Abstract
Understanding how closely related wildlife species and their domesticated counterparts exchange or share parasites, or replace each other in parasite life cycles, is of great interest to veterinary and human public health, and wildlife ecology. Grey wolves (Canis lupus) host and spread endoparasites that can either directly infect canid conspecifics or their prey serving as intermediate hosts of indirectly transmitted species. The wolf recolonization of Central Europe represents an opportunity to study parasite transmission dynamics between wildlife and domestic species for cases when a definitive host returns after local extinction – a situation equivalent to a ‘removal experiment’.
Here we investigate whether the re–appearance of wolves has increased parasite pressure on hunting dogs – a group of companion animals of particular interest as they have a similar diet to wolves and flush wolf habitats when hunting. We compared prevalence (P) and species richness (SR) of helminths and the protozoan Sarcocystis to determine whether they were higher in hunting dogs from wolf areas (ndogs = 49) than a control area (ndogs = 29) without wolves. Of particular interest were S. grueneri and S. taeniata, known as ‘wolf specialists’.
Five helminth and 11 Sarcocystis species were identified, of which all helminths and eight Sarcocystis species were shared between dogs and wolves. Overall prevalence and species richness of helminths (P:38.5% vs. 24.1%; SRmean:0.4 vs. 0.3 species) and Sarcocystis (P:63.3% vs. 65.5%, SRmean:2.1 vs. 1.8 species) did not differ between study sites. However, hunting dogs were significantly more likely to be infected with S. grueneri in wolf areas (P:45.2% vs. 10.5%; p = 0.035). The findings suggest that wolves indirectly increase S. grueneri infection risk for hunting dogs since cervids are intermediate hosts and occasionally fed to dogs. Furthermore, a periodic anthelminthic treatment of hunting dogs may be an effective measure to control helminth infections regardless of wolf presence.
Keywords: Canis lupus, Epidemiology, Helminths, Hunting dogs, Metabarcoding, NGS, Protozoa, Sarcocystis
Graphical abstract
Highlights
-
•
General parasite burden in hunting dogs is not increased by wolves.
-
•
General Sarcocystis burden in hunting dogs is high due to raw feeding.
-
•
‘Wolf specialist’ parasite S. grueneri more prevalent in hunting dogs from wolf areas.
1. Introduction
Many pathogens circulate in multi–host systems and do not depend on one single host species. Understanding the epidemiology of multi–host pathogens is critical to the ‘One Health’ concept as wildlife, domesticated animals and humans may be affected by such pathogens and share and exchange them (Taylor et al., 2001, Aguirre et al., 2002, Haydon et al., 2002, Thompson, 2013). Species or populations that maintain a pathogen and are responsible for its spill–over to a target species of interest are generally defined as “reservoirs” (Haydon et al., 2002, Hatcher and Dunn, 2011). In the context of conserving endangered species (van Kesteren et al., 2015, Millan et al., 2016), and recolonization or reintroduction projects (Almberg et al., 2012), the identification of pathogen reservoirs plays an important role for their success. Although spill–over to wildlife species and its effect on endangered or reintroduced species have received increasing attention, the influence of wildlife on closely related domesticated species has rarely been investigated (Thompson, 2013).
The return of an apex predator such as the grey wolf (Canis lupus) to a human–dominated landscape, from which it was absent for a century, is the equivalent of an (unintended) ‘removal experiment’. Such an event provides an excellent opportunity to study how its close relative, the domestic dog, may be affected by the resurrection of parasite cycles for which returning wolves are definitive hosts. Currently, there 47 recognized wolf packs and 15 scent marking pairs in Germany that belong to the Western part of the Central European lowland wolf population (http://www.wolf-sachsen.de/de/verbreitung-in-deutschland), (Supplementary Fig. 1). According to the German Hunting Association (DJV, 2017b; https://www.jaegermagazin.de/jaeger-praxis/jagdschule/die-jagd-2016-in-zahlen) and the University of Göttingen (Ohr and Zeddies, 2006), the number of dogs in Germany ranges between 4.8 and 5.3 million, of which more than 300,000 are owned by hunters.
Given their similar biology and close relatedness, mesopredators like red foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) and the wolf as an apex predator are known to share several helminth species with domestic dogs (Al-Sabi et al., 2013b, Otranto et al., 2015a). All of these carnivores have been recognized as hosts of the protozoan Sarcocystis (Barutzki and Schaper, 2011, Stronen et al., 2011, Prakas et al., 2015, Moré et al., 2016). However, it is unclear at present to what extent one canid may act as ‘substitute’ host for the other and how close their relationship as ‘joint’ definitive hosts of Sarcocystis is (Otranto et al., 2015b). This lack of information is very likely caused by methodological challenges, as there are no morphological techniques to discriminate Sarcocystis sporocysts or oocysts shed by definitive hosts (Xiang et al., 2009). With current molecular genetic tools such as metabarcoding, species detection from canid faecal samples has become possible, and recently wolves have been described as hosts for 12 Sarcocystis species (Lesniak et al., 2017). Furthermore, epidemiological studies of wolves and their ungulate prey species demonstrated that wolf presence increased the prevalence of sarcocysts in their ungulate prey. Accordingly, the Sarcocystis species S. grueneri and S. taeniata, that were identified as well–adapted to wolves and therefore termed ‘wolf specialists’, were mostly responsible for this increase (Lesniak et al., under review).
In general, hunting activities have been identified as a risk factor altering parasite infection risk, for instance, by the protozoan Sarcocystis (Thompson, 2013). In this context, hunting dogs – domestic dogs trained for hunting ungulates and other game – are of interest for several reasons. They can be considered the most likely source of pathogens or parasites that could be transmitted to wolves, but at the same time are potentially at risk of being exposed to wolf–derived parasites themselves. Transmission could occur when hunting dogs are used for hunting in wolf habitats or when fed with game meat by their owners (ESCCAP, 2010, Otranto et al., 2015b), which usually originates from the same ungulate species that wolves prey on (Wagner et al., 2012). While literature on companion dog parasites is regularly published (Barutzki and Schaper, 2003, Barutzki and Schaper, 2011), little is known about the parasite fauna of hunting dogs, and it is unlikely that they are identical (Al-Sabi et al., 2013a, Gómez-Morales et al., 2016). The current wolf recolonization of Central Europe is therefore an ideal system to investigate the potential link between a wild apex predator and its domesticated equivalent, since hunting dogs can be examined in the presence and absence of wolves in comparable habitats. However, in such ‘field experiments’ several (unknown) factors that potentially influence parasite development, survival and transmission cannot be controlled for. This includes, e.g. the microclimate that might affect the survival of parasite stages in the environment (Randolph and Storey, 1999). Another relevant but uncontrollable factor is the anthelminthic treatment of dogs. Depending on a product's target site, it may selectively clear trematodes, cestodes, nematodes or all helminths, but anthelminthics have no effect on protozoa (Martin et al., 1997). Even though the European Scientific Counsel for Companion Animal Parasites recommends a monthly anthelminthic treatment for pets belonging to risk groups such as hunting dogs, the dewormification routine of dogs falls to their owners and may therefore be inconsistent and strongly differ among individuals (ESCCAP, 2010).
We hypothesized that wolves transmit endoparasites to hunting dogs. Such transmission might either occur directly from wolves via the environment to hunting dogs (no intermediate host required) or indirectly via intermediate hosts. There are several reasons why transmission effects might differ among parasites. In contrast to protozoan Sarcocystis species, transmission of helminth species might be strongly reduced or even absent because dogs usually undergo anthelminthic treatments. In addition, transmission might strongly differ among Sarcocystis species and might be particularly strong for species recognized as wolf specialists (Lesniak et al., under review) because of the similar biology of dogs and wolves. Taken these considerations into account, we predicted that (1) the general prevalence and species richness of Sarcocystis would be higher in hunting dogs from areas affected by wolf recolonization compared to hunting dogs from the control site, and that (2) particularly Sarcocystis species recognized as wolf specialists should show a higher prevalence in hunting dogs from the wolf area. Finally, we also predicted that (3) helminth prevalence and species richness of hunting dogs from wolf areas will not be increased compared to the control area.
