Abstract
Histone Deacetylase 11 (HDAC11) is highly expressed in the central nervous system where it has been reported to have roles in neural differentiation. In contrast with previous studies showing nuclear and cytoplasmic localisation, we observed synaptic enrichment of HDAC11. Knockout mouse models for HDACs 1–9 have been important for guiding the development of isoform specific HDAC inhibitors as effective therapeutics. Given the close relationship between HDAC11 and neural cells in vitro, we examined neural tissue in a previously uncharacterised Hdac11 knockout mouse (Hdac11 KO/KO). Loss of HDAC11 had no obvious impact on brain morphology and neural stem/precursor cells isolated from Hdac11 KO/KO mice had comparable proliferation and differentiation characteristics. However, in differentiating neural cells we observed decreased expression of schizophrenia-associated gene Fez1 (fasciculation and elongation protein zeta 1), a gene previously reported to be regulated by HDAC11 activity. FEZ1 has been associated with the dendritic growth of neurons and risk of schizophrenia via its interaction with DISC1 (disrupted in schizophrenia 1). Examination of cortical, cerebellar and hippocampal tissue reveal decreased Fez1 expression specifically in the hippocampus of adult mice. The results of this study demonstrate that loss of HDAC11 has age dependent and brain-region specific consequences.
Introduction
Histone deacetylase 11 (HDAC11) is the most recently identified member of the HDAC family1, with homologues identified in most species examined2,3. HDAC11 has a single lysine deacetylase domain surrounded by a short N- and C-terminus1. This lysine deacetylase domain is shared by all zinc-dependant HDACs (HDAC1-11) and is predicted to catalyse the removal of acetyl groups from acetylated lysine residues. HDAC11 is similar to both class I (HDAC1, -2, -3, and -8) and class II (HDAC4, -5, -6, -7, -9, and -10) zinc-dependent HDACs3. As there is little justification for assigning HDAC11 specifically to class I or class II HDACs, it is considered exclusively as the first and only ‘class IV’ HDAC identified to date4. The catalytic activity of HDAC11 is inhibited by HDAC inhibitors such as Mocetinostat, Vorinostat, Panobinostat and Quinostat at nanomolar concentrations5,6. Recently there have been reports of an inhibitor predicted to specifically target HDAC117.
The expression of HDAC11 is specific to certain tissues, including the central nervous system1. Compared to the other HDACs, Hdac11 mRNA is particularly abundant in the rat brain with a peculiar pattern of expression in the hippocampus where it is most concentrated in the CA1 (Cornu Ammonis 1) region8. HDAC11 appears to be closely related to cell proliferation/differentiation as its expression is mutually exclusive with proliferative marker Ki-67 and its expression increases as neural cells differentiate in vitro 9–11. Overexpression of HDAC11 in mouse fibroblasts inhibits their proliferation12. The Hdac11 gene and its chromosomal region are associated with variability in regional brain volume of mice13,14 and humans15. Additionally, multiple reports suggest associations between HDAC11 and malignant disease16–21. Two studies have implicated HDAC11 as a negative regulator of cell cycle component DNA replication factor Cdt1 (chromatin licensing and DNA replication factor 1)22,23. Whereas other studies have shown a role for HDAC11 in the response of antigen presenting cells24,25 and the differentiation of neural cells10,26. Watanabe et al., demonstrated that knockdown of HDAC11 inhibits dendrite growth of neurons generated in the adult mouse hippocampus26. This appeared to result from a regulatory role upstream of FEZ1 (fasciculation and elongation protein zeta 1), as they show that HDAC11 deacetylates BUBR1 (Budding uninhibited by benzimidazole 1-related protein kinase), relieving its inhibition of CDC20/APC (cell division cycle 20/anaphase-promoting complex), allowing that to ubiquitinate FEZ1 and therefore its degradation.
Knockout mouse models for HDACs 1–9 have demonstrated the consequences resulting from complete loss of each individual HDAC27,28. As previous studies report a role for HDAC11 in cell proliferation and neural differentiation, the aim of this study was to examine the nervous system in a previously uncharacterised Hdac11 knockout mouse. Hdac11 KO/KO mice were born at Mendelian ratios and brain morphology appeared normal suggesting that HDAC11 does not have a critical role during development. Accordingly, in vitro studies on neural cell lines generated from Hdac11 KO/KO mice showed normal proliferation patterns. However, we found that the expression of Fez1 was decreased in both proliferating and differentiating neural cell cultures. We further observed specific decrease in Fez1 gene expression in the hippocampi of adult Hdac11 KO/KO mice. This suggests that HDAC11 has an age-dependent, brain region specific function in regulating FEZ1, a gene associated with schizophrenia. This is important because it demonstrates that the regulatory role of HDAC11 in the hippocampus is developmentally regulated. Given previous associations between HDAC11, FEZ1 and schizophrenia, a goal of future research should be to examine the performance of homozygous Hdac11 knockout mice in behavioural tests.
Results
HDAC11 Expression Increases with Neural Differentiation with Nuclear and Synaptic Localisation
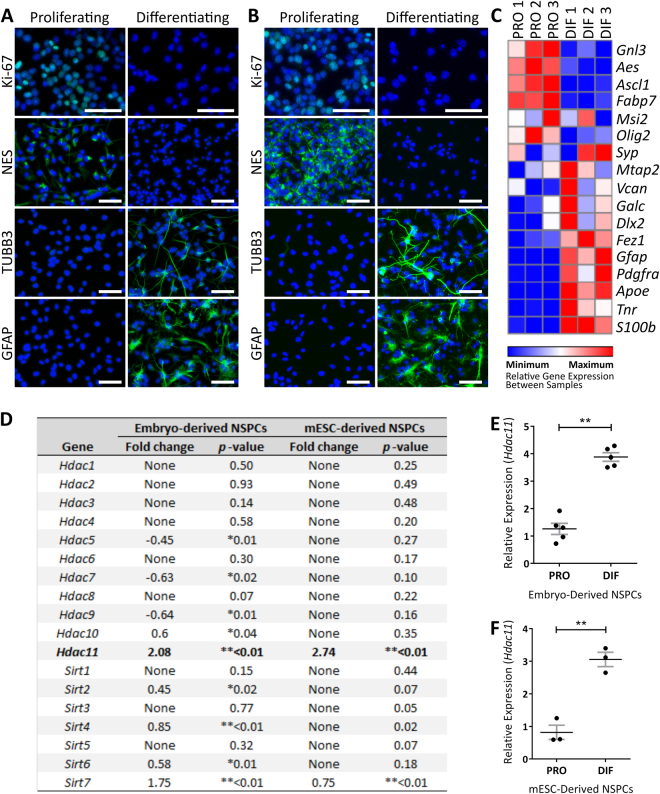
Neural cell lines were generated from the mouse ganglionic eminence (Fig. 1A) and mouse embryonic stem cells (mESCs) (Fig. 1B) to profile HDAC gene expression during differentiation. Following 3 days of differentiation inducing conditions there was a decrease in the expression of proliferation associated Ki-67 and neural stem/precursor cell associated nestin. In contrast, the expression of neuronal associated tubulin beta-3 chain (TUBB3) and astrocyte associated glial fibrillary acidic protein (GFAP) increased (Fig. 1A,B). This transition was further demonstrated by the changing profile of genes associated with neural proliferation and differentiation (Fig. 1C; Supplementary Fig. 1). Examination of Hdac gene expression levels between proliferating and differentiating neural cells revealed that Hdac11 has the largest increase in expression compared to all the other Hdacs and Sirtuins (Fig. 1D; Supplementary Fig. 2). This is similarly observed in mouse embryo-derived and mESC-derived neural cells (Fig. 1D–F).
