activation. One notable finding was the identification of a previously unrecognized subset of mycobacteria-specific CD4+ T cells characterized by the production of interleukin-3. Our results support the use of CD154 expression as a practical and reliable method to isolate live, antigen-specific CD4+ T cells for transcriptomic analysis and potentially for a range of other studies in infected or previously immunized hosts.
Introduction
Commonly used methods to analyze antigen-specific CD4+ T cells have limited compatibility with downstream applications such as isolation of live cells for further manipulation or isolation of high quality RNA from cells for transcriptome analysis. Typically, antigen specific responses are assessed ex vivo either by measuring cytokines secreted into the culture supernatant by ELISA or by enumerating cytokine producing cells by ELISPOT [1]. Neither of these methods permits phenotypic analysis of the cytokine producing cells of interest. Individual cytokine-producing cells can be identified by intracellular cytokine staining (ICS) and subsequent analysis by flow cytometry, however, this technique requires fixation and permeabilization of cells leading to cell death and RNA degradation [2]. Alternate buffer conditions for preservation of RNA integrity during intracellular staining have been suggested, but have yet to be validated [3, 4]. Cell surface cytokine-capture is a promising method that potentially allows identification of live cytokine-producing cells, but this technique requires bi-functional antibodies, and is laborious while suffering from limited sensitivity [5]. In addition, all of the above methods require that a particular cytokine of interest be identified a priori, which may not be appropriate given the plasticity and heterogeneity of T helper (Th) cells [6]. While identification of antigen specific CD4+ T cells using multimers of peptide-loaded MHC II molecules does not require prior knowledge of cytokine production, this technique is limited by the need for epitope discovery and synthesis of peptide-MHC multimers, and by the heterogeneity of MHC II [2].
Alternatively, antigen-specific CD4+ T cells can be identified by surface molecules that are expressed specifically upon antigen stimulation, but this approach remains underexplored [7–10]. CD154, also known as CD40 ligand, is an activation-induced marker on CD4+ T cells and its binding partner CD40 is expressed on a variety of hematopoetic and non-hematopoetic cells [11–15]. Because expression of CD154 on CD4+ T cells after activation is transient, peaking at 6 hours post-activation and then declining due to endocytosis and degradation [16], it is technically challenging to identify populations that upregulate this marker as an indicator of antigen specific activation. To overcome this issue, two modifications of the staining procedure have been previously proposed. In the first approach, inclusion of monensin and fluorochrome-conjugated antibody against CD154 during the stimulation phase resulted in enhanced detection of CD154 by retaining the fluorescent conjugate within the cells after endocytosis [8]. In the second approach, by including an antibody against CD40 that functionally blocks interaction with CD154, greater levels of CD154 were retained on the surface of CD4+ T cells [9]. Both methods were used to identify and isolate antigen-specific CD4+ T cells, with the former method showing potentially greater sensitivity.
In this article, we report the use of activation-induced CD154 expression as a method to identify antigen-specific CD4+ T cells in two experimental models in mice, and show that this method can be used to isolate high quality samples for gene expression analysis by microarray. In one case, we assessed CD154 expression on adoptively transferred transgenic CD4+ T cells from OT-II mice that are specific to a peptide of ovalbumin. In the second model, we assessed CD154 expression on endogenous CD4+ T cells specific to Mycobacterium tuberculosis (Mtb) antigens in mice vaccinated with Mycobacterium bovis Bacillus Calmette-Guerin (BCG). Analysis of both models showed that CD154 expression upon re-stimulation is a valid approach to identify antigen-specific CD4+ T cells, and can be used to facilitate transcriptome analysis of antigen-specific CD4+ T cells. In addition, an in depth analysis of the microarray data from cells obtained by this method resulted in the identification of a previously unrecognized subset of mycobacteria-specific CD4+ T cells that secrete the cytokine interleukin-3 (IL-3).
Materials and Methods
Mice
Six- to 8-wk-old female wild-type C57BL/6 mice were obtained from The Jackson Laboratory. C57BL/6-OT-II TCR-Tg mice expressing GFP were bred in our facility from founders provided by G. Lauvau (Albert Einstein College of Medicine, Bronx, NY). All mice were maintained in specific pathogen-free conditions. All procedures involving the use of animals were in compliance with protocols approved by the Einstein Institutional Animal Use and Biosafety Committees.
Mycobacterial vaccinations
M. bovis Bacillus Calmette-Guérin (BCG)-Danish was obtained from Statens Serum Institute (Copenhagen, Denmark), and was grown in Middlebrook 7H9 medium (Difco Laboratories, BD Diagnostic Systems, Sparks, MD) with oleic acid-albumin-dextrose-catalase (OADC Enrichment; Difco Laboratories, BD Diagnostic Systems, Sparks, MD) and 0.05% tyloxapol (Sigma-Aldrich, St. Louis, MO). Mycobacterium smegmatis (strain mc2155) were grown in liquid cultures in Sauton medium [17]. Bacteria were grown from low passage number frozen stocks, cultured to mid-log phase and then frozen in medium with 5% glycerol at −80°C. Bacteria were thawed, washed, resuspended in PBS containing 0.05% Tween-80, and sonicated to obtain a single-cell suspension prior to infection. Mice were vaccinated with 2 × 106 CFU of bacteria at the base of the tail unless otherwise indicated. Mice were euthanized 4–6 weeks after vaccination to isolate spleen and splenocyte suspensions were prepared by gently forcing spleen through a 70-μm cell strainer. RBC lysis was performed using RBC lysing buffer Hybri-Max (Sigma).
Aerosol infection with M. tuberculosis
Low density freezer stocks were made from M. tuberculosis H37Rv cultured in Middlebrook 7H9 medium containing OADC, 0.5% glycerol, and 0.05% tyloxapol. Prior to infection, bacteria were thawed, sonicated and re-suspended in PBS-Tween containing 0.05% antifoam Y-30 (Sigma). The suspension was loaded into a nebulizer attached to an airborne infection system (University of Wisconsin Mechanical Engineering Workshop). Mice were exposed to aerosolized bacteria for 20 minutes and approximately 100 bacteria were deposited into the lungs of each animal. The inoculum dose was confirmed by plating of whole-lung homogenates at 24 hours post-exposure, with quantification of CFU 4 weeks later. Four weeks after infection, lungs from infected mice were removed and perfused with PBS via the pulmonary artery. Lungs were then treated with Liberase TL (0.3 Wünsch Units/ml; Roche) and DNAse I (10 μg/ml; Sigma) in serum-free RPMI-1640. The lungs were then passed through a 70 μm cell strainer. Mediastinal lymph nodes were passed through a 70 μm cell strainer to make single cell suspension.
Adoptive transfer of OT-II transgenic CD4+ T cells
CD4+ T cells from OT-II TCR-Tg/GFP mice were purified by negative selection using a commercially available kit according to the manufacturer’s instructions (Miltenyi Biotec) and 20,000 cells in 100 μl of PBS were injected intravenously into wild type C57BL/6 mice. Twenty four hours later, ovalbumin and poly (I:C) were administered cutaneously at the base of the tail. On day 7 after vaccination, mice were euthanized to isolate splenocytes.
Re-stimulation and flow cytometry analysis of CD4+ T cells
For CD154 staining, splenocytes were incubated for 6 hours with appropriate antigen, monensin (2 μM; Sigma) and anti-CD154-antibody (0.25 μg/ml) conjugated with PE in 96-well round-bottom plate. For staining intra-cellular IL-3, splenocytes were stimulated with antigen for 6 hours with the last 4 hours in the presence of monensin (5μM) and brefeldin A (5μg/ml; Sigma). Re-stimulation was performed in RPMI medium (Gibco) supplemented with HEPES (Gibco), penicillin-streptomycin (Gibco), fetal bovine serum (10%; Atlanta Biologicals), beta-mercaptoethanol (Gibco), essential amino acids (Gibco) and non-essential amino acids (Gibco). The final concentration of OT-II peptide (ISQAVHAAHAEINEAGR; Mimotopes), ESAT-6 peptide (MTEQQWNFAGIEAAASAIQG; Mimotopes) and P25 peptide (FQDAYNAAGGHNAVF; Mimotopes) was 5 μg/ml. Mtb or M. smegmatis lysate were used at a final protein concentration of 50 μg/ml and was prepared as previously described [18]. After re-stimulation, cells were suspended in PBS and stained with viability dye (LIVE/DEAD Fixable Blue Dead Cell Stain, Molecular Probes), and subsequently with fluorochrome-conjugated antibodies against surface markers in PBS containing fetal bovine serum (2%) and sodium azide (0.05%; Sigma). In some experiments, cells were fixed using paraformaldehyde (2% in PBS; Electron Microscopy Sciences), permeabilized (Fixation/Permeabilization buffer; eBioscience) and stained for intracellular cytokines. Intracellular cytokine staining buffer used was Dulbecco’s PBS with calcium and magnesium (Gibco) containing fetal bovine serum (2%), sodium azide (0.05%), HEPES (1%) and saponin (0.1%; Sigma). The following antibodies were used for staining: CD154-PE (Clone MR1, BD Biosciences), CD4-APC-Cy7 (Clone RM4-5, Tonbo Biosciences), CD44-eFluor 450 (Clone IM7; eBioscience), CD8α-PE-Cy5 (Clone 53-6.7, Tonbo Biosciences), B220-PE-Cy5 (Clone RA3-6B2, BD Biosciences), MHC II-PE-Cy5 (Clone M5/114.15.2, eBioscience), IFNγ-Alexa Fluor 700 (Clone XMG1.2, BD Biosciences), TNF-Alexa Fluor 488 (Clone MP6-XT22, BD Biosciences), IL-2-PE-Cy7 (Clone JES6-5H4, eBioscience), IL-4-APC (Clone 11B11, BD Biosciences), IL-17A-PerCP-Cy5.5 (Clone eBio17B7, eBioscience), IL-21 (FFA21, eBioscience), T-bet (Clone eBio4B10, eBioscience) and IL-3-PE (Clone MP2-8F8, BD Biosciences).
