Summary
A ketogenic diet (KD) recapitulates certain metabolic aspects of dietary restriction such as reliance on fatty acid metabolism and production of ketone bodies. We investigated whether KD might, like dietary restriction, affect longevity and healthspan in C57BL/6 male mice. We find that an isoprotein KD, fed on alternate weeks to prevent obesity (Cyclic KD), reduces mid-life mortality but does not affect maximum lifespan. A non-ketogenic high-fat diet (HF) fed similarly may have an intermediate effect on mortality. Cyclic KD improves memory performance in old age, while modestly improving composite healthspan measures. Gene expression analysis identifies down-regulation of insulin, TOR, and fatty acid synthesis pathways as mechanisms common to KD and HF. However, up-regulation of PPARα target genes is unique to KD, consistent across tissues, and preserved in old age. In all, we show that a non-obesogenic ketogenic diet improves survival, memory, and healthspan in aging mice.
Keywords: ketogenic diet, beta-hydroxybutyrate, longevity, healthspan
eTOC blurb
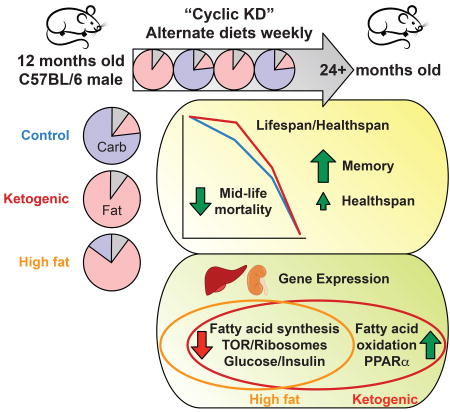
Ketogenic diet is similar in many physiological respects to dietary restriction. Newman et al. show that a cyclic ketogenic diet reduces mortality and improves memory as mice age. Ketogenic diet gene expression pattern is similar to high-fat diet, except activation of PPARα targets. See related paper by Roberts et al.
Introduction
The health benefits of “lifestyle” interventions such as exercise and reduced food intake are increasingly understood to be mediated through networks of specific molecular mechanisms. Unraveling these environmentally-responsive networks has led to the first drug found to extend lifespan in healthy mammals - rapamycin (Harrison et al., 2009) - and the first candidate intervention for testing healthspan extension in a human clinical trial - metformin (Barzilai et al., 2016; Martin-Montalvo et al., 2013). Some components of these networks are normal cellular metabolites that, in addition to their canonical roles in energy metabolism, have additional functions as regulatory or signaling molecules. Acetyl-CoA and protein acetylation may be the best-studied example (Menzies et al., 2016), but many other intermediate metabolites have similar roles (Benayoun et al., 2015; Sabari et al., 2016) including the ketone body beta-hydroxybutyrate (BHB).
BHB is a normal human metabolite that is synthesized in the liver from fat, and then circulates throughout the body as a glucose-sparing energy source. It is intrinsically produced during states such as intermittent fasting and dietary restriction that result in extended longevity, cognitive protection, cancer reduction, and immune rejuvenation (Longo and Panda, 2016; Mattson et al., 2016). Recent work has elucidated an array of signaling functions of BHB: inhibitor of the NLRP3 inflammasome (Youm et al., 2015), inhibitor of histone deacetylases (Shimazu et al., 2013), ligand of G-protein coupled receptors (Offermanns, 2017), and a covalent histone modification associated with fasting-activated genes (Xie et al., 2016). These signaling functions suggest that BHB might itself regulate inflammation and gene expression, with implications for health and longevity. Aberrant inflammasome activity is associated with a variety of age-related diseases (Goldberg and Dixit, 2015), while moderate inhibition of histone deacetylases is associated with enhanced longevity in invertebrates (Woods and Rogina, 2016) as well as amelioration of cognitive decline (Penney and Tsai, 2014) and cardiac dysfunction (Morales et al., 2016) in mice.
A ketogenic diet is one means to deliver high levels of BHB for a prolonged time outside of a fasting or exercise context (Newman and Verdin, 2014). Ketogenic diets generally contain little or no carbohydrate, and stimulate endogenous ketogenesis. We sought to test whether a ketogenic diet carefully matched to comparison diets could enhance the longevity and healthspan of normal mice, and to elucidate potential molecular mechanisms of such effects.
Results
Ketogenic diet reduces mid-life mortality
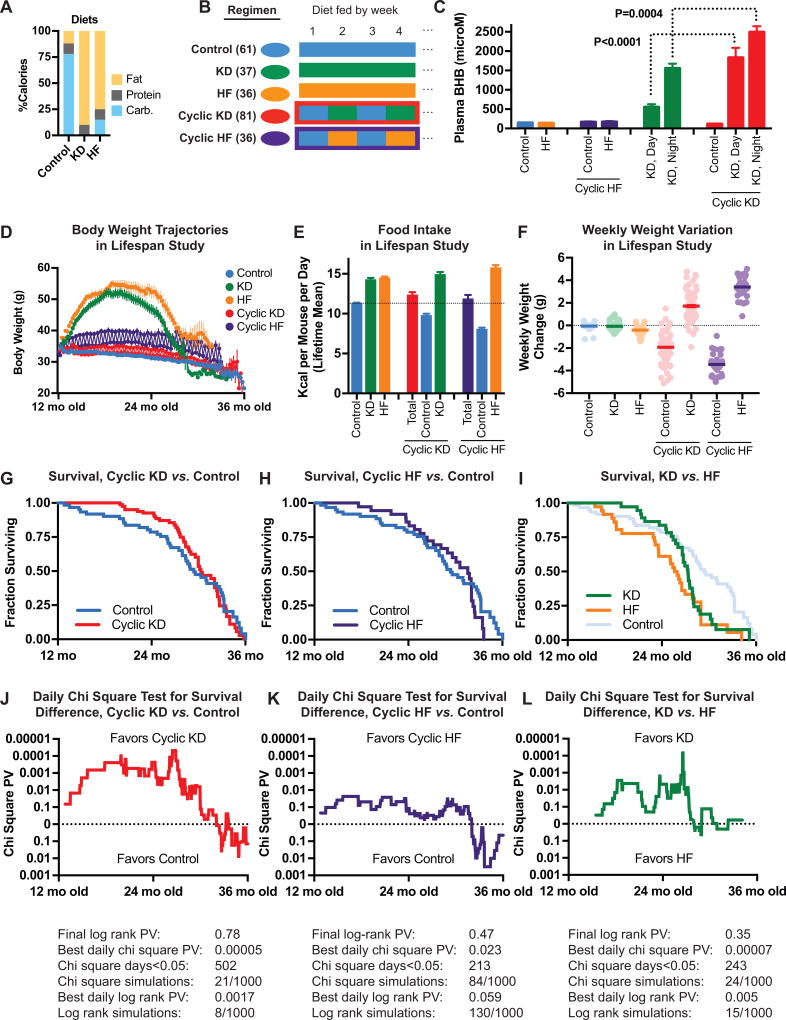
We first set out to develop a diet and feeding regimen that would provide maximum plasma BHB levels with minimum short- and long-term confounding effects. We developed a set of diets based on AIN-93M, with varying fat and carbohydrate content but matched on a per-calorie basis for protein content, micronutrients, and fiber. The diets use similar plant-based fat sources with the minimum saturated fat content that permits a semisolid consistency. Preliminary work showed that zero carbohydrate content generated the highest ketone body levels (Figure S1A), with additional carbohydrates gradually suppressing ketogenesis until reaching baseline plasma BHB levels at 15% of calories from carbohydrates (Figure S1B). Subsequent experiments thus used three diets: zero-carbohydrate ketogenic diet (KD), normal control diet (Control), and 15% carbohydrate high-fat non-ketogenic diet (HF) as a comparison for fat content (Figure 1A). Although mice on HF had normal blood BHB levels, most energy utilization is from fats, similar to KD (Figure S1C–D). Preliminary testing found that both KD and HF were obesogenic when fed continuously, but cycling these diets weekly with the control diet (Figure 1B) resulted in long-term maintenance of normal weight (Figure S1D). KD generated plasma BHB levels similar to fasting (1–2 mM), and highest during the nighttime feeding period (Figure 1C). BHB levels were higher on Cyclic KD than on continuously fed KD, particularly during the daytime (Figure 1C). Based on these preliminary studies, we selected the every-other-week Cyclic KD as the key intervention to test whether long-term exposure to ketone bodies has effects on longevity and healthspan. We organized the three diets into five diet regimens altogether: Control diet, Cyclic KD (alternating KD and control diet weekly), Cyclic HF (alternating HF and control diet weekly), continuously-fed KD, and continuously-fed HF (Figure 1B). C57BL/6 NIA male mice were enrolled into the lifespan study at 12 mo of age.
Figure 1.
Cyclic ketogenic diet (switching weekly between KD and control diet) started at 12 mo old reduces mid-life mortality of C57BL/6 males. A, Diet composition. B, Five diet regimens. C, Plasma BHB levels of 4 mo mice on diets for 10 weeks; blood drawn on consecutive weeks during both day and night (N=6). Cyclic diets indicate the food eaten each week. Data that did not differ between weeks or night/day are combined for clarity. D–L Data from main lifespan study. D, Body weight trajectories. E, Overall mean caloric intake. F, Mean weekly weight change, showing weight cycling on cyclic diets. G–I, Survival curves for Cyclic KD (G) and Cyclic HF (H) vs. Control, and for KD vs. HF (I). J–L, Daily chi square tests of differences in survival for Cyclic KD (J) and Cyclic HF (K) vs. Control, and for KD vs. HF (L). Survival statistics below include the final log rank test, the best daily chi square test (J–L), the best daily log rank test, and the frequency of observed differences occurring among random-data monte carlo simulations. Lifespan study, C57BL/6 NIA males starting 12 mo old: N=61 Control, 37 KD, 26 HF, 81 Cyclic KD, 36 Cyclic HF. See also Figures S1 and S2.
