Abstract
Although outbreaks of highly pathogenic avian influenza in wild and domestic birds have been posing the threat of a new influenza pandemic for the last decade, the first pandemic of the 21st century came from swine viruses. This fact emphasizes the complexity of influenza viral ecology and the difficulty of predicting influenza viral dynamics. Complete control of influenza viruses seems impossible. However, we must minimize the impact of animal and human influenza outbreaks by learning lessons from past experiences and recognizing the current status. Here, we review the most recent influenza virology data in the veterinary field, including aspects of zoonotic agents and recent studies that assessed the pandemic potential of H5N1 highly pathogenic avian influenza viruses.
Keywords: influenza A viruses, host, transmission, pandemic
1 Influenza virus
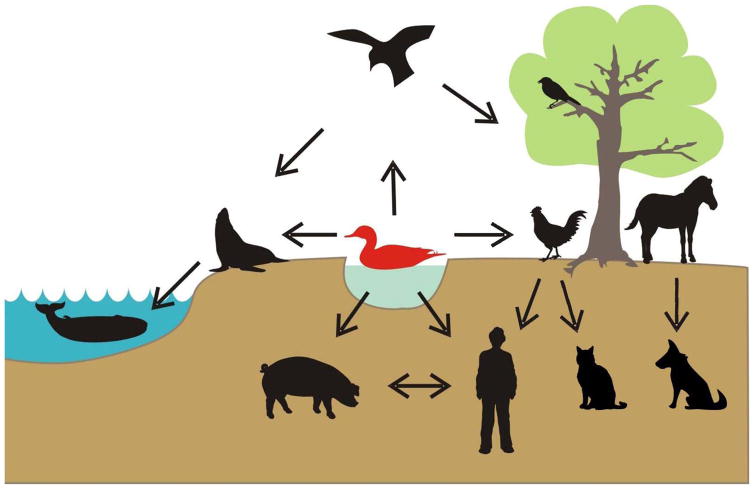
Influenza is a collective term describing infectious diseases caused by highly contagious influenza viruses, a member of the family Orthomyxoviridae. Based on genome composition and encoded proteins, influenza viruses are classified into three types, A, B, and C. Among these three types, only type A virus (influenza A virus) is a major zoonotic agent. Influenza A viruses have been isolated from a variety of animals, including birds, pigs, horses, dogs, sea mammals, and humans (Figure 1).
Figure 1. Hosts of influenza A viruses.
Wild aquatic birds are recognized as a natural reservoir of influenza A viruses, since viruses of all 16 HA and 9 NA subtypes have been isolated from them. In addition, viruses of particular subtypes are circulating in various animals, including birds, pigs, horses, dogs, sea mammals, and humans. Arrows indicate known transmission routes between each animal species. With permission from Wright PF, Neumann G, Kawaoka Y. 2007. In Fields virology, ed. DM Knipe, PM Howley, pp. 1691–740. Philadelphia, Pa.: Lippincott-Raven Publishers.
Based on the antigenicities of two viral glycoproteins, hemagglutinin (HA) and neuraminidase (NA), influenza A viruses are further classified into subtypes; to date, 16 HA subtypes (H1–H16) and 9 NA subtypes (N1–N9) have been identified. Recently identified influenza viruses from bats (1) possess genetically distinct HA genes, possibly H17, although the antigenicity of this HA has not been properly compared with other HA subtypes (2). Wild aquatic birds, particularly of the orders Anseriformes (ducks, geese, swan) and Charadriiformes (gulls, terns, shorebirds) harbor viruses of all of the 16 HA and 9 NA subtypes (3–5) usually without significant symptoms, suggesting that they represent the natural reservoir of influenza A viruses (Figure 1).
The low fidelity of the genome replication machinery, together with the segmented nature of the influenza virus genome, allows the rapid emergence of antigenic variants of these viruses with various pathogenic phenotypes and their transmission and adaptation to new hosts. Humans experienced four global epidemics (pandemics) caused by new influenza viruses in the 20th and 21st centuries (see 3.1 Pandemics). One critical feature of these pandemic viruses is that their HA genes were derived from contemporary avian or swine influenza virus and mutated to allow human-to-human transmission.
2 Influenza in nonhuman animals
Influenza A viruses cause mild to severe diseases in various animals. Among these diseases, avian, swine, and equine influenza circulate globally and are particularly important in veterinary fields due to their economic impact and the potential risk of zoonosis outbreaks.
2.1 Avian influenza
Based on virus pathogenicity in chickens, avian influenza viruses are classified into two groups: highly pathogenic avian influenza (HPAI) viruses and the others [low-pathogenicity avian influenza (LPAI) viruses]. The Office International des Epizooties (OIE) defines highly pathogenic virus strains as follows (6):
any influenza virus that is lethal for six, seven, or eight of eight (>75%) four- to six-week-old susceptible chickens within ten days following intravenous inoculation with 0.2 ml of a 1:10 dilution of a bacteria-free, infectious allantoic fluid,
any H5 or H7 virus that does not meet the criteria in a), but has an amino acid sequence at the HA cleavage site that is compatible with HPAI viruses,
any influenza virus that is not an H5 or H7 subtype and that kills one to five of eight inoculated.
Historically, viruses of either H5 or H7 subtype have caused dreadful HPAI outbreaks in poultry farms (Table 1). Since 1997, multiple transmissions of H5N1 and H7N7 viruses to humans have been reported (see 3.2.1 Avian H5N1 influenza section and 3.2.2 Avian H7N7 influenza section).
Table 1.
Outbreaks of highly pathogenic avian influenza viruses since 1995a.