ELISPOT assay
Purified CD4+ T cells (positive selection, L3T4 microbeads; Miltenyi Biotech) isolated from immunized spleens were incubated in nitrocellulose ELISPOT plates (Millipore) coated with anti-IFNγ (Clone R4-6A2, BD Biosciences), anti-IL-4 (Clone 11B11, BD Biosciences), anti-IL-17A (Clone TC11-18H10, BD Biosciences), or anti-IL-3 (MP2-8F8, purified in-house) antibodies with naïve, T cell-depleted splenocytes and TB9.8 peptide (ESSAAFQAAHARFVAA; 10μg/ml; Mimotopes [19]. Mice used for these experiments received 1 × 107 CFU of BCG subcutaneously at the scruff of the neck. After 20 hours of incubation at 37°C, plates were washed with PBS containing 0.05% Tween 20 and incubated with biotinylated anti-IFNγ (Clone XMG1.2, BD Biosciences), anti-IL-4 (Clone BVD6-24G2, BD Biosciences), anti-IL-17A (Clone TC11-8H4, BD Biosciences), or anti-IL-3 (Clone MP2-43D11, BD Biosciences) antibodies. Bound antibodies were detected using streptavidin-alkaline phosphatase (Life Technologies) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) and nitroblue tetrazolium chloride (NBT) (Sigma). Plates were counted with the aid of computer-assisted image analysis by using an AID ELISPOT reader (Autoimmun Diagnostika GmbH). IL-21 assay was performed using IL-21 mouse ELISPOT kit as per instructions provided by the manufacturer (eBioscience).
Sorting and microarray experiments
CD154 staining was performed with splenocytes isolated from mice vaccinated with BCG and re-stimulated with Mtb lysate as described above. For each mouse, antigen-experienced CD4+ T cells were identified as live cells in the lymphocyte gate that are negative for CD8α, B220 and MHC II and positive for CD4 and CD44. From this population, CD154+ and CD154− cells were sorted separately into tubes containing RNA Protect Cell Reagent (Qiagen). The cells were pelleted and RNA isolation was performed using RNeasy Micro kit (Qiagen). Isolated RNA was quantified using Ribogreen method (Quant-iT Ribogreen RNA Assay Kit, Molecular Probes) with a Nanodrop 3300 instrument (Thermo Fisher Scientific Inc.). RNA quality was assessed using Bioanalyzer (Agilent Technologies). Twenty five nanograms of total RNA was amplified using Ovation Pico WTA Systems (NuGEN; http://www.nugen.com/products/ovation-pico-wta-system-v2) and hybridized onto GeneChip Mouse Gene 1.0 ST arrays [20] (Affymetrix; https://www.thermofisher.com/order/catalog/product/901168). The arrays were then scanned using a GeneChip Scanner 3000 (Affymetrix), and fluorescent intensity for each feature on the array was quantified using GeneChip Operating Software (Affymetrix). Probe summarization and normalization using Robust Multi-array Average (RMA) method is performed using Expression Console software (Affymetrix). Microarray files are available in the GEO database (GEO accession number: GSE98182: https://www.ncbi.nlm.nih.gov/geo).
Analysis of microarray data
Principal Component Analysis (PCA) was performed using top 15% most varying transcripts identified based on standard deviation with R software using the library ‘ggbiplot’ (https://www.rdocumentation.org/packages/ggbiplot). Differentially expressed transcripts were identified by Rank Product analysis performed with R software using the library ‘RankProd’ [21]. Rank product is a non-parametric statistic derived from biological reasoning that detects items that are consistently highly ranked in a number of lists [22]. Heatmaps were generated using TIGR Multiple Experiment Viewer (http://mev.tm4.org/). Pathway analysis was performed using GSEA software obtained from the Molecular Signatures Database (MSigDB) [23]. The KEGG pathway database (http://www.genome.jp/kegg/pathway.html) and TRANSFAC database (http://gene-regulation.com/pub/databases.html) were accessed through GSEA software.
Real-time PCR
The SYBR Green method was used to quantify relative levels of selected transcripts as per the protocols provided by the supplier (Molecular Probes). Single-stranded DNA obtained after amplification of RNA was used as template. Transcripts were selected to cover the entire range of microarray intensity and included genes that are differentially and non-differentially expressed between CD154+ and CD154− population. Primers were designed using NCBI Primer-BLAST software which uses Primer3 to design primers, and BLAST and global alignment algorithm to screen primers against non-specific amplifications [24]. Primer sequences are provided in Supplementary Table 2.
Results
Detection of adoptively transferred antigen-specific CD4+ T cells based on activation-induced expression of CD154
Inclusion of fluorochrome-conjugated anti-CD154 antibody and monensin in the culture medium during stimulation has been shown to enhance the detection of CD154+, antigen-specific CD4+ T cells [8]. In order to evaluate the sensitivity and specificity of this assay, we used a defined monoclonal population of GFP-expressing TCR transgenic CD4+ T cells specific for ovalbumin (OVA) 323–339 peptide (OT-II cells) adoptively transferred into naïve recipient hosts. Following intravenous transfer of OT-II cells, recipient mice were vaccinated with ovalbumin in the presence of Poly (I:C). Seven days after vaccination, we isolated splenocytes and re-stimulated them with ovalbumin peptide in the presence of PE-conjugated anti-CD154 and 2 μM monensin. Flow cytometry analysis showed that re-stimulation with OVA323–339 resulted in specific upregulation of CD154 on GFP+ CD4+ T cells, while the levels of CD154 on endogenous cells remained at background levels in both stimulated and unstimulated conditions (Figure 1A). Over a range of anti-CD154 antibody concentrations, more than 90% of GFP+ OT-II cells were positive for CD154 based on detection of PE by flow cytometry (Figure 1B).
Figure 1. Expression of CD154 on adoptively transferred OT-II transgenic CD4+ T cells.
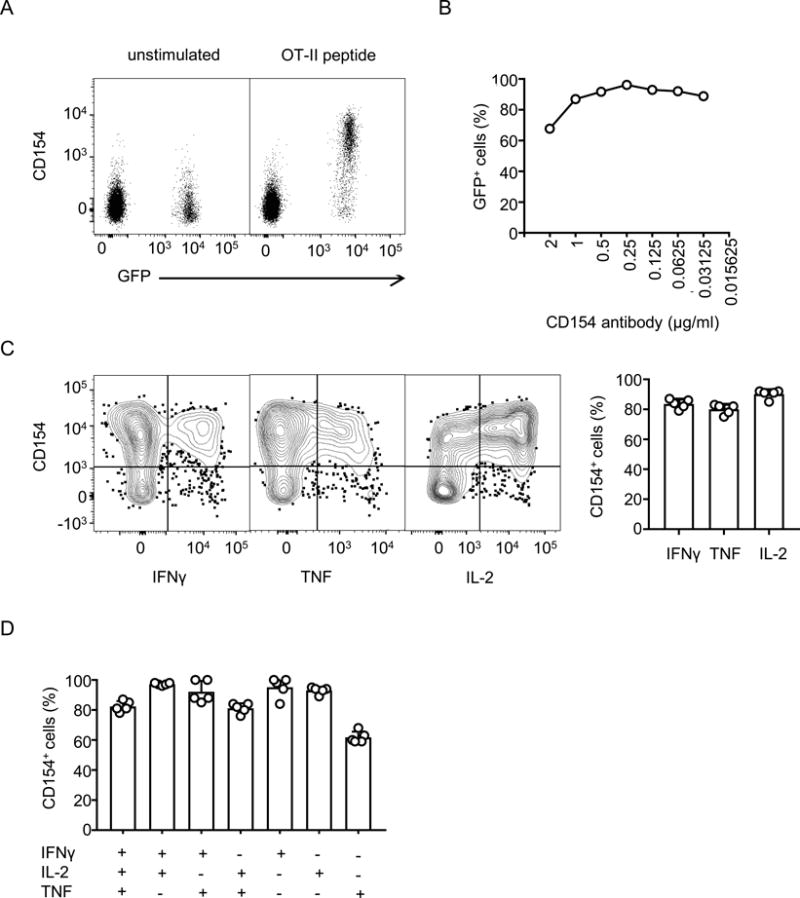
A) Mice were vaccinated with ovalbumin and poly (I:C) one day after adoptive transfer of GFP+ OT-II transgenic CD4+ T cells. On the 7th day after vaccination, splenocytes were isolated and stimulated for 6 hours with ovalbumin peptide in the presence of PE-conjugated anti-CD154 antibody and monensin. Flow cytometry analysis was performed after staining for additional surface markers (anti-CD8α, -B220 and -MHCII) and with viability dye for exclusion of dead cells. The cells shown are singlet events from the lymphocyte gate that are positive for CD4 and negative for Live/Dead stain, CD8α, B220 and MHC-II. Dot plots show levels of CD154 on adoptively transferred OT-II transgenic CD4+ T cells (GFP+) and recipient CD4+ T cells (GFP−) with and without re-stimulation. B) Percentage of GFP+ cells among CD154+ cells when varying concentrations of anti-CD154 antibody were included in the re-stimulation medium. C) Splenocytes, surface-stained for CD154 and other markers as in panel A, were fixed, permeabilized and stained for intra-cellular cytokines. Only GFP+ OT-II CD4+ T cells are shown. The bar chart on right summarizes the percentages of CD154+ cells among total cytokine producing cells. D). Experiments were performed as in Panel C. Cytokine-producing cells were grouped into single, double and triple cytokine-producers using boolean gating strategy and analyzed for the co-expression of CD154. The percentages of cells that co-express CD154 are shown for each group. Results shown represent two independent experiments.
