Abstract
The strong societal urge to reduce the use of experimental animals, and the biological differences between rodent and human skin, have led to the development of alternative models for healthy and diseased human skin. However, the limited availability of primary keratinocytes to generate such models hampers large-scale implementation of skin models in biomedical, toxicological, and pharmaceutical research. Immortalized cell lines may overcome these issues, however, few immortalized human keratinocyte cell lines are available and most do not form a fully stratified epithelium. In this study we compared two immortalized keratinocyte cell lines (N/TERT1, N/TERT2G) to human primary keratinocytes based on epidermal differentiation, response to inflammatory mediators, and the development of normal and inflammatory human epidermal equivalents (HEEs). Stratum corneum permeability, epidermal morphology, and expression of epidermal differentiation and host defence genes and proteins in N/TERT-HEE cultures was similar to that of primary human keratinocytes. We successfully generated N/TERT-HEEs with psoriasis or atopic dermatitis features and validated these models for drug-screening purposes. We conclude that the N/TERT keratinocyte cell lines are useful substitutes for primary human keratinocytes thereby providing a biologically relevant, unlimited cell source for in vitro studies on epidermal biology, inflammatory skin disease pathogenesis and therapeutics.
Introduction
In vitro cultured 3D human skin equivalents (HSEs) and human epidermal equivalents (HEEs) (also known as organotypic skin models) are nowadays often used for studies in the fields of cell biology, tissue engineering, dermatology, and toxicology. These 3D skin models are preferred over the conventional monolayer cultures as they faithfully mimic the in vivo epidermis and skin barrier function and thereby serve as alternatives for experimental animal testing1. To meet the growing demand for these in vitro 3D human organotypic skin cultures, large quantities of primary keratinocytes are required. Human primary keratinocytes are preferably isolated from adult skin excised during plastic surgery (e.g. abdominal wall or breast reduction). Unfortunately, the availability of donor skin is limited and the short in vitro lifespan and inter-donor variation are disadvantages of the use of human primary keratinocytes. Immortalized keratinocyte cell lines could therefore provide an unlimited cell source and solution to the aforementioned issues.
The HaCaT cell line, a spontaneously immortalized human keratinocyte cell line, is widely used in keratinocyte monolayer culture models and also described for the development of organotypic skin models2. HaCaT cells respond to differentiation-promoting stimuli, such as contact inhibition and high calcium concentrations in the culture medium2–4. However, the transcriptional expression pattern of cornified envelope-associated proteins, such as filaggrin, loricrin and involucrin is abnormal compared to normal human primary keratinocytes5,6 and karyotyping of HaCaT cells shows aneuploidy5. Wound regeneration and eczematous organotypic models with HaCaT cells have been reported6–10, however, epidermal stratification is abnormal, aberrant epidermal differentiation protein expression is observed and a stratum corneum is lacking in these HaCaT-derived 3D skin models.
In 2000, the Rheinwald laboratory developed two immortalized keratinocyte cell lines, called the N/TERT cell lines (N/TERT1 and N/TERT2G), generated by transduction of human primary keratinocytes with the human telomerase reverse transcriptase (hTERT) gene and by the spontaneous loss of the pRB/p16INK4a cell cycle control mechanism11. This cell line was shown to have normal differentiation characteristics in monolayer and organotypic skin models. Remarkably, the use of the HaCaT cell line prevailed in experimental dermatological research and to date a limited number of studies have used N/TERT keratinocytes. (See Table 1 for studies using organotypic skin models and Supplemental Table 1 for studies using N/TERT cells in conventional monolayer cultures). Unfortunately, a direct comparison of N/TERT keratinocytes to primary human keratinocytes is lacking and this may have hampered the wide implementation of the N/TERT keratinocyte cell line in biomedical research. In addition, in toxicological research, the HEE models: SkinEthic26, EpiDerm27, and EPISKIN28 are accepted alternatives for skin irritation and corrosion testing. Herein, the N/TERT cell line could be a valuable substitute for primary keratinocytes, however studies on N/TERT cells in HEE models are scarce17,18,21–23 and a thorough characterization with regard to epidermal differentiation and response to inflammatory mediators is lacking.
Table 1.
Studies using in vitro 3D reconstructed skin generated from N/TERT keratinocytes.
| 3D skin model | Dermal matrix | F | Epidermal morphology | Field of study | Reference |
|---|---|---|---|---|---|
| HSE | Collagen type I | Y | Stratified epidermis, sc present | Cell line development | Dickson et al.11 $ |
| HSE | Collagen type I | Y | Stratified epidermis, sc present | Cell line development | Rheinwald et al.12 |
| HSE | DED | Y | Stratified epidermis, sc present | Epidermal biology | Wan et al.6 |
| HSE | DED | Y | Stratified epidermis, sc present | Skin barrier | Man et al.13 |
| HSE | Collagen type I | Y | Stratified epidermis, sc present | Epidermal biology | Dabelsteen et al.14 |
| HEE | None | N | Stratified epidermis, sc present | UV radiation, DNA damage | Bertrand-Vallery et al.15 |
| HSE | Collagen type I | N | Stratified epidermis, sc present | Human papillomavirus | Lazic et al.16 |
| HEE | None | N | Stratified epidermis, sc present | Epidermal biology | Robertson et al.17 |
| HSE | Collagen type I | Y | Stratified epidermis, sc present | Skin barrier | Van Drongelen et al.18 |
| HSE | Collagen type I | Y | Multilayer of cells, sc lacking | UV radiation, DNA damage | Harrison et al.19 |
| HSE | Collagen type I | Y | Multilayer of cells, sc present | Ionizing radiation, DNA damage | Acheva et al.20 |
| HSE | Collagen type I | Y | Stratified epidermis, sc present | Skin barrier | Van Drongelen et al.21 |
| HEE | None | N | Stratified epidermis, sc present | Epidermal biology | Van Drongelen et al.22 |
| HEE | None | N | Stratified epidermis, sc present | Skin sensitization | Alloul-Ramdhani et al.23 |
| HSE | Matriderm | Y | Stratified epidermis, sc present | Epidermal biology | Reijnders et al.24 |
| HSE | Collagen type I | Y | Stratified epidermis, sc present | Ionizing radiation, inflammation | Acheva et al.25 |
$Initial paper describing the N/TERT cell line development; HSE: human skin equivalent; HEE: human epidermal equivalent; DED: de-epidermized dermis; F: Fibroblasts present Yes/No; UV: ultraviolet; sc: stratum corneum.
Earlier studies by our group and others showed that the addition of T-helper-(Th)1 or Th2 cytokines to 3D skin models induce a phenotype in keratinocytes that is similar to psoriasis (PS) or atopic dermatitis (AD), respectively29–31. PS and AD are highly prevalent multifactorial chronic inflammatory skin diseases of which PS is considered to be Th1-Th17 immune cell mediated, while in AD the Th2 cells and cytokines prevail. Current treatment is often aimed at general immunosuppression, which is associated with side effects. Therefore, an unmet medical need exists for novel targeted, therapeutic approaches. In vitro inflammatory skin disease models are considered valuable tools to study disease pathogenesis and can be used in a pre-clinical drug screening setting. However, the large-scale production and implementation of these models is lacking and this may be due to the limited source of primary keratinocytes. The N/TERT keratinocyte cell lines could provide a solution herein, but only if their response to inflammatory mediators such as Th1, Th2, and Th17 cytokines is similar to that of primary keratinocytes.
To illustrate the potential of N/TERT keratinocytes for studies on epidermal biology, inflammatory skin disease pathogenesis and therapeutics, we compared the mRNA and protein expression of two N/TERT keratinocyte cell lines to that of human primary epidermal keratinocytes in monolayer cultures. Next, we generated a N/TERT-HEE model and characterized mRNA and protein expression, and physical barrier properties of this 3D epidermal model by a comparison to HEEs generated from primary keratinocytes. PS-associated (Th1 and Th17) or AD-associated (Th2) cytokines were used to generate 3D human skin inflammation models and we validated disease markers in both models by known anti-inflammatory drugs.
Results
Karyotyping and short tandem repeat analysis of N/TERT keratinocytes
The N/TERT1 and N/TERT2G keratinocytes were karyotyped to assess their ploidy according to standard procedures. N/TERT2G is diploid (46, XY) and N/TERT1 is diploid with an additional chromosome 20 (47, XY, +20) (Supplemental Fig. 1). To assess potential contamination with other cells, short tandem repeat (STR) analysis was performed by quantitative fluorescence (QF)-PCR using 21 specific targets on chromosome 13, 18, 21, and both sex chromosomes X and Y (Supplemental Fig. 2). All targets, except D18S391, are informative and rule out cellular contamination. Only N/TERT1 is shown, as both N/TERT keratinocytes showed identical results.