The overall body weight of mice on Cyclic KD was very similar to controls over the lifespan, with mice on Cyclic HF averaging slightly heavier (Figure 1D). Similarly, HF-fed mice were slightly more obese than KD-fed mice. The KD and HF groups had correspondingly higher lifetime caloric intake than the control or cyclic diets (Figure 1E). Both Cyclic KD and Cyclic HF mice had similar total caloric intake than controls, though caloric intake was markedly higher on KD- or HF-fed weeks and lower on control-fed weeks despite ad libitum feeding at all times. Caloric intake did not change with age (Figure S1E–F), and both the Cyclic KD and Cyclic HF groups maintained a tight weight distribution (Figure S1G). The weekly cycling in caloric intake correlated with weekly weight cycling, with higher amplitude of weight cycling in Cyclic HF than Cyclic KD (Figure 1F). Mice on Cyclic KD and Cyclic HF had gradual weight loss during the first 3–4 days of control diet weeks, with no food aversion or fasting (Figure S1H–J).
A small pilot cohort suggested that Cyclic KD might “square off” the survival curve, with reduced mortality in mid-life but not necessarily an increase in maximum lifespan (Figure S2A). In the main study both Cyclic KD and Cyclic HF indeed appeared to reduce early mortality compared to Control, with a modest increase in median lifespan (Figures 1G and 1H, Table S1). There was no difference in maximum lifespan and the overall survival curves were not different by log rank test. We went on to quantitatively examine the possibility of differing effects on survival at different regions of the lifespan curves. We found that the day-by-day survival of Control and Cyclic KD were significantly different via chi square test up through approximately 30 months of age (Figure 1J). Importantly, beyond 30 months the survival converged. Cyclic HF displayed a similar pattern, albeit of more modest magnitude and significance (Figures 1K and S2C). This suggests that the effect of KD is incremental upon, rather than distinct from, a low-carbohydrate high-fat diet, a concept we explored further in the gene expression analysis below. Unlike the cyclic diets, continuously-fed KD and HF were short-lived, unsurprisingly given the obesity of most mice in these groups. But KD compared to HF showed a pattern of reduced early mortality reminiscent of Cyclic KD versus Control, reaching moderate significance through approximately 26 months of age (Figures 1I and 1L).
We found similar results for all pairwise comparisons (Cyclic KD vs. Control, Cyclic HF vs. Control, KD vs. HF, Cyclic KD vs. Cyclic HF) when comparing daily hazard ratio calculations (Figure S2D) and daily log rank tests (Figure S2E). To guard against the multiple hypothesis problem, and to evaluate the robustness of these results, we performed label-randomization monte carlo simulations. The observed separation of the Control and Cyclic KD survival curves occurred in only 21 of 1000 simulations via daily chi square, and 8 of 1000 simulations via daily log rank. The observed separation of Control and Cyclic HF was present in 84/1000 and 130/1000 of simulations via daily chi square and daily log rank, respectively. Overall, the improved survival of Cyclic KD vs. Control and of KD vs. HF appeared robust, while survival of Cyclic HF lay intermediate between Control and Cyclic KD and less robustly different from either. We interpreted these lifespan data as Cyclic KD improving survival in mid-life but not late-life compared to Control, with Cyclic HF most likely having an intermediate effect.
Necropsy data suggested that the causes of earlier deaths were not distinct from the causes of later deaths. Overall, the prevalence of internal tumors at necropsy was lowest in the Cyclic KD group, although the difference with Control did not reach significance (Figure S2B). Carcinoma of the Harderian gland in the orbit was rare overall but appeared to be more common in the Cyclic KD and KD groups.
Preservation of memory and healthspan with aging
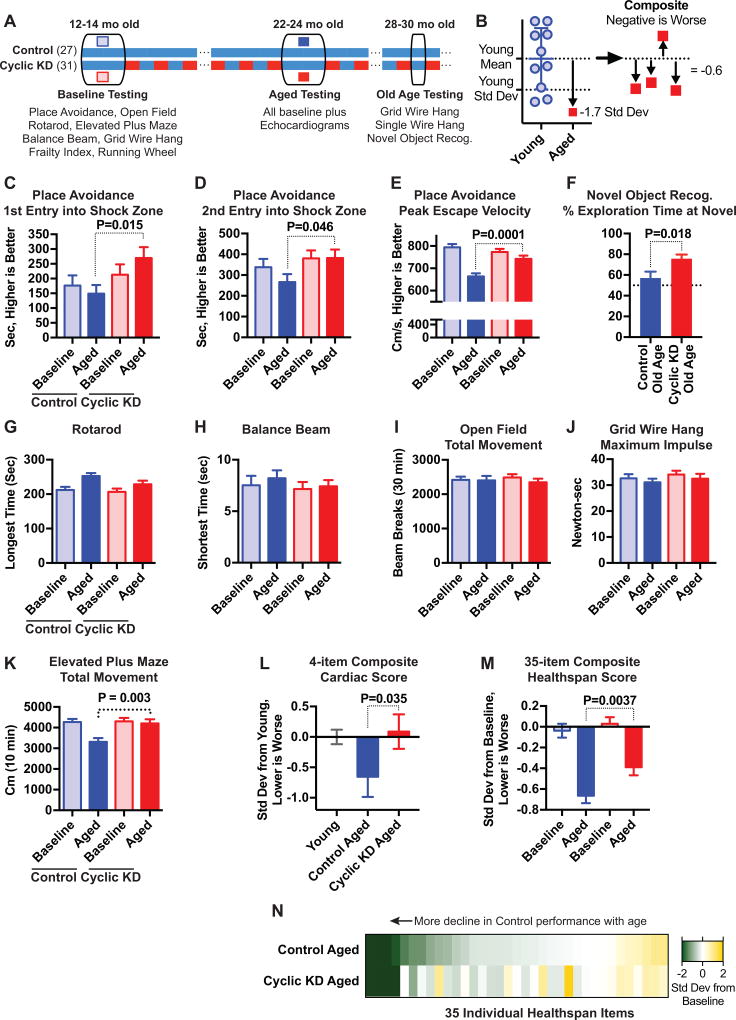
While assessing longevity, we sought in a parallel study to determine if Cyclic KD might have broader effects on healthspan. We enrolled separate cohorts of Cyclic KD and Control diet mice into a longitudinal healthspan testing program, with a Baseline testing period at 12–14 mo old, followed by randomization of half the mice to Cyclic KD, and an Aged testing period at 22–24 mo old (Figure 2A). The testing periods included a diverse set of cognitive and physical function tasks including open field, elevated plus maze, place avoidance, rotarod, balance beam, grid wire hang, clinical frailty scores, and running wheels (complete list in Table S2). The Aged testing period also included, for half the cohort, echocardiograms. All mice were eating the Control diet throughout the Baseline and Aged testing periods, to prevent confounding effects from acute diet intake or weight cycling. Mice in the Cyclic KD group ate KD every-other-week in between the Baseline and Aged testing periods, and after Aged testing was completed. Six months later at 28–30 mo old a more limited set of Old Age testing was performed during weeks that the Cyclic KD group was eating control diet (Figure 2A). The goals of the healthspan studies were to identify any specific phenotypes that are strongly affected by Cyclic KD, and to test whether Cyclic KD broadly affects varied age-related phenotypes (Richardson et al., 2016) since lifespan and healthspan may not always be coupled in mice (Fischer et al., 2016). For the latter purpose, we combined diverse parameters into composite scores normalized as standard deviations below the mean young/baseline performance (Figure 2B).
Figure 2.
Effect of Cyclic KD on memory and other healthspan measures with aging. A, Experimental timeline. All mice ate control diet during Baseline and Aged testing periods. B, Schema of composite scores in L–N. C–D, Memory retrieval tasks of Place Avoidance show better performance in Cyclic KD group. E, Maximum escape velocity following shock in Place Avoidance. F, Old Age Novel Object Recognition test shows improved memory in Cyclic KD group. G–K, Activity and physical function show fewer differences between Cyclic KD and Control groups, but also little age-related decline. L, Normalized composite of four echocardiogram measures using 3 mo old mice as baseline. M–N, 35-item baseline-normalized composite healthspan score (Table S1) shows modestly less age-related decline in Cyclic KD (M), spread across many items (N). Healthspan study, C57BL/6 NIA males starting 12 mo old: N=27 Control, 31 Cyclic KD. See also Figure S3.
We found that the Cyclic KD healthspan group maintained better memory with aging. The primary cognitive test in the healthspan program was place avoidance, a visuospatial learning and memory paradigm conceptually similar to the Morris water maze, but with mice learning to avoid a location where they receive a mild shock. Both groups showed the expected age-associated slowing in the learning/acquisition phase of place avoidance (Figure S2A), but the Cyclic KD group performed markedly better than the Control group in the memory/recall portion (Figures 2C–D). The Cyclic KD group also showed amelioration of age-related decline in a physical performance parameter during this test, the maximum velocity achieved escaping a shock (Figure 2E). Six months later (Old Age, 28–30 mo) we found a similar improvement in memory using a different test paradigm, novel object recognition (Figure 2F).
We found fewer differences between Cyclic KD and Control mice in tests of physical function or spontaneous activity, and only slightly lower frailty scores (Figures 2G–K, S2B–J). However, only one test of spontaneous activity showed a clear decline with age, elevated plus maze exploration, and this was ameliorated in Cyclic KD (Figure 2K). The Old Age tests with single wire hang and grid wire hang showed no difference between groups (Figure S2G–H). Importantly, there were also no differences in anxiety-related behaviors in either the open field or elevated plus maze (Figure S2K–L). The Cyclic KD group maintained a youthful phenotype in a composite cardiac score incorporating heart rate, left ventricular mass, aortic valve pressure gradient, and fractional shortening (Figure 2L). To gauge the overall effect of Cyclic KD on aging, we compiled an unbiased composite score from all 35 quantitative items measured during healthspan testing that had a young or baseline measurement. Cyclic KD modestly ameliorated the age-related decline in overall healthspan performance (Figure 2M), with differences broadly distributed across the 35 items (Figure 2N, Table S2).
Distinct fasting-like gene expression pattern of KD vs. HF
KD has broad effects on gene expression (Badman et al., 2007), and beta-hydroxbutyrate itself can affect gene expression through several mechanisms, including via histone beta-hydroxybutyrylation, by providing acetyl-CoA as a substrate for acetyltransferases, and via inhibiting deacetylases. Gene expression differences between KD, HF, and Control diets might therefore suggest underlying explanations for the effects we observed on survival and healthspan. As a preliminary study, we first examined protein acetylation and fasting-related gene expression in the livers of 9 mo old C57BL/6 mice that had been on KD for 5 months. Plasma BHB reached near-fasting levels over the first week, and was then maintained at lower levels (Figure S4A) as mice gained weight (Figure S4B). We detected increased global protein acetylation in both whole cell lysates and in isolated mitochondria from liver (Figure S4C–D). Quantitative PCR using a panel of fasting-regulated genes (Gut et al., 2013) showed that KD largely activates a fasting-like gene expression pattern (Figure S4E), as well as repressing the key lipid synthesis genes Fasn and Scd1.
