Abstract
Background
Epigenetics, in the broadest sense, governs all aspects of the life of any multicellular organism, as it controls how differentiated cells arrive at their unique phenotype during development and differentiation, despite having a uniform (with some exceptions such as T-cells and germ cells) genetic make-up. The endocrine pancreas is no exception. Transcriptional regulators and epigenetic modifiers shape the differentiation of the five major endocrine cell types from their common precursor in the fetal pancreatic bud. Beyond their role in cell differentiation, interactions of the organism with the environment are also often encoded into permanent or semi-permanent epigenetic marks and affect cellular behavior and organismal health. Epigenetics is defined as any heritable – at least through one mitotic cell division – change in phenotype or trait that is not the result of a change in genomic DNA sequence, and it forms the basis that mediates the environmental impact on diabetes susceptibility and islet function.
Scope of review
We will summarize the impact of epigenetic regulation on islet cell development, maturation, function, and pathophysiology. We will briefly recapitulate the major epigenetic marks and their relationship to gene activity, and outline novel strategies to employ targeted epigenetic modifications as a tool to improve islet cell function.
Major conclusions
The improved understanding of the epigenetic underpinnings of islet cell differentiation, function and breakdown, as well as the development of innovative tools for their manipulation, is key to islet cell biology and the discovery of novel approaches to therapies for islet cell failure.
Keywords: Endocrine pancreas, Islet cells, DNA methylation, Histone marks, Epigenetics
1. Introduction
All cells in a multicellular organism – excluding cell types such as B- and T-cells that undergo genomic rearrangements at specific sites to increase the body's antibody and T-cell receptor diversity – contain an identical set of genomic instructions faithfully reproduced through many cycles of cell division from the zygote to the mature organism. It follows then that diversity between cellular phenotypes and organ function must result from carefully orchestrated regulatory mechanisms that reinterpret the genomic DNA sequence, depending on cues such as developmental morphogens, cell position, hormonal milieu, diurnal cues, and environmental factors to activate and repress specific gene sets within each cell type. While most changes in gene activation status are acute and thus more properly thought of as direct control of gene expression, alterations in the gene activation status that are maintained through at least one mitotic cell division are considered ‘heritable” and thus categorized as ‘epigenetic’ [1]. The Greek word ‘epi’ (επι) means ‘above, over’, thus indicating that epigenetic modifications control biological processes, from gene transcription to complex metabolic phenotypes, without altering the DNA sequence itself.
A striking example of epigenetic control is seen in the hundred or so autosomal loci in the human genome that are expressed in a ‘parent-of-origin’ specific pattern, meaning some are actively transcribed only on the maternally inherited chromosome while others are only transcribed from the paternal chromosome. This process is termed ‘imprinting’ and is mediated by specific patterns of DNA methylation [reviewed in [2]]. Defects in imprinting reveal the crucial role of this type of epigenetic regulation in the development and function of specific tissues. For example, imprinting affects islet cell function, as seen in Beckwith–Wiedemann syndrome, a severe organ overgrowth disorder caused by aberrant gene expression from an imprinted gene cluster located on chromosome 11 [reviewed in [3]]. This disease can result from either specific mutations within the imprinting control regions or from paternal disomy for this chromosomal region. As a result, expression of the tumor suppressor p57 (encoded by CDKN1C) is lost, as it is only transcribed from the maternal chromosome, resulting in impaired cell cycle control. In about half of the Beckwith–Wiedemann patient population, the resulting excess of β-cell mass results in hyperinsulinemia and hypoglycemia [4].
Another imprinted locus affecting insulin secretin is Transient Neonatal Diabetes Mellitus (TNDM). Neonates with TNDM present with hypoinsulinemia, which resolves by three six months of age [5]. This disease can result from paternal hetero- or isodisomy or from aberrant methylation of the maternal allele, suggesting that overexpression of genes within this locus is responsible for the observed phenotype. ZAC (Zinc finger protein that regulates apoptosis and cell cycle) and the non-coding RNA HYMAI (hydatidiform mole-associated and imprinted region) are genes located in the TNDM locus, and murine dual transgenic overexpression recapitulates the human phenotype [6]. Beckwith–Wiedemann syndrome and TNDM thus represent striking examples of the impact of epigenetic control on islet growth and function.
On the molecular level, epigenetic states are ‘encoded’ both by DNA methylation, specifically cytosine methylation of palindromic CpG sequences, and by a multitude of histone modifications, which collectively determine chromatin compaction and accessibility of the transcriptional machinery to the more than 20,000 genes in the mammalian genome [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19]. Frequently, non-coding RNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are also considered to be epigenetic modifiers, as they affect final cellular steady-state mRNA and protein concentrations at the level of translation or mRNA stability, although strictly speaking, the expression of these non-coding RNAs themselves is under epigenetic and genetic control [20], [21], [22], [23].
Epigenetic mechanisms affect organismal function on multiple time-scales: transgenerationally, such as the aforementioned imprinting; within an organism to facilitate permanent or semi-permanent changes to mediate interactions between the environment and the genome [24], [25]; or just a few days, e.g. recurrent cellular differentiation programs such as the generation of intestinal epithelial cells [26]. A unique feature of epigenetic modifications is that they permit adaptation to changing metabolic states while maintaining cellular function and can thus be both flexible and stable. As introduced above through the example of Beckwith–Wiedemann syndrome, epigenetic dysregulation can have dramatic pathophysiological consequences on islet cells and glucose homeostasis (see Table 1 for epigenetic modifiers with known roles in islet development and function).
Table 1.
Chromatin-modifying enzymes required for normal islet development or function.
| Enzyme | Mechanism of action | Effect on transcription | Effect of deletion on pancreas or islets in mice |
|---|---|---|---|
| Brg1 | Part of SWI/SNF complex; nucleosome remodeling; in the pancreas a Pdx1 coactivator | Silencing or activating | Hypoplastic pancreas when deleted early |
| Dicer | ncRNA processing | Silencing or activating | Degranulation and loss of β-cells |
| Dnmt1 | Maintenance of CpG methylation | Usually silencing | Hypoplastic pancreas when deleted early |
| Dnmt3 | De novo CpG methylation | Usually silencing | Dysregulation of glucose-stimulated insulin secretion |
| Ezh22 | Part of PRC2; methylation of lysine 27 on H3 to H3K27 | Silencing | Increased number of endocrine progenitors and mature β-cells |
2. Major classes of epigenetic modifications
2.1. Histone modifications
Genomic material in the cell is packaged into chromatin, which is a DNA-protein complex consisting of linear DNA wrapped around a histone core. A nucleosome is a unit of chromatin defined by 147 base pairs of DNA wound around the histone core, which includes four dimers each of the histones H2A, H2B, H3, and H4. The N-terminal tails of these histones can be covalently modified, which alters chromatin compaction and recruits transcriptional regulators to modulate gene expression in a multitude of ways [27], [28]. Among the modifications to histones are acetylation of lysine and arginine residues, ubiquitination and sumoylation of lysines, and serine and threonine phosphorylation. Genome-wide maps for many of these modifications have been obtained through a technology termed ‘ChIP-Seq’, or chromatin immunoprecipitation followed by high throughput sequencing. For this assay, antibodies directed against specific histone modifications are employed to enrich for those chromatin fragments occupied by histones bearing these marks, and the precipitated DNA is purified and sequenced. Alignment of these sequenced fragments then reveals chromatin regions that are occupied by modified histones of interest.