| Virus | Subtype | Comments |
|---|---|---|
| A/chicken/Scotland/59 | H5N1 | Two chicken flocks |
| A/tern/South Africa/61 | H5N3 | 1,300 Common terns |
| A/turkey/England/63 | H7N3 | 29,000 Breeder turkeys |
| A/turkey/Ontario7732/66 | H5N9 | 8,100 Breeder turkeys |
| A/chicken/Victoria/76 | H7N7 | 25,000 Laying chickens, 17,000 broilers and 16,000 |
| A/chicken/Germany/79 | H7N7 | Outbreaks in former East Germany; numbers unknown |
| A/turkey/England/199/79 | H7N7 | 3 Commercial turkey farms |
| A/chicken/Pennsylvania/1370/83 | H5N2 | 17 Million birds, mostly chickens or turkeys |
| A/turkey/Ireland/1378/83 | H5N8 | 800 Infected turkeys died; 8,640 turkeys, 28,020 chickens, and 270,000 ducks were culled |
| A/chicken/Victoria/85 | H7N7 | 24,000 Broiler breeders, 27,000 laying chickens, 69,000 broilers, and 118,518 unspecified chickens |
| A/turkey/England/50-92/91 | H5N1 | 8,000 Turkeys |
| A/chicken/Victoria/92 | H7N3 | 12,700 Broiler breeders and 5,700 ducks |
| A/chicken/Queensland/95 | H7N3 | 22,000 Laying chickens |
| A/chicken/Puebla/8623-607/94 A/chicken/Queretaro/14588-19/95 | H5N2 | Millions of birds died or were culled; exact numbers are not available |
| A/chicken/Pakistan/447/95 A/chicken/Pakistan/1369-CR2/95 |
H7N3 | 3.2 Million broilers and broiler breeder chicken |
| A/chicken/Hong Kong/220/97 | H5N1 | 1.5 Million chickens and other domestic birds; 6 human fatalities (among 18 infected individuals) |
| A/chicken/New South Wales/1651/97 | H7N4 | 128,000 Broiler breeders, 33,000 broilers, and 261 emus |
| A/chicken/Italy/330/97 | H5N2 | Chickens, turkeys, guinea-fowl, ducks, quail, pigeons, geese, pheasant (all in small numbers) |
| A/turkey/Italy/4580/99 | H7N1 | 8.1 Million laying chickens, 2.7 million meat and breeder turkeys, 2.4 million broiler breeders and broilers, 247,000 guinea-fowl, 260,000 quail, ducks, and pheasants; also backyard poultry and ostriches |
| A/chicken/Chile/4957/2002 | H7N2 | 2 Million birds died or were culled |
| A/grey heron/Hong Kong/861.1/2002 | H5N1 | Outbreak in wild birds in Hong Kong; over 800,000 domestic birds were culled |
| A/chicken/Netherlands/1/2003 | H7N7 | Virus was isolated from 241 poultry farms in The Netherlands, two farms in Belgium, and one farm in Germany; outbreak was controlled by killing more than 30 million birds; one human fatality |
| A/chicken/Canada/AVFV1/2004 A/chicken/Canada/AVFV2/2004 |
H7N3 | Spread to more than 40 commercial poultry farms; outbreak was controlled by culling all 19 million domestic birds in Fraser Valley, British Columbia |
| A/ostrich/South Africa/2004 | H5N2 | Outbreak in ostriches; 26,000 ostriches were culled to control virus spread |
| Various H5N1 viruses (since 2003) | H5N1 | Outbreak started in July 2003 in poultry in Vietnam, Indonesia, and Thailand and has since spread to a number of Southeast Asian, European, Middle Eastern, and African countries; more than 100 million domestic birds have died or have been culled; mortality has also been observed among wild birds; as of March 31, 2011, 318 human fatalities had been reported (among 539 infected individuals) |
| A/chicken/North Korea/2005 | H7N7 | Approximate number of birds affected: 219,000 |
| A/chicken/Saskatchewan/HR-00011/2007 | H7N3 | Outbreak on a broiler breeding farm in Saskatchewan, Canada; depopulation of all animals on the premises (53,000) |
| A/chicken/England/1158-11406/2008 | H7N7 | Outbreak in a laying flock of chicken; death or depopulation of 25,000 animals |
| A/chicken/Spain/6279-2/2009 | H7N7 | Outbreak in a layer hen farm; outbreak was stopped through death (~30,000) and depopulation (~278,000) of all animals on the farm |
| H5N2 viruses | H5N2 | Outbreak in a broiler breeding farm in the Tainan, Changhua, and Pingtung areas of Taiwan |
Modified with permission from Swayne, DE, Halvorson DA. Influenza. In: Saif YM, Barnes HJ, Fadly AM, Glisson JR, McDougald LR, Swayne DE, eds. Diseases of Poultry. 11th ed. Ames: Iowa State University Press; 2003:135–160.
2.1.1 Epizoology
In avian species, especially in waterfowl, influenza A viruses replicate mainly in the intestinal tract. Consequently, huge numbers of viruses are shed in the feces of infected birds (7, 8), resulting in contamination of the lake water inhabited by migratory birds (9). The prevalence of viruses in avian hosts varies depending on seasonality and species. Surveillance studies (5, 10–15) revealed that 60% of mallard ducks were infected with influenza A viruses in the autumn (just before migration) but and less than 10% were infected in spring to summer (during migration). In dabbling ducks, the prevalence of avian influenza viruses ranges from less than 1% to up to 30% during and prior to migration, respectively (5, 15). These findings suggest that juvenile birds that are born in summer are more susceptible to influenza A virus infection and play a critical role in the maintenance of viruses in bird populations.
Most influenza A viruses, including “classical” HPAI strains but excluding the recently circulating H5N1 viruses (see 3.2.1 Avian H5N1 influenza section), typically cause asymptomatic infection in wild aquatic birds. Therefore, virus-infected birds serve not only as a reservoir, but also as a carrier of viruses via global migration. Viruses infecting wild aquatic birds are occasionally introduced into poultry farms. By contrast to the natural hosts, terrestrial birds such as chickens, turkeys, and quails, are susceptible to some of these avian influenza viruses, which cause diseases after repeated replication in these birds. The exceptions are domestic ducks; when these ducks are infected with influenza A viruses they usually exhibit no or limited symptoms, and can, therefore potentially spread the viruses to other poultry.
Viruses of several HA and NA subtypes have been isolated from poultry. Among them, those of the H6, H7, and H9 subtypes appear to readily adapt to chickens (16–18). In addition, the H5 and H7 viruses potentially have a huge impact on the both the poultry industry and public health (see 3.2.1 Avian H5N1 influenza section and 3.2.2 Avian H7N7 influenza section).
2.1.2 Diseases
Poultry infected with LPAI viruses typically exhibit mild respiratory disease, depression, and/or a drop in egg production. In most cases of HPAI strain infection, if the infected poultry die within ~48 h of infection, they show no clinical symptoms; when infected birds live longer than ~48 h, neurological (e.g., tremor and circling) and/or hematological (e.g., edema and cyanosis) symptoms are often present.
2.2 Swine influenza
Swine influenza viruses of different lineages are circulating. In addition, both avian and human viruses can infect and replicate in pigs (19). Therefore, pigs serve as a “mixing vessel” for the generation of reassortant viruses (20).
2.2.1 Epizoology
Swine influenza viruses, as well as human influenza viruses, spread via respiratory droplets. A swine influenza virus that is an ancestor of the recent H1N1 pandemic strain (see 3.1.5 The pandemic (H1N1) 2009 section) was first isolated in 1930 (A/swine/Iowa/15/30 [H1N1]). This swine H1N1 strain and its descendent are referred to as “classical swine viruses” and have circulated worldwide for a long time as a major virus subtype in pig populations. In the 1997–1998 season, H1N2 and H3N2 triple reassortants among human, avian, and swine viruses were isolated in the north-central United States. These viruses possessed human virus-origin HA, NA, and PB1 genes, avian virus-origin PB2 and PA genes, and the remaining genes from classical swine virus (21). These “triple reassortant swine viruses” have rapidly become predominant in North American pig populations, occasionally giving rise to reasssortants possessing HA and/or NA genes derived from other human, avian, or swine virus; their internal gene composition remains fixed.
In Europe, although the classical swine viruses caused outbreaks in pig populations in 1976 (22), they were gradually replaced by entirely avian virus-origin H1N1 viruses that were firstly isolated in 1979 (23). These viruses are referred to as “avian-like swine H1N1 viruses” and are currently circulating in European pig populations.
Human H3N2 viruses were also introduced into worldwide pig populations following the H3N2 pandemic of 1968 (see 3.1.3 Hong Kong influenza in 1968 section). In 1984, reassortant H3N2 swine viruses that possessed human-like HA and NA genes and avian-like swine H1N1 virus-origin internal genes caused severe disease in Italian and French pig populations. These so-called “human-like swine H3N2 viruses” rapidly spread across Europe and established one of the swine virus lineages. Several types of H1N2 swine viruses (e.g., reassortants between the classical swine H1N1 viruses and human H3N2 viruses or triple reassortant swine viruses) have also been isolated globally. In addition, since the swine-origin virus responsible for the (H1N1) 2009 pandemic (see 3.1.5 The (H1N1) 2009 pandemic section) emerged, this virus has been transmitted back to pigs and established a new swine virus lineage.
2.2.2 Diseases
In pigs, influenza A viruses cause decreased appetite, fever, runny nose, coughing, pneumonia, and/or conjunctivitis. However, pigs do not develop severe or lethal infection even with highly virulent viruses such as HPAI (24, 25) or the “Spanish influenza” viruses (26).
2.3 Equine influenza
The first equine influenza virus was isolated from a European horse with a respiratory disorder in 1958 (27). These viruses pose a global threat to horses, especially unvaccinated individuals. Equine influenza is an enzootic disease. Viruses are typically maintained among horse populations, although interspecies transmission occasionally occurs.