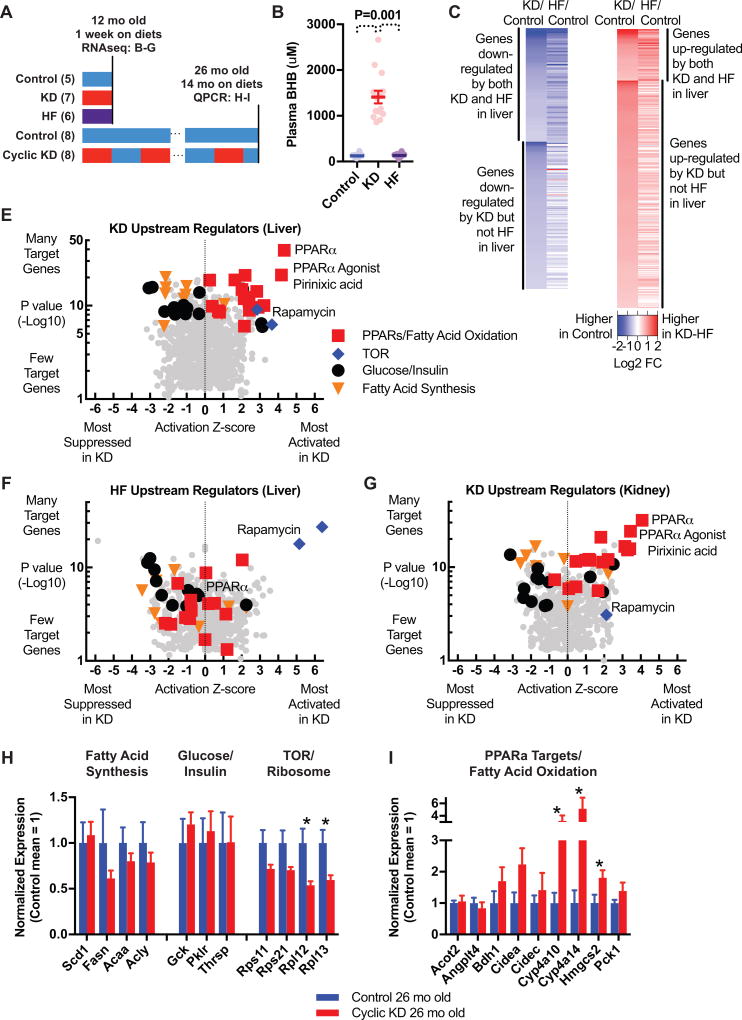
To systematically probe transcriptional changes that might contribute to the mortality and healthspan effects of the Cyclic KD regimen, we performed RNAseq on liver and kidney of 12 mo old mice fed the control diet, KD, or HF for one week (Figure 3A). The goal was to identify patterns of gene expression that might be reproduced each week on the cycling diets in the lifespan study. As expected, KD but not HF increased plasma BHB to near-fasting levels (Figure 3B). We sought to identify gene expression patterns that were common between HF and KD, and those that were unique to KD despite both being high-fat/low-carbohydrate diets that cause weight gain (Figure S4F; complete data set with differentially expressed genes in Table S3). Nearly half of the genes down-regulated by KD were also down-regulated by HF, but surprisingly most genes up-regulated by KD were unique (Figures 3C and S4G). We also found more commonality between liver and kidney in genes up-regulated by KD than in those down-regulated (Figure S4G), suggesting that KD up-regulation includes a coherent transcriptional program shared across tissues, and distinct from HF.
Figure 3.
KD activates PPARα gene expression pattern distinct from HF. A, Experimental timeline. B–G, RNAseq transcriptome analysis of 1 week on diets in 12 mo old mice. B, Plasma BHB levels for study of immediate diet effects. C–D, Gene expression changes in liver, defined permissively for subsequent pathway analysis (Log2FC>0.38, P<0.02), show many genes down-regulated in common by KD and HF but few genes up-regulated in common. E–G, Top ingenuity upstream regulators associated with KD in liver (E), with the same regulators highlighted for HF in liver (F) and KD in kidney (G). PPARα activation pattern is present in both liver and kidney for KD, and not for HF. H–I, Study of 26 mo old Cyclic KD and Control mice. H, Q-PCR of genes involved in fatty acid synthesis, glucose/insulin signaling, and TOR activity. I, Q-PCR of PPARα target genes. B–G, 12 mo mice on diets for 1 week, collected at night, N = 5 Control, 7 KD, 6 HF. H–I, 26 mo mice on diets for 14 mo, collected at night during control-fed week, N = 8 Control, 8 Cyclic KD. See also Figure S4.
Pathway analysis identified PPARα activation as the key element of the KD-specific transcriptional pattern (full data set in Table S3). Ingenuity and Gene Ontology analyses both found three major patterns among genes down-regulated by KD in the liver: suppression of glucose metabolism and insulin/IGF signaling, suppression of fatty acid synthesis, and suppression of TOR activity and ribosomal protein expression (Figure 3E). All three of these patterns were similarly evident in the HF liver data set (Figure 3F and Table 1), consistent with the strong overlap of individual down-regulated genes between KD and HF. Genes up-regulated by KD produced a fourth pattern, including activation of fatty acid oxidation, mitochondrial proteins, and targets of PPAR proteins – with the strongest signal for targets of PPARα (Figure 3E and Table 1). This PPARα activation pattern was conserved in the kidney for KD (Figure 3G), but strikingly absent from the HF liver data set (Figure 3F). Direct comparison of KD and HF revealed another distinct pattern, relative activation of tumor suppressors p53 and p21 by KD (Figure S4H).
Table 1.
Genes and pathways regulated 1 by KD and HF
| Representative Top Gene Ontology Terms (Liver) | Genes Most Up-Regulated by KD but not HF (Liver) | |||||
|---|---|---|---|---|---|---|
| Term | P value |
Fold enrichment |
Gene |
Log2 FC vs. Con |
P value |
Function |
| Genes up-regulated by both KD and HF | Cfd/Adipsin | 3.1 | <0.001 | Adipokine, insulin? | ||
| none | 1300015D01Rik | 1.9 | <0.001 | |||
| Cyp4a14 | 1.8 | <0.001 | Microsome | |||
| Genes up-regulated by KD but not HF | Cidea | 1,7 | <0.001 | Lipase | ||
| Lipid metabolic process | 4.2e-10 | 4.1 | Lamb3 | 1.7 | <0.001 | Basement membrane |
| Mitochondrion | 2.0e-6 | 2.9 | Pck1 | 1.7 | <0.001 | Gluconeogenesis |
| Fatty acid metabolic process | 3.6e-6 | 6.2 | Cyp4a10 | 1.5 | <0.001 | Microsome |
| Peroxisome | 5.0e-6 | 10.8 | Tpm2 | 1.4 | <0.001 | Cytoskeleton |
| Gm15441 | 1.4 | <0.001 | ||||
| Genes down-regulated by both KD and HF | Sema5 | 1.3 | <0.001 | Cell migration | ||
| Cytosolic ribosome | 2.0e-18 | 34.8 | Mfsd2a | 1.3 | <0.001 | Fatty acid transport |
| Translation | 1.7e-5 | 6.2 | Acot2 | 1.2 | <0.001 | Fatty acid oxidation |
| Triglyceride metabolic process | 3.2e-4 | 18.2 | Cntnap1 | 1.2 | <0.001 | Cell adhesion |
| Lipid biosynthetic process | 0.0051 | 5.5 | Themis | 1.2 | <0.001 | T cell development |
| Carbohydrate catabolic process | 0.021 | 12.8 | Cyp4a32 | 1.1 | <0.001 | |
| G0s2 | 1.1 | <0.001 | Apoptosis | |||
| Genes down-regulated by KD but not HF | C330002G04Rik | 1.1 | <0.001 | |||
| Fatty acid biosynthetic process | 0.0021 | 11.2 | Angplt4 | 1.1 | <0.001 | Lipoprotein uptake |
| Slc25a30 | 1.0 | <0.001 | Mitochondrial transport | |||
| Cidec | 1.0 | <0.001 | Lipase | |||
| Upk3b | 1.0 | 0.004 | Cytoskeleton | |||
| Bold = PPARα target (Rakhshandehroo et al., 2010) | ||||||
To determine if these transcriptional patterns were maintained long-term and relevant to the lifespan results, we performed quantitative PCR of selected genes on livers from 26 month old mice that had been on Control or Cyclic KD diet regimens since 12 months of age (Figure 3A). 26 months old corresponds to the maximum separation of the Cyclic KD and Control lifespan curves, and approximates the time of Aged healthspan testing. Mice at this age on Cyclic KD have nighttime plasma BHB levels averaging 1 mM while eating KD (Figure S4I). However, the tissues were collected 4–5 days into a Control feeding week to observe persistent differences unrelated to acute dietary effects or weight changes (Figure S1I). Of the three down-regulation patterns shared by KD and HF, we observed persistent down-regulation of TOR but not of fatty acid synthesis or glucose/insulin signaling (Figure 3H). Mice in the Cyclic KD group also showed persistent up-regulation of PPARα target genes and genes involved in fatty acid oxidation (Figure 3I).
Altogether, this transcriptome analysis revealed many important commonalities between KD and low-carbohydrate HF reflecting common physiological effects that are relevant to longevity, including suppression of insulin/IGF signaling and suppression of TOR activation and protein translation. But KD also has a unique fasting-like pattern of up-regulation of PPARα target genes, common across tissues, and, along with TOR suppression, persistent into old age.
Discussion
We show that long-term exposure to a ketogenic diet, fed every-other-week starting in middle age, reduces mid-life mortality and preserves memory in aging C57BL/6 male mice. Similar feeding of a high-fat/low-carbohydrate non-ketogenic diet appeared to have an intermediate effect on mortality, but survival of this Cyclic HF group could not be definitely distinguished from either the Control or Cyclic KD groups. These results might be interpreted in the broader context of the health effects of dietary restriction (DR) and of segmental DR mimetics such as metformin and rapamycin, and suggests that one or more aspects of a ketogenic diet may similarly act as a segmental DR mimetic. One prior study of KD did not observe a change in survival, but differences included the use of a very low-protein KD formulation (5% of calories), initiation in young mice (8 weeks old), and substantially shorter lifespans in all groups (Douris et al., 2015). Importantly, all of the diets used here had identical per-calorie protein content. Protein-restricted KD may not produce obesity in mice, but our finding that a normal-protein KD is obesogenic is consistent with prior reports (Borghjid and Feinman, 2012).