Staining of intracellular cytokines following antigen stimulation is a standard method used to characterize antigen-specific CD4+ T cells. Since staining for CD154 in the presence of monensin is compatible with subsequent staining for intra-cellular cytokines [8], we combined staining for activation-induced CD154 as described above with staining for intracellular IFNγ, TNF and IL-2. These cytokines were chosen for initial studies because Poly (I:C) is a known Th1 polarizing adjuvant [25]. As expected, most cytokine-producing OT-II cells were co-stained for CD154 (Figure 1C). Similar results were also obtained when the cytokine producing cells were grouped into single, double or triple cytokine producers (Figure 1D). We also observed that approximately 25% of CD154+ cells did not secrete any of these cytokines. It is possible that these cells secrete other cytokines which were not assessed or may represent antigen-specific CD4+ T cells that do not produce detectable cytokines under the conditions used for activation.
Detection of endogenous mycobacteria-specific CD4+ T cells based on activation-induced expression of CD154
We next evaluated the ability of the assay to detect endogenous polyclonal populations of pathogen-specific CD4+ T cells. For this, we tested whether CD154 expression allows identification of mycobacteria-specific CD4+ T cells in a murine model of BCG vaccination. We isolated splenocytes from mice that were previously vaccinated with BCG and stimulated them with P25 peptide of the immunodominant Antigen 85B or lysate of Mtb in the presence of PE-conjugated anti-CD154 antibody and monensin. Splenocytes from unvaccinated mice were used as negative controls. We observed that CD4+ T cells from unvaccinated mice showed minimal background levels of CD154 which did not change across various stimulation conditions (Figure 2A). The background level was slightly higher in the CD44+, antigen-experienced, population compared to the CD44− population, as shown previously [26]. Similar background levels of CD154 were also noted on the unstimulated cells from mice vaccinated with BCG. However, stimulation with P25 peptide or lysate resulted in a significant increase in CD154 levels on cells from vaccinated mice (Figure 2A and B). This increase was seen only in CD44+ cells consistent with stimulation-induced CD154 expression occurring only in antigen-experienced cells. We observed that compared to unvaccinated mice, peptide stimulation in vaccinated mice resulted in 9-fold increase in the percentage of CD154+ CD4+ T cells while the increase was 15 fold with lysate-stimulation. The higher number of antigen-specific cells with lysate-stimulation was consistent with the presence of multiple antigens in the lysate. Based on these fold calculations, we estimated that the percentage of antigen specific cells within the CD154+ population was approximately 90% (peptide stimulation) or 94% (lysate stimulation).
Figure 2. Expression of CD154 on endogenous mycobacteria-specific CD4+ T cells after re-stimulation.
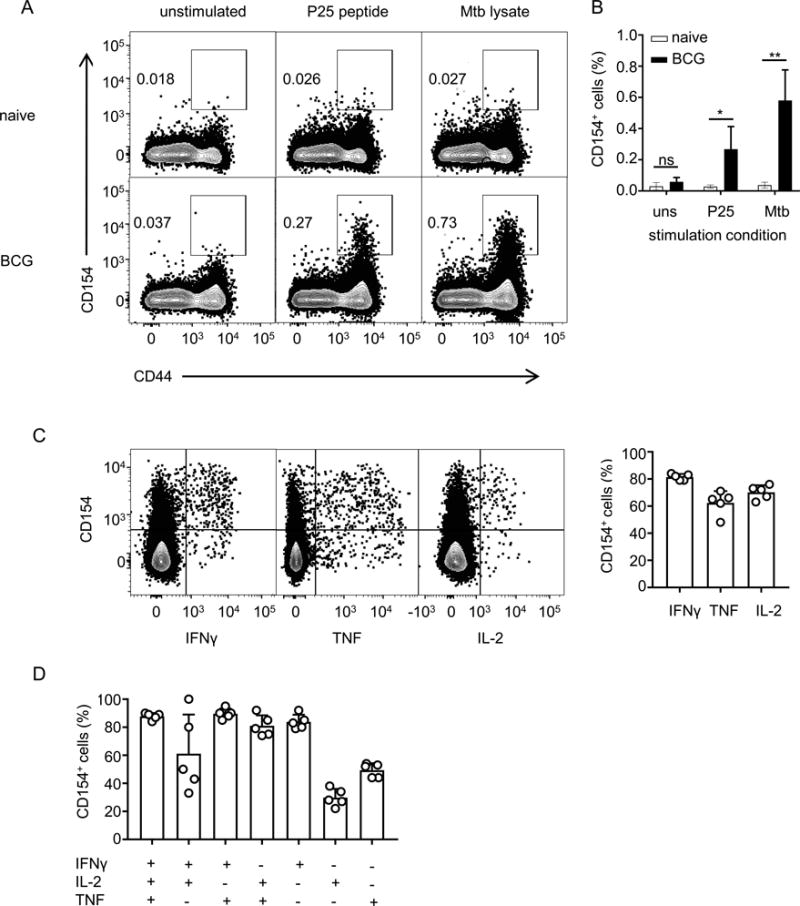
A) Splenocytes from naïve and BCG-vaccinated mice were stimulated ex vivo with Mtb lysate or P25 peptide for 6 hours in the presence of PE-conjugated anti-CD154 antibody and monensin. Flow cytometry analysis was performed after staining for additional surface markers and dead cells. The cells shown are single cells from the lymphocyte gate that are positive for CD4 and negative for Live/Dead stain, CD8α, B220 and MHC-II. Dot plots show levels of CD154 on CD4+ T cells with and without re-stimulation. B) Graph showing the percentages of CD154+ cells in total CD4 cells under various stimulation conditions. uns, unstimulated; P25, P25 peptide; Mtb, Mtb lysate; * p < 0.05; ** p < 0.005 for t-tests comparing naïve and BCG. C) Splenocytes, surface-stained for CD154 as in panel A, were fixed, permeabilized and stained for intracellular cytokines. The bar graph summarizes the percentages of CD154+ cells among total cytokine producing cells. D). Experiments were performed as in Panel C. Cytokine-producing cells were grouped into single, double and triple cytokine-producers using boolean gating strategy and analysed for the co-expression of CD154. The percentages of cells that co-express CD154 are shown for each group. Results shown represent two independent experiments.
We also combined CD154 staining with intracellular cytokine staining (ICS) and found that most cells expressing IFNγ, TNF and IL-2 were positive for CD154, as we observed with adoptively transferred cells (Figure 2C). Similar results were also obtained when the cytokine producing cells were grouped into single, double or triple cytokine producers (Figure 2D). Cells that produced only IL-2 showed the least co-expression with CD154, consistent with findings of a previous study of human PBMCs [8]. We also noted that approximately 75% of the CD154+ endogenous CD4+ T cells did not secrete any of the cytokines that we assayed by ICS (Figure 2C). This percentage of cytokine-negative CD154+ cells was higher than the ~25% that we observed in our adoptive transfer model using OT-II peptide for re-stimulation (Figure 1C). This may be explained in part by secretion of cytokines other than those associated with Th1 cells by a substantial portion of endogenous T cells. In addition, it likely reflects the stronger signal generated by stimulation with optimal concentrations of synthetic peptide epitopes, as in the case of the OT-II experiments. Thus, in the case of Mtb lysate stimulation of endogenous BCG-primed T cells, stimulation was sufficient to induce CD154 expression but not cytokine production. This would be consistent with the known differences that can exist in the thresholds of T cell receptor stimulation for different outcomes of activation [27–29], and is also mirrored by previous studies of human T cells showing a similar higher percentage of cytokine expression by CD154+ cells with peptide stimulation compared with a more polyclonal stimulation using Staphylococcus aureus enterotoxin B [8, 9].