2.3.1 Epizoology
Equine influenza spreads via respiratory droplets. Influenza A viruses of two subtypes, H7N7 and H3N8, are known to cause equine influenza, although no outbreak by a virus of H7N7 subtype, which was the subtype of the first equine influenza virus isolated (28), has occurred during the last two decades in horse populations (29). The H3N8 viruses are considered more pathogenic in horses compared with their H7N7 counterparts and have intermittently caused widespread enzootics in Europe, North America, Africa, and Asia. International transportation of racehorses may contribute to the evolution of equine influenza viruses.
2.3.2 Diseases
The typical symptoms caused by influenza A virus infection in horses are similar to those in humans: fever, cough, loss of appetite, and muscular soreness. In part due to vaccine use (see 2.5 Prevention section), fatal infection is rare in horses. An exceptional enzootic with an avian-origin new lineage of H3N8 viruses occurred in China in 1989; mortality rates were estimated to be as high as 20% (30).
2.4 Diagnosis
Fecal specimens from wild waterfowls or respiratory and/or cloacal swabs from poultry birds are suitable for testing for avian influenza viruses. For swine and equine influenza, nasal swabs are commonly used for diagnostic testing.
The traditional and most reliable method to prove the presence of viruses in a sample is virus isolation. For this purpose, samples are inoculated into embryonated chicken eggs. Avian influenza viruses can be propagate in eggs and readily tested for their HA and NA subtypes by use of haemagglutination-inhibition and neuraminidase-inhibition assays, respectively, with subtype-specific antisera.
Advances in molecular biological technologies have enabled the easy detection of viruses. Reverse transcriptase polymerase chain reaction (RT-PCR) is widely accepted as an alternative method to detect influenza viruses. Although RT-PCR only guarantees the presence of pieces of viral genes, its high sensitivity, speed, and sample capacity makes this method useful for influenza diagnosis, including large-scale surveillance.
Currently, several immunoassay-based influenza rapid diagnostic tests are available in which testing for the presence of viral nucleoprotein in samples is based on an antigen-antibody reaction with a specific monoclonal antibody or antibodies. Although most of these tests are designed mainly for clinical use and their sensitivity may be suboptimal for animal influenza (31), they can facilitate the detection of influenza viruses in clinical samples.
2.5 Prevention
LPAI viruses of the H5 and H7 subtypes have the potential to evolve into HPAI counterparts by accumulating mutations through continuous replication in poultry birds. Therefore, both HPAI and LPAI must be controlled. Because of the potential risk for HPAI virus generation, live attenuated vaccines based on influenza virus are not considered appropriate for avian influenza control. Instead, formalin- or other organic solvent-inactivated vaccines are available in some countries where avian influenza viruses persist. Although inactivated influenza vaccines induce specific immunity against vaccine strains, their efficacy in limited; viruses often replicate in vaccinated animals to some extent, leading to the generation of antigenic variants (32). In fact, many countries that were successful in eradicating HPAI viruses did not use vaccines; rather, they eradicated the viruses by culling infected birds. Some countries however, succeeded in eradicating these viruses by implementing emergency use of inactivated vaccines. In some countries, other virus- (e.g., Newcastle disease and fowl pox viruses) based live recombinant vaccines possessing influenza viral HA genes are approved for animal use (33).
Swine influenza vaccines against the H1N1 and H3N2 subtypes are available in many countries. However, these vaccines are not extensively used to control influenza viruses in pig populations, partly because of the limited risk of swine influenza in pigs (see 2.2.2 Diseases section) and because vaccine efficacy in pig herds remains inconclusive (34).
Contrary to other animal influenza vaccines, equine influenza vaccines are used widely and aggressively, especially in racehorses, and therefore, widespread outbreaks of severe cases in horse populations are limited. Routine use of vaccines, however, allows viruses to circulate in horse populations at subclinical levels, leading to antigenic drift (35). Therefore, constant updates of the vaccine reference strains are essential to maximize vaccine efficacy. A canarypox virus-based live recombinant vaccine expressing equine viral H3 HA is available in several countries (36). In the United States, a cold-adapted, temperature-sensitive modified live equine influenza vaccine is also licensed (37–40).
2.6 Influenza in minor hosts
Viruses closely related to the equine isolate A/canine/Florida/43/04 (H3N8) caused outbreaks of respiratory disease in a racing dog (greyhound) population in 2004 in Florida and spread to the general dog population, although outbreaks in domestic dogs occur mainly in kennels (41). Studies (42, 43) revealed that equine H3N8 viruses that were genetically distinct from the ancestor of the American canine isolates have also transmitted to dogs, indicating that dogs are susceptible to equine H3N8 viruses. Avian H3N2 (44) and the pandemic (H1N1) 2009 viruses (45, 46) have also been transmitted to dogs, although none of them has become established in dog populations.
Influenza A viruses have also been isolated from zoo tigers (H5N1 subtypes) (47, 48), minks (H10N4 subtype) (49), seals (H7N7, H3N2, and H4N6) (50, 51), whales (H13N2, H13N9, and H1N3) (52, 53), and cats (various subtypes including H5N1 and H1N1), sometimes causing lethal infections. Recently, genetic evidence of a new influenza virus lineage was found in fruit bats (1), suggesting that viruses of unknown HA and/or NA subtype may be maintained in as yet unrecognized reservoirs.
3 Zoonotic influenza
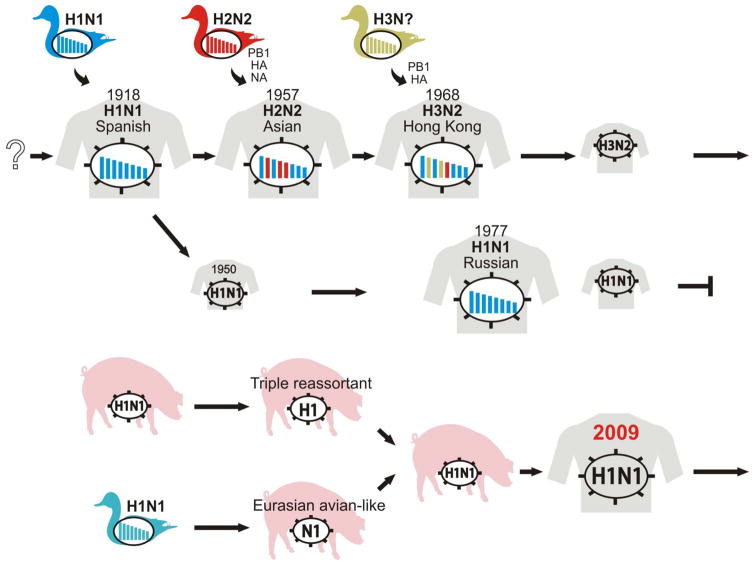
Although many viruses maintained in nonhuman animals have infected humans, most of these infections were self-limiting. However, as exemplified by the “Spanish influenza”, the transmission of influenza A viruses from nonhuman animals to humans can have devastating effects. In the last 150 years, viruses of only three HA subtypes, that is, H1, H2, and H3 viruses, have caused pandemics, although others may have this potential (Figure 2). The key feature of pandemic viruses is the ability to transmit from human to human.
Figure 2. Pandemic influenza viruses and their origins.