2. Material and methods
2.1. Sample collection
Between November 2012 and January 2015, we collected 359 faecal triplicate samples of 78 hunting dogs residing in areas occupied by wolves in the German federal states of Brandenburg and Saxony (50°10′–53°33′ N and 11°14′–15°2′ E; ndogs = 49, nsamples = 230). Hunting dogs were also sampled in a control area in the German federal state of Schleswig–Holstein (53°20′–54°55′ N and 8°36′–11°7′ E; ndogs = 29, nsamples = 129) where no territorial wolves were recognized during the sampling period (Supplementary Fig. 1). Detailed information about the exact collection dates and application of anthelminthics for triplicate samples making up the faecal pools per dog are provided in Supplementary Table S2. If dog owners applied anthelminthics within the intended quarterly sampling schedule, they were asked to collect the triplicate sample beginning with the first faecal dropping after drug application. We intended to avoid a bias towards false negative findings, if samples were collected too late after helminths had been cleared. Additionally, this sampling strategy avoided missing particular parasite taxa, if e.g. a product against only nematodes was applied because we would capture both the flushed species cleared by the drug and the non–affected species due to their ongoing shedding of eggs.
Dog age, breed, function for hunting, information on routine of anthelminthic treatments and feeding habits were supplied by their owners who voluntarily supported the study (Supplementary Table S2). Hence, diet and medical treatments were not controlled by the authors but rely on the voluntary information of participants.
2.2. DNA extraction
Dog faeces were collected on three consecutive days and pooled. DNA was extracted using the NucleoSpin® Soil Kit (Macherey–Nagel, Düren, Germany) according to the manufacturer's protocol. This kit has previously been successfully used for DNA isolation from nematode and cestode eggs from faecal samples (Demeler et al., 2013, Maksimov et al., 2017). For subsequent analyses, DNA from dog faecal pools (n = 359) was again pooled in equimolar ratios per individual (n = 78). Extraction success and DNA concentrations were determined using the NanoDrop® 1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA).
2.3. Metabarcoding
Portions of the helminth and apicomplexan 18S rRNA and cox1 genes were amplified using a set of five primer pairs (Table 1). We restricted the screening for helminths to two primer pairs that were previously used in non–copro–genetic studies (Bowles et al., 1992, Lesniak et al., 2017). As this is the first genetic study of Sarcocystis spp. in dogs, we applied a more redundant study design with three sets of primers to target Sarcocystis species. Each forward and reverse oligonucleotide was tagged with the Fluidigm–specific common sequence CS1 (5′–ACACTGACGACATGGTTCTACA–[TS–For]–3′) or CS2 (5′–TACGGTAGCAGAGACTTGGTCT–[TS–Rev]–3′) to enable subsequent barcoding of the generated PCR products (Fluidigm, San Francisco, California, USA). Target–specific PCRs had a total volume of 12.5 μL containing 1 μL DNA template and were run in 40 cycles in an epGradient S thermocycler (Eppendorf, Hamburg, Germany). The reactions contained 1 × FastStart High Fidelity Reaction Buffer without MgCl2, 0.2 mM dNTPs, 2.5 mM MgCl2, 0.2 μM each primer, 5% DMSO, 0.4 μg/μl BSA (only cox1 PCR) and 0.5 U FastStart High Fidelity Enzyme Blend (all components from Roche, Basel, Switzerland). PCR conditions included an initial denaturation step of 95 °C (10 min); 40 cycles of 95 °C (45 s), 53 °C (18S rRNA PCRs) or 55 °C (cox1 PCR) (45 s), 72 °C (60 s), and a final elongation of 72 °C (10 min). PCR products were purified using Agencourt® AMPure® XP beads (Beckman Coulter GmbH, Krefeld, Germany) in a 1:1 ratio to reduce adapter concatemerization during barcoding. Post–PCR quality control steps included amplicon quality and length check using the 2200 TapeStation Instrument with D1000 ScreenTapes and D1000 Reagents (Agilent Technologies, Santa Clara, California, USA). A 10–fold dilution of the purified amplicon pools was used for the subsequent barcoding PCR with the Access Array Barcode Library for Illumina Sequencers – 384 (Single Direction) (Fluidigm, San Francisco, California, USA) according to the manufacturer's protocol (Access Array™ System for Illumina Sequencing Systems, Chapter 6, pp. 70–72, Fluidigm, San Francisco, California, USA). Barcoded amplicons were pooled and target fragments between 400 and 800 bp were size–selected using the BluePippin® instrument (Sage Science, Inc., Beverly, Massachusetts, USA) with the 1.5% agarose gel cassettes, 250 bp–1.5 kb (Sage Science, Inc., Beverly, Massachusetts, USA). The purified libraries were sequenced on an Illumina MiSeq at the Berlin Center for Genomics in Biodiversity Research (BeGenDiv) using the MiSeq Reagent Kit v3 (600 cycles) (Illumina, Inc., San Diego, California, USA) and a spike–in of 50% PhiX.
Table 1.
Target–specific primers (F: forward direction, R: reverse direction) for the amplification of the variable regions of the helminth and Sarcocystis spp. cox1 and 18S rRNA genes.
| # | target | primer name | primer sequence 5′–3′ | potential target species | ∼product size |
|---|---|---|---|---|---|
| 1 | cox1 | JB3F | TTTTTTGGGCATCCTGAGGTTTATb |
Alaria alatad Echinococcus spp.b,d Mesocestoides litteratusd Taenia spp.d Toxascaris leoninad Toxocara canisd Uncinaria stenocephalad |
396 bp |
| JB4.5R | TAAAGAAAGAACATAATGAAAATGb | ||||
| 2 | 18S | 18S_965Fa | GGCGATCAGATACCGCCCTAGTTc |
Crenosoma vulpisd Capillaria spp.c Trichuris vulpisc |
606 bp |
| 18S_1573Ra | TACAAAGGGCAGGGACGTAATc | ||||
| 3 | 18S | proti15F | TGCCAGTAGTCATATGCTTGTYTd | Sarcocystis spp.d | 378 bp |
| proti440R | CAGGCYCSCTCTCCGGAd | ||||
| 4 | 18S | SarAF | CTGGTTGATCCTGCCAGTAGe | Sarcocystis spp.e | 530 bp |
| SarAR | TTCCCATCATTCCAATCACTe | ||||
| 5 | 18S | SarBF | GGGAGGTAGTGACAAGAAATAACAAe | Sarcocystis spp.e | 467 bp |
| SarBR | GGCAAATGCTTTCGCAGTAGe |
The 18S_965F/18S_1573R primer pair failed to amplify parasite DNA from faecal samples.
2.4. Bioinformatic analysis
As a first step, forward and reverse reads from the Illumina metabarcoding dataset were stratified per sample using bcl2fastq v2.17.1.14 (Illumina, Inc., San Diego, California, USA) with no mismatch tolerance allowed in the barcode. Fastq files were grouped into amplicon sets according to the respective primer used (forward and reverse reads were not merged), and were further processed with USEARCH, a sequence analysis tool that bundles sequence reads into clusters of similarity called ‘operational taxonomic units’ (OTUs) (Edgar, 2010, Edgar, 2013). The USEARCH algorithm performs several quality filtering steps, including a simultaneous chimera filtering. Sequences were trimmed using the USEARCH ‘fastq_filter’ command with the parameter fastq_maxee set to two nucleotides per read, and the amplicon–specific optimal trim length (fastq_trunclen). Singleton reads were discarded with the USEARCH ‘dedup_fulllength’ command, applying a minsize parameter of two reads. Remaining sequences were clustered using the USEARCH ‘cluster_otus’ command, setting the parameter minsize to two reads per cluster and allowing one nucleotide substitution as value for the parameter otu_radius_pct. OTUs were assigned with a similarity of 99% (parameter id set to 0.99) by applying the USEARCH ‘usearch_global’ command.
A custom database to identify Sarcocystis species was constructed from a set of 586,784 sequences for Apicomplexa (Taxonomy ID: 5794) extracted from the NCBI (National Center for Biotechnology Information) database using their taxonomy browser on 18 Oct 2016 (Sayers et al., 2009). Likewise, a custom database to identify helminth species was constructed from a set of 2,948,076 sequences for the taxon of plathelminthes and 1,959,651 sequences for the taxon of nematodes. Identified OTUs were aligned to the custom databases using BLAST® (blastn, Altschul et al., 1990) with an identity threshold of 98%. To assign OTUs to parasite species, only hits with an unambiguous best bit score for one species were collected in a table including the respective parasite species, NCBI accession number, OTU, amplicon, and sample name (Suppl. Tables S3 and S4).
