Abstract
Background
MicroRNAs have emerged as important regulatory non-coding RNAs that tune cellular responses to physiological perturbations and disease conditions. An increasing body of literature underlines the important roles of miRNA function in pancreatic β-cells in response to metabolic, genetic and inflammatory stress. Lessons from genetic loss- and gain-of-function studies have implicated several highly expressed and evolutionary conserved miRNAs in stress signal modulation, resolution and buffering, thereby forming stabilizing miRNA networks that preserve β-cell differentiation, function, proliferation and cell survival.
Scope of Review
This review will summarize our current knowledge of how biologically relevant miRNAs regulate stress responses in pancreatic β-cells, discuss the challenges and opportunities associated with using secreted miRNAs as biomarkers and forecast how mechanistic knowledge of miRNA function can be exploited in developing miRNA-based therapeutics.
Major Conclusions
miRNAs play important roles in the function, differentiation, proliferation, and survival of pancreatic β-cells. Many miRNA families that are regulated by metabolic, genetic, and inflammatory stressors have been found to coordinate the adaptive responses of β-cells in vivo in conditions such as obesity and diabetes.
Keywords: miRNAs, Pancreatic β-cells, Insulin secretion, Diabetes
1. MicroRNA function
MicroRNA (miRNA) molecules are short, non-coding RNAs that mediate RNA silencing and post-transcriptional regulation of gene expression. They are transcribed by RNA polymerase II as long primary transcripts that are capped, polyadenylated, and spliced, thus resembling conventional mRNAs. However, unlike mRNA, the active region of the miRNA is contained within a 70-nucleotide hairpin structure that forms within the RNA transcript. The hairpin is cleaved by the endonuclease enzymes Drosha and Dicer to yield a ≈22-nucleotide double-stranded RNA (dsRNA) [1]. This RNA product, referred to as the ‘mature’ miRNA, resembles a small-interfering RNA (siRNA) and is assembled into the RNA interference (RNAi)-effector complex known as RISC (RNA-induced silencing complex). The miRNA directs RISC to partially complementary mRNA sites by base-pairing [2]. Consequently, the expression of targeted mRNAs is blocked either by translational repression or by mRNA degradation [3], [4]. In this way, a cell can use the RNAi pathway to regulate the expression of many genes simply by transcribing the appropriate miRNA.
In addition to the canonical miRNA pathway, a fundamentally different mode of action has been proposed in which coding transcripts, transcribed pseudogenes, and long non-coding RNAs, collectively referred to as “competing endogenous RNA” (ceRNA), “talk” to each other using miRNA response elements (MREs) [5]. This model proposes that highly expressed MRE-containing RNAs act as “sponges” to titrate miRNAs away from other normal targets and thereby form a large-scale regulatory network across the transcriptome. Experimental evidence for such a ceRNA crosstalk was initially described for the tumor-suppressor gene PTEN, which appears to be regulated by the abundance of its pseudogene (PTENP1) in a Dicer-dependent manner [6]. Additional reports have identified other ceRNAs as physiological regulators, including a long non-coding RNA, which regulates muscle differentiation [7] and an overexpressed 3′ untranslated region (3′ UTR) that induces cancer in transgenic mice [8]. However, quantitative analyses of miR-122 target gene expression in response to increased intracellular target site abundance through controlled overexpression of a miRNA target in hepatocytes and livers showed that profound forced overexpression of a miRNA-binding target is required to increase the target-site abundance sufficiently to achieve derepress the miRNA [9], [10], [11]. Furthermore, even in extreme metabolic stress conditions, global target-site abundance of hepatocytes did not change sufficiently to affect miRNA-mediated repression. By extrapolation, these observations also hold true in pancreatic islets of metabolically stressed mice. These quantitative studies make it unlikely that miRNA target abundance in primary differentiated cells cause significant effects on gene expression and metabolism through a ceRNA effect. One exception of a naturally occurring ceRNA is circular RNA (circRNA) CDR1as/ciRS-7 with >60 closely spaced sites to miR-7 [12], [13]. This non-coding RNA is highly expressed in the hippocampus and other parts of the central nervous system (CNS). However, miR-7 copy numbers per pancreatic β-cell far outnumber its binding sites of CDR1as/ciRS-7 (unpublished data), making it unlikely that this circRNA can act as a physiological sponge in pancreatic β-cells. In addition, very few other circRNAs contain more miRNA-binding sites than expected by chance [14], making endogenous sponges an unlikely scenario of widespread miRNA regulation.
2. The collective role of miRNAs in pancreatic β-cells
The pancreatic β-cell is the sole cell type to store and release insulin in response to metabolic demand and the loss of β-cell function and number underlies much of the pathology of diabetes [15]. In type 1 diabetes (T1D), a deranged immune system leads to an autoimmune attack resulting in destruction of the β-cell population. Type 2 diabetes (T2D) is typically preceded by insulin resistance and accompanied by an inability of liver, muscle and adipose tissue to respond to the normal actions of insulin [16]. To compensate for insulin resistance, pancreatic β-cells increase their production and secretion of insulin; however, β-cell function eventually declines and insulin secretion becomes inadequate. This phenomenon is referred to as β-cell decompensation and involves many molecular defects in glucose sensing, intracellular intermediary metabolism and coupling to membrane potential, insulin synthesis, processing and secretion, β-cell differentiation, proliferation and cell survival [15].
There is increasing evidence that miRNAs in pancreatic β-cells are important posttranscriptional regulators of gene expression that mediate various stress responses in disease. Conditional deletion of the miRNA processing enzyme Dicer has enabled the study of reduced or absent miRNA activity in a cell type-specific and temporal manner. The overall contribution of miRNAs to pancreatic development was investigated by employing Pdx1-Cre mediated deletion of Dicer at ≈e9.5. Mutant mice survived until birth but failed to grow and died postnatally due to a defect in all pancreatic lineages, with the most pronounced reduction in endocrine cell types, including pancreatic β-cells [17]. This developmental defect can at least in part be attributed to abnormal Notch signaling, which is known to inhibit the expression of neurogenin 3 (Ngn3) and thereby constrains endocrine differentiation in the developing pancreas. Indeed Ngn3 levels were dramatically reduced in the pancreas of e15.5 Pdx1-Cre Dicer fl/fl embryos and this downregulation was associated with an increase in the Notch signaling target Hes1 [17]. These data convincingly demonstrate that miRNA expression is required for endocrine pancreatogenesis.
The role of miRNAs during islet cell specification from endocrine Ngn3-positive precursor cells has also been investigated in mice where expression of Cre recombinase was controlled by the Ngn3promoter, leading to conditionally deleted Dicer fl/fl alleles in endocrine progenitor cells. These mice develop a similar but slightly more attenuated phenotype than the Pdx1-Cre Dicer fl/fl, including structural and functional defects in the neonatal period characterized by altered islet morphology, loss of hormone expression and reduced β-cell mass. Interestingly, Dicer1-deficient islet cells expressed many neuronal genes, supporting a model in which miRNA pathways control key transcriptional networks required for suppressing neuronal fate during the maintenance and maturation of newly specified endocrine cells [18].
Two independent studies used a similar genetic approach to investigate the collective role of miRNAs during late embryonic and postnatal development by genetic deletion of a conditional Dicer allele using transgenic mice, in which the Cre recombinase is controlled by the insulin promoter [19], [20]. These mice exhibit normal fetal and postnatal β-cell development and have normal insulin secretion at 2 weeks of age; however, they develop progressive hyperglycemia and overt diabetes in adulthood. Phenotypic characteristics also included altered islet morphology, reduced insulin granules and secretion and decreased β-cell mass. In a third study, Dicer was inactivated in adult mice by the tamoxifen-inducible Pdx1-CreER transgene. These mice develop impaired insulin secretion prior to changes in β-cell mass and pancreatic insulin content. This study also clearly established that β-cell apoptosis at least partially contributes to the decline in β-cell number [21].
Since genetic ablation of Dicer leads to the loss of most miRNAs it is expected that the expression of many gene networks would be affected. Hence these studies are not very informative in determining the role of individual miRNAs in endocrine organ development, cell lineage specification, or β-cell function in adulthood and in response to pathological stress. However, the pancreatic Dicer mutant mice have provided evidence for the notion that miRNAs function to buffer pathway activity by dampening expression of both positive and negative regulators, thereby preventing the expression of ‘disallowed genes’ and inhibiting stochastic fluctuations in signaling pathways [22], [23]. A small group of genes in pancreatic β-cells that are abundantly expressed in most, if not all, other mammalian tissues are highly selectively repressed, among them monocarboxylate transporter-1 (MCT-1/Slc16a1), a carrier of the potent insulin secretagogue pyruvate [24], Maf (cMAF), an enhancer of glucagon expression in α-cells [25], Pdgfra, a receptor tyrosine kinase that regulates β-cell proliferation [26], ornithine aminotransferase (Oat), a mitochondrial enzyme that controls the production of the signaling molecule glutamate [27], Fcgrt, a Fc receptor that mediates the selective uptake of immunoglobulin G, and insulin-like growth factor binding protein (Igfbp4), which inhibits insulin-like growth factor (IGF) signaling [21], [28]. These transcripts are all targeted by miRNAs that are highly expressed in β-cells and were markedly increased in Dicer Pdx1-CteER mice [20]. This analysis highlights the general role of miRNAs as an additional layer of negative gene regulation and specifically how they contribute to the maintenance of pancreatic β-cell function by buffering or even shutting down a subset of ‘disallowed’ genes that would otherwise promote β-cell dedifferentiation and impair insulin secretory capacity and β-cell survival.
3. miRNA expression in pancreatic islet cells of healthy and diabetic subjects
Changes in miRNA levels may reflect physiological or pathological responses of pancreatic islets to constantly changing metabolic environments. miRNA profiling in mouse models of insulin resistance/diabetes and in islets of healthy and type 2 diabetic donors has revealed signature patterns of miRNAs that may be diagnostic for distinct defects of pancreatic islet function.
