Abstract
Thioredoxin (Trx) is one of the two major thiol antioxidants playing essential roles in redox homeostasis and signaling. Despite the importance, there is a lack of method in monitoring Trx redox dynamics in live cells, hindering a better understanding of physiological and pathological roles of the Trx redox system. In this work, we developed the first genetically encoded fluorescent biosensor for Trx redox by engineering a redox relay between the active-site cysteines of human Trx1 and rxRFP1—a redox-sensitive red fluorescent protein. We utilized the resultant biosensor—TrxRFP1—to selectively monitor perturbations of Trx redox in various mammalian cell lines. We subcellularly localized TrxRFP1 to image compartmentalized Trx redox changes. We further combined TrxRFP1 with a green fluorescent Grx1-roGFP2 biosensor to simultaneously monitor Trx and glutathione redox dynamics in live cells in response to chemical and physiologically relevant stimuli.
Along with glutathione, thioredoxin (Trx) is one of the two major thiol-dependent antioxidants in mammals1. The NADPH-dependent reduction of Trx is catalyzed by the flavoenzyme Trx reductase (TrxR), whereas the oxidation of Trx is catalyzed by Trx-dependent peroxiredoxins (Prx) also known as Trx peroxidases (TPx) (Supplementary Results, Supplementary Fig. 1)1. Consequently, the Trx system shuttles electrons from NADPH to TPx, leading to rapid removal of reactive oxygen species (ROS). Trx also plays important roles in modulating protein activities through the redox modulation of cysteine and methionine residues1. Trx can reduce oxidized cysteine residues in proteins, such as ribonucleotide reductase involved in DNA synthesis and repair2, a number of redox-sensitive transcription factors including NF-κB and Ref-1/AP-13,4, apoptosis signal regulating kinase 1 (ASK1)5, and methionine sulfoxide reductases for the reduction of oxidized methionine residues6,7. In addition, the Trx system has been linked to immune responses8, bacterial and viral infection9,10, cell proliferation11, apoptosis12, and pancreatic β-cell functions13. Moreover, the abnormality of the Trx system has been observed in cancer14. The Trx system, especially TrxR, has been considered as a promising target for cancer therapy15.
Our current understanding of redox biology remains largely elusive. Redox signaling, which involves diverse reactive oxygen, nitrogen and sulfur species (ROS, RNS and RSS), small-molecule thiols, proteins, and other types of molecules, is incredibly sophisticated16. Although thermodynamics drives redox reactions in live cells and organisms, the specificity of the reactions is largely determined by the spatial organization of redox signaling components and the kinetics of competitive reactions17. Such complexity is further compounded by the scarcity of research tools that can monitor the redox of specific redox-signaling components in live cells with appropriate specificity and spatiotemporal resolution. Previous studies have reported a family of genetically encoded fluorescent protein (FP)-based biosensors that can sense live-cell redox dynamics18,19. Reversible disulfide bridges have been introduced to the surfaces of a yellow FP (YFP) and green FPs (GFPs), and recently, to the N- and C- termini of a circularly permuted red FP (cpRFP) to derive redox sensitive rxYFP, roGFPs, and rxRFPs20–23. Genetically encoded biosensors (Supplementary Fig. 1) are also available for monitoring various ROS, RNS and RSS such as hydrogen peroxide (H2O2)24–26, organic hydroperoxides (ROOH)27, peroxynitrite (ONOO−)28 and hydrogen sulfide (H2S).29,30 Moreover, rxYFP and roGFPs have been genetically linked to glutaredoxin (Grx), resulting in selective biosensors for the redox dynamics of the oxidized and reduced glutathione (GSSG/GSH) pair31,32. Despite the progress, there is currently no genetically encoded biosensor for probing the redox of the Trx system. Effort has previously been made to fuse rxYFP or roGFPs to Trx but failed to yield any effective biosensor32. The current strategy for probing the redox status of Trx is based on the alkylation of Trx thiols in cell lysates followed by electrophoresis33. This low-throughput method only allows end-point measurements and cannot avoid additional Trx oxidation during sample preparation. The technical difficulty in direct monitoring of the redox status of Trx in live cells has greatly hindered investigations on the physiological and pathological roles of the Trx redox system and the interplays between Trx and other cell signaling components.
Herein we report the first fluorescent probe that can directly monitor the redox status of the active-site cysteines of Trx in live mammalian cells. Our new probe, namely TrxRFP1, was developed by creating and optimizing a redox relay between human Trx1 and rxRFP1—a redox-sensitive red fluorescent protein previously developed in our laboratory22. We have utilized the resultant optimized biosensor, TrxRFP1, to monitor the dynamics of Trx redox induced by various chemicals, such as H2O2, auranofin, and arsenic trioxide. We further localized TrxRFP1 to the cell nucleus and mitochondria, validated TrxRFP1 in a variety of mammalian cell lines, and combined TrxRFP1 with a green fluorescent Grx1-roGFP2 biosensor for simultaneous imaging of Trx and glutathione redox dynamics. We observed Trx redox changes under physiologically relevant conditions, including stimulations by serum or epidermal growth factor (EGF).
RESULTS
Engineering of fluorescent biosensors for Trx redox
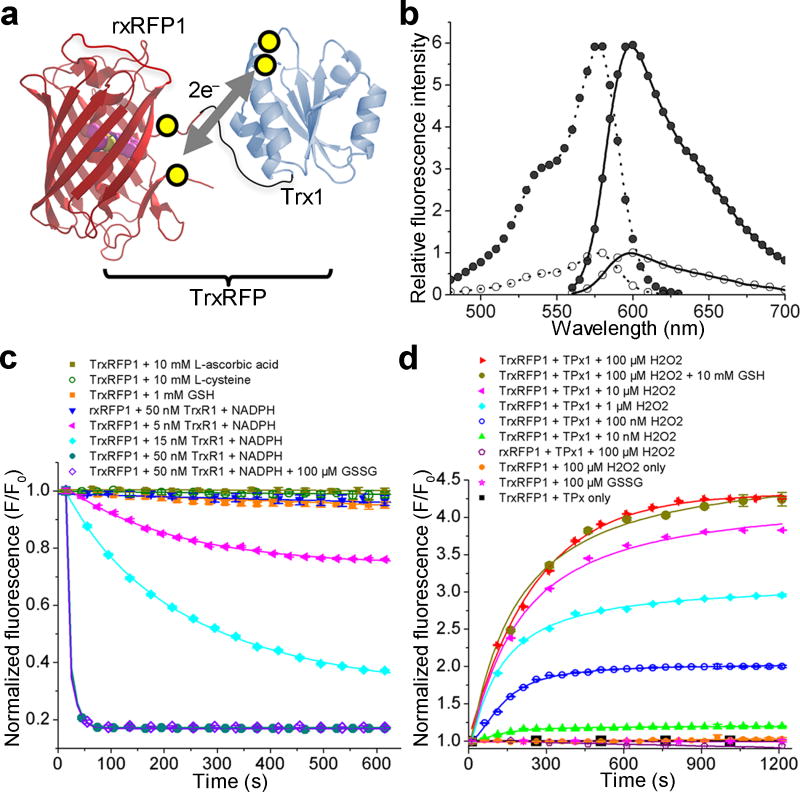
We genetically linked rxRFP1 to Trx1, the human Trx found in cytoplasmic and nuclear compartments. We reasoned that, with an appropriate topological rearrangement, the redox of the active-site cysteine residues (C32 and C35) of Trx1 might be coupled with the redox of the cysteine pair at the termini of rxRFP1 (Fig. 1a). Because Trx1 and rxRFP1 are forced into proximity in the fusion construct, such redox coupling may be kinetically favored. Therefore, the fluorescence of rxRFP1 would become an indicator for the redox of Trx1. We next constructed four chimeric proteins varying in the lengths of the linkers and the relative order of Trx1 and rxRFP1 (Constructs 1–4 in Supplementary Fig. 2). One of the four variants, which has a 30-amino-acid Gly-Ser-rich linker between the C-terminus of Trx1 and the N-terminus of rxRFP1, showed a 4-fold fluorescence change in response to reduction by recombinant human TrxR1 and NADPH (Supplementary Fig. 3). We named this mutant TrxRFP0.1 (Supplementary Fig. 4). Oxidized TrxRFP0.1 was unresponsive to various tested reducing reagents such as L-ascorbic acid and L-cysteine at millimolar concentrations, and reduced glutathione only caused a small, less than 10% fluorescence decrease (Supplementary Fig. 5a). In addition, H2O2 effectively oxidized pre-reduced TrxRFP0.1 in the presence of recombinant human TPx1, leading to a prompt fluorescence increase (Supplementary Fig. 5b). In comparison, rxRFP1 alone in either an oxidized or a reduced state was unresponsive to TrxR1/NADPH and TPx1/H2O2 (Supplementary Fig. 5a,b), further confirming that the observed fluorescence changes of TrxRFP0.1 were caused by the redox of Trx1. Moreover, we mutated each of the two or both active-site cysteine residues of Trx1 to serine and replaced Trx1 in TrxRFP0.1 with one of these three mutants. None of them responded to TrxR1/NADPH or TPx1/H2O2 (Supplementary Fig. 5c,d), supporting that both active-site cysteine residues of Trx1 are directly involved in the redox coupling between Trx1 and rxRFP1.
Figure 1. Design and fluorescence characterization of TrxRFP biosensors.
