Abstract
Background
Genomewide association studies of autoimmune diseases have mapped hundreds of susceptibility regions in the genome. However, only for a few association signals has the causal gene been identified, and for even fewer have the causal variant and underlying mechanism been defined. Coincident associations of DNA variants affecting both the risk of autoimmune disease and quantitative immune variables provide an informative route to explore disease mechanisms and drug-targetable pathways.
Methods
Using case–control samples from Sardinia, Italy, we performed a genomewide association study in multiple sclerosis followed by TNFSF13B locus–specific association testing in systemic lupus erythematosus (SLE). Extensive phenotyping of quantitative immune variables, sequence-based fine mapping, cross-population and cross-phenotype analyses, and gene-expression studies were used to identify the causal variant and elucidate its mechanism of action. Signatures of positive selection were also investigated.
Results
A variant in TNFSF13B, encoding the cytokine and drug target B-cell activating factor (BAFF), was associated with multiple sclerosis as well as SLE. The disease-risk allele was also associated with up-regulated humoral immunity through increased levels of soluble BAFF, B lymphocytes, and immunoglobulins. The causal variant was identified: an insertion–deletion variant, GCTGT→A (in which A is the risk allele), yielded a shorter transcript that escaped microRNA inhibition and increased production of soluble BAFF, which in turn up-regulated humoral immunity. Population genetic signatures indicated that this autoimmunity variant has been evolutionarily advantageous, most likely by augmenting resistance to malaria.
Conclusions
A TNFSF13B variant was associated with multiple sclerosis and SLE, and its effects were clarified at the population, cellular, and molecular levels. (Funded by the Italian Foundation for Multiple Sclerosis and others.)
Multiple sclerosis and systemic lupus erythematosus (SLE) are autoimmune diseases caused by largely unknown environmental factors acting in genetically susceptible persons. Genomewide association studies have provided statistical support for more than 110 independent signals for multiple sclerosis and 43 for SLE.1-3 Although several signals are near genes involved in immunologic processes, the effector mechanisms for most associations remain unknown.
A useful approach to explore disease mechanisms is to search for genetic variants that affect both the risk of autoimmune disease and quantitative immune variables such as circulating levels of immune-cell populations, immunoglobulins, and cytokines.4,5 This approach can reveal disease-related endophenotypes and is especially informative when variables are measured in healthy persons, avoiding secondary effects of the disease and its therapy.4 For such studies, the genetic structure of the Sardinian population,6 with prevalences of multiple sclerosis and SLE that are among the highest in the world,7,8 is likely to facilitate the detection of relevant variants missed in studies of cosmopolitan populations.6,9,10
A genomewide association scan for risk of multiple sclerosis was carried out in Sardinia, Italy. We implemented a strategy to follow up association signals in other populations and assess mechanisms through the characterization of potentially correlated quantitative immune variables and transcript and expression studies. We focused on an association signal in TNFSF13B, which encodes tumor necrosis factor superfamily member 13b (also known as B-cell activating factor [BAFF]). BAFF is a cytokine and drug target that is primarily produced by monocytes and neutrophils and that is essential for B-cell activation, differentiation, and survival.11,12
Methods
Study Sample Sets
Coincident associations were assessed in case–control sets of 2934 patients with multiple sclerosis, 411 patients with SLE, and 3392 controls from across Sardinia, as well as in a population cohort (SardiNIA study) of 6921 volunteers from the Lanusei valley in Sardinia.6,13 Tests of replication involved the use of case–control sets from mainland Italy (2292 patients with multiple sclerosis, 503 patients with SLE, and 2563 controls), Sweden (4548 patients with multiple sclerosis and 3481 controls), the United Kingdom (3176 patients with multiple sclerosis and 2958 controls), and the Iberian Peninsula (1120 patients with SLE and 1300 controls). All participants provided written informed consent.
Genotyping, Imputation, and Variant Characterization
A subgroup of 2273 patients with multiple sclerosis and 2148 controls from Sardinia were genotyped with the Affymetrix SNP Array 6.0 (574,519 single-nucleotide polymorphisms [SNPs]), and 6602 SardiNIA volunteers were genotyped with four Illumina arrays (OmniExpress, ImmunoChip, Cardio-MetaboChip, and ExomeChip; 890,542 SNPs in total).6,14 A much denser genetic map (up to 13.6 million SNPs) was constructed from a reference panel of 2120 Sardinians who underwent whole-genome sequencing and was then propagated (“imputed”) to all genotyped persons.6
Furthermore, at the TNFSF13B locus, we constructed a more refined map, which also included insertion–deletion variants (indels) called by use of the Genome Analysis Tool Kit (GATK) HaplotypeCaller tool. This locus-specific map was used as a reference panel for TNFSF13B locus imputation in the case–control and SardiNIA cohorts. Custom TaqMan assays were designed to genotype variants of interest (rs12874404 and an insertion–deletion variant, GCTGT→A, that we refer to as BAFF-var) in appropriate Sardinian and non-Sardinian samples.
Association Analyses
Genomewide association studies in Sardinian multiple sclerosis case–control sets and locus-specific association studies in the SardiNIA cohort were performed as described previously.6,14 Single-variant follow-up association analyses focusing on rs12874404 and BAFF-var (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org) used PLINK software, version 1.9 (https://www.cog-genomics.org/plink2). P values and effect sizes for SLE were calculated with the use of METAL software (http://genome.sph.umich.edu/wiki/METAL_Program). Bonferroni correction for multiple testing was applied to the nominal P value (P = 0.05). Conditional analyses for risk of multiple sclerosis and for quantitative traits were performed by adding either BAFF-var or rs12874404 as a co-variate to the basic model.
