Abstract
At present, artificial pit mud (APM) is widely used in Chinese liquor-making industry and plays a particular role in the production of Chinese strong flavor liquor (CSFL). However, APM frequently ages during fermentation, thus becoming unsuitable for sustainable use due to its low-quality. The reasons behind, and results of, APM aging during the production of CSFL are not yet understood. Sequencing the V4 region of the 16S rRNA gene shows that prokaryotic diversity is significantly decreased (Shannon’s diversity index, P < 0.01) and community composition is distinctly changed (from 1197 to 865 OTUs) in aging APM. On the phylum level, the increase of Firmicutes and decrease of Proteobacteria are the main consequences of APM aging during the production of CSFL. The counting of cultivatable bacteria confirmed that there was a large increase in Lactobacilli and aerobic spore-forming bacteria in aging low-quality APM (more than twofold). Unexpectedly, the total number of caproic acid-producing bacteria, mainly Clostridia, did not change significantly between the two kinds of APM. Furthermore, biochemical analysis indicates that the pH and the levels of NH4 + and K+ are decreased in aging low-quality APM (P < 0.01). The results obtained in this study support the possibility that environmental factors (pH, nutrients) induce the decrease of prokaryotic diversity, and the changed community composition influences the environmental properties. Therefore, through interfering with the cycle, APM aging can be controlled potentially by adjustment of environmental factors and/or supplementation of diminished or missed microorganisms.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0978-0) contains supplementary material, which is available to authorized users.
Keywords: Artificial pit mud, Prokaryotic diversity, Community composition, Lactobacilli, Clostridia, Biochemical properties
Introduction
Chinese liquor is a traditional alcoholic beverage widely consumed by billions of people annually. Chinese strong flavor liquor (CSFL) accounts for more than 70% of Chinese liquor. It is produced by the solid-state fermentation technique in a mud cellar in which pit mud plays an important role for efficient fermentation during the whole process (Zhao et al. 2012). Natural high-quality pit mud is formed through acclimation of soil microbes during many years of fermentation in a cellar. Some old cellars have been used for several hundreds of years and numerous famous brands, such as Wuliangye and Luzhoulaojiao, are representatives of CSFL produced in this way in China (Song 2016). Each company believes that its inherited natural high-quality pit mud is its unique and pivotal treasure, although the legacy cannot be transferred from one place to another.
At present, the amount of naturally preserved high-quality pit mud is insufficient to keep pace with the rapid development of the industry. Therefore, artificial pit mud (APM) is widely applied in Chinese liquor production (Jing et al. 2010; Zhang et al. 2014a). Usually, artificial pit mud is prepared by mixing high-quality old pit mud or its derivative enriched with microbes primarily by selective cultivation, normal fresh soil and water, and then incubating the mixture for about 2 months in an anaerobic cellar before use (Xu et al. 2010). Matured APM is used for fermentation in new cellars. APM is the habitat of microbes, which act as fermentation starters and producers of various flavor components. Unfortunately, APM ages and this leads to fermentation failure. The adverse change of APM is termed as aging and so the mud is termed as aging low-quality APM. An obvious feature of aging APM is the formation of a large amount of white aggregate comprising calcium lactate and crystal ferrous lactate (Zhang et al. 2014a, b). The common sensory characteristics of aging APM are stiff and weak ester aromas (Zhang 2010; Zhang et al. 2010).
A three-phase model, including domestication, transition, and the maturation period, was proposed for describing the long-term shaping process of the microbial community in high-quality pit mud (Tao et al. 2014). A few studies have reported the difference of the microbial community in pit mud by a combination of polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) and quantitative PCR assays (Zheng et al. 2013; Zhang et al. 2015; Liang et al. 2015). The most recent study using Illumina MiSeq sequencing of the 16S rRNA gene demonstrated that the pit mud anaerobic microbial community was correlated with its quality and environmental factors (Hu et al. 2016). However, there exists no explanation for the aging process, and there are no comprehensive studies of microbial diversity in APM specifically. The relationships between microbial diversity and biogeochemical attributes in aging APM are still poorly understood. Particularly, it is not known which factors are most influential on the quality of pit mud and how to control aging. Considering the large demand for APM and the rapid aging during production, it is necessary to study these relationships.