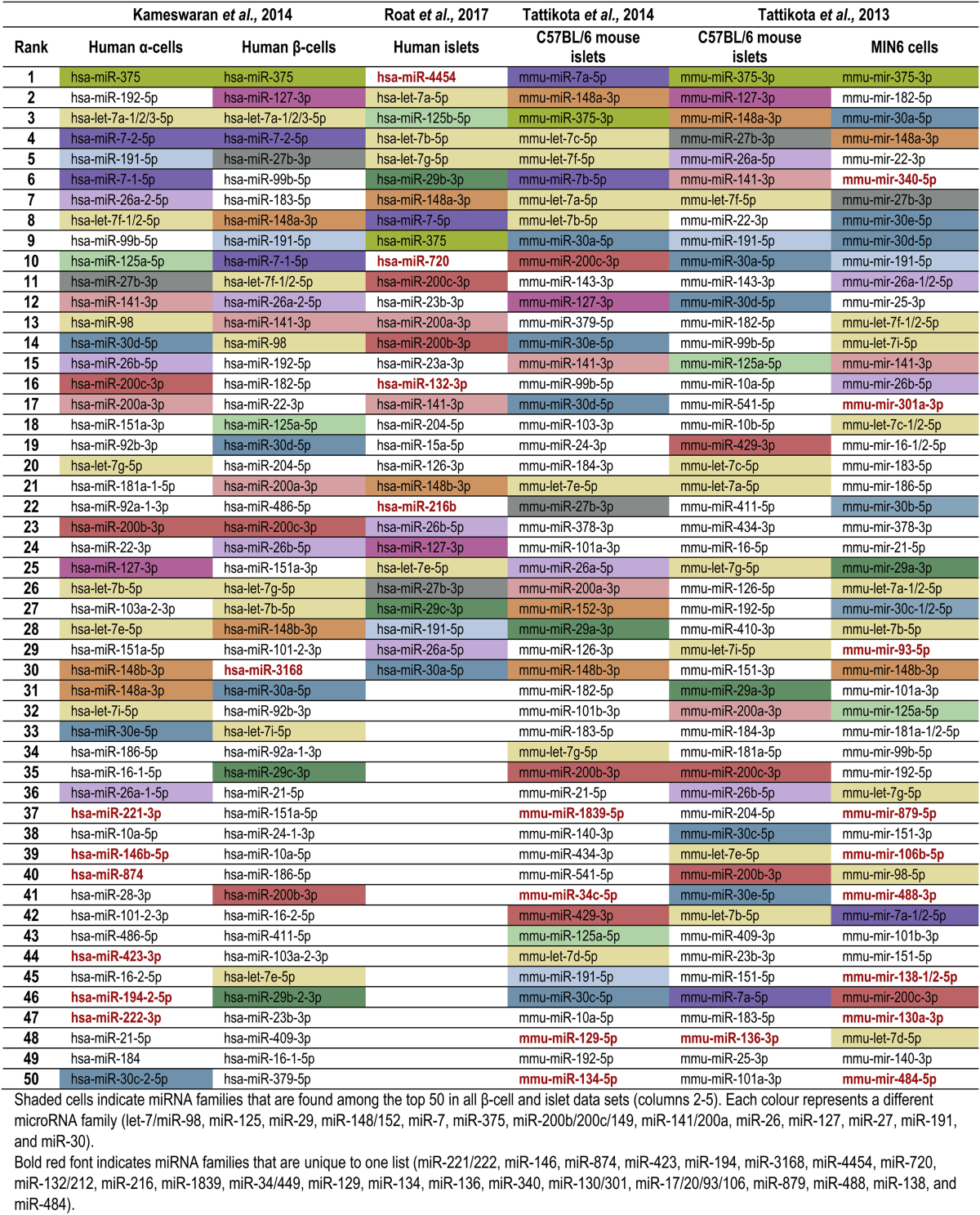
In Table 1, we present the most highly expressed miRNAs in human and mouse pancreatic cells as identified by six profiling studies [29], [30], [31], [32]. The shaded cells indicate miRNA families (identified as miRNAs sharing a common seed sequence, as listed in Targetscan Release 7.1) that are found in at least all of the β-cell and islet data sets. It is important to examine the expression of miRNA families as a functional unit and not only that of individual miRNAs, as a common seed sequence imparts functional homology to the members of a family. Of these 13 consistently identified miRNA families, one (miR-29) is absent in α-cells and another (miR-127) is absent in MIN6 cells. While many miRNA families are consistently detected, their relative expression varies substantially across studies. Furthermore, several miRNA families are found in only a single list (indicated in bold red font). Seven miRNA families are unique to MIN6 cells, five families are detected only in α-cells, and between one and four families are unique to each of the β-cell and islet lists. The fact that MIN6 cells contain the highest number of unique miRNA families suggests that the differential miRNA expression between this cell line and in vivo β-cells may be, in some respects, greater than the difference between α-cells and β-cells.
Table 1.
Most highly expressed miRNAs in profiling studies of human and mouse pancreatic cells.
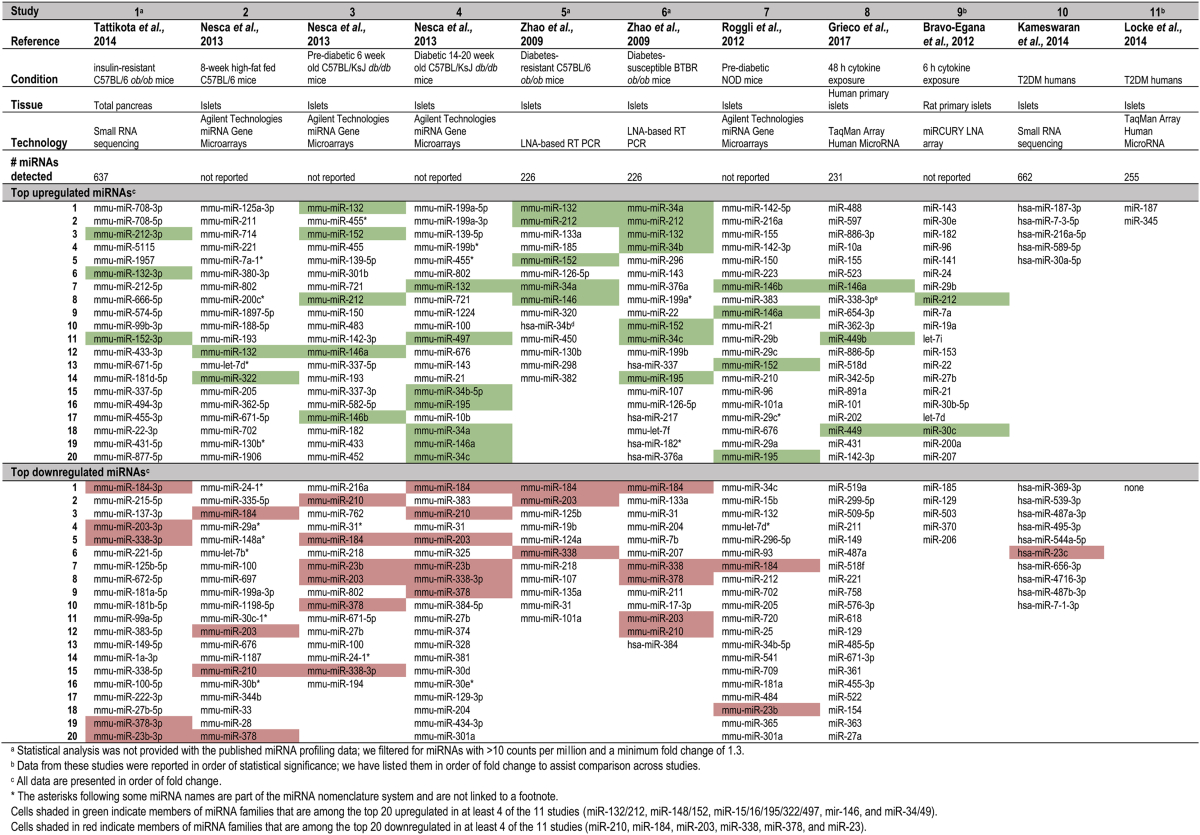
Table 2 summarizes 11 datasets of pancreatic miRNA regulation in a range of conditions, including genetic models of obesity/diabetes and exposure to metabolic stressors in primary cells and in vivo [29], [31], [33], [34], [35], [36], [37], [38]. This is not an exhaustive list and it does not cover the many studies performed in cell lines, as these exhibit abnormal miRNA and transcript levels that may not precisely reflect physiological regulation.
Table 2.
Top regulated pancreatic/islet miRNAs in profiling studies of obesity/diabetes models.
The heterogeneity of results between these studies is striking; while five families (miR-132/212, miR-148/152, miR-15/16/195/322/497, mir-146, and miR-34/49) are upregulated in at least 4 of the 11 studies and 6 families (miR-210, miR-184, miR-203, miR-338, miR-378, and miR-23) are downregulated in at least 4 of the 11 studies, 66 families are unique to a single study. Interestingly, only one of the consistently regulated families (miR-23) was also found in the human islet sample data sets, likely due in part to the limited number miRNAs that were significantly regulated in these studies. This discrepancy of data may be due to distinct patho-physiological mechanisms underlying the range of conditions but other variables such as differences in islet isolation procedures, culturing protocols and miRNA profiling technologies (arrays, qPCR, and small RNA seq) may also account for some of the disparities.
The data in summarized in Table 2 have been obtained via hybridization-based array, quantitative real-time PCR, and small RNA sequencing. The varying attributes of these technologies can greatly influence the reproducibility of data, an issue which has been demonstrated by systematic cross-comparison of results from the same samples across platforms. For example, out of 136 total miRNAs identified as ‘regulated’ in five assays (deep sequencing and four microarrays), only 53 were common to all five [39]. Another study detected 175 miRNAs by microarray, of which only 126 were also detected by small RNA sequencing [40].
It should be noted that Table 2 only includes the top 20 regulated miRNAs from each study, and therefore the above observations do not represent a comprehensive statistical analysis of the data but rather a general illustration of the differences across studies. Nonetheless, if some of the uniquely identified miRNAs were statistically regulated in other studies, the fact that they don't appear in the top 20 still attests to the inconsistency of results. Lastly, it is interesting to note that the most regulated miRNAs are not necessarily the most abundant. For example, miR-184 was downregulated in 7 studies in Table 2, but is only found in the top 50 miRNAs in three of the six studies we examined and is never within the top 20 (Table 1). Even large relative changes in the levels of poorly or modestly expressed miRNAs may produce minor absolute changes in miRNA copy number per cell that are insufficient to impact on target gene expression. Therefore, the significance of relative gene expression changes must be interpreted with caution, and the biological impact be validated experimentally. The miR-148/152 family is notable as it is the only family which appears consistently in the top 50 expressed miRNAs (Table 1) as well as in the top 20 upregulated miRNAs in disease conditions (Table 2). miR-148 regulates insulin synthesis in MIN6 cells and primary islets through regulation of the insulin repressor SOX6 [41]. miR-26, another highly expressed miRNA (Table 1), also regulates insulin transcription in vitro and ex vivo [41]. However, this miRNA was not among the top regulated in any studies in Table 2, leading one to question whether its contribution to β-cell function is relevant in pathophysiological conditions.
