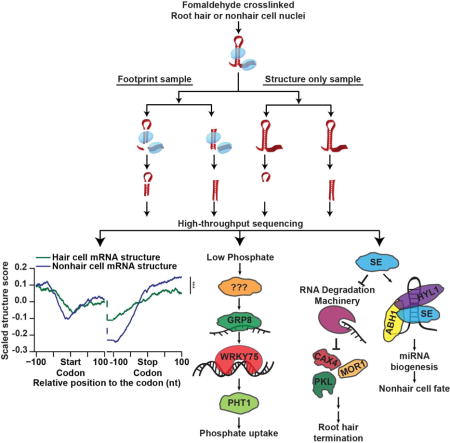
SUMMARY
The Arabidopsis thaliana root epidermis is comprised of two cell types, hair and nonhair cells, which differentiate from the same precursor. Although the transcriptional programs regulating these events are well studied, post-transcriptional factors functioning in this cell fate decision are mostly unknown. Here, we globally identify RNA-protein interactions and RNA secondary structure in hair and nonhair cell nuclei. This analysis reveals distinct structural and protein binding patterns across both transcriptomes, allowing identification of differential RNA binding protein (RBP) recognition sites. Using these sequences, we identify two RBPs that regulate hair cell development. Specifically, we find that SERRATE functions in a microRNA-dependent manner to inhibit hair cell fate, while also terminating growth of root hairs mostly independent of microRNA biogenesis. Additionally, we show that GLYCINE-RICH PROTEIN 8 promotes hair cell fate while alleviating phosphate starvation stress. In total, this global analysis reveals novel post-transcriptional regulators of plant root epidermal cell fate.
eTOC Blurb
Plant root hair cells function in water and nutrient uptake. Foley et al. characterize global RNA-protein interactions and RNA secondary structure in Arabidopsis root hair and non-hair cell nuclei using protein-interaction-profile sequencing. The dataset uncovers cell type-specific RNA secondary structure, RBP binding patterns, and RBP regulators of hair cell fate.
INTRODUCTION
The agricultural industry is responsible for providing food for an ever-growing global population. Currently, population growth is on track to outpace agricultural growth by the year 2050 (OECD and FAO, 2012). This challenge is compounded by climate change, which reduces arable land that can be used for agricultural production, necessitating the development of new technologies to increase crop yield. One method to achieve this is through the study of plant root development, as roots function in the uptake of both water and nutrients from the environment (Grierson et al., 2014). Thus, these studies can result in the engineering of plants that can better tolerate and respond to these environmental stresses, without affecting the development of the agriculturally important aerial tissues.
The plant root epidermis is responsible for absorbing both water and nutrients from the environment (Grierson et al., 2014). During root growth, epidermal precursor cells differentiate (Dolan et al., 1993) into either root hair or nonhair cells. The long hair-like projections of hair cells dramatically increase surface area, allowing uptake of more nutrients from the surrounding soil. Therefore, plants regulate the ratio of root hair to nonhair cells in a manner that is partially dependent on environmental signals (Meisner and Karnok, 1991). More specifically, plants grown under nutrient or water poor conditions develop more hair cells with longer hairs (Bates and Lynch, 1996), thereby greatly increasing the surface area of the root to promote increased absorption.
Phosphate limitation is one of the most common nutrient stresses that plants face when growing in fields for agriculture production. This is because roots can only absorb inorganic phosphates, which are naturally present at very low concentrations in soil (Patrick and Khalid, 1974). Therefore, plants have developed numerous mechanisms by which to maximize the uptake of this nutrient in phosphate poor soil (Niu et al., 2013; Williamson et al., 2001). In fact, researchers have described three major changes in Arabidopsis thaliana (hereafter Arabidopsis) root development during phosphate starvation. First, the primary root ceases downward growth, with a subsequent increase in lateral roots branching away from primary roots (Linkohr et al., 2002). Additionally, the root epidermis dramatically increases the number of root hair cells, while also increasing hair length (Bates and Lynch, 1996). Finally, root epidermal cells secrete acid phosphatases, enzymes that catalyze the conversion of organic into inorganic phosphates, which can be subsequently absorbed (Gilbert et al., 1999). Thus, there is a clear link between response to phosphate starvation and root hair cell fate. However, the molecular mechanisms by which exogenous phosphate levels regulate this cell fate decision are not fully understood.
To better understand this cell fate decision, previous studies have focused primarily on understanding the transcriptional networks present in both hair and nonhair cells (Lee and Schiefelbein, 2002). Two key transcription factors that function in this process are WEREWOLF (WER) and CAPRICE (CPC), which promote nonhair cell (Ryu et al., 2005), and hair cell fate, respectively (Wada et al., 1997). Plants that have null mutations in these genes have dramatic root epidermal phenotypes. However, hair and nonhair cells are never fully absent (Lee and Schiefelbein, 2002). The presence of both cell types, even when these key transcription factors are absent, suggests that there are other pathways that regulate root hair cell fate. In fact, more recent studies have begun to appreciate the numerous post-transcriptional processes that may influence this cell fate decision. Specifically, a recent study identified hair cell specific alternative splicing events (Lan et al., 2013), indicating splicing as one potential post-transcriptional mechanism of cell fate decision regulation.
Beginning with its transcription, each RNA molecule is bound by an ever-changing cohort of RNA-binding proteins (RBPs). These proteins regulate RNA stability, post-transcriptional processing (capping, splicing, etc.), export, localization, and translation (Jangi and Sharp, 2014; Vandivier et al., 2016; Younis et al., 2013). Furthermore, a single RBP can bind to and potentially regulate the transcripts encoded by thousands of different genes (Cruz and Westhof, 2009), allowing these proteins to act as master regulators of developmental switches (Han et al., 2013; Warzecha et al., 2009). However, whether RBPs regulate Arabidopsis root hair cell fate decisions and development is currently unknown.
Like transcription factors, RBPs bind to primary sequence motifs. However, the intricate secondary structures that each RNA forms adds an additional mechanism to regulate RBP-binding (Cruz and Westhof, 2009). More specifically, the structure of an RNA molecule can make RBP recognition sequences inaccessible to a single-stranded RNA (ssRNA) binding protein, or promote binding by a double-stranded RNA (dsRNA) binding protein, or vice versa (Cruz and Westhof, 2009). Therefore, both the RNA sequence and its secondary structure are important cis regulators of RNA-protein interactions.
In this study, we utilized our protein interaction profile sequencing (PIP-seq) technique to simultaneously probe RNA secondary structure and RNA-protein interactions in the nuclei of Arabidopsis root hair and nonhair cells. This analysis revealed cell type specific secondary structure and RBP binding patterns, some of which influence root epidermal cell development. Additionally, these protein-bound sequences were used to identify two RBPs, SERRATE and GLYCINE-RICH PROTEIN 8 (GRP8), that both regulate proper hair cell development. Together, these data elucidate novel post-transcriptional regulators of the plant root epidermal cell fate decision and development.
RESULTS
PIP-seq identifies thousands of cell type specific protein-bound sites
To identify the differences in the nuclear RNA-protein interaction and RNA secondary structure landscapes of root hair and nonhair cells, we used the isolation of nuclei tagged in specific cell types (INTACT) method (Deal and Henikoff, 2010; Wang and Deal, 2015) to obtain highly pure nuclear samples (Figure S1A). This technique utilizes cell type specific promoters to drive expression of a fusion protein that targets a biotin ligase receptor peptide to the nuclear envelope. Therefore, by using plants that expressed this fusion protein under the control of the ADF8 or GL2 promoters we were able to specifically purify nuclei from root hair and nonhair cells, respectively (Figure 1A). In fact, we obtained highly pure nuclei from both cell types that were completely devoid of the cytoplasmic and rough endoplasmic reticulum markers EIF1A, ALDOLASE, and CNX1 (Figure S1A). These highly pure nuclei were then used for subsequent PIP-seq analyses.
Figure 1. Nuclear PIP-seq identifies cell type specific RNA-protein interactions.
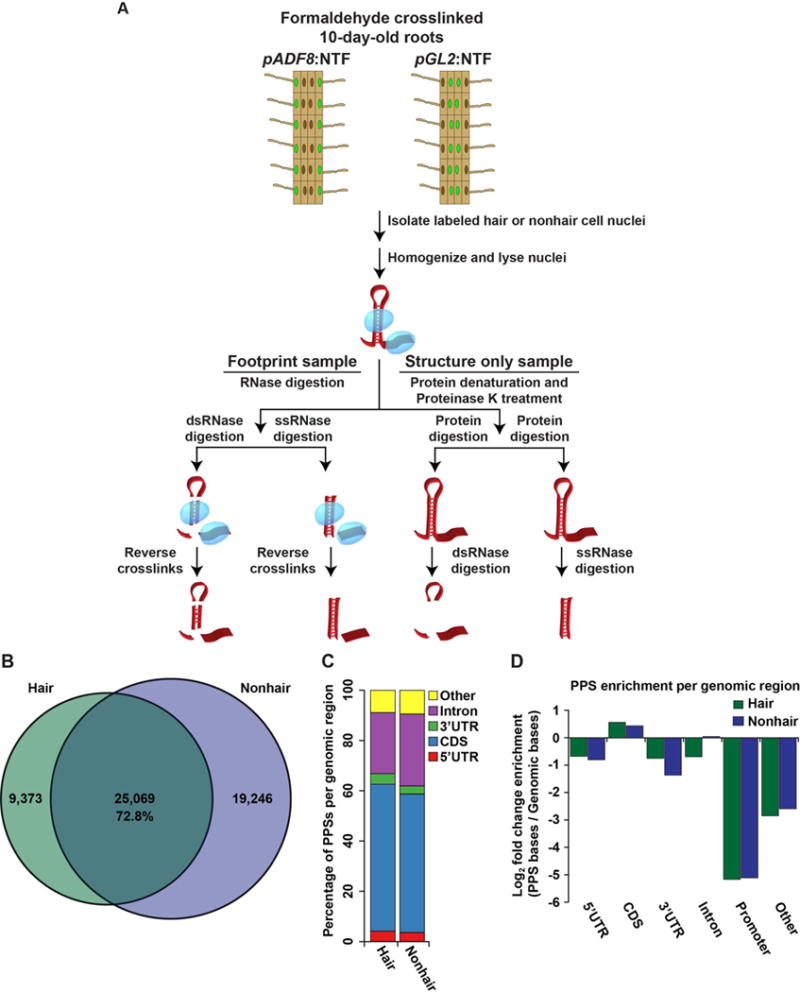
(A) The PIP-seq approach in the nucleus of Arabidopsis root hair and nonhair cells. Fully differentiated root epidermal cells were excised from 10-day-old Arabidopsis plants and crosslinked with a 1% formaldehyde solution. The nuclei of either root hair or nonhair cells (green circles) were then isolated via the INTACT technique. Nuclei were lysed and separated into footprinting and structure only samples. Four total sequencing libraries were then prepared for each replicate experiment as previously described (Gosai et al., 2015). (B) Overlap between protein protected sites (PPSs) identified in hair (green) or nonhair (purple) cell nuclei. The intersection indicates PPSs identified in both cell types that overlap by at least a single nucleotide. (C) Absolute distribution of PPSs throughout regions of mRNA transcripts. (D) Genomic enrichment of PPS density, measured as log2 enrichment of the fraction of PPS base coverage normalized to the fraction of genomic bases covered by indicated nuclear mRNA regions for hair (green bars) and nonhair (purple bars) cells. See also Figures S1–S3.
PIP-seq allows global identification of RNA-protein interaction sites as well as RNA secondary structure (Figure 1A) (Gosai et al., 2015; Silverman et al., 2014). We used ~2 million highly pure nuclei (Figure S1A) for each of two PIP-seq replicates per cell type. These nuclei were lysed, then divided into footprinting and structure only samples (four total libraries per replicate) (Figure 1A). To globally identify RBP-bound RNA sequences, footprinting samples were directly treated with an RNase specific to either ssRNA or dsRNA (ssRNase or dsRNase, respectively), followed by protein denaturation and sequencing library preparation. In contrast, the structure only samples first had proteins denatured in SDS and degraded with Proteinase K prior to RNase digestion. Denaturation of proteins before RNase treatment makes sequences that were RBP-bound in the footprinting sample accessible to RNases in these reactions. Thus, sequences that are enriched in footprinting relative to structure only samples are identified as protein protected sites (PPSs) (Gosai et al., 2015; Silverman et al., 2014) (Figure 1A). Additionally, using the structure only libraries allowed us to determine the native (protein-bound) RNA base-pairing probabilities for the nuclear transcriptomes of Arabidopsis root hair or nonhair cells, as previously described (Gosai et al., 2015; Li et al., 2012a).