Unlike DR, the effect on survival we observed from intermittent feeding of KD was not consistent throughout the lifespan. The mortality in the Cyclic KD group was lowest from 12–30 months old, then converged with the Control group. The lifespan-extending fasting-mimic diet, which involves severe dietary restriction 3–4 days per month, showed a similar pattern (Brandhorst et al., 2015). One explanation may be that changes to cellular physiology in older animals, such as to histone acetylation (Peleg et al., 2016), attenuate the response to the specific mechanisms of DR invoked by Cyclic KD. Alternatively, weight cycling could become a counterproductive stress in old age. In addition to the FMD, a small study of monthly cycling on a high-fat diet also appeared to show increased mortality in old age (List et al., 2013). Finally, there could be an accumulated harm that becomes evident only with prolonged intake of HF or KD, such as, perhaps, liver injury. Understanding the mechanism of this potential harm at old age or long duration will help ensure the safety of any translational applications of cycling or ketogenic diets in humans.
In healthspan testing, we found a striking effect of Cyclic KD on memory as well as more modest effects on a broader range of measures. We saw consistent memory improvement in two distinct tasks over six months. Effects of KD on activity were mixed, with no change in the open field or running wheels but amelioration of the age-related decline in elevated plus maze activity. Physical performance was consistently not affected, even in those tasks with clear age-related declines in controls (balance beam training and single-wire hang) and in those with strong neurocognitive elements (e.g. balance beam). Protein intake was similar in both groups, but it is possible that the periodic weight loss associated with the cyclic regimen limited any improvements in motor strength.
Strengths of this study design for evaluation of a ketogenic diet included the careful matching of protein content, a regimen (every-other-week feeding) that approximated both body weights and caloric intake between with the control-fed group, a comparison group fed a maximally high-fat/low-carbohydrate but non-ketogenic diet, parallel healthspan and lifespan cohorts, and feeding control diet to all mice during healthspan testing to avoid acute confounding effects of ketosis or weight changes. This study also had important limitations, including the use of a single strain, single sex, single start age, single non-obese ketogenic diet regimen, lack of a Cyclic HF group in healthspan testing, and lack of a separate pathology cohort. All of these choices increased the power of the primary comparison but left potentially important variables to be explored in future studies. We selected C57BL/6 males because they are particularly prone to lifespan extension with dietary restriction, even when started in middle age (Pugh et al., 1999). But as most longevity interventions in mice have shown different responses between sexes (Austad and Fischer, 2016) and strains (Liao et al., 2010), it will be critical to test the effect of ketogenic diet or ketone bodies in other backgrounds. Similarly, starting a ketogenic diet intervention at different ages and/or using a variety of durations could determine if the waning benefit of Cyclic KD late in life is due to increasing age, or to increasing duration of treatment. Testing other regimens for feeding ketogenic diets or ketone bodies can ensure that the phenotypes we observed were not due solely to the every-other-week diet cycling, and would provide additional data towards the relevant mechanisms at work. Our study cannot exclude that a cyclic non-ketogenic high-fat/low-carbohydrate diet might have similar effects on memory with aging as the cyclic ketogenic diet. Finally, although we collected necropsy data when animals were euthanized for cause, this could be subject to selection bias and cannot describe age-matched incidence of pathologies as a dedicated pathology cohort could have.
The possible mechanisms of KD in longevity (and cognition) include effects of the low-carbohydrate, high-fat diet composition as well as activities of BHB itself. The former include reduced insulin and IGF signaling, reduced protein synthesis, and suppression of TOR activation. Our gene expression studies helped to distinguish which mechanisms are shared in common with a high-fat/low-carbohydrate diet, and which are unique to KD. KD has been associated in the liver with down-regulation of genes involved in fatty acid synthesis and up-regulation of fatty acid oxidation genes (Douris et al., 2015), including PPARα target genes (Badman et al., 2007). Indeed, PPARα activation is a master switch that is required to fully activate the response to fasting or a ketogenic diet (Badman et al., 2007). A “Western” high-fat diet (not low-carbohydrate) has a distinct profile from KD, with up-regulation of fatty acid synthesis and gluconeogenesis genes, and down-regulation of fatty acid oxidation (Kennedy et al., 2007). Our data show that a high-fat/low-carbohydrate diet is remarkably similar in transcriptional profile to KD. Both suppress fatty acid synthesis, glucose metabolism, and protein synthesis, all of which might provide a mortality benefit common to the cyclic (non-obese) HF and KD used here. The potent up-regulation of PPARα target genes is unique to KD, and might provide one clue to the mechanisms of the incrementally larger effect on survival of Cyclic KD. PPARα activation has been suggested as a calorie restriction mimetic (Barger et al., 2017), but its effects on aging phenotypes have not been deeply explored.
The presence of high concentrations of BHB in plasma defines KD, and BHB has direct activities relevant to aging. These might be categorized as energetics (via metabolism to ATP) or signaling. Increased ATP availability may be highly relevant to the immediate effects of ketone bodies on motor and cognitive function (Murray et al., 2016). However, our behavioral testing was performed during Control diet feeding periods specifically to exclude acute energetic effects of BHB. This suggests a more persistent mechanism, such as a change in mitochondrial number, chromatin structure, inflammatory state, or neuronal architecture and blood supply. The various signaling functions of BHB might generate such persistent effects: reduction of metabolic rate via FFAR3 inhibition (Kimura et al., 2011), reduced lipolysis and immunomodulation via HCAR2 activation (Rahman et al., 2014), inhibition of the NLRP3 inflammasome (Youm et al., 2015), and gene expression changes via deacetylase inhibition (Shimazu et al., 2013) or histone beta-hydroxybutyrylation (Xie et al., 2016).
Direct gene expression effects of histone beta-hydroxybutyrylation are a tempting mechanism to link BHB to the up-regulation of PPARα target genes that we observed – both acutely after one week on KD and persistent in old Cyclic KD mice several days into a control diet week. Indeed, Xie and colleagues reported that BHB-modified H3K9 marks PPARα target genes, and is associated particularly with such genes up-regulated (but not down-regulated) by prolonged fasting (Xie et al., 2016). It is not yet known if histone beta-hydroxybutyrylation is merely a marker of transcriptional activation or is an effector of activation. Our data would fit well into a model where BHB itself provides additional modulation of the basline transcriptional program of a high-fat/low-carbohydrate diet. Whether these transcriptional changes directly modulate the effects of Cyclic KD on mortality or memory remains to be tested. Other models are also plausible. For example, deacetylase inhibition is also associated with many of the effects we observed for Cyclic KD, including preservation of memory (Penney and Tsai, 2014), amelioration of chronic heart remodeling (Morales et al., 2016), and even activation of PPARα (Montgomery et al., 2008).
We have shown that long-term exposure to BHB through a ketogenic diet reduces mid-life mortality and preserves memory in aging normal mice, while activating a distinct transcriptional program involving PPARα target genes. Further work might determine if BHB itself is a segmental mimetic of DR, and could provide candidates for translational therapies of syndromes of aging through either administration of ketone esters (Veech, 2014) or, with a thorough understanding of its downstream signaling mechanisms, activation of BHB’s specific signaling effects. Future work to understand the nature of the persistent effect on memory in particular may lead to therapies to promote cognitive resilience to dementia or illness-associated delirium.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric Verdin (everdin@buckinstitute.org). Certain materials may be subject to Material Transfer Agreements from the Buck Institute, Gladstone Institutes, or the original providing entity.
Experimental Model and Subject Details
Mouse strains, housing, and husbandry
All mice were maintained according to the National Institutes of Health guidelines, and all experimental protocols were approved by the UCSF Institutional Animal Care and Use Committee (IACUC). UCSF is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Mice were maintained in a specific pathogen-free barrier facility on a 7:00 am to 7:00 pm light cycle. The facility is maintained at a temperature of 68–72° F and at >30% humidity. All mice were group housed in the groups in which they arrived, up to 5 mice per cage, unless specified in a particular experiment below. Cages contained absorbent paper chip bedding, and were changed every two weeks. Food was provided in a recess of the metal wire lid at the top of the cage; most experimental diets below were changed weekly, and the wire lid was changed with the food. Water was provided by a centralized system with a metal dispenser extending into the cage from the external rack. Enrichment was provided in the form of a nestlet, and the last surviving mouse in a cage was provided additional enrichment with a Nylabone. Houses or igloos were not used for enrichment.
C57BL/6 male mice were obtained from the National Institute on Aging’s Aged Rodent Colony at 11 months of age. For some preliminary studies, C57BL/6J male mice were obtained at 2–4 mo old from Jackson Labs. The NIA mice are derived from Jackson stock, and were confirmed as Nnt deletion carriers. The age of mice at the time experiments were undertaken is presented in the figures. Briefly, the lifespan study began with 12 mo old mice and followed them until death; the healthspan began with 12 mo old mice and involved behavioral testing at 12–14 mo old, 22–24 mo old, and 28–30 mo old.
Mouse diets and feeding
Food was provided ad libitum at all times. Per-calorie macronutrient content for customized diets (Envigo) is as follows: control, 10% protein, 13% fat, and 77% carbohydrates (TD.150345); KD 10% protein and 90% fat (TD.160153); HF, 10% protein, 75% fat, 15% carbohydrates (TD.160239). The fat sources are Crisco, cocoa butter, and corn oil. All diets are matched on a per-calorie basis for micronutrient content, fiber, and preservatives.
Compositions of the three experimental diets are as follows (g/kg):
| Control TD.150345 |
KD TD.160153 |
HF TD.160239 |
|
|---|---|---|---|
| Casein | 100 | 180 | 160 |
| DL-methionine | 1.6 | 2.88 | 2.5 |
| Corn starch | 512.46 | 0 | 158 |
| Sucrose | 100 | 0 | 30 |
| Maltodextrin | 155 | 0 | 46.5 |
| Crisco | 25 | 440 | 314 |
| Cocoa butter | 0 | 150 | 107 |
| Corn oil | 25 | 85 | 60.7 |
| Cellulose | 35 | 59.19 | 49.3 |
| Calories per gram | 3.7 | 6.7 | 5.8 |
Vitamin supplements included AIN-93-VX vitamin mix (Envigo 110068), thiamin, phylloquinone, choline bitartate, mineral mix (Envigo 98057), calcium phosphate, and calcium carbonate. Crisco is a proprietary blend of partially hydrogenated vegetable oil, with minimal trans-fat content. Fatty acids in KD are, by weight, approximately 24% saturated, 39% monounsaturated, and 37% polyunsaturated.