In the case of some of the most highly expressed miRNAs, whether they play an important role in β-cells remains elusive. For example, miR-27 is one of the consistently top expressed miRNAs in the islets and was downregulated in three disease models (Table 2) but does not have known functions in the β-cells. As miR-27 is known to regulate TGFβ signaling in fibroblasts [42], it could be speculated that this same regulatory network may influence β-cell proliferation and function. miR-127 is another highly expressed miRNA that has been found to correlate with β-cell function in humans [43]. However, its role has yet to be tested in functional studies and it is also not among the top regulated miRNAs in any of the studies in Table 2. The let-7 family is also highly expressed in pancreatic cells and is upregulated in two disease conditions (Table 2). However, its role in β-cells is unclear as global let-7 overexpression in mice has been shown to attenuate glucose-stimulated insulin secretion in one study [44] while a second study observed increased insulin secretion during a glucose tolerance test in mice overexpressing let-7 [45]. In sum, several of the most highly expressed miRNA families in β-cells remain to be thoroughly investigated and their exact roles and mechanisms may be clarified through β-cell-specific manipulation in vivo.
4. miRNAs regulating differentiation and function of pancreatic β-cells
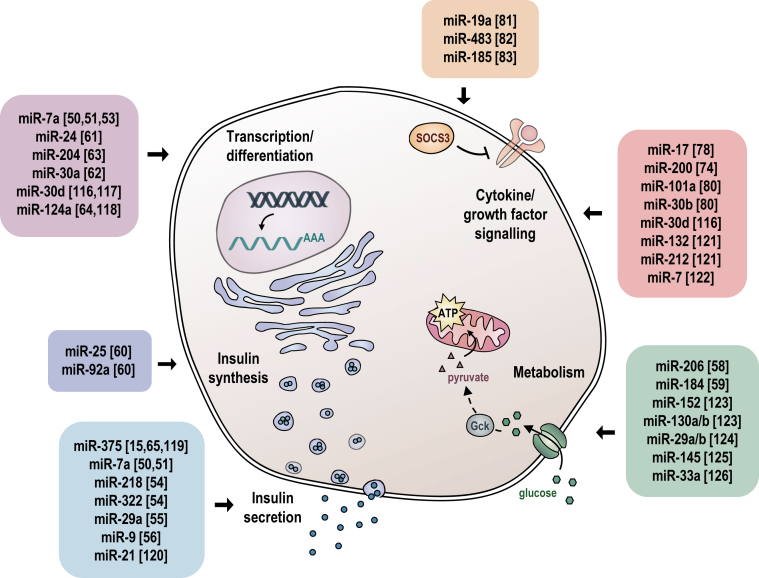
Manipulating expression of β-cell miRNAs in several genetic mouse models has revealed the influence of specific miRNAs on pathways that are known to be crucial for β-cell differentiation and insulin secretion (Figure 1). The majority of studies have attempted to infer miRNA function in pancreatic β-cells from overexpression studies and miRNA silencing experiments by transfecting pancreatic β-cell lines with either synthetic miRNA mimics or anti-miRs, which inhibit endogenous miRNAs by Watson–Crick base pairing. Although these studies may reveal important mechanistic information, they should be treated with caution with respect to inferring miRNA function in primary β-cells in an organismal context for several reasons: 1) Pancreatic cell lines are transformed, constantly replicating cells that have abnormal miRNA and transcript expression profiles; 2) overexpression of synthetic miRNA mimics typically leads to miRNA levels that are orders of magnitude higher than observed in cells; and 3) anti-miRs can have toxic and off-target effects similar to any siRNA and therefore may induce phenotypes that are independent of the miRNA inhibition. The experience of mouse genetic studies in the last 10 years has shown that miRNA loss-of-function rarely results in highly penetrant phenotypes, but rather, influences cellular responses to physiologic and pathophysiologic stresses. One notable exception was revealed upon genetic inactivation of miR-375, the most highly expressed and β-cell-enriched miRNA, which led to overt diabetes. This was due in part to increased and decreased α- and β-cell mass, respectively, and concomitant with altered hormone levels, which form the primary basis for the hyperglycemia [15]. Interestingly, miR-375 also regulates stimulus secretion coupling and exocytosis through mechanisms that are still not entirely understood.
Figure 1.
Schematic illustration of miRNA functions in the pancreatic β-cell. Respective references are shown in brackets [[116], [117], [118], [119], [120], [121], [122], [123], [124], [125], [126]].
Overexpression of miR-375 in vitro reduces insulin secretion by inhibiting insulin exocytosis with no adverse effect on more proximal events such as glucose metabolism. Bioinformatic and experimental target gene analysis of miR-375 has identified several candidates that impact insulin secretion: vesicle transport through interaction with t-SNAREs yeast homologue 1A (Vti1a), V-1/myotrophin (V-1/Mtpn), p38 mitogen-activated protein kinase (Mapk14), monocarboxylic acid transporter member 8 (Slc16A2) and Max interacting protein-1 (Mxi1). Of these, Mtpn and Vti1a showed the strongest regulation upon experimentally altering expression of miR-375. Furthermore, overexpression and silencing of Mtpn correlate with insulin secretion, further strengthening the evidence of this gene as an important miR-375 target in β-cell function (38). miR-375 may also directly target PDK1 transcripts and reduce its protein levels. PDK1 is a key molecule in PI3-kinase signaling and repression results in a decreased ability of the β-cell to upregulate insulin gene expression and DNA synthesis in response to glucose stimulation [46]
While selective re-expression of miR-375 in β-cells of miR-375KO mice normalizes both α- and β-cell phenotypes as well as glucose metabolism, mice with a 2-fold overexpression of miR-375 unexpectedly exhibit normal β-cell mass and function [47]. The discrepancy between the in vitro and in vivo overexpression data may be explained by the higher expression of miR-375 in mouse islets (compared to MIN6 cells and dissociated β-cells), where it may already attain maximal repression of target genes responsible for regulating insulin secretion. Indeed, no downregulation of established targets such as Mtpn or Vtia was observed in islets of mice overexpressing miR-375.
Another important miRNA for β-cell function is the highly expressed miR-7 family, encoded by three unlinked miRNAs (miR-7a1, miR-7a2 and miR-7b) whose mature forms have identical nucleotide sequences in humans. The miR-7 family is a prototypical neuroendocrine miRNA with highest levels in the pituitary, pancreatic islets, hypothalamus and adrenal glands [48], [49]. miR-7 has been shown to be regulated by the circular RNA ‘ciRS-7’, which is most highly expressed in the brain [12], [13]. A recent report indicates that ciRS-7 may also be functional in pancreatic β-cells, where it regulates insulin gene transcription and secretion through changes in miR-7 activity [50]. However, this finding was based upon an in vitro overexpression model and the physiological relevance of ciRS-7 in β-cells should be confirmed using genetic knockout (KO) models.
miR-7 levels are reduced in islets of insulin resistant and obese mice (HFD; ob/ob mice on C57BL/6 background) that are euglycemic due to the compensatory secretion of insulin and increased β-cell mass. These findings were corroborated in prediabetic obese human subjects. In contrast, hyperglycemic rodent models (db/db on BLKS background; non-obese GK diabetic rats), with decompensated β-cell function exhibit increased miR-7 levels [51], [52]. This correlation of miR-7 levels with the diabetogenic state was also confirmed in a transplantation model in which healthy human islets were grafted under the kidney capsule of mice. These animals were maintained either on chow or on HFD to induce β-cell failure. In this experimental setting, human transplants exposed to an obesogenic environment for 12 weeks display clear signs of β-cell failure, with significantly higher glucose and lower circulating human C-peptide levels. Levels of miR-7 were more than 50% downregulated in HFD grafts at 6 weeks of diet, but increased after 10 weeks compared with chow diet transplants – and correlated with the diabetic/decompensation phenotype of these mice [52]. Together, the unique and dynamic regulation of miR-7 in pancreatic islets in response to metabolic stress suggests that it plays a key role in the functional adaptation/decompensation of β-cells and development of T2D.
The functional consequences of miR-7 overexpression in pancreatic β-cells was investigated in two studies that revealed identical phenotypes of β-cell dedifferentiation and overt diabetes [51], [53], demonstrating that this miRNA plays an essentially contrasting role to that of miR-375 in β-cells. Mice expressing miR-7a under the control of the insulin promoter had reduced levels of its target Pax6 and exhibited molecular changes resembling those in haploinsufficient mutant Pax6 mice, such as downregulated expression of endocrine pancreatic identity markers and a marked decrease in insulin content. Conversely, miR-7 knockdown resulted in Pax6 upregulation and promoted β-cell differentiation. Furthermore, Pax6 downregulation reversed the effect of miR-7 knockdown on insulin promoter activity [53]. Studies of β-cell specific miR-7aKO mice also exhibited increased glucose tolerance and insulin secretion in response to an intra-peritoneal glucose challenge due to augmented β-cell function by directly regulating genes that control late stages of insulin granule fusion with the plasma membrane and ternary SNARE complex activity [51]. Direct targets that likely mediate enhanced insulin granule exocytosis included Snca, Cspa, Cplx1, Pkcb, Basp1, Pfn2, Wipf2, and Zdhhc9, all critical components of the exocytotic insulin granule machinery. Electrophysiological studies provided supportive evidence revealing that miR-7a does not affect total numbers of insulin granules per cell or the number of docked granules; rather, it appears to modulate the release competence of insulin granules already in place. These studies collectively demonstrate that miR-7 is a stress responsive miRNA that regulates an interconnecting genomic circuit influencing β-cell differentiation and insulin granule exocytosis, together affirming a role for miR-7 in the adaptation of pancreatic β-cell function in obesity and T2D. Several other studies, performed in pancreatic β-cell lines and primary cells have suggested roles of miRNAs in insulin secretion. miR-218 and miR-322 have been shown to regulate syntaxin binding protein1 (Stxbp1), which modulates the folded conformation of syntaxin 1A that plays an essential role in vesicle trafficking and insulin granule exocytosis [54]. Interestingly, Syntaxin-1a itself and granulophilin are also direct targets of miR-29a and miR-9, respectively, in β-cells [55], [56], suggesting that components of the secretory machinery are under tight control of miRNAs.