Immunologic Phenotyping and Serum Quantifications in the SardiNIA Cohort
Broad leukocyte types, including lymphocytes, neutrophils, and monocytes, were quantified with the use of an automated counter in up to 5937 genotyped volunteers. Flow cytometry was used to profile immune-cell subpopulations in 3669 volunteers, as described previously in a study involving a smaller sample.4 In addition, B-cell and monocyte subsets were assessed cytometrically in 1902 persons (Table S2 in the Supplementary Appendix). Soluble BAFF was measured by enzyme-linked immunosorbent assay (ELISA) (R&D Systems) in serum specimens from 2733 SardiNIA volunteers, and levels of IgA, IgG1, IgG2, IgG3, IgG4, and IgM were measured by Bio-Plex Multiplex Immunoassay (Bio-Rad Laboratories) in serum specimens from 2898 SardiNIA volunteers.
Transcript Studies
Normalized read counts of TNFSF13B (reflecting levels of messenger RNA [mRNA]) and TNFSF13B 3′ untranslated region (3′ UTR) length scores were computed from RNA sequencing data from leukocyte samples from 606 SardiNIA volunteers, and their correlation was assessed with the use of the Pearson coefficient. (See the Methods section in the Supplementary Appendix.) Mapping of expression quantitative trait loci was performed with the use of Merlin software (http://csg.sph.umich.edu//abecasis/Merlin/) and the lmekin() function in R Software.
To determine whether the effect of BAFF-var on levels of soluble BAFF was fully accounted for by effects on transcription, we performed a conditional linear regression analysis of data on 522 persons for whom both soluble BAFF protein levels and RNA sequencing data were available. The effect of BAFF-var on soluble BAFF levels, with and without TNFSF13B 3′ UTR transcript levels, was assessed with a likelihood-ratio test.
MicroRNA Effects on BAFF Expression
Expression studies were performed with primary monocytes and HeLa, HEK293T, and THP1 cells. Primary monocytes were isolated from peripheral-blood mononuclear cells by negative selection. RNA extraction, complementary DNA preparation, and quantitative polymerase-chain-reaction (PCR) analyses were performed with the use of the primers listed in Table S3 in the Supplementary Appendix.
In silico prediction of microRNAs (miRNAs) targeting the wild-type BAFF 3′ UTR and the variant BAFF 3′ UTR (nucleotides 2130 through 2582) was performed with the use of three prediction programs: TargetScan (http://www.targetscan.org/), miRDB (http://mirdb.org/miRDB/), and RNA22 (https://cm.jefferson.edu/rna22/). The miRNAs and related predicted binding sites (Table S4 in the Supplementary Appendix) were considered for further validation. Pull-down assays, miRNA and anti-miRNA transfection, Western blot analysis, ELISAs, and luciferase assays were used to assess miRNA binding and validate BAFF mRNA–miRNA interactions.
Differentiation and Positive Selection Analyses
To assess positive selection, we used standard statistical tests to evaluate allele-frequency differentiation (fixation index15 and population branch statistic16) and haplotype diversity (integrated haplotype score17 and the number of segregating sites by length18) in sequences from 1081 unrelated Sardinians as compared with sequences from European participants in phase 3 of the 1000 Genomes Project.19 For any given statistic, we calculated a “genomic percentile,” ranking BAFF-var relative to 3042 other variants that were sampled across the genome and matched for minor-allele frequency in Sardinians, local recombination rates, and levels of background selection.
Results
Association of a Variant in TNFSF13B with Multiple Sclerosis
Extending previous work14 with more samples and an improved genetic map, we conducted a sequencing-based genomewide association study involving 2273 Sardinian patients with multiple sclerosis and 2148 controls, analyzing approximately 12.2 million SNPs.6 The SNPs that showed a suggestive association (P<5×10−6) (Table S5 in the Supplementary Appendix) included variant rs12874404 near TNFSF13B. This variant is in moderate linkage disequilibrium (r2 = 0.44) with SNP rs9520836, which we previously found to be associated with increased numbers of B cells.4 This locus was thus a strong candidate for evaluating coincident associations between disease risk and endophenotypes and was further investigated.
We confirmed the association with multiple sclerosis at this locus by directly genotyping rs12874404 in a sample of Sardinians that included the discovery set and some additional Sardinians (totaling 2934 patients and 3392 controls) and observing a genomewide significant association (odds ratio, 1.27; P = 1.52×10−9). (See the Results section in the Supplementary Appendix.) However, we found no association between rs12874404 and multiple sclerosis in the case– control sets from mainland Italy, the United Kingdom, or Sweden (the lowest P value was found in the Italian sample [P = 0.51]), even though the risk allele was common and sample sizes provided high statistical power. (See the Results section in the Supplementary Appendix.) Thus, rs12874404 is unlikely to be the variant driving the association.
Because the pattern of linkage disequilibrium around rs12874404 differs between Sardinia and mainland Europe (Fig. S1A in the Supplementary Appendix), we considered whether the signal might come from a variant, such as an indel, that was not assessed in our initial genomewide association study and that was in stronger linkage disequilibrium with rs12874404 in Sardinia than elsewhere. Searching for such variants, we found an indel with GCTGT replaced by A, which had an r2 of 0.76 with rs12874404 in Sardinia but an r2 of only 0.44 among European participants in the 1000 Genomes Project (Fig. S1B in the Supplementary Appendix). Directly genotyping the indel in our Sardinian cohort showed the minor allele (A), which we named “BAFF-var,” as the variant most strongly associated with multiple sclerosis (odds ratio, 1.27; P = 1.23×10−9) (Fig. 1A). Conditional analyses showed BAFF-var or rs12874404 completely explaining the association at TNFSF13B, excluding secondary signals among the approximately 10,000 variants assessed at the locus. (See the Results section in the Supplementary Appendix.)
