Abstract
We investigate the association of MUC1 with castration-resistant prostate cancer (CRPC), bone metastasis, and PC recurrence. MUC1 expression was studied in patient-derived bone metastasis and CRPCs produced by prostate-specific PTEN−/− mice and LNCaP xenografts. Elevations in MUC1 expression occur in CRPC. Among nine patients with hormone-naïve bone metastasis, eight express MUC1 in 61% to 100% of PC cells. Utilizing cBioPortal PC genomic data, we organized a training (n = 300), testing (n = 185), and validation (n = 194) cohort. Using the Cox model, a nine-gene signature was derived, including eight genes from a MUC1-related network (APC, CTNNB1/β-catenin, GALNT10, GRB2, LYN, SIGLEC1, SOS1, and ZAP70) and FAM84B. Genomic alterations in these genes reduce disease-free survival (DFS) in the training (P = .00161), testing (P = .00699), entire (training + testing, P = 5.557e-5), and a validation cohort (P = 3.326e-5). The signature independently predicts PC recurrence [hazard ratio (HR) = 1.731; 95% confidence interval (CI): 1.104-2.712; P = .0167] after adjusting for known clinical factors and stratifies patients with high risk of PC recurrence using the median (HR 2.072; 95% CI: 1.245-3.450, P = .0051) and quartile 3 (HR 3.707, 95% CI: 1.949-7.052, P = 6.51e-5) scores. Several novel β-catenin mutants are identified in PCs leading to a rapid onset of death and recurrence. Genomic alterations in APC and CTNNB1/β-catenin reduce DFS in two independent PC cohorts (n = 485, P = .0369; n = 84, P = .0437). The nine-gene signature also associates with reductions in overall survival (P = .0458) and DFS (P = .0163) in melanoma patients (n = 367). MUC1 upregulation is associated with CRPC and bone metastasis. A nine-gene signature derived from a MUC1 network predicts PC recurrence.
Abbreviations: ADT, androgen deprivation therapy; BR, biochemical recurrence; RP, radical prostatectomy; WHO, the World Health Organization
Introduction
Prostate cancer (PC) is the most common male-specific malignancy in the developed world [1]. The disease progresses with a heterogeneous path. A large proportion of the low-grade [Gleason score 6/WHO grade (group) I] tumors are indolent. Approximately 30% of patients after radical prostatectomy (RP) will experience a rise in serum prostate-specific antigen (PSA) [2]; this biochemical recurrence (BR) significantly increases risk of PC metastasis and the development of castration-resistant prostate cancer (CRPC) [3]. PC predominantly metastasizes to the bone [4]; the standard treatment for this condition is androgen deprivation therapy (ADT). The treatment is palliative, and patients eventually develop CRPC. Although the recent developments have resulted in three sets of mRNA-based signatures, Oncotype DX (Genomic Prostate Score/GPS), Prolaris, and Decipher (Genomic Classifier), that evaluate the risk of PC progression [5], [6], [7], our ability to stratify PCs with high risk of progression remains poor. It is therefore critical to improve our understanding of factors contributing to PC recurrence, metastasis, and CRPC.
Mucin 1 (MUC1) is the most thoroughly studied tumor-associated antigen [8], [9], [10]. This is a cell membrane glycoprotein expressed on the apical surface of most epithelial tissues, including the pancreas, breast, lung, and gastrointestinal tract [11], [12]. The expression plays a protective role for the mucosal epithelial surface [13]. This polarity of apical presence in epithelial cells is lost in cancer cells; MUC1 is upregulated and altered in its pattern of glycosylation in over 70% of cancers [9], [11]. In PC, elevations in the MUC1 protein and aberrant MUC1 glycosylation have been observed [14], [15], [16]. These changes are associated with increases in angiogenesis [17], adverse clinical features, and higher Gleason scores [18]. MUC1 upregulation is weakly related with reductions in disease-free survival (DFS) and overall survival (OS) [18] and associates with adverse histopathology after RP [19]. A panel of three proteins (AZGP1, MUC1, and p53) predicts death in men with local PC [20]. Increases in MUC1 mRNA were observed in PC metastasis, genomic alterations in the MUC1 gene were detected in CRPC, and genomic changes in a 25-factor MUC1 network marginally correlated with PC recurrence [21]. Collectively, MUC1’s involvement in PC recurrence, metastasis, and CRPC development shows great potential that warrants further investigations.
In this study, we examined MUC1 expression in CRPC produced by xenografts and prostate-specific PTEN−/− transgenic mice, and in patients with hormone-naïve PC metastasized to bone. We also determined the association of a MUC1 network with PC recurrence using the PC data sets within the cBioPortal database. We report here 1) a relationship of MUC1 upregulation with CRPC and PC bone metastasis and 2) a nine-gene signature that is strongly associated with PC recurrence.
Materials and Methods
Collecting PC Bone Metastasis
Bone tissues containing metastatic PC were obtained from Hamilton Health Sciences, Hamilton, Ontario, Canada, under approval from the local Research Ethics Board (REB #11-3472).
Generation of CRPC Using Animal Models
LNCaP cells (5 × 106) were used to produce subcutaneous xenograft tumors in 8-week-old male NOD/SCID mice (The Jackson Laboratory); tumor volume was monitored according to our published systems [21]. Tumor growth was measured by serum PSA levels (PSA kit, Abcam). Mice were surgically castrated when tumor reached 100 to 200 mm3. Serum PSA was determined before and following castration. Rise in serum PSA indicates CRPC growth. Animals were sacrificed once tumors reached a volume ≥1000 mm3.
Prostate-specific PTEN−/− mice were produced using PTENloxp/loxp (C;129S4-Ptentm1Hwu/J; the Jackson Laboratory) and PB-Cre4 mice [B6.Cg-Tg(Pbsn-cre)4Prb, the NCI Mouse Repository] following our published conditions [22]. Surgical castration was performed when mice were 23 weeks old and subsequently monitored for 13 weeks. All animal protocols were approved by the McMaster University Animal Research Ethics Board.
Quantitative Real-Time PCR Analysis of MUC1 Expression
Real-time PCR using RNA samples was performed as previously described [21], [22]. All samples were run in triplicate using the following primers: MUC1 (forward): 5′-TGCCGCCGAAAGAACTACG-3′, MUC1 (reverse): 5′-TGGGGTACTCGCTCATAGGAT-3′. β-actin (forward): 5′-ACCGAGCGCGGCTACAG-3′, β-actin (reverse): 5′-CTTAATGTCACGCACGATTTCC-3′.
Immunohistochemistry (IHC)
Slide preparation, processing, and antigen retrieval were carried out according to our established protocols [21], [22]. Slides were blocked in PBS, 1% BSA, and 10% normal goat serum (Vector Laboratories) for 1 hour. MUC1-N (N-terminus) (1:100, BD), MUC1-C (1:50, Fisher Scientific), and prostate acid phosphatase (PAP) (1:300, Abcam) antibodies were added at 4°C overnight. Biotinylated goat anti-mouse IgG or anti-hamster IgG secondary antibodies, and Vector ABC reagent (Vector Laboratories) were incubated according to the manufacturer's instructions. Secondary antibody only was used as negative control. Images were acquired and analyzed using ImageScope software (Leica Microsystems Inc.). Quantification of MUC1-positive PC cells was performed by counting up to 10,000 cells in several regions for MUC1-positve and PAP-positive cells. Percentage of MUC1-postive cells was estimated as MU1 positivity/PAP positivity × 100.
Establishing of a Nine-Gene Genomic Signature from the MUC1 Network
The largest TCGA data set (n = 499), which includes 485 patients with follow-up data, within the cBioPortal database [23], [24] (http://www.cbioportal.org/index.do) was extracted and randomly divided into 10 sets of training (n = 300) and testing (n = 185) cohorts using the RandomizationR package in R. The 25 genes of the MUC1 network [21] plus FAM84B [22] (Supplementary Table 1) were inputted into the Cox model to select for their contributions to hazard ratio (HR) by either forward addition or backward elimination of covariates using SPSS Statistics version 23. The resultant 9 genes were then examined on the 10 testing cohorts and validated on an independent cohort (MSKCC, cBioPortal, n = 194) for effects on DFS, OS, and HR using the Survival package in R.
Statistical analysis
Statistical analysis was performed using Student's t test. Kaplan-Meier surviving curves, log-rank test, receiver-operating characteristic (ROC) curve, and univariate and multivariate Cox proportional hazards regression analyses (Survival package in R and SPSS Statistics version 23). A value of P < .05 is considered statistically significant.