Figure 1.
Hdac11 displays the greatest and most consistent increase in expression as neural cells differentiate. Neural cells were derived from (A) the ganglionic eminence of E14.5 mouse embryos and (B) mESCs then examined for immunoreactivity of Ki-67, Nestin (NES), TUBB3, and GFAP antibodies following 3 days in either proliferation or differentiation inducing conditions. (C) Relative expression of genes associated with neural proliferation in mouse embryo-derived neural cells maintained in proliferative (PRO) conditions or following 3 days of differentiation (DIF) inducing conditions. The heat map displays relative expression values between samples (i.e. across rows). Gene expression was normalised to the mean of Canx and Sdha. The full names of the genes and assays used are listed in Supplementary Table 3. Hdac11 expression increases in (D,E) E14.5 mouse embryo-derived (N = 3, dots = individual sample values, bars = mean ± SE, *p < 0.05 **p < 0.01, two-tailed paired Student’s t-test) and (D,F) mESC-derived neural stem/precursor cells (NSPCs) following 3 days of differentiation inducing conditions (N = 5, dots = individual sample values, bars = mean ± SE, *p < 0.05 **p < 0.01, one-tailed paired Student’s t-test). Gene expression was normalised to the geomean of Ubc, Actb and Eif4a2.
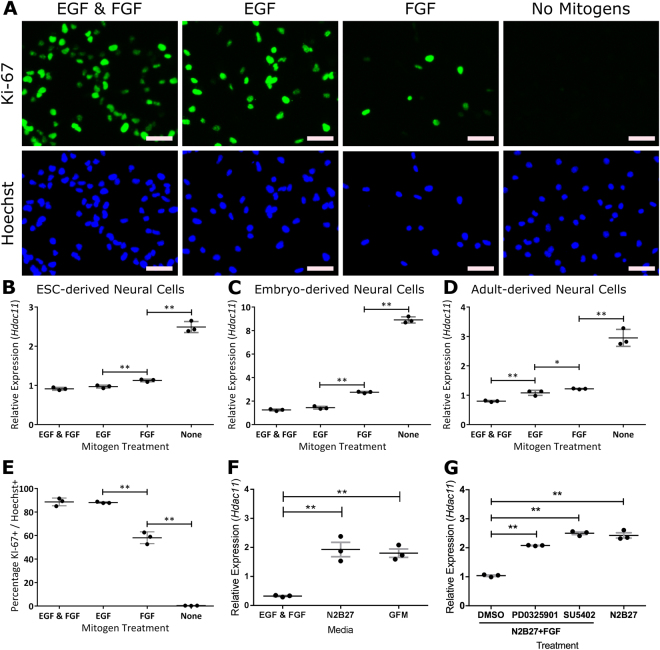
Increasing Hdac11 expression was closely related to progressive removal of mitogens epidermal growth factor (EGF) and fibroblast growth factor (FGF). Removal of mitogens from the culturing media results in fewer proliferating cells (Fig. 2A,E). This mirrored the pattern of increasing Hdac11 expression as the mitogens were removed from mESC-derived (Fig. 2B), embryonic mouse-derived (Fig. 2C) and adult mouse-derived (Fig. 2D) neural cells. When the growth factors that encourage proliferation (EGF & FGF) were replaced with growth factors that encourage differentiation (Brain-derived neurotrophic factor (BDNF), Glial-derived neurotrophic factor (GDNF), Nerve growth factor (NGF) & Neurotrophin-3 (NT-3)) there was an increase in Hdac11 expression (Fig. 2F). Accordingly, small-molecule compounds that are reported to inhibit pathways associated with FGF signalling also increased the expression of Hdac11 (Fig. 2G).
Figure 2.
Hdac11 expression responds to mitogen withdrawal from mESC-, embryonic mouse-, and adult mouse-derived neural cells. (A) Embryo-derived neural cells positive for proliferative marker Ki-62 following withdrawal of mitogens epidermal growth factor (EGF) and/or fibroblast growth factor (FGF) for 3 days. Expression of Hdac11 in (B) embryo-derived neural cells, (C) mESC-derived neural cells and (D) adult mouse-derived neural cells increases following withdrawal of mitogens. (E) Percentage of proliferating cells in each condition as indicated by nuclear Ki-67 immunoreactivity. (F) Hdac11 expression in media containing non-mitogenic growth factors BDNF, GDNF, NGF and NT-3 (collectively GFM “Growth factor media”). (G) Impact of small molecule inhibitors PD0325901 (1 µM) and SU5492 (10 µM) on Hdac11 expression in the presence of FGF. N = 3, dots = individual sample values, bars = mean ± SE, **p < 0.01, *p < 0.05, one-tailed paired Student’s t-test. Gene expression was normalised to the mean of Canx and Sdha.
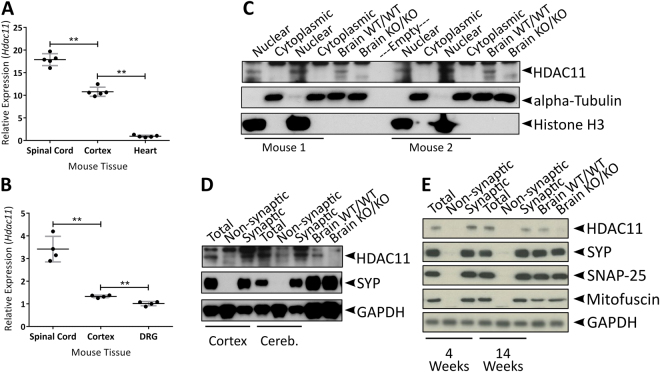
Hdac11 expression was comparatively high in the mouse brain relative to the other HDACs (Supplementary Table 6). Expression was highest in the spinal cord and all neural tissue displayed greater expression than the heart (Fig. 3A,B; Supplementary Fig. 3A). Examination of total brain lysates demonstrated that HDAC11 was enriched in the nuclear fractions (Fig. 3C; Supplementary Figs 3B, 4A,B). However, as there are reports of HDAC11 in the cytoplasm of neural cells26,29, we decided to look more closely at this compartment. Interestingly, HDAC11 was found to be enriched in synaptosomal preparations (Fig. 3D; Supplementary Figs 3C, 4C,D) at both 4 and 14 weeks of age (Fig. 3E; Supplementary Figs 3D, 5A–D). Unfortunately, the HDAC11 antibodies used in this study proved to have a lot of unspecific binding (Supplementary Figs 3 and 6). We assessed HDAC11 expression using western blotting with Hdac11 KO/KO lysates as negative controls. However, we were unable to examine subcellular localisation with histological methods because Hdac11 WT/WT and Hdac11 KO/KO tissue displayed similar patterns of reactivity following the conditions used in this study (Supplementary Fig. 7).
Figure 3.
HDAC11 protein is enriched in the nucleus and at the synapse. (A) Expression of Hdac11 in neural and non-neural tissue of 2-month-old mice. (B) Expression of Hdac11 in tissue prepared from cortex, spinal cord and dorsal root ganglion tissue of P7 mice. (C) HDAC11 was enriched in the nuclear fraction but not in the cytoplasmic fraction of mouse brain lysate. (D) HDAC11 was enriched in synaptic fractions prepared from mouse cortex and cerebellum lysates. (E) HDAC11 was enriched in synaptic fractions prepared from the cortex of 4 week and 14 week-old mice (SYP = Synaptophysin; SNAP-25 = Synaptosomal-associated protein 25). N = 4–5, dots = individual sample values, bars = mean ± STD, **p < 0.01, two-tailed unpaired Student’s t-test.