(a) Schematic representation of the mechanism for TrxRFP sensors. The yellow circles indicate cysteine residues involved in the redox coupling. (b) Excitation (dotted line) and emission (solid line) spectra of reduced (open circle) and oxidized (filled circle) TrxRFP1. Spectra were normalized to the maximal fluorescence of the reduced form. (c) Kinetic traces for the reduction of oxidized proteins. (d) Kinetic traces for the oxidation of reduced proteins. TrxR1 used in these experiments was a commercial rat TrxR1 containing ~ 50% of the full-length selenocysteine (Sec)-containing enzyme.
Trx1 has a total of five cysteine residues. Its three non-active-site cysteine residues (C62, C69, and C73) have been previously reported for regulatory roles and an additional disulfide bond may be formed between C62 and C69 to crosstalk with the glutaredoxin/glutathione system34. To eliminate possible interferences caused by these non-active-site cysteines, we mutated all three to serine to derive TrxRFP0.2 (Supplementary Fig. 2). Next, we randomized the gene fragment encoding rxRFP1 and screened for TrxRFP mutants showing increased dynamic ranges. After three rounds of directed evolution, we arrived at TrxRFP0.9 with three additional mutations (G142S/D228G/V399Q) and a 5.7-fold fluorescence increase from the fully reduced state to the fully oxidized state (Supplementary Fig. 3,4). Compared to TrxRFP0.1, oxidized and reduced TrxRFP0.9 were more reactive toward TrxR1/NADPH and TPx1/H2O2, respectively (Supplementary Fig. 5e,f). Pre-reduced TrxRFP0.9 showed a notable response to as low as 10 nM H2O2 in our TPx1 oxidation assay and remained unreactive to 100 µM H2O2 alone.
In addition to the two cysteine residues forming the reversible disulfide bond, rxRFP1 has two additional cysteine residues. We next created a mutant of TrxRFP0.9 with all nonessential cysteines removed (C217Y/C397S). The C217Y mutation was chosen because mApple35, the parental protein of rxRFP1, has a tyrosine residue at this site, whereas the C397S mutation has previously been shown to have little impact on the spectral and redox properties of rxRFP123. This final mutant was named TrxRFP1 (Supplementary Fig. 4) and subjected to thorough characterization.
Initial characterization of TrxRFP1
Our freshly purified TrxRFP1, which was fully oxidized by molecular oxygen in air during preparation, showed an excitation peak at 576 nm and an emission peak at 600 nm (Fig. 1b), identical to the peaks of rxRFP122. In the presence of dithiothreitol (DTT) or TrxR1/NADPH, TrxRFP1 was reduced quickly to show a nearly 6-fold fluorescence decrease (Fig. 1c, Supplementary Fig. 3, and Supplementary Fig. 6a). The speed and extent of the reduction were dependent of the concentrations of TrxR1. In comparison, rxRFP1 could be reduced by DTT, but not by TrxR1/NADPH. TrxRFP1 inherited excellent specificity from TrxRFP0.1, as millimolar L-cysteine and L-ascorbic acid did not decrease the fluorescence of oxidized TrxRFP1. Interestingly, through this evolution process, the sensitivity of TrxRFP1 to glutathione was further reduced. Millimolar reduced glutathione alone triggered little fluorescence response, and in the presence of 100 µM oxidized glutathione, which is considered an upper concentration limit for typical mammalian cells36, the reduction of TrxRFP1 by TrxR1/NADPH was not impacted (Fig. 1c). The fully reduced TrxRFP1, freshly prepared by DTT reduction and dialysis in a nitrogen (N2)-filled anaerobic chamber, responded to TPx1-catalyzed oxidation by H2O2 (Fig. 1d). The response was dependent of the concentrations of H2O2. In parallel, reduced TrxRFP1 was unreactive to 100 µM H2O2 or oxidized glutathione. In the presence of 10 mM reduced glutathione, oxidation of reduced TrxRFP1 by TPx1/H2O2 was also not affected (Fig. 1d). We further investigated the response of TrxRFP1 to reducing and oxidizing enzymes in the same reaction vessel. As expected, TrxRFP1 reduced by TrxR1/NADPH could be re-oxidized by addition of TPx1/H2O2 (Supplementary Fig. 6b). With the same amount of TrxR1/NADPH, the degree of re-oxidation was dependent of the concentrations of H2O2 and TPx1.
We also tested the responses of TrxRFP1 to Trx-like, redox-sensitive proteins such as glutaredoxin 1 (Grx1) and thioredoxin (Trx)-related protein TRP14. Neither oxidation nor reduction took place (Supplementary Fig. 7), suggesting that the redox states of these proteins are kinetically separated and TrxRFP1 is a specific biosensor. Next, we compared the enzyme kinetics of TrxRFP1 and Trx1 using a commercial rat TrxR1 enzyme (Supplementary Fig. 8). We derived similar Michaelis-Menten constants (KM values, as fit-value ± s.e. of fit, are 2.63±0.49 µM or 2.83±0.69 µM for TrxRFP1 and Trx1, respectively), supporting that TrxRFP1 and Trx1 respond to TrxR1 similarly.
It was reported that the reduction of oxidized Trx1 by glutathione is enzymatically coupled with Grx1, glutathione reductase, and NADPH37, so we further examined our oxidized TrxRFP1 under this condition. In both of our in vitro fluorescence and NADPH absorbance assays, no reaction was detectable for TrxRFP1 (Supplementary Fig. 9). We also created a TrxRFP1 mutant (C32S/C35S/S62C/S69C) in which only the non-active-site disulfide bond can form. This new mutant was reduced under this condition as observed from the NADPH absorbance change at 340 nm. Next, we prepared Trx1 and several Trx1 variants, including Trx1 with an active-site disulfide and three non-active site free thiols, and tested their responses to the glutaredoxin/glutathione reduction (Supplementary Fig. 10). Our results collectively suggest that the glutaredoxin/glutathione system is involved in the reduction of the non-active-site disulfide bond, but not the active-site disulfide bond, of Trx1.
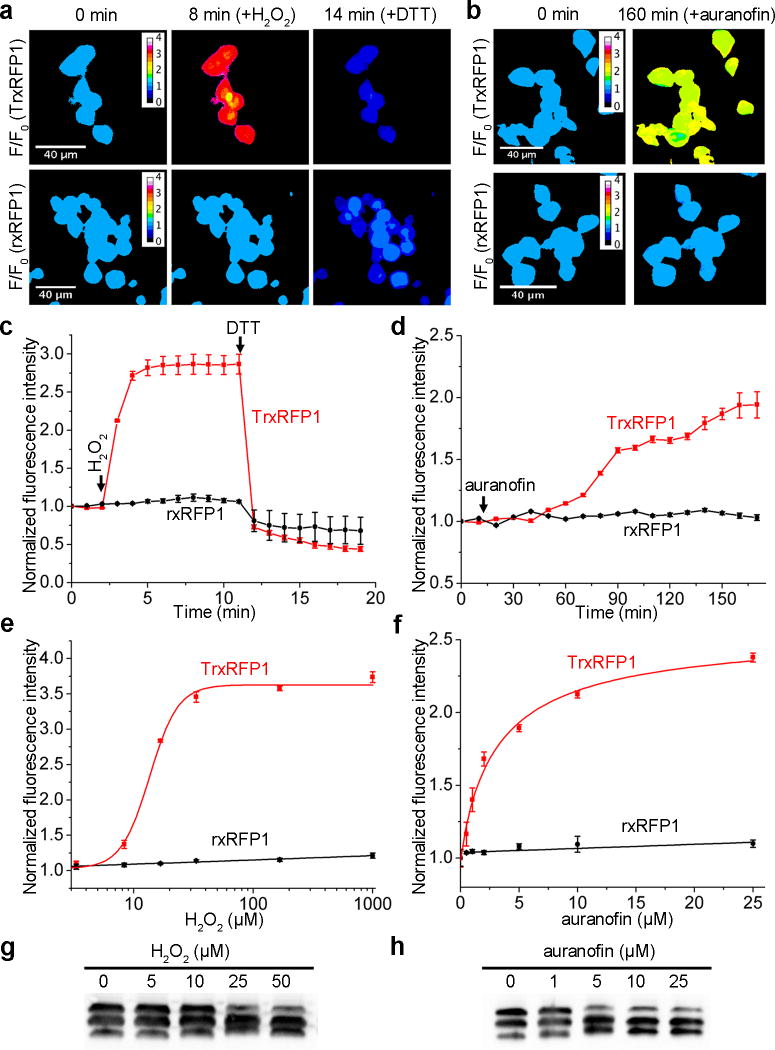
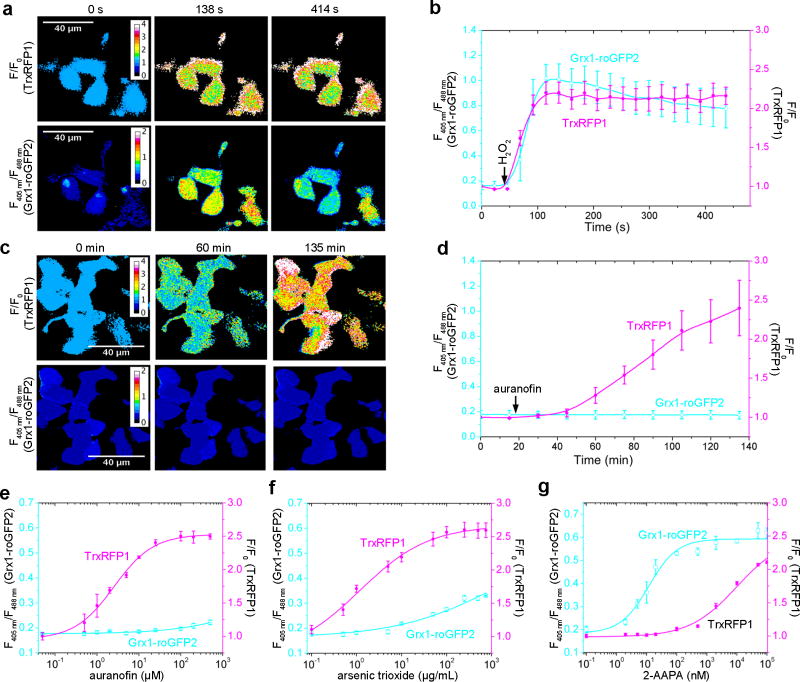
To examine the performance of TrxRFP1 in live cells, we transiently expressed it in human embryonic kidney (HEK) 293T cells. Live mammalian cells express TPx to transform ROS and, at the same time, oxidize Trx (Supplementary Fig. 1). Not surprisingly, addition of a single bolus of 16.7 µM H2O2 to the imaging medium caused an immediate and dramatic fluorescence increase for TrxRFP1-expressing cells (Fig. 2a, Supplementary Fig. 11, and Supplementary Video 1), suggesting the change of Trx to a more oxidized state. The TrxRFP1 sensor remained oxidized during our imaging period until the addition of 10 mM DTT 9 min later. We also imaged H2O2-treated TrxRFP1-expressing cells for a longer period or with less H2O2 and found that the oxidation of TrxRFP1 could be spontaneously reverted (Supplementary Fig. 12), suggesting the presence of an efficient intracellular system to maintain the redox homeostasis of Trx. Furthermore, we treated TrxRFP1-expressing cells with 10 µM auranofin, a previously reported gold(I)-containing TrxR reductase inhibitor38. Like H2O2, auranofin also shifted Trx to a more oxidized state indicated by the gradual fluorescence increase over the monitored period (Fig. 2b, Supplementary Fig. 11, and Supplementary Video 2). In comparison, we did not observe substantial fluorescence increase for H2O2- or auranofin- treated control cells expressing either rxRFP1 or a genetically encoded pH indicator pHRFP22 (Fig. 2c,d and Supplementary Fig. 13).