The resulting PIP-seq libraries were sequenced and provided ~25–35 million raw reads per library. To determine reproducibility, we used a principle component analysis of read coverage in 100 nucleotide (nt) bins. This revealed that biological replicates of each library from the distinct cell types cluster together (Figure S1B), indicating the high quality and reproducibility of our root hair and nonhair nuclear PIP-seq libraries.
To identify PPSs, we used a Poisson distribution model to identify enriched regions in the footprinting compared to the structure only libraries at a false discovery rate (FDR) of 5%, as previously described (Gosai et al., 2015). We identified a total of 34,442 and 44,315 PPSs in root hair and nonhair cell nuclei, respectively (Table S1). To estimate the functional relevance of these nuclear PPSs from both root cell types, we compared flowering plant PhastCons conservation scores (Li et al., 2012a) for PPSs and equal-sized flanking regions. We found that PPS sequences were significantly (p values < 1.2 × 10−71; KS-test) more evolutionarily conserved than flanking regions in both hair and nonhair cells (Figure S1C), indicating that there is evolutionary pressure to constrain these sites, likely due to their ability to interact with RBPs (Gosai et al., 2015).
Additionally, we observed a high overlap of PPSs between biological replicates. Whereas CLIP-seq experiments will often find <35% of protein-bound sites shared between biological replicates (Lebedeva et al., 2011), we observed ~72% of dsRNase identified PPSs, and ~57% of ssRNase identified PPSs found in our first replicate are shared between both biological replicates (Figures S2AB), with ~55–64% of hair cell and 27–36% of nonhair cell PPSs being identified by both ssRNase and dsRNase treatments (Figures S2C–D). When comparing total identified PPSs found in hair cells, we observed 25,069 (72.8%) PPSs are also present in nonhair cells (Figure 1B). Interestingly, we found 16,460 (72.4%) of dsRNase identified hair cell PPSs are common to both cell types (Figure S3A), whereas only 4,323 (34.4%) of ssRNase identified PPSs are common (Figure S3B), with the remaining 4,286 shared PPSs being identified in the dsRNase sample of one cell type and the ssRNase sample of the other cell type. Therefore, these data reveal that many cell type specific protein-bound events are present in lowly-structured, ssRNase-accessible regions.
We next confirmed that these are true differences in protein occupancy at cell type specific PPSs, rather than a representation of differentially expressed mRNAs. To do this, we analyzed PPSs present only in mRNAs expressed in both hair and nonhair cells. We found that the PPSs from both hair and nonhair cells within this subset of transcripts displayed an overlap of 73.4% (Figure S3C), supporting the conclusion that we have identified a subset of cell type specific RNA-protein interactions.
A classification of hair and nonhair cell PPSs revealed that >90% of these sites are localized to mRNAs, with the largest fractions occupying the coding sequence (CDS; ~55%) and introns (~25%) in both cell types (Figure 1C). We then tested the enrichment of PPSs in specific nuclear mRNA regions (e.g., CDS, introns, etc.) by comparing the number of PPS occupied nucleotides to the number of bases annotated as each feature in the TAIR10 Arabidopsis genome. We found that PPSs identified in both cell types were enriched in CDSs, while underrepresented in both untranslated regions (UTRs). Furthermore, introns showed a slight enrichment for PPSs in hair cells, but an underrepresentation in nonhair cells (Figure 1D). These findings are consistent with our previous results using nuclei isolated from whole seedlings (Gosai et al., 2015), both of which indicate that CDSs are highly RBP bound in plant nuclei.
Hair and nonhair cells have distinct RNA-protein interaction and RNA secondary structure profiles in shared mRNAs and lncRNAs
To interrogate the landscape of RBP binding and RNA secondary structure in specific regions of nuclear mRNAs expressed in both hair and nonhair cells, we first calculated their structure scores and PPS densities. The structure score is a generalized log ratio of ds- to ssRNA-seq reads at each nucleotide position. These raw scores are then scaled by generating Z-scores (Berkowitz et al., 2016), with positive and negative scores indicating high likelihood of ds- and ssRNA, respectively (see STAR Methods). To examine the relationship between PPS density and structure score, we focused on the 100 nt up- and downstream of the start and stop codons of nuclear mRNAs expressed in both cell types. From this analysis, we observed the highest PPS density in the CDS with decreased occupancy within the 5′ and 3′ UTRs (Figures 2A–B), consistent with the overall PPS localization and enrichment analysis (Figures 1C–D).
Figure 2. Hair and nonhair cells have distinct RNA-protein interaction and RNA secondary structure profiles.
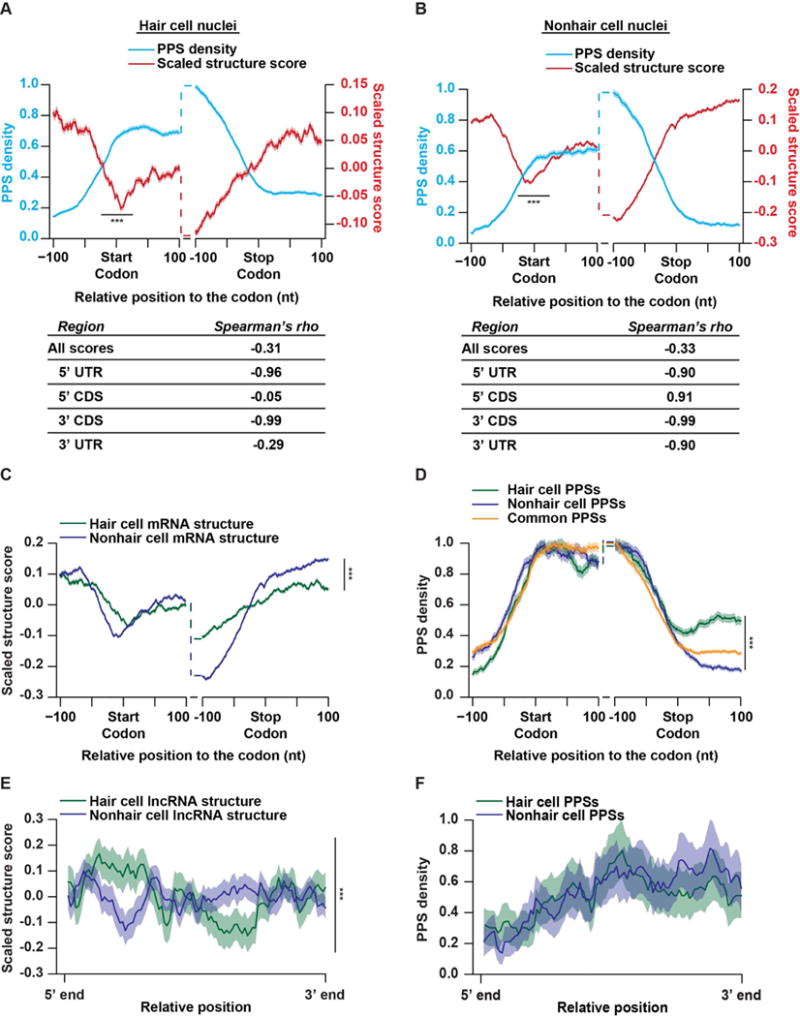
(A–B) PPS density (blue line) and scaled structure score (red line) profiles for nuclear mRNAs at each nucleotide +/− 100 nt from the annotated start or stop codons in hair (A) or nonhair (B) cell nuclei. The tables represent the Spearman’s rho correlations between the PPS density and scaled structure scores across the graphed windows up- and downstream of the start codon, stop codon, or across all detectable mRNA transcripts. (C–D) Scaled structure score (C) or PPS density (D) profiles at each nucleotide +/− 100 nt from the annotated start or stop codons in nuclear mRNAs expressed in both hair (green line) and nonhair (purple line) cells. PPSs are divided into those that are detected in hair cells (green line), nonhair cells (purple line), or common to both cell types (orange line). (E–F) Scaled structure score (E) or PPS density (F) across binned unspliced lncRNAs expressed in root hair (green) or nonhair (purple) cell nuclei. Shading around the solid lines indicates standard error of the mean (SEM) across all detectable transcripts. ***indicates p value < 0.001, Wilcoxon test in all panels. See also Figures S1 and S2.
In contrast to RBP occupancy, we found that secondary structure was higher in both UTRs compared to the CDS within the regions analyzed in both cell types. Additionally, we observed a significant (p values < 6.6 × 10−13; Wilcoxon test) dip in secondary structure directly over start codons, as well as upstream of the stop codon (Figures 2A–B), two characteristics which have been observed in numerous studies of RNA secondary structure across various organisms (Ding et al., 2014; Gosai et al., 2015; Li et al., 2012a, 2012b). Additionally, all of these results are consistent with the patterns observed previously for nuclear mRNA secondary structure from whole seedling nuclei (Gosai et al., 2015). Thus, these structural characteristics across the UTRs and CDS seem to be a consistent feature of the Arabidopsis nuclear mRNA transcriptome.
Consistent with our study of whole seedling nuclei, our combined analyses of RBP binding and RNA secondary structure revealed that these features are anticorrelated across nuclear mRNAs (Spearman’s rho ≤ −0.31; p value < 2.2 × 10−16; asymptotic t approximation) in both root epidermal cell types. In addition to this transcriptome-wide pattern for both cell types, we found even stronger anticorrelations (Spearman’s rho ≤ −0.90; p value < 2.2 × 10−16; asymptotic t approximation) between protein binding and RNA folding within the last 100 nt of 5′ UTRs and CDSs of nuclear mRNAs expressed in both hair and nonhair mRNAs. Interestingly, we observed a discrepancy within the first 100 nt of mRNA 3′ UTRs from root hair and nonhair cells. Specifically, we found a strong negative correlation (Spearman’s rho ≤ −0.99; p value < 2.2 × 10−16; asymptotic t approximation) between protein binding and structure in nonhair cell nuclei, with a much more mild correlation (Spearman’s rho ≤ −0.29; p value < 0.0036; asymptotic t approximation) in hair cell nuclei. This distinct pattern indicates that there may be differential protein binding in the 3′ UTRs of these two cell types.
Conversely, the RBP binding and RNA secondary structure of the first 100 nt of the CDS did not exhibit an anticorrelation. We found no significant correlation in hair cells, as well as a significant positive correlation (Spearman’s rho > 0.91; p value < 2.2 × 10−16; asymptotic t approximation) in nonhair cells. These observations are striking as they oppose the anticorrelation that we found in this same region when profiling mixed nuclei from whole seedlings (Gosai et al., 2015). Taken together, these observations reveal a cell type specific interplay between RNA folding and RBP binding near the start codon of nuclear mRNAs. Given that these results are from highly pure nuclear samples (Figure S1A), the PPSs cannot simply indicate ribosome binding, and are likely caused by cell type specific RBP interactions. Identifying and characterizing these proteins will be the focus of future inquiry.
We next aimed to directly compare the RNA secondary structure patterns in the nuclei of these two cell types by using these scaled structure scores. We found that RNA secondary structure is similar in both cell types within the 200 nt window flanking the start codon (Figure 2C). Conversely, there are significant (p values < 3.1×10−4; Wilcoxon test) differences in RNA secondary structure within the 100 nt windows up- and downstream of the stop codons of mRNAs found in both hair and nonhair cell nuclei. Specifically, we found significantly higher RNA secondary structure in these mRNAs within the last 100 nt of their CDSs in hair compared to nonhair cells, while the opposite pattern was observed for the first 100 nt of their 3′ UTRs (p values < 1.7×10−5 and 2.2×10−16, respectively; Wilcoxon test) (Figure 2C). These differences in secondary structure around the stop codon could provide an intriguing mechanism for regulating RBP binding within these specific transcript regions. Therefore, we also directly compared the density of hair and nonhair cell specific as well as common PPSs (Table S2) in the 200 nt regions surrounding the start and stop codons of mRNAs expressed in both hair and nonhair cells (Figure 2D). Although overall RBP binding had a similar profile across mRNAs from both cell types (Figures 2A–B), there is a significant (p value < 3.4×10−15; Wilcoxon test) increase in hair cell specific RBP binding events within the first 100 nt of the 3′ UTRs of mRNAs expressed in both cell types (Figure 2D). These findings are consistent with the significantly (p value < 2.2×10−16; Wilcoxon test) decreased RNA secondary structure also observed in this transcript region in hair compared to nonhair cells (Figure 2C), given that these features are generally anticorrelated with one another (Figures 2A–B). Thus, this nuclear PIP-seq analysis reveals cell type specific differences in both RNA secondary structure and RBP binding profiles between hair and nonhair cells. In total, our findings suggest that cell type specific RNA folding and RBP binding in protein-coding mRNAs is a likely mechanism for differential regulation of the root hair and nonhair cell transcriptomes, and the resulting cell fate decisions.