To address some of these problems, we first investigated prokaryotic diversities in different APMs collected from the same cellars by amplicon sequencing of the 16S rRNA gene. Second, we selectively cultivated and counted three groups of microbes in APMs on corresponding agar plates. Lastly, we compared the chemical properties of high-quality and aging low-quality APMs. This study aims to (1) characterize the bacterial diversity and community structure in different APMs, (2) identify the main groups of microbes in APMs that are predominantly responsible for high-quality CSFL production, and (3) reveal the relationships between the chemical properties and microbial community composition in APMs.
Materials and methods
APM sample collection
APM samples were collected from Henan Songhe Liquor Corporation located in Zhoukou City (34°02′N, 115°62′E), China. We selected APM samples from three different cellars that had been in use for 5 years. In each cellar, there is a distinct black band on the wall, which indicates aging occurred above the band. High-quality and aging low-quality APM samples were collected at three sites in each cellar below or above the black band, respectively. Samples were taken back to the laboratory on ice and in sealed bags. Before the mixing of the three samples collected in each cellar, all samples were kept at −20 °C. Then, mixed high-quality APM samples were labeled as NS1.1, NS1.2, and NS1.3, and aging low-quality APM samples as OS1.1, OS1.2, and OS1.3.
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted using a PowerSoil DNA isolation kit (MoBio Laboratories, Cat no. 12888). The V4 variable region of the 16S rRNA gene was amplified with universal primers 515F/806R (Zhang et al. 2014b). The detailed PCR conditions were as described (Li et al. 2014b). The barcoded amplicons were pooled with equal molar concentrations of the samples and sequenced using an Illumina MiSeq PE250 (Illumina) by a commercial supplier (Novogene, Beijing).
Processing of sequencing data
The raw sequences were processed using the QIIME pipeline (Caporaso et al. 2010). Briefly, all 16S rRNA sequences with lengths less than 150 bp were removed (excluding primer and barcode region). The remaining good-quality sequences were clustered into operational taxonomic units (OTUs) using a threshold of 97% identity (Edgar 2010). Only OTUs containing at least five reads were considered to be valid in this study.
Culture of different groups of bacteria in APM
For each sample, 1 g of freshly collected wet APM was fully suspended in 100 mL sterile peptone physiological salt (PPS) solution as described (Hu et al. 2015). Then, 1 mL suspension was 10-fold diluted in 10 mL glass tubes. Subsequently, 0.1 mL suspension of these diluents was spread on corresponding agar plate in triplicate. Lactobacilli were cultivated for 3 days on Lactobacilli MRS Agar (US Biological, Cat no. L1021). Aerobic spore-forming bacteria were selected by heating the original suspension at 80 °C for 10 min before dilution, plated on Plate Count Agar (Sigma, Cat no. 70152), and cultivated for 3 days. Clostridia were cultivated anaerobically for 1 week on Reinforced Clostridia Medium (RCM, BD Difco, Cat no. 218081) supplemented with 15 g/L agar (Sigma, Cat no. A1296). Anaerobic conditions were achieved by AnaeroPack-Anaero in sealed jars (Mitsubishi Gas Chemical) for Clostridia. All agar plates were placed in a temperature-constant incubator set at 37 °C.
Biochemical property analysis
APM moisture level was determined by drying soils at 60 °C for 48 h immediately after the sampling. The pH was measured with a pH meter in the slurry (APM to distilled H2O: 1:5, w/v). NH4 + concentration was determined using the sodium salicylate method (Mulvaney 1996). Humus content was determined by the dichromate oxidation method using a spectrophotometer (Mehlich 1984). Available phosphorus (AP) was extracted with ammonium fluoride and hydrochloric acid (Liu et al. 2014), and measured by UV photometer (Spectrum lab 24, Shanghai, China). Available potassium (K+) was measured by flame photometry after NH4OAc neutral extraction (Li et al. 2014a, b).
Statistical analysis
The Shannon diversity index was calculated according to the method described previously (Tao et al. 2014). The cluster analysis was conducted with the unweighted pair group method using average linkages (UPGMA) (Hammer et al. 2001). Principal component analysis (PCA) and hierarchical clustering analysis (CA) were used for sample comparison in Unifrac (http://bmf.colorado.edu/unifrac). The statistical significance of the difference between the means of samples was tested by one-way analysis of variance (ANOVA).
Project registration and nucleotide sequence accession
The original pyrosequencing data are available at the European Nucleotide Archive (accession no. PRJEB19195).
