ABSTRACT
Akkermansia muciniphila has evolved to specialize in the degradation and utilization of host mucus, which it may use as the sole source of carbon and nitrogen. Mucus degradation and fermentation by A. muciniphila are known to result in the liberation of oligosaccharides and subsequent production of acetate, which becomes directly available to microorganisms in the vicinity of the intestinal mucosa. Coculturing experiments of A. muciniphila with non-mucus-degrading butyrate-producing bacteria Anaerostipes caccae, Eubacterium hallii, and Faecalibacterium prausnitzii resulted in syntrophic growth and production of butyrate. In addition, we demonstrate that the production of pseudovitamin B12 by E. hallii results in production of propionate by A. muciniphila, which suggests that this syntrophy is indeed bidirectional. These data are proof of concept for syntrophic and other symbiotic microbe-microbe interactions at the intestinal mucosal interface. The observed metabolic interactions between A. muciniphila and butyrogenic bacterial taxa support the existence of colonic vitamin and butyrate production pathways that are dependent on host glycan production and independent of dietary carbohydrates. We infer that the intestinal symbiont A. muciniphila can indirectly stimulate intestinal butyrate levels in the vicinity of the intestinal epithelial cells with potential health benefits to the host.
KEYWORDS: Akkermansia muciniphila, anaerobes, butyrate, cross-feeding, intestine, microbiome, mucus, syntrophy
IMPORTANCE
The intestinal microbiota is said to be a stable ecosystem where many networks between microorganisms are formed. Here we present a proof of principle study of microbial interaction at the intestinal mucus layer. We show that indigestible oligosaccharide chains within mucus become available for a broad range of intestinal microbes after degradation and liberation of sugars by the species Akkermansia muciniphila. This leads to the microbial synthesis of vitamin B12, 1,2-propanediol, propionate, and butyrate, which are beneficial to the microbial ecosystem and host epithelial cells.
INTRODUCTION
The mammalian intestinal tract harbors complex microbial ecosystems that have been forged by millennia of coevolution between microbes and hosts. It is suggested that the evolution of metabolic interdependencies has led to strong deterministic processes that shape the composition of the microbiota during development (1). The diversity and richness of the gut microbiota within individuals, as well as the similarity in composition between individuals, are governed by several selective pressures within host habitats, such as diet (2, 3). Recent extreme interventions have illustrated the importance of dietary carbohydrates on the intestinal microbial community succession (4, 5). While dietary fibers affect substrate availability for the colonic microbiota, the mucus lining that covers the epithelial cells forms a consistent factor along its internal surface and is proposed to function as an endogenous prebiotic (6–9). The mucosal layer of the intestine is characterized by specific microbiota communities enriched with taxa affiliated with the family Lachnospiraceae (also known as Clostridium cluster XIVa) and the phylum Verrucomicrobia (10–15).
Akkermansia muciniphila is a mucus-colonizing member of the gut microbiota that has evolved to specialize in the degradation and utilization of host mucus, which it may use as the sole source of carbon and nitrogen (16, 17). Its mucin degradation activity leads to the production of 1,2-propanediol, propionate, and acetate (17). In addition, its mucosal foraging results in the availability of sugars liberated from mucus glycans and subsequent acetate production can stimulate coexistence of butyrogenic bacteria within the same mucosal niche (16). Microbe-produced short-chain fatty acids are described as major health-promoting compounds (18, 19). Because of its location close to the host cells, a symbiotic mucobiome could therefore be particularly important in fostering health in terms of nutrient exchange, communication with the host, regulation of the immune system, and resistance against invading pathogens.
Dietary intervention studies (13), in vitro mucosal model studies (20), and microbiota comparisons of gut lumen and epithelial biopsy specimens (11) have revealed strong cooccurrence of specific mucolytic bacteria (A. muciniphila, Bacteroides spp., and Ruminococcus spp.) and second-line butyrate producers (Anaerostipes caccae, Eubacterium spp., Faecalibacterium prausnitzii, and Roseburia intestinalis). This cooccurrence may be indicative of shared metabolic networks among the different microbial groups. In vitro isotope labeling has identified lactate and acetate as important precursors of butyrate production in human fecal samples (21). On top of this, kinetic modeling showed the likelihood for the dominant butyrate producers, such as Anaerostipes coli and Eubacterium hallii, to use short-chain fatty acids for butyrate production by utilizing lactate and acetate via the butyryl coenzyme A (CoA):acetate CoA transferase route, the main metabolic pathway for butyrate synthesis in the human colon (22).