By mapping specific histone marks and the binding profiles of major islet transcription factors, Pasquali and colleagues were able to compile genome-wide maps of distinct chromatin states in human islets, from promoters to active enhancers [29]. Studies such as these have revealed that major chromatin states can be defined by just a handful of histone marks. Activating marks include the trimethylation of the fourth lysine on histone 3 (H3K4me3), which is associated with active promoters at genes enriched in CpG islands (see below), and acetylation of the 27th lysine of histone H3 (H3K27Ac), which is a mark of active enhancers, whereas repressed chromatin states are often marked by H3K27me3 [30]. Interestingly, combinations of specific histone modifications can also be relevant, such as in the case of a ‘bivalent’ chromatin state characterized by the presence of both the ‘repressive’ H3K27me3 and the ‘activating’ H3K4me3 mark at the same gene, a phenomenon first found in embryonic stem cells [31]. It is thought that this bivalent state facilitates the subsequent differentiation process, because genes will become active as soon as the H3K27me3 mark is removed from promoters or stably repressed if the H3K4me3 mark is erased. Together, histone modifications are reflected in transcriptional activation or chromatin compaction.
Chromatin compaction itself regulates accessibility of genetic loci to DNA-binding proteins, and is regulated, in turn, by the action of these sequence-specific transcription factors, especially the so-called ‘pioneer factors’ that can contact their binding sites even when nucleosome-bound within inactive chromatin regions [32]. There are multiple methodologies available for the study of chromatin accessibility, and one of the first systematic applications of these techniques to islet biology was published by Gaulton and colleagues in 2010, who employed “FAIRE-Seq” (formaldehyde-assisted isolation of regulatory elements followed by high-throughput sequencing) to map open chromatin regions in whole human islets [33]. They identified 340 genes with islet-selective open chromatin regions, among them, not surprisingly, the β-cell expressed PDX1, SLC30A8, and NKX6.1 loci, and found that open chromatin regions are enriched for SNPs associated with the genetic risk for type 2 diabetes. More recently, a much-improved technology, termed ‘ATACseq’, for ‘Assay for Transposase-Accessible Chromatin with high throughput sequencing’, became available to map open chromatin with much higher resolution [34], [35]. ATACseq utilizes the hyperactive T5 transposase, which is used to simultaneously cut DNA and ligate adaptors for sequencing [36]. The Greenleaf group, recognizing that the transposase could only access naked or near-naked DNA, adapted this technology to use on intact open chromatin, which was then sequenced, while closed chromatin was inaccessible [34], [35]. This approach has since been used successfully to provide detailed maps of accessible chromatin in sorted human α- and β-cells, which will greatly facilitate the functional characterization of diabetes-associated genetic variants and aid in the integration of global transcription regulatory networks [37].
2.2. DNA methylation
The second major epigenetic modification to our genomes occurs through methylation of nuclear DNA on specific residues, specifically at the C5 position of cytosines to create 5-methylcytosine. This modification occurs almost exclusively in the context of CpG dinucleotides in non-neuronal cells, which through their palindromic nature provide a convenient way to maintain the methylated state of a specific genomic region through mitotic cell divisions. This is so because once a DNA strand is replicated during S-phase, the enzyme DNA methyltransferase 1 (DNMT1) recognizes the now half-methylated sites and methylates the opposite cytosine to restore the original state utilizing S-adenosyl-methionine as the methyl donor (Figure 1). Thus, DNA methylation is the only epigenetic mark for which we understand inheritance through DNA replication. Most parental DNA methylation marks are erased during early embryogenesis and then reestablished and subsequently maintained in a cell type-specific manner. With the exception of the aforementioned imprinting diseases, which are caused by improper DNA methylation status, DNA methylation was considered for a long time to be a ‘boring’ epigenetic mark, because of its apparently invariant nature once established.
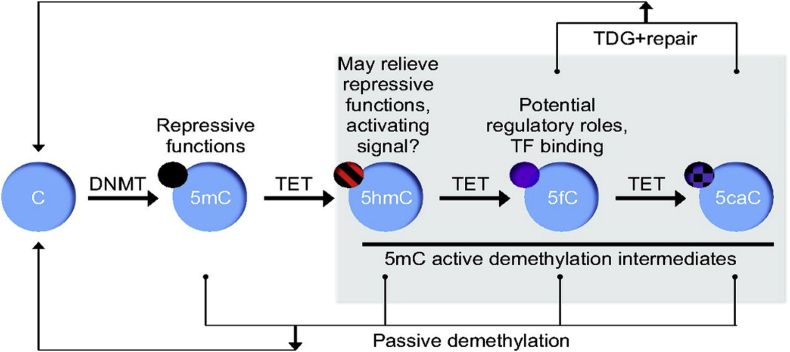
Figure 1.
5mC oxidation and demethylation process. 5′-methyl cytosine (5mC) is generated by the action of DNA methyltransferases (DNMTs), and can be oxidized by TET enzymes to produce 5hmC, 5fC, and 5caC. All oxidative derivatives can be diluted out during replication, due to lack of recognition by the DNMT1 (passive demethylation). Alternatively, 5fC and 5caC can be excised by TDG and repaired by the base excision repair mechanism to C (active demethylation).
However, this all changed with the discovery in 2009 of an enzymatic pathway for targeted removal of DNA methylation (Figure 1). Members of the ‘Ten-eleven translocation’ or ‘Tet’ gene family were shown to be able to catalyze oxidation of 5mC to 5-hydroxymethylcytosine (5hmC) [38], [39], which provides the starting point for two non-mutually exclusive mechanisms leading to demethylation of specific CpG's. In the first model, the Tet enzymes can oxidize 5hmC further to 5-formlyctosine (5fC) and then 5-carboxylcytosine (5caC), although at reduced efficacy compared to oxidation from 5mC to 5hmC [40] (Figure 1). Following further enzymatic steps, 5caC bases can eventually be replaced by un-modified cytosines through the base excision repair mechanism [41]. Second, and perhaps more likely to matter in vivo, hydroxymethylation of specific cytosines results in their passive but targeted demethylation in replicating cells, because the maintenance DNA methyltransferases do not recognize hemimethylated 5 hmC nucleotides as a substrate following DNA replication during S-phase, resulting in an unmethylated newly synthesized strand (Figure 1). Regardless of the molecular mechanism, hydroxymethylation effected by the Tet enzymes at specific loci presents a pathway to decrease DNA methylation even for fully committed cell lineages. Around the same time as the discovery of the role of Tet enzymes in de-methylation, determining CpG methylation and hydroxymethylation genome-wide became feasible [42], [43], [44], quickly establishing that while the fully methylated state of repetitive elements is invariant, methylation levels at promoters and enhancers can be highly divergent between different tissues, and even dynamic within the same cell type, for instance during the aging process [45].
DNA methylation frequently represses transcription and causes gene silencing [6], although it was shown more recently that, on occasion, DNA methylation correlates with gene activation [46]. While in this review, we are focusing on the context of gene regulation and imprinting introduced above, DNA methylation is also critical for X chromosome inactivation in females, and to maintain genome integrity through inactivating transposable elements, many of which retain their ability to ‘jump,’ even once integrated into the human genome [47]. These divergent roles are reflected in the global landscape of DNA methylation, where large stretches of repetitive sequences, including those of transposable elements, are fully methylated, while gene promoters are frequently unmethylated.
The human genome contains about 30 million CpG dinucleotides, constituting approximately 1% of the haploid genome, much less than would be expected by chance, indicating that CpGs were actively selected against during evolution. While the majority of CpGs are dispersed sparsely throughout the genome, a small fraction (1–2%) is clustered into dense arrays termed ‘CpG islands’ (CGIs). CGIs are defined as genomic regions with more than 50% CpG content over a span greater than 500 base-pairs in length [48]. Strikingly, the majority of CGIs occur near RNA polymerase II promoters, suggesting a direct impact on gene transcription depending on their methylation status. CGI methylation can directly regulate gene expression by determining whether or not the basal transcriptional machinery can bind to a gene promoter. Most CpGs in the genome are highly methylated, and, as introduced above, this level of methylation ensures genomic stability by silencing transposable elements. In contrast, gene promoters and distal regulatory elements such as enhancers are often lowly and variably methylated [45], [49], [50].