Transcriptome analysis of mycobacteria-specific CD4+ T cells induced by M. bovis BCG vaccination
To characterize CD154+ CD4+ T cells in a broader, unbiased fashion, we performed transcriptome analysis of these cells. We isolated splenocytes from mice that were previously vaccinated with BCG and stimulated them with lysate of Mtb in the presence of PE-conjugated anti-CD154 antibody and monensin. We then sorted CD154+ and CD154− antigen-experienced (CD44+) CD4+ T cells using high speed cell sorting (FACS). Although we sorted higher numbers of CD154− cells compared to CD154+ cells and subsequently obtained greater amounts of RNA from these cells, both CD154+ and CD154− cells yielded high quality RNA as indicated by their high RNA Integrity Number (RIN) values. (Supplementary Figure 1A and 1B). After amplifying the RNA using standard protocols, microarray experiments were carried out with Affymetrix Gene Chips [30]. Quality of the microarrays was assessed using guidelines adopted by Immunological Genome Project (ImmGen), which is a compendium that characterizes microarray gene expression profiles of cells in the murine immune system (https://www.immgen.org/). Comparison of the quality metrics showed that the microarrays in the current study outperformed the arrays from ImmGen, based on higher dynamic range and area under the curve and lower coefficient of variation (Supplementary Figure 1C). In addition, no significant differences were observed for these metrics between microarrays performed for CD154+ and CD154− population (Supplementary Figure 1C). These results showed that high quality microarray data can be obtained from FACS-sorted CD154+ cells.
CD154+ cells were expected to have a different transcriptome signature than CD154− cells for at least two reasons. First, since they were activated by Mtb lysate, they should preferentially express transcripts associated with cellular activation. Second, CD154+ cells should be enriched for helper populations generated by the original BCG vaccination. We performed Principal Component Analysis (PCA) to test whether CD154+ cells had a unique transcriptome signature. The analysis revealed that the CD154+ cells grouped separately from CD154− cells, indicating differences in transcriptome of these populations (Figure 3A). We next identified genes that were differentially expressed between the CD154+ andCD154− populations using Rank product, a non-parametric method of identifying differentially transcribed genes in microarrays [22]. Compared to CD154− cells, there were 271 genes upregulated in CD154+ cells while 195 genes were downregulated (Figure 3B & Supplementary Table 1). All of these genes were included in the gene set used for PCA. Quantitative PCR analysis of selected transcripts showed that relative transcript levels were highly correlated with microarray intensity (Figure 3C). Many of these differentially expressed transcripts were from genes known to be associated with CD4+ T cell responses. For example, the top 10 transcripts that were more highly expressed in CD154+ compared to CD154− cells were predominantly from genes encoding cytokines and chemokines known to be secreted by CD4+ T cells (Figure 3D). Similarly, compared to CD154+ cells, CD154− cells expressed higher levels of transcripts encoding LRRC32/GARP, a cell surface protein known to be associated with regulatory T cells and FOXP3, the master transcription factor for regulatory T cells. However, there were notable exceptions indicating previously unrecognized features of the transcriptional profiles of CD4+ T cells undergoing activation by cognate antigens. For example, the most highly expressed transcript in CD154+ cells versus CD154− cells encodes a cytoskeletal protein called Ermin (ERMN), which is believed to be restricted to oligodendrocytes [31]. Similarly, the most highly expressed gene for a transcription factor in CD154+ versus CD154− cells encodes Zinc Finger E-Box Binding Homeobox 2 (ZEB2), which has been implicated in CD8+ T cell and myeloid cell function but to our knowledge has not been investigated for its potential functions in CD4+ T cells [32, 33].
Figure 3. Differential gene expression in CD154+ versus CD154− CD4+ T cells.
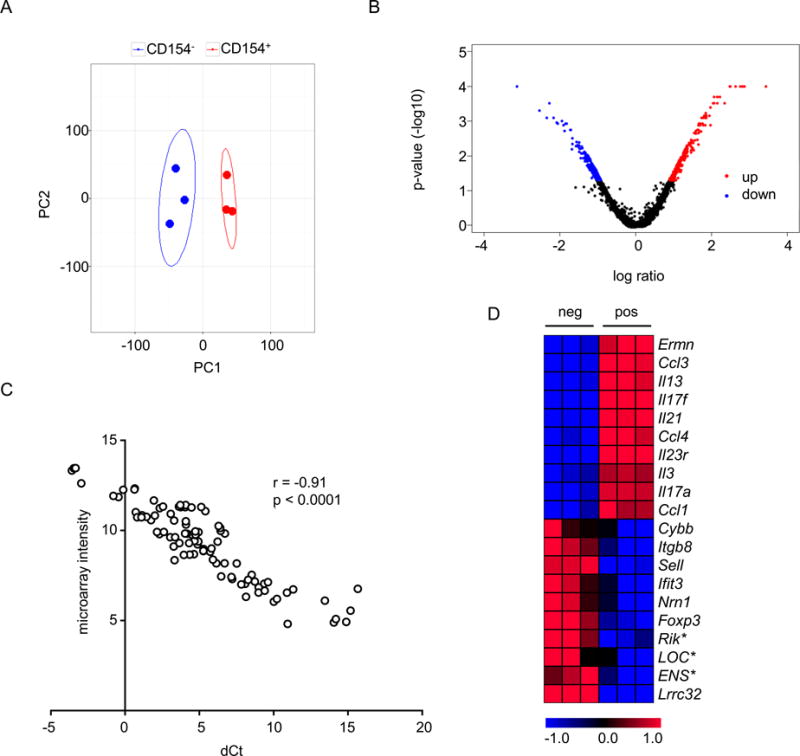
Microarray experiments were performed with RNA isolated by cell sorting from CD154+ and CD154− cells from BCG-vaccinated mice. A) Principal component analysis of microarray data using 15% most varying genes identified based on standard deviation. First two principal components (PC) are shown. Each dot represents a microarray and ellipses represents 95% confidence intervals. B) Volcano plot showing differential gene expression. P-values were obtained from rank product analysis. C) Quantitative PCR analysis of selected transcripts. dCt, delta cycle threshold calculated based on house keeping gene, Ywhaz. Genes analyzed and primers used are listed in Supplementary Table 2. Each spot represents a biological replicate. D. Heatmap showing normalized intensities from microarray analysis of top 10 differentially expressed transcripts. The list is sorted for magnitude for differential expression with Ermn as most highly expressed in CD154+ cells and Lrrc32 as most highly expressed in CD154− cells. Abbreviations for transcript names are derived from the Mouse Genome Informatics website (http://www.informatics.jax.org). The * symbol indicates features representing less defined genes; refer to Supplementary Table 1 for details.
Pathway analysis of transcriptome data and cytokine expression
We hypothesized that those transcriptional changes seen in CD154+ cells should mirror changes associated with cellular activation since these cells are activated by cognate antigen. To explore this possibility, we performed pathway analysis of the microarray data using Gene Set Enrichment Analysis (GSEA) [23]. By using genes sets derived from the KEGG pathway database, we found that the pathways that were significantly enriched in the CD154+ transcriptome represented metabolic changes associated with activation of T cells (Figure 4A). We also investigated whether targets of certain transcription factors were selectively enriched in CD154+ population using sets of genes sharing a transcription factor binding site, as defined in the TRANSFAC database. This analysis showed that the targets of transcription factors known to be associated with cellular activation, growth and proliferation were enriched in the CD154+ population (Figure 4B).
Figure 4. Pathway analysis and cytokine gene expression analysis of microarray data.