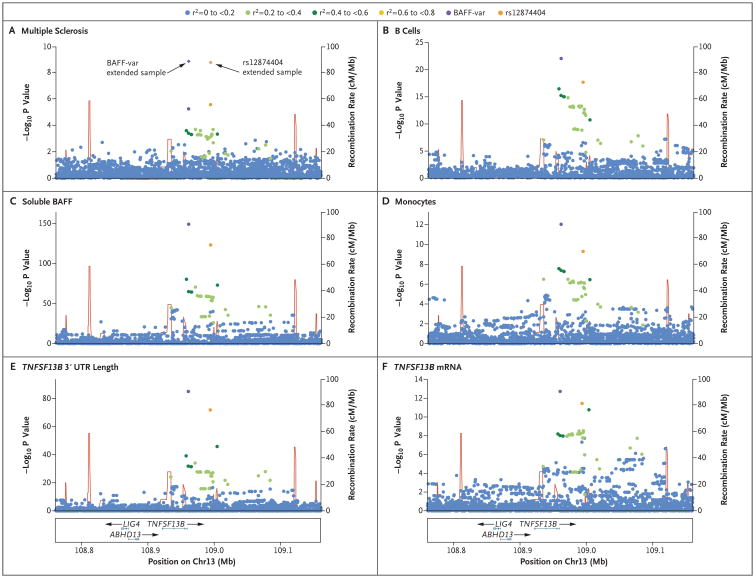
Figure 1. Regional Association Plots.
Shown are association results in the TNFSF13B region for multiple sclerosis (Panel A), B-cell percentage with respect to lymphocytes (Panel B), level of soluble B-cell activating factor (BAFF) (Panel C), monocyte counts (Panel D), length of the 3′ untranslated region (3′ UTR) of TNFSF13B (Panel E), and TNFSF13B messenger RNA (mRNA) (Panel F). Each panel shows the association strength (expressed as negative log10 P values) versus the genomic position (on the hg19/GRCh37 genomic build), with specifications on genes, exons, and direction of transcription shown in the box at the bottom of the column. In each plot, BAFF-var is the variant with the strongest association. Other variants in the region are color-coded to reflect their extent of linkage disequilibrium with BAFF-var (taken from pairwise r2 values calculated for Sardinian haplotypes). Panel A shows results for 2273 patients with multiple sclerosis and 2148 controls and for the extended sample (diamonds) of 2934 patients with the disorder and 3392 controls. Panels B through F refer to the SardiNIA study cohort.
We further fine-mapped the association through cross-population comparisons. BAFF-var is especially common across Sardinia (frequency, 26.5%) and progressively less common in southern Europe (5.7% in Italy and 4.9% in Spain) and northern Europe (1.8% in the United Kingdom and Sweden). We estimated that we would have power to detect an association with BAFF-var only in the available Italian samples. (See the Results section in the Supplementary Appendix.) By genotyping these samples (in which we initially did not replicate the rs12874404 association), we replicated the association of BAFF-var with risk of multiple sclerosis (odds ratio, 1.25; P = 0.01) (Table S6 in the Supplementary Appendix). Together, these findings pinpoint BAFF-var as the variant in TNFSF13B that is most strongly associated with multiple sclerosis.
BAFF-var as a General Autoimmunity Variant
BAFF is also a prime candidate for involvement in other autoimmune diseases, especially SLE20; belimumab, which targets BAFF, has been approved for the treatment of SLE by the Food and Drug Administration.12 We found that BAFF-var was associated with SLE in the case–control sets from Sardinia (odds ratio, 1.38; P = 4.09×10−5), mainland Italy (odds ratio, 1.49; P = 0.002), and the Iberian Peninsula (odds ratio, 1.55; P = 3.05×10−4), with an odds ratio of 1.44 (P = 6.74×10−10) in a combined analysis across all samples (Table S6 in the Supplementary Appendix). These findings indicate that the effect of BAFF-var is not restricted to multiple sclerosis alone.
Associations of BAFF-var with Immune Endophenotypes
We next asked whether BAFF-var might also account for the previously observed associations between variants at the TNFSF13B locus and elevated numbers of B lymphocytes.4 Assessing a much denser set of markers (including indels) in an expanded sample of 3669 persons from the SardiNIA cohort, we found that BAFF-var dominated the association with circulating B cells (B-cell counts, P = 4.23×10−12; B-cell percentage with respect to lymphocytes, P = 9.36×10−23) (Fig. 1B, and Table S7 in the Supplementary Appendix). To refine the target-cell type, we carried out deeper profiling of six B-cell subtypes. Rather than any particular subtype explaining the overall B-cell association, several subtypes were affected, with CD24+CD27+ (comprising unswitched and switched memory) B cells being most strongly associated (Table S8 in the Supplementary Appendix).
Furthermore, BAFF-var dramatically augmented levels of soluble BAFF in SardiNIA volunteers (effect size, 0.77 SD; P = 8.47×10−150) (Fig. 1C, and Table S9 in the Supplementary Appendix). It also increased levels of total IgG (P = 1.68×10−12), IgG1 (P = 2.24×10−14), IgA (P = 7.64×10−9), and IgM (P = 4.70×10−8) (Fig. S2 and Table S9 in the Supplementary Appendix). Finally, we found that BAFF-var was also the lead TNFSF13B variant (P = 9.07×10−13) associated with reduced monocyte counts in SardiNIA volunteers (Fig. 1D, and Table S9 in the Supplementary Appendix), with the largest effect on “classical” CD14+CD16–monocytes (Table S10 in the Supplementary Appendix).
Overall, BAFF-var was the top variant in ge-nomewide significant associations (P<6.9×10−9),6 with 18 immune endophenotypes (Tables S7 through S10 in the Supplementary Appendix). For each of these traits, conditional analysis showed that BAFF-var explained the entire association at the locus (Table S11 in the Supplementary Appendix).