In addition to regulating insulin exocytosis, miRNAs also play a more upstream role in the control of insulin secretion (Figure 1). Intracellular glucose metabolism, a key feature of glucose sensing and insulin secretion, is facilitated by the rate-limiting glucose sensor glucokinase (GCK) and the low-affinity glucose transporter GLUT2, which control intracellular ATP levels by controlling glucose-6-phosphate availability for glycolysis and ATP production. ATP inhibits the KATP channel leading to membrane depolarization, and increased intracellular Ca2+ influx, which then directly triggers the fusion of the insulin granule with the plasma membrane [57]. Interestingly, miR-206 has been detected in GCK-expressing tissues and its expression is increased in response to HFD-feeding. Chow- and HFD-fed miR-206 KO mice have improved glucose tolerance and glucose-stimulated insulin secretion (GSIS) with no changes in insulin sensitivity. The Gck 3′UTR harbors a conserved 8-mer miR-206 binding site which was found to be functional in reporter and GK activity assays [58]. Although data from the global miR-206KO strongly implicates miR-206 as a negative regulator of GCK, there are phenotypic traits that cannot easily be explained by a β-cell autonomous effect, including the reduced body weight of KO mice on chow and HFD. It will be interesting to investigate miR-206 in other tissues such as the CNS and muscle, where expression of this miRNA is higher than in β-cells and liver. It can be speculated that GCK is a disallowed gene in the muscle due to miR-206 inhibition, thereby enabling functional replacement of GCK with hexokinase II (HK2), which differs in its property of end product inhibition by glucose-6-phosphate to prevent uncontrolled glycolysis.
The conserved miR-184 has also been shown to regulate compensatory insulin secretion during insulin resistance. This miRNA is silenced in genetic and diet-induced mouse models of obesity and its expression is rescued upon subjecting mice to a low carbohydrate ketogenic diet [59]. Interestingly, knockdown of glucokinase resulted in increased expression of miR-184, suggesting that β-cells activate the expression of this miRNA to suppress insulin release when either glucose sensing or glycolysis is attenuated. The regulatory function of miR-184 is complex and not completely understood, however, experimental evidence points toward the functional regulation of its target Argonaute2 (Ago2), a component of the miRNA-induced silencing complex, and via mitochondrial substrate flux in the β-cell by suppressing the target Slc25a22 [31].
Other miRNAs have been suggested to regulated the process of insulin synthesis (Figure 1); members of a miRNA family miR-25 and miR-92a have been implicated in insulin expression by gain and loss of function studies in cell lines which respectively resulted in reduced and enhanced glucose-stimulated insulin expression and secretion. This regulation may be mediated through miR-25 and miR-92a binding sites in the insulin transcript that overlap with the site of the RNA binding protein of PTBP1, a known stabilizer of insulin transcripts and enhancer of translational efficacy [60].
Transcription factors represent another opportunity for miRNA-mediated regulation of β-cell functions (Figure 1). These factors play a crucial role in orchestrating the intricate pathways of cellular growth and differentiation by regulating the rate of transcription of an array of genes. Genetic and biochemical studies have begun to unravel the complex cascade of factors that controls the proliferation and differentiation of cells in the developing pancreas and during adult life. Several studies suggest that miRNAs form an additional layer of regulation that contributes to the robustness of these networks. As mentioned earlier, the highly expressed miR-7a is a critical regulator of Pax6 in pancreatic β-cells, thereby maintaining the differentiation state of these cells during metabolic stress [51], [53]. Expression of miR-24 is induced by palmitate in MIN6 cells and isolated primary islets and ectopic expression of miR-24 resulted in failure of β-cell function. This response may have been caused by the direct regulation of the MODY genes TCF1 and NEUROD1, since silencing either of these targets mimicked the cellular phenotype caused by miR-24 overexpression, whereas restoring their expression rescued β-cell function [61].
The transcription factor NeuroD1 has also been reported as a direct target of miR-30a-5p, which is increased in β-cells after exposure to glucotoxic conditions. Experimental overexpression of this miRNA in vitro reduced insulin content and gene expression as well as glucose-stimulated insulin secretion, whereas the restoration of NeuroD1 expression via silencing of miR-30a-5p or overexpression of NeuroD1 rescued β-cell function. Furthermore, glucose tolerance and β-cell dysfunction improved when a recombinant adenovirus expressing si-30a-5p to inhibit miR-30-5p was administered into the coeliac artery of diabetic mice [62]. These studies suggest that NeuroD1 is targeted post-transcriptionally by multiple miRNAs. The basic-leucine zipper transcription factor MafA is also an important β-cell enriched protein that functions as a differentiation factor and potent transactivator of the insulin gene. It is a highly regulated protein, whose expression and DNA binding activities diminish in acute hyperglycemic conditions and diabetes as well as in Thioredoxin-interacting protein (TXNIP) knockout mice [63]. MafA is directly regulated by miR-204, which itself is activated in diabetic conditions. This study reveals a TXNIP-miR-204-MAFA-insulin axis that may contribute to diabetes progression and should be further investigated using genetic models [63].
The neuron-specific miR-124a has been reported to regulate the expression of Foxa2 and its downstream target Pdx-1. Foxa2 is a master regulator of pancreatic endoderm development and of genes involved in glucose metabolism and insulin secretion. Investigations in cell lines indicate that miR-124a-2 modifies basal and glucose- or KCl-stimulated intracellular free Ca2+ concentrations without affecting the secretion of insulin, consistent with an altered sensitivity of the β-cell exocytotic machinery to Ca2+ [64]. However, the physiological role of miR-124 in pancreatic β-cells has been questioned by deep RNA sequencing studies in islets and primary β-cells which revealed that this neuron-enriched miRNA either absent or among the lowest expressed miRNAs in the adult endocrine pancreas and that its higher expression in MIN6 and INS1 cell lines is due to a partial differentiation towards neuronal cell lineages [32], [65].
5. miRNAs and β-cell proliferation
The maintenance of β-cell mass throughout life is a highly regulated process that is essential to preserve normal glucose homeostasis during aging. Defects in pancreatic β-cell proliferation alter islet cell composition over time, and can contribute to the hyperglycemia that characterizes the diabetic state. The dynamic adaptation of β-cell mass in adult life is influenced by various metabolic stresses, which control the balance between proliferation and apoptosis. Several miRNAs have been implicated in this process. For instance, the genetic ablation of the most abundant miR-375 induces progressive hyperglycemia and decreased pancreatic β-cell mass as a result of impaired proliferation [15]. These mice also have increased α-cell numbers and hyperglucagonemia in the fasted and fed state, which likely contributes to the hyperglycemia. Interestingly, the α-cell phenotype can be completely rescued by a transgene expressing miR-375 from the insulin promoter, suggesting that the increased α-cell activity in miR-375KO mice is secondary to a β-cell autonomous defect. The β-cell proliferation phenotype in miR-375KO animals is exaggerated in pancreatic islets of obese mice (ob/ob) in which miR-375 is genetically deleted. These animals exhibit a profoundly diminished proliferative capacity of the endocrine pancreas that results in a severely diabetic state and ultimately death [15]. In silico bioinformatic analysis and transcriptomic RNA sequencing data from 375KO islets revealed that this miRNA regulates a cluster of genes controlling cellular growth and proliferation (caveolin1 (Cav1), inhibitor of DNA binding 3 (Id3), Smarca2, Ras-dexamethasone-induced-1 (Rasd1), regulator of G protein signaling 16 (Rgs16), eukaryotic elongation factor 1 epsilon 1 (Eef1e1), apoptosis-inducing factor, mitochondrion-associated 1 (Aifm1), cell adhesion molecule 1 (Cadm1), HuD antigen (HuD), complement component 1 q subcomponent binding protein (C1qbp), and gephyrin (Gphn)). These data provide genetic evidence that miR-375 is essential for normal glucose homeostasis, α- and β-cell turnover, and adaptive β-cell expansion in response to increasing insulin demand in insulin resistance [15]. Other studies have reported potential pro- and anti-proliferative roles of miRNAs in β-cells [66], [67], [68], [69], [70]. These studies were performed in cell lines or primary β-cells in vitro and thus their impact on pancreatic β-cell compensation in response to chronic metabolic stress will need to be studied in vivo.
6. miRNAs and β-cell survival
Increasing evidence from animal models and humans indicates that apoptosis is the main mode of β-cell death leading to T1D, and that it also leads to decreased β-cell mass in T2D [71], [72]. In vivo and in vitro studies suggest that members of the miR-200 family are involved in pancreatic β-cell apoptosis [73], [74], [75], [76], [77]. The miR-200 family consists of five evolutionarily conserved miRNAs (miR-141, miR-200c, miR-200a, miR-200b and miR-429) and is localized in two polycistronic clusters, mir-141/200c and mir-200a/200b/429 [77]. Based on the homology of their seed sequences, miR-200 family members can be subdivided into 2 distinct functional miRNA subfamilies containing either the miR-141/200a (AACACUG) or miR-200b/200c/429 (AAUACUG) seed, which are predicted to regulate different target genes. All members of the miR-200 family are highly expressed in the endocrine pancreas with miR-200c being the prominent form. The obese db/db mouse model on the C57BLKS/J background, which develops uncontrolled T2DM due to impaired β-cell function and apoptosis, has significantly increased levels of miR-200. Interestingly, miR-200 has a negative impact on β-cell function as transgenic mice with β-cell-specific overexpression of miR-141/200c at similar levels as measured in db/db mice developed overt diabetes due to massive induction of apoptosis. On the other hand, genetic deletion of the two miRNA clusters conferred protection in a gene dosage manner against β-cell apoptosis in three experimental models of apoptosis: the Akita mouse, a multiple low dose streptozotocin (STZ) model, and high-fat diet-fed mice receiving a single dose of STZ. In contrast, complete loss of miR-200 did not affect β-cell function or glucose tolerance in unstressed conditions [77]. Gene expression analysis of islets from gain- and loss-of-function mice identified Dnajc3, Jazf1, Rps6kb1 and Xiap (which encode anti-apoptotic proteins), and Dusp26 (which encodes a pro-survival signaling protein) as direct targets, suggesting that these miRNAs promote β-cell survival. These studies also found that the tumor suppressor, Trp53, is regulated by miR-200, and that ablation of Trp53 or its downstream target, Bax, prevents miR-200-induced apoptosis of β-cells and the resultant T2D phenotype. Importantly, silencing of miR-200c with inhibitory antagomirs reduced the expression of pro-apoptotic genes that are induced by cytokines and associated with T2D in islets [77]. Together, these studies strongly implicate the miR-200 family in programmed cell death of pancreatic islets as a response to oxidative and endoplasmic reticulum stress.