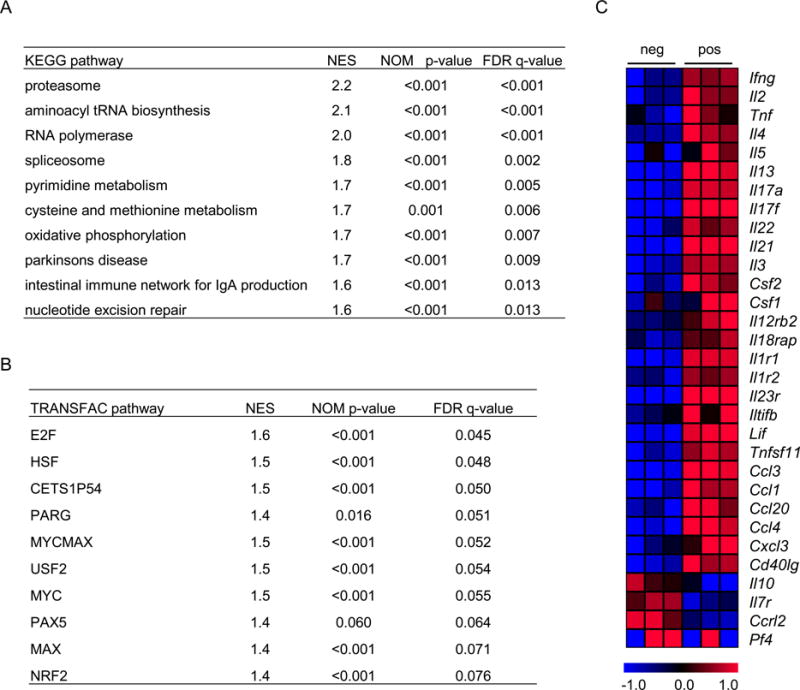
A) Pathway analysis was carried out on data from microarray experiments performed with RNA isolated from CD154+ and CD154− cells from BCG-vaccinated mice. Gene Set Enrichment Analysis (GSEA) software was used, with genes sorted for p-values obtained from rank product analysis. The top 10 pathways from KEGG database that are enriched in the transcriptome of CD154+ cells are shown. B) Analysis was performed as in panel A using groups of transcripts where each group represents genes predicted to bind a common transcription factor as described by the TRANSFAC database. The top 10 transcription factors whose targets are enriched in the transcriptome of CD154+ cells are shown. C) Heat map showing cytokine transcripts differentially expressed between CD154+ and CD154− cells. The list is organized in such a way that cytokines and chemokines are grouped separately and cytokines of similar properties (e.g. belonging to one T helper subset) are grouped together. Abbreviations for transcript names are derived from the Mouse Genome Informatics website (http://www.informatics.jax.org).
We also carried out a focused analysis of expression of cytokines, chemokines and their receptors, based on the microarray data. This identified 31 transcripts that were differentially expressed between CD154+ and CD154− populations. Among these, 27 were more highly expressed in the CD154+ population while only 4 were more highly expressed in the CD154− population (Figure 4C). Transcripts more highly expressed in the CD154+ population included those encoding cytokines known to be associated with various T helper populations, such as the signature cytokines of Th1 (IFNγ), Th2 (IL-4, IL-5 and IL-13), Th17 (IL-17A), and T follicular helper (IL-21) populations (Figure 4C).
While mycobacteria are well known to induce Th1 cells, induction of transcripts associated with other helper cell subsets was unexpected. We selected signature cytokines for Th2 (IL-4), Th17 (IL-17A) and Tfh (IL-21) cells for protein level validation using intra-cellular cytokine staining. While we were able to detect IL-4 and IL-17A secreting cells in BCG-vaccinated mice in some cases (Supplementary Figure 2A and 2B), we were unable to detect them consistently in all experiments. ELISPOT assays were also performed to analyze CD4+ T cells secreting these cytokines, which showed that significantly higher frequencies of cells producing IL-17A but not IL-4 were observed in BCG vaccinated compared to naïve mice (Supplementary Figure 2C). Neither technique detected IL-21 producing CD4+ T cells, possibly reflecting the known difficulties in detecting IL-21 producing germinal center Tfh cells [34, 35]. Based on our results, published reports [36–40] and known caveats in directly comparing RNA and protein levels [41, 42], we surmise that helper cells other than Th1 cells are generated during BCG vaccination but at low frequencies.
Transcripts that were more highly expressed in the CD154− population included that encoding IL-10, a cytokine associated with regulatory T cells. The expression of IL-10 transcript, together with the relative increase in Lrrc32 and Foxp3 transcripts in the CD154− population compared to the CD154+ population (Figure 3D and 4C), was consistent with previous reports showing that regulatory T cells lack pre-formed CD154 and that induced regulatory T cells are unlikely to be included in the CD154+ population [43–45].
Identification of an IL-3 secreting subset of mycobacteria-specific CD4+ T cells
An in depth analysis of our transcriptome data revealed that among the cytokines not classically associated with known T helper subsets, transcripts encoding IL-3 showed the highest differential expression between CD154+ and CD154− populations of CD4+ T cells in BCG vaccinated mice. This was an unexpected finding since IL-3, a cytokine mainly associated with anti-parasite immunity and hematopoiesis, has been seldom considered or studied in the context of mycobacterial immunity [46]. We validated the microarray results by quantitative PCR, which showed that transcripts encoding IL-3 were 9 times more abundant in CD154+ cells compared to CD154− cells (Figure 5A). We then examined whether IL-3 secretion by CD4+ T cells could be detected at the protein level. Mice were vaccinated with BCG, and 4 weeks after vaccination, CD4+ T cells were re-stimulated in vitro with Mtb lysate and stained for intracellular IL-3. We observed that approximately 0.2% of CD4+ T cells secreted IL-3 after re-stimulation, and the secretion was restricted to antigen-experienced (i.e., CD44+) CD4+ T cells (Figure 5B). This was approximately 18 fold less than the frequency of IFNγ producing CD4+ T cells detected by intracellular cytokine staining (Figure 5C) or by ELISPOT assays (Figure 5D). IL-3 secreting CD4+ T cells co-expressed IFNγ (Figure 5E), indicating that IL-3 producing cells were also CD154+ since IFNγ producing cells were almost uniformly CD154+. The co-expression of IFNγ but not IL-4 or IL-17A suggested that these cells were related to Th1 cells (Figure 5E), and this was further reinforced by the finding that they also expressed T-bet (Figure 5F).
Figure 5. Induction of IL-3 producing CD4+ T cells by mycobacteria.
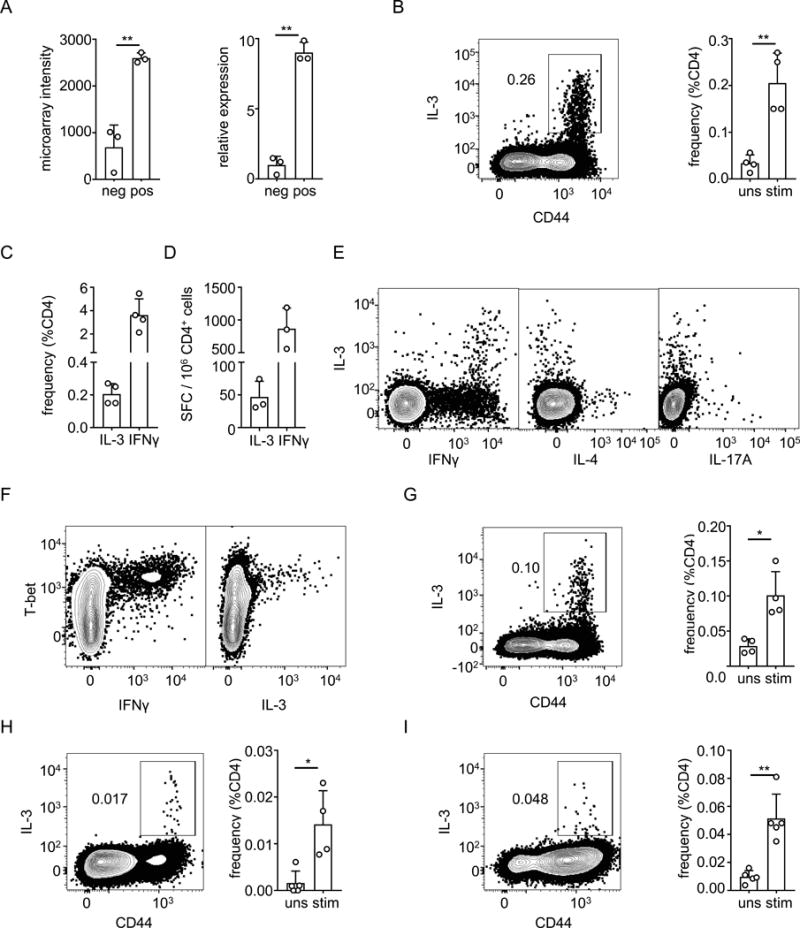
A) Analysis of IL-3 expression at RNA level of CD154− (neg) and CD154+ (pos) CD4+ T cells sorted from spleens of BCG-vaccinated mice. Graph on left shows normalized microarray intensity of IL-3 transcripts, and graph on right shows quantitative PCR analysis of IL-3 transcripts. B) Splenocytes from BCG-vaccinated mice were stimulated with Mtb lysate for 6 hours. Cells were stained with antibodies for surface markers including CD44, fixed and intracellular cytokine staining for IL-3 was performed. The cells shown are single cells from the lymphocyte gate that were positive for CD4 and negative for Live/Dead stain, CD8α, B220 and MHC-II. Barplot on the right summarizes the frequencies of IL-3 producing CD4+ T cells either without stimulation (uns) or with stimulation (stim). C, D). Mice were vaccinated with BCG and stimulated as in Panel B, and the frequencies of IL-3 producing and IFNγ producing CD4+ T cells in spleens were determined by intracellular cytokine staining (C) or by ELISPOT (D). SFC, spot forming cells. E) Analysis of splenocytes from BCG-vaccinated mice performed as in panel B, using intracellular cytokine staining to show co-expression of IL-3 with IFNγ, IL-4 and IL-17A. F). Intracellular staining of splenocytes prepared as in panel B and stained for T-bet in addition to IL-3 and IFNγ. G). Splenocytes from mice were infected with M. smegmatis, re-stimulated for 6 hours in vitro with lysate of M. smegmatis, and then stained and analyzed as in (B). H, I). Mice were infected with M. tuberculosis by aerosol route and four weeks later, single cell suspensions made from mediastinal lymph nodes (H) or lungs (I) were re-stimulaed with ESAT-6 peptide in the presence of brefeldin A and monensin. The cells were then surface stained, fixed, permeabilized and stained for intracellular IL-3. The cells shown are single cells from the lymphocyte gate that were positive for CD4 and negative for Live/Dead stain, CD8α, B220 and MHC-II. Barplot on the right summarizes the frequencies of IL-3 producing CD4+ T cells either without stimulation (uns) or with stimulation (stim). Bars in all plots show mean values for 3-4 replicates, open circles show values for individual mice, and error bars show 1 SD. * p < 0.05; ** p < 0.005 for Student’s t-tests. Results shown are representative of two independent experiments.