Figure 2. Characterization of TrxRFP1 in HEK 293T cells.
(a,b) Time-lapse pseudocolored fluorescence images (F/F0) of HEK 293T cells expressing either TrxRFP1 or rxRFP1 sequentially treated with 16.7 µM H2O2 and 10 mM DTT (a) or with 10 µM auranofin (b), showing H2O2- and auranofin- induced fluorescence changes of TrxRFP1 but not rxRFP1. H2O2 and auranofin were added at t = 2 min and t = 12 min, respectively (Scale bar = 40 µm). (c,d) Fluorescence intensity traces for TrxRFP1 or rxRFP1 in HEK 293T. The intensities were normalized to the value at t = 0 min and shown as the mean and s.d. of randomly selected eight cells from three independent replicates. The red and black lines are for TrxRFP1 and rxRFP1, respectively. The arrows indicate the time points for addition of chemicals. (e,f) Fluorescence responses of TrxRFP1 (red) or rxRFP1 (black) in HEK 293T to various concentrations of H2O2 (e) or auranofin (f), suggesting that TrxRFP1 can selectively sense the redox changes of Trx in live cells. Data are shown as mean and s.d. of three independent experiments. (g,h) Redox urea-PAGE and immunoblotting analysis of TrxRFP1 in HEK 293T cells treated with H2O2 (g) or auranofin (h) at the indicated concentrations, showing the increase of protein oxidation in response to the increase of H2O2 or auranofin (Full gel for 2g is in Supplementary Fig. 21d. The full gel for 2h is in Supplementary Fig. 21e). The three bands from top to bottom are interpreted as the TrxRFP1 protein containing no, one, and two disulfide bonds, respectively.
To further explore the sensitivity of TrxRFP1 in live cells, we characterized the responses of TrxRFP1 to various concentrations of H2O2 and auranofin (Fig. 2e,f). The EC50’s (the concentration to give a half-maximal response) for 10-min H2O2 treatment and 3-h auranofin treatment were 13.5±0.7 µM (fit-value ± s.e. of fit) and 3.2±1.1 µM, respectively. In the tested concentration ranges, H2O2 and auranofin did not appreciably increase the fluorescence of rxRFP1-expressing HEK 293T cells. We further performed redox urea-polyacrylamide gel electrophoresis (PAGE) and immunoblotting analysis34 of TrxRFP1 in HEK 293T cells treated with different concentrations of H2O2 or auranofin. We treat cells with neutral alkylating agent iodoacetamide, subsequently reduced cell lysates, and further treat the lysates with acidic alkylating agent iodoacetic acid. We were able to separate the TrxRFP1 protein based on its redox states. As the concentration of H2O2 or auranofin increases, the oxidation of TrxRFP1 increases (Fig. 2g,h), corroborating our measurements based on the fluorescence of TrxRFP1. These results further confirm that TrxRFP1 is a selective indicator for Trx redox and the engineered redox relay in TrxRFP1 is necessary for sensing Trx redox changes.
It is a general concern that the use of redox sensors in live cells may inevitably perturb intrinsic physiology. Expression of TrxRFP1 may result in the additional Trx1 domain and rxRFP1 which also contains cysteines. Fortunately, Trx1 is a highly abundant protein in mammalian cells39, whereas cysteines in rxRFP1 are not very reactive because rxRFP1 alone does not respond to micromolar and even low millimolar H2O2 in vitro and in live cells (Fig. 1d,2e). We assessed the effect of TrxRFP1 expression in mammalian cells, in terms of cell viabilities, in response to H2O2 and auranofin, which are both modulators of the thioredoxin redox system. After 24-h incubation with H2O2 or auranofin, we did not observe any substantial difference for the viabilities of mammalian cell lines (e.g. HEK 293T, HeLa, MCF-7, and SW620) with or without TrxRFP1 overexpression (Supplementary Fig. 14). We also compared the proliferation of HEK 293T cells expressing or not expressing TrxRFP1 and observed no difference (Supplementary Fig. 15).
Localization of TrxRFP1 to subcellular compartments
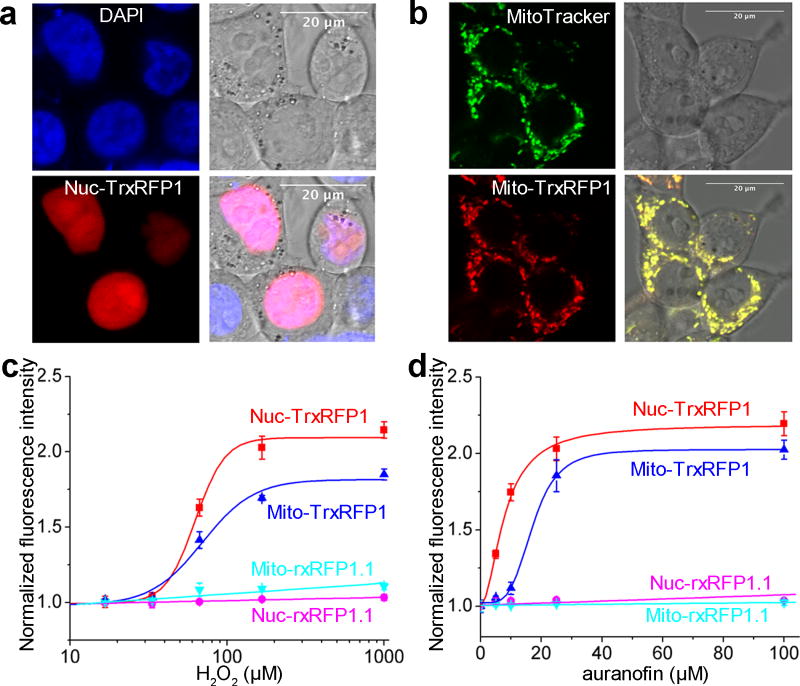
In addition to the cytosol, the Trx redox system exists in the nucleus and mitochondria of mammalian cells, we therefore subcellularly localized TrxRFP1 to monitor compartmentalized Trx redox dynamics. We appended a nuclear localization sequence (NLS) or a mitochondrial targeting sequence (MTS) to TrxRFP1. This led to the proper localization of TrxRFP1 in either the mammalian cell nucleus or mitochondria, as confirmed by the co-localization of TrxRFP1 with a nuclear stain DAPI (Fig. 3a) or a mitochondrial stain MitoTracker Green (Fig. 3b). Treating the cells expressing nuclear or mitochondrial TrxRFP1 with H2O2 or auranofin resulted in robust fluorescence increases (Supplementary Fig. 16). However, both nuclear and mitochondrial TrxRFP1 are less sensitive to exogenous H2O2 and auranofin than unlocalized TrxRFP1, as higher concentrations are needed to trigger comparable fluorescence responses (Fig. 3c,d). In particular, the EC50’s for 10-min H2O2 treatment were 62.6±3.6 µM (fit-value ± s.e. of fit) and 69.6±9.7 µM for nuclear and mitochondrial TrxRFP1, respectively. The EC50’s for 3-h auranofin treatment were 7.9±0.8 and 17.1±1.2 µM for nuclear and mitochondrial TrxRFP1, respectively. These observations are reasonable, since exogenous H2O2 and auranofin have to cross cellular space and layers of lipid membranes to reach the nuclear or mitochondrial Trx redox system. Moreover, compared to the cytosol, the cell nucleus and mitochondria may possess sophisticated systems to quickly degrade ROS. We utilized a HyPer3 biosensor40 to directly monitor cytosolic and nuclear H2O2 and more robust fluorescence changes were observed for cytosolic HyPer3 than nuclear HyPer3 (Supplementary Fig. 17), suggesting that the effective concentrations of H2O2 in the cytosol are indeed higher than in the nucleus upon extracellular addition of H2O2. Furthermore, the Trx redox system in mammalian mitochondria is based on Trx2 and TrxR241, which is homologous to, but distinct from the cytosolic and nuclear Trx redox system based on Trx1 and TrxR1. Therefore, although our Trx1-based TrxRFP1 biosensor displayed cross-reactivity as expected42, it may not have the optimal kinetics to sense the redox of the mitochondrial Trx system.