We also found that soluble BAFF levels were significantly higher in patients with multiple sclerosis or SLE than in controls. Furthermore, using preclinical samples available for SardiNIA volunteers in whom multiple sclerosis developed up to 12 years later, we observed elevated levels of soluble BAFF before disease diagnosis (Table S12 and the Results section in the Supplementary Appendix). We infer that BAFF-var is the causal variant driving an increase in soluble BAFF and a cascade of immune effects leading to increased autoimmunity risk.
Effect of BAFF-var on Levels of Soluble BAFF
The observation that BAFF-var creates an alternative polyadenylation motif, AATAAA, from the sequence AAT[GCTGT/A]AA prompted the hypothesis that the variant 3′ UTR could raise soluble BAFF levels by generating a truncated TNFSF13B transcript in the 3′ UTR. Indeed, in 606 sequenced leukocyte transcriptomes of SardiNIA participants (Fig. 2), BAFF-var correlated with a decrease of approximately 35% in the longer transcript per allele (P = 8.34×10−86) (Fig. 1E, and Fig. S3 in the Supplementary Appendix), an effect confirmed by quantitative PCR analysis of primary monocyte RNA (Fig. S4 in the Supplementary Appendix). Thus, BAFF-var generates a shorter TNFSF13B 3′ U T R.
Figure 2. RNA Sequencing Coverage across the TNFSF13B Gene.
RNA expression levels are plotted as the relative number of reads across the TNFSF13B locus, spanning the last exon (exon 6) and the 3′ UTR. The mean expression levels for the three alternative genotypes (wild type [WT]/WT, BAFF-var/BAFF-var, and WT/BAFF-var) are shown. The 3′ UTRs (long and short forms) are represented with dark gray bars, and the BAFF-var position is specified.
The transcriptome data also implicated BAFF-var as an expression quantitative trait locus that increases TNFSF13B expression (P = 2.37×10−13) (Fig. 1F). However, the elevated mRNA levels could explain only 24 to 27% of the effect on protein (soluble BAFF) levels (Fig. 3A, and the Results section in the Supplementary Appendix), indicating that an increase in translation level was also probably involved. Higher levels of BAFF-var protein could be due to the presence of miRNA sites in the longer, but not in the shorter, 3′ UTR that might reduce protein production from the longer 3′ UTR.21
Figure 3. Post-transcriptional Regulation of BAFF Protein Expression by MicroRNAs.
Panel A shows a box plot of soluble BAFF levels, as determined by enzyme-linked immunosorbent assay (ELISA), in serum specimens from 2733 genotyped SardiNIA volunteers, stratified according to genotype. Panel B shows the results of Western blot analysis of endogenous BAFF protein in THP1 cells 48 hours after transfection of the microRNA (miRNA) precursors. Panel C shows BAFF protein expression normalized to HSP90 levels and plotted relative to the scrambled miRNA control (miR-Ctr). Panel D shows relative miRNA expression in primary monocytes, quantified by reverse transcription and quantitative polymerase-chain-reaction (qPCR) analysis using U6 RNA as the endogenous control. Panel E shows quantification by qPCR of BAFF mRNA levels after miR-15a overexpression in THP1 cells, normalized to GAPDH mRNA levels. Soluble BAFF levels were measured by ELISA after transfection of the locked nucleic acid–anti–miR-15a oligonucleotide into THP1 cells, as shown in Panel F, and into primary monocytes, as shown in Panel G; results were normalized to locked nucleic acid–anti-miR control samples. Data in Panels C through G are the means of at least three independent experiments. I bars represent standard errors. Level of significance, calculated by t-test, is indicated.
We identified in silico candidate miRNA sites that were absent from the truncated 3′ UTR (Table S4 in the Supplementary Appendix) and tested them by pulling down miRNA–mRNA complexes from THP1 lysates using biotinylated RNA probes for the TNFSF13B wild-type 3′ UTR and the variant 3′ UTR (Fig. S5A in the Supplementary Appendix). The miRNAs in the miR-15/107 family22 and miR-494 were more enriched in the probe for the wild-type 3′ UTR than in that for the variant 3′ UTR (Fig. S5B in the Supplementary Appendix), suggesting their preferential interaction with the longer 3′ UTR.
Confirming their inhibitory effect on BAFF expression, overexpression of miR-15a, miR-497, and miR-646, but not miR-16 and miR-494, significantly down-regulated levels of endogenous BAFF protein in THP1 cells (Fig. 3B and 3C). One of these miRNAs, miR-15a, is highly expressed in primary monocytes (Fig. 3D) and thus was used in further validation experiments; its overexpression did not lower levels of endogenous TNFSF13B mRNA (Fig. 3E), consistent with post-transcriptional regulation of BAFF protein production. Further supporting the regulation of BAFF protein levels, suppression of miR-15a with the use of a locked nucleic acid–anti-miR increased production of soluble BAFF, both in THP1 cells and in primary monocytes (Fig. 3F and 3G).
To verify that miR-15a directly targets TNFSF13B, we cloned the wild-type BAFF 3′ UTR and the variant BAFF 3′ UTR into the pmirGLO reporter vector (Fig. 4A) and analyzed luciferase activity in HeLa cells, which express low levels of miR-15a (Fig. S6 in the Supplementary Appendix). Co-transfection of miR-15a with pmirGLO wild-type BAFF 3′ UTR or pmirGLO variant BAFF 3′ UTR substantially reduced reporter levels, whereas reporters bearing mutated miR-15a binding sites were refractory to repression by miR-15a (Fig. 4B).
Figure 4. Differential Regulation of BAFF Expression by miR-15a.