Differentiation kinetics and dynamics of N/TERT keratinocytes in conventional cultures
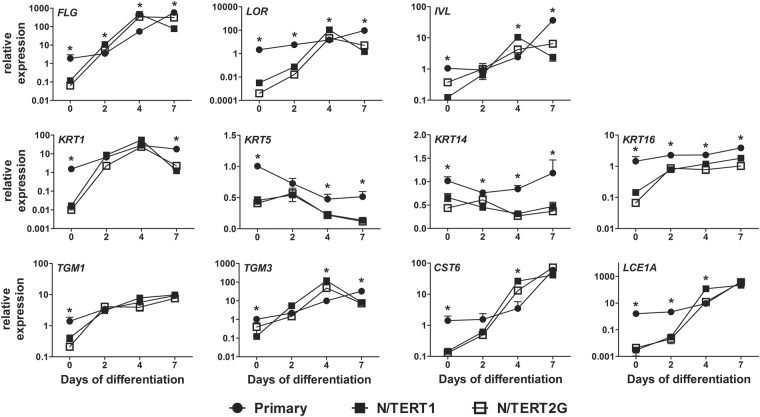
To evaluate the differentiation dynamics of the N/TERT1 and N/TERT2G keratinocytes we performed a time course and compared N/TERT keratinocytes to primary human keratinocytes. We analysed epidermal differentiation gene expression levels in monolayer cultures up to seven days of differentiation. Relative expression levels of N/TERT keratinocytes were normalized to primary keratinocytes, giving an insight into the basal expression levels and expression patterns of N/TERT keratinocytes at different time points (Fig. 1, t = 0, 2, 4 and 7 days of differentiation). At confluency, which is the start of the differentiation phase (t = 0) due to cell-cell contact inhibition, the basal expression level of all investigated epidermal genes was significantly lower in N/TERT keratinocytes (terminal differentiation genes: filaggrin (FLG), loricrin (LOR), involucrin (IVL), late cornified envelope (LCE) 1A; keratins (KRT): KRT1, KRT5, KRT14, KRT16; and genes involved in cornification and desquamation: transglutaminase (TGM) 1, TGM3, cystatin M/E (CST6)). Expression levels increased at day 2 and at day 4 the expression of FLG, LOR, IVL, TGM3, CST6, LCE1A was significantly higher in the N/TERT keratinocytes as compared to primary keratinocytes, indicating a significantly stronger induction rate of N/TERT keratinocytes from day 0 to day 4. The N/TERT keratinocyte gene expression levels reached a maximum at day 4 and dropped below the primary keratinocyte levels at day 7. Primary keratinocytes tend to reach their highest expression levels not earlier than at day 7. These results indicate that the N/TERT keratinocytes respond very strongly to differentiation stimuli and reach similar expression levels of terminal differentiation genes as primary keratinocytes, albeit at an earlier time point in culture.
Figure 1.
N/TERT keratinocytes express terminal differentiation genes. mRNA expression of epidermal differentiation genes by N/TERT keratinocyte monolayer cultures (N = 3 for N/TERT 1 and N/TERT2G) were compared to primary keratinocytes (N = 6 donors). Bars represent mean ± SEM. *p < 0.05 relative to primary keratinocyte at the same time point.
Human epidermal equivalents can be generated from human N/TERT keratinocytes
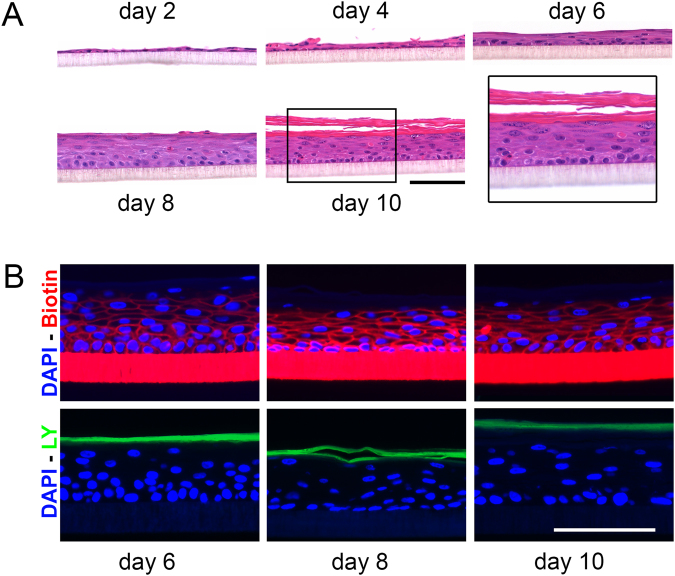
To study the potential of N/TERT keratinocytes for the implementation in 3D skin model development, we optimised N/TERT HEE culture protocols (Supplemental Fig. 3) and studied the epidermal stratification and functional characteristics herein. Briefly, using K-SFM medium (as compared to DMEM which is used in primary keratinocyte HEE cultures) during the proliferation phase of the culture period and CnT-PR-3D medium supplemented with DMEM (60:40) in the differentiation phase resulted in most optimal epidermal morphology (Supplemental Fig. 4A). Furthermore, the seeding of approximately 1·105 N/TERT cells (Supplemental Fig. 4B) directly from liquid nitrogen (passage 0) or cultured cells until passage 4 (Supplemental Fig. 4C) generated high quality HEEs where N/TERT keratinocytes generated a multilayered epidermis with presence of a stratum granulosum and a basket weave stratum corneum in approximately 10 days of air-exposed culture (Fig. 2A). Using immunohistochemistry we studied the expression of epidermal differentiation protein FLG, the expression of the proliferation marker Ki67, and the expression of anti-microbial peptides human beta defensin 2 (hBD2), and skin-derived antileukoprotease (SKALP). We compared the expression patterns with HEE cultures generated from primary human keratinocytes and found highly similar protein expression patterns, apart from a lower expression of FLG and stronger expression of SKALP in the N/TERT-HEEs (Supplemental Fig. 4D,E). To analyse the stratum corneum barrier characteristics of N/TERT keratinocyte-based 3D HEEs we performed ‘outside-in’ (Lucifer Yellow (LY)) and ‘inside-out’ (EZ-Link Sulfo-NHS-LC-LC-Biotin) tracer penetration assays, as previously reported32. We found restricted dye penetration from day 6 of the air-liquid interphase culture onwards (Fig. 2B). These findings correspond to data obtained with HEEs generated from primary human keratinocytes32.
Figure 2.
N/TERT1 keratinocytes are suitable for culture in HEE models. (A) Haematoxylin Eosin (HE) staining of N/TERT1 keratinocytes in the HEE model system while developing from day 2 to day 10 of air exposure. (B) Epidermal barrier properties of the HEE constructs by biotin penetration (red) and Lucifer yellow penetration (green). Scale bar = 100 µm.
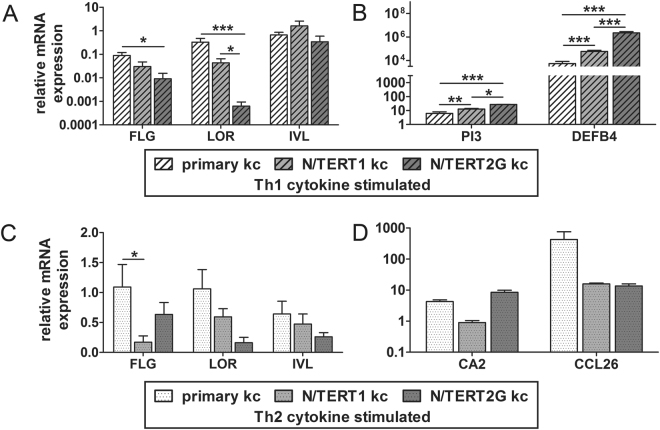
Human N/TERT and primary keratinocytes respond similar to pro-inflammatory cytokine stimulation in conventional monolayer cultures
Next, the expression levels of terminal differentiation genes and inflammatory disease marker genes were analysed in monolayer N/TERT keratinocytes that were stimulated with pro-inflammatory Th1 or Th2 cytokines. Upon stimulation with Th1 cytokines (TNFα, IFNγ, and IL-1α) the expression levels of FLG and LOR decreased (normalized to unstimulated keratinocytes) and in particular in N/TERT2G keratinocytes this downregulation was significantly stronger as compared to primary keratinocytes (Fig. 3A). The expression of psoriasis-related host defence genes PI3 and DEFB4 33–35 (encoding SKALP/elafin and hBD2, respectively) was increased in both primary and N/TERT keratinocytes, but N/TERT keratinocytes showed a significantly stronger upregulation and this was most pronounced in N/TERT2G cultures (Fig. 3B). After stimulation with Th2-cytokines (IL-4 and IL-13), we observed a minor decrease (only statistically significant for FLG) in FLG, LOR and IVL expression in N/TERT keratinocytes (Fig. 3C), while markers for AD, C-C motif chemokine ligand 26 (CCL26) and carbonic anhydrase 2 (CA2)30,36, were found to be upregulated (Fig. 3D). Overall, the N/TERT keratinocytes appear more responsive to Th1 cytokine stimulation than primary keratinocytes. Upon addition of Th2 cytokines, N/TERT1 keratinocytes express less CA2 than primary keratinocytes and N/TERT2G keratinocytes. CCL26 is upregulated by both N/TERT cell lines and the primary keratinocytes upon Th2 cytokine addition, albeit a stronger induction rate of primary keratinocytes. These differences are, however, not statistically significant due to the high interdonor variability of the primary keratinocytes.
Figure 3.
N/TERT keratinocytes and primary keratinocytes respond similar to pro-inflammatory cytokine stimulation in conventional monolayer cultures. (A) Terminal differentiation genes and (B) psoriasis marker mRNA expression of monolayer N/TERT keratinocytes and primary keratinocytes after stimulation with Th1 cytokines (IL-1α, TNFα, and IFNγ). (C) Terminal differentiation genes and (D) AD marker mRNA expression of monolayer N/TERT keratinocytes and primary keratinocytes after stimulation with Th2 cytokines (IL-4 and IL-13). Bars represent mean ± SEM. *p < 0.05 **p < 0.01 ***p < 0.001 relative to primary keratinocytes.