2.3. Non-coding RNAs
A third type of epigenetic regulators are non-coding RNAs (ncRNAs). An interest in ncRNAs began in the 1990s. The long non-coding RNA (lncRNA) Xist, associated with the inactive X chromosome [51], and H19, associated with the imprinted IGF2-locus [52], were first characterized during that decade, and RNA interference through microRNAs (miRNAs) was discovered around the same time [53], [54], [55]. In recent years, there has been an explosion of knowledge and publication about the transcription, processing, and function of ncRNAs (for recent reviews see [56], [57], [58], [59]).
Mature miRNAs are 21–25 nucleotide single-stranded molecules with high sequence homology to the 3′ UTR of one or—more commonly—multiple protein-encoding RNAs. In mammals, they are processed by cleavage of longer double-stranded RNA precursors by the ribonuclease Dicer [60]. They bind their targets in an RNA-induced silencing complex (RISC) complex that includes an Argonaute protein, the enzyme that cleaves miRNA targets (reviewed in [61]). Higher homology mRNAs are targeted for immediate degradation, while for mRNA with lower sequence homology to miRNAs translation is blocked, leading to de-adenylation and ultimate degradation of the mRNA [59].
lncRNAs are defined as having a length of greater than 200 nucleotides, but they are often much longer. They are usually shorter than mRNA, but like mRNA are spliced and polyadenylated. Unlike mRNAs, lncRNAs are enriched in the nuclear fraction of the cell. lncRNAs can be divided into multiple classes [62]. These include: (1) long intergenic ncRNAs (lincRNAs) that do not overlap with any coding gene; (2) antisense lncRNAs; (3) pseudogenes; (4) long intronic ncRNAs; and (5) promoter- and enhancer-associated transcripts.
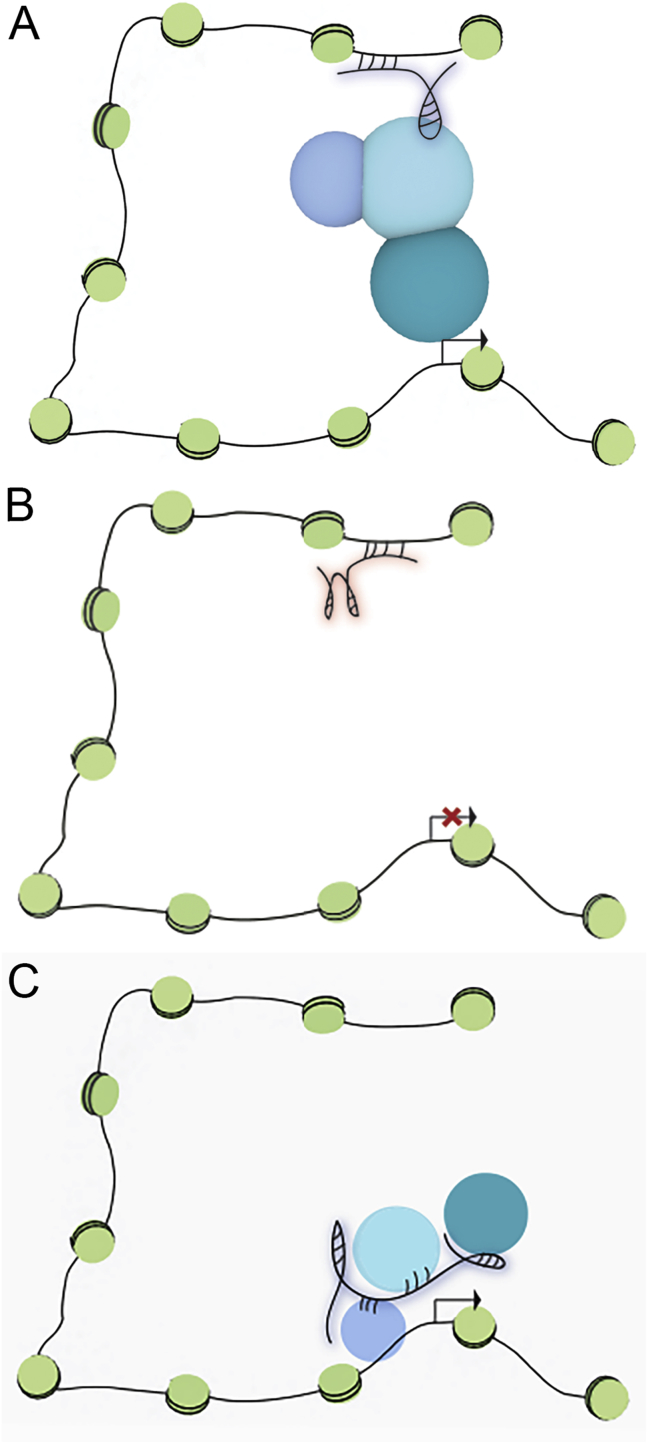
These disparate lncRNAs function through a variety of mechanisms (Figure 2), and lncRNAs can both activate and repress gene expression. lncRNAs such as Xist act as scaffolds and bind both proteins that direct them to specific sites within the genome as well as the chromatin remodeling complexes that either silence or activate gene transcription [63]. At least in the case of Xist, spreading and silencing occurs only in cis, i.e. only on one of the two X-chromosomes, and seems to be controlled by proximity, explaining the major differences in Xist occupancy between the active and inactive X chromosomes [64]. Other ways IncRNAs can influence gene expression include mediating enhancer/promoter looping [65], repression of gene expression by antisense transcription [66], and stabilization/destabilization of mRNA by hybridization [67].
Figure 2.
lncRNAs epigenetically regulate gene transcription through multiple mechanisms. (A) lncRNAs can directly or indirectly bind enhancers and recruit transcription factors to promote chromatin looping and gene activation. (B) lncRNAs can directly or indirectly bind enhancers to block chromatin looping, resulting in gene silencing. (C) lncRNAs can directly or indirectly interact with chromatin while recruiting HATs, HDACs or other chromatin-modifying enzymes to activate or silence gene transcription.
3. Epigenetic impact on islet cell development
3.1. Epigenetic modifiers contribute to pancreas development
The pancreas is derived from the foregut endoderm, from which the liver, lung, esophagus, and stomach also arise. Cells in the foregut endoderm have to “decide” which path of differentiation to follow, and epigenetic processes help to shape this decision. Thus, when Xu and colleagues mapped key chromatic marks in pancreatic versus hepatic cell fate choices, they found that marks of active transcription (histone H3 acetylation) and polycomb-complex mediated repression (H3K27me3) were divergent between genes fated to be expressed in the pancreas versus the liver [68]. Consequently, genetic ablation of the enzymes responsible for these chromatin marks led to a partial redistribution of cells between the two primordia.
Mouse or human embryonic stem cells can be differentiated into foregut endoderm, multipotent pancreatic progenitors, and even immature endocrine cells in vitro using precisely defined but still improving combinations of growth factors and other medium additives [69], [70]. This paradigm by and large recapitulates the sequence of gene activation and repression events that occur during pancreatogenesis in vivo. For example, Wang and colleagues found that the FOXA transcription factors bind their enhancer targets as early as the foregut endoderm stage, while the pancreas-enriched transcription factor PDX1 occupies only a subset of enhancers once the embryonic stem cells are induced to differentiate towards the pancreatic lineage. At this later stage of pancreatic differentiation, FOXA binding at liver-specific enhancers is lost but maintained at pancreas-specific enhancers, and the chromatin at the latter changes from a ‘poised’ to an ‘active’ state, at least as reflected in specific histone modifications [71]. These observations in ES cell culture recapitulate earlier findings using mouse models that identified the Foxa and Pdx1 genes as essential to pancreas development [72], [73], [74].