During the study, the fat composition of Crisco was changed by the manufacturer, with a slight reduction in average chain length and saturation causing a loosening of the consistency of KD and HF. This was compensated for by the additional of cocoa butter to KD and HF to restore a semisolid consistency. No difference in food intake or weight was observed after the adjustment.
KD and HF have a dough-like texture that permits them to be placed in the food well of the cage-top wire lid, in the same manner as pellets. All custom diets were changed weekly for all cages; for cKD and cHF, the diet was switched at this time. Food intake was determined from food remaining in the cage-top. Minimal or no wastage was observed in the cage bottoms.
Mice at the NIA Aged Rodent Colonies are fed NIH31, which contains per-calorie 24% protein, 14% fat, and 62% carbohydrates. Mice would have eaten this diet until shipped from the Aged Rodent Colony to our laboratory at 11 mo old. Once arrived, mice were fed the Gladstone Institutes standard vivarium chow until they were enrolled into a study and started on the custom diets. Gladstone vivarium chow is 5053 PicoLab diet (Ralston Purina Company, St. Louis, MO), and contains per-calorie 24% protein, 13% fat, and 62% carbohydrates. Thus, mice in the lifespan and healthspan studies a 24% kcal protein chow until started on our custom diets at 12 months of age.
Method Details
Lifespan endpoints
Endpoints for healthy lifespan were based on the NIA Interventions Testing Program (Miller et al., 2007) with additional stringent criteria developed with UCSF LARC veterinarians. The goal of the euthanasia criteria was to relieve and prevent any suffering without unnecessarily reducing healthy lifespan. Indications for euthanasia generally involved any obvious discomfort, impending death, systemic signs of unwellness, or any condition that was likely a harbinger of impending discomfort or death. Benign conditions of aging, even if abnormal in young mice, did not prompt euthanasia unless they were associated with these criteria or thought likely to lead to them over days or weeks.
All mice were weighed weekly, and monitored with increasing frequency up to 5x/week after 24 mo old. Specific indications for euthanasia of aged mice included the following:
-
-
moribund
-
-
BCS 2 or less
-
-
poor general health
-
-
poor mobility
-
-
any obvious discomfort
-
-
discoordination, ataxia, or abnormal gait
-
-
progressive weight loss to >15% below normal body weight
-
-
unexplained weight gain >15% above prior body weight
-
-
wounds that do not heal after two weeks of treatment, or worsen despite treatment
-
-
development of new wounds while on treatment for other wounds
-
-
full-thickness wounds that expose underlying fascia or muscle, wounds >2cm in any dimension, or any wound that is obviously causing discomfort
-
-
rectal prolapse, penile prolapse, or paraphimosis
-
-
suspected tumor associated with other signs of systemic unwellness (wound, weight loss, etc.)
-
-
suspected tumor >2cm or ulcerated
Lifespan Study: Start and End
Mice were enrolled into the lifespan study on a rolling basis over a 24 month period as they were obtained from the NIA Aged Rodent Colony, 16–30 mice per month. The first group was enrolled in November 2014, and the final group in March 2016. As above, all mice were fed vivarium chow until enrolled in a study. Mice were group housed, maintaining the same groups in which they arrived from NIA. Cages were pseudorandomly assigned to the five diet conditions, staggered between months, in pairs of cages. This resulted in all diet conditions being spread out across the entire study cohort in time.
Lifespan data collection ended when our laboratory moved from the Gladstone Institutes (San Francisco, CA) to the Buck Institute for Research on Aging (Novato, CA) in March 2017. The move involved shipping mice in boxes, as well as changes to the cage and bedding systems. The move was assumed to potentially confound any further lifespan observations, particularly given the rolling nature of the study. At the time of the move, 76% of enrolled mice had died. All mice are included in the data presented in Figure 1. Mice alive at the time of the move were censored at the date they were packed into boxes. Exclusion of younger cohorts (those mostly still alive at the time of the move) does not affect the shape of the lifespan curves.
Lifespan Study: Necropsies
Gross necropsies were performed on most mice that were euthanized, or that were found within 24 hours of death. “Internal tumors” includes any tumor in the abdomen or thorax, primarily of the liver, spleen, or mesentery. The denominator for internal tumor rate is the total number of necropsies. “Eye tumors” were histologically confirmed as carcinomas of the Harderian gland. As these are externally visible, the denominator is the total number of deaths. Neurological causes of death include head tilt, ataxia, and lower body or unilateral paralysis, all of which prompt euthanasia. The denominator for neurological deaths is all mouse deaths.
Lifespan Study: Statistical Analysis
Based on data from our pilot study, we anticipated that Cyclic KD might result in a squaring of the mortality curve with better survival through mid-life but not necessarily increased maximum lifespan. Therefore, we used methods that could detect and quantify such a time-dependent effect on mortality, in addition to presenting standard lifespan curve characteristics.
The log rank (Mantel-Cox) test assesses overall differences between survival curves. We present pairwise log rank P values, as well as the time to decile survival (including median survival).
There are several methods to assess differences in mortality at different times. One is to use a statistical test that differently weighs events at different times. One such commonly used test is Gehan-Breslow-Wilcoxon, which gives more weight to early deaths and has been used in mouse lifespan studies (Martin-Montalvo et al., 2013). The Fleming-Harrington test also incorporates a customizable weighting function. However, we lack a non-arbitrary rationale for selecting a specific magnitude of weighting. And a critical threshold of weighting magnitude (that which clearly separates groups) is abstract to interpret. Therefore, rather than using weighted statistics to assess the entire lifespan curve, we opted instead to use unweighted tests at different time points across the lifespan curve. This is systematic and unbiased, and could provide a straightforward interpretation – that mortality or survival differ at specific ranges of timepoints.
First, we calculated daily chi square tests to assess differences between pairwise groups on each day of the lifespan. We used Stata “sts list” to generate a table for each diet group with the number of mice at risk and the surviving proportion for each day of the lifespan. We then used custom code to calculate pairwise chi square tests of expected and actual survival: Control vs. Cyclic KD, Control vs. Cyclic HF, HF vs. KD, and Cyclic HF vs. Cyclic KD. For example, the survival proportion of the Control group was used to calculate expected surviving/dead numbers for the cyclic KD group, and compared to the actual surviving/dead numbers of the Cyclic KD group. These data are presented in Figure 1J–L and in Figure S2.
Second, we used daily-truncated data sets to calculate log rank tests and cumulative hazard ratios across the lifespan. For each day of age from when mice were first enrolled (365 days) until the last recorded death (1096 days), we generated a data set as if the study were terminated on that day, with later deaths converted to censorings. We used the same pairwise comparisons: Control vs. Cyclic KD, Control vs. Cyclic HF, HF vs. KD, and Cyclic HF vs. Cyclic KD. We used Stata “sts test” to perform log rank tests. To calculate estimates of the daily hazard ratio, we similarly truncated the survival data, but this time into moving 180-day windows. Each truncated data set started with only mice that were alive on the first day of the window, and converted any deaths beyond 180 days to censorings. We generated a new 180-day window centered on each day. We then used Stata “stcox” to calculate the cumulative hazard ratio. The window was chosen to capture enough of the relatively-rare deaths within each window to reduce noise in the resulting plot. Both log rank and hazard ratio data are presented in Figure S2.
Importantly for interpreting these plots, the chi square test is instantaneous, while the log rank test is cumulative. In other words, the chi square test assesses differences in survival on that day, while the log rank test or hazard ratio assesses for differences in survival up until that day. A chi square test can generate a low P value from very transient survival differences, while log rank requires more sustained differences for a low P value.
Third, in order to address the multiple hypothesis problem inherent in interpreting P values across hundred of days of lifespan, we performed randomized-data monte carlo simulations. We randomly shuffled the diet group of all mice in the data set, and performed the same daily log rank and daily chi square procedures described above. For log rank, we used the best daily log rank P value as the relevant metric. For chi square, since very transient differences between curves can generate a low P value, we instead used “total days with P<0.05”. We performed 1000 simulations for each of the three pairwise comparisons: Control vs. Cyclic KD, Control vs. Cyclic HF, and HF vs. KD. We obtained similar results for the chi square simulations using “most consecutive days with P<0.05” as the metric, and using P<0.01 as the P value threshold (except for Cyclic HF vs. Control, as this chi square P never reached P<0.01). The number of randomized simulations that generated results better than observed in the actual data is shown in Figures 1 and S2.
Healthspan Study
All behavioral testing was performed in consultation with the Gladstone Neurobehavioral Core, incorporating expert recommendations as to the type, sequence, and timing of the various behavioral tests. A key factor was their suitability for use with old mice, compatibility for performing multiple tests on the same animals, and suitability for repeat measurements over time. Preference was given to selecting tests that had been reported in the literature to show declines in performance with age in C57BL/6, or that were analogous to well-validated correlates of aging in humans such as grip strength and gait speed. All tests were carried out during normal daytime hours, in normal room light except where noted. The healthspan cohort was divided into two subcohorts, each half Cyclic KD and half Control, in order to stagger the behavioral testing by one month. Thus, about 30 mice were tested at a time. For tests involving multiple repetitions in a day, all mice underwent the first repetition before proceeding in the same order with the second repetition. Most of the Baseline and Aged testing took place while mice were temporarily housed in the behavioral core holding room. Upon completion of testing (4–6 weeks) they were transferred back to their usual holding room. The only exceptions were the running wheels, echocardiograms, and frailty index, which were performed in/from the usual holding rooms after the mice were transferred back. Old Age testing was performing in the usual holding rooms.
As described in the figures and text, all mice were fed Control diet during the Baseline testing period from 12–14 mo old. Upon completion of Baseline testing, mice were randomly assigned by cages to either the Control group (continue on continuous control diet) or the Cyclic KD group (begin the Cyclic KD regimen, alternating control and KD weekly). Randomization was performed by dice roll. These group assignments were maintained for the remainder of the study.