Psoriasis-like HEE model (PS-HEE) generated from N/TERT keratinocytes
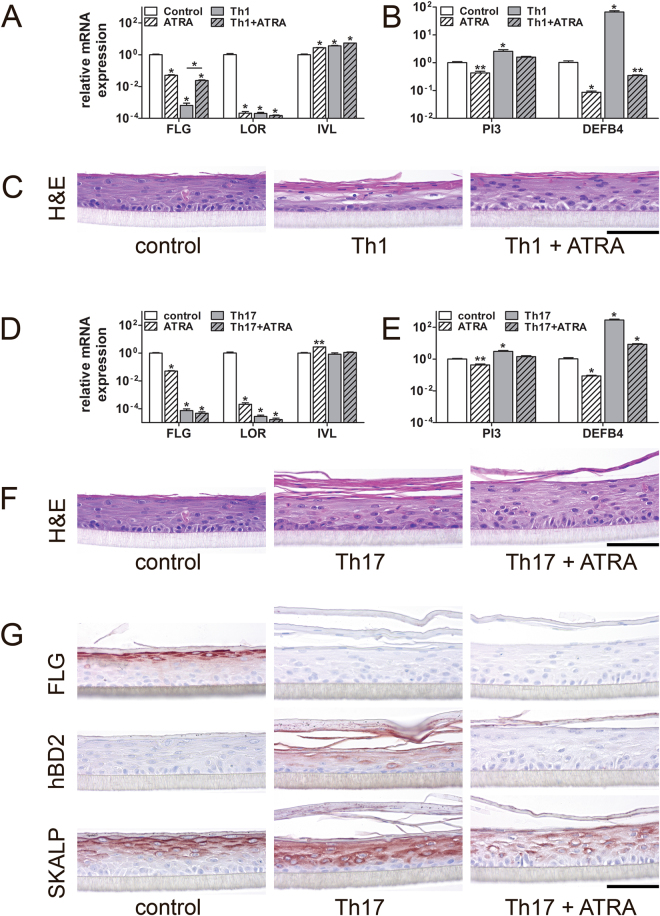
We explored the potential of N/TERT keratinocytes for generating in vitro inflammatory skin models for psoriasis by the addition of pro-inflammatory cytokines to N/TERT-HEEs. We added psoriasis-associated cytokines of Th1 (TNFα, IL-6, and IL-1α) and Th17 (IL-17 and IL-22) origin to N/TERT1 HEEs during the last three days of the air-liquid interphase culture. We furthermore aimed to rescue the disease phenotype by the addition of all trans retinoic acid (ATRA), an anti-psoriatic drug, to the culture medium supplemented with Th1 or Th17 cytokines as previously described for primary keratinocyte 3D skin models31,37.
Upon Th1 and Th17 cytokine stimulation we found a strong and significant decrease in the expression of FLG and LOR, while IVL expression was upregulated by Th1 cytokines (Fig. 4A,B). ATRA treatment successfully rescued FLG expression levels after the Th1 cytokine stimulation, but also significantly altered FLG, LOR and IVL expression alone. Both Th1 and Th17 cytokines significantly induced the expression of the host defence genes PI3 and DEFB4 (Fig. 4B,E). This closely resembles the transcriptional expression patterns that are found in psoriasis lesions in vivo 38–40. ATRA treatment reduced the expression of PI3 and DEFB4 in Th1 and Th17 cytokine stimulated N/TERT-HEEs to levels that are close to the control situation (Fig. 4B,E).
Figure 4.
N/TERT1 keratinocytes are suitable to generate a psoriasis-like HEE (PS-HEE) disease model. (A,B) N/TERT HEEs were harvested for mRNA expression analysis and (C) morphological analysis after stimulation with Th1 cytokines (TNFα, IL-6, and IL-1α) and rescued by addition of all trans retinoic acid (ATRA). (D,E) N/TERT HEEs were harvested for mRNA expression analysis and (F,G) morphological analysis after stimulation with Th17 cytokines (IL-17 and IL-22) and rescued by addition of all trans retinoic acid (ATRA). Images are representative of N = 3 N/TERT HEE experiments. Bars represent mean ± SEM. *p < 0.001 **p < 0.01 relative to control unstimulated keratinocytes. Scale bar = 100 µm.
Th1 and Th17 cytokines affected the epidermal morphology as haematoxylin eosin stainings showed parakeratosis (retained nuclei in the stratum corneum), thickening of the stratum corneum and the absence of a stratum granulosum in these cultures (Fig. 4C,F), all features of psoriatic skin. Treatment with ATRA improved the epidermal morphology in both cytokine models (Fig. 4C,F). The changes in epidermal morphology were accompanied by lowered protein expression levels of the terminal differentiation protein FLG, while psoriasis markers hBD2 and SKALP were upregulated. ATRA treatment lowered hBD2 and SKALP expression but did not restore FLG protein levels (Fig. 4G and Supplemental Fig. 5). These results indicate that N/TERT cells can be used to generated PS-HEEs and we were able to validate the model by the treatment with a known anti-psoriatic drug.
Atopic dermatitis-like HEE model (AD-HEE) generated from N/TERT keratinocytes
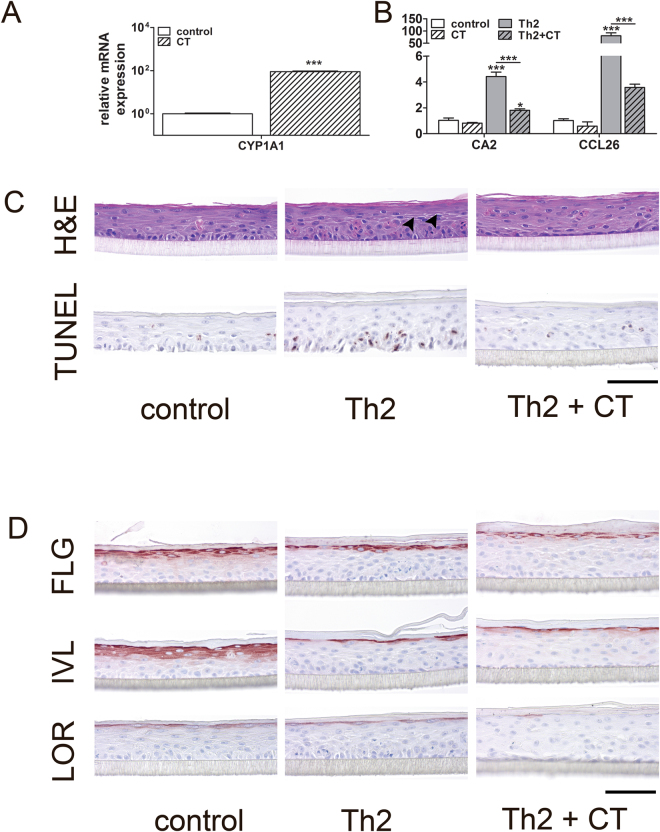
We used the Th2 cytokines IL-4 and IL-13 to induce histopathological and molecular hallmarks of AD. These include acanthosis, spongiosis, and apoptosis41–43 and disease-associated gene and protein expression patterns in HEEs generated from N/TERT1 and N/TERT2G keratinocytes. To explore the potential of the N/TERT atopic dermatitis-like inflammatory HEE model (AD-HEE) model for pre-clinical drug screening, we used coal tar as a therapeutic model agent. Coal tar therapy is an ancient treatment for AD and psoriasis44,45 and we recently uncovered its molecular mechanism of action, namely activation of the aryl hydrocarbon receptor (AHR)46. The well known AHR target gene, cytochrome p450 family member 1A1 (CYP1A1)47 is strongly induced by primary keratinocytes after coal tar exposure48 and N/TERT keratinocytes respond in a similar manner (Fig. 5A, Supplemental Fig. 6A). Other AHR target genes like 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) inducible poly(ADP-ribose) polymerase (TIPARP), cytochrome p450 family member 1B1 (CYP1B1), aryl hydrocarbon receptor repressor (AHRR) (Supplemental Fig. 6A) and epidermal differentiation genes, IVL and HRNR (Supplemental Fig. 6B) were also induced in N/TERT keratinocytes after coal tar stimulation while the expression of AD-related genes30,49 CCL26 and CA2 were significantly upregulated in the N/TERT AD-HEE (Fig. 5B). Epidermal morphology of N/TERT AD-HEEs showed signs of spongiosis in the suprabasal layers (intercellular oedema, seen as white spaces between adjacent cells in the HE staining) (Fig. 5C, panel HE, arrowheads) and apoptotic basal cells (Fig. 5C, panel TUNEL). Coal tar treatment reduced the histopathological AD hallmarks but did not rescue the Th2-cytokine mediated downregulation of epidermal differentiation proteins, FLG, IVL and LOR (Fig. 5D).
Figure 5.
N/TERT1 keratinocytes can be used to generate a atopic dermatitis-like HEE (AD-HEE) disease model. (A) mRNA expression of N/TERT1 HEEs after coal tar stimulation. Activation of the aryl hydrocarbon receptor (AHR) is measured via the induction of the AHR target gene, CYP1A1. (B) N/TERT HEEs were harvested for mRNA and (C) morphological analysis after stimulation with Th2 cytokines (IL-4 and IL-13) and rescued by addition of coal tar extract. (D) Protein expression was visualised by immunohistochemistry. Bars represent mean ± SEM. *p < 0.05 **p < 0.01 ***p < 0.001. Scale bar = 100 µm.