In addition to mRNAs, we examined both RNA secondary structure and RNA-protein interactions in long noncoding RNAs (lncRNAs) that are found in the nucleus. Using a comprehensive list of Arabidopsis lncRNAs (Liu et al., 2012), we first analyzed the secondary structure of these transcripts in root hair and nonhair cell nuclei (Figure 2E). Taking the entire length of the unspliced annotated lncRNAs, we divided each transcript into 100 equally sized bins. Graphing the average scaled structure score of each bin, we found significant (p value < 2.2×10−16; Wilcoxon test) differences between the structure profiles of the lncRNAs found in both root hair and nonhair cells. Specifically, these lncRNAs in root hair cell nuclei exhibited increased structure at the 5′ end of the transcript, while being less structured near the 3′ end than these lncRNAs in nonhair cell nuclei (Figure 2E). As the structural profiles differ dramatically, we next examined PPS binding across lncRNAs. This analysis revealed that a vast majority (>82%) of lncRNA mapping PPSs in hair cells are shared with nonhair cell nuclei. Unsurprisingly, when graphing the PPS density across all lncRNAs identified in root hair or nonhair cells, these profiles were not significantly different (p value > 0.05; Wilcoxon test) (Figure 2F). Therefore, like mRNAs, lncRNAs exhibit cell type specific secondary structure. However, unlike mRNAs, these differences do not result in a significant difference in RBP binding across these transcripts. Although these transcripts are bound by similar numbers of proteins in each cell type, this difference in secondary structure likely indicates that differing cohorts of proteins are binding lncRNAs in hair and nonhair cells.
SERRATE regulates root hair length and hair cell fate in a microRNA-independent and a microRNA-dependent manner, respectively
To determine whether cell type specific RBP binding regulates the root hair and nonhair cell fate decision, we identified RBPs that function in a cell type specific manner. It is worth noting that all PPSs correspond to protein-bound sites in fully differentiated cells, rather than non-terminally differentiated precursor cells. Therefore, only protein binding sites established in these precursor cells, and maintained after differentiation will lead to the identification of RBPs necessary for cell fate decision. To identify those PPSs potentially necessary for differentiation, we subsetted all identified PPSs into those that are hair or nonhair cell specific as well as those common to both cell types (Figure 1B and Table S2). Taking these three subsets of RBP bound sequences, we used the motif finding algorithm MEME (Bailey et al., 2009) to identify enriched protein-bound sequences. We identified a combined 54 significantly (E values < 0.01) enriched motifs using these three subsets (Table S3).
To identify the specific RBPs that interact with a subset of these motifs, we performed RNA affinity chromatography followed by mass spectrometry analysis. In this technique, we covalently attached a synthetic RNA motif or a scrambled sequence control to agarose beads. We then incubated these RNA baits, as well as a bead-only control, with whole root lysate, and stringently washed away any weakly bound proteins. The specifically bound proteins were identified via mass spectrometry. Using this approach, we identified 58 annotated RBPs that are at least 4-fold enriched for interaction with at least one of the twelve tested sequence motifs, as compared to the scrambled sequence and bead-only negative controls (Figure S4 and Table S4). One motif of particular interest, a GGN repeat motif that was enriched in PPSs common to both root hair and nonhair cell nuclei, was found to interact with the RBP SERRATE (SE) (AT2G27100) (Figure 3A). SE is known to function in conjunction with ABA HYPERSENSITIVE 1/CAP-BINDING PROTEIN 80 (ABH1/CPB80, AT2G13540) and HYPONASTIC LEAVES 1 (HYL1, AT1G09700) in microRNA (miRNA) biogenesis, where these three RBPs recruit DICER-LIKE 1 (DCL1, AT1G01040) to primary miRNA transcripts to allow their processing to mature miRNAs (Dong et al., 2008; Yang et al., 2006). Additionally, SE and ABH1/CBP80 regulate alternative splicing across the Arabidopsis transcriptome. This variety of functions indicated that SE was a reasonable candidate as a potential regulator of root hair cell fate. We first confirmed that SE is expressed at similar levels in both root hair and nonhair cell nuclei (Figure S6A), then performed RNA immunoprecipitation (RIP) followed by RT-qPCR to confirm that SE interacts with transcripts containing the GGN repeat motif in vivo. To do this, we incubated lysates from formaldehyde crosslinked roots with polyclonal α-SE, α-ABH1/CBP80, or the negative control rabbit IgG antibody. We first confirmed pulldown of SE and ABH1/CBP80 by these antibodies (Figure S5), then performed RT-qPCR for 13 GGN repeat containing mRNAs (Table S5). We found that all 13 of the transcripts were significantly (all p values < 0.05; Welch’s t-test) enriched >1.5-fold in the α-SE compared to the IgG control RIP samples, as opposed to the ACTIN2 negative control (Figure 3B). Furthermore, none of the 13 transcripts were enriched in α-ABH1/CBP80 compared to the IgG control RIP samples. Taken together, these findings indicate that SE interacts in vivo with GGN motif-containing mRNAs, while ABH1/CBP80 does not.
Figure 3. SERRATE regulates hair cell fate and hair length in a partially microRNA-independent manner.
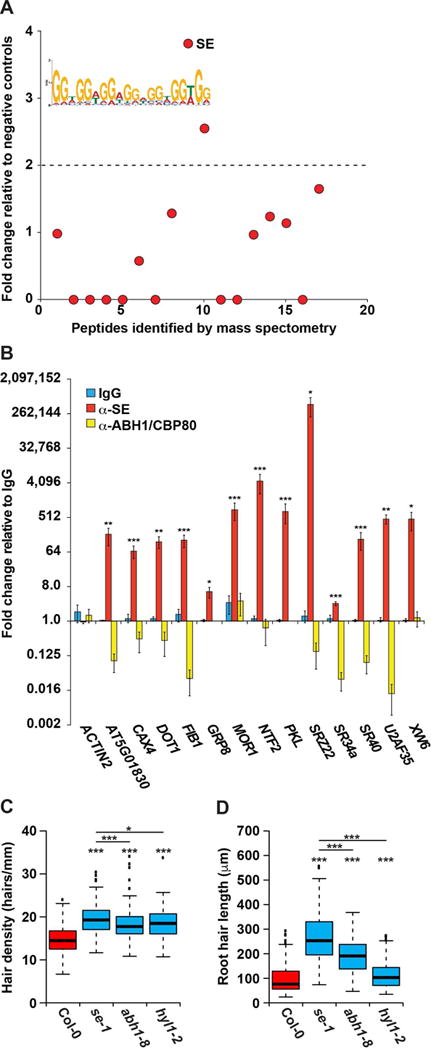
(A) RNA affinity chromatography followed by LC-MS was performed on whole root cell lysate using the MEME identified GGN repeat motif as bait. The number of peptide spectrum matches (PSMs) for each identified peptide was graphed as fold change over the average PSMs of scrambled RNA bait and no RNA controls. Peptides above the dotted line have a more than 2-fold change and correspond to candidate RBPs. SE is denoted as being highly bound by our analysis. (B) RIP-qPCR was performed on whole root lysate using rabbit α-IgG (blue bars), α-SE (red bars), or α-ABH1/CBP80 (yellow bars) antibodies, graphed as fold change relative to the IgG negative control pull down, n = 4. Error bars indicate SEM. *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, Welch’s t-test. (C–D) Root hair cell density (hairs/mm) (C) and root hair length (μm) (D) of Col-0, se-1, abh1-8, and hyl1-2 mutant plants. For analysis of root hair length n=400, and for root hair density n > 135. *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, while N.S. denotes p value > 0.05, Wilcoxon test. See also Figures S1, S2, and S4–S6.
After validating in vivo GGN motif-containing mRNA binding by SE, we next determined whether this protein regulates root hair cell fate and development. To do this, we measured the root hair cell density (hairs/mm) and root hair length in 8-day-old wildtype Col-0 (hereafter WT) and SE hypomorphic (se-1) seedlings (Clarke et al., 1999; Serrano-Cartagena et al., 1999). From this analysis, we found that se-1 mutant seedlings had significantly (p values < 2.2×10−16; Wilcoxon test) more root hair cells that are significantly (p values < 2.2×10−16; Wilcoxon test) longer as compared to WT (Figures 3C–D), indicating that SE functions in both promoting root nonhair cell fate and terminating root hair extension. The difference in hair cell density on se-1 plants could be caused by either promotion of hair cell fate, resulting in ectopic hair cells, or by decreased epidermal cell size, packing hair cells closer together. Therefore, we measured the size of hair cell bodies and found that there is no significant (p value > 0.05; Wilcoxon test) difference in their size in se-1 compared to WT roots (Figure S6B). Combined, these findings demonstrate that SE functions both in precursor epidermal cells to promote nonhair cell fate, as well as in differentiated hair cells to terminate hair growth. This variety of functions is unsurprising as this RBP binds (Figure 3B) and post-transcriptionally regulates many different transcripts (Clarke et al., 1999; Raczynska et al., 2014).
We next tested whether SE influences the cell fate decision by functioning in the same pathway as the canonical transcription factors CAPRICE and WEREWOLF. Taking CAPRICE null (cpc-1) and wildtype WS plants as a control, we measured levels of SE and GL2 in plant roots, while 5S rRNA was used as a control. Given that cpc-1 plants are enriched in nonhair cells, we observed the expected significant (p value < 0.05; Welch’s t-test) increase in abundance of the nonhair cell specific GL2 marker gene. Conversely, no significant difference in SE level was observed between wildtype WS and cpc-1 plants (Figure S6C), which was consistent with our western blot results (Figure S6A). Additionally, we measured the abundance of several transcription factors known to function in the CAPRICE/WEREWOLF pathway in the roots of se-1 plants, and also observed no significant (p value > 0.05; Welch’s t-test) difference in their levels compared to wildtype plants (Figure S6D). Together, these data suggest that SE functions independently of the canonical CAPRICE/WEREWOLF pathway in determining root hair cell fate.
As SE functions in both microRNA biogenesis and alternative splicing, our next goal was to differentiate the effect of these two regulatory mechanisms on its function in root hair cell development. To do this, we looked for root hair length and density phenotypes in null mutants of ABH1/CBP80 (abh1-8) and hypomorphic mutants of HYL1 (hyl1-2), both of which are known to function in conjunction with SE during plant miRNA biogenesis. We measured root hair density for 8-day-old WT, abh1-8, and hyl1-2 seedlings and found significant (p values < 5.6×10−15; Wilcoxon test) increases in the density of root hairs in both abh1-8 and hyl1-2 mutant compared to WT (Figure 3C). These increases were similar in magnitude to those seen in the se-1 mutant seedlings, indicating that this root hair cell fate phenotype is miRNA biogenesis dependent. Additionally, we found the root hair lengths in abh-1 and hyl1-2 seedlings to be significantly (p values < 3.7 × 10−9; Wilcoxon test) longer than those of WT. However, they are also significantly (p values < 2.2×10−16; Wilcoxon test) shorter than those observed for se-1 seedlings (Figure 3D). This mild increase in hair length in abh1-8 and hyl1-2 mutant roots indicates that decreased miRNA biogenesis in se-1 plants accounts for a portion of the root hair length phenotype. However, there are also important SE-specific regulatory functions that add to the increased hair length observed in se-1 mutant seedlings. Taken together, these findings reveal that although the function of SE in the microRNA biogenesis pathway is required for regulating root hair cell fate, this protein also has specific effects on root hair length.
In order to better understand these SE-specific effects on hair length, we investigated the root phenotypes of mutants lacking one of several GGN motif-containing genes that we found were bound by SE (Figure 3B). Although none of these genes are known to function in root hair cell fate, three of them have known roles in root development. CATION EXCHANGER 4 (CAX4, AT5G01490) promotes both primary and lateral root growth in plants subjected to Cd2+ toxicity (Mei et al., 2009). MICROTUBULE ORGANIZATION 1 (MOR1, AT2G35630) regulates microtubule assembly, and when temperature sensitive mor1-1 mutants are grown at the restrictive temperature there is an increase in primary root diameter (Whittington et al., 2001). Additionally, the chromatin-remodeling factor PICKLE (PKL, AT2G25170) is necessary for silencing embryonic genes and promoting lateral root development (Furuta et al., 2011; Ogas et al., 1999). Interestingly, when screening 8-day-old seedlings lacking any one of these proteins (cax4-1, mor1-1, and pkl1-1) we found significantly (all p values < 0.001; Wilcoxon test) aberrant root hair length as compared to WT (Figures 4A–C). Specifically, we observed that cax4-1 and pkl1-1 mutant seedlings had longer root hairs (Figures 4A and 4C), similar to se-1. Conversely, we found that mor1-1 mutant seedlings grown at the restrictive temperature displayed shorter root hairs compared to WT (Figure 4B). Taken together, these data suggest that the increased root hair length observed for se-1 plants is likely due to the additive effects of misregulation of numerous mRNA substrates. We next confirmed that CAX4, MOR1, and PKL transcripts are not regulated by the CAPRICE/WEREWOLF transcription factor network by measuring their abundance in the roots of wildtype WS and cpc-1 plants. From this analysis, we observed no significant (p value > 0.05; Welch’s t-test) difference (Figure S6E) in the levels of these three in the absence of CAPRICE function.