Results
Overall bacterial diversity is decreased
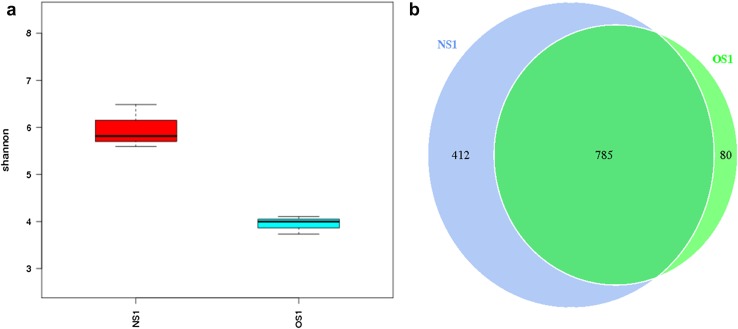
Totally, 323,827 qualified reads with an average length of 253 bp were obtained from all pit mud samples and each sample contained 32,795–63,264 reads (supplementary materials, Table S1). A total of 50,005 tags in the dataset with an average of 1735 unique tags in each sample and an average of 724 OTUs were obtained based on 97% identity in 16S rRNA sequences. Rarefaction analysis indicated that all communities were well represented, since the alpha rarefaction curves reached the saturation plateau (supplementary materials, Fig. S1). The Shannon index suggests that the microbial diversity is significantly decreased in aging APM (Fig. 1a, P < 0.01). As shown in the Venn diagram, the richness of the community in aging APM also decreased (Fig. 1b, from 1197 to 865 OTUs, P < 0.05).
Fig. 1.
Alpha-diversity analysis of the OTUs. a Shannon diversity index; b Venn diagram. NS1 new samples are high-quality APMs; OS1 old samples are aging low-quality APMs
Total phyla and orders are fewer in aging APM
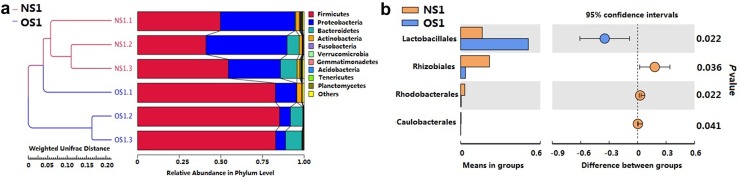
There are a total of 25 bacteria phyla and two Achaea phyla in high-quality APM, compared to 19 bacteria phyla and one Achaea phyla in aging low-quality APM according to the statistical analysis of OTUs (supplementary materials, dataset 1). Among the 20 shared phyla, Firmicutes and Proteobacteria were dominant. Although OS1.1 is a bit closer to the NS1 cluster, phylogenetic analysis indicates these samples are well represented. The two shared dominant phyla represent 89.99% in NS1 and 92.20% in OS1 among the top ten phyla, though they are in different percentages (Fig. 2a). For example, there is 48.40% Firmicutes and 41.59% Proteobacteria in high-quality APM, but 83.62% Firmicutes and 9.06% Proteobacteria in aging low-quality APM. Therefore, the ratio of Firmicutes/Proteobacteria increases by nearly sevenfold (from 1.16 to 9.23). On the class level, there are 93 bacterial classes in high-quality APM and 75 classes in aging low-quality APM (supplementary materials, dataset 2). The class of Bacilli is the most abundant in both high- and low-quality APMs. As shown on the heatmap (supplementary materials, Fig. S2), Lactobacillales is the most abundant order in OS1, while Actinomycetales and Chthoniobacterales are the two most abundant orders in NS1. A further statistical analysis confirms that Lactobacillales is significantly abundant in aging low-quality APM (Fig. 2b, P < 0.05). However, three orders (Rhizobiales, Rhodobacterales, Caulobacterales) are significantly abundant in high-quality APM (P < 0.05).
Fig. 2.