Since BCG is derived from a pathogenic species of mycobacterium, it is possible that it has retained properties that interfere with its ability to induce optimal protective immunity [47]. Generation of IL-3 may be one of the mechanisms, since mast cells and basophils that express IL-3 receptor and mediate Type II immunity, may have adverse effect on the generation of an effective type I immune response [48]. To determine whether IL-3 production by CD4+ T cells is restricted to BCG or is a more general feature of immune responses to mycobacteria, we injected mice with the non-pathogenic mycobacterium M. smegmatis, and examined IL-3 production by CD4+ T cells 4 weeks later. We found that M. smegmatis also induced IL-3 production from CD4+ T cells, albeit at a lower level than BCG, suggesting IL-3 induction is likely to be a conserved feature of host responses to various mycobacterial species (Figure 5G).
We also assessed whether IL-3 producing CD4 T cells are induced after infection with Mtb in a model of low-dose aerosol infection. We assessed IL-3 production by CD4+ T cells 4 weeks after mycobacterial infection and found out that both mediastinal lymph node and lung contained IL-3 producing CD4+ T cells that are specific to mycobacterial peptide ESAT-6. (Figure 5H and 5I). Thus, transcriptome analysis of mycobacteria-specific CD4+ T cells resulted in the identification of a previously unrecognized subset characterized by the production of IL-3.
Discussion
Using two different experimental models, we have shown in the current study that detection of activation-induced CD154 expression is a reliable method to identify and isolate antigen-specific CD4+ T cells for downstream applications. Our approach was based on modifications of a method reported previously that uses inclusion of fluorescent antibodies to CD154 and monensin in the media during in vitro antigen re-stimulation of CD4+ T cells [8]. We initially validated this method using adoptively transferred transgenic CD4+ T cells specific to a peptide of ovalbumin, and then extended and confirmed our results in a second model examining endogenous mycobacteria-specific CD4+ T cells after BCG vaccination. In these models, we found that CD154 expression served as a reliable marker for at least 90% of the antigen-specific CD4+ T cells that could be reactivated by antigen exposure in vitro. Expression microarray analysis performed on mycobacteria-specific CD4+ T cells further supported the validity of using CD154 expression as a surrogate marker for antigen-specificity. In addition, microarray analysis revealed that while multiple functional T helper subsets were present in the CD154+ population, antigen-specific regulatory T cells were likely to be excluded, consistent with previous reports in the literature [43–45].
Transcriptome analysis of antigen-specific CD4+ T cells highlighted several understudied molecules expressed by these cells. One of these is Ermn, the most highly expressed transcript in CD154+ relative to CD154− cells, which encode a cytoskeletal protein Ermin. Expression of ERMN is believed to be restricted to oligodendrocytes, and has been shown to bind and re-organize F-actin in these cells [31]. ERMN belongs to the family of ERM proteins which function upstream and downstream of Rho GTPases, and probably control process outgrowth and morphological changes during oligodendrocyte differentiation [31]. While the function of ERMN in CD4+ T cells or other immune cells is yet to be studied, it is possible that similar to oligodendrocytes, the function of ERMN in these cells is related to cytoskeletal changes during activation. Another transcript that showed high levels of expression in CD154+ cells encoded ZEB2, a two-handed zinc-finger transcription factor that binds DNA at tandem, separated consensus E-box sites [32]. Based on microarray intensities, expression of Zeb2 was highest among all transcription factors in CD154+ versus CD154− cells from BCG-vaccinated mice. Recent studies have shown that ZEB2 expression is important for the differentiation of effector and memory CD8+ T cells after infection with lymphocytic choriomeningitis virus [32, 49]. While ZEB2 was shown to be downstream of T-bet, and these two factors co-regulate many of the same genes, it appears that ZEB2 modulates these downstream genes independently. Given that ZEB2 plays a similar role in the maturation Natural Killer cells [50], and is required for the development of plasmacytoid dendritic cells and monocytes [33], it would be relevant to assess whether ZEB2 has similar functions in the differentiation of CD4+ T cells.
Our experiments also showed that BCG vaccination and M. smegmatis infection induce IL-3 producing CD4+ T cells which are related to Th1 cells. We pursued IL-3 producing CD4+ T cells because, in the microarray analysis, transcripts for this cytokine showed the most differential expression among cytokines that are not associated with a defined T helper population. In addition, this cytokine has not been investigated in detail in the context of anti-mycobacterial immunity. IL-3 was originally described as a cytokine secreted by CD4+ T cells and has been shown to have proliferative effects on various cell types [51, 52]. Studies on IL-3 knock-out mice showed that IL-3 has minor nonredundant effects on hematopoiesis, but plays a significant role in anti-parasite immunity through its action on mast cells and basophils [46, 53–56]. There is increasing interest in this cytokine given recent reports on the involvement of IL-3 in augmenting features of sepsis, systemic lupus erythematosus or the experimental autoimmune encephalomyelitis model of multiple sclerosis [57–59].
Our results showed that Mtb infection by the aerosol route induced IL-3 producing CD4+ T cells in the lung. While we perfused lungs prior to processing the tissue into single cell suspensions, we cannot exclude the possibility that these cells reside within the lung vasculature since simple perfusion may be inefficient at removing intravascular T cells [60]. Further analysis of these cells after intra-vascular staining will allow us to better determine their location within the lung [61]. This is important because it has been shown that Th1 cells in the lung parenchyma are highly protective while those in the lung vasculature are only weakly protective [62]. If they are located within the lung parenchyma, their location within the granuloma and their relationship with other cells, especially those expressing the receptor to IL-3, will be important issues to address in future studies. Additionally, the question of whether IL-3 producing CD4+ T cells contribute significantly to immunity during infection with Mtb, and whether IL-3 producing CD4+ T cells induced by BCG vaccination are recalled during an Mtb challenge, will require further study. Given the role of IL-3 in anti-parasite-immunity, IL-3 producing CD4+ T cells may have effects that are deleterious for anti-tuberculosis immunity, as has been suggested generally for Th2 cytokines [48]. That IL-3 producing CD4+ T cells are induced by M. smegmatis, a non-pathogenic mycobacterium, does not exclude this possibility. This deleterious effect could be mediated by mast cells which are abundant at barrier surfaces including respiratory epithelium [63]. Mast cells have been shown to interact with Mtb triggering the release of histamine, β-hexosaminidase, TNF-α and IL-6 [64]. In addition, activation of mast cells using compound 48/80 in Mtb infected mice can lead to increased bacterial burdens, presumably by decreasing Th1 cytokines and increasing IL-10 [65]. Basophils and eosinophils can also infiltrate inflamed lung and mediate activities that exacerbate pathology and promote mycobacterial growth [66–68].
In summary, our studies support the use of activation-induced expression of CD154 as a method to detect and isolate antigen-specific CD4+ T cells in murine models of infection and vaccination. This method has advantages over intracellular cytokine staining when downstream applications require live cells, or when high quality RNA needs to be isolated for transcriptomic studies. This approach can also be used when tetramers or bi-functional antibodies for cell surface cytokine-capture assays are not readily available. In addition, this method is useful when analyzing responses to complex antigen mixtures such as extracts of whole microorganisms or tumor cells, or when the choice of cytokine to be assayed is uncertain.
Supplementary Material
Acknowledgments
We thank staff from the Einstein Genomics and Flow Cytometry Core facilities for assistance with microarray and cell sorting experiments. We thank Pooja Arora for insightful discussions.
This work was supported by NIH/NIAD grants 1R21AI092448 (awarded to SAP) and 2P01AI063537 (awarded to WRJ, SAP and JC). Core resources for flow cytometry and microarray analysis were supported by the Einstein Cancer Center (CA13330). Support for CTJ was provided by NIH Training Grant GM07491.