Direct evidence for the critical role of epigenetic processes in pancreas development comes from multiple studies in which key epigenetic enzymes were removed in the developing pancreas using Cre/loxP-mediated cell type-specific gene ablation. Thus, without the DNA methyltranferase Dnmt1, pancreas development is halted due to inappropriate activation of the p53 locus [75]. Likewise, a hypoplastic pancreas results when a critical component of the SWI/SNF nucleosomal remodeling complex, Brg1, is ablated in the pancreas primordium, at least in part due its role as a Pdx1 co-activator [76]. In addition, pancreas-wide embryonic deletion of the miRNA-processing ribonuclease Dicer results in a hypoplastic pancreas, likely due to alterations in Notch signaling pathway components [77]. It is perhaps unsurprising that complete ablation of these global regulators of the epigenome results in such dramatic phenotypes, but they still serves as a powerful confirmation of the concept that without the interplay of lineage-specific transcription factors with epigenetic enzymes, normal development cannot occur.
3.2. Differentiation of the endocrine cells types is controlled by epigenetic enzymes
Key evidence for the impact of epigenetic modifying enzymes comes from the use of specific inhibitors and various gene ablation models in the mouse. For instance, when explants of the embryonic pancreas anlage are cultured in the presence of HDAC (histone deacetylase) inhibitors, which would be expected to prevent gene silencing, multipotent progenitor cells produce an excess of endocrine cells at the expense of the exocrine pancreas, resulting in increases in both the β- and δ-cell populations [78]. As would be expected from these results, overexpression of HDAC enzymes led to decreased numbers of β- and δ-cells [79].
A key mediator of gene silencing at the chromatin level is the ‘Polycomb repressive complex’, named after a mutant Drosophila melanogaster fruit fly. During early development, Polycomb group genes function in the silencing of Hox gene clusters in organisms ranging from flies to humans. The Polycomb repressive complex 2 (PRC2) acts by methylating lysine 27 of histone H3 to H3K27me3. This modification is followed by binding of the PRC1 complex, which ubiquitylates histone 2A (H2A), blocking the action of the histone 3 lysine 4 (H3K4)-methylating enzymes needed to achieve an active promoter state [80]. During differentiation of stem cells in vitro, transcriptional regulators tended to lose H3K27 trimethylation, while those that gained H3K27 trimethylation tend be involved in cell function and morphology. When the PRC2 component Ezh2, necessary for the gene silencing process, was deleted in the pancreas anlage, the number of endocrine progenitors and hence mature β-cells was increased [81], which is consistent with the results from the stem cell analyses, since endocrine cells are among the first cells to differentiate within the pancreas.
3.3. Maintaining mature endocrine cell function and islet cell identity by epigenetic means
The major endocrine cell types, i.e. insulin-producing β-cells, glucagon-producing α-cells and somatostatin-secreting δ-cells, are closely related in terms of embryonic lineage and share common functional properties in terms of stimulus-secretion coupling, which is reflected in overall similar gene expression profiles [37], [82]. Nevertheless, specific differences in gene activation programs need to be established and maintained to assure the unique functions of each cell type throughout life. This cell-type discrimination is accomplished by a highly complex network of DNA-binding transcription factors, whose individual impact has been delineated in great detail over the past 20 years through gene ablation studies in the mouse [72], [73], [74], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93]. One particularly intriguing example, which also highlights the coordinated action of DNA-binding transcription factors with chromatin modifying enzymes, is the α-cell transcription factor Arx, or Aristaless-related homeobox. This DNA-binding protein is required for α-cell maintenance [94], [95], but it also needs to be repressed in β-cells to prevent their transdifferentiation into glucagon-producing cells [96]. Arx silencing in β-cells is accomplished, at least in part, by binding of the transcription factor Nkx2.2 to the Arx promoter, followed by recruitment of the DNA methyltransferase Dnmt3, increased CpG methylation, and binding by MeCP2, a methylated DNA binding protein, to establish and maintain a fully repressed state [97], [98]. Consequently, genetic ablation of the genes that establish DNA methylation at the Arx locus in β-cells causes transdifferentiation of the mutant cells towards the α-cell phenotype.
In addition to the manipulation of the enzymes that mediate epigenetic modifications described above, genome-wide studies of chromatin marks and open chromatin states are contributing to elucidate the epigenetic landscape of islet endocrine cell subtypes [29], [37], [99], [100]. Given the remarkable plasticity of the endocrine cell types [94], [96], [101], it is quite satisfying that major histone marks such as the activating H3K4me3 and repressive H3K27me3 histone modifications are similar between human α- and β-cells, while a more dissimilar pattern is observed in the more distantly related exocrine pancreas [100]. Confirmation of the likeness of chromatin states between α- and β-cells was recently obtained by ATACseq analysis [37]. The remarkable cellular plasticity of α-cells as seen in mouse gene and cell ablation models finds its corollary in the fact that human α-cells exhibit many more ‘bivalently’ marked loci than α-cells. Bivalency refers to the simultaneous presence of activating H3K4me3 and repressive H3K27me3 histone modifications at the same promoter, an occurrence first observed in genes that need to be rapidly activated following differentiation from embryonic stem cells [31].
Non-coding RNAs help to maintain the identity and function of islet cells types. One of the first RNAs shown to have a role in pancreas function was miR-375 [102]. Its overexpression suppressed insulin secretion, and it repression conversely enhanced insulin secretion. Mice completely lacking mir-375 display impaired glucose homeostasis, increased α-cell mass, and decreased β-cell mass due to reduced proliferation [103]. Later, a wider role of miRNAs in β-cell function was discovered with a deletion of Dicer within the β-cell domain. These mice lose insulin expression, and the degranulated β-cells slowly disappear from the pancreas [104], [105].
An appreciation of the importance of lncRNAs to islet biology has taken longer to develop than that for miRNAs. In 2012, Jorge Ferrer's group reported the presence of >1,000 islet-specific lncRNAs in mice and examined the expression of lncRNAs during directed differentiation of human ES cells to insulin-producing cells [106]. Moreover, many lncRNAs were differentially regulated in islets from donors with type 2 diabetes compared to controls. In addition, deletion of the lncRNA termed ‘β-cell long intergenic non-coding RNA’ (βlinc) in mice resulted in a reduction of β-cells with a concomitant increase in δ-cells along with impaired glucose homeostasis [107].
A key functional property of the β-cell is its ability to sense a dynamic range of physiological glucose concentrations and react with a graded insulin secretory response. Central to this unique property is the near-exclusive expression of glucokinase, with a high Km for glucose (around 8 mM in humans) [108], [109], as opposed to hexokinase, which is active in neurons, skeletal muscle cells, etc. Because of the very low Km of hexokinase for glucose, it essentially always operates at Vmax. In addition, glucokinase displays cooperativity, with a Hill-coefficient of 1.7, resulting in an inflection point in the glucose response curve of 3.9 mM, which enables it to function as the glucose sensor and to maintain the set-point for blood glucose of 5 mM [108], [109], [110]. Because of the low Km for glucose of hexokinase, it is critical that the β-cell not activate transcription of HK1, the gene which encodes it, as any significant expression of this enzyme would shift the glucose response curve to the left, hence causing hypoglycemia. This consequence has been seen perhaps most strikingly in humans with activating HK1 promoter mutations which lead to congenital hyperinsulinism in children due to inappropriate expression of Hexokinase I [111]. Indeed, multiple enzymes and transporters exist, the expression of which would interfere with β-cell function; this fact has led to the concept that the corresponding genes are specifically repressed, or ‘disallowed’ in β-cells [110], [112], [113], [114], [115], [116]. Multiple mouse models support a role for epigenetics in regulating these disallowed genes. In mice, deletion of the de novo DNA methylatransferase Dnmt3a resulted in promoter hypomethylation and elevated gene expression at Hk1 and Lactase Dehydrogenase A (Ldha), another protein normally suppressed in mature β-cells [117]. Furthermore, Dicer inactivation in adult β-cells led to the same degranulation of β-cells observed when the gene was deleted developmentally and was accompanied by an upregulation of six normally silent genes [104], [105], [118].