Beta-diversity analysis of the community composition. a Phylogenetic tree and corresponding relative abundance in phylum level of different samples. b Significantly different orders in two groups (P < 0.05). NS1 new samples are high-quality APMs, OS1 old samples are aging low-quality APMs
Community composition is simplified in aging APM
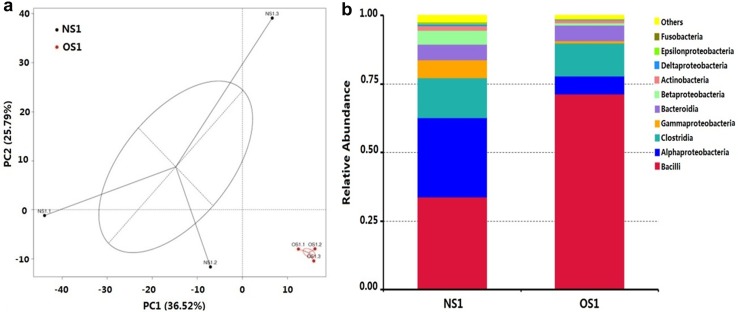
To evaluate similarities in different communities, principal component analysis (PCA) was conducted using Unifrac approaches. The PCA showed that all samples were clustered into high-quality (NS1) and low-quality (OS1) clusters (Fig. 3a). Microbes in the high-quality APM are highly dispersed, suggesting that these three samples are possibly in different transitional states. In contrast, microbes in the aging low-quality APM are extremely centralized. On the class level, there are a total of 62 classes in the high-quality APM and 50 classes in the aging low-quality APM. Seven shared classes were identified as being dominant (relative abundance of more than 0.5%), accounting for between 96.30 and 98.38% in each sample. These included Bacilli, Alphproteobacteria, Clostridia, Gammaproteobacteria, Bacteroidia, Betaproteobacteria, and Actinobacteria (Fig. 3b). After comparison, only the relative abundance of Bacilli was significantly higher, and the relative abundance of Alphproteobacteria was lower in aging low-quality APM (P < 0.05, supplementary materials, dataset 3).
Fig. 3.
Statistical analyses of microbial communities between different samples. a Cluster analyses based on principal component analysis (PCA) using weighted UniFrac. b Relative abundances of different classes in the two types of APMs. Presented taxa are the top ten classes. NS1 high-quality APM, including samples NS1.1, NS1.2, and NS1.3; OS1 aging low-quality APM, including samples OS1.1, OS1.2, and OS1.3
On the genus level, 14 genera were determined as being dominant among a total of 200 observed genera in all high-quality APMs. There are much fewer dominant genera in aging low-quality APM (i.e., only 4 genera making up more than 0.5%, including Lactobacillus, Ruminococcus, Ochrobactrum, and Bacillus among 144 genera). The other ten core genera only existing in high-quality APM are Sporosarcina, Devosia, Achromobacter, Paracoccus, Pigmentiphaga, Pseudomonas, Aeromonas, Denitrobacter, Garciella, and Amorphomonas. A total of 65 genera were absent in aging low-quality APM, accounting for 0.36% of the total classified sequences (supplementary materials, dataset 4). Further analysis on the species level suggests that the abundance of Lactobacillus acidipiscis and Bacillus thermoamylovorans is higher in aging low-quality APM than in high-quality APM, with 21.50% and 0.92%, respectively (supplementary materials, dataset 5).
Cultivation of specific groups agrees with sequencing results
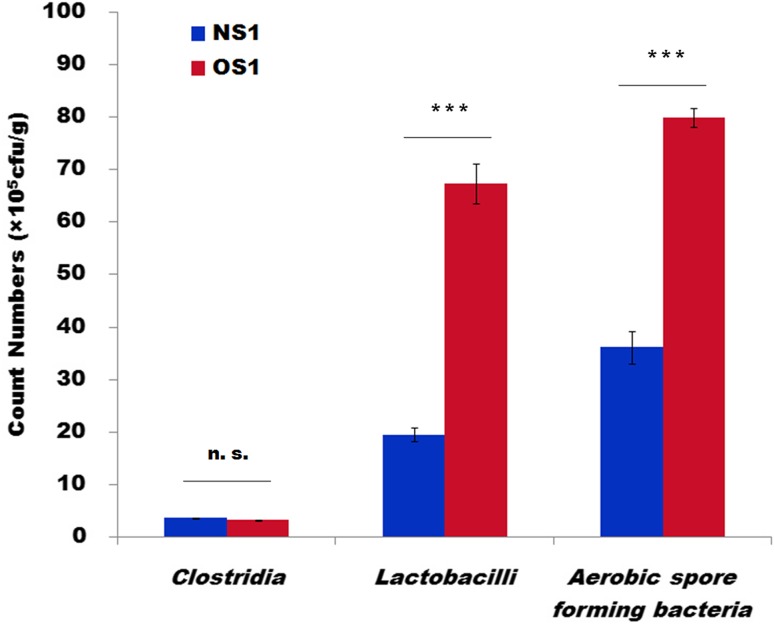
Three representative bacterial groups were selectively cultivated on different agar plates. The viable cell number was counted and calculated for comparison (Table S2). There are more than two times Lactobacilli in aging low-quality APM on average. The number of aerobic spore-forming bacteria in aging low-quality APM is around two times higher than that in high-quality APM. However, anaerobic Clostridia are slightly less abundant in aging low-quality APM and are mostly caproic acid-producing bacteria (Fig. 4). The relative ratio of the cell number from these three bacterial groups is similar to the sequencing data (relative abundance between Lactobacilli:Bacilli:Clostridia), suggesting high confidence of the culture-dependent and culture-independent methods.