Characterisation of Hdac11KO/KO Mice
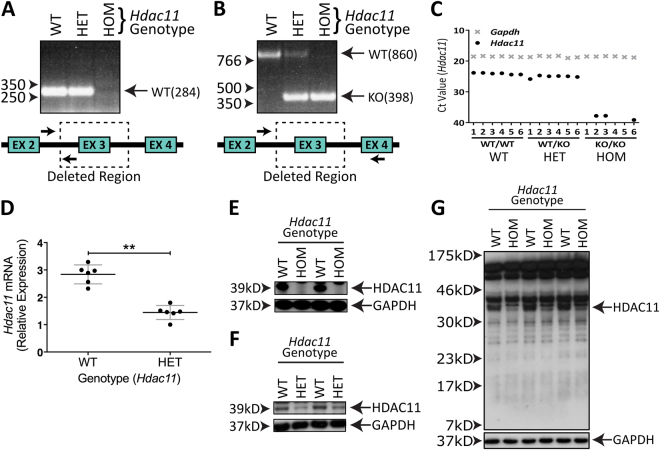
Mice with complete loss of exon 3 from the Hdac11 gene (Fig. 4A,B; Supplementary Fig. 8A,B) were born at Mendelian ratios (Fig. 5A). Accordingly, no transcript was detected by primers complementary to the cDNA sequence within exon 3 (Fig. 4C). The Hdac11 transcript was detected in brain lysates at reduced levels in Hdac11 WT/KO mice compared to Hdac11 WT/WT mice (Fig. 4D). There was no impact on the expression of the other HDAC transcripts (Supplementary Fig. 9). We examined HDAC11 protein using an antibody recognising the amino acids 2–16 at the N-terminus (Fig. 4E,F; Supplementary Figs 6A,E, 8C) and an antibody recognising a more C-terminal part of the protein at amino acid 269 (Supplementary Fig. 6B). Hdac11 KO/KO mice displayed complete loss of the protein (Fig. 4E, Supplementary Figs 6A, 8C) whereas Hdac11 WT/KO mice expressed HDAC11 at a level lower than wild type mice (Fig. 4F, Supplementary Figs 6F, 8D). The antibody targeting the N-terminus of HDAC11 did not detect a remaining protein fragment in the Hdac11 KO/KO brain tissue (Fig. 4G; Supplementary Figs 6G, 8E).
Figure 4.
Mutation of Hdac11 gene results in loss of HDAC11 expression. (A) A primer pair (arrows) requiring a sequence in the deleted region (dotted box) encompassing exon 3 of the Hdac11 gene fails to produce a 284 bp in Hdac11 KO/KO (HOM) mice. (B) A primer pair that covers the deleted region yields a smaller product in mice heterozygous or homozygous for the mutant allele. (C,D) Lower levels of the Hdac11 transcript and (F) HDAC11 protein were detected in Hdac11 WT/KO mice compared to Hdac11 WT/WT mice. Gene expression was normalised to that of Gapdh. (E) No HDAC11 protein was detected in Hdac11 KO/KO mice. (G) A truncated form of HDAC11 was not detected in Hdac11 KO/KO mouse brain. Antibody used in panels 4E, 4F and 4G is anti-HDAC11 (Sigma, Catalogue #H4539). N = 6 (dots = individual sample values), bars = mean ± STD, **p < 0.01, one-tailed unpaired Student’s t-test.
Figure 5.
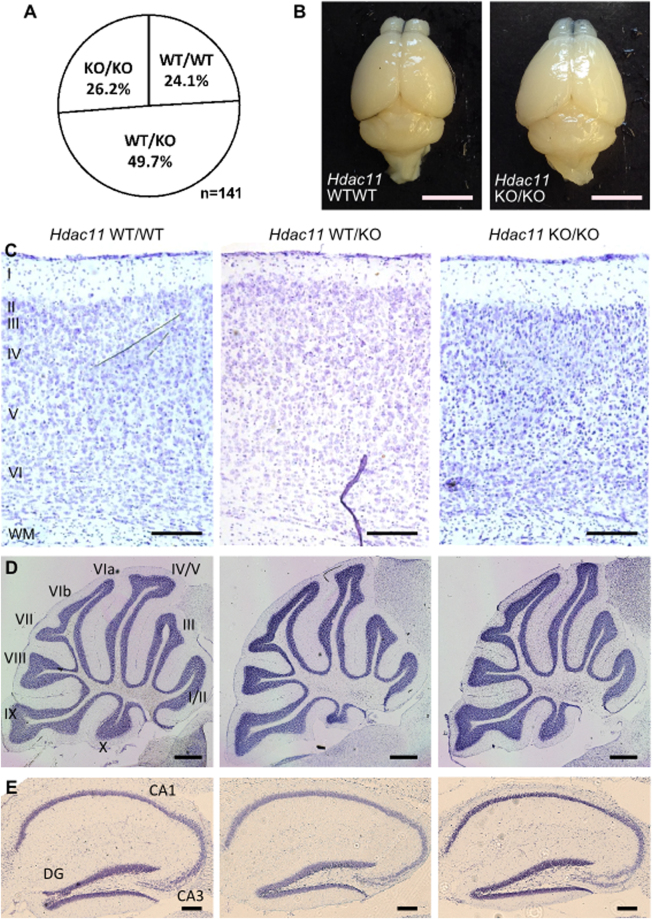
Hdac11 KO/KO mice are born at Mendelian ratios with normal gross brain morphology. (A) Viable mice were born at approximately Mendelian ratios. (B) The gross brain morphology of two-month-old Hdac11 KO/KO mice is similar to Hdac11 WT/WT mice. Cresyl violet (Nissl) staining in (C) cortical (layers I-VI, WM = white matter) (D) cerebellar (lobules I-X) and (E) hippocampal regions (DG = Dentate Gyrus; CA = Cornu Ammonis). Scale bars = (B) 5 mm, (C) 200 µm (D) 500 µm and (E) 200 µm.
Given reports that HDAC11 has roles in proliferation and our observations that HDAC11 was intimately linked with the proliferative status of neural cells we reasoned that loss of HDAC11 may impact development. However, the brains of Hdac11 KO/KO mice appeared similar to those of Hdac11 WT/WT mice at 2 months of age (Fig. 5B). The cortex and cerebellum were of a similar size and there was no obvious regional disruption (Fig. 5C,D). Similarly, there was no disruption to the appearance of the major hippocampal regions (Fig. 5E). For closer examination of neural proliferation and differentiation we derived neural cells from Hdac11 KO/KO mice for in vitro investigations.