2.5. Statistical analyses
Statistical analyses were performed in R version 3.2.1 (R–Development–Core–Team, 2008). To test our predictions, we used five different generalized linear models (GLM). While each model contained a different response variable, all models included the same predictor variables: ‘wolf presence’ (binary: present, absent), ‘dog age’ (0–14 years) and ‘sampling effort’ (1–11 samples). To test whether the general prevalence of Sarcocystis is higher in wolf areas (prediction 1) we used a binomial model with a binary response variable ‘Sarcocystis spp. infection’ (present vs. absent). To test whether Sarcocystis species richness is higher in wolf areas (prediction 1) we used a Poisson model with ‘Sarcocystis species richness’ (the number of species) as a response. To test prediction 2 that particular Sarcocystis species recognized as wolf specialists (Lesniak et al., under review) are more prevalent in hunting dogs from wolf areas, we used two binomial models with the binary response variables: (1) ‘S. grueneri infection’ and (2) ‘S. taeniata infection’ (present vs. absent). To test whether the prevalence of helminths is different between wolf and control areas (prediction 3), we used a binomial model with the binary response variable ‘helminth infection’ (present vs. absent). Finally, to test whether helminth species richness is different between wolf and control areas (prediction 3), we used a Poisson model with ‘helminth species richness’ (the number of species) as a response.
3. Results
3.1. Infection with Sarcocystis spp.
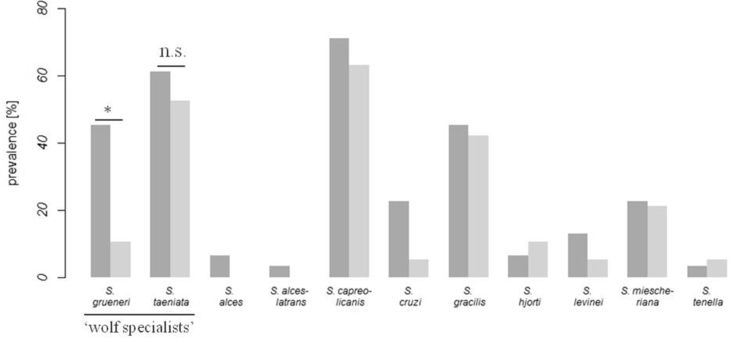
Overall Sarcocystis spp. prevalence was 63.3% in hunting dogs from wolf areas and 65.5% in hunting dogs from control areas. In the dataset, 109 operational taxonomic units (OTUs) were detected (Table 2) which were aligned to 11 Sarcocystis species (Fig. 1). In 22.5% (wolf area) and 37.9% (control area) of all hunting dogs, OTUs were assigned to unknown Sarcocystis sp.
Table 2.
Number of detected operational taxonomic units (OTUs) per Sarcoystis species based on 18S rRNA sequences from six different amplicons analysed with USEARCH.
| species assignment | number of OTUs |
|---|---|
| S. alces | 2 |
| S. alceslatrans | 1 |
| S. capreolicanis | 22 |
| S. cruzi | 6 |
| S. gacilis | 6 |
| S. grueneri | 3 |
| S. hjorti | 4 |
| S. levinei | 2 |
| S. miescheriana | 5 |
| Sarcocystis sp. | 9 |
| S. taeniata | 57 |
| S. tenella | 1 |
Fig. 1.
Normalized Sarcocystis spp. prevalence in hunting dogs from the wolf area (dark grey, n = 49) and control area without wolves (light grey, n = 29). Hunting dogs were infected with 11 distinct Sarcocystis species, of which two species only occurred in wolf inhabited areas. They were significantly more likely to be infected with the ‘wolf–specialized’ parasite S. grueneri when sharing their habitat with wolves (p = 0.035). It was not possible to determine a correlation for an infection with the other ‘wolf specialist’ S. taeniata and wolf presence (n.s. = not significant, p = 0.476). P values were extracted from GLMs.
For general Sarcocystis prevalence (GLM, overall log likelihood ratio test, χ2 = 14.280, df = 4, n = 78, p = 0.003) no significant difference was detected between study sites (p = 0.556), although prevalence significantly increased with sampling effort (p = 0.004) and significantly decreased with dog age (p = 0.014). Similarly, for Sarcocystis species richness (GLM, overall log likelihood ratio test, χ2 = 17.965, df = 4, n = 78, p < 0.001) no significant difference was detected between study sites (p = 0.380), although species richness significantly increased with increasing number of analysed samples (p < 0.001) and significantly decreased with dog age (p = 0.013). In contrast, an infection with S. grueneri (GLM, overall log likelihood ratio test, χ2 = 8.088, df = 4, n = 78, p = 0.044, Fig. 1) was significantly more likely to occur in hunting dogs sharing their habitat with wolves than in hunting dogs from control areas (p = 0.035), while sampling effort (p = 0.779) and dog age (p = 0.157) had no significant effect. For infections with S. taeniata no significant effect for any of the predictors was detected (GLM, overall log likelihood ratio test, χ2 = 2.500, df = 4, n = 78, p = 0.476).
3.2. Infection with helminths
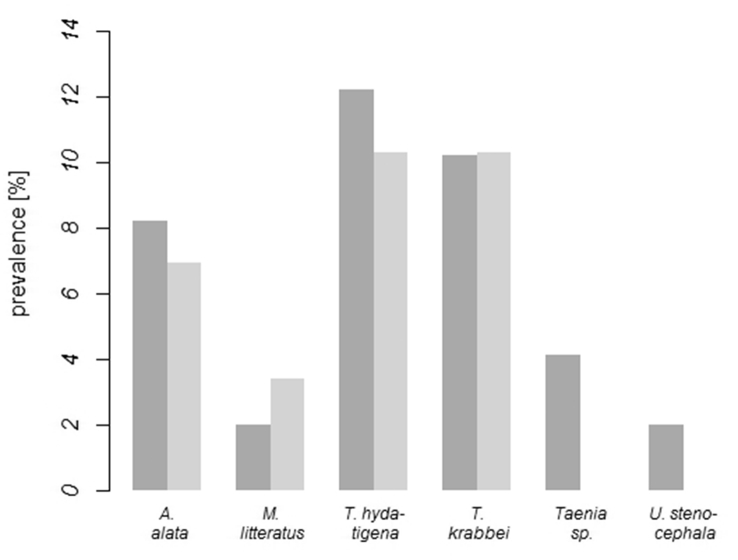
Due to the bioinformatic restrictions we set regarding the taxonomic resolution, the nematode–specific 18S_965F and 18S_1573R primer pair did not produce sequence hits against parasites. All respective reads were discarded from the dataset and further analysis. Based on cox1–amplicons, overall helminth prevalence was 38.5% in hunting dogs from the wolf inhabited area and 24.1% in hunting dogs from the control area. In the dataset, 416 OTUs (Table 3) were detected which were aligned to one known trematode, three cestode and one nematode species (Fig. 2). Sequences were assigned to Taenia sp. in 4.1% of hunting dogs from the wolf area.
Table 3.
Number of detected operational taxonomic units (OTUs) per helminth species based on cox1 sequences from two different amplicons analysed with USEARCH.
| species assignment | number of OTUs |
|---|---|
| Alaria alata | 4 |
| Mesocestoides litteratus | 26 |
| Taenia hydatigena | 76 |
| T. krabbei | 308 |
| Taenia sp. | 1 |
| Uncinaria stenocephala | 2 |
Fig. 2.
Normalized helminth prevalence in hunting dogs from the wolf area (dark grey, n = 49) and control area without wolves (light grey, n = 29). Lack of statistical significance was determined using a GLM.
No significant effect for any of the predictors was detected for overall helminth infection risk of hunting dogs (GLM, overall log likelihood ratio test, χ2 = 1.675, df = 4, n = 78, p = 0.643) or helminth species richness (GLM, overall log likelihood ratio test, χ2 = 2.129, df = 4, n = 78, p = 0.546).
4. Discussion
The recent recolonization of Central Europe by wolves has triggered several scientific studies on their ecology (Ansorge et al., 2010, Nowak et al., 2011, Wagner et al., 2012, Andersen et al., 2015, Reinhardt et al., 2015, Nowak and Mysłajek, 2016, Szentiks et al., 2016). Investigating parasite infections in wildlife has a long history that mainly focused on helminth occurrence (Craig and Craig, 2005). Recently, metabarcoding techniques have enabled the high–throughput species identification of protozoan parasites such as Sarcocystis in ungulates (Lesniak et al., under review) and wolves (Lesniak et al., 2017), representing the intermediate and definitive hosts, respectively. By applying these techniques, we explored the helminth and Sarcocystis fauna of hunting dogs in the context of the ongoing wolf recolonization. We showed that overall prevalence and species richness of Sarcocystis and helminths in hunting dogs did not significantly differ between animals from wolf and control areas. Even though we could not control for unknown variables that may differ between the two study sites, this result indicates that wolves have a minor epidemiological influence on their domesticated conspecifics regarding general parasite burden. The only statistically significant effect detected was an increased prevalence of the wolf specialist parasite S. grueneri in hunting dogs from areas affected by wolf recolonization compared to hunting dogs from the control area. This finding underlines the impact of wolves regarding the spread of multi–host pathogens in a predator–prey system, in which prey species are intermediate hosts that serve as source of infection for alternative definitive host such as dogs.