Discussion
Immortalized human cell lines have been instrumental for scientists to validate data obtained in animal models and for the translation to human biology and disease. Yet, the demand for complex tissue-like cell culture models to better represent the in vivo situation is ever rising. In skin research, organotypic 3D skin models are nowadays considered the gold standard, however, the use of primary human keratinocytes is herein required to obtain optimal tissue morphology. High costs for commercially available primary cells and the large quantity of cells needed to generate 3D skin models have directed research groups to isolate primary keratinocytes from surplus skin. Unfortunately, the availability of donor skin is limited and high inter-donor variability requires a multitude of biological replicates for a good reproducibility of results. Immortalized N/TERT keratinocytes are diploid (Supplemental Figs 1 and 2) with the exception of an additional chromosome 20 in the N/TERT1 keratinocyte cell line. A gain of chromosome 20 is related to increased proliferative potential and often observed in immortalized cells and in several cancer cell lines48,50–54. Nevertheless, both N/TERT keratinocytes mimic major characteristics of primary human keratinocytes and have superior qualities over the most widely used HaCaT (aneuploid5) keratinocyte cell line with regard to epidermal differentiation and the formation of a stratified epithelium. The herein provided literature overview of research utilizing N/TERT keratinocytes (Table 1 and Supplemental Table 1) enables others to determine the suitability of the N/TERT keratinocytes for their research.
Besides the use of HaCaT cells, the use of the Rho kinase inhibitor, Y-27632, was demonstrated to generate immortalized epithelial cell lines55. Although it is possible to generate immortalized primary foreskin keratinocytes, and to prolong the life span of adult keratinocytes through the use of Y-2763255,56, cells require the inhibitor throughout the cell culture. Since Rho kinases are vital for many cellular processes, potential off-target effects may occur. In addition, the adult keratinocytes are not stably immortalized by the Rho kinase inhibition and senescence occurs after five to eight passages. Therefore, this method seems less suitable for experiments that require extensive cell handling, sorting and prolonged subcultures.
The initial study first describing the N/TERT1 and N/TERT2G cell lines11 already showed the potential of N/TERT keratinocytes to retain normal growth and differentiation characteristics in vitro. These data together with the herein presented direct comparison to human primary keratinocytes clearly indicate that N/TERT keratinocytes largely behave like primary keratinocytes. N/TERT keratinocytes tend to differentiate quicker than primary keratinocytes as shown by the gene expression peaks at day 4 of differentiation in N/TERT keratinocytes and at day 7 of differentiation in primary human keratinocytes (Fig. 1). Therefore, when using N/TERT keratinocytes in a monolayer culture system, one should be cautious about the time of harvesting when differentiation status is critical for the experimental outcome.
Since the introduction of the N/TERT cell line seventeen years ago, we only found sixteen papers describing the use of these cells in the development of organotypic human skin models (Table 1). The majority have used a dermal substrate based on collagen, fibroblasts or human decellularized dermis. Only four studies15,17,22,23 have generated an epidermis only model from N/TERT keratinocytes, similar to the commercially available SkinEthic26, EpiDerm27, and EPISKIN28 whom are generated from primary human keratinocytes. A thorough characterization of epidermal morphology, differentiation, and barrier function of N/TERT HEEs was still lacking, hence the current study. We found that both N/TERT cell lines (N/TERT1 and N/TERT2G) generate HEEs that are similar to primary keratinocyte-based HEEs. The only protein that is absent in normal skin, yet detected in N/TERT HEEs is SKALP. This antimicrobial protein is involved in keratinocyte host defence responses and highly upregulated in psoriasis skin57. The expression of SKALP in organotypic skin cultures is however not unusual31. N/TERT keratinocytes are derived from primary neonatal foreskin keratinocytes, which are known for higher expression levels of SKALP in comparison to primary adult keratinocytes31.
We found the basket weave stratum corneum of N/TERT HEEs to form a functional barrier to low molecular weight tracers (Fig. 2A,B) that are widely used for studying the outside-inside and inside-outside barrier function of the in vitro reconstructed epidermis32,58. The stratum corneum lipid composition and ultrastructure of N/TERT HSEs has been studied by the group of El Ghalbzouri21, showing a high similarity to HSEs generated from primary human keratinocytes. Although we cannot exclude potential differences between N/TERT and primary keratinocytes for the penetration of other tracer molecules or microbial components, and differences between the stratum corneum formation in HSE versus HEE models may occur, N/TERT keratinocytes show a great potential substituting primary keratinocytes in 3D reconstructed models for normal skin. Important to note is that the HEE culture model system should not be considered as a full thickness skin model, but as an epidermal equivalent. The inert plastic filter obviously does not replicate the functions of the native basement membrane and the absence of a dermal matrix also precludes investigations on dermal-epidermal interactions. Other groups already showed the feasibility of generating full thickness skin equivalents using N/TERT cells18,21,23, therefore we focussed on the application of the N/TERT cells in the epidermal only models and models for inflammatory skin diseases. Altogether the N/TERT keratinocytes are now described in different culture models for human skin, using diverse culture protocols by various research groups. This clearly indicates the good reproducibility of data obtained from N/TERT cells and strengthens the notion that this cell line is a powerful tool in experimental dermatology research.
To date, no studies investigated the feasibility of using the N/TERT-HEEs for studies on inflammatory skin diseases. Psoriasis and atopic dermatitis are highly prevalent, inflammatory skin disease of multifactorial aetiology but with a distinct disease pathophysiology. Increased proliferation, lack of a stratum granulosum and parakeratosis (retention of nuclei in the stratum corneum resulting from incomplete keratinocyte maturation59) are morphological hallmarks of psoriatic skin60–62. Epithelium-specific genes like DEFB4, PI3, S100A7, and KRT16 and their corresponding proteins are highly expressed in psoriatic epidermis and are considered disease markers57,63. The immunological component of psoriasis is marked by high levels of pro-inflammatory cytokines and chemokines, including interferon-γ, interleukins 1, 6, 17, and 22 and tumor necrosis factor (TNF)α64–69. Addition of the abovementioned pro-inflammatory cytokines to human primary keratinocytes is frequently described to generate in vitro models for psoriatic skin31,70–74. We found that the addition of Th17 cytokines, IL-17 and IL-22, to the N/TERT-HEEs most closely resembled psoriasis skin: presence of parakeratosis, lack of a stratum granulosum (and thus low levels of FLG and LOR), and induced levels of hBD2 and SKALP (Fig. 4G). The addition of Th1 cytokines (TNFα, IL-6, and IL-1α) resulted in similar effects, yet these cytokines had more deleterious effects on the epidermis resulting in less optimal epidermal morphology (Supplemental Fig. 5). As we intended to compare the responses of N/TERT keratinocytes to primary keratinocytes, we applied the cytokine cocktails used for primary keratinocytes and did not optimise concentrations or combinations which may improve the epidermal morphology of the Th1 model. Other psoriasis hallmarks, like the presence of rete ridges, microabcesses of Munro, pustules of Kogoj, and aberrations of dermal capillaries60,75 can obviously not be studied in our model, as it lacks a dermal compartment with immune cells and blood vessels. Of note, the N/TERT PS-HEE does not show acanthosis or hyperproliferation. Since the majority of primary keratinocyte-derived organotypic skin models lack these features, this seems a problem inherent to the skin- or epidermis equivalent model system used, and not the source of keratinocytes. The only in vitro psoriasis model showing acanthosis due to increased proliferation is a commercially available model27 where normal human keratinocytes are seeded on a collagen gel with fibroblasts isolated from lesional psoriatic skin. This suggests that fibroblast-secreted factors, and not T-cell derived cytokines, might be a source of keratinocyte mitogens.
Histological examination of AD skin shows dermal infiltrates that are mainly comprised of Th2-helper cells that produce IL-4 and IL-1376. Like in psoriasis, IL-17 also plays a role in AD pathophysiology77. However, novel systemic therapies are aimed at the Th2-mediated inflammation and phase 3 clinical trials on a human monoclonal antibody (Dupilimab) against IL-4 receptor alpha that inhibits signaling of IL-4 and IL-13 have been completed recently78. IL-4 signaling reduces expression of important skin barrier proteins, like FLG, LOR, and IVL46,79–83, which is thought to be correlated to the impaired skin barrier function in AD84–86. This, however, is of recent controversy since FLG null mutations and a complete absence of LOR and IVL does not diminish barrier properties of HEEs32. We were able to generate an AD-like phenotype in our N/TERT AD-HEE by stimulating with IL-4 and IL-13 during the final three days of the air-liquid interphase culture (Fig. 5). The similar effects on N/TERT keratinocytes as compared to primary keratinocytes on downstream target genes by the herein investigated pro-inflammatory cytokines, suggest that the signalling pathways involved in the Th1, Th17, and Th2 mediated pathogenesis, like the JAK/STAT87 and NF-ĸB88 pathways are functional in both N/TERT keratinocyte cell lines.
In both N/TERT HEE disease models, established drugs targeting the epidermal keratinocyte compartment successfully reduced inflammation hallmarks suggesting a place for N/TERT keratinocytes in drug development or screening pipelines. ATRA is a widely used anti-psoriatic drug and binds to nuclear receptors, retinoic acid receptor (RAR) and retinoid X receptor (RXR) in the epidermis89. Coal tar activates the AHR leading to a dampened Th2 cytokine-mediated inflammatory response and the reduction of spongiosis and apoptosis46. Research in the field of AHR signalling has been rapidly emerging in the past years. This ligand-activated transcription factor is a master regulator of pivotal cell biological processes like xenobiotic metabolism, cell proliferation, differentiation, and migration in a variety of cells and tissues (reviewed by Esser et al.90). In the skin, the AHR regulates epidermal proliferation, differentiation, melanogenesis, wound healing, response to UV radiation, and immune responses91. AHR activation in N/TERT keratinocytes results in upregulated expression of known target genes CYP1A1, CYP1B1, TIPARP (Fig. 5A, Supplemental Fig. 6A) and negative feedback repressor gene AHRR, which opens up avenues to use the N/TERT cells as a convenient and unlimited cell source of human keratinocytes to study AHR-related mechanisms in the skin.