Non-uniform insulin secretion and function by β-cells is a concept that has been revisited over the years [119], [120], [121]. New single-cell assays have provided multiple tools for investigating these different populations, including single-cell RNAseq [122], [123], [124], mass cytometry [125], single-cell calcium imaging [45], [126], and the ability to delineate β-cell subpopulations on the basis of cell-surface antigens [127]. It is currently unclear whether this heterogeneity reflects different subgroups of β-cells, or merely temporary subpopulations that are cycling between states. Whichever is the case, epigenetics likely plays a role in the regulation of these subgroups. However, state-of-the-art single-cell techniques are unlikely to provide a satisfactory answer for any given gene, except for traditional bisulfite sequencing methods that assay one allele at a time [128]. Although single-cell ATACseq has already been reported [129], [130], even for single-cell RNA-seq where most transcripts will have multiple copies, low or even moderate expression can lead to “drop-outs”, or false negatives, for expression of a certain gene in any given cell [131]. A single cell contains only two copies of a certain chromatin region or DNA sequence, resulting in either 0, 1, or 2 reads per cell. Thus, if in single-cell ATACseq a region is not sequenced, determining whether it is truly an area of inaccessible chromatin or if the sequence reads are missing by chance, is difficult. Computational biology tools that take into account thousands of genomic regions simultaneously will have to be devised in order to be able to determine if β-cell subtypes are reflected on the chromatin level.
4. Epigenetics and human metabolic disease
Given what we have learned about the impact of epigenetic factors for the development and maintenance of islet cell identity and function, it is not surprising that alterations in the epigenetic program can have severe pathophysiological consequences. In fact, multiple studies in rodent models and humans have shown that even in utero exposure can lead to multi-generational inheritance of metabolic disease (see below), which clearly has to be mediated by epigenetic processes, as there are no mutations in the DNA sequence evident in these processes. Likewise, the obvious effects of the modern lifestyle of limited physical activity and obesity have a dramatic effect on the incidence of type 2 diabetes in the Western and increasingly also the developing world. The worldwide increase in type 2 diabetes cannot be explained by an altered prevalence of disease risk-conferring mutations. While the precise molecular causality is much more difficult to prove for epigenetic events than for mutations to the DNA, an important contribution of an altered epigenome to T2D susceptibility is supported by multiple lines of evidence.
4.1. Intrauterine development and the transgenerational epigenetic inheritance of metabolic disease
The ‘Dutch hunger winter’ (‘hongerwinter’ in Dutch) is the most famous example of the effects of maternal diet to diabetes risk. At the end of World War II, Nazi occupiers imposed a strict diet on the population of the northwestern Netherlands, with daily rations as low as 600 kcal. Children of the mothers that were pregnant during this time period were not only born with low birth weight as expected, but also had a significantly higher risk of metabolic disease as adults [132]. A separate study following boys with known birth-weight until age 64 found that individuals with low birth weight had a higher incidence of impaired glucose tolerance and impaired β-cell function than others in the same cohort [133].
Rodent models offer the opportunity to study these phenomena in isolation and to find correlates of phenotypic changes in the epigenome. The effects of maternal nutritional status during pregnancy on the long-term metabolic health of offspring can be studied in models of intrauterine growth restriction (IUGR), either by providing very limited maternal nutrition or by performing uterine artery ligation to induce uteroplacental insufficiency [134], [135]. While these paradigms of course affect multiple organ systems, in many cases the activity of crucial β-cell transcription factors such as Pdx1 was altered, evidently due to a modified epigenetic state [136], [137], [138]. In another paradigm, pregnant dams were fed isocaloric diets, but one group was protein-restricted. This alteration in macronutrient intake was sufficient to cause hypermethylation and decreased activation of the Hnf4a promoter, an effect that did not resolve as the offspring aged [138].
In the scenarios described thus far, the fetus itself is exposed to detrimental conditions that affect its metabolic health; thus, they are examples of the ‘developmental origins of health and disease’, or DOHAD, hypothesis [133]. Remarkably, though, in certain detrimental conditions effects are seen in the generation not directly exposed to the stimulus, resulting in transgenerational inheritance not mediated by changes to the DNA sequence.
4.2. Parental transmission of epigenetic risk for metabolic dysfunction
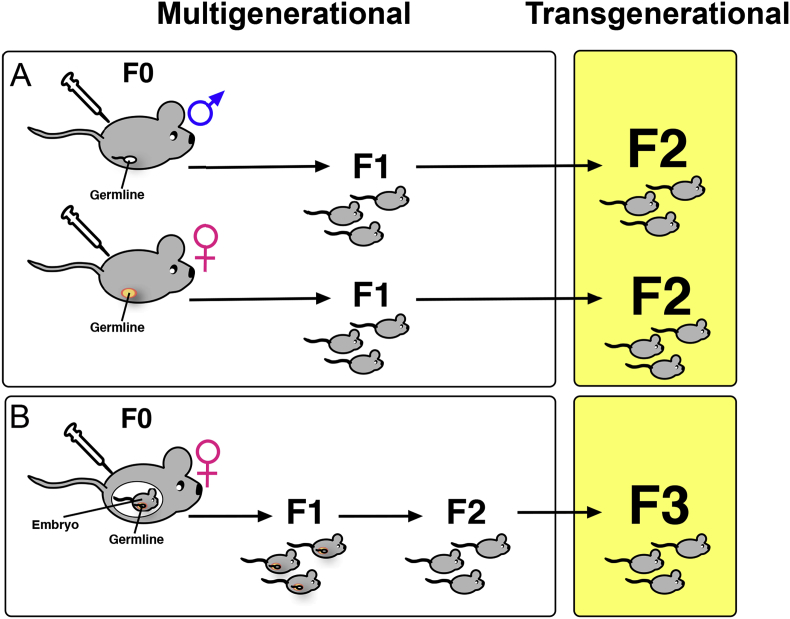
When considering the consequences of the DOHAD hypothesis, it is essential to be aware that aberrant epigenetic alterations can occur by exposure to detrimental conditions in utero, as exemplified by the Dutch hunger winter described above, or through inheritance of epigenetic effects to the next generation via molecular changes to the parental gametes. Finding the molecular evidence for transmission of epigenetic marks through parental gametes is particularly difficult since most epigenetic marks such as DNA methylation and histone modifications undergo near wholesale erasure during gametogenesis and again in the preimplantation embryo post-fertilization to establish and maintain cellular pluripotency. Thus, for true epigenetic transgenerational inheritance, environmental cues must be reflected in permanent epigenetic changes that are exempt from erasure. Therefore, it is essential to carefully consider the timing of environmental exposure to differentiate multigenerational from transgenerational epigenetic effects. Thus, when a F1 fetus is exposed to a detrimental condition such as nutrient deprivation or maternal stress in utero, the gametes of that fetus (F2) will also experience the biochemical consequences of the exposure. Therefore, transmission of an epigenetic effect should only be considered to be transgenerational if its phenotypic consequences are also observed in the F3 generation (see Figure 3).
Figure 3.
Epigenetic inheritance can be multigenerational or transgenerational. A) Multigenerational inheritance refers to a change in a trait or phenotype in the F1 offspring of males or non-pregnant females (F0) exposed to a stimulus that impacts the epigenome without changing the DNA sequence. B) In the case of a pregnant female exposed to an environmental toxin, for instance, the F0 parent, the F1 fetus, and the F2 germline within the fetus are all exposed. Therefore, in this case, only if the F3 generation also shows an epigenetically altered phenotype does transgenerational inheritance occur.