Figure 4. SE bound GGN motif containing genes regulate root hair cell development.
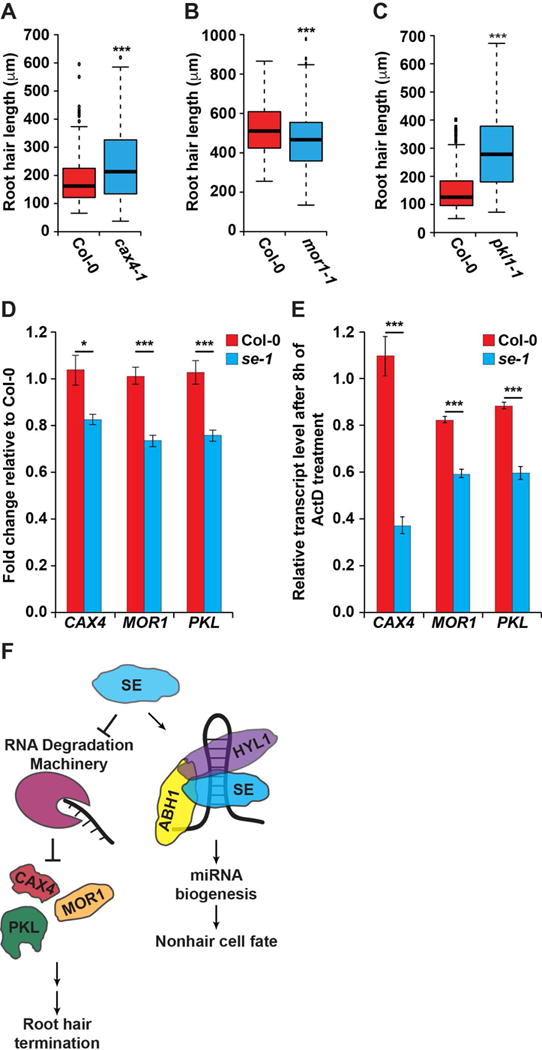
(A–C) Root hair length for null cax4-1 (A), mor1-1 (B), and pkl1-1 (C) mutant plants as compared to wild type Col-0. For root hair length analysis n=200. *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, Wilcoxon test. (D) RT-qPCR of SE bound genes in WT (red) and se-1 (blue) roots, n = 6. (E) Roots from both WT (red) and se-1 (blue) plants were subjected to Actinomycin D treatment for 8 hours to inhibit transcription, followed by RT-qPCR analysis of the mRNAs noted in the figure, n = 3. *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, Welch’s t-test. Error bars indicate SEM. (F) A model of the role of SE in both the microRNA-independent promotion of root hair termination, as well as the microRNA-dependent promotion of the nonhair cell fate. See also Figures S1, S2, and S4–S6.
To test how SE affects the abundance of these RNAs, we measured their levels in the roots of WT and se-1 plants. We found that all three transcripts are significantly (all p values < 0.05; Welch’s t-test) decreased in se-1 roots (Figure 4D), suggesting that SE stabilizes these transcripts. To further test this idea, we excised roots from 8-day-old WT and se-1 plants and incubated them in liquid media containing the transcription inhibitor Actinomycin D (Act D). Specifically, we incubated these roots for 8 hours in this media, and then measured changes in transcript levels in the absence of transcription via RT-qPCR. This analysis revealed significantly (p value < 0.001; Welch’s t-test) decreased transcript levels in se-1 roots compared to WT, indicating decreased transcript stability in the absence of SE function (Figure 4E). In total, our results reveal that SE promotes the nonhair cell fate in a miRNA biogenesis dependent manner, while also terminating root hair growth by stabilizing the mRNA transcripts of proteins involved in specifying hair length in plant roots (Figure 4F).
GRP8 regulates root hair cell fate independently of GRP7
As we had observed a dramatic difference in secondary structure and protein binding in the first 100 nt of 3′ UTRs of root hair and nonhair cell mRNAs (Figures 2C–D), we next examined this region for enriched protein-bound motifs. To do this, we performed MEME on all hair or nonhair cell PPSs located in this area. While we did not observe any significant (E-value < 0.05) motifs in nonhair cell PPSs, we identified three significant (E-value < 0.05) motifs in hair cell PPSs (Figure S7A). Although the two GA-rich motifs are similar to other motifs identified in nonhair cell PPSs and common PPSs (Table S3), the TG-rich motif was only previously identified in hair cell specific PPSs (Table S3). As this motif is enriched in regions of differential protein binding, we next aimed to identify what proteins are able to bind to this sequence, and determine their role in root hair development.
We performed RNA affinity chromatography using this TG-rich motif, and found four annotated RBPs that were >10-fold enriched over our negative controls. In addition to RBP45A (AT4G54900), we found multiple members of the family of GLYCINE-RICH PROTEINs, GRP2 (AT14G13850), GRP7 (AT2G21660), and GRP8 (AT4G39260) interacted with this sequence motif (Figures 5A and S4 and Table S4). GRPs are nuclear localized hnRNP-like proteins (Streitner et al., 2012) that are required for numerous processes in plants, including responses to various biotic and abiotic stresses via their function in regulating both alternative splicing and microRNA biogenesis (Lewinski et al., 2016). Therefore, we were unsurprised when we observed that similar levels of GRP7 and GPR8 were present in the nuclei of both root hair and nonhair cells (Figure S6F). Using an antibody that recognizes both native GRP7 and GRP8, we performed RIP-qPCR to validate in vivo binding of GRP7/8 to TG-rich motif containing transcripts (Table S5) in formaldehyde-crosslinked whole root lysate. Given that both GRP7 and GRP8 are known to bind the GRP8 transcript (Schöning et al., 2008), we used it as a positive control, and identified a significant enrichment of this transcript in the α-GRP7/8 compared to our rabbit IgG negative control pulldown (Figure 5B). Of the eight TG-rich motif containing mRNAs tested, we found six genes to be significantly (all p values < 0.05; Welch’s t-test) enriched in the α-GRP7/8 compared to the IgG negative control pulldown (Figure 5B). These data reveal either GRP7, GRP8, or both proteins bind to TG-rich motif-containing transcripts in vivo.
Figure 5. GRP8 regulates root hair cell fate in a GRP7-independent manner.
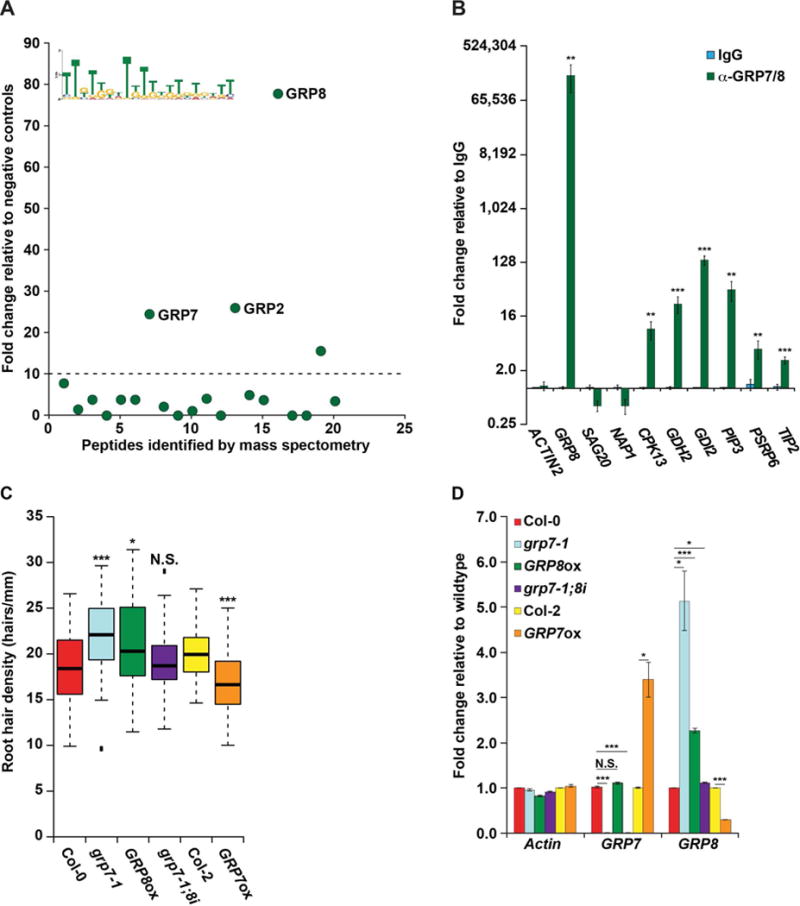
(A) RNA affinity chromatography followed by LC-MS was performed on whole root cell lysate using the MEME identified TG-rich motif as bait. Peptides above the dotted line have a more than 10-fold change and are candidate RBPs, with three GRPs denoted. (B) RIP-qPCR was performed on whole root lysate using rabbit IgG (blue bars) or rabbit serum raised against GRP7 and GRP8 (green bars) graphed as fold change relative to IgG. (C) Root hair cell density was measured in 8-day-old seedlings of WT or plants with decreased or increased GRP7 (grp7-1 or GRP7ox, respectively), increased GRP8 (GRP8ox), or decreased GRP7 with WT levels of GRP8 (grp7-1;8i), n > 50. * and *** denote p value < 0.05 and 0.001, respectively, while N.S. denotes p value > 0.05, Wilcoxon test. (D) RT-qPCR of root tissue from lines with altered GRP7 and/or GRP8 levels, graphed as fold change relative to WT (Col-0 or Col-2). For (B) and (D), *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, Welch’s t-test. Error bars indicate SEM. See also Figures S1, S2, S4, S6, and S7.
As the GRP7/8 bound motif was enriched specifically in hair cell PPSs, we tested plants aberrantly expressing these proteins for root hair cell phenotypes. From this analysis, we found that root hair cell density in the grp7-1 null mutant is significantly (p value < 3.3 × 10−7; Wilcoxon test) increased compared to WT plants (Figure 5C). In accordance, plants overexpressing GRP7 (GRP7ox) demonstrate significantly (p value < 2.8 × 10−8; Wilcoxon test) decreased hair cell density compared to their respective WT plants (Col-2) (Figure 5C). As mentioned previously, GRP7 is known to bind to GRP8 transcripts, thereby decreasing GRP8 expression levels (Schöning et al., 2008), resulting in the grp7-1 and GRP7ox lines exhibiting significantly (p values < 0.05; Welch’s t-test) increased or decreased GRP8 levels as compared to WT plants, respectively (Figure 5D). Thus, to differentiate the effects of each protein in hair cell differentiation, we required additional mutant plant lines. For instance, we identified a mutant line with an insertion in the GRP8 promoter (CS803581/SAIL_75_G05; hereafter referred to as GRP8ox) that resulted in a significant (p value < 0.001; Welch’s t-test) increase in the levels of GRP8 mRNA in these plants relative to WT. Importantly, this increase in GRP8 levels does not cause a concomitant alteration in GRP7 abundance in GRP8ox plants (Figure 5D). We examined root hair cell density in these plants, and revealed a significantly (p value < 0.015; Wilcoxon test) increased root hair density as compared to WT, strongly suggesting that this is a GRP8-dependent phenotype (Figure 5C). To determine the effects of altering GRP7 alone on root hair cell fate, we also measured the density of these cells in a plant line that contains a GRP7 null mutation (grp7-1), as well as an artificial microRNA targeting GRP8, which returns the levels of this mRNA close to those of WT (hereafter grp7-1;8i) (Streitner et al., 2012) (Figure 5D). We found that these plants exhibit a similar root hair density as WT (p value > 0.825; Wilcoxon test) (Figure 5C), indicating that this is indeed a GRP7 independent phenotype. Therefore, the grp7-1 plants only exhibited increased root hair cell density as a result of increased GRP8 levels, not due to the absence of GRP7.