Results
Upregulation of MUC1 in CRPC
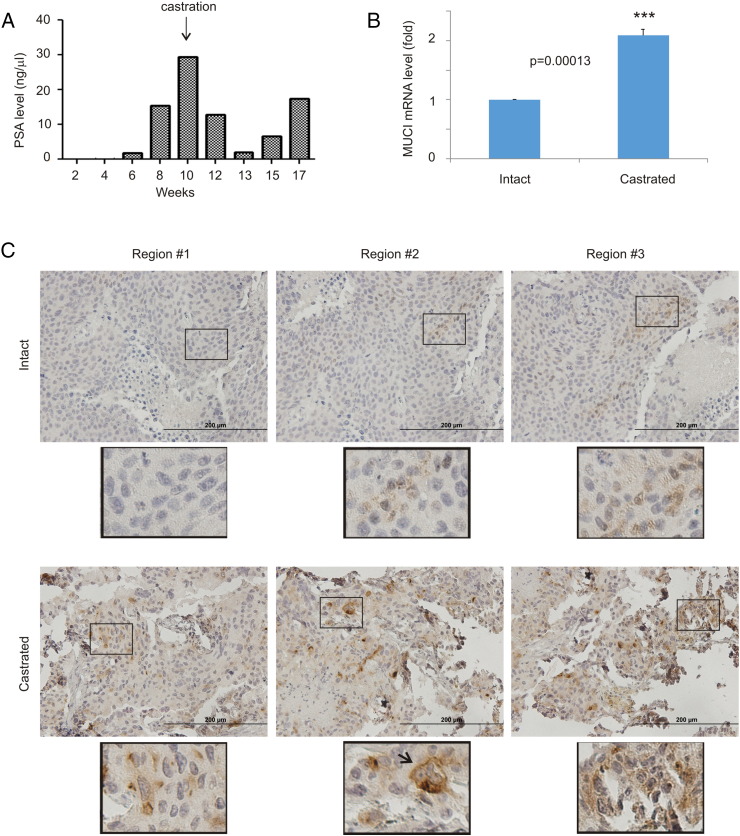
CRPC is the leading cause of PC fatalities; specific genomic alteration in the MUC1 gene was observed in CRPC [21]. To examine MUC1 expression in CRPC, we implanted LNCaP cells, a well-established androgen-dependent PC cell line, into NOD/SCID mice. Surgical castration initially reduced tumor growth, followed by a subsequent tumor regrowth (Figure 1A). The regrown tumors (CRPCs) exhibited a significant increase in MUC1 mRNA compared to the xenografts developed in intact mice (Figure 1B). Elevations in MUC1 protein in CRPC were also demonstrated; cell surface expression and a special clustering pattern for MUC1 were observed (Figure 1C), which is consistent with MUC1 being a cell surface protein [11], [12] and its detection in prostate cancer stem-like cells [21].
Figure 1.
MUC1 upregulation in animal models of CRPC. (A) PSA levels in NOD/SCID mice bearing LNCaP cell-derived xenograft tumors prior to and after castration. (B) Real-time PCR analysis of MUC1 mRNA in hormone-naive (n = 3) and castration-resistant (n = 3) LNCaP xenograft tumors. Statistical analysis was performed using Student's t test (two-tailed). (C) IHC staining of MUC1 in LNCaP xenograft tumors produced in intact (n = 3) and castrated mice (n = 3). Typical images from three different regions of individual tumors are shown. The indicated regions were enlarged three-fold.
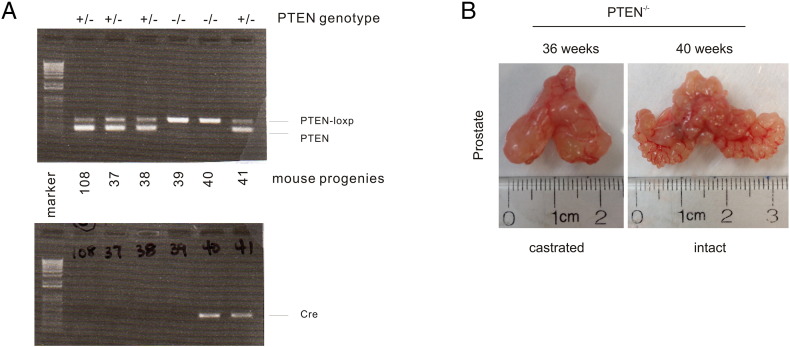
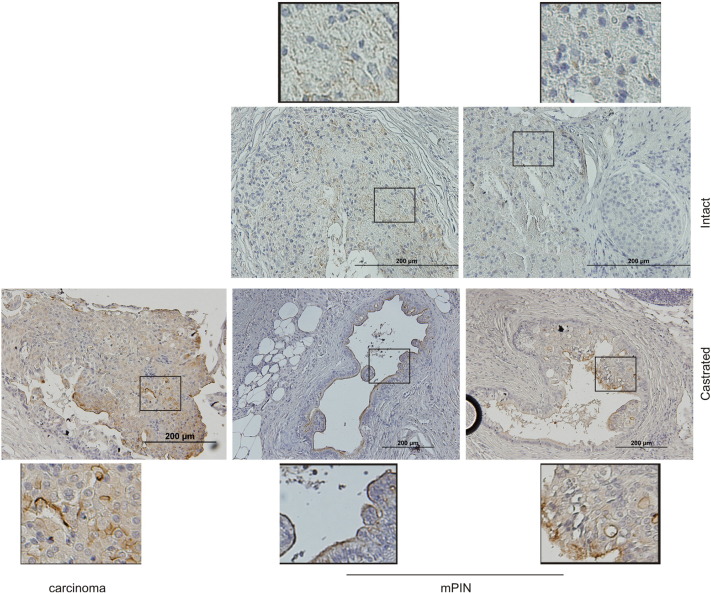
Furthermore, we have generated prostate-specific PTEN−/− mice (Supplementary Figure 1A), castrated the animals at 23 weeks, and euthanized the mice 13 weeks later. CRPCs were clearly developed (Supplementary Figure 1B). Compared to tumors in intact mice, tumors in castrated mice express an increased level of MUC1 (Figure 2). Intriguingly, the MUC1 protein shows a preference of expression in the luminal layer epithelial cells in mouse PINs produced in castrated PTEN−/− mice (Figure 2), and this preference is lost in carcinoma (Figure 2), resembling the MUC1’s expression pattern in nontumor [13] and tumor tissues [9], [11]. Collectively, we provide direct evidence for MUC1 upregulation in CRPC.
Supplementary Figure 1.
Production of CRPC in prostate-specific PTEN−/− mice. (A) Prostate-specific PTEN−/− mice were produced by crossing PTEN-loxp/loxp mice with PB-Cre4 (prostate-specific expression of Cre) mice. Genotyping of six mouse progenies for the PTEN locus (top panel) and the Cre transgene locus (bottom panel) using tail genomic DNA. (B) PTEN−/− mice were castrated at 23 weeks old. Typical images of age-matched partial urogenital systems (bladder, prostatic lobes, seminal vesicles) from an intact and a castrated mouse are shown.
Figure 2.
Increases in MUC1 expression in CRPC generated from prostate-specific PTEN−/− mice. Generation of prostate-specific PTEN−/− mice is detailed in Supplementary Figure 1. PTEN−/− mice were castrated at 23 weeks old and monitored for 13 weeks, followed by IHC straining for MUC1. Typical images of MUC1 staining in intact (n = 3) and castrated (n = 3) mouse prostate. The indicated regions were enlarged three-fold. Note: MUC1 is largely detected in the luminal surface of mouse PINs in castrated mice. mPIN, mouse PIN.