A worrisome example of detrimental environmental exposures that have multigenerational effects on β-cell function is parental exposure to endocrine disruptors. While malnutrition can be easily diagnosed and prevented at trivial costs, the effects of environmental toxins are mostly unknown and can take decades to be discovered. One such industrial chemical is Bisphenol A (BPA), which is among the most commonly produced chemicals in the world today, with an annual production of 4 million metric tons. BPA is used for the synthesis of epoxy resins and plastics, and finds its way into ubiquitous consumer goods such as water bottles. BPA and related compounds mimic the action of estrogens, and fetal exposure to BPA causes impaired glucose handling in rodents, which appears to be caused by altering the epigenetic state of β-cell expressed genes Pdx1 and Igf2, resulting in impaired insulin secretion [139], [140]. Sperm of the F1 generation after BPA exposure exhibited similar changes to the DNA methylation state as the F0 generation, suggesting that in this case, and likely other, epigenetic transmission can occur via the male lineage [141]. Given the plethora of new chemical structures and xenobiotics being produced world-wide, it is extremely likely that many other detrimental effects of environmental toxins will be discovered in the future, a subset of which will have multigenerational and transgenerational effects.
When males are exposed to detrimental or diabetogenic conditions, the paternal contribution can be isolated to epigenetic information carried in sperm and observed in phenotypes of the F2 generation. Thus, when male rats where fed a chronic high fat diet (HFD), their female offspring showed glucose intolerance and reduced β-cell function that worsened with age, which clearly had to be the consequence of epigenetic changes carried in the sperm. In fact, many genes known to function in insulin exocytosis and β-cell survival were differentially expressed in β-cells of females born to an obese father compared to those of females that were offspring of a lean male [142]. Remarkably, new evidence is accumulating that these epigenetic effects on metabolic health are not confined to rodents, but can also occur in humans, as the sperm DNA methylome was found to be significantly different between lean and obese men [143], suggesting, though not proving, that a predisposition to obesity could be transmitted epigenetically.
4.3. The methylome is dynamic during ontogeny
When comparing genome-wide profiles and maps of various histone marks to those of DNA methylation among cell types and between individuals, it is quite clear that DNA methylation is the more uniform and stable mark. In fact, originally it was thought that once the methylome is established in the various cell lineages during early fetal development, the identical methylation pattern is maintained in each cell type, because the maintenance DNA methylase DNMT1, which is associated with the DNA replication fork, immediately remethylates the hemimethylated CpGs that result from DNA replication. As introduced above, this view is clearly an over-simplification, and the methylation state can be highly dynamic even within the same cell lineage, as shown by genome-wide methylation studies comparing, for instance, young and old β-cells [45]. The discovery of the Tet hydroxymethylation enzymes provided the molecular underpinnings for a targeted demethylation pathway (Figure 1).
Consequently, for many key β-cell factors, changes in the methylation status of their promoter have been determined and correlated with changes in gene activity and islet function. For instance, it was found that in islets from people with type 2 diabetes, the promoter of the key transcriptional coactivator PGC-1α (peroxisome proliferator activated receptor gamma coactivator-1 alpha) is hypermethylated compared to controls and also exhibits reduced steady-state transcript levels [144], providing a possible explanation for reduced mitochondrial function in diabetic β-cells. In a different study, increases in body mass index and hemoglobin A1c correlated with increased islet cell DNA methylation and reduced expression of the glucagon-like peptide 1 receptor, which suggests a reduced responsiveness of β-cells to incretins in obese and/or diabetic patients [145]. While it is impossible in these cross-sectional studies to prove that aberrant DNA methylation of any specific gene causes diabetes, they nevertheless suggest that impaired epigenetic regulation of islet cell genes could factor into disease development.
Today, it is possible to determine the DNA methylation status genome-wide, either by ‘bisulfite sequencing’ or by array-bases methodologies. While array-based techniques offer methylation analysis at lower costs than bisulfite sequencing, they only cover about 0.5% of all CpG sites in the genome and therefore can miss important changes to the methylome. Array-based methods were used to find 273 differentially methylated regions in whole islets when comparing the promoter methylation of five donors with type 2 diabetes to eleven controls [146]. However, genome-wide methylation analysis employed in a similar experiment with six controls and eight donors with type 2 discovered 25,820 differentially methylated regions [147]. Using this same technology, Avrahami and colleagues showed that the mouse β-cell methylome is dynamic with age, especially at distal enhancers, and that alterations frequently occur in genes that control β-cell function [45].
5. Conclusions
The interpretation of the genome—with a few exceptions the same in all our cell types—is essential to enable the development of multicellular organisms with a division of function among the multiple organ systems and cell types. The micro-organ of the pancreatic islet is no exception, and epigenetic regulators play key roles in endocrine cell development, differentiation, and function. While over the past 20 years, the focus of researchers world-wide has been on the determination of the function of individual epigenetic factors, and more recently, the genome-wide mapping of epigenetic marks with increasing breadth and depth of coverage, the future promises to allow for direct, targeted control of gene function through epigenetic control. In addition to broad-acting drugs, such as inhibitors of histone deacetylase, which can reduce the sensitivity of islets to apoptosis-inducing cytokines [148], current and future approaches will employ mutated TALE and CRISPR-Cas proteins tethered to epigenetic enzymes such as DNA methyltransferases, a combination which can target these modifiers to a single selected locus in the genome [149], [150]. These approaches will thus enable control of islet cell function and proliferation without any changes to the DNA sequence. When combined with cell-type specific delivery systems, these methods will be able to elevate precision medicine for diabetes to a whole new level.
Conflict of interest
None declared.