Pancreatic β-cell death can be caused by direct interaction with activated macrophages and T-cells, and/or exposure to soluble mediators secreted by inflammatory cells, including cytokines, nitric oxide (NO), and oxygen free radicals (75). In vitro exposure of β-cells to IL-1β or to IL-1β plus interferon (IFN)-γ causes functional changes in pancreatic function similar to those observed in pre-diabetic patients. Prolonged exposure to of a combination of IL-1β plus IFN-γ and/or tumor necrosis factor (TNF)-α consistently leads to β-cell death [73]. Our increasing knowledge of cytokine signaling pathways that trigger β-cell apoptosis and experimentally robust readouts have led to several studies investigating the influence of miRNAs on apoptotic signaling pathways. A miRNA expression profile on human islet preparations exposed to the cytokines IL-1β plus IFN-γ identified 57 miRNAs that were modulated by cytokine exposure. Three of them, miR-23a-3p, miR-23b-3p, and miR-149-5p, were downregulated by cytokines and found to regulate the expression of the proapoptotic Bcl-2 proteins DP5 and PUMA and human β-cell apoptosis [36]. These results identify a novel cross talk between a key family of miRNAs and proapoptotic Bcl-2 proteins in human pancreatic β-cells, broadening our understanding of cytokine-induced β-cell apoptosis in early T1D.
Thioredoxin-interacting Protein (TXNIP) is a key regulator of diabetic β-cell apoptosis as TXNIP inhibition prevents diabetes in mouse models of T1D and T2D. TXNIP expression is differentially regulated by cytokines, with TNFα having no effect, IL-1β leading to downregulation of TXNIP transcript levels, and IFN-γ inducing expression in INS-1 β-cells and primary islets [78]. The distinct and partly opposing effects of these pro-inflammatory cytokines on β-cell TXNIP expression are in part regulated by miR-17 and miR-200 [77], [78], [79]. Another example of cytokine-mediated miRNA regulation includes the stimulatory action of IL-1β on miR-101a and miR-30b expression in MIN6 cells, which decreases insulin content and gene expression and increases β-cell death. The miR-101a and miR-30b mediated β-cell apoptosis is associated with diminished expression level of the antiapoptotic protein Bcl2 and suggests that these miRNAs could contribute to cytokine-mediated β-cell dysfunction occurring during the development and progression of T1D [80].
Lastly, several miRNAs have been shown to regulate key components of signaling pathways and may modulate the response of β-cells to stress stimuli by titrating a critical component of a signal transduction pathway (Figure 1). Thus, miRNAs may mediate negative feedback to prevent pathologic hyperactivity and to restore homeostasis once stress resolves. Alternatively, activation of a given miRNA may inhibit negative regulators of a signaling pathway and could thereby promote an irreversible activation of a signaling pathway and contribute to disease. For instance, miR-19a-3p has been shown to be decreased in diabetic patients. Overexpression of miR-19a-3p promoted cell proliferation and insulin secretion, while it suppressed the apoptosis of pancreatic β-cells. Furthermore, the levels of suppressor of cytokine signaling 3 (SOCS3) are increased in diabetic patients, and are inversely correlated with the miR-19a-3p level, suggesting that the downregulation of miR-19a-3p during metabolic stress leads to the upregulation of SOCS3, which may contribute to the dysfunction of pancreatic β-cells [81]. SOCS3 is also targeted by miR-483, which protects β-cells against proinflammatory cytokine-induced apoptosis. This effect is corroborated by the correlation between increased miR-483 expression and the expanded β-cell mass observed in the islets of prediabetic db/db mice. This study also suggests that miR-483 has opposite effects in α- and β-cells by targeting SOCS3, and the imbalance of miR-483 and its targets may therefore play a role in the etiology of diabetes [82]. A third miRNA that has been reported to directly regulate SOCS3 via its 3′-UTR is miR-185. Overexpression of miR-185 enhanced insulin secretion from pancreatic β-cells, promoted cell proliferation and protected cells from apoptosis. Contrary to miR-185's protective effects, SOCS3 significantly suppressed functions of β-cell and inactivated STAT3 pathway. When treating cells with miR-185 mimics in combination with SOCS3 overexpression, the inhibitory effects of SOCS3 were reversed [83]. These studies highlight the tight control of this inhibitory cytokine signaling molecule by multiple conserved miRNAs, thereby preventing its hyperactivity and/or contributing to signal resolution following activation by inflammatory signals.
7. Are miRNAs part of a thrifty genotype?
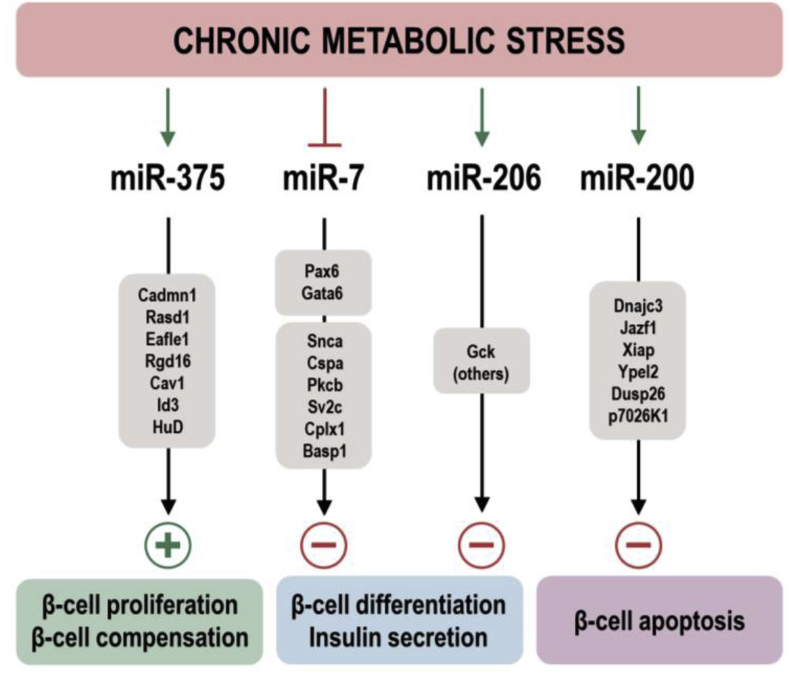
As discussed above, the ablation of all miRNAs in pancreatic β-cells by genetic deletion of Dicer results in progressive hyperglycemia through impaired insulin secretion leading to glucose intolerance and ultimately the development of diabetes [18], [19], [20], [21]. Interestingly, genetic inactivation of miR-375, the most abundant miRNA in the developing and adult endocrine pancreas, revealed that it is essential for maintaining normal β-cell mass in unstressed conditions and for the hypertrophic response of β-cells during metabolic stress in obesity [15]. This phenotype therefore represented a partial phenocopy of the pancreatic β-cell-specific DicerKO mice and suggested that the Dicer defect could be explained by the synergistic action of several β-cell miRNAs, each contributing to the defect in pancreatic β-cells when inactivated. Interestingly, this additive or synergistic model of ‘positive miRNAs’ did not hold up when miR-7a and miR-200 function was studied in β-cells. miR-7a2KO mice exhibit enhanced insulin secretion upon glucose stimulation whereas overexpression impairs insulin gene expression and secretion [51] and genetic deletion of the miR-200 family protects β-cells from apoptosis during metabolic stress whereas overexpression initiated massive apoptosis [77]. As illustrated in Figure 2, these genetic studies provide strong evidence that miRNAs in β-cells exert both positive and negative regulatory functions, but it also raises the question of why the endocrine pancreas expresses two evolutionarily conserved miRNA families, each comprising several unlinked paralogues, that exert strong negative influence on β-cells. A possible explanation is that these miRNAs evolved and their expression in β-cells was maintained during evolution because they served the advantageous function of protecting humans and other mammals from increased insulin secretion (and therefore hypoglycemia) during historical periods of famine [84]. It is worth recollecting that, during evolution, hypoinsulinemic states would have a stronger adverse impact on reproduction and survival than hyperinsulinemic conditions. Scientific proof of this concept is difficult to obtain, although the potentially advantageous functions of a seemingly detrimental miRNA could be tested in the case of miR-200KO mice, which are resistant to apoptosis following metabolic stress. Pregnancy imposes a state of metabolic stress that is known to temporarily increase β-cell mass during gestation, followed by a return to non-pregnant levels through apoptosis. The expectation would be that, as apoptosis is impaired in miR-200KO mice following pregnancy, these mice would have progressively increasing pancreatic β-cell mass leading to the development of relative hyperinsulinemia after multiple pregnancies, which may result in smaller litter sizes when exposed to a calorie restriction compared to dams that express miR-200. In order to provide support for a ‘thrifty miRNA hypothesis’ it will also be important to investigate if certain miRNAs (i.e. miR-7, miR-200) are upregulated and their targets repressed in islets of populations that are evolutionarily considered survivors of historical famine (i.e. indigenous African populations). The concept could also be addressed by comparing miRNA and target gene expression in homeostatic and stress conditions of certain rodents such as the desert spiny mouse (Acomys cahirinus) that have adapted to conditions of intermediate famine, and mice that evolved in the absence of this genetic pressure. Spiny mice exhibit metabolic pathways that result in diabetes without marked insulin resistance due to low supply of insulin on abundant nutrition. These animals respond with obesity and glucose intolerance, β-cell hyperplasia, and hypertrophy on a high-fat diet. The resulting hyperglycemia and hyperinsulinemia are initially mild, after a few weeks turn into a β-cell decompensation phase with loss of insulin synthesis and secretion and ketone acidosis [85]. Together, such studies may shed light if miRNAs with negative influence on β-cell function could have played an advantageous role in historical famines.