We next confirmed that this phenotype was due to ectopic hair cell production, rather than changes in the size of epidermal hair cells (Figure S6G). Like our analysis of SE, we probed GRP7 and GPR8 for a potential role in the CAPRICE/WEREWOLF pathway using qPCR analyses. Similar to the results for SE, we observed no significant (p value > 0.05; Welch’s t-test) difference in GRP7 or GRP8 abundance in cpc-1 roots compared to the wildtype control (Figure S6H). Additionally, we found no significant (p value > 0.05; Welch’s t-test) difference in the levels of transcription factor transcripts that function in the CAPRICE/WEREWOLF pathway in the GRP8ox line compared to control (Figure S6I). Combined, these data reveal that GRP8 promotes root hair cell fate in a GRP7 independent manner, uncovering another novel post-transcriptional regulator of this important plant developmental process that is not affected by the CAPRICE/WEREWOLF pathway.
GRP8 promotes phosphate starvation stress response
One of the major factors regulating root hair cell fate is environmental signaling, such as nutrient deprivation. Therefore, a regulator of root hair cell fate may play a role in nutrient stress response. In fact, a recent microarray analysis of phosphate starved Arabidopsis roots revealed a mild increase in GRP8 levels during the phosphate starvation response (Woo et al., 2012). Given this observation, in conjunction with our identification of GRP8 as a regulator of root hair cell fate (Figure 5), we next investigated the role of this RBP in the phosphate starvation stress response pathway. To begin, we performed RT-qPCR on the roots of WT plants grown on control and low phosphate media and validated that GRP8 abundance is significantly (p value < 1.1×10−9; Welch’s t-test) upregulated upon phosphate starvation (Figure 6A), thereby verifying that this gene does respond to phosphate deprivation. We then examined the response of WT, GRP8ox, and grp7-1;8i plants to phosphate starvation. Using these plants, we first measured the levels of acid phosphatase activity from their roots under control and 3-day phosphate starvation conditions. This analysis revealed acid phosphatase levels to be significantly (p value < 0.05; Wilcoxon test) increased in the GRP8ox plants as compared to WT (Figure 6B) with no significant (p value < 0.05; Wilcoxon test) difference between grp7-1;8i and WT plants (Figure 6B). These results indicate that there is a GRP8-dependent and GRP7-independent increase in acid phosphatase activity in Arabidopsis roots upon phosphate starvation.
Figure 6. GRP8 functions in the phosphate starvation response pathway.
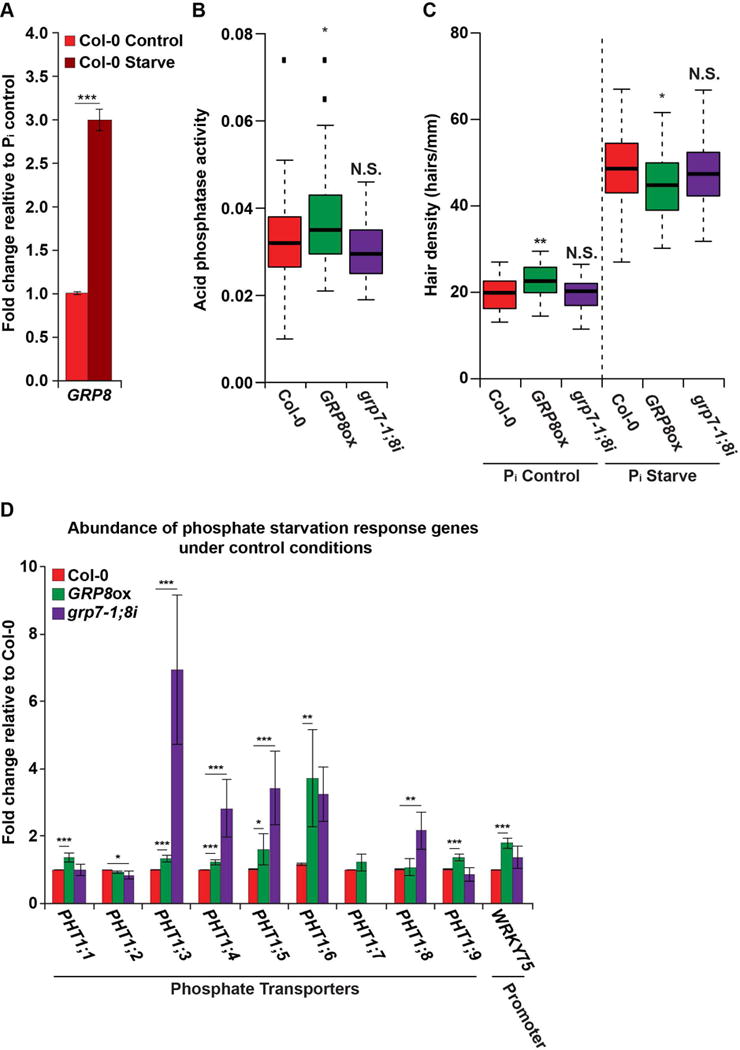
(A) RT-qPCR measuring GRP8 levels in Col-0 plants after three days of phosphate deprivation (dark red bar) or control treatment (light red bar). (B) Acid phosphatase activity in the roots of phosphate starved Col-0 and GRP7/8 mutant 8-day-old seedlings, n > 40. (C) Root hair cell density (hairs/mm) in 8-day-old seedlings after three days of phosphate starvation. (D) Levels of phosphate starvation response genes as measured by RT-qPCR in roots from Col-0 (blue), GRP8ox (green), and grp7-1;8i (purple) grown under control conditions. For (A) and (D), * and ** denote p value < 0.05 and 0.01, respectively, Welch’s t-test. For (B) and (C), * and ** denote p value < 0.05 and 0.01, respectively, Wilcoxon test. Error bars indicate SEM. See also Figures S1, S2, S4, S6–S7.
Acid phosphatases are secreted from the root epidermis, therefore phosphatase activity corresponds to root surface area (Gilbert et al., 1999). To determine whether increased phosphatase activity is a consequence of increased root hair cell number, we measured hair cell density under both normal and phosphate deprivation conditions. From this analysis we observed that GRP8ox plants exhibited significantly (p value < 0.05; Wilcoxon test) decreased hair cell density under the starved conditions as compared to WT, while there was no change in grp7-1;8i plants (Figure 6C), indicating that there is an uncoupling of GRP8-dependent regulation of cell fate decision from phosphate starvation response. Furthermore, these findings demonstrate that the increase in acid phosphatase activity is especially sizeable in GRP8ox plants (Figure 6B), as there are fewer hair cells to secrete these enzymes during phosphate deprivation.
In order to better understand the roles of GRP8 and GRP7 in phosphate deprivation response, we measured the expression of numerous phosphate starvation response genes in the roots of GRP8ox and grp7-1;8i plants (Péret et al., 2011). To do this, we collected RNA from the roots of 8-day-old WT, GRP8ox, and grp7-1;8i seedlings under both control and phosphate starvation conditions and performed RT-qPCR on a panel of phosphate response genes (Figures S7B–C). From this analysis, we observed a significant (all p values < 0.05; Welch’s t-test) increase in the levels of several PHOSPHATE TRANSPORTER 1 (PHT1) family genes in the roots of GRP8ox plants under normal growth conditions (Figure 6D). Most of the PHT1 family genes are upregulated under phosphate starvation (Figure S7C), providing a mechanism to maximize the uptake of phosphate when it is most scarce, allowing alleviation of the stress that the plant undergoes (Muchhal et al., 1996). Specifically, PHT1;1 (AT5G43350) expression, which we found was increased in GRP8ox plants, has been linked to increased phosphate uptake and increased plant survival under phosphate starvation (Wang et al., 2014). In addition to heightened PHT1 levels, we found significantly (all p values < 0.05; Welch’s t-test) increased levels of the WRKY-domain containing transcription factor WRKY75 (AT5G13080) in GRP8ox as compared to WT roots (Figure 6D). This is notable because WRKY75 is known to promote PHT1;1 transcription during phosphate starvation, and may be involved in the transcription of other PHT1 family genes (Wang et al., 2014). Interestingly, the grp7-1;8i plants exhibit upregulation of several PHT1 family genes (PHT1;3, PHT1;4, PHT1;5, PHT1;8) (Figure 6D), indicating that there is a GRP7-dependent inhibition of several of these genes. Therefore, these data indicate that there is a GRP8-dependent increase in the levels of most PHT1 family transcripts, while GRP7 also affects several of these mRNAs.
We next aimed to determine if GRP8 directly binds to any of these phosphate deprivation response transcripts. As the α-GRP7/8 antibody binds to both GRP7 and GRP8 proteins, testing direct binding of GRP8 required performing RIP-qPCR in the roots of grp7-1 plants grown under phosphate deprivation. Using this assay, we tested for GRP8 binding to PHT1 family genes and WRKY75. Although there is no significant (all p values > 0.05; Welch’s t-test) enrichment of PHT1 family transcripts in GRP8 pulldown samples, we did observe a significant (all p values < 0.05; Welch’s t-test) enrichment of WRKY75 (Figure 7A; p value < 0.05; Welch’s t-test) specifically in our α-GRP8 samples as compared to our IgG negative contol. These data reveal that GRP8 binds to WRKY75 in vivo, leading to its altered transcript level. Thus, the GRP8-dependent regulation of WRKY75 results in increased PHT1 family phosphate transporter mRNA expression in the roots of 8-day-old seedlings.
Figure 7. GRP8 alleviates phosphate deprivation stress.
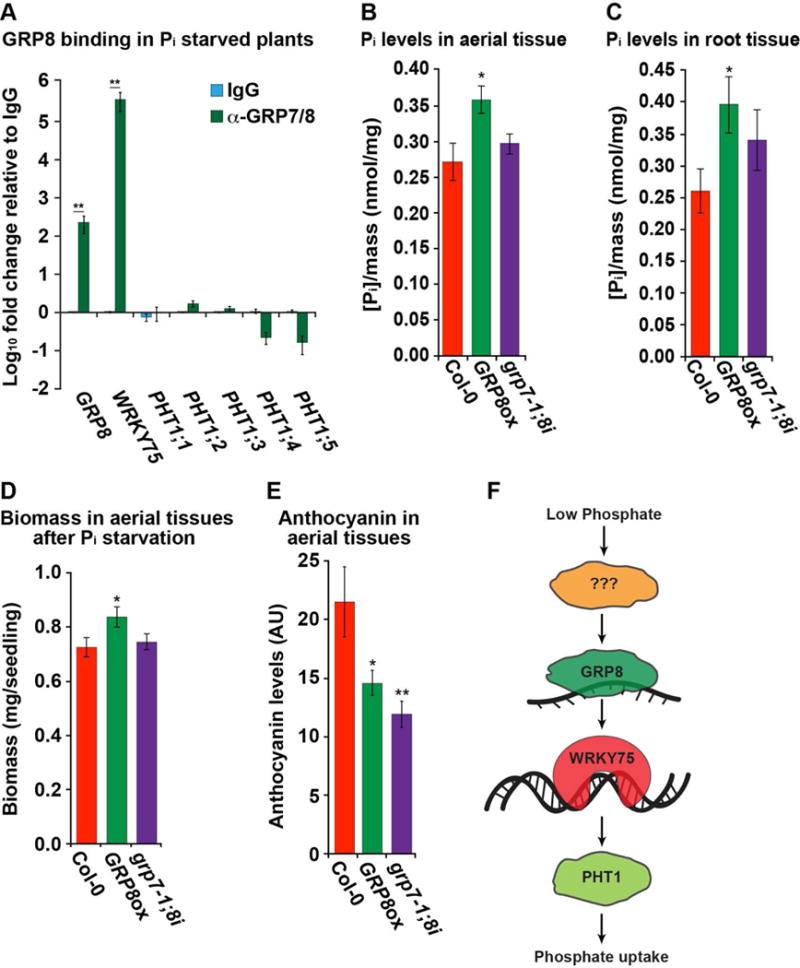
(A) RIP-qPCR of root tissue from grp7-1 plants grown under phosphate starvation. RIP-qPCR was performed with a rabbit IgG (blue) or rabbit serum raised against GRP7 and GRP8 (green) graphed as fold change relative to α-IgG, n = 4 (B–C) Measurement of phosphate levels normalized to mass after 3-days of phosphate starvation in the shoots (B) or roots (C) of 8-day-old seedlings, n = 12. (D–E) Biomass (D) or anthocyanin levels (E) for 18-day-old seedlings after 2 weeks of phosphate deprivation, n = 12. For (A)-(E), *, **, and *** denote p value < 0.05, 0.01, and 0.001, respectively, Welch’s t-test. Error bars indicate SEM. (F) A model of the role of GRP8 on the plant phosphate starvation response. See also Figures S1, S2, S4, S6–S7.