Extensive Expression of the MUC1 Protein in Hormone-Naïve PCs Metastasized to the Bone
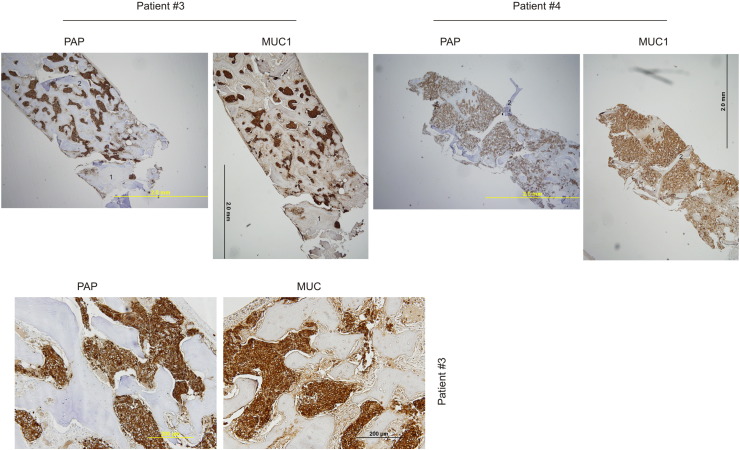
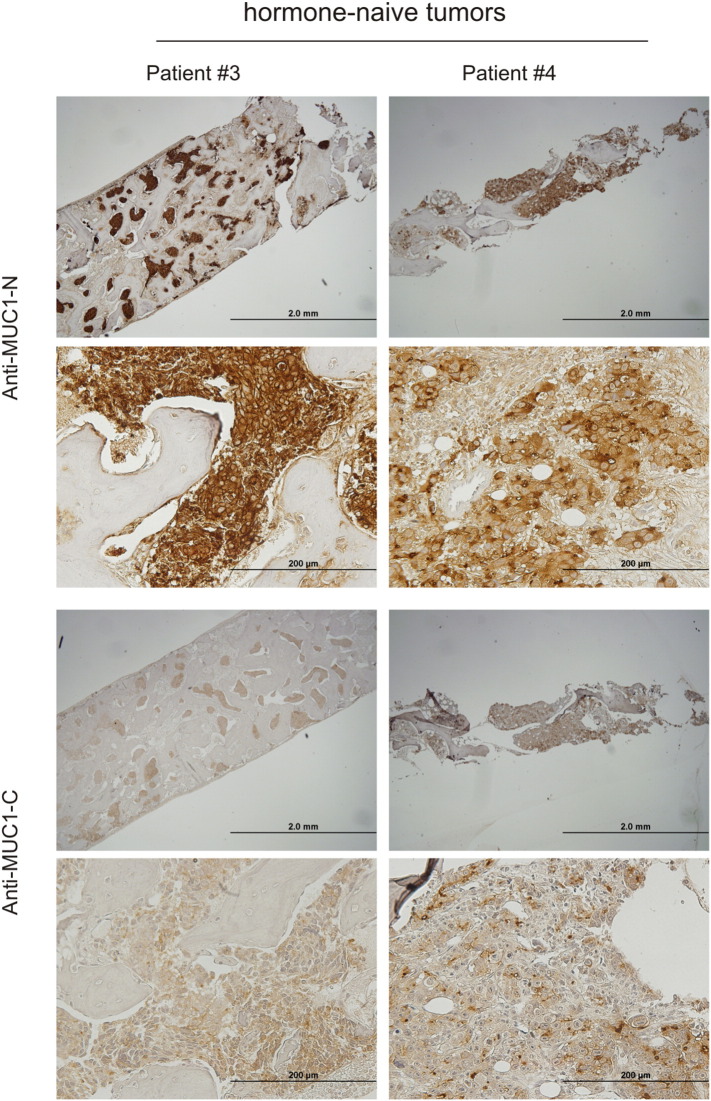
Metastasis to the bone is a major cause of morbidity in patients with PC. We recently observed increases in MUC1 mRNA in metastatic PCs [21]. To examine the MUC1 protein in PC metastasized to the bone, we obtained nine bone biopsy samples from patients who presented with PC metastatic to bone at diagnosis; the tumors were thus hormone naïve (Table 1). The metastatic prostate origin was confirmed by prostate acid phosphatase (PAP) staining, a widely used surrogate marker of PC cells in the clinic. Based on PAP-positive cells, extensive MUC1 expression in PC cells was demonstrated (Figure 3, Table 1). A good coordinate expression of MUC1 detected by antibodies against MUC1 N- and C-terminus was observed (Supplementary Figure 2). Collectively, we show the first evidence for extensive MUC1 expression in hormone-naïve PC bone metastasis in humans. These observations are novel and clinically significant (see Discussion for details).
Table 1.
MUC1 Expression in Bone Metastases of PC Derived from Hormone-Naïve Patientsa
| Patient | Ageb | PSAc | Hormone Status | MUC1/PAP (%)d |
|---|---|---|---|---|
| 2 | 71 | 26 | Naive | 0e |
| 3 | 77 | 522 | Naive | 89.6% |
| 4 | 81 | 649 | Naive | 84.1% |
| 6 | 61 | 205 | Naive | 64.8% |
| 9 | 71 | 21 | Naive | 61.1% |
| 10 | 80 | 10 | Naive | 100% |
| 11 | 73 | 164 | Naive | 74.8% |
| 12 | 62 | 34 | Naive | 97.8% |
| 13 | 72 | 163 | Naive | 75.7% |
Prostate adenocarcinoma in bone was confirmed by PSA and PAP staining.
Age at diagnosis.
PSA at diagnosis.
Ratio of MUC1-positive cells in PAP-positive cells.
MUC1 protein undetectable.
Figure 3.
Extensive expression of MUC1 in PC-derived bone metastases from hormone-naïve patients. Bone tissues with hormone-naïve PC were obtained from patients #3 and #4 and were IHC stained for PAP and MUC1 using an antibody to a MUC1 N-terminal region. Matched images of a low magnification for PAP and MUC1 staining are shown (top panels). Regions marked with the same number in the PAP and MUC1 image of individual patients are matched. Please note that the #2 region in the PAP image of patient 4 was dislocated during IHC staining. Matched images of PAP and MUC1 staining for patient #3 in a higher magnification are also included (bottom panel).
Supplementary Figure 2.
Expression of MUC1 in bone PC metastases. IHC staining of PC bone metastases (n = 9) for both MUC1-N (N-terminal fragment) and MUC1-C (C-terminal fragment). Typical images for the indicated patients at a low- (scale bar: 2 mm) and high- (scale bar: 200 mm) power magnification are provided.
Derivation of a Nine-Gene Signature from a MUC1 Network with a Robust Predicting Value of PC Recurrence
BR is associated with significant increases in risk for PC metastasis and CRPC [3]. Despite extensive research effort searching for BR-associated biomarkers, effective biomarkers are not available. Our observed MUC1 upregulation in CRPC and bone metastasis suggests a relationship between MUC1 and BR. Indeed, a weak association of MUC1 and PC recurrence was recently reported [18]; MUC1 associates with adverse pathology after RP [19]; and MUC1 along with AZGP1 and p53 predicts death in patients with local prostate tumor [20]. We reasoned that our recently identified MUC1 network consisting of 25 genes [21] would enhance the effectiveness of BR prediction.
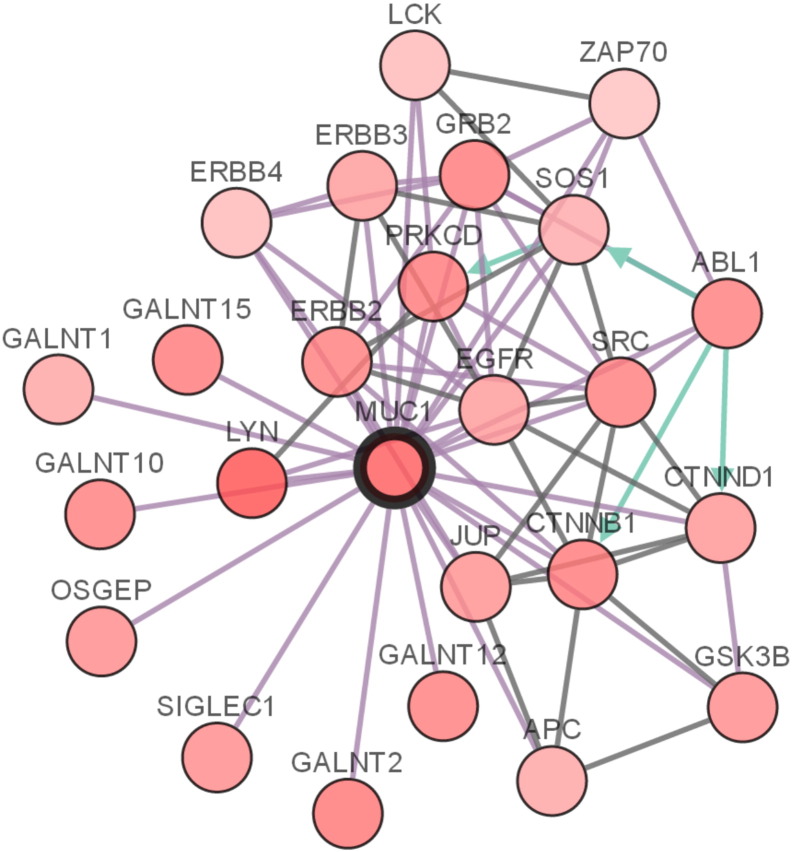
To address this possibility, we extracted a population (n = 485) with genomic alterations and pathological data from the TCGA data set (n = 499) within the cBioPortal database. Ten random sets of training (n = 300) and testing (n = 185) cohorts were generated (Table 2). FAM84B is a novel factor associated with PC progression [22]. A systematic variable selection from the 25 genes of the MUC1 network (Supplementary Figure 3) and FAM84B for their specific genomic alterations (Supplementary Table 1) using the Cox model yielded 16 candidate genes (Table 3). The top eight genes appeared in at least two training cohorts (Table 3); LYN and GALNT15 have P values approaching the significant level (P < .05) and HR > 1 (Table 3); the top eight genes (GALNT10, SOS1, ZAP70, FAM84B, GRB2, SIGLEC1, CTNNB1, and APC) plus LYN and GALNT15 (Table 3) were thus further considered. The 95% confidence interval (CI) for GALNT15 was large (Table 3); its removal from this 10-gene list improved HR and P values in 6 training cohorts (Supplementary Table 2). We thus defined the nine-gene signature for their specific genomic alterations (Table 3).
Table 2.