One other important field for which the N/TERT keratinocytes could be an immensely valuable asset is genome editing. The CRISPR/Cas9 system, first introduced for mammalian cell genome editing in 201392, now facilitates virtually unlimited genetic manipulation and is described in studies on cell biology, disease pathophysiology, drug screening, and therapeutic interventions. In vitro technologies based on CRISPR/Cas9 require serial passaging of transfected cells and clonal cultures for the selection of your desired genotype. Primary cells with a limited life span are therefore not suitable for genome editing strategies and studies with CRISPR/Cas9 in skin biology or diseases remain scarce93,94. The unlimited lifespan, diploid nature and high similarity to primary keratinocytes make N/TERT keratinocytes the ideal cell source for genome editing studies.
Altogether, this work directs towards an expanded utilization of the N/TERT cell lines as a substitute for primary cells in biologically relevant applications in the fields of cell biology, tissue engineering, dermatology, and toxicology.
Methods
N/TERT keratinocytes karyotyping and short tandem repeat (STR) analysis
The N/TERT keratinocytes were karyotyped to assess chromosome abnormalities. In short, colcemid (Invitrogen, New York, USA) treated, hypotonic N/TERT keratinocytes were fixed by methanol:acetic acid (3:1) before Giemsa-staining on glass slides. Metaphases were analysed under light microscope by Clinical Assistant (RVC, Baarn, The Netherlands). Additional short tandem repeat (STR) analysis, to assess the cell lines purity, was performed by QF-PCR using the Aneufast QF-PCR kit (molGENTIX, Barcelona, Spain) according to the manufacturers protocol. Electrophoretograms were generated by use of GeneMarker (SoftGenetics LLC, Stage College, USA).
Isolation of primary human keratinocytes
Human abdominal or breast skin was obtained from plastic surgery procedures after informed consent and in line with the principles and guidelines of the Declaration of Helsinki. Skin biopsies were taken and human primary keratinocytes were isolated as previously described31 and stored in liquid nitrogen until further use.
Monolayer cultures of human primary keratinocytes and human N/TERT keratinocytes
Human primary keratinocytes (P1) were cultured in a 24 wells plate in keratinocyte growth medium (Lonza, Walkersville, USA) until near 100% cell confluence before stimulation with either Th1 (TNFα (30 ng/mL), IFNγ (500 U/mL) and IL-1α (30 ng/mL)), Th2 (IL-4 (50 ng/mL) and IL-13 (50 ng/mL)), or Th17 (IL-17 (50 ng/mL) and IL-22 (50 ng/mL)) cytokines. All cytokines were obtained from Preprotech, London, UK. Two human N/TERT keratinocyte cell lines (N/TERT-1 and N/TERT-2G), purchased from J. Rheinwald laboratory (Harvard Medical School, Boston, USA), were cultured in a 24 well plate in keratinocyte-serum free medium (K-SFM, Gibco) until 50% confluence when switched to experiment medium (50% K-SFM and 50% DF-K (DMEM:Ham’s F12, 1:1 (vol/vol) with added bovine pituitary extract (BPE, 25 µg/mL, Invitrogen,) L-glutamine (2mM, Invitrogen), EGF (0.2 ng/mL, Invitrogen) and CaCl2 (300 µM, Sigma-Aldrich, Saint Louis, USA), as described by Dickson et al.11. Stimulation with Th1, Th2 and Th17 cytokines (in concentrations mentioned above) was performed near 100% cell confluence. Cells were harvested 24, 48, and 72 hours post stimulation for quantitative gene expression analysis.
Human epidermal equivalent (HEE) generation
N/TERT-HEEs were generated as previously described32,95 with few modifications (Supplemental Fig. 3). Briefly, inert transwells (ThinCerts, Greiner Bio-One GmbH) were coated with rat tail collagen (100 µg/mL, BD Biosciences, Bedford, USA) at 4 °C for 1 hour. 105 N/TERT1 or N/TERT2G keratinocytes were seeded on the transwells in 100 µL K-SFM medium (Gibco) in a 24 wells format. After 48 hours, cultures were switched to a mixture of CnT-PR-3D medium (CELLnTEC, Bern, Switzerland) and DMEM medium (60:40 (v/v)) for 24 hours and then cultured at the air-liquid interface for 10 days. Culture medium was refreshed every other day. Optimisation of the culture protocol is shown in Supplemental Fig. 4A–C.
Psoriasis (PS) and atopic dermatitis (AD) HEE development
N/TERT-HEEs were generated as described above (Supplemental Fig. 3A). From day 7 to 10 of the air-liquid interphase culture, PS-associated or AD-associated cytokines were added to the cultures (Supplemental Fig. 3B). For the PS model, Th1 cytokines, TNFα (5 ng/mL), IL-6 (5 ng/mL) and IL-1α (10 ng/mL) or Th17 cytokines IL-17 (30 ng/mL), and IL-22 (30 ng/mL) were used. The AD model was generated by adding the Th2 cytokine IL-4 (10 ng/mL) to the culture media. The PS or AD phenotype in HEEs was rescued by addition of all trans retinoic acid (ATRA, 1 µM, Sigma-Aldrich) or a coal tar extract (prepared from crude pix lithanthracis, Fagron BV, Capelle aan den IJssel, Netherlands), respectively at day 8 of the culture experiment (Supplemental Fig. 3C). At day 10, N/TERT-HEEs were harvested for gene expression analysis and immunohistochemistry.
Total RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was isolated using the Favorprep total tissue RNA kit (Favorgen Biotech, Taiwan), according to the manufacturers protocol. cDNA was generated, after DNase treatment, and used for quantitative real-time PCR (qPCR) by use of the MyiQ Single-Colour Real-Time Detection System (Bio-Rad laboratories, Hercules, USA) for quantification with Sybr Green and melting curve analysis. All primers (Table 2) were obtained from Biolegio (Nijmegen, The Netherlands). Target gene expression levels were normalized to the expression of human acidic ribosomal phosphoprotein P0 (RPLP0). The relative expression levels of all genes of interest were measured using the 2−ΔΔCT method96.
Table 2.
Primer sequences of all primers.
| Gene | Name | Forward primer (5′–3′) | Reverse primer (5′–3′) | E * |
|---|---|---|---|---|
| FLG | Filaggrin | acttcactgagtttcttctgatggtatt | tccagacttgagggtctttttctg | 1.89 |
| LOR | Loricrin | aggttaagacatgaaggatttgcaa | ggcaccgatgggcttagag | 2.08 |
| IVL | Involucrin | acttatttcgggtccgctaggt | gagacatgtagagggacagagtcaag | 1.93 |
| HRNR | Hornerin | tacaaggcgtcatcactgtcatc | atctggatcgtttggattcttcag | 2.12 |
| LCE1A | Late cornified envelope 1 A | tgcaagagtggctgagatgc | agacaacacagttggtgtcagg | 2.18 |
| KRT1 | Keratin 1 | gatgaaatcaacaagcggacaa | tggtagagtgctgtaaggaaatcaatt | 2.24 |
| KRT5 | Keratin 5 | atctctgagatgaaccggatgatc | cagattggcgcactgtttctt | 2.26 |
| KRT14 | Keratin 14 | ggcctgctgagatcaaagactac | cactgtggctgtgagaatcttgtt | 1.93 |
| KRT16 | Keratin 16 | gatcattgcggccaccat | tgctcatacttggtcctgaagtca | 2.01 |
| TGM1 | Transglutaminase 1 | cccccgcaatgagatctaca | atcctcatggtccacgtacaca | 1.99 |
| TGM3 | Transglutaminase 3 | ggaaggactctgccacaatgtc | tgtctgacttcaggtacttctcatactg | 2.06 |
| CST6 | Cystatin M/E | tccgagacacgcacatcatc | ccatctccatcgtcaggaagtac | 1.98 |
| CA2 | Carbonic anhydrase II | aacaatggtcatgctttcaacg | tgtccatcaagtgaaccccag | 2.02 |
| CCL26 | C-C motif chemokine ligand 26 | tcattcagtaaagaggcgaagtattatc | cagttttttggagggcatctg | 1.89 |
| DEFB4 | Human beta defensin 2 | gatgcctcttccaggtgttttt | ggatgacatatggctccactctt | 1.99 |
| PI3 | Skin-derived antileukoprotease | catgagggccagcagctt | tttaacaggaactcccgtgaca | 2.02 |
| CYP1A1 | Cytochrome p450 family 1A1 | ctggagaccttccgacactctt | gtaaaagcctttcaaacttgtgtctct | 2.02 |
| CYP1B1 | Cytochrome p450 family 1B1 | tggctgctcctcctcttcac | ccacgacctgatccaattctg | 2.02 |
| TIPARP | TCDD inducible polymerase | aagctcctccacctcttgaa | tctgcagaaacagggacttg | 2.05 |
| AHRR | Aryl hydrocarbon receptor repressor | aaggctgctgttggagtctcttaat | gatgtagtcataaatgttctggtgcat | 2.15 |
| RPLP0 | Ribosomal protein P0 | caccattgaaatcctgagtgatgt | tgaccagcccaaaggagaag | 2.02 |
*E is efficiency as fold increase in fluorescence per PCR cycle.