As GRP8 promotes phosphate transporters, we next tested its role in alleviating both short-term and long-term phosphate starvation. We first measured phosphate levels in the aerial and root tissue of WT, GRP8ox, and grp7-1;8i seedlings after three days of phosphate starvation. This assay revealed significantly (p value < 0.05; Welch’s t-test) increased phosphate levels in both tissues in GRP8ox plants as compared to WT and grp7-1;8i seedlings (Figures 7B–C). These results indicated that both phosphate uptake and phosphate efflux to the shoots are upregulated in plants with higher GRP8 levels. Additionally, we subjected plants to long-term (12-day) phosphate starvation and assayed both biomass and anthocyanin levels in the shoots of WT, GRP8ox, and grp7-1;8i seedlings, since phosphate starvation inhibits plant growth while promoting production of anthocyanin. We observed significantly (p value < 0.05; Welch’s t-test) greater biomass in the shoots of GRP8ox as compared to WT and grp7-1;8i plants (Figure 7D). We also found significantly (p value < 0.05; Welch’s t-test) decreased anthocyanin accumulation in the aerial tissue of both GRP8ox and grp7-1;8i as compared to the WT plants (Figure 7E). These data indicate that GRP8 is required for alleviating this plant stress by promoting increased phosphate uptake and biomass accumulation, while both GRP7 and GRP8 function in the reduction of the anthocyanin accumulation associated with phosphate starvation.
DISCUSSION
Here, we use PIP-seq to examine both the RNA-protein interaction and RNA secondary structure landscapes of nuclei from root hair and nonhair cells, which comprise the Arabidopsis root epidermis. Analyzing highly pure populations of hair or nonhair cell nuclei (Figure S1) revealed thousands of cell type specific protein-bound sites as well as many shared sites, which are enriched in the coding sequence of the mRNA transcriptomes of both cell types (Figure 1).
This study compares global patterns of RNA secondary structure and RNA-protein interactions across the nuclear transcriptomes of two distinct cell types. This large-scale analysis identifies distinct profiles in specific regions of mRNA transcripts. For instance, mRNAs found in both cell types exhibit an increase in protein binding in the CDS, which corresponds to a relative decrease in secondary structure (Figures 2A–B). Interestingly, both RNA secondary structure and protein binding exhibit distinct patterns in the 3′UTR of root hair and nonhair cell nuclei (Figures 2C–D). RNA-protein interactions are known to be key regulators of numerous developmental events across various species. Therefore, our observation of cell type specific RNA-protein interactions is not very surprising. More interestingly, the main driver of RNA secondary structure has always been believed to be the primary sequence of a transcript. Thus, when analyzing the same RNA populations in two different cell types, one would expect to see virtually identical RNA folding patterns. Here, we describe distinct secondary structural profiles for mRNAs passing our expression threshold in both root hair and nonhair cell nuclei. These data reveal that there are further drivers of RNA folding in eukaryotic transcriptomes than simply the primary sequence, such as RNA-protein interactions, covalent modifications, and/or other factors. Furthermore, we use the cell type specific patterns of RNA secondary structure and RNA protein interactions to identify the preferred interacting motif of GRP8 (Figures 2A–D and S7A), which we then demonstrate is a novel regulator of root hair cell fate and plant phosphate stress response (Figures 5–7). Thus, our study exemplifies the use of a genome-wide screen to identify novel regulators of a biological process.
It is of note that we observe the greatest difference in both protein occupancy and RNA folding in the 3′ UTRs of mRNAs expressed in both hair and nonhair cells (Figures 2C–D). These complementary profiles provide two potential models to explain this phenomenon. First, cell type specific protein binding could regulate the folding of RNA transcripts, resulting in distinct folding patterns. Conversely, the distinct RNA folding patterns could in fact regulate protein binding. This latter model is supported by our findings that lncRNAs exhibit similar overall protein binding profiles while displaying distinct patterns of RNA secondary structure between root hair and nonhair cells (Figures 2E–F), suggesting that a different array of ssRNA- and dsRNA-binding RBPs are interacting with the distinctly structured lncRNAs found in these two cell types. Future studies will further investigate this type of cell-type specific RNA folding and RNA-protein interactions, determining the mechanisms involved in their feedback regulation.
This study also reveals an interesting pattern in nuclear RNA folding. Specifically, our analysis reveals that in both root hair and nonhair cell nuclei the CDS is less structured than both UTRs (Figures 2A–B), which is consistent with our nuclear PIP-seq performed in mixed nuclei from whole seedlings (Gosai et al., 2015). Although this pattern is consistent between all three nuclear PIP-seq datasets, the opposite pattern has been observed in studies performed on whole cell (mostly cytoplasmic) RNA populations. These whole cell studies have been performed on unopened flower buds utilizing ds/ssRNA-seq (Li et al., 2012a), as well as on whole seedlings with structure-seq (Ding et al., 2014). Although these studies were performed using different techniques in a variety of Arabidopsis tissues, these data support the idea that the nuclear and cytoplasmic transcriptomes may in fact have distinct RNA secondary structure profiles. As with cell type specific RNA folding, these distinct folding patterns could be due to different cohorts of RBPs in the nucleus and cytoplasm, and/or distinct post-transcriptional covalent modifications present in these cellular compartments. However, these consistent results across various studies and structure probing techniques warrant additional analyses to better understand this phenomenon.
In addition to describing global patterns, we used our PPS data to identify enriched protein-bound sequences and identify the RBPs that interact with a number of these sequences. More specifically, using RNA affinity chromatography we first identified SE as a candidate regulator of root hair cell development, while providing evidence of its preferred binding motif, a GGN repeat, in target RNAs (Figures 3A–B). Phenotypic analyses reveal that SE inhibits root hair cell fate in a miRNA biogenesis-dependent manner (Figure 3C), while also terminating root hair tip growth in differentiated cells primarily (Figure 3D). We found that several SE-bound transcripts are necessary for proper root hair length (Figures 4A–C), and exhibit reduced abundance and stability in the absence of SE (Figures 4D–E). Combined, these data provide a working model in which SE functions with other microRNA biogenesis regulators to promote the nonhair cell fate, while also functioning independently to promote the stability of several mRNAs necessary for proper root hair termination (Figure 4F). Therefore, determining the mechanism by which SE positively regulates the abundance of these mRNAs that are required for proper root hair development is an area of future study.
Analyzing the first 100 nt of 3′ UTRs, where the greatest difference in protein binding was observed, led to the identification of a hair cell specific TG-rich motif (Table S3 and Figure S7A). Through both RNA affinity chromatography and phenotypic analyses we found that GRP8 binds this TG-rich motif, and promotes root hair cell fate in a GRP7-independent manner (Figure 5). This finding is of particular interest since plants overexpressing GRP8 do not exhibit the deleterious aerial phenotypes described for se-1 (Clarke et al., 1999; Serrano-Cartagena et al., 1999), making this a potential candidate for engineering more stress resistant crop plants. This idea is further supported by our observations that GRP8 is upregulated upon phosphate starvation, and promotes increased acid phosphatase activity (Figure 6). Additionally, we found that GRP8 alone has substantial effects in promoting phosphate uptake, efflux, and biomass accumulation while simultaneously alleviating anthocyanin production during phosphate starvation (Figure 7). In fact, our findings indicate the presence of a novel model of plant phosphate starvation response (Figure 7F). Specifically, we demonstrated that GRP8 is dramatically upregulated during phosphate starvation (Figures 6A and S7C), and promotes the abundance of mRNAs encoding phosphate transporters as well as WRKY75, which regulates transcription of several PHT1 transporters, while also binding directly to WRKY75 (Figures 7A–B). The increase in PHT1 mRNA abundance likely explains the increased phosphate levels and biomass accumulation in GRP8 overexpressing plants (Figures 7C–D), as well as decreased anthocyanin accumulation in the aerial tissues (Figure 7E). Thus, our working model suggests that phosphate deprivation initiates a signaling pathway that promotes GRP8 abundance, which in turn binds to WRKY75 mRNA thereby promoting its abundance. Increased WRKY75 then promotes phosphate transporter expression, resulting in increased phosphate levels in the plant (Figure 7F). Although the specific signaling pathways and mechanisms by which GRP8 leads to increased WRKY75 levels must be further elucidated, our findings point to the exciting possibility that GRP8 overexpression is a viable target for engineering more stress resistant crop plants, which is a hypothesis that will be addressed with future research.
In total, our findings have revealed the power of PIP-seq in identifying biologically significant RBPs through a genome-wide screen. In the future, these newly described hair cell regulatory proteins will be further studied to better understand the mechanisms by which they regulate this agriculturally important plant phenotype.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact/Corresponding Author, Dr. Brian D. Gregory (bdgregor@sas.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Plant Materials
Seedlings were grown on 0.5× MS plates with 1% sucrose and 0.8% Phytoblend, grown vertically at 20°C, in a 16 h light/8 h dark cycle. The purified nuclei used in this study were extracted from 10-day-old seedlings of ADF8:NTF/ACT2p:BirA or GL2:NTF/ACT2p:BirA Columbia-0 (Col-0) ecotype of Arabidopsis thaliana using the INTACT methodology (Wang and Deal, 2015). Additionally, the lysates for all western blots were from these same 10-day-old seedlings.
The lysates used for RNA immunoprecipitation (RIP) RT-qPCR and motif-interacting protein analyses were from the whole roots of 8-day-old seedlings of the genotypes as noted.
METHOD DETAILS
Cross-linking and INTACT purification
Immediately before nuclei purification, 10-day-old seedlings of ADF8:NTF/ACT2p:BirA or GL2:NTF/ACT2p:BirA were crosslinked in nuclear purification buffer (20 mM MOPS (pH 7), 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA) plus 1% (vol/vol) formaldehyde under vacuum for 10 minutes, followed by a five minute quench with 125 mM Glycine under vacuum for an additional five minutes. Crosslinked seedlings then underwent INTACT purification as previously described (Wang and Deal, 2015).
Briefly, 3 grams of root tissue were pulverized in liquid nitrogen, then resuspended in 10 mL of nuclear purification buffer. The resuspension was passed through a 0.45 μm nylon filter, and incubated on ice for 10 min. The samples were then centrifuged at 1,200 rcf for 10 min, and the pelleted nuclei were resuspended in 1 mL of nuclear purification buffer. Following resuspension, 25 μL of streptavidin coated M-280 Dynabeads (Life Technologies; Carlsbad, CA, USA) were washed twice with nuclear purification buffer, and then combined with the samples in nuclear purification buffer. The samples were allowed to rotate end over end at 4°C for 30 min. Samples were then transferred to 15 mL conical tubes, and washed 4 times with 12 mL of nuclear purification buffer plus 0.1% Tween20. After the last wash the beads were resuspended in 1 mL of nuclear purification buffer and transferred to a 1.7 mL tube and washed twice more. The final samples were resuspended in 20 μL of nuclear purification buffer, snap frozen in liquid nitrogen, and stored at −80°C until processing.
Western blotting
Western blots using lysates from INTACT purified nuclei or 10-day-old roots were performed using α-CNX1 (1:1,000; AS12 2365; Agrisera; Vännäs, Sweden), α-EIF1A (1:1,000; AS10 934; Agrisera; Vännäs, Sweden), α-ALDOLASE (1:1,000; AS08 294; Agrisera; Vännäs, Sweden), or α-H3 (1:1,000; ab1791; Abcam; Cambridge, MA, USA) antibodies were performed as previously described (Kupsch et al., 2012). Briefly, lysates were fractionated on a 4–12% SDS NuPage gel in MES at 100 V for 2 h. Transfer to PVDF was performed at 200 mA at 4°C for 2 h. The membrane was then briefly washed in water, and allowed to block at 4°C overnight in 5% milk in TBST. The membrane was blotted with primary antibody diluted in 5% milk at RT for 2 h, then underwent 3× 10 minute washes in TBST. The secondary antibody was diluted 1:5,000 in TBST and the membrane was blotted at RT for 2 h. Three 15 minute washes with TBST were performed, followed by one 5 minute wash in TBS. The membrane was then removed from liquid and ECL Prime Western Blotting Detection Reagent (GE Healthcare; Little Chalfont, UK) was applied to the membrane for one minute. Images were taken incrementally every 10 seconds until saturation of the images.