Fig. 4.
Colony number of three representative bacterial groups cultivated on different selective agar plates. NS1 high-quality APM, including samples NS1.1, NS1.2, and NS1.3; OS1 aging low-quality APM, including samples OS1.1, OS1.2, and OS1.3. All data are presented as mean ± standard deviations (n = 3). Significance was determined by ANOVA (P < 0.05). ***P < 0.01; n. s. nonsignificant
Biochemical properties are changed in aging APM
The pH is an important factor that has complex impacts on the properties of APM. The pH value decreased from around 6.50 in high-quality APM to 4.54 in aging low-quality APM. As the water-holding capacity is normally reduced in aging low-quality APM, the relative content of water, as another factor (moisture), was assayed. However, there is no obvious difference in moisture in the two kinds of APM. Nutrients are also important contributors to the biochemical properties of soil. Concentrations of NH4 + and available K+ are considerably lower in aging APM. In contrast, the concentration of available phosphorus (AP) is significantly higher in aging APM (Table 1, P < 0.01). Moreover, the content of humus is slightly higher in aging APM.
Table 1.
Comparison of biochemical properties among different samples
| Items/samples | NS1.1 | NS1.2 | NS1.3 | OS1.1 | OS1.2 | OS1.3 |
|---|---|---|---|---|---|---|
| pH* | 6.25 ± 0.1 | 6.50 ± 0.2 | 6.88 ± 0.1 | 4.50 ± 0.2 | 4.68 ± 0.1 | 4.49 ± 0.1 |
| Moisture (%) | 37.81 ± 3.4 | 38.02 ± 2.9 | 38.12 ± 2.6 | 37.66 ± 1.3 | 38.14 ± 2.5 | 38.25 ± 4.2 |
| Humus (%)* | 21.05 ± 1.5 | 23.88 ± 1.3 | 24.41 ± 2.1 | 30.15 ± 2.6 | 33.10 ± 3.1 | 29.84 ± 0.8 |
| NH4 + (mg/kg)* | 183.1 ± 1.3 | 180.8 ± 1.9 | 170.6 ± 0.8 | 129.2 ± 2.4 | 130.8 ± 3.2 | 131.1 ± 1.5 |
| AP (mg/kg)* | 191.2 ± 9.6 | 205.3 ± 10.5 | 214.5 ± 12.3 | 825.0 ± 32.1 | 846.1 ± 64.5 | 802.4 ± 57.8 |
| Available K+ (mg/kg)* | 112.5 ± 3.2 | 102.4 ± 1.5 | 107.8 ± 2.6 | 59.8 ± 0.8 | 60.1 ± 1.2 | 62.5 ± 0.7 |
All data are presented as mean ± standard deviations (n = 3)
AP available phosphorus
* Significant differences in a row between NS1 and OS1 as determined by ANOVA (P < 0.05)
Discussion
The standard production of Chinese liquor is through four steps, including cooking, saccharification, fermentation, and distillation, using grain as the main raw material. The final product is colorless or transparently yellowish, giving off a pure and pleasant fragrance of esters as the main fragrant composites. APM plays leading role in the production of Chinese liquor, especially of CSFLs that are produced through fermentation in cellars. However, APM aging is common in the production process and aging APM fails to produce high-quality liquor (Zhang et al. 2014a, b). The aging of APM is manageable but unavoidable, as this process is affected by complex biochemical reactions during continued fermentation (Cai et al. 2009). To control the process of aging effectively, it is necessary to compare the difference between high-quality APM and aging low-quality APM, from the perspective of both chemistry and biology.