References
- 1.Klinman D. ELISPOT assay to detect cytokine-secreting murine and human cells. Curr Protoc Immunol. 2008 doi: 10.1002/0471142735.im0619s83. Chapter 6: p. Unit 6 19. [DOI] [PubMed] [Google Scholar]
- 2.Bercovici N, Duffour MT, Agrawal S, Salcedo M, Abastado JP. New methods for assessing T-cell responses. Clin Diagn Lab Immunol. 2000;7(6):859–64. doi: 10.1128/cdli.7.6.859-864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, Shehata SI, Nelson AM, Shapovalova NV, Levi BP, Ramanathan S. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat Methods. 2016;13(1):87–93. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klemm S, Semrau S, Wiebrands K, Mooijman D, Faddah DA, Jaenisch R, van Oudenaarden A. Transcriptional profiling of cells sorted by RNA abundance. Nat Methods. 2014;11(5):549–51. doi: 10.1038/nmeth.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manz R, Assenmacher M, Pfluger E, Miltenyi S, Radbruch A. Analysis and sorting of live cells according to secreted molecules, relocated to a cell-surface affinity matrix. Proc Natl Acad Sci U S A. 1995;92(6):1921–5. doi: 10.1073/pnas.92.6.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20(1):4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havenar-Daughton C, Reiss SM, Carnathan DG, Wu JE, Kendric K, Torrents de la Pena A, Kasturi SP, Dan JM, Bothwell M, Sanders RW, Pulendran B, Silvestri G, Crotty S. Cytokine-Independent Detection of Antigen-Specific Germinal Center T Follicular Helper Cells in Immunized Nonhuman Primates Using a Live Cell Activation-Induced Marker Technique. J Immunol. 2016;197(3):994–1002. doi: 10.4049/jimmunol.1600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med. 2005;11(10):1113–7. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 9.Frentsch M, Arbach O, Kirchhoff D, Moewes B, Worm M, Rothe M, Scheffold A, Thiel A. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat Med. 2005;11(10):1118–24. doi: 10.1038/nm1292. [DOI] [PubMed] [Google Scholar]
- 10.Kirchhoff D, Frentsch M, Leclerk P, Bumann D, Rausch S, Hartmann S, Thiel A, Scheffold A. Identification and isolation of murine antigen-reactive T cells according to CD154 expression. Eur J Immunol. 2007;37(9):2370–7. doi: 10.1002/eji.200737322. [DOI] [PubMed] [Google Scholar]
- 11.Lederman S, Yellin MJ, Krichevsky A, Belko J, Lee JJ, Chess L. Identification of a novel surface protein on activated CD4+ T cells that induces contact-dependent B cell differentiation (help) J Exp Med. 1992;175(4):1091–101. doi: 10.1084/jem.175.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noelle RJ, Roy M, Shepherd DM, Stamenkovic I, Ledbetter JA, Aruffo A. A 39-kDa protein on activated helper T cells binds CD40 and transduces the signal for cognate activation of B cells. Proc Natl Acad Sci U S A. 1992;89(14):6550–4. doi: 10.1073/pnas.89.14.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1(3):167–78. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 14.Brines RD, Klaus GG. Polyclonal activation of immature B cells by preactivated T cells: the role of IL-4 and CD40 ligand. Int Immunol. 1993;5(11):1445–50. doi: 10.1093/intimm/5.11.1445. [DOI] [PubMed] [Google Scholar]
- 15.Grewal IS, Flavell RA. The role of CD40 ligand in costimulation and T-cell activation. Immunol Rev. 1996;153:85–106. doi: 10.1111/j.1600-065x.1996.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 16.Yellin MJ, Sippel K, Inghirami G, Covey LR, Lee JJ, Sinning J, Clark EA, Chess L, Lederman S. CD40 molecules induce down-modulation and endocytosis of T cell surface T cell-B cell activating molecule/CD40-L. Potential role in regulating helper effector function. J Immunol. 1994;152(2):598–608. [PubMed] [Google Scholar]
- 17.Hatfull GF. The molecular genetics of Mycobacterium tuberculosis. Curr Top Microbiol Immunol. 1996;215:29–47. doi: 10.1007/978-3-642-80166-2_2. [DOI] [PubMed] [Google Scholar]
- 18.Johnson AJ, Kennedy SC, Lindestam Arlehamn CS, Goldberg MF, Saini NK, Xu J, Paul S, Hegde SS, Blanchard JS, Chan J, Jacobs WR, Jr, Sette A, Porcelli SA. Identification of Mycobacterial RplJ/L10 and RpsA/S1 Proteins as Novel Targets for CD4+ T Cells. Infect Immun. 2017;85(4) doi: 10.1128/IAI.01023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweeney KA, Dao DN, Goldberg MF, Hsu T, Venkataswamy MM, Henao-Tamayo M, Ordway D, Sellers RS, Jain P, Chen B, Chen M, Kim J, Lukose R, Chan J, Orme IM, Porcelli SA, Jacobs WR., Jr A recombinant Mycobacterium smegmatis induces potent bactericidal immunity against Mycobacterium tuberculosis. Nat Med. 2011;17(10):1261–8. doi: 10.1038/nm.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schinke-Braun M, Couget JA. Expression profiling using affymetrix genechip probe arrays. Methods Mol Biol. 2007;366:13–40. doi: 10.1007/978-1-59745-030-0_2. [DOI] [PubMed] [Google Scholar]
- 21.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22(22):2825–7. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 22.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1–3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trumpfheller C, Caskey M, Nchinda G, Longhi MP, Mizenina O, Huang Y, Schlesinger SJ, Colonna M, Steinman RM. The microbial mimic poly IC induces durable and protective CD4+ T cell immunity together with a dendritic cell targeted vaccine. Proc Natl Acad Sci U S A. 2008;105(7):2574–9. doi: 10.1073/pnas.0711976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koguchi Y, Thauland TJ, Slifka MK, Parker DC. Preformed CD40 ligand exists in secretory lysosomes in effector and memory CD4+ T cells and is quickly expressed on the cell surface in an antigen-specific manner. Blood. 2007;110(7):2520–7. doi: 10.1182/blood-2007-03-081299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waldrop SL, Davis KA, Maino VC, Picker LJ. Normal human CD4+ memory T cells display broad heterogeneity in their activation threshold for cytokine synthesis. J Immunol. 1998;161(10):5284–95. [PubMed] [Google Scholar]
- 28.Itoh Y, Germain RN. Single cell analysis reveals regulated hierarchical T cell antigen receptor signaling thresholds and intraclonal heterogeneity for individual cytokine responses of CD4+ T cells. J Exp Med. 1997;186(5):757–66. doi: 10.1084/jem.186.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auphan-Anezin N, Verdeil G, Schmitt-Verhulst AM. Distinct thresholds for CD8 T cell activation lead to functional heterogeneity: CD8 T cell priming can occur independently of cell division. J Immunol. 2003;170(5):2442–8. doi: 10.4049/jimmunol.170.5.2442. [DOI] [PubMed] [Google Scholar]
- 30.Dafforn A, Chen P, Deng G, Herrler M, Iglehart D, Koritala S, Lato S, Pillarisetty S, Purohit R, Wang M, Wang S, Kurn N. Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. Biotechniques. 2004;37(5):854–7. doi: 10.2144/04375PF01. [DOI] [PubMed] [Google Scholar]
- 31.Brockschnieder D, Sabanay H, Riethmacher D, Peles E. Ermin, a myelinating oligodendrocyte-specific protein that regulates cell morphology. J Neurosci. 2006;26(3):757–62. doi: 10.1523/JNEUROSCI.4317-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, Seuntjens E, Stryjewska A, Zweier C, Roychoudhuri R, Gattinoni L, Bird LM, Higashi Y, Kondoh H, Huylebroeck D, Haigh J, Goldrath AW. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med. 2015;212(12):2027–39. doi: 10.1084/jem.20150194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Briseno CG, Grajales-Reyes GE, Haldar M, Iwata A, Kretzer NM, Kc W, Tussiwand R, Higashi Y, Murphy TL, Murphy KM. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc Natl Acad Sci U S A. 2016;113(51):14775–14780. doi: 10.1073/pnas.1611408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar-Daughton C, Reiss SM, Brigger M, Bothwell M, Sette A, Crotty S. A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. J Immunol. 2016;197(3):983–93. doi: 10.4049/jimmunol.1600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suto A, Kashiwakuma D, Kagami S, Hirose K, Watanabe N, Yokote K, Saito Y, Nakayama T, Grusby MJ, Iwamoto I, Nakajima H. Development and characterization of IL-21-producing CD4+ T cells. J Exp Med. 2008;205(6):1369–79. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huygen K, Abramowicz D, Vandenbussche P, Jacobs F, De Bruyn J, Kentos A, Drowart A, Van Vooren JP, Goldman M. Spleen cell cytokine secretion in Mycobacterium bovis BCG-infected mice. Infect Immun. 1992;60(7):2880–6. doi: 10.1128/iai.60.7.2880-2886.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar M, Behera AK, Matsuse H, Lockey RF, Mohapatra SS. A recombinant BCG vaccine generates a Th1-like response and inhibits IgE synthesis in BALB/c mice. Immunology. 