Figure 2.
Schematic illustration of metabolic stress-regulated miRNAs and their primary targets and cellular function as observed in genetic animal models. Plus (+) and minus (−) signs indicate positive and negative regulation, respectively.
8. Circulating miRNAs from endocrine pancreatic cells
miRNAs have also been detected in blood, where they are present in plasma, platelets, erythrocytes, and nucleated blood cells. Plasma miRNAs are remarkably stable, even under long-term storage conditions at room temperature, multiple freeze–thaw cycles, boiling, and a broad spectrum of basic and acidic pH [86], [87]. Plasma miRNAs are packaged in microparticles (exosomes, microvesicles, and apoptotic bodies) [88], [89] or associated with RNA-binding proteins (Argonaute2 (Ago2)) [90] or lipoprotein complexes (high-density lipoprotein [HDL]) [91] that protect them from degradation by RNases.
Recent studies have suggested that miRNAs are selectively secreted from tissues in response to specific stimuli and are taken up by a distant target cell, where they regulate gene expression by canonical miRNA signaling and contribute to inter-organ communication [92]. However, this concept of ‘miRNA hormonal signaling’ has not been proven unequivocally. It would be important to substantiate any claim of miRNAs as secreted messengers by demonstrating circulating miRNA uptake in recipient organs and enrichment of Ago engagement using unbiased approaches such as RNA Seq. Furthermore, the physiological relevance of such signaling is unproven considering the low plasma concentrations, rapid renal clearance and minute uptake of miRNAs in tissues [93], [94].
Another area of study proposes that circulating miRNAs may serve as serum biomarkers because of their characteristic expression in different tissues and disease states [95]. For example, high dose STZ administration leads to increased circulating miR-375 levels prior to the onset of hyperglycemia. Levels of miR-375 were also increased before the onset of diabetes in the NOD mouse, a model of autoimmune diabetes [96] and in human T1D subjects, but not in maturity onset diabetes of the young (MODY) and T2D patients that only exhibit mild β-cell demise over a long period of time [47]. These observations therefore suggest that circulating miR-375 can be used as a marker for β-cell death and as a potential predictor of diabetes in conditions with acute and profound β-cell destruction.
However, although miR-375 is highly enriched in the endocrine pancreas, it has also been detected in neuroendocrine cells of the intestine, skin and lung, as well as goblet cells, thyroid and adrenal glands, which may therefore be significant sources of circulating miR-375. Indeed, Latreille at al. found that plasma miR-375 levels in mice that exclusively express miR-375 in β-cells correspond to approximately 1% of miR-375 levels found in wildtype mice [47]. Furthermore, in apoE-deficient mice, miR-375 belonged to the most highly differentially regulated blood miRNAs [91]. Conversely, plasma miR-375 levels are reduced in obese mouse models (HFD and ob/ob on C57BL/6 background) that exhibit increased pancreatic β-cell function, proliferation and absence of apoptosis. Together, this indicates that miR-375 levels in the blood are influenced by multiple processes, including production/secretion by multiple cell types, renal clearance, metabolic stress and possibly inflammatory responses, thus making it challenging to exploit circulating miR-375 levels as a biomarker for β-cell pathology.
9. miRNA therapeutics
The inappropriate miRNA expression that has been linked to a variety of diseases forms a reasonable rationale for the pharmacological interference with miRNA expression. Various approaches to increase and decrease miRNA function or abundance have been developed; expression of small RNAs in cells has become a routine procedure through transient or stable transfection or viral transduction of pri-miRNA transgenes, pre-miRNAs, mature miRNA/miRNA∗, small interfering RNA (siRNA) or short hairpin RNAs [97], [98].
Replenishing miRNA as modified dsRNAs in tissues where expression is lost or reduced has been attempted in vivo as single and multiple dose administrations. However, data from these studies showed that sustained target regulation was difficult to achieve even when high doses were given, due to inefficient delivery or degradation of the synthetic RNA [99].
miRNAs can be inhibited by short, single-stranded RNA or DNA molecules that bind to miRNAs by Watson–Crick base pairing or miRNA ‘decoys’ [100] or ‘sponges’ [101] to reduce the activity of specific miRNAs [102]. Antagomirs were the first miRNA inhibitors that were shown to have activity in mammals [103]. They are synthetic antisense oligonucleotides (ASOs) that contain 2′-O-methyl-modified ribose sugars, terminal phosphorothioates and are conjugated at the 3′ end with a cholesterol molecule, which facilitates delivery in cells by associating with lipoproteins and binding to cell surface membrane receptors such as the scavenger receptor BI for high-density lipoproteins and low-density lipoprotein receptor (LDLR) for low-density lipoproteins [104], [105]. Alternative RNA chemistries have also been successfully used to inhibit miRNAs in vitro and in vivo. These chemistries include 2′-O-methoxyethyl, 2′-fluoro and 2′,4′-methylene (locked nucleic acids (LNAs)), and have been shown to exhibit greater affinity to miRNAs and stability than 2′-O-methyl RNA oligonucleotides [106], [107], [108]. Antagomirs localize to the cytoplasm and do not affect pre-miRNA levels, suggesting that they target the mature miRNA. Their mode of action is through high-affinity base pairing with miRNAs, which prevents the binding of miRNAs to their target transcripts. Antagomirs can also induce miRNA degradation by the ‘tailing-and-trimming pathway’, in which highly complementary target RNAs trigger both tailing and trimming of miRNAs, which ultimately decreases their abundance [106], [107], [108], [109], [110].
Antisense oligonucleotides have been studied in vitro and in vivo for their ability to silence miRNAs in pancreatic islets. Unpublished studies from our own group indicate that the endocrine pancreas does not readily take up conjugated or unconjugated miRNAs when injected at high doses (3 × 80 mg/kg) in mice. Uptake of ASOs is also quite inefficient in isolated pancreatic islets in vitro and miRNA inhibition could only be achieved at high ASO concentrations and prolonged exposure, which frequently resulted in toxicity and enhanced apoptosis.
One application for pharmacological intervention in miRNAs involved in metabolism may be for the generation of endoderm and insulin-producing cells through differentiation of induced pluripotent stem cells that can be generated from the reprogramming of skin fibroblasts [111]. Substitution of highly expressed β-cell miRNAs and/or silencing of dominant non-β-cell miRNAs (i.e. the liver miR-122 or neuronal miR-124) may affect the ability of multipotent stem cells to differentiate into insulin-positive cells. Evidence that such approaches indeed promote β-cell differentiation come from virus-mediated overexpression of miR-375 in human skin fibroblast-derived induced pluripotent stem cells (iPSCs), which was sufficient to trigger their differentiation into insulin-expressing cells and allow glucose-dependent insulin secretion in vitro [112]. Overexpression of miR-186 and miR-375 by chemical transfection of human iPSCs promoted islet-like cell cluster formation and induced β-cell lineage markers [113]. Furthermore, human embryonic stem cells with induced overexpression of miR-410 and miR-495, prior to differentiation into endoderm, led to improved glycemic control when transplanted in mouse models of gestational or T2D [114], [115]. These studies demonstrate that miRNAs can direct developmental decisions and promote differentiation of distinct cell lineages and may be exploited therapeutically in stem-cell-based approaches aiming to replace differentiated endocrine cells for the treatment of diabetes.
In conclusion, miRNAs play essential roles in the function, differentiation, proliferation, and survival of pancreatic β-cells. Many miRNA families that are regulated by metabolic, genetic, and inflammatory stressors have been found to coordinate the adaptive responses of β-cells in vivo in conditions such as obesity and diabetes. Others have only been examined through in vitro approaches and remain to be manipulated in vivo in order to assess their physiological relevance, while some other stress-regulated miRNAs have no identified function in β-cells to date. It is interesting to note that some highly conserved miRNAs have detrimental effects in the β-cells, which may lead one to hypothesize that these were evolutionarily “thrifty miRNAs” whose actions were once important for survival but have now become unfavourable in the face of metabolic diseases. Continued in vivo studies of specific miRNA families in pancreatic β-cells may serve to advance their use in therapeutic approaches, either as biomarkers or as targets of pharmacological interventions, for the treatment of metabolic diseases.
Acknowledgement
This work was funded by the Swiss National Science Foundation and a ‘KFSP_SmallRNAs’ grant from the Unispital Zurich. M.P.L. is supported by a Doctoral Foreign Study Award from the Canadian Institutes of Health Research.
Conflicts of interest
None declared.