Disrupted Fez1 Expression in the Hippocampus of Adult Hdac11KO/KO Mice
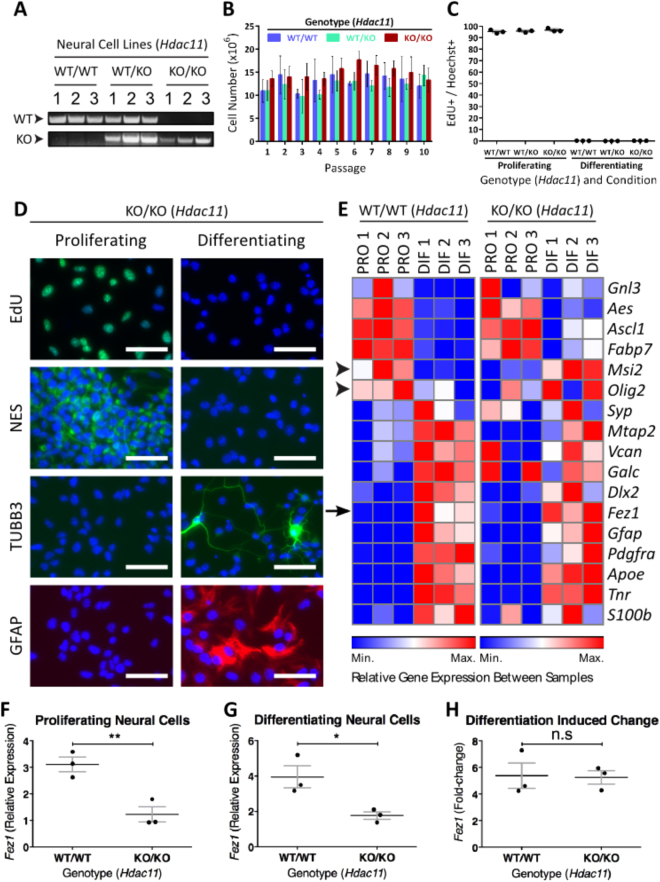
Neural cell lines generated from Hdac11 KO/KO mice (Fig. 6A; Supplementary Fig. 8F) displayed similar proliferation rates to those generated from Hdac11 WT/WT mice (Fig. 6B). 5-ethynyl-2′deoxyuridine (EdU) is a thymidine analogue which is incorporated into the DNA of dividing cells. All neural cells generated from Hdac11 WT/WT and Hdac11 KO/KO mice displayed no EdU incorporation over a 24 hour period following 2 days of induced differentiation (Fig. 6C). During this process the expression of neural stem/precursor cell and neural differentiation associated markers appeared largely normal (Fig. 6D,E; Supplementary Figs 10, 11, 12, 13). We observed similar levels of expression of all Hdac genes (except Hdac11) in Hdac11 KO/KO and Hdac11 WT/WT neural cell lines (Supplementary Fig. 9). Furthermore, the expression profile of neural associated genes was similar in neural cells generated from Hdac11 KO/KO and Hdac11 WT/WT mice (Fig. 6E). The exception being the expression of proliferation associated gene Msi2 (Musashi 2) which displays a small decrease (−0.27-fold, p < 0.01) during the differentiation of Hdac11 WT/WT cells but an increase (0.46-fold, p < 0.01) during the differentiation of cells derived from Hdac11 KO/KO mice (Fig. 6E, arrow head). However, there was not a statistically significant difference in the level of Msi2 expression when comparing Hdac11 KO/KO and Hdac11 WT/WT cells with each other in proliferating (p = 0.1) or differentiating (p = 0.24) conditions (Supplementary Table 7). Olig2 displayed a similar pattern to Msi2 (Fig. 6E, arrow head), and this too was not statistically different between Hdac11 KO/KO and Hdac11 WT/WT cells (Supplementary Table 7).
Figure 6.
Reduced Fez1 expression in neural cells derived from Hdac11 KO/KO mice. (A) Three neural cell lines were generated from Hdac11 WT/WT, Hdac11 WT/KO and Hdac11 KO/KO mice. (B) Cells were counted following each passage every 4 days for 10 passages. (C) Cells derived from all mice exited the cell cycle following 3 days of differentiation inducing conditions. (D) Presence of EdU, Nestin (NES), TUBB3, and GFAP in cells derived from the ganglionic eminence of E14.5 Hdac11 KO/KO mouse embryos following 3 days of proliferation or differentiation inducing conditions. (E) Relative expression of genes associated with neural proliferation in mouse embryo-derived neural cells given 3 days of either proliferation (PRO) or differentiation (DIF) associated conditions. Msi2 and Olig2 expression display opposing changes during the differentiation of Hdac11 WT/WT and Hdac11 KO/KO cells (arrow heads). The expression of Fez1 (arrow) was decreased in neural cells derived from Hdac11 KO/KO mice following 3 days of either (F) proliferating or (G) differentiating inducing conditions. (H) Fez1 expression increased in neural cells derived from Hdac11 WT/WT and Hdac11 KO/KO mice at similar levels following 3 days differentiation. N = 3 (dots = individual sample values), bars = mean ± SE, **p < 0.01, *p < 0.05, two-tailed unpaired Student’s t-test. Gene expression was normalised to the mean of Canx and Sdha.
Comparison of gene expression profiles of proliferating neural cells derived from Hdac11 KO/KO and Hdac11 WT/WT revealed an impact on Fez1 expression (Fig. 6F). Fez1 expression increases following the differentiation of both Hdac11 KO/KO and Hdac11 WT/WT cells (Fig. 6E arrow, 6H). However, its expression remains lower in differentiating Hdac11 KO/KO cells compared to Hdac11 WT/WT cells (Fig. 6G). This was not due to a reduced increase of Fez1 expression during the differentiation of Hdac11 KO/KO neural cells (Fig. 6H). The impact on Fez1 caught our attention because of Watanabe et al.’s reports of the regulatory activity of HDAC11 on FEZ126. Additionally, it was the only gene on the gene expression panel to be affected in both proliferating and differentiating neural cells with statistical significance (Supplementary Table 7).
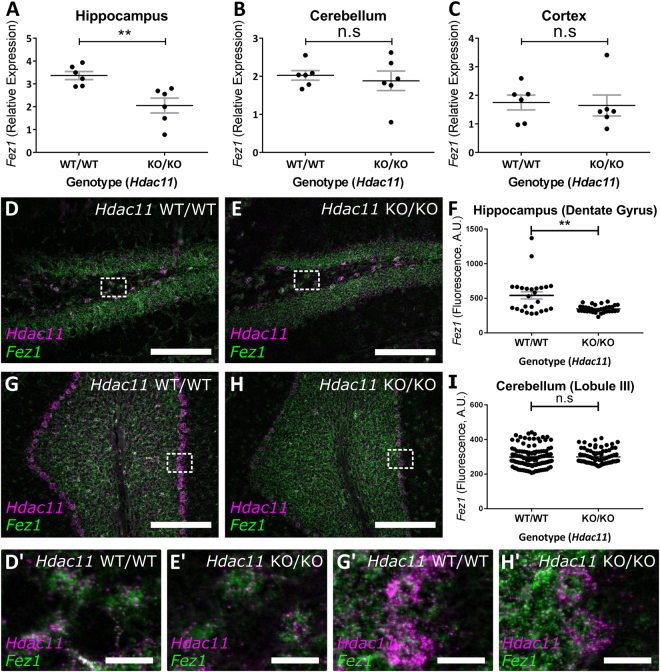
We sought to corroborate observations from our in vitro studies and therefore examined Fez1 expression in brain lysates from Hdac11 KO/KO mice. This did not reveal an impact of HDAC11 loss on Fez1 expression (Supplementary Fig. 14). To explore the possibility that the loss of HDAC11 has a region specific impact on the expression Fez1 we examined the cortex, hippocampus and cerebellum of Hdac11 KO/KO. These experiments were done on juvenile (P10/“10 day old”) and adult (P60/“2 month old”) mice. This revealed that Fez1 expression was specifically downregulated in the hippocampus of adult Hdac11 KO/KO mice (Fig. 7A). In contrast, expression of Fez1 in the hippocampus was unaffected in juvenile Hdac11 KO/KO mice (Supplementary Fig. 15A). This age-dependent effect on Fez1 is region specific because it was expressed at similar levels in the cortex and cerebellum of Hdac11 KO/KO juvenile (Supplementary Fig. 15B,C) and adult mice (Fig. 7B,C).