References
- 1.Wu C., Morris J.R. Genes, genetics, and epigenetics: a correspondence. Science. 2001;293(5532):1103–1105. doi: 10.1126/science.293.5532.1103. [DOI] [PubMed] [Google Scholar]
- 2.Elhamamsy A.R. Role of DNA methylation in imprinting disorders: an updated review. Journal of Assisted Reproduction and Genetics. 2017;34(5):549–562. doi: 10.1007/s10815-017-0895-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuman C., Beckwith J.B., Weksberg R. Beckwith-Wiedemann syndrome. In: Pagon R.A., editor. GeneReviews(R) 1993. Seattle (WA) [Google Scholar]
- 4.DeBaun M.R., King A.A., White N. Hypoglycemia in Beckwith-Wiedemann syndrome. Seminars in Perinatology. 2000;24(2):164–171. doi: 10.1053/sp.2000.6366. [DOI] [PubMed] [Google Scholar]
- 5.Temple I.K., Mackay D.J.G., Docherty L.E. Diabetes mellitus, 6q24-related transient neonatal. In: Pagon R.A., editor. GeneReviews(R) 1993. Seattle (WA) [Google Scholar]
- 6.Ma D. Impaired glucose homeostasis in transgenic mice expressing the human transient neonatal diabetes mellitus locus, TNDM. Journal of Clinical Investigation. 2004;114(3):339–348. doi: 10.1172/JCI19876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambrosi C., Manzo M., Baubec T. Dynamics and context-dependent roles of DNA methylation. Journal of Molecular Biology. 2017;429(10):1459–1475. doi: 10.1016/j.jmb.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 9.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Research. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luger K. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 11.Parthun M.R. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26(37):5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- 12.Allfrey V.G., Faulkner R., Mirsky A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng S.S. Dynamic protein methylation in chromatin biology. Cellular and Molecular Life Sciences. 2009;66(3):407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan F., Shi Y. Epigenetic regulation: methylation of histone and non-histone proteins. Science in China – Series C: Life Sciences. 2009;52(4):311–322. doi: 10.1007/s11427-009-0054-z. [DOI] [PubMed] [Google Scholar]
- 15.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews Genetics. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 16.Lennartsson A., Ekwall K. Histone modification patterns and epigenetic codes . Biochimica et Biophysica Acta (BBA) – General Subjects. 2009;1790(9):863–868. doi: 10.1016/j.bbagen.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Handy D.E., Castro R., Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123(19):2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.K., Samaranayake M., Pradhan S. Epigenetic mechanisms in mammals. Cellular and Molecular Life Sciences. 2009;66(4):596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner B.M. Defining an epigenetic code. Nature Cell Biology. 2007;9(1):2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 20.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peschansky V.J., Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9(1):3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holoch D., Moazed D. RNA-mediated epigenetic regulation of gene expression. Nature Reviews Genetics. 2015;16(2):71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa F.F. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410(1):9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Feil R., Fraga M.F. Epigenetics and the environment: emerging patterns and implications. Nature Reviews Genetics. 2012;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 25.Vineis P. Epigenetic memory in response to environmental stressors. FASEB Journal. 2017;31(6):2241–2251. doi: 10.1096/fj.201601059RR. [DOI] [PubMed] [Google Scholar]
- 26.Roostaee A. Epigenetics in intestinal epithelial cell renewal. Journal of Cellular Physiology. 2016;231(11):2361–2367. doi: 10.1002/jcp.25401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 28.Gurard-Levin Z.A., Almouzni G. Histone modifications and a choice of variant: a language that helps the genome express itself. F1000Prime Reports. 2014;6:76. doi: 10.12703/P6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquali L. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nature Genetics. 2014;46(2):136. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein B.E. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 32.Iwafuchi-Doi M., Zaret K.S. Pioneer transcription factors in cell reprogramming. Genes & Development. 2014;28(24):2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaulton K.J. A map of open chromatin in human pancreatic islets. Nature Genetics. 2010;42(3):255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buenrostro J.D. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buenrostro J.D. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Current Issues in Molecular Biology. 2015;109 doi: 10.1002/0471142727.mb2129s109. 21 29 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adey A. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biology. 2010;11(12):R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ackermann A.M. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Molecular Metabolism. 2016;5(3):233–244. doi: 10.1016/j.molmet.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahiliani M. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito S. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber A.R. Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nature Communication. 2016;7:10806. doi: 10.1038/ncomms10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi I. 5-hydroxymethylcytosine represses the activity of enhancers in embryonic stem cells: a new epigenetic signature for gene regulation. BMC Genomics. 2014;15:670. doi: 10.1186/1471-2164-15-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu M. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nature Protocol. 2012;7(12):2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu M. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Avrahami D. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved beta cell function. Cell Metabolism. 2015;22(4):619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hark A.T. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405(6785):486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 47.Hancks D.C., Kazazian H.H., Jr. Active human retrotransposons: variation and disease. Current Opinion in Genetics & Development. 2012;22(3):191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zampieri M. Reconfiguration of DNA methylation in aging. Mechanisms of Ageing and Development. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Sheaffer K.L. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes & Development. 2014;28(6):652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stadler M.B. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 51.Clemson C.M. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. Journal of Cell Biology. 1996;132(3):259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brannan C.I. The product of the H19 gene may function as an RNA. Molecular and Cellular Biology. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Metzlaff M. RNA-mediated RNA degradation and chalcone synthase A silencing in petunia. Cell. 1997;88(6):845–854. doi: 10.1016/s0092-8674(00)81930-3. [DOI] [PubMed] [Google Scholar]
- 54.van der Krol A.R. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2(4):291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fire A. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 56.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 57.Beermann J. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiological Reviews. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 58.Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cellular and Molecular Life Sciences. 2016;73(13):2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson R.C., Doudna J.A. Molecular mechanisms of RNA interference. Annual Review of Biophysics. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernstein E. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 61.Hutvagner G., Simard M.J. Argonaute proteins: key players in RNA silencing. Nature Reviews Molecular Cell Biology. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 62.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spitale R.C., Tsai M.C., Chang H.Y. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6(5):539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown C.J. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 65.Bohmdorfer G., Wierzbicki A.T. Control of chromatin structure by long noncoding RNA. Trends in Cell Biology. 2015;25(10):623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pelechano V., Steinmetz L.M. Gene regulation by antisense transcription. Nature Reviews Genetics. 2013;14(12):880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 67.Yuan S.X. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Letters. 2014;349(1):87–94. doi: 10.1016/j.canlet.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 68.Xu C.R. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332(6032):963–966. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kroon E. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nature Biotechnology. 2008;26(4):443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 70.Rezania A. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature Biotechnology. 2014;32(11):1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 71.Wang A. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16(4):386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao N. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes & Development. 2008;22(24):3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jonsson J. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371(6498):606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 74.Offield M.F. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122(3):983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 75.Georgia S., Kanji M., Bhushan A. DNMT1 represses p53 to maintain progenitor cell survival during pancreatic organogenesis. Genes & Development. 2013;27(4):372–377. doi: 10.1101/gad.207001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McKenna B. Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet beta cells. Cell Reports. 2015;10(12):2032–2042. doi: 10.1016/j.celrep.2015.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lynn F.C. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56(12):2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 78.Haumaitre C., Lenoir O., Scharfmann R. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Molecular and Cellular Biology. 2008;28(20):6373–6383. doi: 10.1128/MCB.00413-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lenoir O. Specific control of pancreatic endocrine beta- and delta-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes. 2011;60(11):2861–2871. doi: 10.2337/db11-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Kruijsbergen I., Hontelez S., Veenstra G.J.C. Recruiting polycomb to chromatin. The International Journal of Biochemistry & Cell Biology. 2015;67:177–187. doi: 10.1016/j.biocel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu C.R. Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. EMBO Journal. 2014;33(19):2157–2170. doi: 10.15252/embj.201488671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dorrell C. Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Research. 2008;1(3):183–194. doi: 10.1016/j.scr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 83.De Vas M.G. Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development. 2015;142(5):871–882. doi: 10.1242/dev.110759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Decker K. Gata6 is an important regulator of mouse pancreas development. Developmental Biology. 2006;298(2):415–429. doi: 10.1016/j.ydbio.2006.