References
- 1.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang L., Du W.W., Yang X., Chen K., Ghanekar A., Levy G. Versican 3′-untranslated region (3′-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB Journal. 2013;27:907–919. doi: 10.1096/fj.12-220905. [DOI] [PubMed] [Google Scholar]
- 9.Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Molecular Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosson A.D., Zamudio J.R., Sharp P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Molecular Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M. Impact of MicroRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Molecular Cell. 2016;64:565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 14.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biology. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahn S.E., Hull R.L., Utzschneider K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 17.Lynn F.C., Skewes-Cox P., Kosaka Y., McManus M.T., Harfe B.D., German M.S. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 18.Kanji M.S., Martin M.G., Bhushan A. Dicer1 is required to repress neuronal fate during endocrine cell maturation. Diabetes. 2013;62:1602–1611. doi: 10.2337/db12-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalis M., Bolmeson C., Esguerra J.L., Gupta S., Edlund A., Tormo-Badia N. Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandelbaum A.D., Melkman-Zehavi T., Oren R., Kredo-Russo S., Nir T., Dor Y. Dysregulation of Dicer1 in beta cells impairs islet architecture and glucose metabolism. Experimental Diabetes Research. 2012;2012 doi: 10.1155/2012/470302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Sanchez A., Nguyen-Tu M.-S., Rutter G.A. DICER inactivation identifies pancreatic β-cell “disallowed” genes targeted by microRNAs. Molecular Endocrinology. 2015;29:1067–1079. doi: 10.1210/me.2015-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen S.M., Brennecke J., Stark A. Denoising feedback loops by thresholding – a new role for microRNAs. Genes & Development. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- 23.Pullen T.J., Khan A.M., Barton G., Butcher S.A., Sun G., Rutter G.A. Identification of genes selectively disallowed in the pancreatic islet. Islets. 2010;2:89–95. doi: 10.4161/isl.2.2.11025. [DOI] [PubMed] [Google Scholar]
- 24.Ishihara H., Wang H., Drewes L.R., Wollheim C.B. Overexpression of monocarboxylate transporter and lactate dehydrogenase alters insulin secretory responses to pyruvate and lactate in beta cells. Journal of Clinical Investigation. 1999;104:1621–1629. doi: 10.1172/JCI7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kataoka K., Shioda S., Ando K., Sakagami K., Handa H., Yasuda K. Differentially expressed Maf family transcription factors, c-Maf and MafA, activate glucagon and insulin gene expression in pancreatic islet alpha- and beta-cells. Journal of Molecular Endocrinology. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- 26.Chen H., Gu X., Liu Y., Wang J., Wirt S.E., Bottino R. PDGF signalling controls age-dependent proliferation in pancreatic β-cells. Nature. 2011;478:349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginguay A., Cynober L., Curis E., Nicolis I. Ornithine aminotransferase, an important glutamate-metabolizing enzyme at the crossroads of multiple metabolic pathways. Biology (Basel) 2017 Mar 7;6(1) doi: 10.3390/biology6010018. pii: E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sitar T., Popowicz G.M., Siwanowicz I., Huber R., Holak T.A. Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13028–13033. doi: 10.1073/pnas.0605652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kameswaran V., Bramswig N.C., McKenna L.B., Penn M., Schug J., Hand N.J. Epigenetic regulation of the DLK1-MEG3 MicroRNA cluster in human type 2 diabetic islets. Cell Metabolism. 2014;19:135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roat R., Hossain M.M., Christopherson J., Free C., Jain S., Guay C. Identification and characterization of microRNAs associated with human β-cell loss in a mouse model. American Journal of Transplantation. 2017;17:992–1007. doi: 10.1111/ajt.14073. [DOI] [PubMed] [Google Scholar]
- 31.Tattikota S.G., Rathjen T., McAnulty S.J., Wessels H.-H., Akerman I., van de Bunt M. Argonaute2 mediates compensatory expansion of the pancreatic β cell. Cell Metabolism. 2014;19:122–134. doi: 10.1016/j.cmet.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tattikota S.G., Sury M.D., Rathjen T., Wessels H.-H., Pandey A.K., You X. Argonaute2 regulates the pancreatic β-cell secretome. Molecular & Cellular Proteomics. 2013;12:1214–1225. doi: 10.1074/mcp.M112.024786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nesca V., Guay C., Jacovetti C., Menoud V., Peyot M.-L., Laybutt D. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia. 2013;56(10):2203–2212. doi: 10.1007/s00125-013-2993-y. [DOI] [PubMed] [Google Scholar]
- 34.Zhao E., Keller M.P., Rabaglia M.E., Oler A.T., Stapleton D.S., Schueler K.L. Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mammalian Genome. 2009;20:476–485. doi: 10.1007/s00335-009-9217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roggli E., Gattesco S., Caille D., Briet C., Boitard C., Meda P. Changes in MicroRNA expression contribute to pancreatic b-cell dysfunction in prediabetic NOD mice. Diabetes. 2012;61(7):1742–1751. doi: 10.2337/db11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grieco F.A., Sebastiani G., Juan-Mateu J., Villate O., Marroqui L., Ladrière L. MicroRNAs miR-23a-3p, miR-23b-3p, and miR-149 5p regulate the expression of proapoptotic BH3 only proteins DP5 and PUMA in human pancreatic β-cells. Diabetes. 2017;66:100–112. doi: 10.2337/db16-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bravo-Egana V., Rosero S., Klein D., Jiang Z., Vargas N., Tsinoremas N. Inflammation-mediated regulation of MicroRNA expression in transplanted pancreatic islets. Journal of Transplantation. 2012;2012:1–15. doi: 10.1155/2012/723614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Locke J.M., Da Silva Xavier G., Dawe H.R., Rutter G.A., Harries L.W. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia. 2014;57:122–128. doi: 10.1007/s00125-013-3089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Git A., Dvinge H., Salmon-Divon M., Osborne M., Kutter C., Hadfield J. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buitrago D.H., Patnaik S.K., Kadota K., Kannisto E., Jone D.R., Adusumilli P.S. Small RNA sequencing for profiling microRNAs in long-term preserved formalin-fixed and paraffin-embedded non-small cell lung cancer tumor specimens. PLoS One. 2015;10:1–12. doi: 10.1371/journal.pone.0121521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melkman-Zehavi T., Oren R., Kredo-Russo S., Shapira T., Mandelbaum A.D., Rivkin N. miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. The EMBO Journal. 2011;30:835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng X., Huang C., Senavirathna L., Wang P., Liu L. miR-27b inhibits fibroblast activation via targeting TGFβ signaling pathway. BMC Cell Biology. 2017;18:9. doi: 10.1186/s12860-016-0123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolmeson C., Esguerra J.L., Salehi A., Speidel D., Eliasson L., Cilio C.M. Differences in islet-enriched miRNAs in healthy and glucose intolerant human subjects. Biochemical and Biophysical Research. 2011;404:16–22. doi: 10.1016/j.bbrc.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Frost R.J., Olson E.M. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu H., Shyh-Chang N., Segrè A.V., Shinoda G., Shah S.P., Einhorn W.S. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouaamari E.I., Baroukh N., Martens G.A., Lebrun P., Pipeleers D., van Obberghen E. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Latreille M., Herrmanns K., Renwick N., Tuschl T., Malecki M.T., McCarthy M.I. miR-375 gene dosage in pancreatic β-cells: implications for regulation of β-cell mass and biomarker development. Journal of Molecular Medicine (Berlin) 2015;93:1159–1169. doi: 10.1007/s00109-015-1296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed K., LaPierre M.P., Gasser E., Denzler R., Yang Y., Rülicke T. Loss of microRNA-7a2 induces hypogonadotropic hypogonadism and infertility. Journal of Clinical Investigation. 2017;127:1061–1074. doi: 10.1172/JCI90031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Scientific Reports. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Latreille M., Hausser J., Stützer I., Zhang Q., Hastoy B., Gargani S. MicroRNA-7a regulates pancreatic β cell function. Journal of Clinical Investigation. 2014;124:2722–2735. doi: 10.1172/JCI73066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esguerra J.L., Bolmeson C., Cilio C.M., Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kredo-Russo S., Mandelbaum A.D., Ness A., Alon I., Lennox K.A., Behlke M.A. Pancreas enriched miRNA refines endocrine cell differentiation. Development. 2012;139:3021–3031. doi: 10.1242/dev.080127. [DOI] [PubMed] [Google Scholar]
- 54.Lang H., Ai Z., You Z., Wan Y., Guo W., Xiao J. Characterization of miR-218/322-Stxbp1 pathway in the process of insulin secretion. Journal of Molecular Endocrinology. 2015;54:65–73. doi: 10.1530/JME-14-0305. [DOI] [PubMed] [Google Scholar]
- 55.Bagge A., Dahmcke C.M., Dalgaard L.T. Syntaxin-1a is a direct target of miR-29a in insulin-producing β-cells. Syntaxin-1a is a direct target of miR-29a in insulin-producing β-cells. Hormone and Metabolic Research. 2013;45:463–466. doi: 10.1055/s-0032-1333238. [DOI] [PubMed] [Google Scholar]
- 56.Plaisance V., Abderrahmani A., Perret-Menoud V., Jacquemin P., Lemaigre F., Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. Journal of Biological Chemistry. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 57.Porte D., Jr., Kahn S.E. β-Cell dysfunction and failure in type 2 diabetes: potential mechanisms. Diabetes. 2001;50(Suppl. 1):S160–S163. doi: 10.2337/diabetes.50.2007.s160. [DOI] [PubMed] [Google Scholar]
- 58.Vinod M., Patankar J.V., Sachdev V., Frank S., Graier W.F., Kratky D. MiR-206 is expressed in pancreatic islets and regulates glucokinase activity. American Journal of Physiology Endocrinology and Metabolism. 2016;311:E175–E185. doi: 10.1152/ajpendo.00510.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tattikota S.G., Rathjen T., Hausser J., Khedkar A., Kabra U.D., Pandey V. miR-184 regulates pancreatic β-cell function according to glucose metabolism. Journal of Biological Chemistry. 2015;290:20284–20294. doi: 10.1074/jbc.M115.658625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Setyowati Karolina D., Sepramaniam S., Tan H.Z., Armugam A., Jeyaseelan K. miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biology. 2013;10:1365–1378. doi: 10.4161/rna.25557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu Y., You W., Wang H., Li Y., Qiao N., Shi Y. MicroRNA-24/MODY gene regulatory pathway mediates pancreatic β-cell dysfunction. Diabetes. 2013;62:3194–3206. doi: 10.2337/db13-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J.W., You Y.H., Jung S., Suh-Kim H., Lee I.K., Cho J.H. miRNA-30a-5p-mediated silencing of Beta2/NeuroD expression is an important initial event of glucotoxicity-induced beta cell dysfunction in rodent models. Diabetologia. 2013;56:847–855. doi: 10.1007/s00125-012-2812-x. [DOI] [PubMed] [Google Scholar]