Figure 7.
Reduced Fez1 expression in the hippocampus of Hdac11 KO/KO adult mice. The expression of Fez1 was examined in the hippocampus, cortex, cerebellum and spinal cord of 2-month-old Hdac11 KO/KO mice. (A) Only the hippocampus of Hdac11 KO/KO mice had lower relative Fez1 expression compared to Hdac11 WT/WT mice. The expression of Fez1 was unaffected in the (B) cerebellum or (C) cortex in Hdac11 KO/KO mice. Gene expression was normalised to the mean of Canx and Sdha (A–C). (D–F) Decreased Fez1 expression was observed in selected Hdac11 positive cells within the dentate gyrus of 2-month-old mouse hippocampus. Regions indicated with a perforated box are displayed in panels D’ and E’. (G–I) There was no impact on Fez1 expression in selected Hdac11 positive cells within the lobule III of 2-month-old mouse cerebellum. Regions indicated with a perforated box are displayed in panels G’ and H’. N = 6 (A–C), N = 3(n ≥ 25) (F,H) (dots = individual sample values), bars = mean ± SE, **p < 0.01, two-tailed unpaired Student’s t-test. Fez1 = Green, Hdac11 = Magenta, scale bar = 200 µm (D,E,G,H) and 25 µm (D’,E’,G’,H’).
Using in situ hybridisation, we observed reduced Fez1 expression in Hdac11 positive cells within the hippocampus but not the cerebellum (Fig. 7D–I; Supplementary Fig. 15D,E). In contrast, Hdac11 was reduced in both the hippocampus and the cerebellum (Supplementary Fig. 15F,G). Using qPCR, we confirmed the presence of an Hdac11 transcript in Hdac11 KO/KO tissue with an assay that targets the cDNA sequence which has not been deleted in the mutant. This assay detects RNA expression, albeit at lower levels in Hdac11 KO/KO mice which is likely a result of nonsense mediated decay of the mutant transcript (Supplementary Table 8). Also for comparison, we examined oligodendrocyte associated genes Mbp (Myelin basic protein) and Plp1 (Proteolipid protein 1) because they had previously been reported to be decreased following loss of Hdac11 10. However, we did not see a statistically significant impact on the expression of either of these genes (Supplementary Table 8).
Discussion
Our observations are consistent with previous work demonstrating a particularly high abundance of HDAC11 in neural tissue1,8,9. Reports that its expression increases as neural cells differentiate were replicated using multiple in vitro approaches10,11. These observations may reflect emerging roles for HDAC11 as a regulator of the cell cycle and neural differentiation10,12,22,23,26. Previous studies had examined the consequence of partial loss or increase in HDAC11 expression10,12,26. However, the consequence of complete Hdac11 knockout in a neural context had not yet been reported. The aim of this study was to examine the neural tissue of homozygous Hdac11 knockout mice.
Fertile, homozygous Hdac11 knockout mice were born at Mendelian ratios with no obvious phenotypes. This contrasts with the homozygous knockout mice of all class I HDACs (Hdac1, -2, -3, and -8) and certain class II HDACs (Hdac4 and -7)30,31 which display either complete or partial embryonic/perinatal lethality27,28. Hdac11 expression increases during the first 2 months of postnatal mouse brain development9. However, we found that the gross anatomy of the brain appeared normal in 2-month-old homozygous Hdac11 knockout mice. Three of the previously generated Hdac knockout mice (Hdac5, -6, and -9) displayed only subcellular or age-dependent phenotypes32–34. Similarly, Hdac11 knockout mice do not display an obvious phenotype but rather have an age dependent brain region specific decrease in Fez1. It is interesting to note that HDAC11 and HDAC6 appear to be recruited to the same protein complex1,26 and both are reported to regulate CDC20, a component of the pathway that controls FEZ1 ubiquitination26,35.
Partial loss of Hdac11 expression has been reported to impact on the proliferation of fibroblasts in vitro 12. Together with reports of roles for HDAC11 in the cell cycle22,23 and its association with cancer and brain volume13–20 we predicted that there would be an impact on the proliferation rate of cells derived from the nervous system of homozygous Hdac11 knockout mice. However, we did not find any effect from HDAC11 loss on neural proliferation and these cells also exited the cell cycle when directed. Accordingly, the gross brain morphology of homozygous Hdac11 knockout mice appeared normal suggesting these processes had at least generally been regulated appropriately. One possibility is that HDAC11 has roles specific to cancer cells. Supporting this idea is a report that HDAC11 knockdown selectively impacts carcinogenic cell lines, leaving normal cell lines unharmed20.
Previous studies have reported that knockdown of Hdac11 results in decreased expression of IL-10 (Interleukin 10)24, Mbp (Myelin basic protein) and Plp (Proteolipid protein 1) genes10. Additionally, Deubzer et al. report unpublished results that HDAC11 represses the transcription of Bmp4 (Bone morphogenetic protein 4) in a neuroblastoma cell line20. We found that the expression of oligodendrocyte associated genes Mbp and Plp were expressed at reduced levels in the adult Hdac11 KO/KO hippocampus but this was not statistically significant. It is likely that loss of HDAC11 has cell type specific effects on gene expression which should be considered carefully when investigating its function. The previous studies on HDAC11 function had examined macrophages24, oligodendrocyte precursor cell lines10, fibroblast cells12 and a neuroblastoma cell line20. We propose that the Hdac11 knockout mouse line can be used for investigation of tissues and cell types relevant to these experiments.
HDAC11 was initially reported to be localised to the nucleus1 where it is understood to regulate gene expression10,24. Indeed, the results of this study show that HDAC11 is enriched in nuclear fractions. It has been reported that HDAC11 becomes increasingly localised in cytoplasmic compartments during the differentiation of neural progenitors in the hippocampus26. Despite limitations preventing us using HDAC11 antibodies to examine HDAC11 localisation using immunocytochemistry, we did observe HDAC11 expression in cytoplasmic fractions and more specifically in synaptosomes. This provides additional insight into the subcellular localisation of HDAC11 and offers the possibility that it has a regulatory role in the synaptic compartment. Like class IIa HDACs, HDAC11 may shuttle between cellular compartments in response to stimuli. HDAC11 has previously been reported to regulate dendrite development26 and enrichment of HDAC11 at the synapse supports the enticing possibility that regulating its activity will impact synaptic plasticity and memory.
We observed decreased Fez1 expression in both proliferating and differentiating cells derived from homozygous Hdac11 knockout mice so we investigated this in more detail. Examination of different brain regions revealed that this effect was specific to the hippocampus of adult mice which was surprising given that we also observed decreased Fez1 expression in proliferating neural cells in vitro. This may reflect an acquired identity of these in vitro lines and reinforces the importance of follow up studies on appropriate tissue.
A previous study showed that FEZ1 ubiquitination is downstream of a pathway activated by HDAC11 activity26. Given their results, it would be predicted that loss of HDAC11 activity would increase FEZ1 protein expression. Regrettably, we do not report FEZ1 protein levels in response to loss of HDAC11 as we were unable to detect the expression of FEZ1 using commercially available antibodies. The decreased level of Fez1 transcript in response to loss of HDAC11 may be due to a self-regulatory function of FEZ1. This self-regulatory function may involve interaction with DISC1 (Disrupted in schizophrenia 1)36. Patients with schizophrenia carrying a high risk allele for the disease in the DISC1 gene show reduced FEZ1 expression in the hippocampus37.