To study the causes and consequences of aging, we first investigated the prokaryotic diversity and community composition in different APMs by the high-throughput sequencing technique. Our results demonstrate that Firmicutes and Proteobacteria are the two most dominant phyla in APM. Linear discriminant analysis effect size (LEfSe) shows that the majority of different biomarkers also belong to these two phyla (Fig. S3). It has also been reported that Firmicutes were selected by pit mud conditions as the most acclimated prokaryotes, e.g., they dominated at 95.7% of prokaryotes after 1 year of domestication, though their abundance decreased yearly (Tao et al. 2014). This is a little different from our results, as the relative abundance of Firmicutes is about 48.4 and 83.6% in our high- and low-quality APM, respectively. However, Bacteroidetes other than Proteobacteria were identified from pit mud earlier by both DGGE and analysis of the 16S rRNA from the clone library as the second dominant phylum in pit mud (Wang et al. 2013). In our study, Bacteroidetes accounting for around 5.78–5.91% are still a prominent phylum (in third position), but are much less abundant than Proteobacteria. The differences between these dominant phyla are quite possibly caused by soil conditions and other environmental factors, as all Proteobacteria, Bacteroidetes, and Firmicutes were widely reported as the main bacterial community in aerobic soil (Lauber et al. 2009; Ferrenberg et al. 2013).
Previous studies have identified members of Clostridium, Bacillus, and Sporolactobacillus genera in pit mud as well (Yue et al. 2007). However, there is no information on the status or quality of pit mud used in these studies. A comparison of the diversity and composition between the high- and low-quality APM suggests that significant diversity decrease and composition imbalance are the main results of aging (as shown in Fig. 1). Particularly, the relative ratio between Bacilli and Alphaproteobacteria is greatly changed in different kinds of APMs (Fig. 3b, and Table S3, 0.34/0.29 in high-quality APM, contrasting with 0.71/0.07 in aging low-quality APM). Analysis on the order level suggests that Lactobacillales are markedly increased in aging low-quality APM (Fig. 2b). A further investigation on the family level indicates that both Lactobacillales and Sporolactobacillaceae are increased, though only the increase of the former is significant in aging low-quality APM. The same result was obtained by combined PCR-DGGE and quantitative PCR assay of aging pit mud used in Chinese Luzhou-flavor liquor (Liang et al. 2015).
Many studies have demonstrated the community changes by the culture-independent methods. However, there is no correlation analysis by the culture-dependent method. As only cultivatable microbes can be supplemented artificially for bioaugmentation, three bacterial groups were cultivated and counted for confirmation of 16S rRNA results and potential application (Fig. 4 and dataset 2). Analysis of their colony-forming units validated the relative abundance or ratio of these microbes in aging APM. Among them, the main lactic acid producers, Lactobacilli, are most abundant, which can restrain the production of caproic acid (Yao et al. 2010). In contrast to traditional notions, the abundance of Clostridia that are core attributes for the production of caproic acid is not changed in aging low-quality APM (Hu et al. 2015). The result suggests that the role of Clostridia may not be as important as previously believed. At least, the cell number of Clostridia is not a major factor that influences the aging of APM.
Although aging is a complex process, it is believed that environmental factors are the main determinants that influence the shift of the prokaryotic community and structure in the soil and pit mud (Yao et al. 2010; Hu et al. 2016). This study revealed that the pH and the content of NH4 +, two of the most important environmental factors influencing the prokaryotic community structure, were lower in aging APM. This is easy to understand as lactic acid accumulation results in the decline of pH as reported (Belenguer et al. 2007; Sun et al. 2010). Low pH reduces prokaryotic diversity and changes the community composition (Hartman et al. 2008; Wang et al. 2012). Low NH4 + reduces the prokaryotic community, perhaps because of lowering pH and/or lowering available nitrogen (Tao et al. 2014). The same correlations are revealed in this study in APM. Meanwhile, nutrients, such as AP and humus, are more abundant in aging low-quality APM. They may promote the growth of specific microbes, as bacterial diversity and composition were primarily driven by the total C/N/P ratios in soil (Delgado-Baquerizo et al. 2016).