Genetic evidence suggests that the “Spanish influenza” viruses originated from an avian influenza virus of the H1N1 subtype in 1918. The introduction of the HA, NA, and PB1 genes from contemporary avian viruses of the H2N2 subtype into the genetic background of the H1N1 viruses in 1957 led to the emergence of the “Asian influenza” viruses. The H2N2 viruses were replaced in 1968 by a new reassortant between an avian virus (which provided the H3 HA and PB1 genes) and a human virus (which provided the remaining genes), that is, the H3N2 “Hong Kong influenza” virus. Although re-emerging H1N1 viruses (the “Russian influenza” viruses) have co-circulated with the H3N2 viruses in human populations since 1977, they became extinct after the emergence of the “pandemic (H1N1) 2009” viruses whose gene segments were derived from contemporary swine viruses (see Figure 3). Avian influenza viruses of the H5N1, H7N7, and H9N2 subtypes have been sporadically isolated from humans over the last 15 years, suggesting their potential to transmit to humans and cause future influenza pandemics. Modified with permission from Neumann et al. (125).
3.1 Pandemics
Influenza pandemics are caused by viruses that possess HAs to which most humans are immunologically naïve. Once a pandemic occurs, 20% to 40% of the world population could be infected within a short period of time, resulting in social and economic consequences. Since 1918, we have experienced four influenza pandemics with three HA subtype viruses.
3.1.1 Spanish influenza in 1918
Although the first outbreak of this pandemic is believed to have occurred in the United States during World War I, Spain, which was neutral during this war, was the first to report cases and therefore this pandemic was referred to as the “Spanish influenza.” Genetic analysis of viral RNAs recovered from lung tissues of victims of the 1918 pandemic (54, 55) revealed that the causative agent was an avian-origin virus of the H1N1 subtype.
The “Spanish influenza” was one of the most catastrophic infectious diseases in human history: one-third (approximately 500 million) of the world’s population at that time are estimated to have been infected with the causative agent and 2.5% (approximately 50 million) of the world’s population are estimated to have killed by the virus. Although patients exhibited typical “flu-like symptoms”, such as fever, runny nose, cough, headache, and muscular soreness, the course of the disease in some patients was remarkably quick and severe compared with contemporary seasonal influenza infection. In contrast to HPAI viruses, 1918 pandemic viruses did not cause systemic viral infection (56). The extraordinary pathogenicity of these viruses was tested in animal models (mice, ferrets, and nonhuman primates) with “Spanish influenza” viruses reconstructed by using the molecular biological technique of reverse genetics (57–60).
3.1.2 Asian influenza in 1957
Influenza viruses of the H2N2 subtype suddenly started spreading among humans in Southern China and quickly spread to neighboring Asian countries in 1957. This “Asian influenza” then attacked the entire world by the end of that year. Simultaneously, human H1N1 viruses became extinct likely because their niche in humans was occupied by the newly emerged pandemic viruses. Approximately one million people were estimated to be killed by the “Asian influenza” (61). Since the pathogenicity of the original 1957 pandemic virus was not high, the novelty of the HA and/or NA antigenicity to the human population was thought to be responsible for the mortality and morbidity of the “Asian influenza”. Genetic information suggests that the 1957 pandemic virus is a reassortant between human and avian viruses: the HA, NA, and PB1 genes were from avian viruses, and the remaining genes were from descendants of the 1918 pandemic virus (62, 63).
3.1.3 Hong Kong influenza in 1968
Another pandemic virus with H3 HA and N2 NA genes also emerged in South Asia. This so-called “Hong Kong influenza” was estimated to kill one million people by the winter of 1969–1970. Again, the H2N2 virus in humans was extinct when the H3N2 virus emerged. Both the HA and PB1 genes were of avian-origin, while the rest of its genes were from a descendant of the former pandemic virus of the H2N2 subtype (62, 64). Because the N2 NA genes of the 1957 and 1968 pandemic viruses were shared, immunity against N2 NA was prevalent in the human population and the impact of the “Hong Kong influenza” was moderate.
3.1.4 Russian Influenza in 1977
H1N1 viruses closely resembling those that were circulating in humans in the 1950’s (65) re-emerged and spread worldwide causing the so-called “Russian influenza”. Russian Influenza mainly affected the young, with mortality rates of greater than 50% among school-aged children. Because influenza viruses do not remain genetically unchanged during repeated replication, this unusual pandemic is thought to have been caused by a virus that was maintained in a laboratory.
3.1.5 The (H1N1) 2009 pandemic
In mid-April, the Centers for Disease Control and Prevention (CDC) isolated influenza A viruses of the H1N1 subtype that were genetically distinct from H1N1 viruses that was circulating at that time from two children in Southern California. The Public Health Agency of Canada also detected similar viruses in specimens received from Mexico on April 23. The causative agents efficiently transmitted among humans rapidly spread worldwide. On June 12, the World Health Organization declared the first influenza pandemic of the 21st century; the causative agent was referred to as the “pandemic (H1N1) 2009” virus.
The virus did not belong to any known lineages; its PB2 and PA genes were from North American avian viruses, its PB1 gene was from a contemporary human H3N2 virus, its HA, NP, and NS genes were from the “classical swine virus” (see 2.2 Swine influenza section), and its NA and M genes were from “avian-like swine viruses” (Figure 3). Because the immediate ancestors of the pandemic (H1N1) 2009 virus were swine viruses, its origin is believed to have been pigs. The direct ancestor of this pandemic virus, however, has not been found in pigs or any other animals to date.
Figure 3. Genetic origins of the “pandemic (H1N1) 2009” virus.
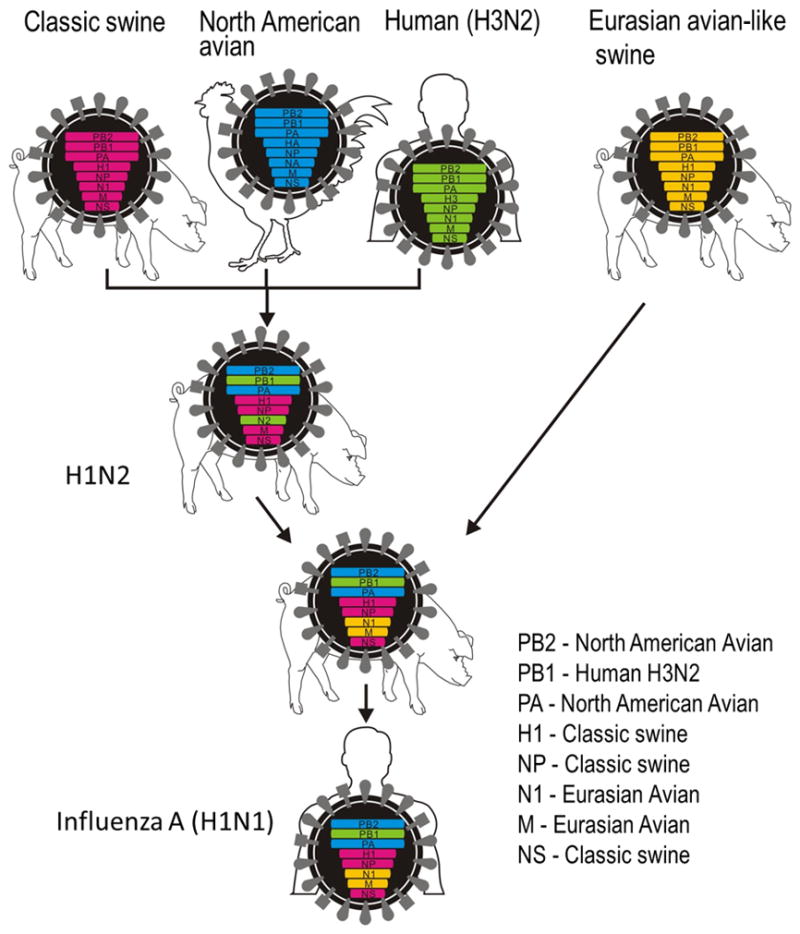
The H1N2 triple reassortant among human, avian, and swine viruses in pigs was firstly recognized in the 1997–1998 ‘flu season in the north-central United States (21). The North American “triple reassortant swine viruses” are thought to have reassorted with a Eurasian “avian-like swine H1N1 viruses” in pigs somewhere, resulting in the emergence of an H1N1 virus that was highly transmissible to and among humans. With permission from Neumann et al. (125).