4.1. Sarcocystis infection of hunting dogs and impact of wolves
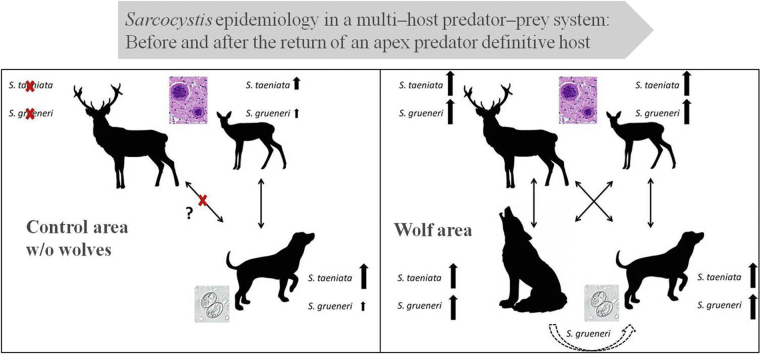
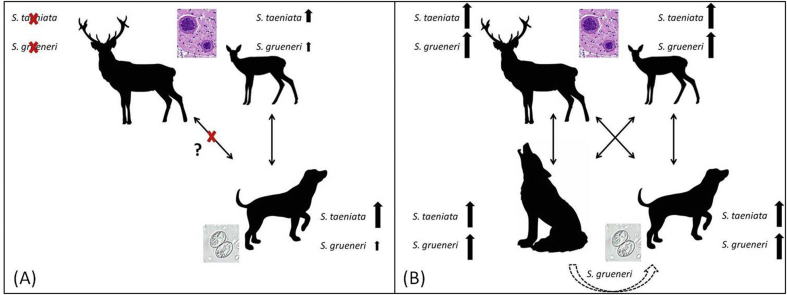
We identified 11 known Sarcocystis species in hunting dogs. Wolves have been previously shown to host as well 11 known Sarcocystis species (Lesniak et al., 2017) of which eight were shared with hunting dogs in this study. These species included the two wolf specialists S. grueneri and S. taeniata that are known to occur in German roe deer (Capreolus capreolus) and red deer (Cervus elaphus) (Lesniak et al., under review), and hence serve as the most likely source of infection for hunting dogs. In contrast to our first prediction, overall Sarcocystis prevalence in hunting dogs did not significantly differ between wolf (63.3%) and control areas (65.5%). However, it is considerably lower than the overall Sarcocystis prevalence of 95% in wolves from Germany (Lesniak et al., 2017), and much higher than previously reported prevalence between 2% and 9% in companion dogs from Germany (Barutzki and Schaper, 2003, Barutzki and Schaper, 2011). A likely explanation for the lack of a wolf–associated increase in overall Sarcocystis prevalence is that in another study (1) a substantial increase was only documented in red deer, whereas in other ungulates the increase was only slight and limited to a non–significant trend (Lesniak et al., under review), and that (2) this overall increase in Sarcocystis prevalence was mainly driven by the two wolf specialist species (Lesniak et al., under review). The current study demonstrates that hunting dog infection with S. grueneri – but not with S. taeniata – was more likely to occur when wolves were present. Consistent with the above mentioned study, wolves most likely increased the prevalence of S. grueneri in their ungulate prey species, thereby being indirectly responsible for an increase in prevalence in hunting dogs because Sarcocystis cannot be directly transmitted between canids but require an intermediate host (Fig. 3B). Hunting dogs had a lower prevalence of S. grueneri in control areas where wolves were absent. Until now, no other wild canids have been described as definitive hosts of S. grueneri and S. taeniata, which were not detected in genetic studies in red foxes and raccoon dogs (Prakas et al., 2015, Moré et al., 2016).
Fig. 3.
S. grueneri and S. taeniata developmental cycles with their intermediate and definitive host in areas without wolves (A) and with wolves (B). In wolf habitats, wolves increase S. grueneri prevalence in their prey, in turn leading to a higher infection rate in hunting dogs. S. grueneri and S. taeniata strains spread by wolves are well–adapted to both ungulate species, while S. grueneri and S. taeniata strains spread by hunting dogs from the control area are restricted to roe deer (right ungulate pictogram). The epidemiological influence of wolves regarding the spread of Sarcocystis in comparison to hunting dogs has a higher impact on red deer (left ungulate pictogram) than on roe deer. Sarcocystis strains in hunting dogs from the wolf area are likely to be a mixture of both dog and wolf strains. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Here, as elsewhere, hunting dogs and mesopredators are the most likely source of infection for grazing ungulates. A study of ungulates from the control area suggested that both S. grueneri and S. taeniata occur in roe deer but not in red deer (Lesniak et al., under review). In order to explain these findings we propose the following conceptual model of Sarcocystis transmission dynamics: parasite strains of species that are spread by hunting dogs in control areas are only well–adapted to roe deer but not to red deer (Fig. 3A), whereas strains spread by wolves are well–adapted to both cervids (Fig. 3B). Investigation of other definitive hosts, including studies using more variable genome sequences in order to identify parasite strains, would be required to either increase confidence in this model or reject it. Additionally, further (unknown) factors – other than wolf absence or presence and dog age or sampling effort for which we had controlled in the respective linear models – may potentially alter the parasite infection risk for hunting dogs and may vary between the two different study regions. Such limitations cannot be ruled out in field studies like this, and may therefore limit the conclusions that can be drawn from the results. For example, microclimate (Randolph and Storey, 1999) or habitat structure, including husbandry types and (wildlife) host density and distribution (Sousa and Grosholz, 1991) may influence survival and transmission of infectious parasite stages in the environment but have not been considered in this study. Nevertheless, our findings indicate that the presence of a returning definitive host, the grey wolf, seems to represent a low parasite infection risk for hunting dogs and is currently limited to one particular protozoan parasite.
Dog owners reported that their dogs receive remains of game such as roe deer, red deer, wild boar and moufflon as well as of farm animals such as cattle, horse and sheep (see Supplementary Table S1). According to a Canadian study, the feeding of commercial meat according to the Biologically Appropriate Raw Food (BARF) method has become an increasingly popular trend among pet owners (Schlesinger and Joffe, 2011). Both raw–feeding diets are potential sources of the high Sarcocystis prevalence and diversity detected in hunting dogs and may explain the lack of a systematic link between wolf presence and overall Sarcocystis prevalence or species richness in hunting dogs.
4.2. Hunting dogs as potential definitive host of Sarcocystis
Apart from the above mentioned ‘wolf specialists’ S. grueneri and S. taeniata, nine other known Sarcocystis species and Sarcocystis parasites of uncharacterized species (Sarcocystis sp.) were detected in the faecal samples of the dogs. In general, the detection of (rare) parasite species in this study could be an artefact of technical limitations such as sequencing errors that cannot be totally excluded despite the stringent quality filtering (Edgar, 2010, Edgar, 2013) and might lead to a potential misidentification of species. This could be the case for species like S. alces and S. alceslatrans that were detected, even though their life cycles were not expected to be present in the study areas as moose (Alces alces) that function as intermediate hosts are absent. However, if these sequences were correctly identified, an explanation for their presence in hunting dog faeces could be that hunting dogs, unlike wolves, have access to ungulates rarely seen in Germany such as moose (Dahlgren and Gjerde, 2008, Gjerde, 2014), most likely because of potential hunting tourism of their owners. In this case, this was the first description of domestic dogs being a suitable definitive host for S. alces, previously only known from red foxes and arctic foxes (Vulpes lagopus) (Dahlgren and Gjerde, 2010). S. alceslatrans has been experimentally shown to reproduce in dogs and coyotes (Canis latrans, Dubey, 1980). However, since moose are rare in Germany (Niedzialkowska et al., 2014) the import of these parasite species by domestic dogs can be considered a minor threat to wolves because their developmental cycles cannot be completed without the appropriate intermediate host(s).