PIP-seq library preparation
~Two million INTACT purified nuclei were lysed in 850 μl RIP buffer (25 mM Tris-HCl, pH = 7.4; 150 mM KCl, 5 mM EDTA, pH = 7.5; 0.5% NP40; 10 μM DTT; 1 tablet protease inhbitors and 0.5μl/ml RNaseOUT (Life Technologies; Carlsbad, CA, USA)) by manual grinding. The resulting cell lysate was treated with RNase-free DNase (Qiagen; Valencia, CA, USA). The lysates were then split and treated with either 100 U/ml of a single-stranded RNase (ssRNase) (RNaseONE (Promega; Madison, WI, USA)) with 200 μg/ml BSA in 1X RNaseONE buffer for 1 hour at room temperature (RT), or 2.5 U/ml of a double-stranded RNase (dsRNase) (RNaseV1 (Ambion; Austin, TX, USA)) in 1X RNA structure buffer for 1 hour at 37°C as previously described (Silverman et al., 2014). See Figure 1A for a schematic representation of library preparation. Proteins were then denatured and digested by treatment with 1% SDS and 0.1 mg/ml Proteinase K (Roche; Basel, Switzerland) for 15 minutes at RT. Proteinase digestion was followed by a 2 hour incubation at 65°C to reverse the RNA-protein cross-links.
To determine whether nuclease resistant regions in RNAs are due to protein binding or specific secondary structures, we also determined the digestion patterns of ds- and ssRNases immediately following protein digestion. To do this, we performed the identical treatments as described above except that the cross-linked nuclear lysates were treated with 1% SDS and 0.1 mg/ml Proteinase K (Roche; Basel, Switzerland) and ethanol precipitated prior to being treated with the two RNases. In this way, the SDS and Proteinase K solubilized and digested the proteins allowing us to deduce PPSs within all detectable RNAs in the cells of interest (see Figure 1A for schematic).
The digested RNA was then isolated using the Qiagen miRNeasy RNA isolation kit following the included protocol (Qiagen; Valencia, CA, USA). To ensure that only high quality RNA samples were used for PIP-seq library preparation, the purified RNA was run on a Eukaryotic Total RNA Pico Series II chip (5067-1513; Agilent Technologies; Wilmington, DE, USA) using a BioAnalyzer 2100 system. Finally, the purified RNA was used as the substrate for strand-specific sequencing library preparation as previously described (Silverman et al., 2014). All of the RNase footprinting libraries (a total of 4 for each replicate: ss- and dsRNase treatments, footprint and structure only) were sequenced on an Illumina HiSeq2000 using the standard protocol for 50 base pair single read sequencing.
RNA affinity chromatography
We used motifs identified within PPS sequences (Figure S4 and Table S5) as baits to isolate interacting proteins by affinity ‘pulldown’ studies. Specifically, RNA baits (covalently-linked to agarose beads) containing the identified motif of interest (IDT; Coralville, IA, USA) were incubated in a binding reaction (3.2 mM MgCl2, 20 mM creatine phosphate, 1 mM ATP, 1.3% polyvinyl alcohol, 25 ng of yeast tRNA, 70 mM KCl, 10 mM Tris, pH 7.5, 0.1 mM EDTA) with ~56 μg of 10-day-old Arabidopsis whole root lysate at RT for 30 minutes. Beads were washed four times with GFB-200 (20 mM TE, 200 mM KCl) plus 6 mM MgCl2 and once with 20 mM Tris-HCl (pH 7.4). The RNA-bound proteins were then directly trypsinized on the beads.
MS-ready sample preparation
Multiple independent samples for the selected motifs and their corresponding controls were used to average out experimental variability, optimize detection limits, and improve signal to noise ratio for robust specific identification. MS sample preparations and analyses were performed as described previously (Onder et al., 2008; Onder et al., 2006). Briefly, RNA-bound proteins were treated directly on the beads with 100 mM NH4HCO3 containing ~6 ng/μl of MS-grade trypsin (Promega; Madison, WI, USA) and incubated at 37°C for 12–18 hrs. These samples were extracted first with 1%HCOOH/2%CH3CN, and several times with 50% CH3CN; combined peptide extracts were vacuum dried and desalted using a ZipTip procedure before resuspending in ~5–10 μL LC buffer A (0.1% HCOOH (v/v) in 5:95 CH3CN:H2O) for MS analysis.
RIP-RT-qPCR
RNA immunoprecipitaion (RIP) was performed on whole root tissue from Col-0 or grp7-1 as described previously. To begin, fresh roots were submerged in PBS plus 1% (vol/vol) formaldehyde and vacuum infiltrated at room temperature (RT) for 10 minutes. One molar Glycine (Sigma-Aldrich; St. Louis, MO, USA) was added to a final concentration of 125 mM before an additional five minutes of vacuum infiltration. The root tissue was then washed five times with distilled water, patted dry, and snap frozen in liquid nitrogen.
On the day of the RIP, the roots were ground into a fine powder in liquid nitrogen using a mortar and pestle, and resuspended in RIP buffer (150 mM NaCl, 20 mM Tris (pH=8.6), 1 mM EDTA, 5 mM MgCl2, 0.5% NP40, 1 tablet/10 ml protease inhibitor (Roche; Basel, Switzerland), 0.5 μl/ml RNaseOUT RNaseOUT (Life Technologies; Carlsbad, CA, USA) at ~1 g / 1.2 mL. This lysate was then subjected to 30 min of sonication (30 s on and 2 min off) and centrifuged twice for 15 min at max speed to remove any pelleted debris.
While the tissue is being prepared, 50 μL of Protein A beads (Life Technologies; Carlsbad, CA, USA) were washed twice with PBS then resuspended in 400 μL. Antibodies were then added to the beads at 5–10 μg per reaction, and allowed to rotated at 4°C for >2 hour s. The antibodies used were α-SE (AS09 532, Agrisera; Vännäs, Sweden), α-ABH1/CBP80 (AS09 531, Agrisera; Vännäs, Sweden), rabbit serum raised against native recombinant Sinapis alba GRP10, which recognizes Arabidopsis GRP7 and GRP8 or normal rabbit IgG (3125, Cell Signaling Technology; Danvers, MA, USA). The beads were then washed twice with RIP buffer, and resuspended in whole root lysate, followed by a 90 min rotation at 4°C. The RIP was t hen washed six times with RIP buffer, and resuspended in QIAzol. Immunoprecipitated RNA was then isolated using the miRNeasy mini kit (Qiagen; Valencia, CA, USA), and an RT was performed on 100–200 ng of RNA using Superscript II (18064014, Life Technologies; Carlsbad, CA, USA) with random hexamer priming following the manufacturers protocol. The cDNA was then subjected to 15 cycles of preamplification using the SsoAdvanced PreAmp Supermix (172–5160, BioRad; Hercules, CA, USA) kit, following the manufacturer’s protocol. The preamplified template DNA was then used to perform qPCR using the 2× SYBR Green Master Mix (B21202, Bimake, Houston, TX, USA) and following the manufacturer’s protocol. All primers used in RT-qPCR are listed in Table S5.
Measurement of root hair density and root hair length
Seeds were sterilized in a 30% Clorox solution for 15 min followed by five washes with autoclaved water. After the last wash seeds were resuspended in 0.15% sterile agarose and stratified at 4°C for at least 48 hours. Seedlings were grown on 0.5X MS plates with 1% sucrose and 0.8% Phytoblend, grown vertically at 20°C, in a 16 h light/8 h dark cycle. Measurements of basal root hair density and length were performed on 8-day-old seedlings by imagine with a dissecting microscope and measuring root hair length using JBrowse. Root hair density was calculated by measuring a length of primary root and counting all visible hairs along that length.
For phosphate starved plants, all seeds were planted on the described 0.5X MS plates and incubated for 5 days. On the fifth day the seedling on each plate were transplanted to two new plates, one identical 0.5X MS plate and one 0.5X MS plate without phosphate. The control and starved plates were then replaced in the incubator for another three days. The root hair cell density and root hair length were then measured as described above.
For the temperature sensitive mor1-1 plants, the plants were grown at 20°C for four days, then transferred to 31°C for another two days before imaging and phenotyping.
Measurement of RNA stability
To measure the stability of mRNA transcripts we took 8-day-old plants grown on 0.5X MS plates and excised the roots below the hypocotyl. Taking 30 roots per biological replicate, we then submerged these into 6 mL of liquid 0.5X MS media supplemented with 1% sucrose and 10 μM Actinomycin D. To account for any immediate changes to the transcriptome we allowed these to incubate for 4 hours before taking our baseline 0 hour time point. We then collected roots after 8 hours of treatment, dabbing them dry of media, and snap freezing them in liquid nitrogen. We then proceeded to perform RNA extraction and RT-qPCR as previously described.
Measurement of acid phosphatase activity
To measure acid phosphatase activity, plants that had been phosphate starved were taken and the primary root was excised and placed in 300 μL of assay buffer (3.4 mM 4-naphthyl phosphate, 2.5 mM FastRed TR) and incubated at RT for 15 min. Then 150 μL of assay buffer was taken and absorbance at 405 nm was measured.
Measurement of phosphate concentration
Seedlings were germinated on 0.5X MS plates, and 5-day-old seedlings were transplanted to control or phosphate starved plates for three days. After phosphate starvation, the hypocotyl was cut to separate the seedlings into roots and shoots, and the tissue from five seedlings was pooled and weighed. This tissue was immediately placed into 1 mL of 1% glacial acetic acid and frozen in liquid nitrogen. The tissue underwent 8 rounds of freezing and thawing in liquid nitrogen and a room temperature water bath. After the eighth round of thawing, 100 μL of supernatant was taken and placed into 200 μL of water and 700 μL phosphate assay buffer (A: 2.85% H2SO4, 0.85% NH4MoO4, B: 10% ascorbic acid, A:B = 6:1). The samples were then incubated at 37°C for 60 minutes, and absorbance was measured at 810 nm (Zhang et al., 2014). A standard curve was generated and the concentration of soluble phosphate per milligram of tissue was reported.
Measurement of anthocyanin
Seedlings were germinated on 0.5X MS plates, and 3-day-old seedlings were transplanted to control or phosphate starved plates for 14 days. After phosphate starvation, the hypocotyl was cut to separate the seedlings into roots and shoots, and the aerial tissue from five seedlings was pooled and weighed. The tissue was then submerged in a 18:1:81 solution of propanol:HCl:water, before incubation at 100°C for 3 min. Samples were then centrifuged at >20,000 × g for 15 min. The supernatant was taken and absorbance was measured at 535 nm and 650 nm. The absorbance due to anthocyanin was calculated as:
QUANTIFICATION AND STATISTICAL ANALYSIS
Experiment specific information
The measurement precision, number of biological replicates (n), statistical tests performed, and significance for each experiment can be found in the figure legend of the experiment, as well as the RESULTS section of this document.
Read processing and alignment
PIP-seq reads were first trimmed to remove 3′ sequencing adapters using cutadapt (version 1.2.1 with parameters -e 0.06 -O 6 -m 14). The resulting trimmed sequences were collapsed to unique reads and aligned to the TAIR10 Arabidopsis genome sequence using TopHat (version 2.0.10 with parameters –library-type fr-secondstrand –read-mismatches 2 –read-edit-dist 2 –max-multihits 10 –b2-very-sensitive –transcriptome-max-hits 10 –no-coverage-search –no-novel-juncs). PCR duplicates were collapsed to single reads for all subsequent analyses.
Identification of PPSs
PPSs were identified using a modified version of the CSAR software package (Muiño et al., 2011). Specifically, read coverage values were calculated for each base position in the genome and a Poisson test was used to compute an enrichment score for footprint versus structure only libraries. PPSs were then called with a false discovery rate of 5% as previously described (Gosai et al., 2015; Silverman et al., 2014).
Calculating the structure score statistic
For every base of detectable transcripts, we calculated the dsRNA-seq and ssRNA-seq coverages from the structure only samples, then calculated the structure score as described previously (Gosai et al., 2015). Briefly, when given the dsRNA-seq and ssRNA-seq coverages (nds, nss) of a given base i, the structure score is determined as:
where Si is the structure score, dsi and ssi are the normalized read coverages, and Lds, Lss are the total covered length by mapped dsRNA-seq and ssRNA-seq reads, respectively. The total coverage length was used as the normalization constant instead of the total number of mapped reads used previously, because we believe it is a more reasonable assumption for the transcriptome to have comparable levels of paired/unpaired regions. It is of note that we used a generalized log ratio (glog) instead of normal log-odds because it can tolerate 0 values (positions with no dsRNA or ssRNA read coverage) as well as being asymptotically equivalent to the standard log ratio when the coverage values are large. Only sense-mapping reads were used, as we are entirely concerned with the intra-molecular interactions contributing to the self-folding secondary structure.