Most patients without underlying conditions exhibited mild clinical symptoms and did not require hospitalization, although animal experiments revealed that the pathogenic potential of this virus was higher than that of seasonal influenza viruses (13, 66–70). The “pandemic (H1N1) 2009” viruses have now completely replaced the “Russian Influenza” viruses and are considered seasonal influenza viruses.
3.2 Animal influenza
Avian viruses had long been thought not to cause lethal infections in humans. Since 1997, however, human infections with avian viruses of three HA subtypes (i.e., H5, H7, and H9) have been reported. Fortunately, human infections with avian viruses have been self-limiting and human-to-human transmission has been infrequent.
3.2.1 Avian H5N1 influenza
The first human cases of H5N1 HPAI virus infection occurred in 1997 in Hong Kong; 18 patients, including 6 lethal cases, were infected with this virus that was circulating in birds in poultry markets (71–73). This outbreak was quickly eliminated likely due to the extensive slaughter of birds. Genetic analysis revealed that all of the genes of the causative virus were derived from avian H5N1 viruses.
This H5N1 virus re-emerged in 2003 and infected humans in Asian countries, such as Vietnam, Indonesia, and Thailand. The virus then spread to Europe, The Middle East, and Africa, causing numerous poultry outbreaks and sporadic human infections with fatal outcomes. As of July 6, 2012, 607 confirmed cases with 358 fatalities have been reported since 2003. The H5N1 viruses occasionally cause systemic infections (74–78), that is, their replication sites are not restricted to respiratory organs, but also include intestine, heart, spleen, kidney, and liver. In addition, recent studies (79–81) using ferrets, which are the gold standard for assessing human-to-human transmissibility of influenza viruses, demonstrated that viruses possessing the H5 HA have the potential to efficiently transmit among humans (see 3.3 Human-to-human transmission).
During replication in birds and infection of mammals, the H5N1 viruses acquired a remarkable ability: unlike other avian influenza viruses, including HPAI strains, they became lethal to wild aquatic birds, such as geese, gulls, and shelducks. The first outbreak of H5N1 viruses among wild aquatic birds occurred at Qinghai Lake, China in 2005 (82, 83). H5N1 viruses closely related to the Qinghai Lake isolates rapidly spread West and, as a result, all of the recent H5N1 isolates in Europe and Africa are descendants of the Qinghai Lake viruses.
3.2.2 Avian H7N7 influenza
Another subtype of HPAI viruses that has been associated with sporadic human infections is the H7N7 subtype. In 2003, H7N7 HPAI viruses attacked layer farms in The Netherlands and spread to neighboring countries, causing poultry outbreaks. These wholly avian influenza viruses also transmitted to humans: among 89 individuals positive for viral RNAs, 83 developed conjunctivitis and one fatal case was reported (84, 85).
3.2.3 Avian H9N2 influenza
Avian H9N2 influenza viruses, which cause LPAI, have circulated in poultry worldwide since the mid-1980s. In South China, viruses of this subtype are frequently isolated from pigs (86–88). In addition, seven human cases with mild symptoms were reported in South China and Hong Kong between 1998 and 2003 (89–92). These viruses have been shown experimentally to have mammalian-like characteristics, for example in their receptor-binding specificity (88, 93–95). The H9N2 viruses are, therefore, candidates for the next pandemic strain.
3.2.4 Other animal influenza
As exemplified by the (H1N1) 2009 pandemic, nonhuman animal viruses whose HA subtypes are the same as those of contemporary human viruses have the potential to cause influenza pandemics. Therefore, viruses of both the H2 and H3 subtypes that have already proven their pandemic potential and are constantly detected in avian species and/or pig populations may be responsible for the next pandemics.
3.3 Human-to-human transmission
Even prior to the human cases of H5N1 HPAI in 1997, avian influenza viruses of the H7 subtype were known to directly transmit to humans (96, 97). Similarly, it was known that pig-to-human transmission of swine influenza viruses could occasionally occur (98), with the earliest cases recorded in 1958. These findings suggest that these viruses can infect and replicate efficiently enough to exhibit influenza symptoms in humans. However, none of these viruses, except for the swine-origin “pandemic (H1N1) 2009” virus (see 3.1.5 The pandemic (H1N1) 2009 section), established sustained human-to-human transmission. Therefore, sustained transmissibility of viruses is fundamentally different from infectivity or replicability and is essential for a virus to adapt to a new host, leading to an influenza pandemic.
The receptor-binding specificity of viral HA is believed to be one of the key factors for avian viruses to transmit efficiently among humans. At the initial step of the viral infection cycle, HA, a glycoprotein on the virions, binds to sugar chains terminating in sialic acid that are expressed on the cell surface. The sialic acid-terminating sugar chain, therefore, serve as cellular receptor for influenza virus infection. Human and avian viruses have different receptor-binding specificity: the former and latter preferentially bind to sialic acids linked to downside galactose by α2,6-linkages (Siaα2,6Gal) and by α2,3-linkages (Siaα2,3Gal), respectively (99–102). In the human upper airway, epithelial cells mainly express Siaα2,6Gal, although some cells in the lung express Siaα2,3Gal (103, 104). By contrast, Siaα2,3Gal is abundant in avian intestine, a major replication site of avian influenza viruses (105–108). This receptor distribution in hosts and viral receptor specificity account for the molecular basis for the species barrier between humans and birds. In fact, all known pandemic viruses preferentially bind to Siaα2,6Gal (100–102, 109).
Some human isolates of H5 HPAI viruses exhibited increased binding to Siaα2,6Gal (110, 111). Nevertheless, importantly, evidence for human-to human transmission of H5 viruses is limited. In addition, although classical and recent swine viruses exhibited preferential binding to Siaα2,6Gal (112), the “triple reassortant swine viruses” (see 2.2.1 Epizoolog section) that sporadically transmitted to humans (113) did not cause a pandemic until they reassorted with another swine virus, resulting in the “pandemic (H1N1) 2009” strain (see 3.1.5 The (H1N1) 2009 pandemic section). These facts suggest that receptor-binding specificity conversion is necessary, but not sufficient to confer human-to-human transmissibility to nonhuman animal viruses.
Because transmissibility is one of the most complex virologic events, no appropriate in vitro assessment system has yet been established. To assess human-to-human transmissibility of influenza viruses, therefore, we rely on animal models. Ferrets, which develop human-like influenza symptoms (e.g., fever, coughing, sneezing, and runny nose) upon infection with human influenza viruses are considered to be the most reliable animal model to study human-to-human transmissibility. Indeed, the distribution patterns of Siaα2,3Gal and Siaα2,6Gal in their respiratory organs resemble those of humans: human-type receptors (Siaα2,6Gal) predominate in the upper respiratory tract (114). Recent studies suggest that guinea pigs could serve as an alternative transmission model for human influenza viruses (115, 116); however, since this small mammal does not support efficient transmission of some human viruses via the respiratory droplets (117) and both avian- and human-type receptors are found on its nasal and tracheal epithelial cells (117–119), ferrets remain the first choice for influenza virus transmission studies.
Several influenza viruses possessing avian-origin HA whose subtype is distinct from that of past pandemic strains have been tested in ferrets for their potential to cause human-to-human transmission. Although some avian H5N1 isolates or their mutants partially or preferentially recognized human-type receptors (Siaα2,6Gal) (110, 120), they did not cause ferret-to-ferret transmission via respiratory droplets (120, 121). Similarly, avian H7 and H9 viruses were not transmissible via respiratory droplets despite their human virus-like receptor-binding specificity (122, 123).