The report of reduced FEZ1 gene expression in the hippocampus of patients with schizophrenia is particularly relevant as this was seen in the homozygous Hdac11 knockout mice37. The relationship between HDAC11 activity and schizophrenia-associated phenotypes is strengthened by Watanabe et al.’s evidence for a signalling cascade implicating HDAC11, FEZ1 and DISC126. Sheng et al. examined the CA1 and CA2/3 regions of the hippocampus from post-mortem schizophrenia patient tissue38. They identified copy number increases in the HDAC11 gene which resulted in increased HDAC11 expression specifically in the CA2/3 region. In contrast, post-mortem tissue from patients with a different psychiatric disease (bipolar disorder) displayed no difference in the copy number or expression of HDAC11 compared to healthy controls. A more recent study reports an association between schizophrenia and single nucleotide polymorphisms (SNPs) in the human HDAC11 gene39.
The emerging association between HDAC11 and schizophrenia is interesting as it should be considered during the development of therapeutic drugs. The promise of treating schizophrenia by selective inhibition of certain HDACs has previously been discussed in a review by Weïwer et al.40. In their review they note that Hdac11 is expressed in the hippocampus and such patterns provide a spatial association with schizophrenia but emphasise that functional inferences cannot be drawn from this. The results of this study demonstrate that Hdac11 knockout mice provide a means to examine the functional association between HDAC11 and schizophrenia-associated FEZ1.
Our findings are specific to the adult-mouse hippocampus, suggesting that this area will be particularly important to following investigations, particularly for investigating the molecular pathways regulated by HDAC11. Disrupted performance on hippocampal dependent tasks such as the Morris water maze, T-maze and radial arm maze are reported in mouse models of schizophrenia41. Furthermore, it would be interesting to investigate the response of Hdac11 knockout mice to phencyclidine, which is reported to result in a variety of phenotypes associated with schizophrenia including hyperlocomotion41. Hdac11 knockout mice may share similar characterisics to Fez1 knockout mice which do not display any anatomical abnormalities but do display a hyperlocomotion phenotype in a variety of tasks42.
Genetics is important in the etiology of schizophrenia but it remains difficult to determine the precise causative genetic factors in this complex polygenic disorder43. HDAC11 may have a more general association with psychiatric disease. Studies have found links between HDAC11, the administration and reinforcement of drugs. An impact on Hdac11 expression is reported in the brain of rodents administered with cocaine44, alcohol45 and methamphetamine46,47. Incidentally Fez1 KO/KO mice have an enhanced response to methamphetamine treatment, at least in part due to increased mesolimbic dopaminergic transmission42. Understanding how these observations converge on HDAC11 may provide an important insight into the neural mechanisms disrupted in a variety of disorders. Furthermore, it is important that we understand the consequences of losing HDAC11 activity as this will be valuable for guiding the development of isoform specific HDAC inhibitors as therapeutics for disease.
Methods
Cell culture media
Mouse Embryonic Stem Cell (mESC) proliferation media was formulated in a basal media of Dulbecco’s modified Eagle medium (DMEM), High Glucose, GlutaMAX™ (Life Technologies, Catalog #61965-026) with 20% embryonic stem cell grade FBS (Source BioScience LifeSciences, Catalog #S0451). The media contained 1× Non-Essential Amino Acids (Life Technologies, Catalog #11140-050), 50 μM beta-mercaptoethanol (Life Technologies, Catalog #31350-010) and 10 μg/mL (Leukemia inhibitory factor) LIF (Millipore, Catalog #ESG1106).
Neural proliferation media was formulated in a NeuroCult™ NSC Basal Medium (Mouse) (STEMCELL Technologies, Catalog #05700) with NeuroCult™ NSC Proliferation Supplement (Mouse) (STEMCELL Technologies, Catalog #05701). The medium contained 20 ng/mL recombinant mouse epidermal growth factor (rmEGF) (Peprotech Inc, Catalog #315-09) and 10 ng/mL recombinant mouse fibroblast growth factor (rmFGF) (Peprotech Inc, Catalog #450-33).
Neural differentiation media (N2B27) is a formulation of 50% DMEM/F-12, GlutaMAX™ (Life Technologies, Catalog #10565-018) and 50% Neurobasal® Medium (Life Technologies, Catalog #21103-049). This contained 0.5x N-2 Supplement (Life Technologies, Catalog #17502-048), 0.5x B-27® Serum-free Supplement (Life Technologies, Catalog #17504-044), 0.5x GlutaMAX™ Supplement (Life Technologies, Catalog #35050-087), 1x Non-Essential Amino Acids (Life Technologies, Catalog #11140-050) and 50 μM beta-mercaptoethanol (Life Technologies, Catalog #31350-010).
Generation and expansion of mESC-derived neural cells
Generation of mESC-derived neural cells was carried out according to Conti et al.48 before they were maintained in the neural proliferation media described above.
Isolation and expansion of mouse embryo-derived neural cells
The mouse embryonic ganglionic eminence was dissected and cultured according to Chojnacki and Weiss’s protocol49. The cells were cultured in suspension on untreated plastic in neural proliferation media (see cell culture media). They were passaged every 4 days by dissociation with Accutase® (Sigma, Catalog #A6964) and reseeded at a density of 10,000 cells/cm2.
Experimental proliferation and differentiation of neural cell cultures
For consistency between approaches, all proliferating neural cell lines were seeded as an adhered monolayer on 0.1 mg/mL poly-DL-ornithine (Sigma, Catalog #P8638) followed by 5 μg/mL laminin (Life Technologies, Catalog #23017-015) coated plastic before experimental procedures began. Following a typical passage, proliferating neural cells were dissociated into a single cell suspension ensured by passing the suspension through a sterile 40 μm cell strainer (SLS, Catalog #352340) into a 50 mL tube. Cells destined for proliferative conditions were seeded at 20,000/cm2, whereas those for differentiation inducing conditions were seeded at 100,000/cm2. After 24 hours, the cells had adhered to the surface and the media was replaced with the experimental media. For proliferative conditions, the media was replaced with neural proliferation media (see cell culture media). For differentiation inducing conditions, the media was replaced with neural differentiation media (see cell culture media). The media was replaced with fresh media every day until the cells were harvested.
5-ethynyl-2’-deoxyuridine (EdU) labelling
Incorporation of the thymidine analogue EdU into the DNA of proliferating cells over a 24 hour period was detected using the Click-IT® EdU Alexa Fluor® 488 Imaging kit (Life Technologies, Catalog #C10337) according to the manufacturer’s instructions.
Immunocytochemistry
Cells for immunocytochemistry assays were seeded into black walled and optical bottomed 96 well plates (Fisher Scientific, #10281092). They were fixed for 20 minutes in a 4% formaldehyde solution (Sigma, Catalog #F1635). Primary and secondary antibodies were prepared in Phosphate buffered solution (PBS) (Life Technologies, Catalog #14190-169) made with 10% donkey serum (Sigma, Catalog #D9663), 0.1% Triton-X 100 (Sigma, Catalog #T8717) as described in supplementary Table 1. After each step the samples were washed three times with PBS. All assays were counterstained for 10 minutes with Hoechst 33342 (Life Technologies, Catalog #H3570) prepared in PBS to a concentration 1 μg/mL.
Microscopy and Image Analysis
Fluorescent images were acquired using an inverted microscope (Axio Observer, ZEISS). Analyses of Ki-67 or EdU positive cells were quantified using Fiji/ImageJ (http://imagej.nih.gov/ij/). Cell nuclei were set as regions of interest (ROI) as defined by the nuclear counter stain (i.e. Hoechst 33342). Then the average signal intensity (i.e. Ki-67 and EdU) was measured for each ROI using the Analyze Particles command. A cell was considered positive if the average signal intensity for its ROI was greater than the ROI with the maximum value from staining with the secondary antibody alone (Ki-67 experiment) or differentiation inducing (i.e. proliferation preventing) condition (EdU experiment).