Structure score profile analysis of mRNAs
The structure score for every base of each detected transcript was first calculated using all mapped and spliced reads. In addition to the minimum dsRNA-seq plus ssRNA-seq read coverage requirement discussed above, we only considered mRNAs with intact CDS regions, ≥ 45 nt 5′UTRs, ≥ 140 nt 3′UTRs and a minimum coverage of 50 reads across the entire transcript. To generate profiles, the Z-score of the structure score was calculated for each nucleotide with respect to the graphed window as previously described (Berkowitz et al., 2016).
To analyze profiles across detectable lncRNAs, we divided the length of the transcript into 100 equally sized bins. Taking the average scaled structure score across each bin, we then graphed the profile of these scores.
PPS profile analysis of mRNAs
PPS occupancy was converted to a score at each nucleotide, with a 1 indicating that a protein was bound and a 0 indicating that the nucleotide was unbound. The average PPS occupancy was calculated for all transcripts passing the expression criteria described above. PPS density was then graphed such that the region of highest occupancy was normalized to a density of 1.0.
Mass Spectrometry Analyses
Tryptic peptide extracts were analyzed using nLC-MS/MS (Dionex/LCPackings Ultimate nano-LC coupled to a Thermo LCQ Deca XP+ ion trap mass spectrometer) in duplicate. 1 μl of the peptide sample (in LC buffer A, 0.1% HCOOH (v/v) in 5:95 CH3CN:H2O) was first loaded onto a μ-Precolumn (PepMapTM C18, LC-Packings), washed for 4 min at a flow rate of 25 μl/min with LC buffer A, then transferred onto an analytical C18-nanocapillary HPLC column (PepMapAcclaim100). Peptides were eluted at 280 nl/min flow rate with a 120 minute gradient of LC buffers A and B (0.1% (v/v) formic acid in 80:20 acetonitrile:water) ranging from 5%-95% B. A fused silica emitter tip with 8-μm aperture (FS360-75-8-N-5-C12; New Objective) mounted to a Thermo nanospray ionization (NSI) source at 1.8 kV was used for positive ionization of peptides. Mass spectra were collected using Thermo Xcalibur 2.0 software. The top 3 principal ions from each MS scan were trapped and fragmented during the chromatographic gradient, using dynamic exclusion to maximize detection of ions (range 200–2000 m/z). The trapped ions were subjected to collision-induced dissociation (CID) with He, and ~4000 spectra (MS/MS) were collected to cover the entire chromatography elution profile.
Spectral Data Analyses and Protein ID
Experimentally collected MS/MS tandem data were searched against the Arabidopsis Proteome Database (NCBI, latest version) using Thermo Proteome Discoverer 1.4 software. The search was restricted to full trypsin digestion with a maximum of 3 missed cleavages and potential modifications for methionine (oxidation) and cysteine (carbamidomethylation); other parameters were standard for LCQ Deca XP+ instrumentation. Peptide filters were set to standard Xcorr vs charge state values; X corr = (1.5, 2.0, 2.25, 2.5) for charges (+1,+2,+3,+4), respectively. Spectral assignments were manually scrutinized to validate the reliability of the protein identifications. Mass spectral data are summarized in Supplemental Table 4.
DATA AND SOFTWARE AVAILABILITY
Accession numbers
The raw and processed data for PIP-seq from our analyses have been deposited into the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE86459.
Genome browser availability
The sequencing data presented here is also available through the EPIC-CoGe genome browser (Lyons and Freeling, 2008): https://genomevolution.org/coge/NotebookView.pl?nid=1767.
Supplementary Material
Highlights.
Root epidermal cell types exhibit distinct patterns of RNA folding and RBP binding
SERRATE regulates root hair length independent of its roles in microRNA biogenesis
GRP8 is a RBP regulator of root hair cell fate
GRP8 promotes phosphate uptake, thereby alleviating phosphate starvation stress
Acknowledgments
The authors would like to thank Dr. Kimberly L. Gallagher, Dr. Richard S. Poethig, and members of the B.D.G. lab both past and present for helpful discussions. Also, Ruthsabel O’Lexy for discussion of the phosphate stress analyses. This work was funded by NSF grants MCB-1243947 and IOS-1444490 to B.D.G., DFG grants STA653 and SPP1530 to D.S., and NIGMS T32GM008216-29 to S.W.F.
Abbreviations
- RBP
RNA binding protein
- ssRNA
single-stranded RNA
- dsRNA
double-stranded RNA
- PIP-seq
protein interaction profile sequencing
- SE
SERRATE
- GRP8
GLYCINE RICH PROTEIN 8
- INTACT
isolation of nuclei in tagged specific cell types
- dsRNase
double-stranded ribonuclease
- ssRNase
single-stranded ribonuclease
- PPS
protein protected site
- nt
nucleotide
- CDS
coding sequence
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Notes to reviewers: We have created an interactive public web browser for visualizing all of the data presented in this manuscript through EPIC-CoGe (https://genomevolution.org/coge/NotebookView.pl?nid=1767). To protect the anonymity of the reviewers, we suggest using a proxy server, such as the free anonymous browsing provided by http://www.anonymizer.com/. The sequencing data herein is also available through GenBank. (GEO accession numbers are below.) GEO data is available to reviewers through the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=qxohkmmablixfep&acc=GSE86459
AUTHOR CONTRIBUTIONS
R.B.D. and B.D.G. conceived the study. S.W.F., R.B.D., and B.D.G. designed the experiments. S.W.F., S.J.G., D.W., N.S., A.C.S., T.K., A.S., E.L., F.D., B.A.G., D.S., R.B.D., and B.D.G. performed experiments. S.W.F. and B.D.G. analyzed the data. S.W.F., R.B.D., and B.D.G. wrote the paper with assistance from all authors. The authors have read and approved the manuscript for publication
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acid Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;19:529–538. [Google Scholar]
- Berkowitz ND, Silverman IM, Childress DM, Kazan H, Wang LS, Gregory BD. A comprehensive database of high-throughput sequencing-based RNA secondary structure probing data (Structure Surfer) BMC Bioinformatics. 2016;17:215. doi: 10.1186/s12859-016-1071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JH, Tack D, Findlay K, Van Montagu M, Van Lijsebettens M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J Cell Mol Biol. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- Cruz JA, Westhof E. The Dynamic Landscapes of RNA Architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. A Simple Method for Gene Expression and Chromatin Profiling of Individual Cell Types within a Tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K, Kubo M, Sano K, Demura T, Fukuda H, Liu YG, Shibata D, Kakimoto T. The CKH2/PKL Chromatin Remodeling Factor Negatively Regulates Cytokinin Responses in Arabidopsis Calli. Pland Cell Physiol. 2011;52:618–628. doi: 10.1093/pcp/pcr022. [DOI] [PubMed] [Google Scholar]
- Gilbert GA, Knight JD, Vance CP, Allan DL. Acid phosphatase activity in phosphorus-deficient white lupin roots. Plant Cell Environ. 1999;22:801–810. [Google Scholar]
- Gosai SJ, Foley SW, Wang D, Silverman IM, Selamoglu N, Nelson ADL, Beilstein MA, Daldal F, Deal RB, Gregory BD. Global Analysis of the RNA-Protein Interaction and RNA Secondary Structure Landscapes of the Arabidopsis Nucleus. Mol Cell. 2015;57:376–388. doi: 10.1016/j.molcel.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J. The Arabidopsis Book. The American Society of Plant Biologists; 2014. Root Hairs; p. e0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi M, Sharp PA. Building robust transcriptomes with master splicing factors. Cell. 2014;159:487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Lin WD, Santi S, Schmidt W. Mapping gene activity of Arabidopsis root hairs. Genome Biol. 2013;14:R67. doi: 10.1186/gb-2013-14-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell. 2002;14:611–618. doi: 10.1105/tpc.010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski M, Hallmann A, Staiger D. Genome-wide identification and phylogenetic analysis of plant RNA binding proteins comprising both RNA recognition motifs and contiguous glycine residues. Mol Genet Genomics MGG. 2016;291:763–773. doi: 10.1007/s00438-015-1144-1. [DOI] [PubMed] [Google Scholar]
- Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, Gregory BD. Regulatory Impact of RNA Secondary Structure across the Arabidopsis Transcriptome. Plant Cell. 2012a;24:4346–4359. doi: 10.1105/tpc.112.104232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, Aiyer S, Valladares O, Yang J, Bambina S, Sabin LR, et al. Global Analysis of RNA Secondary Structure in Two Metazoans. Cell Rep. 2012b;1:69–82. doi: 10.1016/j.celrep.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002;29:751–760. doi: 10.1046/j.1365-313x.2002.01251.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. Genome-Wide Analysis Uncovers Regulation of Long Intergenic Noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- Mei H, Cheng NH, Zhao J, Park S, Escareno RA, Pittman JK, Hirschi KD. Root development under metal stress in Arabidopsis thaliana requires the H+/cation antiporter CAX4. New Phytol. 2009;183:95–105. doi: 10.1111/j.1469-8137.2009.02831.x. [DOI] [PubMed] [Google Scholar]
- Meisner CA, Karnok KJ. Root Hair Occurrence anld Variation with Environment. Agron J. 1991;83:814. [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS. Responses of root architecture development to low phosphorus availability: a review. Ann Bot. 2013;112:391–408. doi: 10.1093/aob/mcs285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD, and FAO. OECD-FAO Agricultural Outlook. Paris: Organisation for Economic Co-operation and Development; 2012. p. 2012. [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. PNAS. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick WH, Khalid RA. Phosphate Release and Sorption by Soils and Sediments: Effect of Aerobic and Anaerobic Conditions. Science. 1974;186:53–55. doi: 10.1126/science.186.4158.53. [DOI] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 2011;16:442–450. doi: 10.1016/j.tplants.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Raczynska KD, Stepien A, Kierzkowski D, Kalak M, Bajczyk M, McNicol J, Simpson CG, Szweykowska-Kulinska Z, Brown JWS, Jarmolowski A. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2014;42:1224–1244. doi: 10.1093/nar/gkt894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KH, Kang YH, Park Y, Hwang I, Schiefelbein J, Lee MM. The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Dev Camb Engl. 2005;132:4765–4775. doi: 10.1242/dev.02055. [DOI] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 2008;36:6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Cartagena J, Robles P, Ponce MR, Micol JL. Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol Gen Genet MGG. 1999;261:725–739. doi: 10.1007/s004380050016. [DOI] [PubMed] [Google Scholar]
- Silverman IM, Li F, Alexander A, Goff L, Trapnell C, Rinn JL, Gregory BD. RNase-mediated protein footprint sequencing reveals protein-binding sites throughout the human transcriptome. Genome Biol. 2014;15:R3. doi: 10.1186/gb-2014-15-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitner C, Köster T, Simpson CG, Shaw P, Danisman S, Brown JWS, Staiger D. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res. 2012;40:11240–11255. doi: 10.1093/nar/gks873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandivier LE, Anderson SJ, Foley SW, Gregory BD. The Conservation and Function of RNA Secondary Structure in Plants. Annu Rev Plant Biol. 2016;67:463–488. doi: 10.1146/annurev-arplant-043015-111754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Wang D, Deal RB. Epigenome profiling of specific plant cell types using a streamlined INTACT protocol and ChIP-seq. Methods Mol Biol Clifton NJ. 2015;1284:3–25. doi: 10.1007/978-1-4939-2444-8_1. [DOI] [PubMed] [Google Scholar]
- Wang H, Xu Q, Kong YH, Chen Y, Duan JY, Wu WH, Chen YF. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 2014;164:2020–2029. doi: 10.1104/pp.113.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO. MOR1 is essential for organizing cortical microtubules in plants. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SPCP, Fitter AH, Leyser HMO. Phosphate Availability Regulates Root System Architecture in Arabidopsis. Plant Physiol. 2001;126:875–882. doi: 10.1104/pp.126.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, MacPherson CR, Liu J, Wang H, Kiba T, Hannah MA, Wang XJ, Bajic VB, Chua NH. The response and recovery of the Arabidopsis thalianatranscriptome to phosphate starvation. BMC Plant Biol. 2012;12:62. doi: 10.1186/1471-2229-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J Cell Mol Biol. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- Younis I, Dittmar K, Wang W, Foley SW, Berg MG, Hu KY, Wei Z, Wan L, Dreyfuss G. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife. 2013;2:e00780. doi: 10.7554/eLife.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Lu S, Liu D. A major root-associated acid phosphatase in Arabidopsis, AtPAP10, is regulated by both local and systemic signals under phosphate starvation. J Exp Bot. 2014;65:6577–6588. doi: 10.1093/jxb/eru377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