Panel A shows a representation of luciferase reporter constructs. Wild-type (WT) BAFF 3′ UTR and variant (var) BAFF 3′ UTR sequences were cloned into the pmirGLO vector. Numbers 1 and 2 correspond to the miR-15a binding sites. Panel B shows the cotransfection of BAFF reporter constructs (WT or var) with miRNA-15a precursor or with a negative control into HeLa cells. “Mut. 1” and “Mut. 2” refer to the reporters mutated in the first and second miRNA-15a binding sites depicted in Panel A. Firefly luciferase levels were normalized to renilla luciferase activity for each sample, and all values were plotted relative to the WT construct. The graph is representative of four independent experiments. Level of significance, calculated by t-test, is indicated.
Importantly, cotransfection of miR-15a with the variant BAFF 3′ UTR reduced reporter activity to levels similar to those seen for the wild-type BAFF 3′ UTR with one of the two miR-15a sites mutated, probably because one miR-15a site is absent from the variant BAFF 3′ UTR. These data indicate that BAFF-var raises soluble BAFF levels by favoring the production of a shorter transcript that is less inhibited by miR-15a. However, the higher luciferase activity from short reporters than from long reporters — either basal or cotransfected with miR-15a after mutating miR-15a binding sites — suggests that miR-15a does not fully explain the BAFF-var regulatory effects, and other miRNAs or RNA-binding proteins may also influence soluble BAFF levels.
Evidence of Positive Selection on BAFF-var
We next assessed whether the high frequency of BAFF-var in the isolated Sardinian population (Fig. S8A in the Supplementary Appendix) is consistent with the effects of random genetic drift acting across the genome or whether it is more likely the outcome of selection specifically favoring BAFF-var in Sardinia (positive selection). We found that BAFF-var has greater differences in frequency between Sardinians and Europeans than the vast majority of variants matched for allelic frequency in Sardinia and for other pertinent genetic features (99.97th genomic percentile for both fixation index and population branch statistic) (Table S13 and Fig. S8B in the Supplementary Appendix). In addition, the core haplotype carrying BAFF-var is remarkably longer than haplotypes carrying variants with matched genetic features (99.91th genomic percentile for integrated haplotype score and >99.99th genomic percentile for the number of segregating sites by length) (Figs. S8C and S9A in the Supplementary Appendix), a finding consistent with the hypothesis of positive selection for BAFF-var in Sardinia. The combination of extreme frequency differences and long core haplotype suggests that BAFF-var has been positively selected,23 most likely as an adaptation to a selective pressure particularly prevalent in Sardinia.
Discussion
Since its discovery, BAFF has been studied in animal models,24,25 with an indication for a role in autoimmunity from mice that overexpress BAFF and have symptoms resembling those of human SLE and Sjögren's syndrome.11 In contrast, case–control studies of human autoimmune disease that compared levels of soluble BAFF in biologic fluids have overall been inconclusive. (See the Discussion section in the Supplementary Appendix.) Furthermore, previous genomewide association studies provided no genetic evidence (in large consortium efforts2,26) or weak genetic evidence (in a small study27) for its role in autoimmune disease. The convergent genetic, transcriptomic, and functional evidence now supports its effects in autoimmunity. A schematic depiction of the effects of BAFF-var is shown in Figure 5. Our findings of naturally overexpressed BAFF in the general population are reminiscent of the large increases in IgM, IgG, and IgA in constitutive Baff-transgenic mice.11 Furthermore, the lack of association of BAFF-var with any of the 153 T-cell traits that we assessed (Table S7 in the Supplementary Appendix) is consistent with the development of auto-immunity to the same extent in T-cell–sufficient and T-cell–deficient Baff-transgenic mice.28 We infer that the increased risk of multiple sclerosis and SLE associated with BAFF overexpression mainly involves humoral immunity, consistent with the efficacy of B-cell–depleting therapies.29,30
Figure 5. Effects of BAFF-var at the Levels of Transcript Type, Protein Production, and Cellular Response.
Shown is a schematic depiction of the location of BAFF-var within TNFSF13B and its effects on the generation of mRNAs with different 3′ UTR lengths. The number and location of miRNA sites, production of soluble BAFF, endophenotypes, and autoimmunity risk are indicated. BAFF-var creates an alternative polyadenylation signal that generates a shorter 3′ UTR transcript lacking a miRNA binding site. In contrast to the wild-type allele, which is associated with long 3′ UTR only, BAFF-var leads to a mixed population of mRNAs with long and short 3′ UTRs, resulting in higher production of soluble BAFF. In turn, the increased levels of soluble BAFF lead to higher numbers of B cells and immunoglobulins, reduced levels of monocytes, and an increased risk of autoimmunity.
However, the effects of BAFF-var are broad, and the involvement of specific B-cell and immunoglobulin subtypes in the pathogenesis of multiple sclerosis and SLE remains unclear and requires further explication. Moreover, how the observed BAFF-var–driven changes in peripheral blood relate to effects in mucosal and target organ compartments remains unresolved. For instance, the reduction in circulating monocytes reported here could result from their soluble BAFF–driven mobilization or transformation into macrophages or dendritic cells at sites of inflammation, consistent with findings in mice.31 Alternatively, it may reflect negative feedback in response to the enhanced production of soluble BAFF in monocytes.