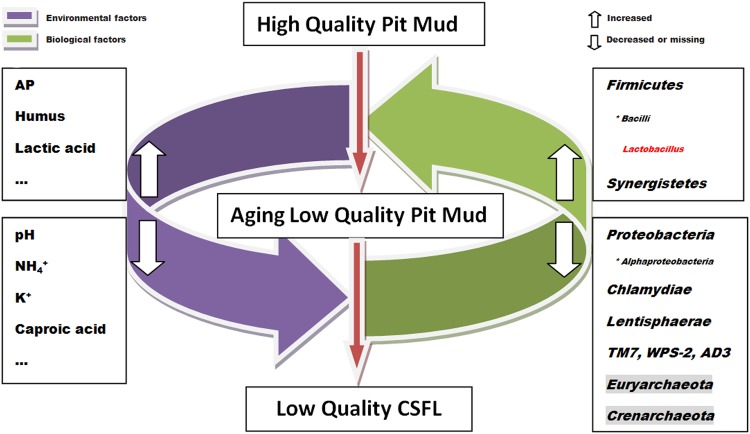
Finally, a possible mechanism of APM aging based on this study was proposed (Fig. 5). First, there are some unqualified reasons, such as changes in temperature, moisture, nutrients accumulation of metabolites, contamination, as well as degeneration, for APM to age during fermentation (Cai et al. 2009). Then, environmental factors are changed in aging APM. These include the increase of AP, humus, and lactic acid, and the decrease in pH, NH4 +, K+, and caproic acid. Subsequently or even simultaneously, biological factors are altered. On the phylum level, the abundance of Firmicutes and Synergistetes is increased. At the same time, more phyla decrease in number or are even missing in aging APM, including two Achaea phyla (labeled by shading). The decrease of pH corresponded to the increase of lactic acid and the high abundance of Lactobacilli, which produce lactic acid as the major end product during fermentation. On the other hand, Lactobacilli increased in response to the increase of lactic acid, but with the decrease of pH and caproic acid. The interaction of these two kinds of factors drives the aging process of pit mud to a severe degree until it becomes completely non-fermentative. Further research is needed to verify the main factors that cause aging and to elucidate how this information can be applied to control the aging process of APM during CSFL production.
Fig. 5.
A possible mechanism of pit mud aging. Achaea phyla are shaded; the most significantly increased genus is shown in red
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The samples were kindly supplied by the chief engineer Xuesi Li from Henan Songhe Liquor Corporation. This study was partially supported by a grant from Zhoukou Normal University (No. ZKNU2015107) to Z. Sun. C. Chen (No. 152300410229) and F. Tian (No. 172102310626) received grants from the Agency of Science and Technology of Henan Province. All founders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with ethical standards
Conflict of interest
There is no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0978-0) contains supplementary material, which is available to authorized users.
References
- Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol. 2007;73:6526–6533. doi: 10.1128/AEM.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Liu G, Qi Y, Li F. The degradation cause of man-made aged pit-mud in Northern China & the related prevention measures. Liquor Mak Sci Technol. 2009;180(6):67–69. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Baquerizo M, Reich PB, Khachane AN, Campbell CD, Thomas N, Freitag TE, Abu Al-Soud W, Sørensen S, Bardgett RD, Singh BK. It is elemental: soil nutrient stoichiometry drives bacterial diversity. Environ Microbiol. 2016;19(3):1176–1188. doi: 10.1111/1462-2920.13642. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Ferrenberg S, O’Neill SP, Knelman JE, Todd B, Duggan S, Bradley D, Robinson T, Schmidt SK, Townsend AR, Williams MW, Cleveland CC, Melbourne BA, Jiang L, Nemergut DR. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 2013;7:1102–1111. doi: 10.1038/ismej.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):1–9. [Google Scholar]
- Hartman WH, Richardson CJ, Vilgalys R, Bruland GL. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc Natl Acad Sci USA. 2008;105:17842–17847. doi: 10.1073/pnas.0808254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XL, Du H, Xu Y. Identification and quantification of the caproic acid-producing bacterium Clostridium kluyveri in the fermentation of pit-mud used for Chinese strong-aroma type liquor production. Int J Food Microbiol. 2015;214:116–122. doi: 10.1016/j.ijfoodmicro.2015.07.032. [DOI] [PubMed] [Google Scholar]
- Hu X, Du H, Ren C, Xu Y. Illuminating anaerobic microbial community and cooccurrence Patterns across a quality gradient in Chinese liquor fermentation pit-muds. Appl Environ Microbiol. 2016;82(8):2506–2515. doi: 10.1128/AEM.03409-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X, Tang Y, Ren D, Yao W. Development and present research status of manmade pit-mud. Liquor Mak Sci Technol. 2010;9(195):77–80. [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lu J, Li X, Ren T, Cong R, Zhou L. Dynamics of potassium release and adsorption on rice straw residue. PLoS One. 2014;9(2):e90440. doi: 10.1371/journal.pone.0090440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rui J, Mao Y, Yannarell A, Mackie R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol Biochem. 2014;68:392–401. doi: 10.1016/j.soilbio.2013.10.017. [DOI] [Google Scholar]