Figure 3. Subcellularly localized TrxRFP1.
(a,b) Co-localization of nuclear (a) and mitochondrial (b) TrxRFP1 with a nuclear stain DAPI and a mitochondrial stain MitoTracker Green, respectively (Scale bar = 20 µm). (c,d) Fluorescence responses of nuclear (red) and mitochondrial (blue) TrxRFP1 in HEK 293T to various concentrations of H2O2 (c) or auranofin (d), suggesting that TrxRFP1 can selectively sense the subcellular redox changes of Trx in live cells. Fluorescence responses of nuclear (cyan) and mitochondrial (magenta) rxRFP1.1 are also shown as the controls. Data are represented as mean and s.d. of three independent experiments.
Chemical-induced Trx redox dynamics in mammalian cells
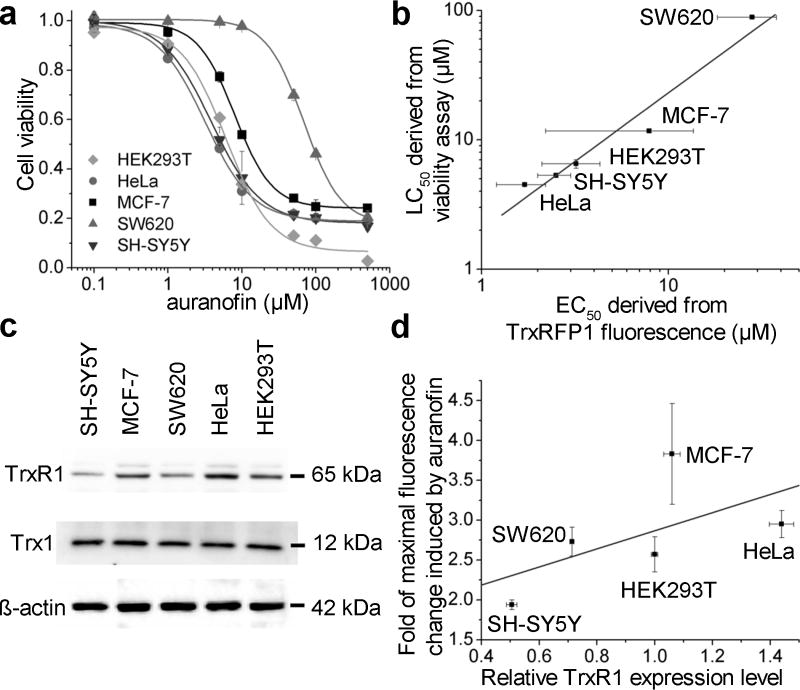
To demonstrate the general applicability of TrxRFP1, we expressed TrxRFP1 in several additional mammalian cell lines, including cervical cancer HeLa cells, breast cancer MCF-7 cells, colon carcinoma SW620 cells, and neuroblastoma SH-SY5Y cells. Fluorescence responses to H2O2- and auranofin- induced Trx oxidation were observed in all conditions (Supplementary Fig. 18, Supplementary Fig. 19, and Supplementary Videos 3,4). We further compared the responses of TrxRFP1 in different cell lines to various concentrations of auranofin after a 3-h incubation period. Each showed different sensitivity to auranofin with an EC50 of 1.7±0.5 µM (fit-value ± s.e. of fit) for HeLa, 7.9±5.7 µM for MCF-7, 28.1±9.8 µM for SW620, and 2.5±0.5 µM for SH-SY5Y (Supplementary Fig. 18). Moreover, the magnitudes of their fluorescence responses were different with approximately 2.95-fold, 3.83-fold, 2.73-fold, 1.94-fold maximal changes for HeLa, MCF-7, SW620, and SH-SY5Y, respectively. To understand the biological meanings of the differences across cell lines, we further determined the viability responses of various cell lines to auranofin after 24-h incubation. The LC50’s (the concentration to kill half cells) were determined to be ~ 6.5, 4.5, 11.7, 88.4, 5.3 µM for HEK 293T, HeLa, MCF-7, SW620, and SH-SY5Y, respectively (Fig. 4a). These LC50 values roughly correlate with the EC50 values derived from the fluorescence measurements of TrxRFP1 (Fig. 4b), suggesting that TrxR is indeed a cellular target of auranofin to cause cell growth inhibition and death. The differences in EC50 and LC50 values across various cell lines were likely due to differences in bioavailability and drug permeability43. On the other hand, it is difficult to rationalize the differences in the magnitudes of auranofin-induced fluorescence changes across various cell lines. We probed the expression levels of Trx1 and TrxR1 in various cell types (Fig. 4c), and did not observe much difference for the expression of Trx1 but observed dramatic differences for TrxR1 across various cell lines. SH-SY5Y, which has a low level of TrxR1, indeed showed a relatively small fluorescence change in response to high concentrations of auranofin, suggesting that Trx1 in untreated SH-SY5Y is likely more oxidized than Trx1 in other tested cell lines. However, when all tested cell lines are considered together, the correlation between the maximal fluorescence changes and the TrxR1 levels is weak (Fig. 4d). We reason that the redox of Trx1 is determined by the interplay between Trx1 oxidation and reduction and that the TrxR1 expression level is not the only factor. Moreover, intracellular TrxRFP1 was not fully oxidized by high concentrations of auranofin (Fig. 2h), suggesting that cells have complicated systems to regulate Trx redox in response to auranofin and such systems may be different in various types of cells. This may also contribute to the observed differences in TrxRFP1 responses across various cell lines.
Figure 4. Use of TrxRFP1 in various mammalian cell lines.
(a) The viabilities of indicated cell lines in response to 24-h auranofin treatment. (b) The correlation between EC50 values derived from TrxRFP1 fluorescence and LC50 values derived from viability assays of various cell lines (R2 = 0.95). (c) Western blots of endogenous TrxR1, Trx1 and β-actin in various cell lines (Full gel is in Supplementary Fig. 21a–c). (d) A plot for relative TrxR1 expression levels and the fold of fluorescence changes induced by auranofin across various cell lines (R2 = 0.35). Data in panel a are shown as mean and s.d. of three independent experiments. Error bars in panels b and d are s.e.m. from curve fitting or s.d. from quantification of Western blot bands of three independent replicates.
Simultaneous monitoring of Trx and glutathione redox
TrxRFP1 is a red fluorescent biosensor spectrally orthogonal to common green fluorescent biosensors. A green-fluorescent excitation-ratiometric Grx1-roGFP2 biosensor was previously reported for monitoring glutathione redox dynamics in mammalian cells32. Since glutathione and Trx are the two major thiol-dependent antioxidants in mammals, we next examined the use of TrxRFP1 and Grx1-roGFP2 to simultaneously monitor the two redox systems. We expressed both probes in HEK 293T cells and treated the cells with H2O2. Because H2O2 is coupled to the redox of both glutathione and Trx (Supplementary Fig. 1), we observed concurrent fluorescence changes for both biosensors (Fig. 5a,b and Supplementary Video 5). We next treated cells co-expressing the two biosensors with auranofin, a small molecule preferably targeting TrxR. A prominent fluorescent change was observed for TrxRFP1 but not for Grx1-roGFP2 (Fig. 5c,d and Supplementary Video 6). We further examined the responses of the cells co-expressing the two biosensors to various concentrations of auranofin, arsenic trioxide, and 2-AAPA (Fig. 5e–g). No change in Grx1-roGFP2 fluorescence was observed for a wide range of auranofin concentrations (Fig. 5e), further supporting that auranofin is a selective inhibitor of TrxR over the glutathione redox system. Arsenic trioxide also preferably triggered the response of TrxRFP1, but at high concentrations (> 10 µg/ml), the fluorescence of Grx1-roGFP2, represented as fluorescence excitation ratios (F405nm/F485nm), also started to change (Fig. 5f). This result corroborates the previous finding that TrxR is a target for arsenic trioxide44. 2-AAPA is a glutathione reductase inhibitor45. At low nanomolar concentrations, it induced the oxidation of Grx1-roGFP2, but not TrxRFP1 (Fig. 5g). When its concentration increased above 100 nM, the fluorescence of TrxRFP1 started to rise, indicating the oxidation of both Trx and glutathione induced by high concentrations of 2-AAPA. All these results collectively suggest that TrxRFP1 and Grx1-roGFP2 can be utilized simultaneously to monitor the redox dynamics of Trx and glutathione in mammalian cells, that the Trx and glutathione redox systems can be individually perturbed, and that a thermodynamic equilibrium for Trx1 and glutathione is not reached in live mammalian cells.
Figure 5. Simultaneous monitoring of thioredoxin and glutathione redox dynamics using TrxRFP1 and Grx1-roGFP2.