Demographics of Patient Populations
| Characteristics | Training Seta (n = 300) | Testing Seta (n = 185) |
|---|---|---|
| Age (years) | ||
| Mean median ± SD | 61.2 ± 0.4 | 61.5 ± 0.67 |
| Q1(SD)-Q3(SD) | 56.1 (0.32)-65.9 (0.3) | 56.3 (0.64)-66 (0.63) |
| Follow-up (months) | ||
| Mean median ± SD | 27 ± 1.33 | 27.2 ± 2.07 |
| Q1(SD)-Q3(SD) | 14.7 (0.7)-45.1 (1.2) | 14 (1.5)-44.5 (2.1) |
| Recurred | ||
| No (mean ± SD, %) | 245.5 ± 4.8, 82% | 149.5 ± 4.8, 81% |
| Yes (mean ± SD, %) | 53.5 ± 4.8, 18% | 35.5 ± 4.8, 19% |
| Tumor stages | ||
| T2a (mean ± SD, %) | 7.7 ± 2, 2.6% | 5.3 ± 2, 2.9% |
| T2b (mean ± SD, %) | 5.5 ± 1.2, 1.7% | 1.8 ± 1.2, 1% |
| T2c (mean ± SD, %) | 102.1 ± 4.2, 34% | 61.9 ± 4.2, 33.5% |
| T3a (mean ± SD, %) | 94.7 ± 2.8, 31.6% | 60.4 ± 2.8, 32.6% |
| T3b (mean ± SD, %) | 81.6 ± 5.4, 27.2% | 48.4 ± 5.4, 26.2% |
| T4 (mean ± SD, %) | 5.7 ± 1.3, 1.9% | 4.3 ± 1.3, 2.3% |
| Surgical margin | ||
| R0 (mean ± SD, %) | 188 ± 4.5, 62.7% | 121 ± 4.5, 65.4% |
| R1 (mean ± SD, %) | 90.8 ± 4.2, 30.3% | 51.2 ± 4.2, 27.7% |
| R2 (mean ± SD, %) | 2.5 ± 0.7, 0.8% | 2.2 ± 0.8, 1.2% |
| Rx (mean ± SD, %) | 9.8 ± 2.6, 3.3% | 5.2 ± 2.6, 2.8% |
Ten random pairs of training and testing sets were generated; all numbers here are the respective means ± SD.
Supplementary Figure 3.
The MUC1 network. The network was generated as described in our recent publication [17]. Briefly, the interactions were derived using the pathway and interaction data from HPRD, Reactome, NCI-Nature Cell Map. The network was constructed using the system provided by cBioPortal (http://www.cbioportal.org/).
Table 3.
Candidate Genes Selected Using the Training Sets
| Gene | Gen Alta | HRb | 95% CIb | P Valueb | Frequencyc |
|---|---|---|---|---|---|
| GALNT10d, e | Amp Mut | 5.177 | 1.269-21.311 | .022* | 4 |
| SOS1d, e | Amp Mut | 23 | 5.181-98.222 | <.0001* | 10 |
| ZAP70d, e | Amp Mut | 4.524 | 1.765-11.598 | .002* | 5 |
| FAM84Bd, e | Amp | 2.236 | 1.053-4.746 | .036* | 2 |
| GRB2d, e | Amp Mut | 7.696 | 1.031-57.462 | .047* | 3 |
| SIGLEC1d, e | Amp Mut | 6.46 | 1.98-21.074 | .002* | 4 |
| CTNNB1d, e | Amp Mut | 6.553 | 1.883-22.808 | .003* | 5 |
| APCd, e | Homdel Mut | 3.084 | 1.2-7.924 | .019* | 5 |
| LYNd, e | Amp Mut | 2.535 | 0.993-6.471 | .052 | 1 |
| GALNT15d | 7.645 | 1.013-57.701 | .049* | 1 | |
| ERBB2 | 2.464 | 0.614-9.889 | .203 | 1 | |
| CTNND1 | 6.139 | 0.839-44.933 | .074 | 2 | |
| GALNT2 | 1.776 | 0.536-5.888 | .348 | 1 | |
| MUC1 | 0.365 | 0.037-3.597 | .388 | 1 | |
| JUP | 0.017 | 0.009-0.586 | .024* | 1 | |
| ABL1 | 0 | 0-0.005 | 0 | 1 |
Genomic alterations are included only for the final set of genes. Amp, amplification; Mut, mutation; Homdel, homdeletion.
Best HR, CI, and P value produced in 10 training sets.
Number of training sets from which the candidates were selected.
Candidates that are evaluated in 10 training sets.
The final set of genes or the nine-gene set.
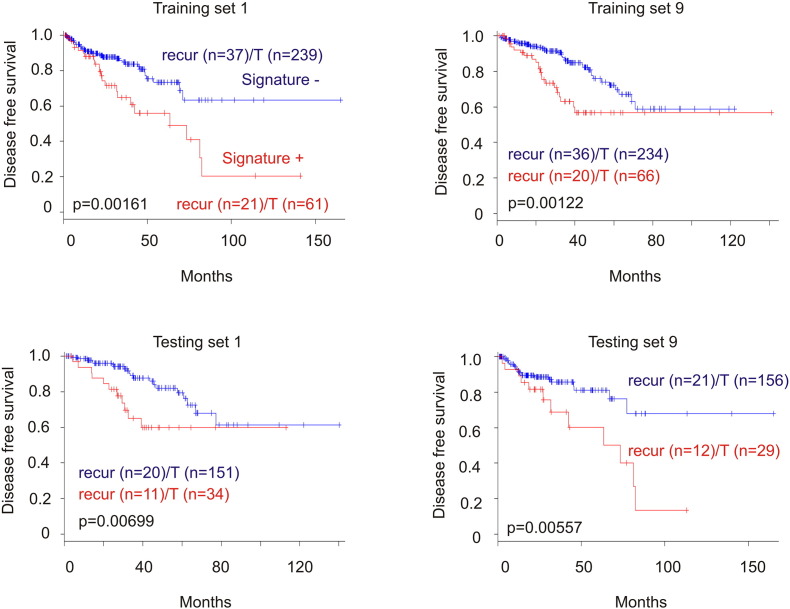
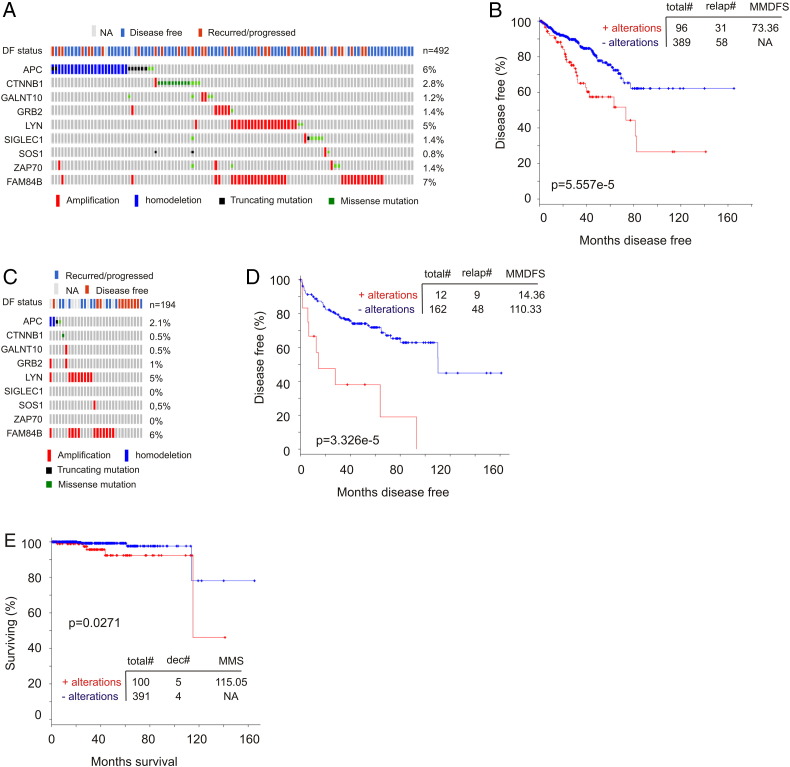
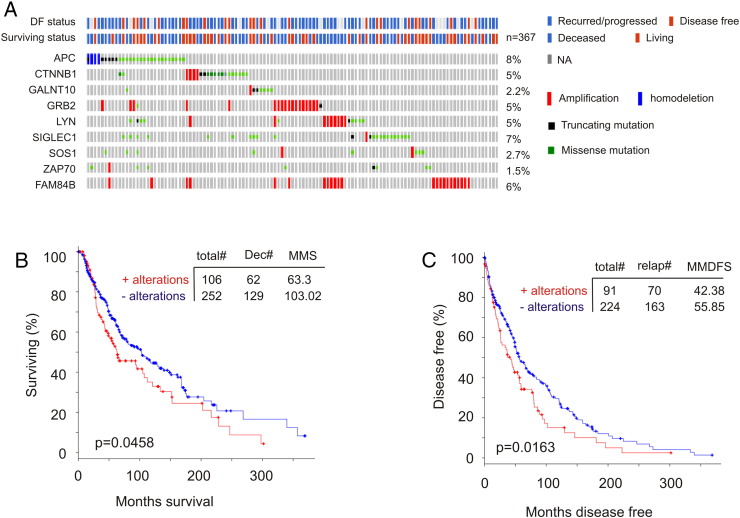
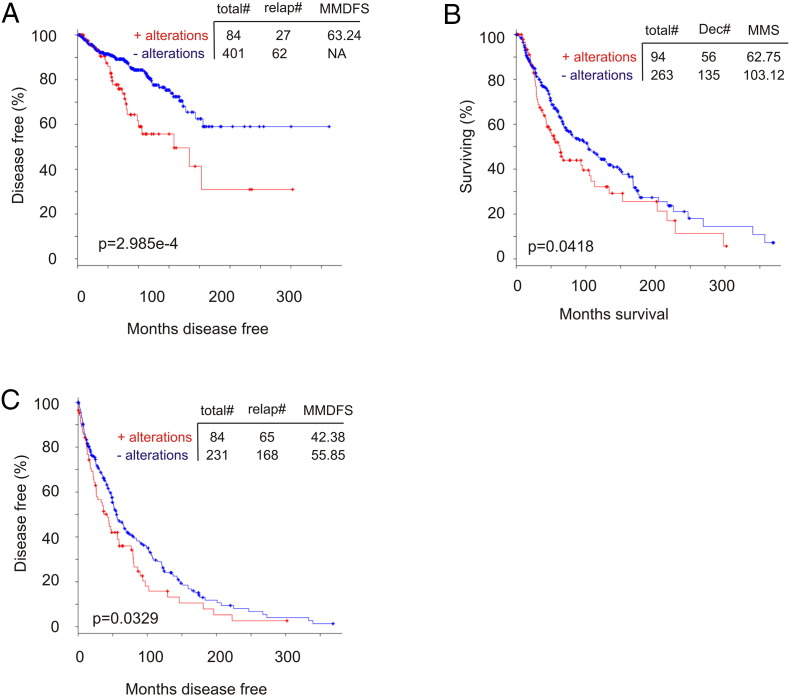
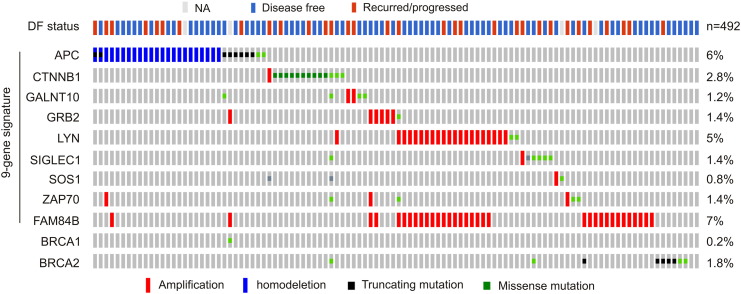
Reanalysis of the 9-gene signature revealed a robust association with reductions of DFS in all 10 training cohorts (smallest P = 6.11e-7) and 8 of 10 testing cohorts (smallest P = 5.75e-5) (Figure 4, data not shown). Of note, the 9-gene signature was a risk factor for BR in all training and 8 of 10 testing populations based on HR (Supplementary Table 3). The signature was not associated with reductions in DFS (data not shown), and neither was it a risk factor in the same two testing cohorts (Supplementary Table 3), which likely resulted from patient randomization. It is thus important to use multiple sets of randomized training and testing cohorts. We further examined this nine-gene signature in the entire TCGA cohort (n = 492) (Figure 5A) and demonstrated its robust association with decreases in DFS (Figure 5B). This association was also revealed in an independent cohort of primary PC (n = 194) (cBioPortal) [25] (Figure 5, C and D). Furthermore, among the 9 PC-related fatalities in the TCGA population (n = 491; 9/491 = 1.8%), 5 were in the 9-gene signature-positive population (n = 100; 5/100 = 5%; P = .0271) (Figure 5E). Additionally, the 9-gene signature is also detected in a patient population of cutaneous melanoma (n = 367) (TCGA, cBioPortal) (Figure 6A) and associates with reductions in OS and DFS in these patients (Figure 6, B and C).
Figure 4.
The nine-gene signature associates with reductions in DFS in training and testing cohorts. See Table 3 for the identities of nine genes and the types of genomic alterations being tested in the signature. The signature was evaluated in 10 individual sets of training and testing populations; typical results in 2 cohort sets are shown. Kaplan-Meier and log-rank tests were performed using the R Survival Package. Recurr, recurrence; T, total. For training cohort, n = 300; for testing cohort, n = 185.