Place Avoidance
Place avoidance is an interactive visuospatial learning and memory task, similar in conception to the Morris water maze. The apparatus is from Biosignal, and the tracking and control software is Biosignal Tracker. The room is kept at low light. Mice are place in a clear plastic cylinder (40 cm diameter, 25 cm high) sitting on a rotating platform whose floor is made up of parallel metal bars (4 mm diameter, spaced every 1 cm). The platform rotates at 1 rpm. A region encompassing roughly a 60 degree wedge of the platform is designated the shock zone. If the mouse enters the shock zone, it receives a non-painful but startling shock delivered through the bars that make up the floor (0.2 mA current, duration 0.5 sec, latency from zone entrance 0.5 sec, intershock latency 1.5 sec). The shock zone is fixed to the orientation of the room, so as the platform rotates the mouse would rotate into the shock zone once per minute if it did not move. There are no visual indications of where the shock zone is; it is defined only in the videotracking software that determines when to deliver a shock. The mouse learns where the shock zone is relative to visual cues in the room, and must frequently move around the platform to avoid rotating into it. Like the water maze, this is a multi-day protocol. Each daily session is 10 minutes long. Day 1, habituation to the apparatus with no shocks. Days 2–4/5, learning/training sessions with shocks delivered. Day 5 or 6, memory/probe trial with shocks turned off for the first 5 minutes, followed by retraining with the shocks turned back on for the second 5 minutes. The primary outcome in the training trials is number of shock-entries (unique entries into the shock zone resulting in a shock) per session. The primary outcome in the probe trial is the time until first entry into the shock zone, with a secondary outcome of time until the second entry (to account for an oblique or transient first entry). We found that mice learned more slowly in the Aged test than they did at Baseline, in the sense of the number of entries per training trial declining more slowly. Since the key outcomes was probe trial performance, we trained the Aged mice an extra day to reach the same final training performance as the final baseline training. In addition to the learning and memory data, the videotracking provides measures of neuromuscular performance. Mice are generally startled by the shock, and jump horizontally to escape the area. We extracted the peak instantaneous velocity (measured in 1/30 sec increments) during the 1 second before and after each shock, as well as the time required to reach this peak velocity. These measures, maximum escape velocity and minimum reaction time, were interpreted as healthspan items. They are the only shock-motivated physical performance measures we collected, and might reasonably represent peak possible physical performance.
Open Field
The open field apparatus (automated Flex-Field/Open-Field Photobeam Activity System; San Diego Instruments) consisted of four identical clear plastic chambers (40 × 40 × 30 cm) with two 16 × 16 photobeam arrays to detect horizontal and vertical (rearing) movements. Total movements were reported, and then stratified as peripheral (4 beams on either flank of each array; 8 beams total) or central (middle 8 beams of each array). Mice were recorded for 30 minutes, in low light conditions.
Rotarod
Rotarod was performed with a Med Associates ENV-575(M) apparatus, using a 3-day protocol. Day 1 involved three training runs at fixed 16 rpm. Days 2 and 3 each involved 6 data runs, 3 in the morning and 3 in the afternoon. The data runs on days 2 and 3 used steady acceleration of the rod from 4–40 rpm over 5 minutes. The time was stopped at 5 minutes or when the mouse fell. Mice were removed if they rotated around the rod twice.
Elevated Plus Maze
The EPM 2000 apparatus (Kinder Scientific) consists of two platforms joined in a plus+ shape. Each platform is 78 c × 5 cm, with a 5 cm square region where they meet. One platform is flat and open. The other platform has 10 cm high walls. The entire apparatus is 62 cm above the floor. Infrared beams in the platform base track mouse movement through the apparatus, reporting both distance moved and dwell time. The room is kept at low light. Mice are permitted to explore for 10 minutes.
Balance Beam
A 15” long plastic beam is suspended about 20” above the table, with an open platform at one end and a dark box with 1.5” square opening at the other end. Mice were tested one at a time, placed on the beam near the open platform. Testing was done over three days. Day 1 involved two untimed trials where mice were gently guided across the beam into the box, followed by two timed trials. Day 2 involved two additional times trials. Day 3 used a narrower beam, and involved two untimed guided trials followed by two timed trials. Days 1–2 used a ½ inch diameter beam with circular cross section, while day 3 used a ¼ wide square beam. Time was recorded by stopwatch from the initial placement on the beam until the torso entered the dark box. Foot slips were recorded by the observer. Mice that fell were immediately placed back on the beam at the spot of the fall. The primary outcome was fastest time on the narrow/test beam. Secondary outcomes included fastest time on the wide/training beam, and numbers of slips and falls.
Grid Wire Hang
The apparatus consists of a 15×25 cm grid of fine wire mesh suspended 34 cm above the table on supports. The walls and rear of the supports are covered with smooth plastic boards to prevent escape, and a larger plastic board is placed atop the wire mesh to prevent a mouse from climbing around the edge. The mouse is placed on the wire mesh, gently shaken to ensure a grip, and the mesh then inverted onto the supports. The test lasts for 180 seconds, or until the mouse falls. The table top is covered with layers of soft towels to prevent injury from a fall. A mouse that immediately reached and fell was replaced on the mesh, but only once. Three trials were run. The primary outcome is maximum hanging impulse, defined as (hang time (sec) × body weight (kg) × 9.8 m/ŝ2).
In-Cage Running Wheels
Med Associates Wireless Running Wheels (ENV-044) were placed one per cage into the normal group-housed cages for one week. To fit the wheels into the cages, the usual wire cage-top was replaced with a flat one, and food placed on the cage floor in sterile glass jars. Additional food was added halfway through the week. The primary outcome was total distance traveled in 7 days, divided by the number of mice in that cage. Note that this outcome may be non-linear to number of mice per cage, and does not represent individual mouse activity; it is therefore only suitable to compare groups.
Single Wire Hang
A 2 mm diameter metal bar is suspended between two supports about 40 cm apart, and 34 cm above the table top. The table top is covered with layers of soft towels to prevent injury from a fall. The mouse is gently lowered onto the wire until it grips the wire with both forepaws, then it is let go. The timer is started. If the mouse falls, it is immediately replaced onto the wire and a fall is recorded. If it crawls to the end of the wire and climbs onto the supports, it is immediately replaced onto the wire and an escape is recorded. The test lasts for 180 seconds, or until the mouse falls ten times. The primary outcomes are the number of falls in the first 60 seconds, and the total time elapsed (until the 10th fall or 180 seconds, whichever is first).
Novel Object Recognition
An enlarged version of the usual home cage was used as the arena, 56×28×21 cm L×W×H, with normal bedding on the floor. This familiar environment was intended to focus attention on the two objects placed in the cage, equidistant from the corners and each other. The test protocol was designed to be adaptable, since it was not clear beforehand how different the objects would have to be to generate adequate recognition in 30 month-old mice that had age-related cognitive and sensory impairments. All objects were about 10 cm high and upright, supported by a flat base hidden under the bedding. The initial training used two relatively similar objects, tissue culture flasks either filled with orange sand or half-filled with white sand - thus differing in color and in opacity but identical in size and outline. Each trial lasted 5 minutes, with exploration time manually scored by the observer using two stopwatches. A mouse was counted as exploring an object if its nose was oriented towards the object and within 2 cm. Rearing onto the object was counted as exploration unless the face/nose was clearly oriented away from the object while rearing. On day 1, the first training trial was performed with two identical objects (alternating the orange and white objects between mice). On day 2, a second training trial was performed in the morning, during which total exploratory time decreased. We proceeded to the first probe trial in the afternoon, during which one of the objects was swapped for the different one (alternating which side was novel between mice). In this first probe trial we observed increased total exploration time, but minimal preference for the novel object in either group. We interpreted this as the novel object being insufficiently distinct. We proceeded to a second probe trial on day 3, replacing the novel object from the first probe trial with a different object and swapping the position of the familiar and novel objects. This third object was roughly the same size and shape, but made of legos and with a more distinct outline. In this second probe trial we observed a decrease in total exploratory time in the Con group, still with little preference for the novel object; but the cKD group showed less decline in exploration and a strong preference for the novel object. We confirmed with an independent group of 12 month-old mice that there was no intrinsic preference for the lego object. The primary outcome was % of exploratory time at the novel object. To remove random effects from low-exploring mice, we excluded mice with <5 seconds total exploratory time from the analysis. Ideally, the outcome measure should be more smoothly weighted towards animals with more absolute exploratory time at the novel object. Three different methods of weighting all reached similar, statistically significant results: (% novel time × novel time), (novel time – familiar time), and (novel time).
Mouse Clinical Frailty Index
We used the previously described (Whitehead et al., 2014) and validated (Kane et al., 2016) 31-item mouse clinical frailty index. Body temperatures were obtained from the abdomen with a Braun Thermoscan infrared thermometer, using the highest of three readings. Reference normal for body weight and temperature were obtained from 12 month-old C57BL/6 male mice also obtained from NIA. All observations were performed by the same experimenter.
Echocardiograms
Mouse transthoracic echocardiograms were performed as previously described (Mohamed et al., 2016). Briefly, mice were anesthetized with isoflurane, placed on a heating pad, and ventral fur shaved with clippers. Echocardiography was performed with the Vevo 770 High-Resolution Micro-Imaging System (VisualSonics) with a 15-MHz linear-array ultrasound transducer. The left ventricle was assessed in both parasternal long-axis and short-axis views at a frame rate of 120?Hz. End-systole or end-diastole were defined as the phases in which the left ventricle appeared the smallest and largest, respectively, and used for ejection-fraction measurements. To calculate the shortening fraction, left ventricular end-systolic and end-diastolic diameters were measured from the left ventricular M-mode tracing with a sweep speed of 50 mm/s at the papillary muscle. Bmode was used for two-dimensional measurements of end-systolic and end-diastolic dimensions. The echocardiographer was blinded to the study groups.