Nevertheless, besides supporting BAFF as a target in multiple sclerosis and SLE, our results have implications for clinical research. A pertinent one, to be assessed in further studies, would be that patients stratified according to BAFF-var status might show a differential benefit from anti-BAFF therapies. One might also anticipate that patients carrying BAFF-var would have a weaker response to B-cell–depleting therapies owing to a faster resurgence of memory B cells.32 In fact, patients with SLE with higher basal levels of BAFF have been found to have poorer or shorter clinical responses to rituximab than those with lower levels.33,34
Our findings also underline the critical balance between regulation of immune responses to defend against infection and possible tissue damage from the elicited responses. Such evolutionary trade-offs might be a more general phenomenon, consistent with previous work.4,35
BAFF-var may have been positively selected in Sardinia by providing resistance to Plasmodium falciparum or P. vivax malaria, both of which were strikingly prevalent in Sardinia until their eradication in the 1950s.36 The malaria-driven selective pressure to maintain high BAFF levels is supported by models of malaria in mice, in which BAFF overexpression confers protection against lethal malaria infection.37 In addition, as shown here, BAFF-var increases antibody production, and classic findings showed that antibody transfer from adults with immunity to malaria to acutely infected children reduced blood-stage parasitemia and disease severity.38
Overall, the evolutionary scenario we propose is that BAFF-var was selected as an adaptive response to malaria infection, resulting in an increased present-day risk of autoimmunity.
Supplementary Material
Acknowledgments
Supported by grants (2011/R/13 and 2015/R/09, to Dr. Cucca) from the Italian Foundation for Multiple Sclerosis; contracts (N01-AG-1-2109 and HHSN271201100005C, to Dr. Cucca) from the Intramural Research Program of the National Institute on Aging, National Institutes of Health (NIH); a grant (FaReBio2011 “Farmaci e Reti Biotecnologiche di Qualità,” to Dr. Cucca) from the Italian Ministry of Economy and Finance; a grant (633964, to Dr. Cucca) from the Horizon 2020 Research and Innovation Program of the European Union; a grant (U1301.2015/AI.1157. BE Prat. 2015-1651, to Dr. Cucca) from Fondazione di Sardegna; grants (“Centro per la ricerca di nuovi farmaci per malattie rare, trascurate e della povertà” and “Progetto collezione di composti chimici ed attività di screening,” to Dr. Cucca) from Ministero dell'Istruzione, dell'Università e della Ricerca; grants (HG005581, HG005552, HG006513, and HG007022, to Dr. Abecasis) from the National Human Genome Research Institute; a grant (9-2011-253, to Dr. Todd) from JDRF; a grant (091157, to Dr. Todd) from the Wellcome Trust; a grant (to Dr. Todd) from the National Institute for Health Research (NIHR); and the NIHR Cambridge Biomedical Research Centre. Dr. Idda was a recipient of a Master and Back fellowship from the Autonomous Region of Sardinia.
We thank all the volunteers who generously participated in this study; Charleston Chiang of the University of California, Los Angeles, for advice on selection tests; Debbie Smyth of the University of Cambridge, Franco Galimi, Giuseppe Delogu, and Valeria Lodde of the University of Sassari, Gianna Costa of the University of Cagliari, Arya Biragyn, Nan-ping Weng, and Monica Bodogai of the NIH, and Cristian Caria of Istituto di Ricerca Genetica e Biomedica–Consiglio Nazionale delle Ricerche for technical assistance; Marcella Devoto for helpful suggestions; Lidia Leoni, Carlo Podda, and the high-performance computing group of the Center for Advanced Studies, Research, and Development in Sardinia (CRS4) for information technology support; the staff of Myriam Gorospe's laboratory at the NIH and Chris Jones and Paolo Zanella of CRS4 for continuous help and port; and the volunteers and staff at the NIHR Cambridge Biomedical Research Centre. Extended acknowledgments are presented in the Supplementary Appendix.
This article is dedicated to the memory of Dr. Giulio Rosati, who died on August 15, 2016.
Appendix.
Maristella Steri, Ph.D., Valeria Orrù, Ph.D., M. Laura Idda, Ph.D., Maristella Pitzalis, Ph.D., Mauro Pala, Ph.D., Ilenia Zara, B.Eng., Carlo Sidore, Ph.D., Valeria Faà, M.S., Matteo Floris, Ph.D., Manila Deiana, Isadora Asunis, Eleonora Porcu, Ph.D., Antonella Mulas, Ph.D., Maria G. Piras, M.S., Monia Lobina, Ph.D., Sandra Lai, M.S., Mara Marongiu, Ph.D., Valentina Serra, Ph.D., Michele Marongiu, B.S., Gabriella Sole, M.S., Fabio Busonero, Ph.D., Andrea Maschio, M.S., Roberto Cusano, M.S., Gianmauro Cuccuru, M.S., Francesca Deidda, Ph.D., Fausto Poddie, M.S., Gabriele Farina, M.D., Mariano Dei, M.S., Francesca Virdis, Ph.D., Stefania Olla, Ph.D., Maria A. Satta, M.D., Mario Pani, M.D., Alessandro Delitala, M.D., Eleonora Cocco, M.D., Jessica Frau, M.D., Giancarlo Coghe, M.D., Lorena Lorefice, M.D., Giuseppe Fenu, M.D., Paola Ferrigno, M.D., Maria Ban, Ph.D., Nadia Barizzone, Ph.D., Maurizio Leone, M.D., Franca R. Guerini, M.D., Matteo Piga, M.D., Davide Firinu, M.D., Ingrid Kockum, Ph.D., Izaura Lima Bomfim, Ph.D., Tomas Olsson, Ph.D., Lars Alfredsson, Ph.D., Ana Suarez, M.D., Patricia E. Carreira, M.D., Maria J. Castillo-Palma, M.D., Joseph H. Marcus, B.S., Mauro Congia, M.D., Andrea Angius, Ph.D., Maurizio Melis, M.D., Antonio Gonzalez, Ph.D., Marta E. Alarcón Riquelme, Ph.D., Berta M. da Silva, Ph.D., Maurizio Marchini, Ph.D., Maria G. Danieli, M.D., Stefano Del Giacco, M.D., Alessandro Mathieu, M.D., Antonello Pani, M.D., Stephen B. Montgomery, Ph.D., Giulio Rosati, M.D., Jan Hillert, M.D., Ph.D., Stephen Sawcer, Ph.D., Sandra D'Alfonso, Ph.D., John A. Todd, Ph.D., John Novembre, Ph.D., Gonçalo R. Abecasis, Ph.D., Michael B. Whalen, Ph.D., Maria G. Marrosu, M.D., Alessandra Meloni, Serena Sanna, Ph.D., Myriam Gorospe, Ph.D., David Schlessinger, Ph.D., Edoardo Fiorillo, Ph.D., Magdalena Zoledziewska, Ph.D., Francesco Cucca, M.D.