Figure 5.
The nine-gene signature robustly correlates with decreases in DFS and OS in PC. (A) The indicated types of genomic alterations for the nine genes in the TCGA data set (n = 492) within the cBioPortal database [23], [24] are shown; only the proportion of cohorts containing the nine-gene signature are included. Each column is for individual tumor. Note: This cohort was used to generate the training and testing subcohorts. DF, disease free; NA, not available. (B) Analysis of DFS using the TCGA cohort. Total#, total number of cases; relap#, number of relapsed cases; MMDFS, median months disease-free survival. (C, D) Genomic alterations for the nine genes in a subcohort (n = 194) within a MSKCC data set (cBioPortal) [25] (C) and the effects of the nine-gene signature on DFS in this cohort (D). (E) Analysis of patients with the nine-gene signature–positive or –negative tumors for their overall survival using the TCGA data set. Dec#, number of deceased cases; MMS, median months survival.
Figure 6.
The nine-gene signature associates with reductions in DFS and OS in patients with cutaneous melanoma. Data were extracted from the TCGA data set of cutaneous melanoma within the cBioPortal database. Analysis of genomic alterations in the nine genes in the TCGA cohort (n = 367); the DF and survival status for these patients are also included (A). The nine-gene signature associates with a significant reduction in OS (B, n = 358) and DFS (C, n = 315) in these patients.
Although the nine-gene list includes a non-MUC1 network member FAM84B, the eight-gene signature of the MUC1 network after removing FAM84B retains the core association with decreases in DFS and OS in PC and cutaneous melanoma (Supplementary Figure 4, A-C). Comparing Figure 5B to Supplementary Figure 4A, FAM84B clearly contributes to the association. Furthermore, removal of any individual gene from the nine-gene list decreased its association with shortening of DFS in PC (the TCGA cohort, cBioPortal; data not shown), validating their necessity in the signature set.
Supplementary Figure 4.
Genomic alterations in the eight MUC1 network genes within the nine-gene list associate with reductions in DSF and OS in patients with either PC or skin cutaneous melanoma. FAM84B gene was removed from the nine-gene set (APP, CTNNB1, GALNT10, GRB2, LYN, SIGLEC1, SOS1, ZAP70, and FAM84B). The specific genomic alterations (see Table 3 for details) in the remaining eight MUC1 network gene associate with decreases in DFS in PC (A) and cutaneous melanoma (C) as well as OS (B) in cutaneous melanoma. Genomic data were extracted from the TCGA cohorts of PC and skin cutaneous melanoma within the cBioPortal database.
APC and β-Catenin Contributions to PC Recurrence
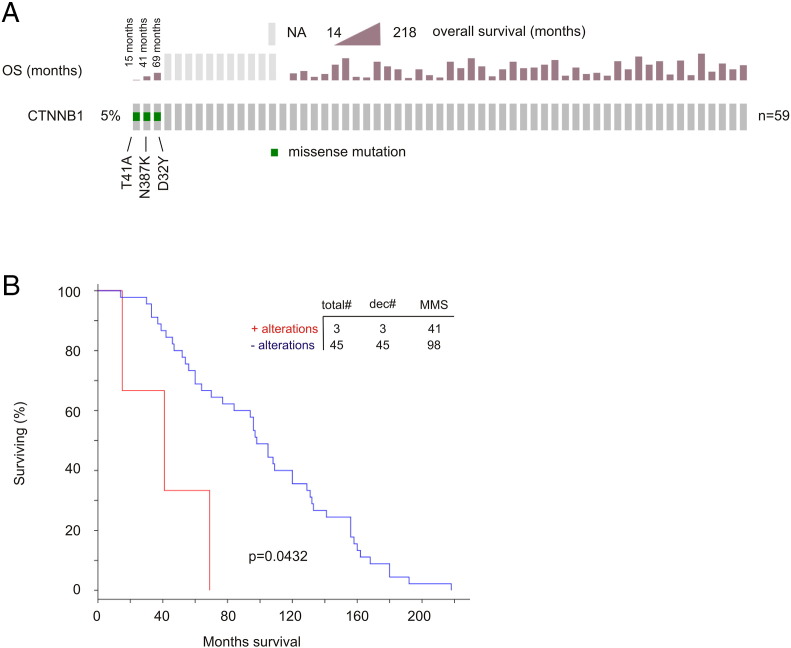
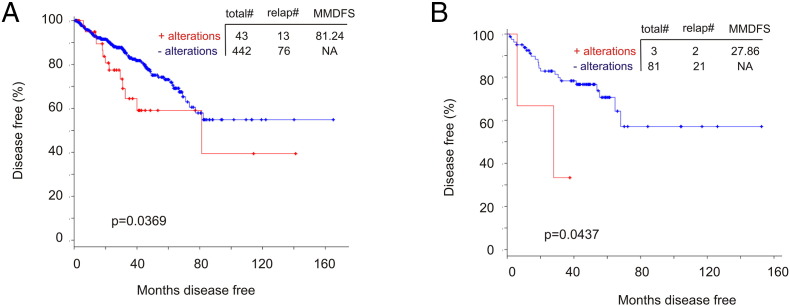
The major type of genomic alterations detected in CTNNB1 (encoding β-catenin) was missense mutations (Figure 5A). We thus examined these mutations in details. cBioPortal contains nine independent PC populations; missense mutations in β-catenin were extracted from seven cohorts (Table 4). T41A was found in six populations (Table 4). In cohorts with follow-up data, T41A-positive tumors caused PC death (1 for 1, the Michigan cohort) and recurrence (1 for 2, the TCGA cohort) (Table 4). Missense mutations at D32 were detected in four cohorts (Table 4). Intriguingly, in the metastatic PC cohort of Michigan [26] with 100% fatality (n = 48), patients with the missense mutations in β-catenin had a rapid onset of death (Supplementary Figure 5). The residues D32, S33, S37, T41, and S45 are within the destruction box of β-catenin and are required for β-catenin degradation, a process mediated by the degradation complex including APC, GSK-3β, and casein kinase 1α [27], [28], [29]. Missense mutations in these residues stabilize β-catenin, contribute to tumorigenesis, and occur in multiple tumor types, including D32Y, S33F, and T41A in PC [28], [30]. Nonetheless, we describe for the first time a set of missense mutations at D32, S33, S37, T41, and S45 in PC (Table 4) and novel missense mutations detected outside of the destruction box [28], N387K, W383G, and R225H. These novel mutations are observed in tumors causing either PC death or recurrence (Table 4). While whether all mutations described here promote PC progression remains unknown, their mutual exclusivity with genomic alterations of the APC tumor suppressor (Figure 5A), a well-demonstrated theme in colon cancer [28], suggests their contributions together with APC in PC recurrence. Indeed, we provide the first evidence that genomic alterations in APC and β-catenin significantly shorten DFS in the TCGA (n = 492, P = .0369) and MSKCC (n = 103, P = .0437) cohorts (cBioPortal) (Supplementary Figure 6).