(a, c) Time-lapse pseudocolored fluorescence images of HEK 293T cells expressing both TrxRFP1 and Grx1-roGFP2 treated with 13.3 µM H2O2 (a) or 15 µM auranofin (c), indicating that H2O2 induces changes in both thioredoxin and glutathione redox systems, whereas auranofin induces the oxidation of thioredoxin but not glutathione (Scale bar = 40 µm). In the top row are pseudocolored ratiometric images (F/F0) for TrxRFP1, and in the bottom row are pseudocolored ratiometric images (405 nm excitation/488 nm excitation) for Grx1-roGFP2. H2O2 and auranofin were added at t = 46 s and t =16 min, respectively. (b,d) Ratio traces for TrxRFP1 or and Grx1-roGFP2 in HEK 293T cells in panels a and c, shown as mean and s.d. of six individual cells from three independent replicates. The magenta and cyan lines are for TrxRFP1 and Grx1-roGFP2, respectively. The arrows indicate the time points for addition of H2O2 or auranofin. (e–g) Fluorescence responses of TrxRFP1 (red) and Grx1-roGFP2 in HEK 293T to various concentrations of auranofin (e), arsenic trioxide (f), or 2-AAPA (g), suggesting that the Trx redox system and the glutathione redox system can be individually perturbed. Data are shown as mean and s.d. of three independent experiments.
Responses of TrxRFP1 to physiological stimuli
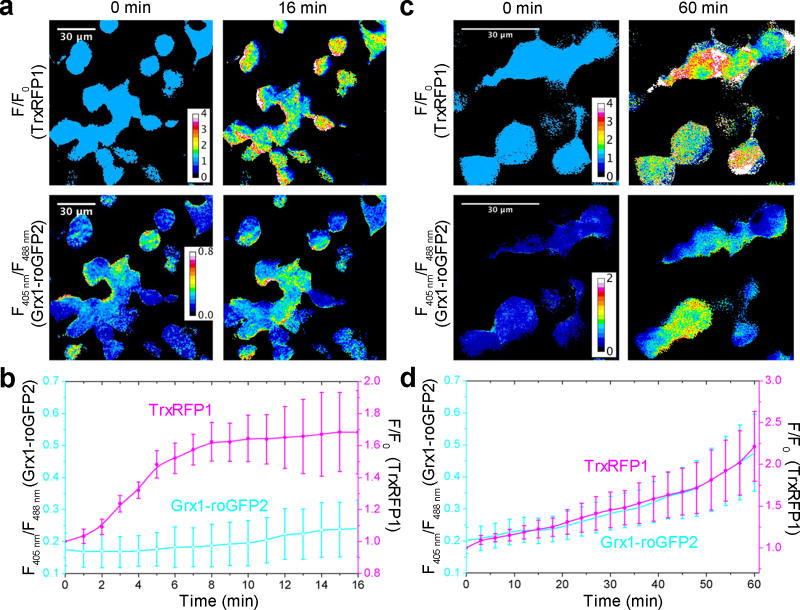
The sensitivity of TrxRFP1 to nanomolar H2O2 in vitro suggests that the Trx system in live cells could be oxidized by low concentrations of H2O2 generated under physiologically relevant conditions. To test this, we treated HEK 293T cells co-expressing TrxRFP1 and Grx1-roGFP2 with 10% fetal bovine serum (FBS) post a 6-h serum starvation. FBS contains a mixture of growth factors and hormones, which are expected to stimulate various signaling pathways. A robust oxidation of Trx1 was observed, as the fluorescence of TrxRFP1 increased rapidly in the first 10 min post stimulation (Fig. 6a,b). In contrast, we only observed a very small fluorescence change for Grx1-roGFP2. We further tested the response of HEK 293T cells to epidermal growth factor (EGF), which is known to induce the biological production of H2O246. We observed fluorescence changes for both TrxRFP1 and Grx1-roGFP2, suggesting the oxidation of both Trx1 and glutathione under this condition (Fig. 6c,d and Supplementary Video 7). In comparison to FBS stimulation after serum starvation, the EGF-induced TrxRFP1 response was relatively slow. It is not yet possible for us to fully rationalize the observations. Potentially, these different processes can activate different compartmentalized sources of H2O2 or induce different antioxidant systems. These results, however, at least support that TrxRFP1 could detect Trx redox changes in live cells under physiologically relevant conditions. We further repeated our serum and EGF stimulation experiments with control HEK 293T cells expressing the pH indicator pHRFP22. None of these conditions increased the fluorescence of pHRFP (Supplementary Fig. 20), suggesting that the observed fluorescence increase of TrxRFP1 was not caused by pH changes.
Figure 6. Responses of TrxRFP1 to physiological stimuli in HEK 293T cells.
(a, b) Time-lapse responses of TrxRFP1 and Grx1-roGFP2 to serum stimulation at t = 0 min, indicating a robust oxidation of Trx (Scale bar = 30 µm). Prior to the 10% FBS treatment, cells were subjected to 6-h serum starvation. (c, d) Time-lapse responses of TrxRFP1 and Grx1-roGFP2 to epidermal growth factor (EGF, 500 ng/mL) treatment at t = 0 min, indicating the oxidation of both Trx and glutathione (Scale bar = 30 µm). Pseudocolored ratiometric images (F/F0) for TrxRFP1 and pseudocolored ratiometric images (405 nm excitation/488 nm excitation) for Grx1-roGFP2 are shown in panels a and c. Ratio traces for TrxRFP1 or and Grx1-roGFP2 are presented in panels b and d as mean and s.d. of eight individual cells from three independent replicates. The magenta and cyan lines are for TrxRFP1 and Grx1-roGFP2, respectively.
DISCUSSION
Trx is a cellular protein containing highly reactive cysteines important for redox homeostasis and signaling. Prior to our work, there were no genetically encoded biosensors for monitoring Trx redox in live cells. To develop a selective biosensor for Trx redox, we linked a redox sensitive rxRFP1 protein with Trx1. A similar fusion strategy based on roGFPs and rxYFP has been previously utilized to create biosensors for glutathione and H2O225,32, but it failed for Trx17,32,47. Different from roGFPs and rxYFP, rxRFP1 adopts a circularly permuted topology to allow a new fusion geometry. This indeed resulted in novel biosensors in which the disulfide exchange between rxRFP1 and Trx1 is possible. We localized TrxRFP1 to the cell nucleus and mitochondria and successfully monitored compartmentalized Trx redox dynamics in live mammalian cells. We also utilized TrxRFP1 to monitor Trx redox dynamics induced by H2O2 and auranofin in various cell lines. In particular, the fluorescence responses of TrxRFP1 to 3-h auranofin treatment were indicative of cell viabilities determined post 24-h auranofin treatment. Such correlation is due to the fact that TrxR is a major cellular target of auranofin. On the basis of this, our TrxRFP1 biosensor may be used for mechanistic studies on the interactions between interested molecules and the Trx redox system. It may also be utilized in high-throughput screening assays to identify selective molecular modulators of the Trx redox system. Considering the importance of the Trx system and its implications in various diseases, TrxRFP1 may open the door for a large array of exciting studies.
Trx1 is typically considered as a reductant for oxidized cysteines in proteins1. It was not surprising that the fluorescence of fully oxidized TrxRFP1 decreased quickly in response to TrxR/NADPH, because the disulfide in Trx1 reduced by TrxR/NaDPH can subsequently exchange with the disulfide in rxRFP1. Furthermore, fully reduced TrxRFP1 responded to TPx/H2O2 whereas rxRFP1 alone was unresponsive, suggesting that Trx1 oxidized by TPx/H2O2 can further oxidize the two cysteine residues in reduced rxRFP1. Chemically, such reversibility is not surprising because oxidation is an exact reverse of the reduction reaction. Biologically, the result indicates that the reversibility may also be important in vivo and Trx may be able to modulate the redox of its interacting proteins in both directions.
The red fluorescent color of TrxRFP1 makes it highly attractive. Not only because it enhances tissue penetration and minimizes phototoxicity, but also TrxRFP1 can be paired with many existing GFP-based biosensors for monitoring more than one parameters. We have demonstrated the use of TrxRFP1 and Grx1-roGFP2 to simultaneously monitor glutathione and Trx redox dynamics in live cells. Furthermore, we show that the redox of glutathione and the Trx1 active site can be individually perturbed without shifting the other. This aligns with our in vitro enzymes assays, which indicate that the glutaredoxin/glutathione system reduces the non-active site disulfide bond, but not the active-site disulfide bond, of Trx1. This finding also corroborates the emerging perspective that various cellular redox couples are quasi-independent from each other and a thermodynamic equilibrium is not reached in live cells48.
The specificity of TrxRFP1 is partially due to the low reactivity of rxRFP1 toward H2O2 and its slow reaction with the glutathione couple. There is essentially no response for rxRFP1 to micromolar and low millimolar H2O222. In contrast, roGFP2 is more reactive toward H2O225,32. Therefore, rxRFP1 is a more preferable scaffold than roGFP2 for construction of redox-relay based biosensors. This unique circularly permuted topology of rxRFP1 may also be advantageous, as shown in this study. Moreover, despite the high specificity of TrxRFP1, rxRFP1 serves as an excellent control to further confirm the connection between the observed fluorescence changes of TrxRFP1 and the redox dynamics of Trx.
Human expresses several Trx isozymes. Although we demonstrated that the Trx1-based TrxRFP1 biosensor could be utilized to monitor the dynamic of the mitochondrial Trx redox system, a better mitochondrial biosensor may be engineered from Trx2 and rxRFP1. Furthermore, although the data presented here were mainly based on experiments in live human cell lines, TrxRFP1 or similar biosensor may be applicable in other mammalian organisms, other eukaryotic systems, or even prokaryotic cells, because of the strong homology between the Trx redox systems across species. There is significant interest to monitor Trx redox dynamics in pathogenic bacteria or parasites, because inhibition of bacterial or parasitic thioredoxin reductase has been consider as new therapeutic mechanisms10,49. This work may be extended to these studies.