Healthspan Composite Scores
Composite scores were calculated as described in the text and figure. A young baseline was chosen as the reference performance – for most tests, this was the pooled result of all mice during Baseline testing. For echocardiogram parameters, this was data from separate 3 mo old C57BL/6 male mice. For each parameter, the mean and standard deviation was calculated from the reference group. Then, each individual mouse result in the aged groups was normalized as the number of baseline standard deviations from the baseline mean. Each parameter was assigned a directionality, with positive (+) representing more youthful or better performance. This simple normalization permits the combination of results across tests with widely varying absolute values and distributions, maintaining equal weighting of each test result. The normalized values were averaged across multiple tests, and all mice in each group. A nonparametric test (Mann-Whitney) was used to compare group means as the performance distribution in aged mice was not necessary expected to follow a Gaussian distribution, and tests of normality of the composite score data showed they did not. The 4-item cardiac score includes: heart rate, weight-normalize left ventricular mass, aortic valve pressure gradient, and fractional shortening. The 35-item healthspan score includes all meaningful quantitative measurements that were obtained during healthspan testing, including the four echocardiogram items (listed in full in Table S2). Tests done in the Old Age testing period could not be included, as they lacked a young or baseline reference group for comparison (Grid Wire Hand was done at all three testing periods, but on a slightly different apparatus in Old Age and so may not be directly comparable).
Plasma Beta-Hydroxybutyrate Measurements
Blood was obtained via distal tail-snip (10–40 uL) or, if the animal was being euthanized for tissue collection, by cardiac puncture immediately following euthanasia. Blood was collected into lithium-heparin coated microvettes (Sarstedt CB 300 LH), and plasma separated by centrifugation at 1500 × g for 15 minutes at 4 degC. Plasma was frozen at −20 degC until thawed for assay. We confirmed that freeze-thawing had no effect on assay results. We measured BHB using the StanBio LiquiColor Beta-Hydroxybutyrate colorimetric test (2440-058), run using 3 uL sample volumes in triplicate. We found it was critical to subtract baseline absorbance of the sample-enzyme mixture prior to adding catalyst, in order to account for any sample hemolysis.
Western Blots
Mice used for the Western blot data (Figure S4) were a preliminary cohort of mice, C57BL/6 males obtained at 4 m old from NIA. They were then fed either control diet or KD for 21 weeks. Tissues were collected in the morning (9a-noon), snap-frozen in liquid nitrogen immediately following euthanasia then transferred to −80 degC for storage. Mitochondrial isolation was performed as previously described (Hirschey et al., 2009). Briefly: A small quantity of liver was placed into homogenization buffer containing protease and deacetylase inhibitors, finely diced, and then homogenized in a glass tube homogenizer. Differential centrifugation first pelleted debris, then intact mitochondria. Whole cell lysates were prepared by placing a small quantity of liver directly in 1% SDS sample buffer, and homogenizing with a handheld rotary homogenizer. Protein concentrations were assayed with a BCA kit prior to loading onto 10% polyacrylamide gels. Approximately 20 ug were loaded per lane. Antibodies included anti-acetyllysine (Cell Signaling 9441), anti-ATP5α (Mitoscience MS502), and anti-histone H3 (Upstate 07-690). We used the LiCor Odyssey system for secondary antibodies, blot scanning, and quantitation. The presented blots are representative of multiple replicates, including from different animals following different lengths of KD treatment.
Quantitative PCR
Mice cohort and tissue collection for the QPCR experiment in Figure S4 (a preliminary cohort of mice) was identical to that described for Western blots, above. Frozen tissue samples were stabilized in RNAlater-ICE (Qiagen), then homogenized in Trizol (Ambion) using a handheld rotary homogenizer. RNA isolation proceeded by the manufacturer’s protocol. Integrity of the RNA was verified on a denaturing agarose gel. cDNA synthesis was carried out with Superscript (ThermoFisher). Realtime PCR was performed in a SYBR-green system using the methods and primer sets previously described (Gut et al., 2013).
Mice for the QPCR studies in Figure 3 were C57BL/6 males from NIA, obtained at 11 mo old, and at 12 months old placed on the Control (8) or Cyclic KD (8) regimen. They were treated identically to mice in the lifespan study. At 26 mo old the mice were euthanized across two consecutive nights (midnight-3am). Tissues were collected immediately upon euthanasia and snap-frozen in liquid nitrogen, then transferred to −80 degC for storage. For each mouse 30mg of liver tissue was homogenized using a Bullet Blender (speed 8 and time 3 at 4C) in RNA STAT 60 (Tel-Test) and RNA was isolated per manufacturers protocol. cDNA synthesis was performed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed using a Bio-Rad CFX384 and gene expression was measured by the delta/delta CT method using the CFX manager software. Gene expression in liver samples were normalized to a basket of four reference genes: Albumin, Transthyretin, Fibrinogen, and GAPDH. Primer sequences are listed in Table S4.
RNAseq
Mice for the RNAseq data were C57BL/6 males from NIA, obtained at 11 mo old, and at 12 months old placed on control diet, KD, or HF for one week. Mice were individually housed in order to collect individual food intake data. Blood and tissues were both collected at night (midnight-3am), both to maximize plasma BHB levels and ensure synchrony of the feeding state for all mice. One blood sample was collected the night before euthanasia via distal tail-snip; a second blood sample was obtained by cardiac puncture immediately following euthanasia. Tissues were collected immediately upon euthanasia and snap-frozen in liquid nitrogen, then transferred to −80 degC for storage. Frozen tissue samples were stabilized in RNAlater-ICE (Qiagen) prior to homogenization in RNeasy buffer RLT using a using a handheld rotary homogenizer. RNA isolation proceeded using the RNeasy Mini kit (Qiagen) per the manufacturer’s protocol. The Gladstone Institutes Genomics Core carried out the downstream processing of the RNA samples. RNA integrity and concentrations were confirmed with an Agilent Bioanalyzer. The standard core protocol uses Ovation RNAseq System V2 (NuGesn) for cDNA synthesis and amplification. This system features chimeric primers for first-strand synthesis with a unique 5’ RNA sequence and 3’ random hexamer or poly-T DNA, and single primer isothermal amplification of the resulting cDNA. A library is created from the amplified cDNA using the Ovation Ultralow DR Multiplex System (NuGen), then analyzed for quality via Bioanalyzer, and quantified by quantitative PCR (KAPA). Next-generation sequencing is done with a HiSeq 2000 (Illumina), with the 18 barcoded samples run in pools over 4 lanes in total. The total yield per sample varied from approximately 1500 to 3000 megabases.
The resulting single-end 50 base pair reads were trimmed of adaptors and low-quality reads using Fastq-mcf, then aligned to mouse reference sequences using TopHat 2.0.11 software. This resulted in approximately 40 million aligned reads per sample. For the liver data set, we excluded genes with <500 reads in total, and the software USeq DefinedRegionDifferentialSeq was used both to tally up per-gene counts, normalize these counts, and compute differential expression based using a negative binomial distribution for gene expression performed by DEseq version 1. For the kidney data set, per-gene counts were arrived at using Subread FeatureCounts, and then edgeR was used to both normalize the counts as CPM (counts per million) and to compute differential expression between samples. In this dataset, we excluding genes that did not have at least two samples (of 18) with initial CPM between 0.5 and 5. For both data sets, false discovery rate was calculated using the Benjamini-Hochberg method. The criteria for selection of a list of differentially expressed genes for use in subsequent pathway analysis included P=0.02 and Log2FC>0.379 (fold change>30%), which generated lists of several hundred genes per comparison. These criteria were chosen based on 1) the biological plausibility of the genes that met these criteria, including those previously confirmed as regulated by KD via Q-PCR; 2) the expectation from prior published work that KD or HF diet should alter the expression of hundreds or thousands of genes in the liver; and 3) to provide adequate power for pathway analysis while accepting caution in interpreting any individual gene. Our biological interpretation of the RNAseq data is based solely on list/pathway analysis, which is robust to uncertainties in individual genes.
Pathway analysis was carried out using Ingenuity Pathway Analysis and the Gene Ontology AmiGO Term Enrichment tool. For the Ingenuity analysis, the entire data set was uploaded, and a two-direction analysis carried using filters to restrict the analysis to differentially expressed genes as above. For Gene Ontology, individual lists of genes were uploaded and analyzed separately. For the presentation of Upstream Regulator data in Figure 3, we manually identified the four patterns that represented the majority of the top hits (by Z-score and P value) in the Liver/KD data set. We then highlighted that same set of regulators in the Kidney/KD and Liver/HF data sets, in order to show commonality (or lack thereof) with upstream regulators predicted for Liver/KD. We manually reviewed the Liver/HF and Kidney/KD data sets to ensure that there were no other major patterns present among the top predicted regulators in these data sets. Heat maps and scatter plots were generated with Graph Pad Prism.
Quantification and Statistical Analysis
Unless otherwise noted, figures are presented as mean +/− SEM, P values are calculated by unpaired two-tailed T test with the assumption of Gaussian distributions, and statistical calculations are carried out in Prism 7.0a (GraphPad Software, La Jolla CA). Exact “N” are specified in the figure legends for each experiment, and refer to the number of individual mice. Statistical details specific to a particular experiment are included in the description of that experiment in Method Details above. Although healthspan testing involved the longitudinal comparison of groups, the use of paired T tests was generally not possible due to small numbers of animal deaths between the Baseline and Aged behavioral testing. In order to limit multiple hypothesis testing, statistical tests were generally limited to the comparison of interest; in healthspan testing this was Control Aged vs. Cyclic KD Aged. As described above, the analysis of lifespan curves included calculations performed in Stata/SE 14.2 (StataCorp, College Station TX). Custom analysis programs were run with Perl v5.16.1 on OS X 10.11.
Data and Software Availability
The RNAseq data described in Figure 3 has been deposited in GEO with accession GSE101657.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-acetyllysine | Cell Signaling | 9441 |
| Anti-ATP5α | Mitoscience | MS502 |
| Anti-histone H3 | Upstate | 07-690 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Control diet (custom) | Envigo | TD.150345 |
| High-fat diet (75% kcal fat, custom) | Envigo | TD.160153 |
| Ketogenic diet (90% kcal fat, custom) | Envigo | TD.160239 |
| Critical Commercial Assays | ||
| LiquiColor Beta-Hydroxybutyrate colorimetric test | StanBio | 2440-058 |
| Deposited Data | ||
| RNAseq fastq files and processed expression data | GEO | GSE101657 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 male mice | NIA Aged Rodent Colony | N/A |
| Oligonucleotides | ||
| See Table S4 | ||
| Software and Algorithms | ||
| Prism | GraphPad | 7.0a |
| Stata/SE | Stata Corp | 14.2 |
| Perl | Perl.org | 5.16.1 |
| Ingenuity Pathway Analysis | Ingenuity | 2017 |
| AmiGO | Gene Ontology | 1.8 |
Highlights.