Table 4.
β-Catenin Mutations in PC and Their Impact on PC Death and Recurrence.
| Patient Cohort | n | Mutation (n); Patient | Survival or Recurrence |
|---|---|---|---|
| Michigan, Nature 2012 | 59 | T41A (1) | Deceased (OS 15 months) |
| D32Y (1) | Deceased (OS 69 months) | ||
| N387 K (1) | Deceased (OS 41 months) | ||
| Robinson et al., Cell 2015 | 150 | T41A (2) | NA |
| S37C (1) | NA | ||
| S45C (1) | NA | ||
| S45F (1) | NA | ||
| FH, Nat Med 2016 | 56 | T41A (1) | NA |
| D32V (1) | NA | ||
| M14 V;S37Y (1) | NA | ||
| S45P (1) | NA | ||
| M763I (1) | NA | ||
| Trento/Cornell/Broad 2016 | 81 | T41A (1) | NA |
| D32N (1) | NA | ||
| S33A (1) | NA | ||
| MSKCC, Cancer cell 2010 | 103 | A522T;S45P | NA |
| W383G (1) | Recurred | ||
| CPC-GENE, Nature 2017 | 449 | T41A (1) | NA |
| TCGA provisional | 489 | T41A (1); HC-7738a | Recurred, DF:13.8 months |
| T41A (1); EJ-A65Da | DF, censored: 12.9 months | ||
| D32Y (1), HC-8262a | DF, censored: 22.3 months | ||
| D32V (1), EJ-5494a | DF, censored: 48.5 months | ||
| D32H (1), EJ-5525a | Recurred, DF: 17.9 months | ||
| S33A (1) | DF; censored: 41 months | ||
| S33Y (1) | DF; censored: 21.8 months | ||
| S33C (1) | Recurred; DF: 18.4 months | ||
| S45P (1) | DF | ||
| R225H (1) | Recurred | ||
| R342K (1) | DF | ||
| T75I (1) | DF |
All cohorts were extracted from cBioPortal.
Frequently mutated resides are bolded.
n, number of patients; Mutation (n), number of cases with the indicated mutations.
Patient ID is included only for patients with PC containing either T41A or missense mutations in D32 within the TCGA dataset.
Supplementary Figure 5.
Mutations in β-catenin in D32, T41, and N387 significantly shorten overall survival in patients with metastatic prostate cancer. Data were extracted from the Michigan data set [22] (cBioPortal). (A) The three missense mutations and their associated OS are indicated. (B) Kaplan-Meier analysis of OS time in metastatic PCs with and without the missense mutations in β-catenin. Statistical analysis was performed using log-rank test. Both analyses were carried out using the tools provided by cBioPortal. Total#, total number of cases; dec#, number of deceased cases; MMS, median months survival.
Supplementary Figure 6.
Genomic alterations in APC and β-catenin significantly associate with reductions in DFS in patients with PC. (A) Please see Figure 5A for the types of genomic alterations that occurred in APC and β-catenin in the TCGA data set (cBioPortal). Kaplan-Meier curve was constructed and log-rank test was performed using tools provided by cBioPortal. (B) Data were extracted from a subcohort from the MSKCC data set within the cBioPortal database [21] and analyzed for DFS using Kaplan-Meier curve and log-rank test. Total#, total number of cases; relap#, number of relapsed cases; MMDFS, median months disease-free survival.
The Nine-Gene Signature Stratifies Patients with High Risk of PC Recurrence and Is an Independent Risk Factor for PC Recurrence
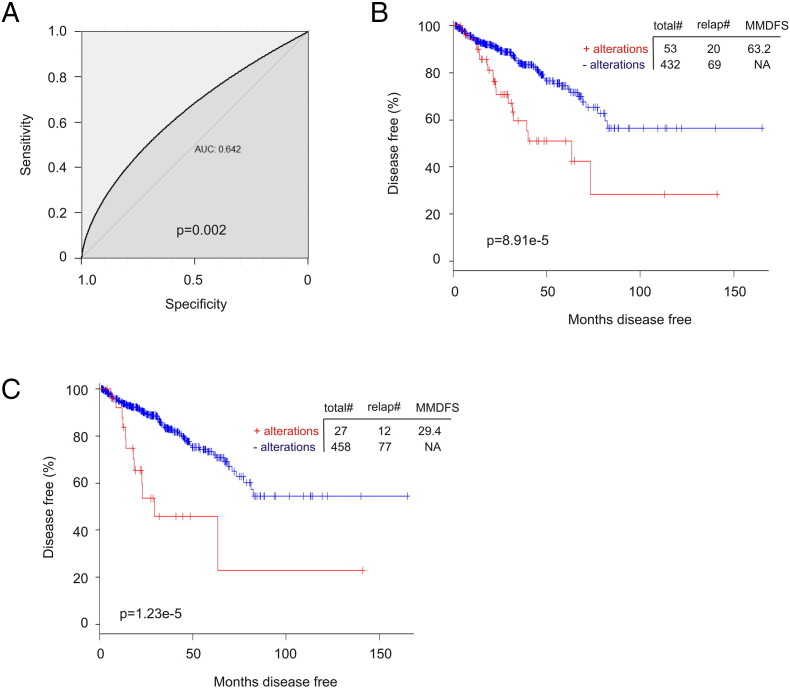
To examine whether the nine-gene signature stratify patients with elevated risk in PC recurrence, we scored each patients based on their nine-gene signature using ∑(fi)n (fi: Cox coefficient of genei, n = 9) (Supplementary Table 4). Scores derived from the signature have an AUC value of 0.64 (P = .002) in predicting PC recurrence (Supplementary Figure 7A). The median (≥0.657) and Q3 (≥1.089) scores classify patients into high- and low-risk group of PC recurrence (Supplementary Figure 7, B and C). While the 9-gene signature has DFS = 73.4 months (95% CI: 37.1-109.5, P = 5.557e-5), the median and Q3 scores (Supplementary Table 4) show respective DFS = 63.2 months (95% CI: 23.2-103.2, P = 8.91e-5) and 29.4 months (95% CI: 3.5-55.2, P = 1.23e-5).
Supplementary Figure 7.
The nine-gene signature stratifies high- and low-risk groups of prostate cancer patients with respect to recurrence. (A) Scores of the nine-gene signature were assigned to individual patients based on the formula ∑(fi)n, (fi: Cox coefficient of individual factors, n = 9) (see Supplementary Table 4 for details). ROC curve was constructed using the pROC package in R. The data set used is the TCGA within cBioPortal. (B, C) Based on the scores (Supplementary Table 4), the median (B) and Q3 scores (C) was used to stratify patients into high-risk (score positive) and low-risk groups (score negative), which were analyzed for the associated DFS times (Kaplan-Meier curve and log-rank test) using R statistics.
We further demonstrated the 9-gene signature being an independent risk factor for PC recurrence (HR = 1.731, 95% CI: 1.104-2.712, P = .0167) after adjusting for TMN tumor stage, age at diagnosis, radical prostatectomy (total) GS, and surgical margin (Table 5). Instead of total GS, we also analyzed the World Health Organization (WHO) PC grading system [WHO grade (group) I-V, see Supplementary Table 4 for details]; the signature continues to predict PC recurrence (HR = 1.769, 95% CI: 1.130-2.768, P = .0126) after adjusting for the WHO grades, TMN tumor stage, age, and surgical margin. Of note, the high-risk group of patients stratified by the median and Q3 scores is associated with an elevated and independent HR = 2.07 (95% CI: 1.245-3.450, P = .0051)/total GS or HR = 2.054 (95% CI: 1.234-3.417, P = .0056)/WHO grades and HR = 3.707 (95% CI: 1.949-7.052, P = 6.51e-5)/total GS or HR = 3.832 (95% CI: 2.015-7.286, P = 4.18e-5)/WHO grades, respectively. Additionally, by performing similar analysis on the nine genes individually, we observed a general worsening in individual HR and P values in multivariate Cox analysis compared to univariate analysis only in the eight MUC1 network genes (Supplementary Table 5), which validates the eight genes belonging to the MUC1 network.