In conclusion, by engineering a redox relay between Trx1 and rxRFP1, we developed the first genetically encoded fluorescent biosensor that allows the observation of real-time dynamics of the Trx redox system in live mammalian cells. We further combined the resultant TrxRFP1 sensor with a green fluorescent Grx1-roGFP2 biosensor to simultaneously monitor Trx and glutathione redox dynamics in live cells in response to chemical and physiologically relevant stimuli. With these new capabilities, a large array of new studies will be enabled to further our understanding of redox biology.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
ONLINE METHODS
Reagents and general methods
The human Trx1 gene fragment and all other synthetic DNA oligonucleotides were purchased from Integrated DNA Technologies. The genes for human TrxR1 and TPx1 were purchased from DNASU Plasmid Repository. Restriction endonucleases were purchased from Thermo Scientific Fermentas. Accura high-fidelity DNA polymerase and EconoTaq DNA polymerase were purchased from Lucigen. Products of PCR and restriction digestion were purified using gel electrophoresis and Syd Laboratories Gel Extraction columns. Plasmid DNA was purified using Syd Laboratories Miniprep columns. DNA sequences were analyzed by Retrogen. All other chemicals were purchased from Sigma-Aldrich, Fisher Scientific, and VWR, unless otherwise noted. Anti-human Trx1 antibody (Cat # 11538) and recombinant rat TrxR1 (Cat # 14638) were purchased from Cayman Chemical. Anti-human/mouse/rat TrxR1 antibody (Cat # MAB7428-SP) was purchased from R&D Systems. Secondary antibodies and the anti-β-actin antibody (Cat # sc-130656) were purchased from Santa Cruz Biotechnology. Human recombinant EGF (Cat # 354052) was purchased from Corning. Glutathione reductase (Cat # G3664) was purchased from Sigma-Aldrich.
Plasmid construction
To build TrxRFP0.1, oligonucleotide Trx-F was paired with 0.1-R1, 0.1-R2, 0.1-R3, or 0.1-R4 (Supplementary Table 1) stepwise in four individual polymerase chain reactions (PCR) using a purchased human Trx1 gene as the template in the first reaction and the products of previous reactions as the templates in the subsequent reactions. Oligonucleotides 0.1-F and pBAD-R were used to amplify an rxRFP1 gene fragment from a previously reported pBAD-rxRFP1 plasmid22. Next, an overlap PCR reaction was performed to assemble the two fragments using Trx-F and pBAD-R to afford a full-length TrxRFP0.1 gene. TrxRFP0.2, TrxRFP0.3, TrxRFP0.4 were constructed similarly using Trx-F/2-R and 2-F/pBAD-R, Trx-F/3-R and 3-F/TrxRFP3-R, and Trx-F/4-R and 4-F/pBAD-R oligonucleotide pairs, respectively. To introduce the C32S mutation to TrxRFP0.1, pBAD-F/C32S-R and C32S-F/pBAD-R were used to amplify two fragments from TrxRFP 0.1, and subsequently, an overlap PCR reaction was performed using pBAD-F and pBAD-R. Similarly, the C35S mutant and the C32S/ C35S double mutant of TrxRFP0.1 were made using C35S-F, C35S-R, C32C35S-F and C32C35S-R. To construct TrxRFP0.2, we utilized Trx-F/C62S-R, or C69/73S-F/pBAD-R to amplify two fragments from TrxRFP 0.1, which were next linked together by using an overlap PCR reaction with Trx-F and pBAD-R. To introduce the C217Y mutation to TrxRFP0.9, we utilized a similar overlap PCR strategy based on Trx-F, C217S-R, C217S-F and pBAD-R. Because C397 is close to the N-terminus, C397-R was used to replace pBAD-R to introduce the C397S mutation. To create the TrxRFP1SSCC (C32S/C35S/S62C/S69C) mutant, oligonucleotide pairs pBAD-F and 32/35S-R, 32/35S-F and 62/69C-R, and 62/69C-F and pBAD-R were utilized to amplify three individual fragments from TrxRFP1, which were next assembled in an overlap PCR with pBAD-F and pBAD-R. To construct the plasmid for bacterial expression of human TrxR1, a purchased gene was amplified with oligonucleotides TRND-F and TRND-R. Similarly, PRX-F and PRX-R were used to amplify the gene of human TPx1. To construct the plasmids for human Trx1, Trx1CCSS, and Trx1SSCC, we used Trx-F and Trx-R to amplify the corresponding gene fragments from TrxRFP0.1, TrxRFP1 and TrxRFPSSCC, respectively. To construct the plasmid for bacterial expression of human Grx1, Grx-F and Grx-R were used to amplify the gene from a HEK 293 cDNA library (Biosettia). All aforementioned assembled or amplified gene fragments were digested with Xho I and Hind III, and ligated into a pre-treated compatible pBAD/His B plasmid (Life Technologies). Oligonucleotides CMV-F1 and CMV-TrxRFP1-R were used to amplify the gene of TrxRFP1 from its corresponding pBAD plasmid. To construct a plasmid for nuclear expression of TrxRFP1, we used CMV-F1, Nuc-R1, Nuc-R2, and Nuc-R3 to amplify the gene of TrxRFP1 and further append three repeats of a nuclear localization sequence (DPKKKRKV). To construct the plasmid for mitochondrial expression of TrxRFP1, oligonucleotides Mito-F and Mito-TrxRFP-R were used amplify a mitochondrial targeting sequence (MLSLRQSIRFFKPATRTLCSSRYLL) from pMito-rxRFP1.123, and Mito-TrxRFP-F and CMV-TrxRFP1-R were used amplify the TrxRFP1 fragment. The products were assembled in an overlap PCR with Mito-F and CMV-TrxRFP1-R. These assembled or amplified gene fragments were next digested with Hind III and Xba I, and ligated into a pre-digested pcDNA3 plasmid (Life Technologies). To add a nuclear exclusion sequence (NES) to TrxRFP1, NES-F1 and NES-F2 were paired with NES-R for PCR in two steps and the resultant gene was digested with BamH I and Xba I, and ligated into a pre-digested pNES-rxRFP1 plasmid23. Using similar procedures, we created plasmids for the cytosolic or nuclear expression of HyPer3 from a synthetic HyPer3 gene fragment and oligonucleotides HyPer-NES-F, HyPer-NES-R, HyPer-F, HyPer-Nuc-R1, Nuc-R2 and Nuc-R3. All ligation products were used to transform DH10B Escherichia coli cells, which were next plated on LB agar plates supplemented with ampicillin (100 µg/mL). Individual colonies were used to seed liquid culture for preparation of DNA. All constructs were confirmed by DNA sequencing.
Random mutagenesis and library screening
Randomization was limited to the gene fragment encoding the GS-rich linker and rxRFP1. First, rxRFP0.2 was amplified with oligonucleotides Trx-F/0.2-R and 0.2_F/pBAD_R. The fragments were digested with Xho I and BamH I, and BamH I/Hind III, respectively, and ligated into a pre-treated compatible pBAD/His B plasmid. This procedure inserted a new BamH I restriction site downstream to Trx1 without change the protein sequence. Error-prone PCR reactions were next carried out with oligonucleotides 0.2_F and pBAD_R according to a reported procedure50. The DNA was digested with BamH I and Hind III and ligated into the pre-digested, aforementioned plasmid. The resultant gene library was introduced into DH10B cells and plated onto LB agar plate supplemented with 100 µg/mL ampicillin and 0.1% (w/v) l-arabinose. Colonies were imaged using a previously described setup22. Fluorescence before and after spraying of DTT (50 mM) was quantified and the fluorescence ratios of individual colonies were evaluated using the Fiji image analysis software51. Colonies showing large fluorescence changes were selected and cultured in 96-well plates containing 2YT medium supplemented with 100 µg/mL ampicillin and 0.2% (w/v) l-arabinose. Bacterial cells were first shaken at 37°C and 250 rpm for 24 h, and next, at room temperature for another 12 h. Cells were pelleted and lysed using B-PER Bacterial Protein Extraction Reagents (Pierce). Cell debris was removed by centrifugation. Fluorescence emission of clear cell lysates was quantified on a BioTek Synergy Mx Microplate Reader before and after treatments of TrxR1 (10 µM)/NADPH (200 µM). Three clones with largest dynamic ranges were selected and corresponding plasmids were minipreped. Their mixture was used as the template for the next round of random mutagenesis.
Protein expression and purification
All proteins were expressed and purified as previously described22. The purities of the prepared proteins were analyzed by SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis, see Supplementary Fig. 21). To prepare reduced sensor proteins or the reduced forms of Grx1 and TRP-14, we incubated the proteins with 100 molar equivalent of DTT at room temperature overnight, which were next dialyzed into 1× phosphate buffered saline (PBS) using Thermo Scientific Snakeskin dialysis tubing (7k molecular weight cutoff) in an N2-filled anaerobic chamber. Purified recombinant TrxR1 and TPx1 proteins were also buffer-exchanged into 1× PBS, aliquoted to 10 µL each at 50 µM, and stored at −80°C for long-term use.