The two Sarcocystis spp. known from domesticated ungulate intermediate hosts, S. cruzi from cattle (Bos taurus) and S. tenella from sheep (Ovis spp.), are known to reproduce in domestic dogs (Dubey, 1976, Erber, 1982). Both parasites were also identified in our sample of hunting dogs, indicating that, besides game, hunting dogs also have access to meat of domesticated ungulates. Here, we demonstrate for the first time that S. levinei, native to North Africa (El-Dakhly et al., 2011) and Asia (Huong, 1999, Claveria and Cruz, 2000), also circulates in a domestic cycle in Germany. This species uses water buffaloes (Bubalus spp.) as intermediate hosts and can reproduce in domestic dogs as shown by experimental infection (Ghosal et al., 1987). Previously, there were no reports of S. levinei from Europe. We assume that this Sarcocystis species was either imported via infected domestic dogs that were adopted from the original range countries, or by feeding infected meat to resident domestic dogs. Due to an increasing popularity of water buffaloes for pasturing purposes and meat production, S. levinei is able to circulate in Germany (Braun and Preuss, 2008).
To the best of our knowledge, this is the first report of domestic dogs being a possible definitive host for the six Sarcocystis species using wild ungulate intermediate hosts –S. capreolicanis, S. gracilis, S. grueneri, S. hjorti, S. miescheriana, and S. taeniata. Domestic dogs share all of these species with wolves (Lesniak et al., 2017) but only S. capreolicanis, S. gracilis and S. miescheriana with red foxes and raccoon dogs (Moré et al., 2016), and S. hjorti with red and arctic foxes, respectively (Dahlgren and Gjerde, 2010).
Some faecal samples analyzed in this study also contained Sarcocystis parasites for which no species has been described to date and that previous authors had deposited as Sarcocystis sp. under six different entries in GenBank (for more details see Suppl. Table S3). In two separate cases, single dogs were infected with Sarcocystis parasites that shared highest sequence similarity with an isolate from the diaphragm of a red deer from Lithuania (JN256125) and an Australian feral goat (Capra hircus) from Malaysia (KR155226). In the case of the four other GenBank entries for which several hunting dogs were positive, raccoon dogs (KT873744: 10 positive hunting dogs) and red foxes (KT873770: four positive hunting dogs; KT873771: two positive hunting dogs; KT873774: nine positive hunting dogs) from Germany had been previously described as definitive hosts (Moré et al., 2016). The two hits against the isolates from Lithuania and Malaysia were rather questionable due to their geographical origin and host species, respectively. However, the sequence hits against Sarcocystis sp. isolated from wild mesopredators from Germany are more likely to be real signals instead of artefacts.
4.3. Helminth fauna of hunting dogs and impact of wolves
Overall helminth prevalence of hunting dogs was similar between wolf (38.5%) and control areas (24.1%), and considerably lower than in wolves (89%, Lesniak et al., 2017). Even though our study potentially missed some nematode taxa such as Toxocara or Toxascaris due the bioinformatic filtering, it is unlikely that the detection of these species would considerably increase the overall prevalence. Based on the described methods, we were not able to detect any link between wolf presence and helminth infection risk or helminth species richness in hunting dogs. This is consistent with our prediction that dog anthelminthic treatments prevent an increase of helminth burden, even if wolf areas might be more contaminated with helminths than control areas. This finding is also consistent with the lack of a link between indirectly transmitted metacestodes (Taenia spp.) and wolf presence as shown by a previous study (Lesniak et al., 2017).
A second explanation for similar helminth prevalences in both study sites is that hunting dogs represent equally good hosts for the detected helminths and that during the absence of wolves hunting dogs may have replaced wolves as definitive host, keeping those parasite life cycles alive, even though their ‘main’ host went locally extinct.
Thirdly, red foxes and raccoon dogs are as well known as definitive hosts of – amongst others – A. alata, Mesocestoides sp., Taenia spp. and U. stenocephala (Loos-Frank and Zeyhle, 1982, Al-Sabi et al., 2013b). In contrast to wolves, foxes have never been extinct in Central Europe and the neozoan raccoon dog has been present since the 1960s (Stubbe, 1977). According to the hunting bag published by the German Hunting Association almost 470.000 foxes and 28.000 raccoon dogs were legally killed in 2015/16 (DJV, 2017a), indicating that both mesopredators occur in higher numbers than wolves. Assuming an average pack size of 7.5 individuals (http://www.wolf-sachsen.de/de/leben-im-rudel) the number of wolves currently living in Germany can roughly be estimated between 350 and 400 individuals.
Out of five helminth species detected, four have an indirect life cycle involving at least one intermediate host. Alaria alata requires snails, then frogs and then wild boars (Sus scrofa) (Möhl et al., 2009), the latter being the most likely source of infection for hunting dogs. Even though the feeding of raw wild boar meat should be avoided due to the potential presence of the pseudorabies virus (PRV) (Chiari et al., 2015) – causing the fatal Aujetzky's disease in dogs (Schoniger et al., 2012) – 24% of the hunters reported to feed wild boar to their dogs (Supplementary Table S1). Mesocestoides sp. requires an unknown first intermediate host (Voge, 1967), then small mammals or birds (Zalesny and Hildebrand, 2012) as second intermediate hosts, respectively, with rodents being a likely source of infection for hunting dogs (Krucken et al., 2017).
Taenia spp. require ungulates which hunting dogs, unlike companion dogs, have access to. In the study area (Germany) roe deer, red deer and wild boar are the most likely wild intermediate hosts of T. hydatigena and T. krabbei (Lesniak et al., 2017), whereas their cysticerci are also known from farm animals like sheep (Priemer et al., 2002), pigs and cattle (Nguyen et al., 2016). Whereas in our study we found that helminth prevalence in hunting dogs amounted to 2.0% for Uncinaria stenocephala, 3.4% for Mesocestoides litteratus, 8.2% for A. alata and up to 12.2% for Taenia spp., previous studies in companion dogs found that helminth prevalence ranges between 0.0% for A. alata and U. stenocephala, 0.2% for Mesocestoides spp., and 1.2% for Taeniids (Barutzki and Schaper, 2003, Barutzki and Schaper, 2011). The divergent feeding (and hunting) behaviour of companion dogs and hunting dogs probably explains the higher prevalence of these helminths in the latter. Even though none of the detected helminths has a high zoonotic potential, it should be pointed out that apart from wild carnivores, hunting dogs function as hosts of A. alata and M. litteratus. Hence, they contribute to the spread of these parasites which may have a health impact on humans, e.g. if larvae are ingested from undercooked frog legs or meat of small vertebrates (Freeman et al., 1976, Möhl et al., 2009, Zalesny and Hildebrand, 2012).
4.4. Operational taxonomic units (OTUs) as measure of diversity and study limitations
High–throughput sequencing (HTS) approaches like metabarcoding have revolutionized many research fields including (veterinary) parasitology (Mardis, 2008, Menke et al., 2014, Hino et al., 2016). The simultaneous molecular–genetic identification of species from a diverse sample (e.g. faeces) via HTS and a subsequent bioinformatic analysis are increasingly supplementing or substituting labour–intensive microscopy, culturing and cloning experiments that were required in the past. Commonly, amplicon reads are clustered into units by nucleotide similarity forming so called OTUs (Blaxter et al., 2005). The more diverse the nucleotide sequences are, the more OTUs will be distinguished per amplicon. In this paper, we compared the OTUs generated from faecal samples of hunting dogs to apicomplexan and helminth sequences from NCBI GenBank to assign parasite species. According to our BLAST search, 109 OTUs matched Sarcocystis entries of at least 11 different species with a similarity of 98% or better. The number of OTUs per Sarcocystis species ranged from one (e.g. S. tenella) to 57 OTUs (S. taeniata) indicating a wide range of intraspecific nucleotide diversity within the analyzed gene fragments of the 18S rRNA gene. Similarly for helminths, OTUs within a range between one (Taenia sp.) and 308 OTUs (T. krabbei) per species were clustered based on two amplicons covering parts of the cox1 gene. A major advantage of generating genetic sequence data is the possible assignment of species of morphologically indistinguishable parasite stages such as oocysts. Furthermore, depending on the patent period and parasite load, genetic approaches have been demonstrated to be more sensitive than other copro–diagnostic methods (microscopy or coproantigen–ELISA), which impedes the comparison to other studies (Al-Sabi’ et al., 2007, Salant et al., 2010).