Morphological and immunohistochemical analysis
HEEs were fixed in 4% formalin solution for 4 hours and subsequently embedded in paraffin. 6 µm sections were stained with hematoxylin and eosin (H&E, Sigma-Aldrich) or processed for immunohistochemical analysis. Sections were blocked for 15 minutes with 5% normal goat, rabbit, or horse serum in phosphate-buffered saline (PBS) and subsequently incubated with the antibodies mouse anti-FLG, rabbit anti-LOR, mouse anti-IVL, mouse anti-Ki67, goat anti-hBD2, or rabbit anti-SKALP/Elafin for 1 hour at room temperature. Next, a 30 minute incubation step with biotinylated rabbit anti-goat, horse anti-mouse, or goat anti-rabbit (Vector Laboratories, Burlingame, USA) was performed, followed by a 30 minute incubation with avidin-biotin complex (Vector laboratories). The peroxidase activity of 3-Amino-9-ethylcarbazole (AEC) was used to visualize the protein expression and the sections were mounted using glycerol gelatin (Sigma-Aldrich). See Table 3 for details on the antibodies used.
Table 3.
Antibodies used for immunohistochemistry.
| Antibody; clone | Manufacturer | Dilution |
|---|---|---|
| FLG; clone 15C10 | Leica Biosystems, Newcastle, UK | 1:100 |
| LOR; Covance 145P-100 | Covance Inc., Princeton, USA | 1:4000 |
| IVL; Mon150 | Van Duijnhoven et al.97 | 1:20 |
| Ki67; MIB-1 | DAKO, Heverlee, Belgium | 1:50 |
| pAb to beta 2 defensin | Abcam, Cambridge, UK | 1:100 |
| SKALP/Elafin; 92-1 | Schalkwijk et al.33 | 1:500 |
Lucifer yellow dye penetration assay
To study the outside-in stratum corneum barrier function, Lucifer Yellow (1 mM, Sigma-Aldrich) was applied on top of the HEEs and was allowed to incubate for 60 minutes, in the dark, at room temperature32,58. HEEs were fixated in buffered 4% formalin solution, embedded in paraffin and sectioned. 6 µm sections were deparaffinised and mounted with Fluoromount-G, containing DAPI (eBioscience Inc. San Diego, USA). SDS treatment of the HEEs as a positive control for the Lucifer Yellow penetration upon skin barrier disruption was shown in a previous study by our group32.
Biotin penetration assay
To study the inside-out stratum corneum barrier function, the HEEs were turned upside down and EZ-link sulfo-NHS-LC-biotin (3.3 mg/mL, Thermo Fisher Scientific, Waltham, USA) was applied on the bottom of the filters and allowed to incubate for 60 minutes at room temperature32. HEEs were fixated in buffered 4% formalin solution, embedded in paraffin and sectioned. 6 µm sections were deparaffinised and incubated for 30 minutes, in the dark, with 1:200 Alexa Fluor 594 streptavidin (Thermo Fisher Scientific) conjugate. The sections were mounted with Fluoromount-G containing DAPI.
Statistics
Data are represented as mean ± SEM of at least three technical (N/TERT keratinocytes) or biological (primary keratinocytes) replicates. Raw ΔCt values were used to statistically analyze the quantitative PCR results using the commercially available SPSS software (IBM SPSS Statistics 22). One-way analysis of variance followed by Bonferroni post hoc testing was performed. P < 0.05 was considered to be of statistical significance.
Literature search
A search of relevant literature that reports the use of N/TERT1 and/or N/TERT2G cell lines has been conducted in April 2017. A first extensive literature search was performed by investigating all publications that cited the original N/TERT paper by Dickson et al.11. From these 229 publications, the abstract and materials and methods section was evaluated for the use of N/TERT1 and N/TERT2G cell lines. This led to a total of 58 papers, of which 42 studies used the cells in conventional monolayer culture systems (Supplemental Table 1) and 16 studies used the cells to generate skin- or epidermal equivalents (Table 1). Furthermore, we used Pubmed and Google Scholar and the search terms included ‘N/TERT’, ‘N/TERT1’, ‘N/TERT2G’, ‘TERT’, ‘TRT’, ‘keratinocytes’, ‘epidermal equivalent’, and ‘skin equivalent’.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on request.
Electronic supplementary material
Acknowledgements
This work was funded by a grant from the Radboud Institute for Molecular Life Science (RIMLS) and a ZonMw TOP grant 91211052. E.B was financially supported by a VENI-Grant 91616054 from ZonMw, The Netherlands Organization for Health Research and Development.
Author Contributions
J.Sm. and E.B. conceived the study and wrote the manuscript. J.Sm., H.N., G.R., I.V., and G.Z. performed the experiments. P.Z. and J.S. critically revised the manuscript and supervised the study.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12041-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Eaglstein WH, Falanga V. Tissue engineering and the development of Apligraf a human skin equivalent. Advances in wound care: the journal for prevention and healing. 1998;11:1–8. [PubMed] [Google Scholar]
- 2.Boelsma E, Verhoeven MC, Ponec M. Reconstruction of a human skin equivalent using a spontaneously transformed keratinocyte cell line (HaCaT) The Journal of investigative dermatology. 1999;112:489–498. doi: 10.1046/j.1523-1747.1999.00545.x. [DOI] [PubMed] [Google Scholar]
- 3.Engelhart K, El Hindi T, Biesalski HK, et al. In vitro reproduction of clinical hallmarks of eczematous dermatitis in organotypic skin models. Archives of dermatological research. 2005;297:1–9. doi: 10.1007/s00403-005-0575-7. [DOI] [PubMed] [Google Scholar]
- 4.Berning M, Pratzel-Wunder S, Bickenbach JR, et al. Three-Dimensional In Vitro Skin and Skin Cancer Models Based on Human Fibroblast-Derived Matrix. Tissue engineering. Part C, Methods. 2015;21:958–970. doi: 10.1089/ten.tec.2014.0698. [DOI] [PubMed] [Google Scholar]
- 5.Boukamp P, Petrussevska RT, Breitkreutz D, et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. The Journal of cell biology. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan H, Yuan M, Simpson C, et al. Stem/progenitor cell-like properties of desmoglein 3dim cells in primary and immortalized keratinocyte lines. Stem cells. 2007;25:1286–1297. doi: 10.1634/stemcells.2006-0304. [DOI] [PubMed] [Google Scholar]
- 7.Bikle DD, Xie Z, Tu CL. Calcium regulation of keratinocyte differentiation. Expert review of endocrinology & metabolism. 2012;7:461–472. doi: 10.1586/eem.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo EY, Piao YJ, Kim JS, et al. Identification of calcium-induced genes in HaCaT keratinocytes by polymerase chain reaction-based subtractive hybridization. Archives of dermatological research. 2002;294:411–418. doi: 10.1007/s00403-002-0355-6. [DOI] [PubMed] [Google Scholar]
- 9.Wilson VG. Growth and differentiation of HaCaT keratinocytes. Methods in molecular biology. 2014;1195:33–41. doi: 10.1007/7651_2013_42. [DOI] [PubMed] [Google Scholar]
- 10.Seo MD, Kang TJ, Lee CH, et al. HaCaT Keratinocytes and Primary Epidermal Keratinocytes Have Different Transcriptional Profiles of Cornified Envelope-Associated Genes to T Helper Cell Cytokines. Biomolecules & therapeutics. 2012;20:171–176. doi: 10.4062/biomolther.2012.20.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson MA, Hahn WC, Ino Y, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Molecular and cellular biology. 2000;20:1436–1447. doi: 10.1128/MCB.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rheinwald JG, Hahn WC, Ramsey MR, et al. A two-stage, p16(INK4A)- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Molecular and cellular biology. 2002;22:5157–5172. doi: 10.1128/MCB.22.14.5157-5172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Man YK, Trolove C, Tattersall D, et al. A deafness-associated mutant human connexin 26 improves the epithelial barrier in vitro. The Journal of membrane biology. 2007;218:29–37. doi: 10.1007/s00232-007-9025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabelsteen S, Hercule P, Barron P, et al. Epithelial cells derived from human embryonic stem cells display p16INK4A senescence, hypermotility, and differentiation properties shared by many P63 + somatic cell types. Stem cells. 2009;27:1388–1399. doi: 10.1002/stem.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertrand-Vallery V, Belot N, Dieu M, et al. Proteomic profiling of human keratinocytes undergoing UVB-induced alternative differentiation reveals TRIpartite Motif Protein 29 as a survival factor. PloS one. 2010;5:e10462. doi: 10.1371/journal.pone.0010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazic D, Hufbauer M, Zigrino P, et al. Human papillomavirus type 8 E6 oncoprotein inhibits transcription of the PDZ protein syntenin-2. Journal of virology. 2012;86:7943–7952. doi: 10.1128/JVI.00132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson ED, Weir L, Romanowska M, et al. ARNT controls the expression of epidermal differentiation genes through HDAC- and EGFR-dependent pathways. Journal of cell science. 2012;125:3320–3332. doi: 10.1242/jcs.095125. [DOI] [PubMed] [Google Scholar]
- 18.van Drongelen V, Alloul-Ramdhani M, Danso MO, et al. Knock-down of filaggrin does not affect lipid organization and composition in stratum corneum of reconstructed human skin equivalents. Experimental dermatology. 2013;22:807–812. doi: 10.1111/exd.12271. [DOI] [PubMed] [Google Scholar]
- 19.Harrison W, Cochrane B, Neill G, et al. The oncogenic GLI transcription factors facilitate keratinocyte survival and transformation upon exposure to genotoxic agents. Oncogene. 2014;33:2432–2440. doi: 10.1038/onc.2013.199. [DOI] [PubMed] [Google Scholar]