In this study, we test the hypothesis that A. muciniphila can serve as the keystone species supporting a syntrophic network in a mucosal environment. Therefore, we studied the metabolic interactions between A. muciniphila and representative intestinal butyrate-producing bacteria; F. prausnitzii (representative of the family Ruminococcaceae also known as Clostridium cluster IV) and A. caccae and E. hallii (representatives of Lachnospiraceae also known as Clostridium cluster XIVa). The results indicate the existence of trophic chains on mucus between A. muciniphila and the butyrate-producing F. prausnitzii and A. caccae, while true bidirectional metabolic cross-feeding dependent on vitamin B12 was observed between A. muciniphila and E. hallii, indicative of a mutualistic symbiosis.
RESULTS
Growth and metabolism of intestinal butyrate producers on mucus or mucus-derived sugars.
In order to test whether Akkermansia muciniphila can serve as a keystone species in an environment where mucus is the main nutrient source, we first tested the ability of butyrate-producing mucosal colonizers to grow on mucus and mucus-derived sugars in the absence of A. muciniphila. When incubated in culture media with mucus as the sole carbon and nitrogen source, none of the butyrate-producing strains tested, Anaerostipes caccae, Eubacterium hallii, and Faecalibacterium prausnitzii, were able to grow or produce metabolites (see Table S2A in the supplemental material).
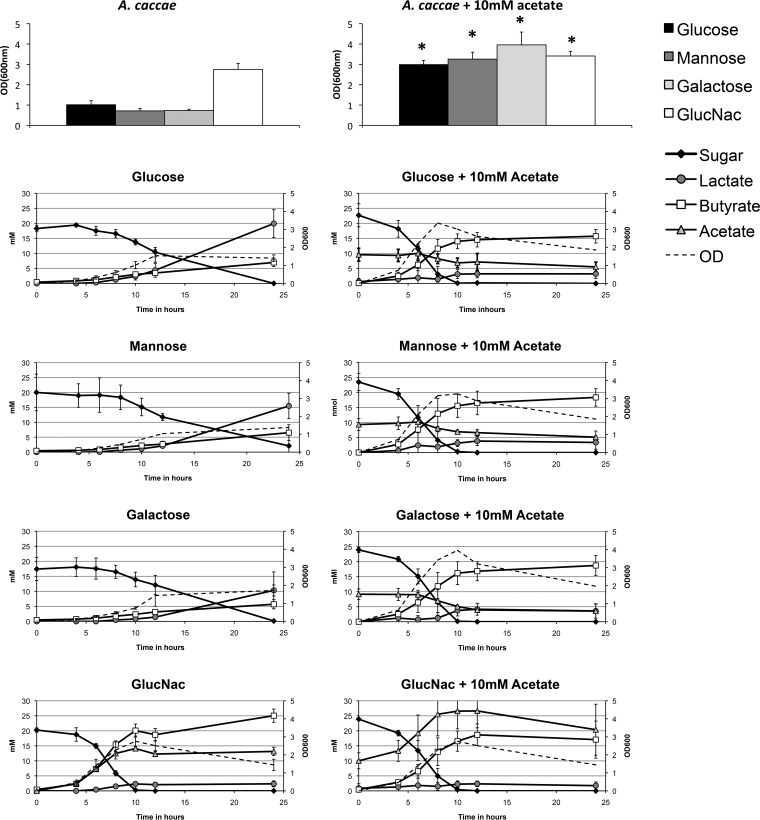
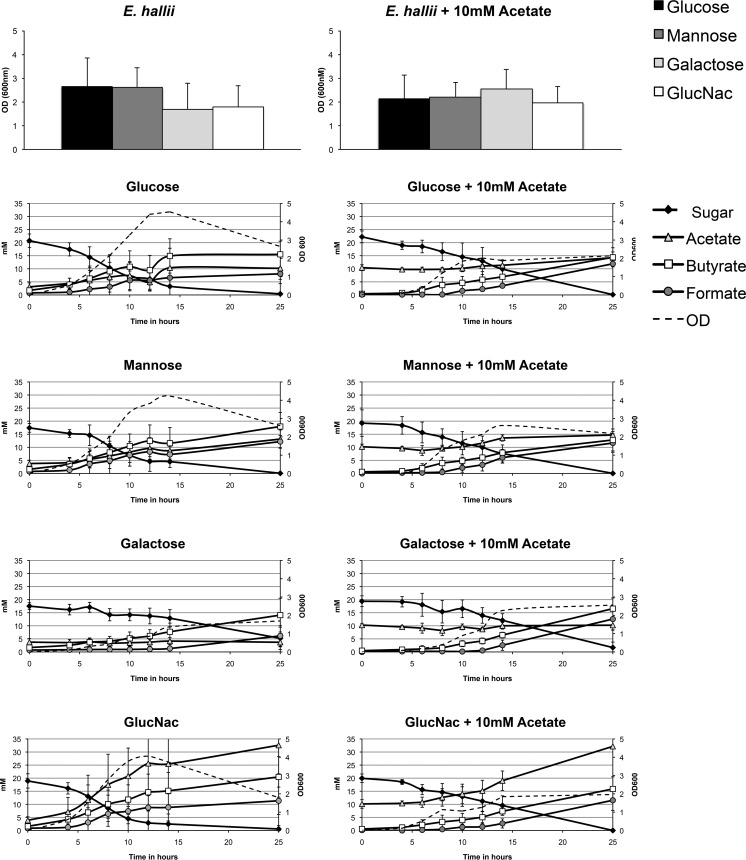
The mucin sugars d-galactose, d-mannose, GlcNAc, GalNAc, and l-fucose and the non-mucin sugar glucose were subsequently tested as possible carbon sources for each butyrate-producing species. Minimal media used for the bacteria differed as a result of different minimal requirements for protein and spore elements (see Materials and Methods for details on the composition of the media). F. prausnitzii is known to be able to grow on GlcNAc and galactose (23). In addition, we tested the growth of F. prausnitzii on mannose and GalNAc, but no growth was observed (Table S2B). A. caccae was observed to use glucose, d-mannose, d-galactose, and GlcNAc for growth, and the main fermentation products were acetate, butyrate, and lactate (Fig. 1). The highest A. caccae cell numbers and acetate production were reached with GlcNAc, possibly due to the fact that fermentation of this amino sugar can replace the need for acetate in the medium (Fig. 1). E. hallii showed the same preference for sugars as A. caccae did, resulting in growth on glucose, d-mannose, d-galactose, and GlcNAc (Fig. 2). The main fermentation products of E. hallii were observed to be acetate, butyrate, and formate. Again GlcNAc resulted in the highest production of acetate and butyrate compared to the other sugars, but this was not accompanied with increased cell numbers of E. hallii (Fig. 2).