Despite several advantages, the copro–genetic approach applied in this study has its drawbacks that affect all sequences isolated in this study: firstly, the detection of parasite DNA in faeces does not necessarily imply that the excreting organism functions as definitive host in which the parasite reproduces since intestinal passage of parasites due to coprophagia or presence of non–infectious stages in the diet cannot be excluded (Boes et al., 1998). Secondly, a potential incomplete taxonomic resolution of the target genes at the species level (Kim et al., 1999) as well as database incompleteness are potential sources of false negative or false positive results, respectively. Also sequencing errors could lead to a misidentification, e.g. if two species are genetically similar. These methodological limitations might particularly apply to the less commonly detected parasite species that might furthermore be preferentially missed due to another known problem of metagenomic approaches – the PCR amplification bias towards more abundant DNA molecules (Brooks et al., 2015).
Above that, the direction of transmission in our field experiment cannot be inferred from the molecular methods we apply. The epidemiological conclusions we draw are solely based on theoretical thoughts and not molecular evidence.
5. Conclusion
Using current molecular genetic tools, we characterized endoparasites from faecal samples of hunting dogs and investigated whether Sarcocystis and helminth transmission dynamics change when a large carnivore host returns as an apex predator to its former habitat. Our study provides valuable knowledge in the context of the ongoing stakeholder discussion related to the wolf recolonization of Central Europe. Our general findings indicate that the return of wolves has a negligible impact on dog health regarding parasites and consequently on the health of dog owners since, e.g. highly zoonotic species such as the fox tapeworm Echinococcus multilocularis are absent in the investigated sample of hunting dogs and rarely occur in wolves as previously shown (Lesniak et al., 2017). Regarding parasites that are considered ‘wolf specialists’, this study supports the idea that hunting dogs from the wolf area experience an increase in infection risk of S. grueneri. Hunting dogs most likely acquire this parasite because the game that they are fed showed an increase in prevalence of this species when wolves are present (Lesniak et al., under review). Furthermore, due to the high prevalence of Sarcocystis itself in dog faeces we recommend reducing or abandoning the feeding of raw meat to dogs. Simply by cooking the remains or commercially available BARF meat, sarcocysts – but also other parasites and pathogens – will be inactivated. Such a measure is furthermore advisable to prevent imported, non–endemic parasites from establishing their life cycles outside of their original distribution ranges.
For helminths, this study suggests that hunting dogs may have substituted wolves as hosts for specific helminth species during the period of wolf extinction in Central Europe. Our findings support the recommendation to periodically deworm hunting dogs in order to prevent wolves from increasing their helminth loads.
Acknowledgements
We are grateful to Nina Hartmann, Susanne Kosanke, Susan Mbedi, Kirsten Richter and Mandy Wolfram for technical support, to José Grau and Dorina Lenz for bioinformatic help, to the German Wolf Documentation and Information Center for supplying the wolf distribution map, to the hunters associated with the forest ministries of Brandenburg, Saxony and Schleswig–Holstein for supplying dog faeces and background information, to two anonymous reviewers for their input regarding the manuscript. The study was funded by the German Federal Environmental Foundation (Az 20012/200), the state forestry services of Brandenburg (Az 35-2130/7+5-58/13), Saxony (Az 51-9210.71/107) and Schleswig–Holstein (Az 7461.9), the Open Access Fund of the Leibniz Association, and the Leibniz Institute for Zoo and Wildlife Research.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ijppaw.2017.09.001.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Distribution of grey wolves in Germany and hunting dog sampling sites. First wolf packs recolonized the region Lusatia along the federal state borders of Brandenburg (BB) and Saxony (SN) in the year 2000, from where wolves expanded their range into northwesterly direction. Until today, no resident wolves have been recognized in the federal state of Schleswig–Holstein (SH). The blue (‘control site’ without wolves) and red (‘wolf area’) dotted lines frame the study sites where the sampled hunting dogs live and work.
References
- Aguirre A.A., Ostfeld R.S., Tabor G.M., House C., Pearl M.C. Oxford University Press; New York: 2002. Conservation Medicine: Ecological Health in Practice. [Google Scholar]
- Al-Sabi M.N., Kapel C.M., Johansson A., Espersen M.C., Koch J., Willesen J.L. A coprological investigation of gastrointestinal and cardiopulmonary parasites in hunting dogs in Denmark. Vet. Parasitol. 2013;196:366–372. doi: 10.1016/j.vetpar.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Al-Sabi M.N.S., Chriél M., Jensen T.H., Enemark H.L. Endoparasites of the raccoon dog (Nyctereutes procyonoides) and the red fox (Vulpes vulpes) in Denmark 2009–2012 – a comparative study. Int. J. Parasitol. Parasites Wildl. 2013;2:144–151. doi: 10.1016/j.ijppaw.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sabi M.N.S., Kapel C.M.O., Deplazes P., Mathis A. Comparative copro-diagnosis of Echinococcus multilocularis in experimentally infected foxes. Parasitol. Res. 2007;101:731–736. doi: 10.1007/s00436-007-0537-4. [DOI] [PubMed] [Google Scholar]
- Almberg E.S., Cross P.C., Dobson A.P., Smith D.W., Hudson P.J. Parasite invasion following host reintroduction: a case study of Yellowstone's wolves. Philosophical Trans. R. Soc. Lond. Ser. B, Biol. Sci. 2012;367:2840–2851. doi: 10.1098/rstb.2011.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersen L.W., Harms V., Caniglia R., Czarnomska S.D., Fabbri E., Jedrzejewska B., Kluth G., Madsen A.B., Nowak C., Pertoldi C., Randi E., Reinhardt I., Stronen A.V. Long-distance dispersal of a wolf, Canis lupus, in northwestern Europe. Mammal. Res. 2015;60:163–168. [Google Scholar]
- Ansorge H., Holzapfel M., Kluth G., Reinhardt I., Wagner C. Die Rückkehr der Wölfe. Das erste Jahrzehnt. Biol. unserer Zeit. 2010;40:244–253. [Google Scholar]
- Barutzki D., Schaper R. Endoparasites in dogs and cats in Germany 1999-2002. Parasitol. Res. 2003;90(Suppl. 3):S148–S150. doi: 10.1007/s00436-003-0922-6. [DOI] [PubMed] [Google Scholar]
- Barutzki D., Schaper R. Results of parasitological examinations of faecal samples from cats and dogs in Germany between 2003 and 2010. Parasitol. Res. 2011;109(Suppl. 1):S45–S60. doi: 10.1007/s00436-011-2402-8. [DOI] [PubMed] [Google Scholar]
- Blaxter M., Mann J., Chapman T., Thomas F., Whitton C., Floyd R., Abebe E. Defining operational taxonomic units using DNA barcode data. Philosophical Trans. R. Soc. B Biol. Sci. 2005;360:1935–1943. doi: 10.1098/rstb.2005.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes J., Johansen M.V., Eriksen L., Bøgh H.O., Nansen P., Stephenson L.S. False-positive Trichuris suis egg counts in pigs in relation to coprophagia. Parasite. 1998;5:91–93. doi: 10.1051/parasite/1998051091. [DOI] [PubMed] [Google Scholar]
- Bowles J., Blair D., McManus D.P. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 1992;54:165–173. doi: 10.1016/0166-6851(92)90109-w. [DOI] [PubMed] [Google Scholar]
- Braun P.G., Preuss S.E. Nutritional composition and chemico-physical parameters of water buffalo milk and milk products in Germany. Milchwissenschaft. 2008;63:70–72. [Google Scholar]
- Brooks J.P., Edwards D.J., Harwich M.D., Rivera M.C., Fettweis J.M., Serrano M.G., Reris R.A., Sheth N.U., Huang B., Girerd P., Vaginal Microbiome C., Strauss J.F., Jefferson K.K., Buck G.A. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiari M., Ferrari N., Bertoletti M., Avisani D., Cerioli M., Zanoni M., Alborali L.G., Lanfranchi P., Lelli D., Martin A.M., Antonio L. Long-term surveillance of Aujeszky's disease in the alpine wild boar (Sus scrofa) EcoHealth. 2015;12:563–570. doi: 10.1007/s10393-015-1064-x. [DOI] [PubMed] [Google Scholar]
- Claveria F.G., Cruz M.J. Sarcocystis levinei infection in Philippine water buffaloes (Bubalus bubalis) Parasitol. Int. 2000;48:243–247. doi: 10.1016/s1383-5769(99)00025-2. [DOI] [PubMed] [Google Scholar]
- Craig H.L., Craig P.S. Helminth parasites of wolves (Canis lupus): a species list and an analysis of published prevalence studies in Nearctic and Palaearctic populations. J. Helminthol. 2005;79:95–103. doi: 10.1079/joh2005282. [DOI] [PubMed] [Google Scholar]
- Dahlgren S.S., Gjerde B. Sarcocystis in moose (Alces alces): molecular identification and phylogeny of six Sarcocystis species in moose, and a morphological description of three new species. Parasitol. Res. 2008;103:93–110. doi: 10.1007/s00436-008-0936-1. [DOI] [PubMed] [Google Scholar]
- Dahlgren S.S., Gjerde B. The red fox (Vulpes vulpes) and the arctic fox (Vulpes lagopus) are definitive hosts of Sarcocystis alces and Sarcocystis hjorti from moose (Alces alces) Parasitology. 2010;137:1547–1557. doi: 10.1017/S0031182010000399. [DOI] [PubMed] [Google Scholar]
- Demeler J., Ramünke S., Wolken S., Ianiello D., Rinaldi L., Gahutu J.B., Cringoli G., von Samson-Himmelstjerna G., Krücken J. Discrimination of gastrointestinal nematode eggs from crude fecal egg preparations by inhibitor-resistant conventional and real-time PCR. PloS One. 2013;8:e61285. doi: 10.1371/journal.pone.0061285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DJV, 2017a. https://www.jagdverband.de/jagdstatistik.