RNA Extraction and Real Time PCR
Real time PCR was carried out using the 2−ΔΔCt method as described previously50. The reverse transcription was carried out using the High-capacity RNA-to-cDNATM Kit (Life Technologies, Catalog #4387406) according to the manufacturer’s instructions. TaqMan® assays (Supplementary Tables 2, 3, 4) and TaqMan® Gene Expression Master Mix (Life Technologies, Catalog #4369016) were used according to the manufacturer’s instructions. Two Hdac11 TaqMan® assays were used in this study as one covered the exon deleted in the Hdac11 mice Hdac11 KO/KO (Exons 2–3; Mm01183513_m1) and the other does not (Exons 5–6; Mm00523422). Data were collected on the C100TM Thermal Cycler and CFX96TM Real-Time System (BIO-RAD). Data collection from TaqMan® Low Density Arrays (TLDA) was carried out on the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems).
Preparation of nuclear, cytoplasmic and synaptosome fractions
Nuclear and cytoplasmic fractionation was carried out as described in Davies et al.51. Synaptosome fractions were prepared as described in Valencia et al.52.
Western Blotting
Western blotting was carried out as described by Landles et al.53. The antibody incubation details are described in Supplementary Table 5. Following washes with PBS/0.1% Tween® 20 to remove residual antibody, the membranes were developed using an Amersham ECLTM Western Blotting Kit (GE Lifesciences, Catalog #RPN2109) and Amersham Hyperfilm ECLTM (GE Lifesciences, Catalog #28906837).
Hdac11 knockout mice
Details of the Hdac11 knockout mice can be found on the Taconic website (http://www.taconic.com/mouse-model/hdac11-ko-6978 ). The gene was targeted in C57BL/6 ESCs by deletion of exon 3 leading to a premature stop codon. To identify the genotype, a PCR reaction with the forward primer (TGCTGCCTGTGAGCCACTGC) and reverse primer (AGAATGGCTGTCTCCCTAGG to detect the wild type allele; or CCTTGGAATAGCATCTCAGG to detect the knockout allele) were used. The reaction was run on a program of 4 minutes at 95 °C, before 36 cycles of 25 seconds at 95 °C, 20 seconds at 60 °C and 45 seconds at 72 °C. The program finished with a single final step for 6 minutes at 72 °C.
Nissl staining
Mice were anesthetised with an injection of 100 μL sodium pentobarbital (J.M.L, Catalog #103-9130). The mice were transcardially perfused with 20 mL of PBS followed by perfusion with 20 mL of 10% formalin (i.e. 4% PFA) (PRC, Catalog #PRC/R/38). After the perfusion was complete, the brain was stored in 10% formalin at 4 °C overnight before equilibration in 30% sucrose (Fisher Scientific, Catalog #10744771) made in PBS at 4 °C. The brain was then snap frozen in 2-methylbutane (Sigma, Catalog #277258) that had been cooled on dry ice. The brains were sectioned every 30 μm on a cryostat, cooled to −17 °C. The sections were attached to glass adhesion SuperFrost® plus slides (VWR, Catalog #631-0446). They were stained in a 1% cresyl violet (Acros Organics, Catalog #405760025), 1% Glacial acetic acid solution for 10 minutes at room temperature (cerebellum and hippocampus images) or 60 minutes at 37 °C (cortex images). The slides were then dehydrated in increasing amounts of ethanol with only a few seconds incubation in the final 100% ethanol step to avoid removing too much stain. They were then incubated in Xylene (Sigma, Catalog #33817) for 15 minutes before mounting with DPX mounting medium (Fisher, Catalog #D/5319/05).
In situ hybridisation
In situ hybridisation was carried out using RNAscope® Technology (Advanced Cell Diagnostics) according to the manufacturer’s instructions. The Hdac11 probe targeted the region 35-981 of NM-1449.9.2. The Fez1 probe targeted the region 2-1010 of NM-183171.4. To ensure that comparative numbers of cells were included in the quantification, single, individual cells expressing Hdac11 were selected within the region of interest (dentate gyrus of the hippocampus) (see Supplementary Fig. 15D,E).
Electronic supplementary material
Acknowledgements
We wish to thank Edward Smith and Parul Choudhary for technical advice. We thank Edward Seto and colleagues for providing the Hdac11 knock-out mice from their colony at the University of South Florida. This work was funded by the Medical Research Council. DB was a Medical Research Council CASE student (G1000393).
Author Contributions
D.T.B., C.L.B. and G.P.B. designed the study. D.T.B., C.L., A.S.P., A.C.B. and J.K.D. performed the experiments. D.T.B., analysed the data and prepared the figures. T.R. contributed to the materials for the study. D.T.B., C.L.B. and G.P.B. wrote the manuscript. All authors discussed and interpreted the data and revised the manuscript.
Competing Interests
J.K.D. and C.L.B. were employees of Neusentis, Pfizer Ltd. and T.R. is an employee of Merck & Co. All other authors declare no competing financial interests.
Footnotes
Caroline L. Benn and Gillian P. Bates contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11630-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol. Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 2.Pandey R, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fournel M, et al. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol. Cancer Ther. 2008;7:759–768. doi: 10.1158/1535-7163.MCT-07-2026. [DOI] [PubMed] [Google Scholar]
- 6.Arts J, et al. JNJ-26481585, a novel “second-generation” oral histone deacetylase inhibitor, shows broad-spectrum preclinical antitumoral activity. Clin. Cancer Res. 2009;15:6841–6851. doi: 10.1158/1078-0432.CCR-09-0547. [DOI] [PubMed] [Google Scholar]
- 7.Tian, Y. et al. A sub-milligram-synthesis protocol for in vitro screening of HDAC11 inhibitors. Bioorg. Med. Chem. Lett., 10.1016/j.bmcl.2016.03.116 (2016). [DOI] [PubMed]
- 8.Broide RS, et al. Distribution of histone deacetylases 1–11 in the rat brain. J. Mol. Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 9.Liu, H., Hu, Q., Kaufman, A., D’Ercole, A. J. & Ye, P. Developmental expression of histone deacetylase 11 in the murine brain. J. Neurosci. Res. 86, 537–543, 10.1002/jnr.21521 (2008). [DOI] [PMC free article] [PubMed]
- 10.Liu, H., Hu, Q., D’Ercole A. J. & Ye, P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia57, 1–12, 10.1002/glia.20729 (2009). [DOI] [PMC free article] [PubMed]
- 11.Androutsellis-Theotokis A, Chrousos GP, McKay RD, DeCherney AH, Kino T. Expression profiles of the nuclear receptors and their transcriptional coregulators during differentiation of neural stem cells. Horm. Metab. Res. 2013;45:159–168. doi: 10.1055/s-0032-1321789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagui TK, Sharma SS, Ma L, Pledger WJ. Proliferative status regulates HDAC11 mRNA abundance in nontransformed fibroblasts. Cell Cycle. 2013;12:3433–3441. doi: 10.4161/cc.26433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosen GD, et al. Genetic modulation of striatal volume by loci on Chrs 6 and 17 in BXD recombinant inbred mice. Genes Brain Behav. 2009;8:296–308. doi: 10.1111/j.1601-183X.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaglani SM, Lu L, Williams RW, Rosen GD. The genetic control of neocortex volume and covariation with neocortical gene expression in mice. BMC Neurosci. 2009;10:44. doi: 10.1186/1471-2202-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inkster B, et al. Histone deacetylase gene variants predict brain volume changes in multiple sclerosis. Neurobiol. Aging. 2013;34:238–247. doi: 10.1016/j.neurobiolaging.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Voelter-Mahlknecht S, Ho AD, Mahlknecht U. Chromosomal organization and localization of the novel class IV human histone deacetylase 11 gene. Int. J. Mol. Med. 2005;16:589–598. [PubMed] [Google Scholar]