- 20.Acheva A, Ghita M, Patel G, et al. Mechanisms of DNA damage response to targeted irradiation in organotypic 3D skin cultures. PloS one. 2014;9:e86092. doi: 10.1371/journal.pone.0086092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Drongelen V, Danso MO, Mulder A, et al. Barrier properties of an N/TERT-based human skin equivalent. Tissue engineering. Part A. 2014;20:3041–3049. doi: 10.1089/ten.tea.2014.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Drongelen V, Haisma EM, Out-Luiting JJ, et al. Reduced filaggrin expression is accompanied by increased Staphylococcus aureus colonization of epidermal skin models. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2014;44:1515–1524. doi: 10.1111/cea.12443. [DOI] [PubMed] [Google Scholar]
- 23.Alloul-Ramdhani M, Tensen CP, El Ghalbzouri A. Performance of the N/TERT epidermal model for skin sensitizer identification via Nrf2-Keap1-ARE pathway activation. Toxicology in vitro: an international journal published in association with BIBRA. 2014;28:982–989. doi: 10.1016/j.tiv.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Reijnders CM, van Lier A, Roffel S, et al. Development of a Full-Thickness Human Skin Equivalent In Vitro Model Derived from TERT-Immortalized Keratinocytes and Fibroblasts. Tissue engineering. Part A. 2015;21:2448–2459. doi: 10.1089/ten.tea.2015.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acheva A, Schettino G, Prise KM. Pro-inflammatory Signaling in a 3D Organotypic Skin Model after Low LET Irradiation-NF-kappaB, COX-2 Activation, and Impact on Cell Differentiation. Frontiers in immunology. 2017;8:82. doi: 10.3389/fimmu.2017.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Brugerolle de F, Picarles V, Chibout S, et al. Predictivity of an in vitro model for acute and chronic skin irritation (SkinEthic) applied to the testing of topical vehicles. Cell biology and toxicology. 1999;15:121–135. doi: 10.1023/A:1007577515215. [DOI] [PubMed] [Google Scholar]
- 27.Cannon CL, Neal PJ, Southee JA, et al. New epidermal model for dermal irritancy testing. Toxicology in vitro: an international journal published in association with BIBRA. 1994;8:889–891. doi: 10.1016/0887-2333(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 28.Tinois E, Tiollier J, Gaucherand M, et al. In vitro and post-transplantation differentiation of human keratinocytes grown on the human type IV collagen film of a bilayered dermal substitute. Experimental cell research. 1991;193:310–319. doi: 10.1016/0014-4827(91)90102-Z. [DOI] [PubMed] [Google Scholar]
- 29.Kamsteeg M, Bergers M, de Boer R, et al. Type 2 helper T-cell cytokines induce morphologic and molecular characteristics of atopic dermatitis in human skin equivalent. The American journal of pathology. 2011;178:2091–2099. doi: 10.1016/j.ajpath.2011.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamsteeg M, Zeeuwen PL, de Jongh GJ, et al. Increased expression of carbonic anhydrase II (CA II) in lesional skin of atopic dermatitis: regulation by Th2 cytokines. The Journal of investigative dermatology. 2007;127:1786–1789. doi: 10.1038/sj.jid.5700752. [DOI] [PubMed] [Google Scholar]
- 31.Tjabringa G, Bergers M, van Rens D, et al. Development and validation of human psoriatic skin equivalents. The American journal of pathology. 2008;173:815–823. doi: 10.2353/ajpath.2008.080173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niehues, H., Schalkwijk, J., van Vlijmen-Willems, I. M. et al. Epidermal equivalents of filaggrin null keratinocytes do not show impaired skin barrier function. The Journal of allergy and clinical immunology, doi:10.1016/j.jaci.2016.09.016 (2016). [DOI] [PubMed]
- 33.Schalkwijk J, van Vlijmen IM, Alkemade JA, et al. Immunohistochemical localization of SKALP/elafin in psoriatic epidermis. The Journal of investigative dermatology. 1993;100:390–393. doi: 10.1111/1523-1747.ep12471990. [DOI] [PubMed] [Google Scholar]
- 34.Harder J, Bartels J, Christophers E, et al. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 35.Ong PY, Ohtake T, Brandt C, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. The New England journal of medicine. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 36.Bao L, Shi VY, Chan LS. IL-4 regulates chemokine CCL26 in keratinocytes through the Jak1, 2/Stat6 signal transduction pathway: Implication for atopic dermatitis. Molecular immunology. 2012;50:91–97. doi: 10.1016/j.molimm.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 37.van den Bogaard EH, Tjabringa GS, Joosten I, et al. Crosstalk between keratinocytes and T cells in a 3D microenvironment: a model to study inflammatory skin diseases. The Journal of investigative dermatology. 2014;134:719–727. doi: 10.1038/jid.2013.417. [DOI] [PubMed] [Google Scholar]
- 38.Nonomura K, Yamanishi K, Yasuno H, et al. Up-regulation of elafin/SKALP gene expression in psoriatic epidermis. The Journal of investigative dermatology. 1994;103:88–91. doi: 10.1111/1523-1747.ep12391802. [DOI] [PubMed] [Google Scholar]
- 39.Madsen P, Rasmussen HH, Leffers H, et al. Molecular cloning, occurrence, and expression of a novel partially secreted protein “psoriasin” that is highly up-regulated in psoriatic skin. The Journal of investigative dermatology. 1991;97:701–712. doi: 10.1111/1523-1747.ep12484041. [DOI] [PubMed] [Google Scholar]
- 40.Leigh IM, Navsaria H, Purkis PE, et al. Keratins (K16 and K17) as markers of keratinocyte hyperproliferation in psoriasis in vivo and in vitro. The British journal of dermatology. 1995;133:501–511. doi: 10.1111/j.1365-2133.1995.tb02696.x. [DOI] [PubMed] [Google Scholar]
- 41.Hamid Q, Boguniewicz M, Leung DY. Differential in situ cytokine gene expression in acute versus chronic atopic dermatitis. The Journal of clinical investigation. 1994;94:870–876. doi: 10.1172/JCI117408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Yoo HM, Choi I, et al. Interleukin 4-induced proliferation in normal human keratinocytes is associated with c-myc gene expression and inhibited by genistein. The Journal of investigative dermatology. 1996;107:367–372. doi: 10.1111/1523-1747.ep12363346. [DOI] [PubMed] [Google Scholar]
- 43.Junghans V, Jung T, Neumann C. Human keratinocytes constitutively express IL-4 receptor molecules and respond to IL-4 with an increase in B7/BB1 expression. Experimental dermatology. 1996;5:316–324. doi: 10.1111/j.1600-0625.1996.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 44.Slutsky JB, Clark RA, Remedios AA, et al. An evidence-based review of the efficacy of coal tar preparations in the treatment of psoriasis and atopic dermatitis. Journal of drugs in dermatology: JDD. 2010;9:1258–1264. [PubMed] [Google Scholar]
- 45.Thami G, Sarkar R. Coal tar: past, present and future. Clinical and experimental dermatology. 2002;27:99–103. doi: 10.1046/j.1365-2230.2002.00995.x. [DOI] [PubMed] [Google Scholar]
- 46.van den Bogaard EH, Bergboer JG, Vonk-Bergers M, et al. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. The Journal of clinical investigation. 2013;123:917–927. doi: 10.1172/JCI65642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma Q. Induction of CYP1A1. The AhR/DRE paradigm: transcription, receptor regulation, and expanding biological roles. Current drug metabolism. 2001;2:149–164. doi: 10.2174/1389200013338603. [DOI] [PubMed] [Google Scholar]
- 48.Kallioniemi OP, Kallioniemi A, Piper J, et al. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- 49.Kagami S, Kakinuma T, Saeki H, et al. Significant elevation of serum levels of eotaxin-3/CCL26, but not of eotaxin-2/CCL24, in patients with atopic dermatitis: serum eotaxin-3/CCL26 levels reflect the disease activity of atopic dermatitis. Clinical and experimental immunology. 2003;134:309–313. doi: 10.1046/j.1365-2249.2003.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muleris M, Almeida A, Gerbault-Seureau M, et al. Detection of DNA amplification in 17 primary breast carcinomas with homogeneously staining regions by a modified comparative genomic hybridization technique. Genes Chromosomes Cancer. 1994;10:160–170. doi: 10.1002/gcc.2870100303. [DOI] [PubMed] [Google Scholar]
- 51.Kallioniemi A, Kallioniemi OP, Citro G, et al. Identification of gains and losses of DNA sequences in primary bladder cancer by comparative genomic hybridization. Genes Chromosomes Cancer. 1995;12:213–219. doi: 10.1002/gcc.2870120309. [DOI] [PubMed] [Google Scholar]
- 52.Schlegel J, Stumm G, Scherthan H, et al. Comparative genomic in situ hybridization of colon carcinomas with replication error. Cancer research. 1995;55:6002–6005. [PubMed] [Google Scholar]
- 53.Iwabuchi H, Sakamoto M, Sakunaga H, et al. Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer research. 1995;55:6172–6180. [PubMed] [Google Scholar]
- 54.Savelieva E, Belair CD, Newton MA, et al. 20q gain associates with immortalization: 20q13.2 amplification correlates with genome instability in human papillomavirus 16 E7 transformed human uroepithelial cells. Oncogene. 1997;14:551–560. doi: 10.1038/sj.onc.1200868. [DOI] [PubMed] [Google Scholar]