The first virus with avian virus-derived HA that was transmissible among ferrets was obtained by serial passage of a reassortant virus between an avian H9N2 virus (which supplied the HA and NA genes) and a human H3N2 virus (which supplied the remaining six genes) (95). Reassortants possessing avian H9 HA and N9 NA genes and the remaining genes from a “pandemic (H1N1) 2009” strain were also transmissible in ferrets (124). Among viruses of the H5 subtype, a reassortant with avian H5 HA that recognizes human receptors and seasonal virus-derived N2 NA was transmissible via respiratory droplets in one of two contact ferrets (79). More recent studies revealed that following serial passage in ferrets, viruses possessing H5 HAs that recognize human receptors in either a wild-type avian H5N1 virus (81) or a “pandemic (H1N1) 2009” virus background (80) were highly transmissible in a ferret model. One of these studies also demonstrated that in addition to the receptor recognition, HA stability is important for avian virus HA to support efficient transmission in ferrets (79). These results underscore the potential of viruses with HA subtypes to which humans are immunologically naïve to cause human-to-human transmission, leading to influenza pandemics.
4 Concluding remarks
Because of the great their threat to public health and economies worldwide, influenza A viruses must be controlled. However, their rapid evolution hampers the development of effective vaccines that confer permanent and universal immunity to hosts. In addition, these viruses can infect and replicate in numerous animal species. Furthermore, recent studies have demonstrated the pandemic potential of viruses of HA subtypes other than H1, H2, and H3. Economic and social globalization has prompted the active flow of people and products, allowing viruses to spread rapidly all over the world. To minimize the threat posed by zoonotic influenza, early detection and local containment are essential. To this end, global surveillance programs and systems to monitor influenza in animals, especially wild and domestic birds and pigs, are urgently needed.
LITERATURE CITED
- 1.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, et al. A distinct lineage of influenza A virus from bats. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4269–74. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowdle WR, Galphin JC, Coleman MT, Schild GC. A simple double immunodiffusion test for typing influenza viruses. Bulletin of the World Health Organization. 1974;51:213–5. [PMC free article] [PubMed] [Google Scholar]
- 3.Downie JC, Laver WG. Isolation of a type A influenza virus from an Australian pelagic bird. Virology. 1973;51:259–69. doi: 10.1016/0042-6822(73)90426-1. [DOI] [PubMed] [Google Scholar]
- 4.Slemons RD, Johnson DC, Osborn JS, Hayes F. Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Diseases. 1974;18:119–24. [PubMed] [Google Scholar]
- 5.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiological Reviews. 1992;56:152–79. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swayne DE, Suarez DL. Highly pathogenic avian influenza. Revue Scientifique et Technique. 2000;19:463–82. doi: 10.20506/rst.19.2.1230. [DOI] [PubMed] [Google Scholar]
- 7.Kawaoka Y, Webster RG. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:324–8. doi: 10.1073/pnas.85.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webster RG, Yakhno M, Hinshaw VS, Bean WJ, Murti KG. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978;84:268–78. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinshaw VS, Webster RG, Turner B. Water-bone transmission of influenza A viruses? Intervirology. 1979;11:66–8. doi: 10.1159/000149014. [DOI] [PubMed] [Google Scholar]
- 10.Hinshaw VS, Webster RG, Turner B. The perpetuation of orthomyxoviruses and paramyxoviruses in Canadian waterfowl. Canadian Journal of Microbiology. 1980;26:622–9. doi: 10.1139/m80-108. [DOI] [PubMed] [Google Scholar]
- 11.Hinshaw VS, Wood JM, Webster RG, Deibel R, Turner B. Circulation of influenza viruses and paramyxoviruses in waterfowl originating from two different areas of North America. Bulletin of the World Health Organization. 1985;63:711–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, et al. Influenza A viruses of migrating wild aquatic birds in North America. Vector borne and zoonotic diseases. 2004;4:177–89. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- 13.Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325:481–3. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okazaki K, Takada A, Ito T, Imai M, Takakuwa H, et al. Precursor genes of future pandemic influenza viruses are perpetuated in ducks nesting in Siberia. Archives of Virology. 2000;145:885–93. doi: 10.1007/s007050050681. [DOI] [PubMed] [Google Scholar]
- 15.Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS pathogens. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webster RG, Hulse DJ. Microbial adaptation and change: avian influenza. Revue Scientifique et Technique. 2004;23:453–65. doi: 10.20506/rst.23.2.1493. [DOI] [PubMed] [Google Scholar]
- 17.Domenech J, Dauphin G, Rushton J, McGrane J, Lubroth J, et al. Experiences with vaccination in countries endemically infected with highly pathogenic avian influenza: the Food and Agriculture Organization perspective. Revue Scientifique et Technique. 2009;28:293–305. doi: 10.20506/rst.28.1.1865. [DOI] [PubMed] [Google Scholar]
- 18.Lee YJ, Shin JY, Song MS, Lee YM, Choi JG, et al. Continuing evolution of H9 influenza viruses in Korean poultry. Virology. 2007;359:313–23. doi: 10.1016/j.virol.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 19.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, et al. Potential for transmission of avian influenza viruses to pigs. The Journal of general virology. 1994;75(Pt 9):2183–8. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 20.Scholtissek C, Burger H, Kistner O, Shortridge KF. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–94. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 21.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–6. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nardelli L, Pascucci S, Gualandi GL, Loda P. Outbreaks of classical swine influenza in Italy in 1976. Zentralblatt fur Veterinarmedizin. Reihe B. Journal of veterinary medicine. Series B. 1978;25:853–7. doi: 10.1111/j.1439-0450.1978.tb01062.x. [DOI] [PubMed] [Google Scholar]
- 23.Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bulletin of the World Health Organization. 1981;59:75–8. [PMC free article] [PubMed] [Google Scholar]
- 24.Choi YK, Nguyen TD, Ozaki H, Webby RJ, Puthavathana P, et al. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. Journal of Virology. 2005;79:10821–5. doi: 10.1128/JVI.79.16.10821-10825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipatov AS, Kwon YK, Sarmento LV, Lager KM, Spackman E, et al. Domestic pigs have low susceptibility to H5N1 highly pathogenic avian influenza viruses. PLoS pathogens. 2008;4:e1000102. doi: 10.1371/journal.ppat.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weingartl HM, Albrecht RA, Lager KM, Babiuk S, Marszal P, et al. Experimental infection of pigs with the human 1918 pandemic influenza virus. Journal of Virology. 2009;83:4287–96. doi: 10.1128/JVI.02399-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sovinova O, Tumova B, Pouska F, Nemec J. Isolation of a virus causing respiratory disease in horses. Acta Virologica. 1958;2:52–61. [PubMed] [Google Scholar]
- 28.Mumford J, Wood JM, Scott AM, Folkers C, Schild GC. Studies with inactivated equine influenza vaccine. 2. Protection against experimental infection with influenza virus A/equine/Newmarket/79 (H3N8) The Journal of hygiene. 1983;90:385–95. doi: 10.1017/s0022172400029016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webster RG. Are equine 1 influenza viruses still present in horses? Equine Veterinary Journal. 1993;25:537–8. doi: 10.1111/j.2042-3306.1993.tb03009.x. [DOI] [PubMed] [Google Scholar]
- 30.Guo YJ, Wang M, Zheng SL, Wang P, Ji WJ, Chen QH. Aetiologic study on an influenza-like epidemic in horses in China. Acta Virologica. 1991;35:190–5. [PubMed] [Google Scholar]
- 31.Sakai-Tagawa Y, Ozawa M, Tamura D, Le MQ, Nidom CA, et al. Sensitivity of influenza rapid diagnostic tests to H5N1 and 2009 pandemic H1N1 viruses. J Clin Microbiol. 2010;48:2872–7. doi: 10.1128/JCM.00439-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CW, Senne DA, Suarez DL. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. Journal of Virology. 2004;78:8372–81. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuchs W, Romer-Oberdorfer A, Veits J, Mettenleiter TC. Novel avian influenza virus vaccines. Revue Scientifique et Technique. 2009;28:319–32. doi: 10.20506/rst.28.1.1862. [DOI] [PubMed] [Google Scholar]