In vitro characterization
Protein concentrations were determined using the Bradford assay. The fluorescence of proteins was measured using a BioTek Synergy Mx Microplate Reader with the excitation wavelength at 540 nm and the emission wavelength at 600 nm. Sensor proteins were diluted to desired concentrations with 1× PBS (pH 7.4). Reduction and oxidation kinetics was monitored at room temperature. The wild-type human TrxR1 has a selenocysteine (Sec) residue encoded by a TGA codon upstream to a Sec insertion sequence (SECIS). In our plasmid for the expression of TrxR1, the Sec residue has been replaced with Cys to afford a mutant retaining partial activity52. We used this TrxR1 mutant or a commercial rat TrxR1 (Cayman Chemical) that contains ~ 50% of the full-length Sec-containing protein in our in vitro assays. Unless otherwise stated, oxidized sensor proteins (1 µM) were reduced with our prepared recombinant TrxR1 (10 µM) or commercial rat TrxR1 (50 nM) and NADPH (200 µM) or GSH; reduced sensor proteins (0.5 µM) were mixed with our purified recombinant TPx1 (0.5 µM) and H2O2 at desired concentrations. For reduction and re-oxidation in the same reaction vessel, oxidized TrxRFP1 (1 µM) was first reduced with our prepared TrxR1 (10 µM) and NADPH (200 µM), and next, TPx1 and H2O2 at the indicated concentrations were added. For absorbance based kinetic assays, TrxRFP1 (60 µM) or TrxRFP1sscc (60 µM) were treated with either NADPH (200 µM) and TrxR (10 µM), or a mixture containing NADPH (200 µM), glutathione reductase (45 nM), Grx1 (5 µM), and GSH (10 mM). In our effort to re-examine the previously reported result37, different Trx1 mutants (20 µM) were treated with a mixture containing NADPH (200 µM), glutathione reductase (60 nM), Grx1 (1 µM), and GSH (10 mM). The consumption of NADPH was determined by monitoring the absorbance at 340 nm. To determine the KM values of TrxRFP1 and Trx1 for rat TrxR1, we conducted assays at room temperature with reaction mixtures consisting of 10 nM rat TrxR, 2 mM EDTA, 200 µM NADPH, and 100 mM sodium phosphate (pH 7.4). The concentrations of Trx1 or TrxRFP1 varied from 1 µM to 50 µM. NADPH oxidation was spectrophotometrically monitored at 340 nm to determine reaction rates. KM values were obtained by fitting the data with the Michaelis-Menten equation.
Cell culture and live-cell imaging
HEK 293T, SW620, MCF-7, and HeLa cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) FBS, and SH-SY5Y cells were cultured in DMEM/F12 (1:1) supplemented with 10% (v/v) FBS. All cells were kept at 37°C in humidified air containing 5% CO2. Cells were seeded and transfected as previously described22, and analyzed 72 h post transfection. Cells were rinsed twice with Dulbecco’ s Phosphate Buffered Saline (DPBS), and left in the imaging medium containing DPBS supplemented with 1 mM Ca2+, 1 mM Mg2+ and 1% (w/v) BSA. For the serum stimulation experiment, cells were first rinsed three times with DMEM containing no FBS, maintained in DMEM containing no FBS for 6 h at 37°C, and next stimulated by adding 10% (v/v) FBS. All single-color images were taken with a Motic AE31 inverted epi-fluorescence microscope as previously described22. A Leica SP5 confocal microscope was used to simultaneously image TrxRFP1 and Grx1-roGFP2. TrxRFP1 was excited with a 543 nm HeNe laser, and the red fluorescence emission was collected between 580–630 nm with a hybrid detector (HyD). Grx1-roGFP2 was excited with a 405 nm UV laser and a 488-nm argon-ion laser sequentially, and the green fluorescence emission was collected between 500–530 nm with a PMT (photomultiplier tube) detector. Time-lapse series was acquired at the indicated interval and fluorescence emissions from all channels were scanned sequentially with the “between frames” mode. All images were analyzed using the Fiji image analysis software51 and the ratios of the images were processed according to a published procedure53.
Cell responses to chemical treatments, and cell viability and proliferation assays
To study the responses of cells expressing fluorescent biosensors to H2O2, transfected cells were rinsed twice with DPBS and left in the abovementioned imaging medium. Fluorescence was quantified after 10 min incubation at 37°C. For auranofin, arsenic trioxide, and 2-AAPA, chemicals were added into the fresh culture medium and cells were incubated in the medium for 3 h at 37°C. Cells were collected and cell suspensions were used for quantification. All samples were prepared and measured in triplicates to derive averages and standard deviations. To determine cell viabilities, cells were treated with chemicals for 24 h at 37°C, and we followed the manufacturer’s protocol and used RealTime-Glo™ MT cell viability assays (Promega) to determine cell viability. All fluorescence response and viability data were fitted with a modified Hill equation:
in which k is the EC50 or LC50 reported in the manuscript, n is the Hill coefficient, A is the initial value, and B is the magnitude of change (positive for growth and negative for decay). To compare the growth of cells expressing or not expressing TrxRFP1, we seeded HEK 293T cells in 35-mm culture dishes. They were divided into two groups and one group of cells were transfected with pcDNA3-TrxRFP1 at 70% confluency. Cells were next collected at different time points to determine cell numbers. Briefly, cells were washed with DPBS, trypsinized, and resuspended in DPBS. Cells were further diluted with DPBS and stained with 0.4% trypan blue. Viable cell numbers were determined using a hemocytometer.
Western blotting
To examine the expression levels of Trx1 and TrxR1, we prepared cell lysates from the same number (6×106) of HEK 293T, SW620, MCF-7, Hela and SH-SY5Y cells using a CelLytic M cell lysis buffer (Sigma-Aldrich) supplemented with cOmplete™ Protease Inhibitor Cocktail (Roche). The Bradford assay was used to determine total protein concentrations in the cell lysates. 5 µg of total proteins were loaded to each lane and resolved on a 12% SDS-PAGE. Proteins were next transferred to a nitrocellulose membrane. The membrane was first blocked with 5% skim milk in TBST, and next, incubated with one of the anti-Trx1 (1:5000), anti-TrxR (1:10000) and anti-β-actin (1:10000) antibodies. After washing with TBST, the corresponding horseradish peroxidase (HRP) conjugated secondary antibodies were added. The membrane was visualized with the SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher) on a Bio-Rad Gel Doc XR System.
Redox immunoblotting
We modified a reported procedure34 to performed redox immunoblotting on TrxRFP1 in HEK 293T cells. Untreated, H2O2-treated, or auranofin-treated cells (6×106) were washed twice quickly with ice-cold 1× PBS, pelleted by a brief centrifugation, and lysed with 200 µL urea lysis buffer (1 mM EDTA, 8 M urea, and 50 mM Tris-HCl, pH 8.3) freshly supplemented with 10 mM iodoacetamide. Free thiols in cells were alkylated with iodoacetamide at 37°C for 30 min. Cell debris was next removed by centrifugation and proteins in clear cell lysates were precipitated with ice-cold acetone containing 20 mM HCl. The resultant precipitates were washed twice with ice-cold acetone containing 20 mM HCl and re-suspended in 50 µL urea lysis buffer containing 3.5 mM DTT. After incubation at 37°C for 30 min, 2.5 µL of freshly prepared 600 mM iodoacetic acid was added, followed by another 30-min incubation at 37°C to alkylate additional free thiols resulting from DTT reduction. Total protein concentrations were determined using a bicinchoninic acid (BCA) assay and 10 µg each of total proteins were separated on a 12% urea gel at constant 100 V and 50°C. Next, protein bands were transferred to a nitrocellulose membrane, which was blocked with 5% skim milk in TBST, incubated with the anti-Trx antibody (1:5000) overnight. After washing with TBST, an HRP-conjugated anti-goat secondary antibody was added. The SuperSignal West Pico Chemiluminescent substrate was added to visualize the membrane on a Bio-Rad Gel Doc XR System.
Statistical analysis
An unpaired Student's t-test was used to determine all P values. No statistical method was used to pre-determine the sample size. This study utilized in vitro enzyme assays and cell lines, so biological variation was not a consideration. Experiments were independently repeated at least three times to minimize technical variation. No sample was excluded from data analysis, and no randomization or blinding was employed. For time-lapse imaging data, we randomly selected cells with sufficient signals to quantify the intensities or ratios of individual cells from three independent duplicates. Unless otherwise indicated, data are shown as mean ± s.d., and error bars in figures represent s.d..
Data availability
The DNA sequence of TrxRFP1 has been deposited into GenBank under the accession number KX981912. Materials, associated protocols, and other supporting data are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We acknowledge funding from the National Institutes of Health (R01GM118675 and R21EB021651) and the National Science Foundation (CHE-1351933).