- DJV, 2017b. https://www.jagdverband.de/sites/default/files/2015-16%20Jagdscheininhaber_0.pdf.
- Dubey J.P. A review of Sarcocystis of domestic animals and of other coccidia of cats and dogs. J. Am. Vet. Med. Assoc. 1976;169:1061–1078. [PubMed] [Google Scholar]
- Dubey J.P. Sarcocystis species in moose (Alces alces), bison (Bison bison), and pronghorn (Antilocapra americana) in Montana. Am. J. Vet. Res. 1980;41:2063–2065. [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- El-Dakhly K.M., El-Nesr K.A., El-Nahass E.-S., Hirata A., Sakai H., Yanai T. Prevalence and distribution patterns of Sarcocystis spp. in buffaloes in Beni-Suef, Egypt. Trop. Animal Health Prod. 2011;43:1549–1554. doi: 10.1007/s11250-011-9840-2. [DOI] [PubMed] [Google Scholar]
- Erber M. Life cycle of Sarcocystis tenella in sheep and dog. Z. fur Parasitenkd. 1982;68:171–180. doi: 10.1007/BF00935058. [DOI] [PubMed] [Google Scholar]
- ESCCAP . In: Worm Control in Cats and Dogs. Parasites E.S.C.f.C.A., editor. 2010. [Google Scholar]
- Freeman R.S., Stuart P.F., Cullen J.B., Ritchie A.C., Mildon A., Fernandes B.J., Bonin R. Fatal human infection with mesocercariae of the trematode alaria americana*. Am. J. Trop. Med. Hyg. 1976;25:803–807. doi: 10.4269/ajtmh.1976.25.803. [DOI] [PubMed] [Google Scholar]
- Ghosal S.B., Joshi S.C., Shah H.L. Development of sarcocysts of Sarcocystis levinei in water buffalo infected with sporocysts from dogs. Vet. Parasitol. 1987;26:165–167. doi: 10.1016/0304-4017(87)90086-0. [DOI] [PubMed] [Google Scholar]
- Gjerde B. Morphological and molecular characteristics of four Sarcocystis spp. in Canadian moose (Alces alces), including Sarcocystis taeniata n. sp. Parasitol. Res. 2014;113:1591–1604. doi: 10.1007/s00436-014-3806-z. [DOI] [PubMed] [Google Scholar]
- Gómez-Morales M.A., Selmi M., Ludovisi A., Amati M., Fiorentino E., Breviglieri L., Poglayen G., Pozio E. Hunting dogs as sentinel animals for monitoring infections with Trichinella spp. in wildlife. Parasites vectors. 2016;9:154. doi: 10.1186/s13071-016-1437-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardone L., Deplazes P., Macchioni F., Magi M., Mathis A. Ribosomal and mitochondrial DNA analysis of Trichuridae nematodes of carnivores and small mammals. Vet. Parasitol. 2013;197:364–369. doi: 10.1016/j.vetpar.2013.06.022. [DOI] [PubMed] [Google Scholar]
- Hatcher M.J., Dunn A.f. Cambridge University Press; Cambridge: 2011. Parasites in Ecological Communities : from Interactions to Ecosystems. [Google Scholar]
- Haydon D.T., Cleaveland S., Taylor L.H., Laurenson M.K. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg. Infect. Dis. 2002;8:1468–1473. doi: 10.3201/eid0812.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino A., Maruyama H., Kikuchi T. A novel method to assess the biodiversity of parasites using 18S rDNA Illumina sequencing; parasitome analysis method. Parasitol. Int. 2016;65:572–575. doi: 10.1016/j.parint.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Huong L.T.T. Prevalence of Sarcocystis spp. in water buffaloes in Vietnam. Vet. Parasitol. 1999;86:33–39. doi: 10.1016/s0304-4017(99)00135-1. [DOI] [PubMed] [Google Scholar]
- Kim J., Kim W., Cunningham C.W. A new perspective on lower metazoan relationships from 18S rDNA sequences. Mol. Biol. Evol. 1999;16:423–427. doi: 10.1093/oxfordjournals.molbev.a026124. [DOI] [PubMed] [Google Scholar]
- Krucken J., Blumke J., Maaz D., Demeler J., Ramunke S., Antolova D., Schaper R., von Samson-Himmelstjerna G. Small rodents as paratenic or intermediate hosts of carnivore parasites in Berlin, Germany. PloS One. 2017;12:e0172829. doi: 10.1371/journal.pone.0172829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutkienė L., Prakas P., Sruoga A., Butkauskas D. The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis) Parasitol. Res. 2010;107:879–888. doi: 10.1007/s00436-010-1945-4. [DOI] [PubMed] [Google Scholar]
- Lesniak I., Heckmann I., Heitlinger E., Szentiks C.A., Nowak C., Harms V., Jarausch A., Reinhardt I., Kluth G., Hofer H., Krone O. Population expansion and individual age affect endoparasite richness and diversity in a recolonising large carnivore population. Sci. Rep. 2017;7:41730. doi: 10.1038/srep41730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos-Frank B., Zeyhle E. The intestinal helminths of the red fox and some other carnivores in Southwest Germany. Z. fur Parasitenkd. 1982;67:99–113. doi: 10.1007/BF00929518. [DOI] [PubMed] [Google Scholar]
- Maksimov P., Schares G., Press S., Fröhlich A., Basso W., Herzig M., Conraths F.J. Comparison of different commercial DNA extraction kits and PCR protocols for the detection of Echinococcus multilocularis eggs in faecal samples from foxes. Vet. Parasitol. 2017;237:83–93. doi: 10.1016/j.vetpar.2017.02.015. [DOI] [PubMed] [Google Scholar]
- Mardis E.R. The impact of next-generation sequencing technology on genetics. Trends Genet.TIG. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Martin R.J., Robertson A.P., Bjorn H. Target sites of anthelmintics. Parasitology. 1997;114:111–124. [PubMed] [Google Scholar]
- Menke S., Wasimuddin Meier M., Melzheimer J., Mfune J.K., Heinrich S., Thalwitzer S., Wachter B., Sommer S. Oligotyping reveals differences between gut microbiomes of free-ranging sympatric Namibian carnivores (Acinonyx jubatus, Canis mesomelas) on a bacterial species-like level. Front. Microbiol. 2014;5:526. doi: 10.3389/fmicb.2014.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan J., Lopez-Bao J.V., Garcia E.J., Oleaga A., Llaneza L., Palacios V., de la Torre A., Rodriguez A., Dubovi E.J., Esperon F. Patterns of exposure of iberian wolves (Canis lupus) to canine viruses in human-dominated landscapes. EcoHealth. 2016;13:123–134. doi: 10.1007/s10393-015-1074-8. [DOI] [PubMed] [Google Scholar]
- Möhl K., Große K., Hamedy A., Wüste T., Kabelitz P., Lücker E. Biology of Alaria spp. and human exposition risk to Alaria mesocercariae—a review. Parasitol. Res. 2009;105:1–15. doi: 10.1007/s00436-009-1444-7. [DOI] [PubMed] [Google Scholar]
- Moré G., Maksimov A., Conraths F.J., Schares G. Molecular identification of Sarcocystis spp. in foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) from Germany. Vet. Parasitol. 2016;220:9–14. doi: 10.1016/j.vetpar.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Nguyen M.T., Gabriel S., Abatih E.N., Dorny P. A systematic review on the global occurrence of Taenia hydatigena in pigs and cattle. Vet. Parasitol. 2016;226:97–103. doi: 10.1016/j.vetpar.2016.06.034. [DOI] [PubMed] [Google Scholar]