- 17.Dai W, et al. Promoter CpG island methylation of genes in key cancer pathways associates with clinical outcome in high-grade serous ovarian cancer. Clin. Cancer Res. 2013;19:5788–5797. doi: 10.1158/1078-0432.CCR-13-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruhn B, et al. The expression of histone deacetylase 4 is associated with prednisone poor-response in childhood acute lymphoblastic leukemia. Leuk. Res. 2013;37:1200–1207. doi: 10.1016/j.leukres.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Skov V, et al. Increased gene expression of histone deacetylases in patients with Philadelphia-negative chronic myeloproliferative neoplasms. Leuk. Lymphoma. 2012;53:123–129. doi: 10.3109/10428194.2011.597905. [DOI] [PubMed] [Google Scholar]
- 20.Deubzer HE, et al. HDAC11 is a novel drug target in carcinomas. Int. J. Cancer. 2013;132:2200–2208. doi: 10.1002/ijc.27876. [DOI] [PubMed] [Google Scholar]
- 21.Thole TM, et al. Neuroblastoma cells depend on HDAC11 for mitotic cell cycle progression and survival. Cell Death Dis. 2017;8:e2635. doi: 10.1038/cddis.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong PG, et al. Chromatin unfolding by Cdt1 regulates MCM loading via opposing functions of HBO1 and HDAC11-geminin. Cell Cycle. 2010;9:4351–4363. doi: 10.4161/cc.9.21.13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glozak MA, Seto E. Acetylation/deacetylation modulates the stability of DNA replication licensing factor Cdt1. J Biol. Chem. 2009;284:11446–11453. doi: 10.1074/jbc.M809394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villagra A, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat. Immunol. 2009;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng F, et al. Divergent roles of histone deacetylase 6 (HDAC6) and histone deacetylase 11 (HDAC11) on the transcriptional regulation of IL10 in antigen presenting cells. Mol. Immunol. 2014;60:44–53. doi: 10.1016/j.molimm.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe Y, Khodosevich K, Monyer H. Dendrite development regulated by the schizophrenia-associated gene FEZ1 involves the ubiquitin proteasome system. Cell Rep. 2014;7:552–564. doi: 10.1016/j.celrep.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takase, K., Oda, S., Kuroda, M. & Funato, H. Monoaminergic and Neuropeptidergic Neurons Have Distinct Expression Profiles of Histone Deacetylases. PLoS One8, 10.1371/journal.pone.0058473 (2013). [DOI] [PMC free article] [PubMed]
- 30.Vega RB, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 31.Chang S, et al. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 32.Chang S, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang CL, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/S0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim AH, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang E, et al. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72:559–571. doi: 10.1016/j.neuron.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipska BK, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum. Mol. Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 38.Sheng G, Demers M, Subburaju S, Benes FM. Differences in the circuitry-based association of copy numbers and gene expression between the hippocampi of patients with schizophrenia and the hippocampi of patients with bipolar disorder. Arch. Gen. Psychiatry. 2012;69:550–561. doi: 10.1001/archgenpsychiatry.2011.1882. [DOI] [PubMed] [Google Scholar]
- 39.Kebir O, et al. Family-based association study of common variants, rare mutation study and epistatic interaction detection in HDAC genes in schizophrenia. Schizophr. Res. 2014;160:97–103. doi: 10.1016/j.schres.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 40.Weiwer M, Lewis MC, Wagner FF, Holson EB. Therapeutic potential of isoform selective HDAC inhibitors for the treatment of schizophrenia. Future Med. Chem. 2013;5:1491–1508. doi: 10.4155/fmc.13.141. [DOI] [PubMed] [Google Scholar]
- 41.Jones CA, Watson DJ, Fone KC. Animal models of schizophrenia. Br. J. Pharmacol. 2011;164:1162–1194. doi: 10.1111/j.1476-5381.2011.01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakae N, et al. Mice lacking the schizophrenia-associated protein FEZ1 manifest hyperactivity and enhanced responsiveness to psychostimulants. Hum. Mol. Genet. 2008;17:3191–3203. doi: 10.1093/hmg/ddn215. [DOI] [PubMed] [Google Scholar]
- 43.Kavanagh DH, Tansey KE, O’Donovan MC, Owen MJ. Schizophrenia genetics: emerging themes for a complex disorder. Mol. Psychiatry. 2015;20:72–76. doi: 10.1038/mp.2014.148. [DOI] [PubMed] [Google Scholar]
- 44.Host L, Dietrich JB, Carouge D, Aunis D, Zwiller J. Cocaine self-administration alters the expression of chromatin-remodelling proteins; modulation by histone deacetylase inhibition. J. Psychopharmacol. 2011;25:222–229. doi: 10.1177/0269881109348173. [DOI] [PubMed] [Google Scholar]
- 45.Finegersh A, Homanics GE. Acute ethanol alters multiple histone modifications at model gene promoters in the cerebral cortex. Alcohol Clin. Exp. Res. 2014;38:1865–1873. doi: 10.1111/acer.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omonijo O, et al. Differential effects of binge methamphetamine injections on the mRNA expression of histone deacetylases (HDACs) in the rat striatum. Neurotoxicology. 2014;45:178–184. doi: 10.1016/j.neuro.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres, O. V. et al. An Acute Methamphetamine Injection Downregulates the Expression of Several Histone Deacetylases (HDACs) in the Mouse Nucleus Accumbens: Potential Regulatory Role of HDAC2 Expression. Neurotox. Res., 10.1007/s12640-015-9591-3 (2015). [DOI] [PMC free article] [PubMed]
- 48.Conti, L. et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. Plos Biol. 3, 1594–1606, 10.1371/journal.pbio.0030283 (2005). [DOI] [PMC free article] [PubMed]
- 49.Chojnacki A, Weiss S. Production of neurons, astrocytes and oligodendrocytes from mammalian CNS stem cells. Nat. Protoc. 2008;3:935–940. doi: 10.1038/nprot.2008.55. [DOI] [PubMed] [Google Scholar]
- 50.Benn, C. L., Fox, H. & Bates, G. P. Optimisation of region-specific reference gene selection and relative gene expression analysis methods for pre-clinical trials of Huntington’s disease. Mol. Neurodegener. 3, 10.1186/1750-1326-3-17 (2008). [DOI] [PMC free article] [PubMed]
- 51.Davies SW, et al. Detection of polyglutamine aggregation in mouse models. Methods Enzymol. 1999;309:687–701. doi: 10.1016/S0076-6879(99)09045-X. [DOI] [PubMed] [Google Scholar]
- 52.Valencia A, et al. Striatal synaptosomes from Hdh140Q/140Q knock-in mice have altered protein levels, novel sites of methionine oxidation, and excess glutamate release after stimulation. J. Huntingtons Dis. 2013;2:459–475. doi: 10.3233/JHD-130080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landles C, et al. Proteolysis of Mutant Huntingtin Produces an Exon 1 Fragment That Accumulates as an Aggregated Protein in Neuronal Nuclei in Huntington Disease. J. Biol. Chem. 2010;285:8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.