Table 5.
Univariate and Multivariate Cox Analysis of the Nine-Gene Signature for PC Recurrence
| Univariate |
Multivariate |
||||||
|---|---|---|---|---|---|---|---|
| Clinical Variables | na | HR | 95% CI | P Value | HR | CI (95%) | P Value |
| The 9-gene sigb | |||||||
| 0 | 390 | ||||||
| 1 | 95 | 2.381 | 1.547-3.684 | 3.47e-5* | 1.731 | 1.104-2.712 | .0167* |
| Tumor stagec | |||||||
| ≤ T2 | 184 | ||||||
| T3 and T4 | 295 | 3.93 | 2.178-7.102 | 5.57e-6* | 1.919 | 0.994-3.683 | .0502 |
| Age at diagnosis | 485 | 1.025 | 0.993-1.058 | .128 | 1.002 | 0.970-1.036 | .8879 |
| RP GSd | 485 | 2.209 | 1.773-2.752 | 1.57e-12* | 1.874 | 1.430-2.360 | 1.93e-6* |
| Margin statuse | |||||||
| 0 | 208 | ||||||
| 1 | 147 | 2.162 | 1.418-3.296 | .00034* | 1.271 | 0.798-1.993 | .3203 |
Number of cases.
The nine-gene signature. 0: signature-negative; 1: signature-positive.
Six cases without stage information.
Radical prostatectomy Gleason score.
Surgical margin status: 0, R0; 1, R1 and R2; NA, case without surgical margin information and Rx (n = 30).
In view of the recent demonstration with respect to increased prostate cancer risk in men with germline mutations in BRCA1 and BRCA2 [31], we reasoned whether there is a relationship between the nine-gene signature and these mutations. Somatic mutations in BRCA1 (1 in 492) and BRCA2 (7 in 492) are not a frequent event in the TCGA cohort (Supplementary Figure 8). Of these mutations, the BRCA1 and three BRCA2 mutations co-occur with the nine-gene signature (Supplementary Figure 8). Importantly, addition of BRCA1 and BRCA2 mutations (positive cases 102/recurred cases 32, median DFS months 73.36; negative cases 383/recurred cases 57, median DFS months not reached; P = 1.099e-4) does not enhance the 9-gene's predictive potency (positive cases 96/recurred cases 31, median DFS months 73.36; negative cases 389/recurred cases 58, median DFS months not reached; P = 5.575e-5). Collectively, these observations provide indirect support for the importance of the intercomponent connections in the association of the nine-gene signature with reductions in DFS.
Supplementary Figure 8.
Relationship of the nine-gene signature with BRCA1 and BRCA2 mutations. The indicated types of genomic alterations for the nine genes, BRCA1, and BRCA2 in the TCGA data set (n = 492) within the cBioPortal database are shown; only the proportion of cohorts containing the genomic alterations are included. Each column is for individual tumor. DF, disease free; NA, not available.
Discussion
MUC1 expression is commonly altered in multiple tumor types [9], [11], [12] and promotes tumor progression through activation of the EGFR, β-catenin, NF-κB, PKM2, and other pathways [9], [13], [32]. MUC1 thus has applications in diagnosis and therapy. However, both applications need further investigations.
We provide the first evidence for elevations of the MUC1 proteins in CRPC (Figure 1, Figure 2). Of note, a recent phase II clinical trial reported that a MUC1-based dendritic cells (DC) vaccination delayed PC progression in patients with nonmetastatic CRPC [33]. Our study provides further support for DC-MUC1 vaccine in treating CRPC.
We also observed for the first time the extensive expression of MUC1 protein in hormone-naïve PC metastasized to bone (Figure 3, Table 1). Our research thus strongly suggests that targeting MUC1 should be considered either as a monotherapy or in combination with ADT in treating patients with hormone-naïve bone metastasis.
We further pioneered a thorough investigation of MUC1’s biomarker value in the context of a network setting, an effort supported by the need of multiple factors to effectively predict cancer progression. Our nine-gene signature associates robustly with PC recurrence. Currently, there are 3 sets of mRNA-based signatures to evaluate PC progression: the 17-gene Oncotype DX (GPS) [34], the 31-gene signature of CCP (cell cycle progression; Prolaris) [35], and the 22-gene Decipher signature [5], [6], [7], [36]. Adding to this resource is our novel nine-gene genomic signature. Interestingly, this signature also correlates with reductions in DFS and OS in patients with cutaneous melanoma (Figure 6).
In this signature, individual genes of the MUC1 network display overlaps or connections in association with PC recurrence evidenced by the worsening of their HR and P values in multivariate Cox analysis (Supplementary Table 5). These connections explain why MUC1 is not present in the signature gene list, as the eight genes mediate the functional contributions of MUC1. This scenario is supported by the multiple processes covered by the eight MUC1 network genes in the nine-gene list (Table 3), including tyrosine kinase signaling (LYN and ZAP70); signaling transduction (GRB2 and SOS1) [37]; protein glycosylation (GALNT10 and SIGLEC1), a process that contributes to PC progression [38]; and the Wnt pathway (APC and CTNNB1) (Table 3).
Mutations in APC are mutually exclusive from those occurring in β-catenin in colon cancer [28]. Nonetheless, their contributions to PC recurrence remain unknown. We demonstrate here that genomic alterations in APC and β-catenin associate with PC recurrence (Supplementary Figure 6). In addition to previously documented missense mutations located in the destruction box of β-catenin in PC, D32Y, S33F, and T41A [28], [30], we describe an array of missense mutations in D32, S33, S37, T41, and S45 in PC that has been reported in other tumor types, including hepatocellular carcinoma, colon cancer, ovarian cancer, medulloblastoma, uterine tumors [28], glomangiopericytoma [39], and desmoid-type fibromatosis [40], [41], [42]. T41A was detected in up to 64% of pediatric fibromatosis and contributes to the aggressiveness of the disease [43], [44]. Among three patients harboring T41A PC with follow-up data, two experienced disease progression (Table 4); it is intriguing whether, with a longer follow-up period, patient EJ-A65D will have PC recurrence (Table 4). It is thus tempting to propose a causative role of T41A in PC recurrence. Despite the fact that mutations in D32, S33, S37, T41, and S45 do not occur frequently in PC (Table 4), other mechanisms may be present to compensate for these mutations. Additionally, three new β-catenin mutations were detected on residues outside of the destruction box, N387K, W383G, and R225H, and they occurred together with PC recurrence and fatality. Although the impact of all β-catenin mutations detected here (Table 4) on PC oncogenesis needs additional studies, the observed mutual exclusiveness between APC mutations and β-catenin mutations strongly suggests their roles in PC promotion (Table 4).
The following are the supplementary data related to this article.
List of the MUC1 Network Genes and FAM84B Used in Variable Selection
Optimization of the Initial Selected 10 Genesa
Prediction of PC Recurrence Using the Nine-Gene Signature
Events Analyzed by the Cox Model
Univariate and Multivariate Cox Regression Analysis of Individual Genes within the Nine-Gene List for PC Recurrence
Acknowledgments
Acknowledgements
The results shown here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). We like to thank Dr. Nicholas Wong for providing tissues derived from prostate-specific PTEN−/− mice. X. L. is a recipient of Chinese Government Award for Outstanding Self-Financed Student Abroad. This work was supported in part by a GAP funding from McMaster University and St. Joseph's Hospital in Hamilton, a grant from the Cancer Research Society to D. T., as well as grants from The Plan Project of Shenzhen Science and Technology (grant no. JCYJ20150403094227974) and the Natural Science Foundation of Guangdong Province (grant No. 2014A030313749; grant no. 2013010016559) to F.W.
Acknowledgments
Conflict of Interest Statement
All authors declare no conflict of interest.
Contributor Information
Huixiang Yang, Email: haowei727499@163.com.
Pierre Major, Email: majorp@HHSC.ca.