- 34.Poljak Z, Dewey CE, Martin SW, Christensen J, Friendship RM. Field efficacy of an inactivated bivalent influenza vaccine in a multi-site swine production system during an outbreak of systemic porcine circovirus associated disease. Canadian journal of veterinary research = Revue canadienne de recherche veterinaire. 2010;74:108–17. [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis NS, Daly JM, Russell CA, Horton DL, Skepner E, et al. Antigenic and genetic evolution of equine influenza A (H3N8) virus from 1968 to 2007. Journal of Virology. 2011;85:12742–9. doi: 10.1128/JVI.05319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edlund Toulemonde C, Daly J, Sindle T, Guigal PM, Audonnet JC, Minke JM. Efficacy of a recombinant equine influenza vaccine against challenge with an American lineage H3N8 influenza virus responsible for the 2003 outbreak in the United Kingdom. The Veterinary record. 2005;156:367–71. doi: 10.1136/vr.156.12.367. [DOI] [PubMed] [Google Scholar]
- 37.Youngner JS, Whitaker-Dowling P, Chambers TM, Rushlow KE, Sebring R. Derivation and characterization of a live attenuated equine influenza vaccine virus. American Journal of Veterinary Research. 2001;62:1290–4. doi: 10.2460/ajvr.2001.62.1290. [DOI] [PubMed] [Google Scholar]
- 38.Chambers TM, Holland RE, Tudor LR, Townsend HG, Cook A, et al. A new modified live equine influenza virus vaccine: phenotypic stability, restricted spread and efficacy against heterologous virus challenge. Equine Veterinary Journal. 2001;33:630–6. doi: 10.2746/042516401776249291. [DOI] [PubMed] [Google Scholar]
- 39.Lunn DP, Hussey S, Sebing R, Rushlow KE, Radecki SV, et al. Safety, efficacy, and immunogenicity of a modified-live equine influenza virus vaccine in ponies after induction of exercise-induced immunosuppression. Journal of the American Veterinary Medical Association. 2001;218:900–6. doi: 10.2460/javma.2001.218.900. [DOI] [PubMed] [Google Scholar]
- 40.Townsend HG, Penner SJ, Watts TC, Cook A, Bogdan J, et al. Efficacy of a cold-adapted, intranasal, equine influenza vaccine: challenge trials. Equine Veterinary Journal. 2001;33:637–43. doi: 10.2746/042516401776249354. [DOI] [PubMed] [Google Scholar]
- 41.Crawford PC, Dubovi EJ, Castleman WL, Stephenson I, Gibbs EP, et al. Transmission of equine influenza virus to dogs. Science. 2005;310:482–5. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 42.Daly JM, Blunden AS, Macrae S, Miller J, Bowman SJ, et al. Transmission of equine influenza virus to English foxhounds. Emerging Infectious Diseases. 2008;14:461–4. doi: 10.3201/eid1403.070643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirkland PD, Finlaison DS, Crispe E, Hurt AC. Influenza virus transmission from horses to dogs, Australia. Emerging Infectious Diseases. 2010;16:699–702. doi: 10.3201/eid1604.091489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song D, Kang B, Lee C, Jung K, Ha G, et al. Transmission of avian influenza virus (H3N2) to dogs. Emerging Infectious Diseases. 2008;14:741–6. doi: 10.3201/eid1405.071471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dundon WG, De Benedictis P, Viale E, Capua I. Serologic evidence of pandemic (H1N1) 2009 infection in dogs, Italy. Emerging Infectious Diseases. 2010;16:2019–21. doi: 10.3201/eid1612.100514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin D, Sun S, Du L, Ma J, Fan L, et al. Natural and experimental infection of dogs with pandemic H1N1/2009 influenza virus. The Journal of general virology. 2012;93:119–23. doi: 10.1099/vir.0.037358-0. [DOI] [PubMed] [Google Scholar]
- 47.Amonsin A, Payungporn S, Theamboonlers A, Thanawongnuwech R, Suradhat S, et al. Genetic characterization of H5N1 influenza A viruses isolated from zoo tigers in Thailand. Virology. 2006;344:480–91. doi: 10.1016/j.virol.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 48.Keawcharoen J, Oraveerakul K, Kuiken T, Fouchier RA, Amonsin A, et al. Avian influenza H5N1 in tigers and leopards. Emerging Infectious Diseases. 2004;10:2189–91. doi: 10.3201/eid1012.040759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klingeborn B, Englund L, Rott R, Juntti N, Rockborn G. An avian influenza A virus killing a mammalian species--the mink. Brief report. Arch Virol. 1985;86:347–51. doi: 10.1007/BF01309839. [DOI] [PubMed] [Google Scholar]
- 50.Webster RG, Hinshaw VS, Bean WJ, Van Wyke KL, Geraci JR, et al. Characterization of an influenza A virus from seals. Virology. 1981;113:712–24. doi: 10.1016/0042-6822(81)90200-2. [DOI] [PubMed] [Google Scholar]
- 51.Callan RJ, Early G, Kida H, Hinshaw VS. The appearance of H3 influenza viruses in seals. The Journal of general virology. 1995;76(Pt 1):199–203. doi: 10.1099/0022-1317-76-1-199. [DOI] [PubMed] [Google Scholar]
- 52.Hinshaw VS, Bean WJ, Geraci J, Fiorelli P, Early G, Webster RG. Characterization of two influenza A viruses from a pilot whale. J Virol. 1986;58:655–6. doi: 10.1128/jvi.58.2.655-656.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lvov DK, Zdanov VM, Sazonov AA, Braude NA, Vladimirtceva EA, et al. Comparison of influenza viruses isolated from man and from whales. Bulletin of the World Health Organization. 1978;56:923–30. [PMC free article] [PubMed] [Google Scholar]
- 54.Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1651–6. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taubenberger JK, Reid AH, Krafft AE, Bijwaard KE, Fanning TG. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–6. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 56.Winternitz MC, Wason IM, McNamara FP. The pathology of influenza. New Haven: Yale university press; etc; Yale university. Anthony N. Brady memorial laboratory., Yale university. Anthony N. Brady memorial foundation. 1920; p. 61.p. 11. [Google Scholar]
- 57.Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–23. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 58.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 59.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–9. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe T, Watanabe S, Shinya K, Kim JH, Hatta M, Kawaoka Y. Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc Natl Acad Sci U S A. 2009;106:588–92. doi: 10.1073/pnas.0806959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glezen WP. Emerging infections: pandemic influenza. Epidemiologic Reviews. 1996;18:64–76. doi: 10.1093/oxfordjournals.epirev.a017917. [DOI] [PubMed] [Google Scholar]
- 62.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 63.Schafer JR, Kawaoka Y, Bean WJ, Suss J, Senne D, Webster RG. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–8. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 64.Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–8. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakajima K, Desselberger U, Palese P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature. 1978;274:334–9. doi: 10.1038/274334a0. [DOI] [PubMed] [Google Scholar]
- 66.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–5. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–7. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez DR, Sorrell E, Angel M, Ye J, Hickman D, et al. Fitness of Pandemic H1N1 and Seasonal influenza A viruses during Co-infection: Evidence of competitive advantage of pandemic H1N1 influenza versus seasonal influenza. PLoS currents. 2009;1 doi: 10.1371/currents.RRN1011. RRN1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Safronetz D, Rockx B, Feldmann F, Belisle SE, Palermo RE, et al. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. Journal of Virology. 2011;85:1214–23. doi: 10.1128/JVI.01848-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van den Brand JM, Stittelaar KJ, van Amerongen G, Rimmelzwaan GF, Simon J, et al. Severity of pneumonia due to new H1N1 influenza virus in ferrets is intermediate between that due to seasonal H1N1 virus and highly pathogenic avian influenza H5N1 virus. The Journal of infectious diseases. 2010;201:993–9. doi: 10.1086/651132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–6. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 72.Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–7. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 73.Claas EC, de Jong JC, van Beek R, Rimmelzwaan GF, Osterhaus AD. Human influenza virus A/HongKong/156/97 (H5N1) infection. Vaccine. 1998;16:977–8. doi: 10.1016/s0264-410x(98)00005-x. [DOI] [PubMed] [Google Scholar]