1999;97(3):515–21. doi: 10.1046/j.1365-2567.1999.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Pelayo MC, Bachy VS, Kaveh DA, Hogarth PJ. BALB/c mice display more enhanced BCG vaccine induced Th1 and Th17 response than C57BL/6 mice but have equivalent protection. Tuberculosis (Edinb) 2015;95(1):48–53. doi: 10.1016/j.tube.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Derrick SC, Kolibab K, Yang A, Morris SL. Intranasal administration of Mycobacterium bovis BCG induces superior protection against aerosol infection with Mycobacterium tuberculosis in mice. Clin Vaccine Immunol. 2014;21(10):1443–51. doi: 10.1128/CVI.00394-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lalor MK, Smith SG, Floyd S, Gorak-Stolinska P, Weir RE, Blitz R, Branson K, Fine PE, Dockrell HM. Complex cytokine profiles induced by BCG vaccination in UK infants. Vaccine. 2010;28(6):1635–41. doi: 10.1016/j.vaccine.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen PZ, Brewer H, Weitz K, Camp DG, 2nd, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7(6):e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 43.Koguchi Y, Buenafe AC, Thauland TJ, Gardell JL, Bivins-Smith ER, Jacoby DB, Slifka MK, Parker DC. Preformed CD40L is stored in Th1, Th2, Th17, and T follicular helper cells as well as CD4+ 8- thymocytes and invariant NKT cells but not in Treg cells. PLoS One. 2012;7(2):e31296. doi: 10.1371/journal.pone.0031296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi JY, Eo SK. Detection of Foreign Antigen-specific CD4(+)Foxp3(+) Regulatory T Cells by MHC Class II Tetramer and Intracellular CD154 Staining. Immune Netw. 2013;13(6):264–74. doi: 10.4110/in.2013.13.6.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noyan F, Lee YS, Zimmermann K, Hardtke-Wolenski M, Taubert R, Warnecke G, Knoefel AK, Schulde E, Olek S, Manns MP, Jaeckel E. Isolation of human antigen-specific regulatory T cells with high suppressive function. Eur J Immunol. 2014;44(9):2592–602. doi: 10.1002/eji.201344381. [DOI] [PubMed] [Google Scholar]
- 46.Lantz CS, Boesiger J, Song CH, Mach N, Kobayashi T, Mulligan RC, Nawa Y, Dranoff G, Galli SJ. Role for interleukin-3 in mast-cell and basophil development and in immunity to parasites. Nature. 1998;392(6671):90–3. doi: 10.1038/32190. [DOI] [PubMed] [Google Scholar]
- 47.Baena A, Porcelli SA. Evasion and subversion of antigen presentation by Mycobacterium tuberculosis. Tissue Antigens. 2009;74(3):189–204. doi: 10.1111/j.1399-0039.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salgame P, Yap GS, Gause WC. Effect of helminth-induced immunity on infections with microbial pathogens. Nat Immunol. 2013;14(11):1118–26. doi: 10.1038/ni.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, Kaech SM. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med. 2015;212(12):2041–56. doi: 10.1084/jem.20150186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Helden MJ, Goossens S, Daussy C, Mathieu AL, Faure F, Marcais A, Vandamme N, Farla N, Mayol K, Viel S, Degouve S, Debien E, Seuntjens E, Conidi A, Chaix J, Mangeot P, de Bernard S, Buffat L, Haigh JJ, Huylebroeck D, Lambrecht BN, Berx G, Walzer T. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med. 2015;212(12):2015–25. doi: 10.1084/jem.20150809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ihle JN, Pepersack L, Rebar L. Regulation of T cell differentiation: in vitro induction of 20 alpha-hydroxysteroid dehydrogenase in splenic lymphocytes from athymic mice by a unique lymphokine. J Immunol. 1981;126(6):2184–9. [PubMed] [Google Scholar]
- 52.Yang YC, Ciarletta AB, Temple PA, Chung MP, Kovacic S, Witek-Giannotti JS, Leary AC, Kriz R, Donahue RE, Wong GG, et al. Human IL-3 (multi-CSF): identification by expression cloning of a novel hematopoietic growth factor related to murine IL-3. Cell. 1986;47(1):3–10. doi: 10.1016/0092-8674(86)90360-0. [DOI] [PubMed] [Google Scholar]
- 53.Kim S, Prout M, Ramshaw H, Lopez AF, LeGros G, Min B. Cutting edge: basophils are transiently recruited into the draining lymph nodes during helminth infection via IL-3, but infection-induced Th2 immunity can develop without basophil lymph node recruitment or IL-3. J Immunol. 2010;184(3):1143–7. doi: 10.4049/jimmunol.0902447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lantz CS, Min B, Tsai M, Chatterjea D, Dranoff G, Galli SJ. IL-3 is required for increases in blood basophils in nematode infection in mice and can enhance IgE-dependent IL-4 production by basophils in vitro. Lab Invest. 2008;88(11):1134–42. doi: 10.1038/labinvest.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen T, Kim S, Do JS, Wang L, Lantz C, Urban JF, Le Gros G, Min B. T cell-derived IL-3 plays key role in parasite infection-induced basophil production but is dispensable for in vivo basophil survival. Int Immunol. 2008;20(9):1201–9. doi: 10.1093/intimm/dxn077. [DOI] [PubMed] [Google Scholar]
- 56.Mach N, Lantz CS, Galli SJ, Reznikoff G, Mihm M, Small C, Granstein R, Beissert S, Sadelain M, Mulligan RC, Dranoff G. Involvement of interleukin-3 in delayed-type hypersensitivity. Blood. 1998;91(3):778–83. [PubMed] [Google Scholar]
- 57.Weber GF, Chousterman BG, He S, Fenn AM, Nairz M, Anzai A, Brenner T, Uhle F, Iwamoto Y, Robbins CS, Noiret L, Maier SL, Zonnchen T, Rahbari NN, Scholch S, Klotzsche-von Ameln A, Chavakis T, Weitz J, Hofer S, Weigand MA, Nahrendorf M, Weissleder R, Swirski FK. Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science. 2015;347(6227):1260–5. doi: 10.1126/science.aaa4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Renner K, Hermann FJ, Schmidbauer K, Talke Y, Rodriguez Gomez M, Schiechl G, Schlossmann J, Bruhl H, Anders HJ, Mack M. IL-3 contributes to development of lupus nephritis in MRL/lpr mice. Kidney Int. 2015;88(5):1088–98. doi: 10.1038/ki.2015.196. [DOI] [PubMed] [Google Scholar]
- 59.Renner K, Hellerbrand S, Hermann F, Riedhammer C, Talke Y, Schiechl G, Gomez MR, Kutzi S, Halbritter D, Goebel N, Bruhl H, Weissert R, Mack M. IL-3 promotes the development of experimental autoimmune encephalitis. JCI Insight. 2016;1(16):e87157. doi: 10.1172/jci.insight.87157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson KG, Sung H, Skon CN, Lefrancois L, Deisinger A, Vezys V, Masopust D. Cutting edge: intravascular staining redefines lung CD8 T cell responses. J Immunol. 2012;189(6):2702–6. doi: 10.4049/jimmunol.1201682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, Masopust D. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9(1):209–22. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakai S, Kauffman KD, Schenkel JM, McBerry CC, Mayer-Barber KD, Masopust D, Barber DL. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J Immunol. 2014;192(7):2965–9. doi: 10.4049/jimmunol.1400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wernersson S, Pejler G. Mast cell secretory granules: armed for battle. Nat Rev Immunol. 2014;14(7):478–94. doi: 10.1038/nri3690. [DOI] [PubMed] [Google Scholar]
- 64.Munoz S, Hernandez-Pando R, Abraham SN, Enciso JA. Mast cell activation by Mycobacterium tuberculosis: mediator release and role of CD48. J Immunol. 2003;170(11):5590–6. doi: 10.4049/jimmunol.170.11.5590. [DOI] [PubMed] [Google Scholar]
- 65.Carlos D, de Souza Junior DA, de Paula L, Jamur MC, Oliver C, Ramos SG, Silva CL, Faccioli LH. Mast cells modulate pulmonary acute inflammation and host defense in a murine model of tuberculosis. J Infect Dis. 2007;196(9):1361–8. doi: 10.1086/521830. [DOI] [PubMed] [Google Scholar]
- 66.Wakahara K, Van VQ, Baba N, Begin P, Rubio M, Delespesse G, Sarfati M. Basophils are recruited to inflamed lungs and exacerbate memory Th2 responses in mice and humans. Allergy. 2013;68(2):180–9. doi: 10.1111/all.12072. [DOI] [PubMed] [Google Scholar]
- 67.Nabe T, Matsuya K, Akamizu K, Fujita M, Nakagawa T, Shioe M, Kida H, Takiguchi A, Wakamori H, Fujii M, Ishihara K, Akiba S, Mizutani N, Yoshino S, Chaplin DD. Roles of basophils and mast cells infiltrating the lung by multiple antigen challenges in asthmatic responses of mice. Br J Pharmacol. 2013;169(2):462–76. doi: 10.1111/bph.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirman J, Zakaria Z, McCoy K, Delahunt B, Le Gros G. Role of eosinophils in the pathogenesis of Mycobacterium bovis BCG infection in gamma interferon receptor-deficient mice. Infect Immun. 2000;68(5):2976–8. doi: 10.1128/iai.68.5.2976-2978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