Istituto di Ricerca Genetica e Biomedica, Consiglio Nazionale delle Ricerche Monserrato (M.S., V.O., M.L.I., M. Pitzalis, M. Pala, C.S., V.F., M.F., M. Deiana, I.A., E.P., A. Mulas, M.G.P., M. Lobina, S.L., Mara Marongiu, V.S., Michele Marongiu, G.S., F.B., A. Maschio, F.D., M. Dei, F.V., S.O., A.A., M.B.W., A. Meloni, S. Sanna, E.F., M.Z., F.C.), Center for Advanced Studies, Research, and Development in Sardinia, Parco Scientifico e Tecnologico della Sardegna (I.Z., M.F., R.C., G. Cuccuru), Struttura Complessa Disciplina di Ematologia e Centro Trapianto Cellule Staminali Emopoietiche Wilma Deplano, Ospedale Oncologico di Riferimento Regionale Armando Businco (M. Pani), Dipartimento di Sanità Pubblica, Medicina Clinica e Molecolare, Università di Cagliari (E.C., J.F., G. Coghe, L.L., G. Fenu), Azienda Ospedaliera Brotzu, S.C. Neurologia (P.F., M. Melis), Division of Rheumatology, University and University Hospital of Cagliari (M. Piga, A. Mathieu), Department of Medical Sciences M. Aresu, University of Cagliari (D.F., S.D.G., M.G.M.), Azienda Ospedaliera Brotzu, U.S. Gastroenterologia Pediatrica Ospedale Pediatrico Microcitemico A. Cao (M.C.), and Nephrology, Dialysis, and Transplantation Unit, Giuseppe Brotzu Hospital (A.P.), Cagliari, Dipartimento di Scienze Biomediche, Università degli Studi di Sassari (M.F., F.P., F.C.), Unit of Neurology, Department of Clinical and Experimental Medicine, University of Sassari (G. Farina, G.R.), and Servizio Trasfusionale (M.A.S.) and Clinica Medica (A.D.), Azienda Ospedaliero Universitaria di Sassari, Sassari, Neurology B, Department of Neurological, Biomedical, and Movement Sciences, University of Verona, Verona (G. Farina), Department of Health Sciences, Interdisciplinary Research Center of Autoimmune Diseases, University of Eastern Piedmont, Novara (N.B., S.D.), SC Neurologia, Dipartimento di Scienze Mediche, Istituti di Ricovero e Cura a Carattere Scientifico (IRCCS) Casa Sollievo della Sofferenza, San Giovanni Rotondo (M. Leone), Don C. Gnocchi Foundation IRCCS (F.R.G.), and Referral Center for Systemic Autoimmune Diseases Fondazione IRCCS Cá Granda Ospedale Maggiore Policlinico and University of Milan (M. Marchini), Milan, and Dipartimento di Scienze Cliniche e Molecolari, Università Politecnica delle Marche e Ospedali Riuniti, Ancona (M.G.D.) — all in Italy; Laboratory of Genetics and Genomics, National Institute on Aging, National Institutes of Health, Baltimore (M.L.I., M.G., D.S.); the Department of Clinical Neurosciences (M.B., S. Sawcer) and JDRF–Wellcome Trust Diabetes and Inflammation Laboratory, National Institute for Health Research Cambridge Biomedical Research Centre, Cambridge Institute for Medical Research (J.A.T.), University of Cambridge, Cambridge, United Kingdom; Department of Clinical Neuroscience, Karolinska Institutet at Karolinska University Hospital Solna (I.K., I.L.B., T.O., J.H.), Institute of Environmental Medicine (L.A.) and Institute of Environmental Medicine, Unit of Immunology and Chronic Disease (M.E.A.R.), Karolinska Institute, and Center for Occupational and Environmental Medicine, Stockholm County Council (L.A.), Stockholm; Department of Functional Biology, University of Oviedo, Oviedo (A.S.), Rheumatology Department, Hospital Universitario 12 de Octubre, Madrid (P.E.C.), Department of Internal Medicine, Hospital Universitario Virgen del Rocío, Seville (M.J.C.-P.), Laboratorio de Investigacion 10 and Rheumatology Unit, Instituto de Investigacion Sanitaria–Hospital Clinico Universitario de Santiago, Santiago de Compostela (A.G.), and Centro de Genómica e Investigación Oncológica, Pfizer–Universidad de Granada–Junta de Andalucía, Granada (M.E.A.R.) — all in Spain; Department of Human Genetics, University of Chicago, Chicago (J.H.M., J.N.); Centro Hospitalar do Porto–Hospital Santo Antonio and Unit for Multidisciplinary Research in Biomedicine–Unidade Multidisciplinar de Investigação Biomédica, Porto, Portugal (B.M.S.); Departments of Pathology and Genetics, Stanford University, Stanford, CA (S.B.M.); and Center for Statistical Genetics, University of Michigan, Ann Arbor (G.R.A.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Sawcer S, Franklin RJM, Ban M. Multiple sclerosis genetics. Lancet Neurol. 2014;13:700–9. doi: 10.1016/S1474-4422(14)70041-9. [DOI] [PubMed] [Google Scholar]
- 2.Beecham AH, Patsopoulos NA, Xifara DK, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentham J, Morris DL, Cunninghame Graham DS, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–64. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orrù V, Steri M, Sole G, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–56. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zavattari P, Deidda E, Whalen M, et al. Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: demography, chromosome recombination frequency and selection. Hum Mol Genet. 2000;9:2947–57. doi: 10.1093/hmg/9.20.2947. [DOI] [PubMed] [Google Scholar]
- 6.Sidore C, Busonero F, Maschio A, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 2015;47:1272–81. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosati G. The prevalence of multiple sclerosis in the world: an update. Neurol Sci. 2001;22:117–39. doi: 10.1007/s100720170011. [DOI] [PubMed] [Google Scholar]