Footnotes
ACCESSION CODES
GenBank: KX981912 (rxRFP1)
Author contributions
H.A. conceived and supervised the project. Y.F. performed all experiments, except for that M.M. assisted Y.F. with cell culture and transfection and M.X.W. assisted Y.F. with plasmid preparation and SDS-PAGE. H.A. and Y.F. analyzed the data and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 2.Sengupta R, Holmgren A. Thioredoxin and glutaredoxin-mediated redox regulation of ribonucleotide reductase. World J. Biol. Chem. 2014;5:68–74. doi: 10.4331/wjbc.v5.i1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthews JR, Wakasugi N, Virelizier JL, Yodoi J, Hay RT. Thioredoxin regulates the DNA binding activity of NF-κB by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20:3821–30. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei SJ, et al. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–95. [PubMed] [Google Scholar]
- 5.Saitoh M, et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. Embo J. 1998;17:2596–606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma XX, et al. Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1. J. Biol. Chem. 2011;286:13430–7. doi: 10.1074/jbc.M110.205161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porqué PG, Baldesten A, Reichard P. The involvement of the thioredoxin system in the reduction of methionine sulfoxide and sulfate. J. Biol. Chem. 1970;245:2371–2374. [PubMed] [Google Scholar]
- 8.Sido B, et al. Potential role of thioredoxin in immune responses in intestinal lamina propria T lymphocytes. Eur. J. Immunol. 2005;35:408–17. doi: 10.1002/eji.200424500. [DOI] [PubMed] [Google Scholar]
- 9.Masutani H, Ueda S, Yodoi J. The thioredoxin system in retroviral infection and apoptosis. Cell Death Differ. 2005;12(Suppl 1):991–8. doi: 10.1038/sj.cdd.4401625. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, et al. Inhibition of bacterial thioredoxin reductase: an antibiotic mechanism targeting bacteria lacking glutathione. FASEB J. 2013;27:1394–403. doi: 10.1096/fj.12-223305. [DOI] [PubMed] [Google Scholar]
- 11.Gallegos A, et al. Transfection with human thioredoxin increases cell proliferation and a dominant-negative mutant thioredoxin reverses the transformed phenotype of human breast cancer cells. Cancer Res. 1996;56:5765–70. [PubMed] [Google Scholar]
- 12.Tonissen KF, Di Trapani G. Thioredoxin system inhibitors as mediators of apoptosis for cancer therapy. Mol. Nutr. Food Res. 2009;53:87–103. doi: 10.1002/mnfr.200700492. [DOI] [PubMed] [Google Scholar]
- 13.Shalev A. Minireview: Thioredoxin-interacting protein: regulation and function in the pancreatic β-cell. Mol. Endocrinol. 2014;28:1211–20. doi: 10.1210/me.2014-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arner ES, Holmgren A. The thioredoxin system in cancer. Semin. Cancer Biol. 2006;16:420–6. doi: 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Mahmood DF, Abderrazak A, El Hadri K, Simmet T, Rouis M. The thioredoxin system as a therapeutic target in human health and disease. Antioxid. Redox Signal. 2013;19:1266–303. doi: 10.1089/ars.2012.4757. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Garcia A, Zavala-Flores L, Rodriguez-Rocha H, Franco R. Thiol-redox signaling, dopaminergic cell death, and Parkinson's disease. Antioxid. Redox Signal. 2012;17:1764–84. doi: 10.1089/ars.2011.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarzlander M, Dick TP, Meyer AJ, Morgan B. Dissecting redox biology using fluorescent protein sensors. Antioxid. Redox Signal. 2016;24:680–712. doi: 10.1089/ars.2015.6266. [DOI] [PubMed] [Google Scholar]
- 18.Ren W, Ai HW. Genetically encoded fluorescent redox probes. Sensors. 2013;13:15422–33. doi: 10.3390/s131115422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 2010;13:621–50. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 20.Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: Engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–62. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson GT, et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 2004;279:13044–53. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 22.Fan Y, Chen Z, Ai HW. Monitoring redox dynamics in living cells with a redox-sensitive red fluorescent protein. Anal. Chem. 2015;87:2802–2810. doi: 10.1021/ac5041988. [DOI] [PubMed] [Google Scholar]
- 23.Fan Y, Ai HW. Development of redox-sensitive red fluorescent proteins for imaging redox dynamics in cellular compartments. Anal. Bioanal. Chem. 2016;408:2901–11. doi: 10.1007/s00216-015-9280-3. [DOI] [PubMed] [Google Scholar]
- 24.Belousov VV, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3:281–6. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 25.Gutscher M, et al. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009;284:31532–40. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan B, et al. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat. Chem. Biol. 2016;12:437–43. doi: 10.1038/nchembio.2067. [DOI] [PubMed] [Google Scholar]
- 27.Zhao BS, et al. A highly selective fluorescent probe for visualization of organic hydroperoxides in living cells. J. Am. Chem. Soc. 2010;132:17065–7. doi: 10.1021/ja1071114. [DOI] [PubMed] [Google Scholar]
- 28.Chen ZJ, Ren W, Wright QE, Ai HW. Genetically encoded fluorescent probe for the selective detection of peroxynitrite. J. Am. Chem. Soc. 2013;135:14940–3. doi: 10.1021/ja408011q. [DOI] [PubMed] [Google Scholar]
- 29.Chen S, Chen ZJ, Ren W, Ai HW. Reaction-based genetically encoded fluorescent hydrogen sulfide sensors. J. Am. Chem. Soc. 2012;134:9589–92. doi: 10.1021/ja303261d. [DOI] [PubMed] [Google Scholar]
- 30.Chen ZJ, Ai HW. A highly responsive and selective fluorescent probe for imaging physiological hydrogen sulfide. Biochemistry. 2014;53:5966–5974. doi: 10.1021/bi500830d. [DOI] [PubMed] [Google Scholar]
- 31.Bjornberg O, Ostergaard H, Winther JR. Mechanistic insight provided by glutaredoxin within a fusion to redox-sensitive yellow fluorescent protein. Biochemistry. 2006;45:2362–71. doi: 10.1021/bi0522495. [DOI] [PubMed] [Google Scholar]
- 32.Gutscher M, et al. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5:553–9. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 33.Ungerstedt J, Du Y, Zhang H, Nair D, Holmgren A. In vivo redox state of human thioredoxin and redox shift by the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) Free Radic. Biol. Med. 2012;53:2002–2007. doi: 10.1016/j.freeradbiomed.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 34.Du Y, Zhang H, Zhang X, Lu J, Holmgren A. Thioredoxin 1 is inactivated due to oxidation induced by peroxiredoxin under oxidative stress and reactivated by the glutaredoxin system. J. Biol. Chem. 2013;288:32241–7. doi: 10.1074/jbc.M113.495150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–51. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giustarini D, Dalle-Donne I, Milzani A, Fanti P, Rossi R. Analysis of GSH and GSSG after derivatization with N-ethylmaleimide. Nat. Protoc. 2013;8:1660–9. doi: 10.1038/nprot.2013.095. [DOI] [PubMed] [Google Scholar]
- 37.Du Y, Zhang H, Lu J, Holmgren A. Glutathione and glutaredoxin act as a backup of human thioredoxin reductase 1 to reduce thioredoxin 1 preventing cell death by aurothioglucose. J. Biol. Chem. 2012;287:38210–9. doi: 10.1074/jbc.M112.392225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill KE, McCollum GW, Boeglin ME, Burk RF. Thioredoxin reductase activity is decreased by selenium deficiency. Biochem. Biophys. Res. Commun. 1997;234:293–5. doi: 10.1006/bbrc.1997.6618. [DOI] [PubMed] [Google Scholar]
- 39.Jurado J, Prieto-Alamo MJ, Madrid-Risquez J, Pueyo C. Absolute gene expression patterns of thioredoxin and glutaredoxin redox systems in mouse. J. Biol. Chem. 2003;278:45546–54. doi: 10.1074/jbc.M307866200. [DOI] [PubMed] [Google Scholar]
- 40.Bilan DS, et al. HyPer-3: a genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem. Biol. 2013;8:535–42. doi: 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- 41.Spyrou G, Enmark E, Miranda-Vizuete A, Gustafsson J. Cloning and expression of a novel mammalian thioredoxin. J. Biol. Chem. 1997;272:2936–41. doi: 10.1074/jbc.272.5.2936. [DOI] [PubMed] [Google Scholar]
- 42.Rackham O, et al. Substrate and inhibitor specificities differ between human cytosolic and mitochondrial thioredoxin reductases: Implications for development of specific inhibitors. Free Radic. Biol. Med. 2011;50:689–699. doi: 10.1016/j.freeradbiomed.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 43.Kumar KK, Karnati S, Reddy MB, Chandramouli R. CACO-2 cell lines in drug discovery- an updated perspective. J Basic Clin Pharm. 2010;1:63–9. [PMC free article] [PubMed] [Google Scholar]
- 44.Lu J, Chew EH, Holmgren A. Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12288–93. doi: 10.1073/pnas.0701549104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seefeldt T, et al. Characterization of a novel dithiocarbamate glutathione reductase inhibitor and its use as a tool to modulate intracellular glutathione. J. Biol. Chem. 2009;284:2729–37. doi: 10.1074/jbc.M802683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bae YS, et al. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 1997;272:217–21. [PubMed] [Google Scholar]
- 47.Ostergaard H, Tachibana C, Winther JR. Monitoring disulfide bond formation in the eukaryotic cytosol. J. Cell. Biol. 2004;166:337–45. doi: 10.1083/jcb.200402120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic. Biol. Med. 2008;44:921–37. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biot C, Dessolin J, Grellier P, Davioud-Charvet E. Double-drug development against antioxidant enzymes from Plasmodium falciparum. Redox Rep. 2003;8:280–3. doi: 10.1179/135100003225002916. [DOI] [PubMed] [Google Scholar]
References
- 50.Wilson DS, Keefe AD. Random mutagenesis by PCR. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb0803s51. Chapter 8, Unit8.3. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bar-Noy S, Gorlatov SN, Stadtman TC. Overexpression of wild type and SeCys/Cys mutant of human thioredoxin reductase in E. coli: the role of selenocysteine in the catalytic activity. Free Radic. Biol. Med. 2001;30:51–61. doi: 10.1016/s0891-5849(00)00448-2. [DOI] [PubMed] [Google Scholar]
- 53.Kardash E, Bandemer J, Raz E. Imaging protein activity in live embryos using fluorescence resonance energy transfer biosensors. Nat. Protoc. 2011;6:1835–46. doi: 10.1038/nprot.2011.395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequence of TrxRFP1 has been deposited into GenBank under the accession number KX981912. Materials, associated protocols, and other supporting data are available from the corresponding author upon request.