- Niedzialkowska M., Hundertmark K.J., Jedrzejewska B., Niedzialkowski K., Sidorovich V.E., Gorny M., Veeroja R., Solberg E.J., Laaksonen S., Sand H., Solovyev V.A., Shkvyria M., Tiainen J., Okhlopkov I.M., Juskaitis R., Done G., Borodulin V.A., Tulandin E.A., Jedrzejewski W. Spatial structure in European moose (Alces alces): genetic data reveal a complex population history. J. Biogeogr. 2014;41:2173–2184. [Google Scholar]
- Nowak S., Mysłajek R.W. Wolf recovery and population dynamics in Western Poland, 2001–2012. Mammal. Res. 2016;61:83–98. [Google Scholar]
- Nowak S., Mysłajek R.W., Kłosińska A., Gabryś G. Diet and prey selection of wolves (Canis lupus) recolonising Western and Central Poland. Mamm. Biol. - Z. für Säugetierkd. 2011;76:709–715. [Google Scholar]
- Ohr R., Zeddies G. Universität Göttingen; Göttingen: 2006. Ökonomische Gesamtbetrachtung der Hundehaltung in Deutschland. Studie der Wirtschaftswissenschaften; pp. 1–35. [Google Scholar]
- Otranto D., Cantacessi C., Dantas-Torres F., Brianti E., Pfeffer M., Genchi C., Guberti V., Capelli G., Deplazes P. The role of wild canids and felids in spreading parasites to dogs and cats in Europe. Part II: helminths and arthropods. Vet. Parasitol. 2015;213:24–37. doi: 10.1016/j.vetpar.2015.04.020. [DOI] [PubMed] [Google Scholar]
- Otranto D., Cantacessi C., Pfeffer M., Dantas-Torres F., Brianti E., Deplazes P., Genchi C., Guberti V., Capelli G. The role of wild canids and felids in spreading parasites to dogs and cats in Europe. Part I: Protozoa and tick-borne agents. Vet. Parasitol. 2015;213:12–23. doi: 10.1016/j.vetpar.2015.04.022. [DOI] [PubMed] [Google Scholar]
- Prakas P., Liaugaudaitė S., Kutkienė L., Sruoga A., Švažas S. Molecular identification of Sarcocystis rileyi sporocysts in red foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) in Lithuania. Parasitol. Res. 2015;114:1671–1676. doi: 10.1007/s00436-015-4348-8. [DOI] [PubMed] [Google Scholar]
- Priemer J., Krone O., Schuster R. Taenia krabbei (cestoda : cyclophyllidea) in Germany and its delimitation from T. ovis. Zool. Anz. 2002;241:333–337. [Google Scholar]
- R–Development–Core–Team . 2008. R: a Language and Environment for Statistical Computing. [Google Scholar]
- Randolph S.E., Storey K. Impact of microclimate on immature tick-rodent host interactions (Acari: ixodidae): implications for parasite transmission. J. Med. Entomology. 1999;36:741–748. doi: 10.1093/jmedent/36.6.741. [DOI] [PubMed] [Google Scholar]
- Reinhardt I., Kluth G., Nowak S., Myslajek R. German Federal Agency for Nature Conservation; 2015. Standards for the Monitoring of the Central European Wolf Population in Germany and Poland. BfN-Skripten 398 43. [Google Scholar]
- Salant H., Spira D.T., Hamburger J. A comparative analysis of coprologic diagnostic methods for detection of toxoplama gondii in cats. Am. J. Trop. Med. Hyg. 2010;82:865–870. doi: 10.4269/ajtmh.2010.09-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Feolo M., Geer L.Y., Helmberg W., Kapustin Y., Landsman D., Lipman D.J., Madden T.L., Maglott D.R., Miller V., Mizrachi I., Ostell J., Pruitt K.D., Schuler G.D., Sequeira E., Sherry S.T., Shumway M., Sirotkin K., Souvorov A., Starchenko G., Tatusova T.A., Wagner L., Yaschenko E., Ye J. Database resources of the national center for Biotechnology information. Nucleic acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger D.P., Joffe D.J. Raw food diets in companion animals: a critical review. Can. Veterinary J. 2011;52:50–54. [PMC free article] [PubMed] [Google Scholar]
- Schoniger S., Klose K., Werner H., Schwarz B.A., Muller T., Schoon H.A. Nonsuppurative encephalitis in a dog. Vet. Pathol. 2012;49:731–734. doi: 10.1177/0300985811432349. [DOI] [PubMed] [Google Scholar]
- Sousa W.P., Grosholz E.D. The influence of habitat structure on the transmission of parasites. In: Bell S.S., McCoy E.D., Mushinsky H.R., editors. Habitat Structure: the Physical Arrangement of Objects in Space. Springer Netherlands; Dordrecht: 1991. pp. 300–324. [Google Scholar]
- Stronen A.V., Sallows T., Forbes G.J., Wagner B., Paquet P.C. Diseases and parasites in wolves of the riding mountain national park region, manitoba, Canada. J. Wildl. Dis. 2011;47:222–227. doi: 10.7589/0090-3558-47.1.222. [DOI] [PubMed] [Google Scholar]
- Stubbe M. vol. 14. Hercynia-Ökologie und Umwelt in Mitteleuropa; 1977. pp. 1–10. (Der Marderhund Nyctereutes procyonoides (GRAY, 1834) in der DDR). [Google Scholar]
- Szentiks C.A., Fritsch G., Lesniak I., Galateanu G., Mühldorfer K., Hildebrandt T.B., Greenwood A.D., Krone O. 12th Conference of the European Wildlife Disease Association (EWDA) 2016. One decade of health monitoring in German free-ranging wolves (Canis lupus) p. 81. Berlin, Germany. [Google Scholar]
- Taylor L.H., Latham S.M., woolhouse M.E.J. Risk factors for human disease emergence. Philosophical Trans. R. Soc. Lond. Ser. B Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.C.A. Parasite zoonoses and wildlife: one health, spillover and human activity. Int. J. Parasitol. 2013;43:1079–1088. doi: 10.1016/j.ijpara.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kesteren F., Piggott K.J., Bengui T., Kubri S.B., Mastin A., Sillero-Zubiri C., Paris M., Millar R.P., Macdonald D.W., Shiferaw F., Craig P.S. Helminth parasites in the endangered Ethiopian wolf, Canis simensis. J. Helminthol. 2015;89:487–495. doi: 10.1017/S0022149X14000534. [DOI] [PubMed] [Google Scholar]
- Voge M. Development in vitro of Mesocestoides (cestoda) from oncosphere to young tetrathyridium. J. Parasitol. 1967;53:78–82. [PubMed] [Google Scholar]
- Wagner C., Holzapfel M., Kluth G., Reinhardt I., Ansorge H. Wolf (Canis lupus) feeding habits during the first eight years of its occurrence in Germany. Mamm. Biol. 2012;77:196–203. [Google Scholar]
- Xiang Z., Chen X., Yang L., He Y., Jiang R., Rosenthal B.M., Luan P., Attwood S.W., Zuo Y., Zhang Y.-p., Yang Z. Non-invasive methods for identifying oocysts of Sarcocystis spp. from definitive hosts. Parasitol. Int. 2009;58:293–296. doi: 10.1016/j.parint.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Zalesny G., Hildebrand J. Molecular identification of Mesocestoides spp. from intermediate hosts (rodents) in central Europe (Poland) Parasitol. Res. 2012;110:1055–1061. doi: 10.1007/s00436-011-2598-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of grey wolves in Germany and hunting dog sampling sites. First wolf packs recolonized the region Lusatia along the federal state borders of Brandenburg (BB) and Saxony (SN) in the year 2000, from where wolves expanded their range into northwesterly direction. Until today, no resident wolves have been recognized in the federal state of Schleswig–Holstein (SH). The blue (‘control site’ without wolves) and red (‘wolf area’) dotted lines frame the study sites where the sampled hunting dogs live and work.