- Liang H, Li W, Luo Q, Liu C, Wu Z, Zhang W. Analysis of the bacterial community in aged and aging pit-mud of Chinese Luzhou-flavour liquor by combined PCR-DGGE and quantitative PCR assay. J Sci Food Agric. 2015;95(13):2729–2735. doi: 10.1002/jsfa.7013. [DOI] [PubMed] [Google Scholar]
- Liu X, Meng W, Liang G, Li K, Xu W, Huang L, Yan J. Available phosphorus in forest soil increases with soil nitrogen but not total phosphorus: evidence from subtropical forests and a pot experiment. PLoS One. 2014;9(2):e88070. doi: 10.1371/journal.pone.0088070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlich A. Photometric determination of humic matter in soils, a proposed method. Commun Soil Sci Plant Anal. 1984;15:1417–1422. doi: 10.1080/00103628409367569. [DOI] [Google Scholar]
- Mulvaney RL. Nitrogeninorganic forms. In: Sparks DL, editor. Methods of soil analysis. Madison: SSSA and ASA Inc; 1996. pp. 1123–11184. [Google Scholar]
- Song S. 2015 annual working report of Baijiu Branch, Market Committee, Famous Baijiu Collection Committee, and Design & Equipment Committee of China Alcoholic Drinks Association. Liquor Mak Sci Technol. 2016;6(264):17–28. [Google Scholar]
- Sun YZ, Mao SY, Zhu WY. Rumen chemical and bacterial changes during stepwise adaptation to a high-concentrate diet in goats. Animal. 2010;4:210–217. doi: 10.1017/S175173110999111X. [DOI] [PubMed] [Google Scholar]
- Tao Y, Li J, Rui J, Xu Z, Zhou Y, Hu X, Wang X, Liu M, Li D, Li X. Prokaryotic communities in pit-mud from different-aged cellars used for the production of Chinese strong-flavored liquor. Appl Environ Microbiol. 2014;80:2254–2260. doi: 10.1128/AEM.04070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu M, Xia Y, Wen X, Ding K. Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl Environ Microbiol. 2012;78:7042–7047. doi: 10.1128/AEM.01617-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhang W, Wang H, Liu C. Analysis of bacterial phylogenetic diversity of pit-muds with different cellar ages. Food Sci. 2013;34:177–181. [Google Scholar]
- Xu Y, Wang D, Fan W, Mu X, Chen J. Traditional Chinese biotechnology. Adv Biochem Eng Biotechnol. 2010;122:189–233. doi: 10.1007/10_2008_36. [DOI] [PubMed] [Google Scholar]
- Yao W, Chen M, Zhen D, Guo Y. Isolation of lactate-producing microbes from fermented grains of Luzhou-flavor liquor and their effect on simulative solid-state fermentation. Liquor Mak. 2010;37:37–41. [Google Scholar]
- Yue Y, Zhang W, Liu X, Hu C, Zhang S. Isolation and identification of facultative anaerobes in the pit-mud of Chinese Luzhou-flavor liquor. Microbiol Chin. 2007;34:251–255. [Google Scholar]
- Zhang X. Research on the rejuvenation of aged cellar mud. Liquor Mak. 2010;37:53–54. [Google Scholar]
- Zhang X, Wu Z, Zhang S, Cheng H, Xue Z. Phylogenetic analysis of cultured bacteria in Luzhou-flavor liquor pits in Guizhou. Liquor Mak Sci Technol. 2010;198:23–27. [Google Scholar]
- Zhang J, Song R, Cao J, Xu G, Sun H, Tu X, Fang S. Preliminary analysis of the pit-mud crystal in artificial old cellar. China Brew. 2014;33(3):21–23. [Google Scholar]
- Zhang Y, Cong J, Lu H, Yang C, Yang Y, Zhou J, Li D. An integrated study to analyze soil microbial community structure and metabolic potential in two forest types. PLoS One. 2014;9:e93773. doi: 10.1371/journal.pone.0093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou R, Niu M, Zheng J, Wu C. Difference of microbial community stressed in artificial pit-muds for Luzhou-flavour liquor brewing revealed by multiphase culture-independent technology. J Appl Microbiol. 2015;119(5):1345–1356. doi: 10.1111/jam.12943. [DOI] [PubMed] [Google Scholar]
- Zhao S, Yang C, Xu M, Dou S, Liao Y. A review of research progress about Luzhou-flavor liquor making microbes in liquor production. Food Ferment Technol. 2012;1:24–29. [Google Scholar]
- Zheng J, Liang R, Zhang L, Wu C, Zhou R, Liao X. Characterization of microbial communities in strong aromatic liquor fermentation pit-muds of different ages assessed by combined DGGE and PLFA analyses. Food Res Int. 2013;54:660–666. doi: 10.1016/j.foodres.2013.07.058. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.