- 63.Xu G., Chen J., Jing G., Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nature Medicine. 2013;19:1141–1146. doi: 10.1038/nm.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baroukh N., Ravier M.A., Loder M.K., Hill E.V., Bounacer A., Scharfmann R. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. Journal of Biological Chemistry. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 65.Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 66.Backe M.B., Novotny G.W., Christensen D.P., Grunnet L.G., Mandrup-Poulsen T. Altering β-cell number through stable alteration of miR-21 and miR-34a expression. Islets. 2014;6 doi: 10.4161/isl.27754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsukita S., Yamada T., Takahashi K., Munakata Y., Hosaka S., Takahashi H. MicroRNAs 106b and 222 improve hyperglycemia in a mouse model of insulin-deficient diabetes via pancreatic β-cell proliferation. EBioMedicine. 2017;15:163–172. doi: 10.1016/j.ebiom.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tao H., Wang M.M., Zhang M., Zhang S.P., Wang C.H., Yuan W.J. MiR-126 suppresses the glucose-stimulated proliferation via IRS-2 in INS-1 β cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu Y., Fei X.Q., Yang S.F., Xu B.K., Li Y.Y. Glucose-induced microRNA-17 promotes pancreatic beta cell proliferation through down-regulation of Menin. European Review for Medical and Pharmacological Sciences. 2015;19:624–629. [PubMed] [Google Scholar]
- 70.Jacovetti C., Jimenez V., Ayuso E., Laybutt R., Peyot M.L., Prentki M. Contribution of intronic miR-338-3p and its hosting gene AATK to compensatory β-cell mass expansion. Molecular Endocrinology. 2015;29:693–702. doi: 10.1210/me.2014-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cnop M., Welsh N., Jonas J.C., Jörns A., Lenzen S., Eizirik D.L. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;54(Suppl. 2):S97–S107. doi: 10.2337/diabetes.54.suppl_2.s97. [DOI] [PubMed] [Google Scholar]
- 72.Halban P.A., Polonsky K.S., Bowden D.W., Hawkins M.A., Ling C., Mather K.J. β-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Journal of Clinical Endocrinology & Metabolism. 2014;99:1983–1992. doi: 10.1210/jc.2014-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schickel R., Park S.M., Murmann A.E., Peter M.E. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Molecular Cell. 2010;38:908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Filios S.R., Xu G., Chen J., Hong K., Jing G., Shalev A. MicroRNA-200 is induced by thioredoxin-interacting protein and regulates Zeb1 protein signaling and beta cell apoptosis. J Biol Chem. 2014;289:36275–36283. doi: 10.1074/jbc.M114.592360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y., Li D., Luo J., Tian G., Zhao L.Y., Liao D. Intrinsic cellular signaling mechanisms determine the sensitivity of cancer cells to virus-induced apoptosis. Scientific Reports. 2016;6 doi: 10.1038/srep37213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao Y., Yan W., Lu L., Wang Y., Lu W., Cao Y. p38/p53/miR-200a-3p feedback loop promotes oxidative stress-mediated liver cell death. Cell Cycle. 2015;14:1548–1558. doi: 10.1080/15384101.2015.1026491. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Belgardt B.F., Ahmed K., Spranger M., Latreille M., Denzler R., Kondratiuk N. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nature Medicine. 2015;21:619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 78.Hong K., Xu G., Grayson T.B., Shalev A. Cytokines regulate β-cell thioredoxin-interacting protein (TXNIP) via distinct mechanisms and pathways. Journal of Biological Chemistry. 2016;291:8428–8439. doi: 10.1074/jbc.M115.698365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lerner A.G., Upton J.P., Praveen P.V., Ghosh R., Nakagawa Y., Igbaria A. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metabolism. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng Y., Wang Z., Tu Y., Shen H., Dai Z., Lin J. miR-101a and miR-30b contribute to inflammatory cytokine-mediated β-cell dysfunction. Laboratory Investigation. 2015;95:1387–1397. doi: 10.1038/labinvest.2015.112. [DOI] [PubMed] [Google Scholar]
- 81.Li Y., Luo T., Wang L., Wu J., Guo S. MicroRNA-19a-3p enhances the proliferation and insulin secretion, while it inhibits the apoptosis of pancreatic β cells via the inhibition of SOCS3. International Journal of Molecular Medicine. 2016;38:1515–1524. doi: 10.3892/ijmm.2016.2748. [DOI] [PubMed] [Google Scholar]
- 82.Mohan R., Mao Y., Zhang S., Zhang Y.W., Xu C.R., Gradwohl G. Differentially expressed MicroRNA-483 confers distinct functions in pancreatic β- and α-cells. Journal of Biological Chemistry. 2015;290:19955–19966. doi: 10.1074/jbc.M115.650705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bao L., Fu X., Si M., Wang Y., Ma R., Ren X. MicroRNA-185 targets SOCS3 to inhibit beta-cell dysfunction in diabetes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Neel J.V. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? American Journal of Human Genetics. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 85.Shafrir E., Ziv E., Kalman R. Nutritionally induced diabetes in desert rodents as models of type 2 diabetes: Acomys cahirinus (spiny mice) and Psammomys obesus (desert gerbil) ILAR Journal. 2006;47:212–224. doi: 10.1093/ilar.47.3.212. [DOI] [PubMed] [Google Scholar]
- 86.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 88.Zernecke A., Bidzhekov K., Noels H., Shagdarsuren E., Gan L., Denecke B. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Science Signalling. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 89.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 90.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nature Cell Biology. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams Z., Ben-Dov I.Z., Elias R., Mihailovic A., Brown M., Rosenwaks Z. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4255–4260. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neal C.S., Michael M.Z., Pimlott L.K., Yong T.Y., Li J.Y.Z., Gleadle J.M. Circulating microRNA expression is reduced in chronic kidney disease. Nephrology Dialysis Transplantation. 2011;26:3794–3802. doi: 10.1093/ndt/gfr485. [DOI] [PubMed] [Google Scholar]
- 95.Guay C., Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nature Reviews Endocrinology. 2013;9:513–521. doi: 10.1038/nrendo.2013.86. [DOI] [PubMed] [Google Scholar]
- 96.Erener S., Mojibian M., Fox J.K., Denroche H.C., Kieffer T.J. Circulating miR-375 as a biomarker of β-cell death and diabetes in mice. Endocrinology. 2013;154:603–608. doi: 10.1210/en.2012-1744. [DOI] [PubMed] [Google Scholar]
- 97.Chappell J., Watters K.E., Takahashi M.K., Lucks J.B. A renaissance in RNA synthetic biology: new mechanisms, applications and tools for the future. Current Opinion in Chemical Biology. 2015;28:47–56. doi: 10.1016/j.cbpa.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 98.Cullen B.R. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–425. doi: 10.1038/nature07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Molecular Therapy. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P. MicroRNA-133 controls cardiac hypertrophy. Nature Medicine. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 101.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 103.Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 104.Krützfeldt J., Kuwajima S., Braich R., Rajeev K.G., Pena J., Tuschl T. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Research. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature Biotechnology. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 106.Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ameres S.L., Phillip D., Zamore P. Diversifying microRNA sequence and function. Nature Reviews Molecular Cell Biology. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 108.Davis S., Lollo B., Freier S., Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Research. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ørom U.A., Kauppinen S., Lund A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 110.Lennox K.A., Behlke M.A. A direct comparison of anti-microRNA oligonucleotide potency. Journal of Pharmacy Research. 2010;27:1788–1799. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 111.Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 112.Lahmy R., Soleimani M., Sanati M.H., Behmanesh M., Kouhkan F., Mobarra N. MiRNA-375 promotes beta pancreatic differentiation in human induced pluripotent stem (hiPS) cells. Molecular Biology Reports. 2014;41:2055–2066. doi: 10.1007/s11033-014-3054-4. [DOI] [PubMed] [Google Scholar]
- 113.Shaer A., Azarpira N., Karimi M.H. Differentiation of human induced pluripotent stem cells into insulin-like cell clusters with miR-186 and miR-375 by using chemical transfection. Applied Biochemistry and Biotechnology. 2014;174:242–258. doi: 10.1007/s12010-014-1045-5. [DOI] [PubMed] [Google Scholar]
- 114.Liang D., Zhang Y., Han J., Wang W., Liu Y., Li J. Embryonic stem cell-derived pancreatic endoderm transplant with MCT1-suppressing miR-495 attenuates type II diabetes in mice. Endocrine Journal. 2015;62:907–920. doi: 10.1507/endocrj.EJ15-0186. [DOI] [PubMed] [Google Scholar]
- 115.Mi Y., Guo N., He T., Ji J., Li Z., Huang P. miR-410 enhanced hESC-derived pancreatic endoderm transplant to alleviate gestational diabetes mellitus. Journal of Molecular Endocrinology. 2015;55:219–229. doi: 10.1530/JME-15-0100. [DOI] [PubMed] [Google Scholar]
- 116.Zhao X., Mohan R., Özcan S., Tang X. MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic β-cells. Journal of Biological Chemistry. 2012;287:31155–31164. doi: 10.1074/jbc.M112.362632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tang X., Muniappan L., Tang G., Özcan S. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–293. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sebastiani G., Po A., Miele E., Ventriglia G., Ceccarelli E., Bugliani M. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetologica. 2015;52:523–530. doi: 10.1007/s00592-014-0675-y. [DOI] [PubMed] [Google Scholar]
- 119.Salunkhe V.A., Esguerra J.L., Ofori J.K., Mollet I.G., Braun M., Stoffel M. Modulation of microRNA-375 expression alters voltage-gated Na(+) channel properties and exocytosis in insulin-secreting cells. Acta Physiologica (Oxford) 2015;213:882–892. doi: 10.1111/apha.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roggli E., Britan A., Gattesco S., Lin-Marg N., Abderrahmani A., Meda P. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shang J., Li J., Keller M.P., Hohmeier H.E., Wang Y., Feng Y. Induction of miR-132 and miR-212 expression by glucagon-like peptide 1 (GLP-1) in rodent and human pancreatic β-cells. Molecular Endocrinology. 2015;29:1243–1253. doi: 10.1210/me.2014-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Y., Liu J., Liu C., Naji A., Stoffers D.A. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes. 2013;62:887–895. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ofori J.K., Salunkhe V.A., Bagge A., Vishnu N., Nagao M., Mulder H. Elevated miR-130a/miR130b/miR-152 expression reduces intracellular ATP levels in the pancreatic beta cell. Scientific Reports. 2017;7 doi: 10.1038/srep44986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pullen T.J., da Silva Xavier G., Kelsey G., Rutter G.A. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1) Molecular and Cellular Biology. 2011;31:3182–3194. doi: 10.1128/MCB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kang M.H., Zhang L.H., Wijesekara N., de Haan W., Butland S., Bhattacharjee A. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arteriosclerosis Thrombosis and Vascular Biology. 2013;33:2724–2732. doi: 10.1161/ATVBAHA.113.302004. [DOI] [PubMed] [Google Scholar]
- 126.Wijesekara N., Zhang L.H., Kang M.H., Abraham T., Bhattacharjee A., Warnock G.L. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes. 2012;61:653–658. doi: 10.2337/db11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]