Damu Tang, Email: damut@mcmaster.ca.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Zaorsky NG, Raj GV, Trabulsi EJ, Lin J, Den RB. The dilemma of a rising prostate-specific antigen level after local therapy: what are our options? Semin Oncol. 2013;40:322–336. doi: 10.1053/j.seminoncol.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Semenas J, Allegrucci C, Boorjian SA, Mongan NP, Persson JL. Overcoming drug resistance and treating advanced prostate cancer. Curr Drug Targets. 2012;13:1308–1323. doi: 10.2174/138945012802429615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T, Zattoni F. EAU guidelines on prostate cancer. Part II: treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur Urol. 2014;65:467–479. doi: 10.1016/j.eururo.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Martin NE. New developments in prostate cancer biomarkers. Curr Opin Oncol. 2016;28:248–252. doi: 10.1097/CCO.0000000000000279. [DOI] [PubMed] [Google Scholar]
- 6.McGrath S, Christidis D, Perera M, Hong SK, Manning T, Vela I, Lawrentschuk N. Prostate cancer biomarkers: are we hitting the mark? Prostate Int. 2016;4:130–135. doi: 10.1016/j.prnil.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel KM, Gnanapragasam VJ. Novel concepts for risk stratification in prostate cancer. J Clin Urol. 2016;9:18–23. doi: 10.1177/2051415816673502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apostolopoulos V, Stojanovska L, Gargosky SE. MUC1 (CD227): a multi-tasked molecule. Cell Mol Life Sci. 2015;72:4475–4500. doi: 10.1007/s00018-015-2014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Paula Peres L, da Luz FA, Dos Anjos Pultz B, Brigido PC, de Araujo RA, Goulart LR, Silva MJ. Peptide vaccines in breast cancer: the immunological basis for clinical response. Biotechnol Adv. 2015;33:1868–1877. doi: 10.1016/j.biotechadv.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Wurz GT, Kao CJ, Wolf M, DeGregorio MW. Tecemotide: an antigen-specific cancer immunotherapy. Hum Vaccin Immunother. 2014;10:3383–3393. doi: 10.4161/hv.29836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16:467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Cozzi PJ, Wang J, Delprado W, Perkins AC, Allen BJ, Russell PJ, Li Y. MUC1, MUC2, MUC4, MUC5AC and MUC6 expression in the progression of prostate cancer. Clin Exp Metastasis. 2005;22:565–573. doi: 10.1007/s10585-005-5376-z. [DOI] [PubMed] [Google Scholar]
- 15.Rabiau N, Dechelotte P, Guy L, Satih S, Bosviel R, Fontana L, Kemeny JL, Boiteux JP, Bignon YJ, Bernard-Gallon D. Immunohistochemical staining of mucin 1 in prostate tissues. In Vivo. 2009;23:203–207. [PubMed] [Google Scholar]
- 16.Arai T, Fujita K, Fujime M, Irimura T. Expression of sialylated MUC1 in prostate cancer: relationship to clinical stage and prognosis. Int J Urol. 2005;12:654–661. doi: 10.1111/j.1442-2042.2005.01112.x. [DOI] [PubMed] [Google Scholar]
- 17.Papadopoulos I, Sivridis E, Giatromanolaki A, Koukourakis MI. Tumor angiogenesis is associated with MUC1 overexpression and loss of prostate-specific antigen expression in prostate cancer. Clin Cancer Res. 2001;7:1533–1538. [PubMed] [Google Scholar]
- 18.Eminaga O, Wei W, Hawley SJ, Auman H, Newcomb LF, Simko J, Hurtado-Coll A, Troyer DA, Carroll PR, Gleave ME. MUC1 expression by immunohistochemistry is associated with adverse pathologic features in prostate cancer: a multi-institutional study. PLoS One. 2016;11:e0165236. doi: 10.1371/journal.pone.0165236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durrani N, Waldron M, Chae C, Harewood L, Frydenberg M, Pedersen J, Mills J. Assessing expression of MUC1 and ZAG protein biomarkers in prostate biopsies improves prediction of adverse pathology following radical prostatectomy The Open Prostate. Cancer J. 2015;8:1–9. [Google Scholar]
- 20.Severi G, FitzGerald LM, Muller DC, Pedersen J, Longano A, Southey MC, Hopper JL, English DR, Giles GG, Mills J. A three-protein biomarker panel assessed in diagnostic tissue predicts death from prostate cancer for men with localized disease. Cancer Med. 2014;3:1266–1274. doi: 10.1002/cam4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong N, Major P, Kapoor A, Wei F, Yan J, Aziz T, Zheng M, Jayasekera D, Cutz JC, Chow MJ. Amplification of MUC1 in prostate cancer metastasis and CRPC development. Oncotarget. 2016;7:83115–83133. doi: 10.18632/oncotarget.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong N, Gu Y, Kapoor A, Lin X, Ojo D, Wei F, Yan J, de Melo J, Major P, Wood G. Upregulation of FAM84B during prostate cancer progression. Oncotarget. 2017 doi: 10.18632/oncotarget.15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6 doi: 10.1126/scisignal.2004088. [pp. l1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 28.Polakis P. The oncogenic activation of beta-catenin. Curr Opin Genet Dev. 1999;9:15–21. doi: 10.1016/s0959-437x(99)80003-3. [DOI] [PubMed] [Google Scholar]
- 29.Fu Y, Zheng S, An N, Athanasopoulos T, Popplewell L, Liang A, Li K, Hu C, Zhu Y. Beta-catenin as a potential key target for tumor suppression. Int J Cancer. 2011;129:1541–1551. doi: 10.1002/ijc.26102. [DOI] [PubMed] [Google Scholar]
- 30.Voeller HJ, Truica CI, Gelmann EP. Beta-catenin mutations in human prostate cancer. Cancer Res. 1998;58:2520–2523. [PubMed] [Google Scholar]
- 31.Lecarpentier J, Silvestri V, Kuchenbaecker KB, Barrowdale D, Dennis J, McGuffog L, Soucy P, Leslie G, Rizzolo P, Navazio AS. Prediction of breast and prostate cancer risks in male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J Clin Oncol. 2017;35:2240–2250. doi: 10.1200/JCO.2016.69.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 33.Scheid E, Major P, Bergeron A, Finn OJ, Salter RD, Eady R, Yassine-Diab B, Favre D, Peretz Y, Landry C. Tn-MUC1 DC vaccination of Rhesus macaques and a phase I/II trial in patients with nonmetastatic castrate-resistant prostate cancer. Cancer Immunol Res. 2016;4:881–892. doi: 10.1158/2326-6066.CIR-15-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knezevic D, Goddard AD, Natraj N, Cherbavaz DB, Clark-Langone KM, Snable J, Watson D, Falzarano SM, Magi-Galluzzi C, Klein EA. Analytical validation of the Oncotype DX prostate cancer assay — a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. doi: 10.1186/1471-2164-14-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuzick J, Berney DM, Fisher G, Mesher D, Moller H, Reid JE, Perry M, Park J, Younus A, Gutin A. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106:1095–1099. doi: 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein EA, Haddad Z, Yousefi K, Lam LL, Wang Q, Choeurng V, Palmer-Aronsten B, Buerki C, Davicioni E, Li J. Decipher genomic classifier measured on prostate biopsy predicts metastasis risk. Urology. 2016;90:148–152. doi: 10.1016/j.urology.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Ichaso N, Dilworth SM. Cell transformation by the middle T-antigen of polyoma virus. Oncogene. 2001;20:7908–7916. doi: 10.1038/sj.onc.1204859. [DOI] [PubMed] [Google Scholar]
- 38.Munkley J, Vodak D, Livermore KE, James K, Wilson BT, Knight B, McCullagh P, McGrath J, Crundwell M, Harries LW. Glycosylation is an androgen-regulated process essential for prostate cancer cell viability. EBioMedicine. 2016;8:103–116. doi: 10.1016/j.ebiom.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lasota J, Felisiak-Golabek A, Aly FZ, Wang ZF, Thompson LD, Miettinen M. Nuclear expression and gain-of-function beta-catenin mutation in glomangiopericytoma (sinonasal-type hemangiopericytoma): insight into pathogenesis and a diagnostic marker. Mod Pathol. 2015;28:715–720. doi: 10.1038/modpathol.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tejpar S, Nollet F, Li C, Wunder JS, Michils G, dal Cin P, Van Cutsem E, Bapat B, van Roy F, Cassiman JJ. Predominance of beta-catenin mutations and beta-catenin dysregulation in sporadic aggressive fibromatosis (desmoid tumor) Oncogene. 1999;18:6615–6620. doi: 10.1038/sj.onc.1203041. [DOI] [PubMed] [Google Scholar]
- 41.Huss S, Nehles J, Binot E, Wardelmann E, Mittler J, Kleine MA, Kunstlinger H, Hartmann W, Hohenberger P, Merkelbach-Bruse S. Beta-catenin (CTNNB1) mutations and clinicopathological features of mesenteric desmoid-type fibromatosis. Histopathology. 2013;62:294–304. doi: 10.1111/j.1365-2559.2012.04355.x. [DOI] [PubMed] [Google Scholar]
- 42.Meazza C, Belfiore A, Busico A, Settanni G, Paielli N, Cesana L, Ferrari A, Chiaravalli S, Massimino M, Gronchi A. AKT1 and BRAF mutations in pediatric aggressive fibromatosis. Cancer Med. 2016;5:1204–1213. doi: 10.1002/cam4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang WL, Nero C, Pappo A, Lev D, Lazar AJ, Lopez-Terrada D. CTNNB1 genotyping and APC screening in pediatric desmoid tumors: a proposed algorithm. Pediatr Dev Pathol. 2012;15:361–367. doi: 10.2350/11-07-1064-OA.1. [DOI] [PubMed] [Google Scholar]
- 44.Bo N, Wang D, Wu B, Chen L, Ruixue M. Analysis of beta-catenin expression and exon 3 mutations in pediatric sporadic aggressive fibromatosis. Pediatr Dev Pathol. 2012;15:173–178. doi: 10.2350/10-07-0866-OA.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the MUC1 Network Genes and FAM84B Used in Variable Selection
Optimization of the Initial Selected 10 Genesa
Prediction of PC Recurrence Using the Nine-Gene Signature
Events Analyzed by the Cox Model
Univariate and Multivariate Cox Regression Analysis of Individual Genes within the Nine-Gene List for PC Recurrence