- 8.Piga M, Casula L, Perra D, et al. Population-based analysis of hospitalizations in a West-European region revealed major changes in hospital utilization for patients with systemic lupus erythematosus over the period 2001-2012. Lupus. 2016;25:28–37. doi: 10.1177/0961203315596597. [DOI] [PubMed] [Google Scholar]
- 9.Danjou F, Zoledziewska M, Sidore C, et al. Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat Genet. 2015;47:1264–71. doi: 10.1038/ng.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zoledziewska M, Sidore C, Chiang CWK, et al. Height-reducing variants and selection for short stature in Sardinia. Nat Genet. 2015;47:1352–6. doi: 10.1038/ng.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackay F, Schneider P, Rennert P, Browning J. BAFF AND APRIL: a tutorial on B cell survival. Annu Rev Immunol. 2003;21:231–64. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 12.Navarra SV, Guzmán RM, Gallacher AE, et al. Efficacy and safety of belimumab in patients with active systemic lupus erythematosus: a randomised, placebo-controlled, phase 3 trial. Lancet. 2011;377:721–31. doi: 10.1016/S0140-6736(10)61354-2. [DOI] [PubMed] [Google Scholar]
- 13.Pilia G, Chen WM, Scuteri A, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2(8):e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanna S, Pitzalis M, Zoledziewska M, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet. 2010;42:495–7. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10:639–50. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi X, Liang Y, Huerta-Sanchez E, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–8. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol Biol Evol. 2014;31:1275–91. doi: 10.1093/molbev/msu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium. Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tipton CM, Fucile CF, Darce J, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16:755–65. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afonso-Grunz F, Müller S. Principles of miRNA-mRNA interactions: beyond sequence complementarity. Cell Mol Life Sci. 2015;72:3127–41. doi: 10.1007/s00018-015-1922-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finnerty JR, Wang WX, Hébert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 24.Moore PA, Belvedere O, Orr A, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285:260–3. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 25.Mackay F, Woodcock SA, Lawton P, et al. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med. 1999;190:1697–710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The International Multiple Sclerosis Genetics Consortium (IMSGC), Wellcome Trust Case Control Consortium 2 (WTCCC2) Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandit L, Ban M, Beecham AH, et al. European multiple sclerosis risk variants in the south Asian population. Mult Scler. 2016;22:1536–40. doi: 10.1177/1352458515624270. [DOI] [PubMed] [Google Scholar]
- 28.Groom JR, Fletcher CA, Walters SN, et al. BAFF and MyD88 signals promote a lupuslike disease independent of T cells. J Exp Med. 2007;204:1959–71. doi: 10.1084/jem.20062567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauser SL, Waubant E, Arnold DL, et al. B-cell depletion with rituximab in relapsing–remitting multiple sclerosis. N Engl J Med. 2008;358:676–88. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 30.Kappos L, Li D, Calabresi PA, et al. Ocrelizumab in relapsing-remitting multiple sclerosis: a phase 2, randomised, placebo-controlled, multicentre trial. Lancet. 2011;378:1779–87. doi: 10.1016/S0140-6736(11)61649-8. [DOI] [PubMed] [Google Scholar]
- 31.Zouggari Y, Ait-Oufella H, Bonnin P, et al. B lymphocytes trigger monocyte mobilization and impair heart function after acute myocardial infarction. Nat Med. 2013;19:1273–80. doi: 10.1038/nm.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anolik JH, Barnard J, Owen T, et al. Delayed memory B cell recovery in peripheral blood and lymphoid tissue in systemic lupus erythematosus after B cell depletion therapy. Arthritis Rheum. 2007;56:3044–56. doi: 10.1002/art.22810. [DOI] [PubMed] [Google Scholar]
- 33.Cambridge G, Isenberg DA, Edwards JCW, et al. B cell depletion therapy in systemic lupus erythematosus: relationships among serum B lymphocyte stimulator levels, autoantibody profile and clinical response. Ann Rheum Dis. 2008;67:1011–6. doi: 10.1136/ard.2007.079418. [DOI] [PubMed] [Google Scholar]
- 34.Sanz I. Connective tissue diseases: the conundrum of B cell depletion in SLE. Nat Rev Rheumatol. 2009;5:304–5. doi: 10.1038/nrrheum.2009.100. [DOI] [PubMed] [Google Scholar]
- 35.Raj T, Kuchroo M, Replogle JM, Raychaudhuri S, Stranger BE, De Jager PL. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am J Hum Genet. 2013;92:517–29. doi: 10.1016/j.ajhg.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tognotti E. Program to eradicate malaria in Sardinia, 1946-1950. Emerg Infect Dis. 2009;15:1460–6. doi: 10.3201/eid1509.081317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu XQ, Stacey KJ, Horne-Debets JM, et al. Malaria infection alters the expression of B-cell activating factor resulting in diminished memory antibody responses and survival. Eur J Immunol. 2012;42:3291–301. doi: 10.1002/eji.201242689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen S, McGregor IA, Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature. 1961;192:733–7. doi: 10.1038/192733a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.