- 55.Chapman S, Liu X, Meyers C, et al. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. The Journal of clinical investigation. 2010;120:2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Bogaard EH, Rodijk-Olthuis D, Jansen PA, et al. Rho kinase inhibitor Y-27632 prolongs the life span of adult human keratinocytes, enhances skin equivalent development, and facilitates lentiviral transduction. Tissue engineering. Part A. 2012;18:1827–1836. doi: 10.1089/ten.tea.2011.0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Jongh GJ, Zeeuwen PL, Kucharekova M, et al. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. The Journal of investigative dermatology. 2005;125:1163–1173. doi: 10.1111/j.0022-202X.2005.23935.x. [DOI] [PubMed] [Google Scholar]
- 58.Mildner M, Jin J, Eckhart L, et al. Knockdown of filaggrin impairs diffusion barrier function and increases UV sensitivity in a human skin model. The Journal of investigative dermatology. 2010;130:2286–2294. doi: 10.1038/jid.2010.115. [DOI] [PubMed] [Google Scholar]
- 59.Weinstein GD, McCullough JL, Ross PA. Cell kinetic basis for pathophysiology of psoriasis. The Journal of investigative dermatology. 1985;85:579–583. doi: 10.1111/1523-1747.ep12283594. [DOI] [PubMed] [Google Scholar]
- 60.Schon MP, Boehncke WH. Psoriasis. The New England journal of medicine. 2005;352:1899–1912. doi: 10.1056/NEJMra041320. [DOI] [PubMed] [Google Scholar]
- 61.Boehncke WH, Schon MP. Psoriasis. Lancet. 2015;386:983–994. doi: 10.1016/S0140-6736(14)61909-7. [DOI] [PubMed] [Google Scholar]
- 62.Bowcock AM, Krueger JG. Getting under the skin: the immunogenetics of psoriasis. Nature reviews. Immunology. 2005;5:699–711. doi: 10.1038/nri1689. [DOI] [PubMed] [Google Scholar]
- 63.Glaser R, Harder J, Lange H, et al. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nature immunology. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 64.Bonifati C, Ameglio F. Cytokines in psoriasis. International journal of dermatology. 1999;38:241–251. doi: 10.1046/j.1365-4362.1999.00622.x. [DOI] [PubMed] [Google Scholar]
- 65.Wolk K, Kunz S, Witte E, et al. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 66.Sabat R, Wolk K. Research in practice: IL-22 and IL-20: significance for epithelial homeostasis and psoriasis pathogenesis. Journal der Deutschen Dermatologischen Gesellschaft=Journal of the German Society of Dermatology: JDDG. 2011;9:518–523. doi: 10.1111/j.1610-0387.2011.07611.x. [DOI] [PubMed] [Google Scholar]
- 67.Grossman RM, Krueger J, Yourish D, et al. Interleukin 6 is expressed in high levels in psoriatic skin and stimulates proliferation of cultured human keratinocytes. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:6367–6371. doi: 10.1073/pnas.86.16.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uyemura K, Yamamura M, Fivenson DF, et al. The cytokine network in lesional and lesion-free psoriatic skin is characterized by a T-helper type 1 cell-mediated response. The Journal of investigative dermatology. 1993;101:701–705. doi: 10.1111/1523-1747.ep12371679. [DOI] [PubMed] [Google Scholar]
- 69.Ettehadi P, Greaves MW, Wallach D, et al. Elevated tumour necrosis factor-alpha (TNF-alpha) biological activity in psoriatic skin lesions. Clinical and experimental immunology. 1994;96:146–151. doi: 10.1111/j.1365-2249.1994.tb06244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pouliot-Berube C, Zaniolo K, Guerin SL, et al. Tissue-engineered human psoriatic skin supplemented with cytokines as an in vitro model to study plaque psoriasis. Regenerative medicine. 2016;11:545–557. doi: 10.2217/rme-2016-0037. [DOI] [PubMed] [Google Scholar]
- 71.Chiricozzi A, Nograles KE, Johnson-Huang LM, et al. IL-17 induces an expanded range of downstream genes in reconstituted human epidermis model. PloS one. 2014;9:e90284. doi: 10.1371/journal.pone.0090284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harvey A, Cole LM, Day R, et al. MALDI-MSI for the analysis of a 3D tissue-engineered psoriatic skin model. Proteomics. 2016;16:1718–1725. doi: 10.1002/pmic.201600036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yuki T, Tobiishi M, Kusaka-Kikushima A, et al. Impaired Tight Junctions in Atopic Dermatitis Skin and in a Skin-Equivalent Model Treated with Interleukin-17. PloS one. 2016;11:e0161759. doi: 10.1371/journal.pone.0161759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danso MO, van Drongelen V, Mulder A, et al. TNF-alpha and Th2 cytokines induce atopic dermatitis-like features on epidermal differentiation proteins and stratum corneum lipids in human skin equivalents. The Journal of investigative dermatology. 2014;134:1941–1950. doi: 10.1038/jid.2014.83. [DOI] [PubMed] [Google Scholar]
- 75.Griffiths CE, Barker JN. Pathogenesis and clinical features of psoriasis. Lancet. 2007;370:263–271. doi: 10.1016/S0140-6736(07)61128-3. [DOI] [PubMed] [Google Scholar]
- 76.Werfel T. The Role of Leukocytes, Keratinocytes, and Allergen-Specific IgE in the Development of Atopic Dermatitis. Journal of Investigative Dermatology. 2009;129:1878–1891. doi: 10.1038/jid.2009.71. [DOI] [PubMed] [Google Scholar]
- 77.Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, et al. Low expression of the IL-23/Th17 pathway in atopic dermatitis compared to psoriasis. Journal of immunology. 2008;181:7420–7427. doi: 10.4049/jimmunol.181.10.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simpson EL, Bieber T, Guttman-Yassky E, et al. Two Phase 3 Trials of Dupilumab versus Placebo in Atopic Dermatitis. The New England journal of medicine. 2016;375:2335–2348. doi: 10.1056/NEJMoa1610020. [DOI] [PubMed] [Google Scholar]
- 79.Brown SJ, Kroboth K, Sandilands A, et al. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. The Journal of investigative dermatology. 2012;132:98–104. doi: 10.1038/jid.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim BE, Leung DYM, Boguniewicz M, et al. Loricrin and involucrin expression is down-regulated by Th2 cytokines through STAT-6. Clinical immunology. 2008;126:332–337. doi: 10.1016/j.clim.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jakasa I, Koster ES, Calkoen F, et al. Skin barrier function in healthy subjects and patients with atopic dermatitis in relation to filaggrin loss-of-function mutations. The Journal of investigative dermatology. 2011;131:540–542. doi: 10.1038/jid.2010.307. [DOI] [PubMed] [Google Scholar]
- 82.Henry J, Hsu CY, Haftek M, et al. Hornerin is a component of the epidermal cornified cell envelopes. Faseb Journal. 2011;25:1567–1576. doi: 10.1096/fj.10-168658. [DOI] [PubMed] [Google Scholar]
- 83.Kim BE, Leung DY, Boguniewicz M, et al. Loricrin and involucrin expression is down-regulated by Th2 cytokines through STAT-6. Clinical immunology. 2008;126:332–337. doi: 10.1016/j.clim.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palmer CNA, Irvine AD, Terron-Kwiatkowski A, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nature genetics. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 85.Brown SJ, McLean WHI. Eczema Genetics: Current State of Knowledge and Future Goals. Journal of Investigative Dermatology. 2009;129:543–552. doi: 10.1038/jid.2008.413. [DOI] [PubMed] [Google Scholar]
- 86.Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. The New England journal of medicine. 2011;365:1315–1327. doi: 10.1056/NEJMra1011040. [DOI] [PubMed] [Google Scholar]
- 87.Shen CH, Stavnezer J. Interaction of stat6 and NF-kappaB: direct association and synergistic activation of interleukin-4-induced transcription. Molecular and cellular biology. 1998;18:3395–3404. doi: 10.1128/MCB.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annual review of immunology. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jho SH, Vouthounis C, Lee B, et al. The book of opposites: the role of the nuclear receptor co-regulators in the suppression of epidermal genes by retinoic acid and thyroid hormone receptors. The Journal of investigative dermatology. 2005;124:1034–1043. doi: 10.1111/j.0022-202X.2005.23691.x. [DOI] [PubMed] [Google Scholar]
- 90.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacological reviews. 2015;67:259–279. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 91.Esser C, Bargen I, Weighardt H, et al. Functions of the aryl hydrocarbon receptor in the skin. Seminars in immunopathology. 2013;35:677–691. doi: 10.1007/s00281-013-0394-4. [DOI] [PubMed] [Google Scholar]
- 92.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Webber, B. R., Osborn, M. J. & McElroy, A. N. et al. CRISPR/Cas9-based genetic correction for recessive dystrophic epidermolysis bullosa. NPJ Regen Med1, doi:10.1038/npjregenmed.2016.14 (2016). [DOI] [PMC free article] [PubMed]
- 94.Liu YC, Cai ZM, Zhang XJ. Reprogrammed CRISPR-Cas9 targeting the conserved regions of HPV6/11 E7 genes inhibits proliferation and induces apoptosis in E7-transformed keratinocytes. Asian J Androl. 2016;18:475–479. doi: 10.4103/1008-682X.157399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nygaard UH, Niehues H, Rikken G, et al. Antibiotics in cell culture: friend or foe? Suppression of keratinocyte growth and differentiation in monolayer cultures and 3D skin models. Experimental dermatology. 2015;24:964–965. doi: 10.1111/exd.12834. [DOI] [PubMed] [Google Scholar]
- 96.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 97.van Duijnhoven JL, Schalkwijk J, Kranenborg MH, et al. MON-150, a versatile monoclonal antibody against involucrin: characterization and applications. Archives of dermatological research. 1992;284:167–172. doi: 10.1007/BF00372711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on request.