Feeding isoprotein ketogenic diet to mice every-other-week (Cyclic KD) avoids obesity
Cyclic KD reduces midlife mortality with no change in maximum lifespan
Cyclic KD prevents memory decline with modest other healthspan effects
Gene expression of KD is similar to high fat diet except activation of PPARα targets
Acknowledgments
Behavioral data were obtained with the help of the Gladstone Institutes’ Neurobehavioral Core (supported by NIH grant P30NS065780). Statistical analysis of RNA sequence data was conducted by Alex Williams at the Gladstone Bioinformatics Core. This work was supported by US National Institutes of Health grants K08AG048354 (JCN) and R24DK085610 (EV); Gladstone Institutes intramural funds (EV); Buck Institute intramural funds (EV); The Buck Impact Circle (JCN); and funds from the Larry L. Hillblom Foundation (JCN), Glenn Foundation for Medical Research (JCN), and American Federation for Aging Research (JCN).
Abbreviations
- BHB
beta-hydroxybutyrate
- KD
ketogenic diet
- HF
high-fat/low-carbohydrate (non-ketogenic) diet
- DR
dietary restriction
- PPAR
peroxisome proliferator-activated receptor
- TOR
target of rapamycin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, J.C.N. and E.V.; Methodology, J.C.N. and E.V.; Software, J.C.N.; Formal Analysis, J.C.N., A.J.C, and J.H.; Investigation, J.C.N. (all except where noted), M.Z. (healthspan testing), X.Y. (healthspan testing), A.J.C. (QPCRs), and J.H. (echocardiograms); Resources, S.H.; Writing – Original Draft, J.C.N. and E.V.; Writing – Review and Editing, J.C.N. and E.V.; Visualization, J.C.N. and E.V.; Supervision, E.V.; Funding Acquisition, J.C.N. and E.V.
References
- Austad SN, Fischer KE. Sex Differences in Lifespan. Cell Metab. 2016;23:1022–1033. doi: 10.1016/j.cmet.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Barger JL, Vann JM, Cray NL, Pugh TD, Mastaloudis A, Hester SN, Wood SM, Newton MA, Weindruch R, Prolla TA. Identification of tissue-specific transcriptional markers of caloric restriction in the mouse and their use to evaluate caloric restriction mimetics. Aging Cell. 2017 doi: 10.1111/acel.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai N, Crandall JP, Kritchevsky SB, Espeland MA. Metformin as a Tool to Target Aging. Cell Metab. 2016;23:1060–1065. doi: 10.1016/j.cmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghjid S, Feinman RD. Response of C57Bl/6 mice to a carbohydrate-free diet. Nutr Metab (Lond) 2012;9:69. doi: 10.1186/1743-7075-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G, Dubeau L, Yap LP, Park R, Vinciguerra M, et al. A Periodic Diet that Mimics Fasting Promotes Multi-System Regeneration, Enhanced Cognitive Performance, and Healthspan. Cell Metab. 2015;22:86–99. doi: 10.1016/j.cmet.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douris N, Melman T, Pecherer JM, Pissios P, Flier JS, Cantley LC, Locasale JW, Maratos-Flier E. Adaptive changes in amino acid metabolism permit normal longevity in mice consuming a low-carbohydrate ketogenic diet. Biochim Biophys Acta. 2015;1852:2056–2065. doi: 10.1016/j.bbadis.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KE, Hoffman JM, Sloane LB, Gelfond JA, Soto VY, Richardson AG, Austad SN. A cross-sectional study of male and female C57BL/6Nia mice suggests lifespan and healthspan are not necessarily correlated. Aging (Albany NY) 2016;8:2370–2391. doi: 10.18632/aging.101059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EL, Dixit VD. Drivers of age-related inflammation and strategies for healthspan extension. Immunol Rev. 2015;265:63–74. doi: 10.1111/imr.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut P, Baeza-Raja B, Andersson O, Hasenkamp L, Hsiao J, Hesselson D, Akassoglou K, Verdin E, Hirschey MD, Stainier DY. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat Chem Biol. 2013;9:97–104. doi: 10.1038/nchembio.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Huang JY, Verdin E. Acetylation of mitochondrial proteins. Methods Enzymol. 2009;457:137–147. doi: 10.1016/S0076-6879(09)05008-3. [DOI] [PubMed] [Google Scholar]
- Kane AE, Hilmer SN, Boyer D, Gavin K, Nines D, Howlett SE, de Cabo R, Mitchell SJ. Impact of Longevity Interventions on a Validated Mouse Clinical Frailty Index. J Gerontol A Biol Sci Med Sci. 2016;71:333–339. doi: 10.1093/gerona/glu315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AR, Pissios P, Otu H, Roberson R, Xue B, Asakura K, Furukawa N, Marino FE, Liu FF, Kahn BB, et al. A high-fat, ketogenic diet induces a unique metabolic state in mice. Am J Physiol Endocrinol Metab. 2007;292:E1724–1739. doi: 10.1152/ajpendo.00717.2006. [DOI] [PubMed] [Google Scholar]
- Kimura I, Inoue D, Maeda T, Hara T, Ichimura A, Miyauchi S, Kobayashi M, Hirasawa A, Tsujimoto G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41) Proc Natl Acad Sci U S A. 2011;108:8030–8035. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 2010;9:92–95. doi: 10.1111/j.1474-9726.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List EO, Berryman DE, Wright-Piekarski J, Jara A, Funk K, Kopchick JJ. The effects of weight cycling on lifespan in male C57BL/6J mice. Int J Obes (Lond) 2013;37:1088–1094. doi: 10.1038/ijo.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Panda S. Fasting, Circadian Rhythms, and Time-Restricted Feeding in Healthy Lifespan. Cell Metab. 2016;23:1048–1059. doi: 10.1016/j.cmet.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, et al. Metformin improves healthspan and lifespan in mice. Nat Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Longo V, Harvie M. Impact of intermittent fasting on health and disease processes. Ageing Res Rev. 2016 doi: 10.1016/j.arr.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies KJ, Zhang H, Katsyuba E, Auwerx J. Protein acetylation in metabolism - metabolites and cofactors. Nat Rev Endocrinol. 2016;12:43–60. doi: 10.1038/nrendo.2015.181. [DOI] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Floyd RA, Flurkey K, Hensley KL, Javors MA, Leeuwenburgh C, Nelson JF, Ongini E, et al. An Aging Interventions Testing Program: study design and interim report. Aging Cell. 2007;6:565–575. doi: 10.1111/j.1474-9726.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- Mohamed TM, Stone NR, Berry EC, Radzinsky E, Huang Y, Pratt K, Ang YS, Yu P, Wang H, Tang S, et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation. 2016 doi: 10.1161/CIRCULATIONAHA.116.024692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales CR, Li DL, Pedrozo Z, May HI, Jiang N, Kyrychenko V, Cho GW, Kim SY, Wang ZV, Rotter D, et al. Inhibition of class I histone deacetylases blunts cardiac hypertrophy through TSC2-dependent mTOR repression. Sci Signal. 2016;9:ra34. doi: 10.1126/scisignal.aad5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AJ, Knight NS, Cole MA, Cochlin LE, Carter E, Tchabanenko K, Pichulik T, Gulston MK, Atherton HJ, Schroeder MA, et al. Novel ketone diet enhances physical and cognitive performance. FASEB J. 2016;30:4021–4032. doi: 10.1096/fj.201600773R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Verdin E. Ketone bodies as signaling metabolites. Trends Endocrinol Metab. 2014;25:42–52. doi: 10.1016/j.tem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermanns S. Hydroxy-Carboxylic Acid Receptor Actions in Metabolism. Trends Endocrinol Metab. 2017 doi: 10.1016/j.tem.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Peleg S, Feller C, Forne I, Schiller E, Sevin DC, Schauer T, Regnard C, Straub T, Prestel M, Klima C, et al. Life span extension by targeting a link between metabolism and histone acetylation in Drosophila. EMBO Rep. 2016;17:455–469. doi: 10.15252/embr.201541132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney J, Tsai LH. Histone deacetylases in memory and cognition. Sci Signal. 2014;7:re12. doi: 10.1126/scisignal.aaa0069. [DOI] [PubMed] [Google Scholar]
- Pugh TD, Oberley TD, Weindruch R. Dietary intervention at middle age: caloric restriction but not dehydroepiandrosterone sulfate increases lifespan and lifetime cancer incidence in mice. Cancer Res. 1999;59:1642–1648. [PubMed] [Google Scholar]
- Rahman M, Muhammad S, Khan MA, Chen H, Ridder DA, Muller-Fielitz H, Pokorna B, Vollbrandt T, Stolting I, Nadrowitz R, et al. The beta-hydroxybutyrate receptor HCA2 activates a neuroprotective subset of macrophages. Nat Commun. 2014;5:3944. doi: 10.1038/ncomms4944. [DOI] [PubMed] [Google Scholar]
- Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010;2010 doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A, Fischer KE, Speakman JR, de Cabo R, Mitchell SJ, Peterson CA, Rabinovitch P, Chiao YA, Taffet G, Miller RA, et al. Measures of Healthspan as Indices of Aging in Mice-A Recommendation. J Gerontol A Biol Sci Med Sci. 2016;71:427–430. doi: 10.1093/gerona/glv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veech RL. Ketone ester effects on metabolism and transcription. J Lipid Res. 2014;55:2004–2006. doi: 10.1194/jlr.R046292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead JC, Hildebrand BA, Sun M, Rockwood MR, Rose RA, Rockwood K, Howlett SE. A clinical frailty index in aging mice: comparisons with frailty index data in humans. J Gerontol A Biol Sci Med Sci. 2014;69:621–632. doi: 10.1093/gerona/glt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods JK, Rogina B. The effects of Rpd3 on fly metabolism, health, and longevity. Exp Gerontol. 2016;86:124–128. doi: 10.1016/j.exger.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, Qi S, Li J, Colak G, Chen Y, et al. Metabolic Regulation of Gene Expression by Histone Lysine beta-Hydroxybutyrylation. Mol Cell. 2016;62:194–206. doi: 10.1016/j.molcel.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm YH, Nguyen KY, Grant RW, Goldberg EL, Bodogai M, Kim D, D'Agostino D, Planavsky N, Lupfer C, Kanneganti TD, et al. The ketone metabolite beta-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat Med. 2015;21:263–269. doi: 10.1038/nm.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.