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gradwohl G. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(4):1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haumaitre C. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(5):1490–1495. doi: 10.1073/pnas.0405776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jensen J. Control of endodermal endocrine development by Hes-1. Nature Genetics. 2000;24(1):36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 88.Krapp A. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes & Development. 1998;12(23):3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larsen H.L., Grapin-Botton A. The molecular and morphogenetic basis of pancreas organogenesis. Seminars in Cell & Developmental Biology. 2017 doi: 10.1016/j.semcdb.2017.01.005. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 90.Martin M. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Developmental Biology. 2005;284(2):399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 91.Pierreux C.E. The transcription factor hepatocyte nuclear factor-6 controls the development of pancreatic ducts in the mouse. Gastroenterology. 2006;130(2):532–541. doi: 10.1053/j.gastro.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 92.Seymour P.A. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(6):1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Soyer J. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development. 2010;137(2):203–212. doi: 10.1242/dev.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Collombat P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes & Development. 2003;17(20):2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilcox C.L. Pancreatic alpha-cell specific deletion of mouse Arx leads to alpha-cell identity loss. PLoS One. 2013;8(6):e66214. doi: 10.1371/journal.pone.0066214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Courtney M. The inactivation of Arx in pancreatic alpha-cells triggers their neogenesis and conversion into functional beta-like cells. PLoS Genetics. 2013;9(10):e1003934. doi: 10.1371/journal.pgen.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dhawan S. Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Developmental Cell. 2011;20(4):419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Papizan J.B. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes & Development. 2011;25(21):2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arda H.E. Age-dependent pancreatic gene regulation reveals mechanisms governing human beta cell function. Cell Metabolism. 2016;23(5):909–920. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bramswig N.C. Epigenomic plasticity enables human pancreatic alpha to beta cell reprogramming. Journal of Clinical Investigation. 2013;123(3):1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thorel F. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464(7292):1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Poy M.N. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 103.Poy M.N. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(14):5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kalis M. Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS One. 2011;6(12):e29166. doi: 10.1371/journal.pone.0029166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mandelbaum A.D. Dysregulation of Dicer1 in beta cells impairs islet architecture and glucose metabolism. Experimental Diabetes Research. 2012;2012:470302. doi: 10.1155/2012/470302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moran I. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metabolism. 2012;16(4):435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arnes L. betalinc1 encodes a long noncoding RNA that regulates islet beta-cell formation and function. Genes & Development. 2016;30(5):502–507. doi: 10.1101/gad.273821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matschinsky F.M. Banting Lecture 1995. A lesson in metabolic regulation inspired by the glucokinase glucose sensor paradigm. Diabetes. 1996;45(2):223–241. doi: 10.2337/diab.45.2.223. [DOI] [PubMed] [Google Scholar]
- 109.Matschinsky F.M., Glaser B., Magnuson M.A. Pancreatic beta-cell glucokinase: closing the gap between theoretical concepts and experimental realities. Diabetes. 1998;47(3):307–315. doi: 10.2337/diabetes.47.3.307. [DOI] [PubMed] [Google Scholar]
- 110.Sekine N. Low lactate dehydrogenase and high mitochondrial glycerol phosphate dehydrogenase in pancreatic beta-cells. Potential role in nutrient sensing. Journal of Biological Chemistry. 1994;269(7):4895–4902. [PubMed] [Google Scholar]
- 111.Stanley C.A. Perspective on the genetics and diagnosis of congenital hyperinsulinism disorders. Journal of Clinical Endocrinology & Metabolism. 2016;101(3):815–826. doi: 10.1210/jc.2015-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lemaire K., Thorrez L., Schuit F. Disallowed and allowed gene expression: two faces of mature islet beta cells. Annual Review of Nutrition. 2016;36:45–71. doi: 10.1146/annurev-nutr-071715-050808. [DOI] [PubMed] [Google Scholar]
- 113.Thorrez L. Tissue-specific disallowance of housekeeping genes: the other face of cell differentiation. Genome Research. 2011;21(1):95–105. doi: 10.1101/gr.109173.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Quintens R. Why expression of some genes is disallowed in beta-cells. Biochemical Society Transactions. 2008;36(Pt 3):300–305. doi: 10.1042/BST0360300. [DOI] [PubMed] [Google Scholar]
- 115.Pullen T.J. Identification of genes selectively disallowed in the pancreatic islet. Islets. 2010;2(2):89–95. doi: 10.4161/isl.2.2.11025. [DOI] [PubMed] [Google Scholar]
- 116.Pullen T.J., Rutter G.A. When less is more: the forbidden fruits of gene repression in the adult beta-cell. Diabetes, Obesity and Metabolism. 2013;15(6):503–512. doi: 10.1111/dom.12029. [DOI] [PubMed] [Google Scholar]
- 117.Dhawan S. DNA methylation directs functional maturation of pancreatic beta cells. Journal of Clinical Investigation. 2015;125(7):2851–2860. doi: 10.1172/JCI79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martinez-Sanchez A., Nguyen-Tu M.S., Rutter G.A. DICER inactivation identifies pancreatic beta-cell “disallowed” genes targeted by MicroRNAs. Mol Endocrinol. 2015;29(7):1067–1079. doi: 10.1210/me.2015-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wojtusciszyn A. Insulin secretion from human beta cells is heterogeneous and dependent on cell-to-cell contacts. Diabetologia. 2008;51(10):1843–1852. doi: 10.1007/s00125-008-1103-z. [DOI] [PubMed] [Google Scholar]
- 120.Salomon D., Meda P. Heterogeneity and contact-dependent regulation of hormone secretion by individual B cells. Experimental Cell Research. 1986;162(2):507–520. doi: 10.1016/0014-4827(86)90354-x. [DOI] [PubMed] [Google Scholar]
- 121.Bosco D. Heterogeneous secretion of individual B cells in response to D-glucose and to nonglucidic nutrient secretagogues. American Journal of Physiology. 1995;268(3 Pt 1):C611–C618. doi: 10.1152/ajpcell.1995.268.3.C611. [DOI] [PubMed] [Google Scholar]
- 122.Wang Y.J. Single-cell transcriptomics of the human endocrine pancreas. Diabetes. 2016;65(10):3028–3038. doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Segerstolpe A. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism. 2016;24(4):593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xin Y. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metabolism. 2016;24(4):608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 125.Wang Y.J. Single-cell mass cytometry analysis of the human endocrine pancreas. Cell Metabolism. 2016;24(4):616–626. doi: 10.1016/j.cmet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dadi P.K. TASK-1 potassium channels limit pancreatic alpha-cell calcium influx and glucagon secretion. Molecular Endocrinology. 2015;29(5):777–787. doi: 10.1210/me.2014-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dorrell C. Human islets contain four distinct subtypes of beta cells. Nature Communication. 2016;7:11756. doi: 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frommer M. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Buenrostro J.D. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523(7561):486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cusanovich D.A. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348(6237):910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kharchenko P.V., Silberstein L., Scadden D.T. Bayesian approach to single-cell differential expression analysis. Nature Methods. 2014;11(7):740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ravelli A.C. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351(9097):173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- 133.Hales C.N., Barker D.J. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35(7):595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 134.Constancia M. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417(6892):945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 135.Petry C.J. Diabetes in old male offspring of rat dams fed a reduced protein diet. International Journal of Experimental Diabetes Research. 2001;2(2):139–143. doi: 10.1155/EDR.2001.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Park J.H. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. Journal of Clinical Investigation. 2008;118(6):2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pinney S.E. Exendin-4 increases histone acetylase activity and reverses epigenetic modifications that silence Pdx1 in the intrauterine growth retarded rat. Diabetologia. 2011;54(10):2606–2614. doi: 10.1007/s00125-011-2250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sandovici I. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(13):5449–5454. doi: 10.1073/pnas.1019007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alonso-Magdalena P. Bisphenol A exposure during pregnancy disrupts glucose homeostasis in mothers and adult male offspring. Environmental Health Perspectives. 2010;118(9):1243–1250. doi: 10.1289/ehp.1001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li G.Q. F0 maternal BPA exposure induced glucose intolerance of F2 generation through DNA methylation change in Gck. Toxicology Letters. 2014;228(3):192–199. doi: 10.1016/j.toxlet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 141.Radford E.J. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345(6198):1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ng S.-F. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467(7318) doi: 10.1038/nature09491. 963–U103. [DOI] [PubMed] [Google Scholar]
- 143.Donkin I. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metabolism. 2016;23(2):369–378. doi: 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 144.Ling C. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51(4):615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hall E. DNA methylation of the glucagon-like peptide 1 receptor (GLP1R) in human pancreatic islets. BMC Medical Genetics. 2013;14 doi: 10.1186/1471-2350-14-76. 76–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Volkmar M. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO Journal. 2012;31(6):1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Volkov P. Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes. 2017;66(4):1074–1085. doi: 10.2337/db16-0996. [DOI] [PubMed] [Google Scholar]
- 148.Chou D.H. Inhibition of histone deacetylase 3 protects beta cells from cytokine-induced apoptosis. Chemistry & Biology. 2012;19(6):669–673. doi: 10.1016/j.chembiol.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bernstein D.L. TALE-mediated epigenetic suppression of CDKN2A increases replication in human. Journal of Clinical Investigation. 2015;125(5):1998–2006. doi: 10.1172/JCI77321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu X.S. Editing DNA methylation in the mammalian genome. Cell. 2016;167(1) doi: 10.1016/j.cell.2016.08.056. 233–247 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]