- 74.de Jong MD, Bach VC, Phan TQ, Vo MH, Tran TT, et al. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. The New England journal of medicine. 2005;352:686–91. doi: 10.1056/NEJMoa044307. [DOI] [PubMed] [Google Scholar]
- 75.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nature Medicine. 2006;12:1203–7. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gu J, Xie Z, Gao Z, Liu J, Korteweg C, et al. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet. 2007;370:1137–45. doi: 10.1016/S0140-6736(07)61515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uiprasertkul M, Puthavathana P, Sangsiriwut K, Pooruk P, Srisook K, et al. Influenza A H5N1 replication sites in humans. Emerging Infectious Diseases. 2005;11:1036–41. doi: 10.3201/eid1107.041313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uiprasertkul M, Kitphati R, Puthavathana P, Kriwong R, Kongchanagul A, et al. Apoptosis and pathogenesis of avian influenza A (H5N1) virus in humans. Emerging Infectious Diseases. 2007;13:708–12. doi: 10.3201/eid1305.060572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen LM, Blixt O, Stevens J, Lipatov AS, Davis CT, et al. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology. 2012;422:105–13. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–8. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–41. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen H, Smith GJ, Zhang SY, Qin K, Wang J, et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–2. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 83.Liu J, Xiao H, Lei F, Zhu Q, Qin K, et al. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- 84.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–61. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–93. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- 86.Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? Journal of Virology. 2001;75:9679–86. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cong YL, Wang CF, Yan CM, Peng JS, Jiang ZL, Liu JH. Swine infection with H9N2 influenza viruses in China in 2004. Virus Genes. 2008;36:461–9. doi: 10.1007/s11262-008-0227-z. [DOI] [PubMed] [Google Scholar]
- 88.Yu H, Hua RH, Wei TC, Zhou YJ, Tian ZJ, et al. Isolation and genetic characterization of avian origin H9N2 influenza viruses from pigs in China. Veterinary Microbiology. 2008;131:82–92. doi: 10.1016/j.vetmic.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 89.Guo Y, Li J, Cheng X. Discovery of men infected by avian influenza A (H9N2) virus. Zhonghua shi yan he lin chuang bing du xue za zhi = Zhonghua shiyan he linchuang bingduxue zazhi = Chinese journal of experimental and clinical virology. 1999;13:105–8. [PubMed] [Google Scholar]
- 90.Lin YP, Shaw M, Gregory V, Cameron K, Lim W, et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9654–8. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, et al. Human infection with influenza H9N2. Lancet. 1999;354:916–7. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 92.Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. Journal of Clinical Microbiology. 2005;43:5760–7. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–62. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 94.Choi YK, Ozaki H, Webby RJ, Webster RG, Peiris JS, et al. Continuing evolution of H9N2 influenza viruses in Southeastern China. Journal of Virology. 2004;78:8609–14. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sorrell EM, Wan H, Araya Y, Song H, Perez DR. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7565–70. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.DeLay PD, Casey HL, Tubiash HS. Comparative study of fowl plague virus and a virus isolated from man. Public Health Reports. 1967;82:615–20. [PMC free article] [PubMed] [Google Scholar]
- 97.Taylor HR, Turner AJ. A case report of fowl plague keratoconjunctivitis. The British journal of ophthalmology. 1977;61:86–8. doi: 10.1136/bjo.61.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007;44:1084–8. doi: 10.1086/512813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–73. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 100.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 101.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–12. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–55. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 103.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–6. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 104.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, et al. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 105.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–73. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuchipudi SV, Nelli R, White GA, Bain M, Chang KC, Dunham S. Differences in influenza virus receptors in chickens and ducks: Implications for interspecies transmission. Journal of molecular and genetic medicine : an international journal of biomedical research. 2009;3:143–51. doi: 10.4172/1747-0862.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kimble B, Nieto GR, Perez DR. Characterization of influenza virus sialic acid receptors in minor poultry species. Virology journal. 2010;7:365. doi: 10.1186/1743-422X-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu JE, Yoon H, Lee HJ, Lee JH, Chang BJ, et al. Expression patterns of influenza virus receptors in the respiratory tracts of four species of poultry. Journal of veterinary science. 2011;12:7–13. doi: 10.4142/jvs.2011.12.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Childs RA, Palma AS, Wharton S, Matrosovich T, Liu Y, et al. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nature Biotechnology. 2009;27:797–9. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gambaryan A, Tuzikov A, Pazynina G, Bovin N, Balish A, Klimov A. Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology. 2006;344:432–8. doi: 10.1016/j.virol.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 111.Yamada S, Suzuki Y, Suzuki T, Le MQ, Nidom CA, et al. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–82. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 112.Bradley KC, Jones CA, Tompkins SM, Tripp RA, Russell RJ, et al. Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (Novel 2009 H1N1) Virology. 2011;413:169–82. doi: 10.1016/j.virol.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 113.Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, et al. Triple-Reassortant Swine Influenza A (H1) in Humans in the United States, 2005–2009. N Engl J Med. 2009 doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 114.Leigh MW, Connor RJ, Kelm S, Baum LG, Paulson JC. Receptor specificity of influenza virus influences severity of illness in ferrets. Vaccine. 1995;13:1468–73. doi: 10.1016/0264-410x(95)00004-k. [DOI] [PubMed] [Google Scholar]
- 115.Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A. 2006;103:9988–92. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lowen AC, Mubareka S, Steel J, Palese P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007;3:1470–6. doi: 10.1371/journal.ppat.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sun Y, Bi Y, Pu J, Hu Y, Wang J, et al. Guinea pig model for evaluating the potential public health risk of swine and avian influenza viruses. PloS one. 2010;5:e15537. doi: 10.1371/journal.pone.0015537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, et al. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. The American journal of pathology. 2007;171:1215–23. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu Q, Wang W, Cheng X, Zengel J, Jin H. Influenza H1N1 A/Solomon Island/3/06 virus receptor binding specificity correlates with virus pathogenicity, antigenicity, and immunogenicity in ferrets. Journal of Virology. 2010;84:4936–45. doi: 10.1128/JVI.02489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maines TR, Chen LM, Van Hoeven N, Tumpey TM, Blixt O, et al. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011;413:139–47. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103:12121–6. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, et al. Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7558–63. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, et al. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PloS one. 2008;3:e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kimble JB, Sorrell E, Shao H, Martin PL, Perez DR. Compatibility of H9N2 avian influenza surface genes and 2009 pandemic H1N1 internal genes for transmission in the ferret model. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12084–8. doi: 10.1073/pnas.1108058108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–9. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]