FIG 1 .
Metabolic activity of A. caccae on mucin-derived sugars. A. caccae was grown on monosaccharide present in the glycan chain of mucin. The OD600 values and HPLC profiles are shown for the sugars that resulted in positive growth. The sugars that gave positive test results were also used to perform experiments with the addition of 10 mM acetate. The graphs show the mean values for the experiments performed a minimum of three times in duplicate. Values that are significantly different (P < 0.05) in the presence of 10 mM acetate or absence of acetate are indicated by an asterisk. GlucNac, N-acetylglucosamine.
FIG 2 .
Metabolic activity of E. hallii on mucin-derived sugars. E. hallii was grown on monosaccharide present in the glycan chain of mucin. The OD600 value and HPLC profiles are shown for sugars that resulted in positive growth. The sugars that gave positive test results were also used to perform experiments with the addition of 10 mM acetate. The graphs show the mean values for the experiments performed a minimum of three times in duplicate.
Acetate enhances growth of A. caccae but not E. hallii on mucin-derived sugars.
The average production of 10 mM acetate by A. muciniphila grown in medium containing mucin could serve as the substrate for growth of butyrogens. Therefore, we added 10 mM acetate to cultures growing on glucose, d-mannose, d-galactose, and GlcNAc. In the case of A. caccae, this did indeed lead to the production of butyrate, acetate, lactate, and formate as measured in a minimal medium. Furthermore, these butyrate production levels were significantly higher than the observed butyrate production without added acetate (Fig. 1).
Weak growth of A. caccae on l-fucose was observed after the addition of acetate but without detected metabolite production. Acetate alone did not support growth (Table S2C). The addition of acetate to the growth media of E. hallii did not result in differences in growth or metabolite profile, possibly due to its own production of acetate (Fig. 2).
The overall fermentation efficiency was determined by calculating the carbon balance at each monosaccharide condition. The recovery of carbon atoms varied in between 70 and 100%, depending on the biomass produced that explains the loss (Tables 1 and 2).
TABLE 1 .
Carbon balance of A. caccae on mucin-derived sugars with or without acetate
| Sugar | No. of carbons (mM) |
Carbon recovery (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Substrates |
Products |
||||||||
| Sugar | Acetate | Lactate | Acetate | Butyrate | Formate | CO2 | Avg | SD | |
| Glucose | 110 | 60 | 26 | 24 | 101 | 13 | |||
| Glucose + 10 mM acetate | 136 | 8 | 8 | 62 | 2 | 82 | 71 | 0 | |
| Mannose | 121 | 55 | 27 | 27 | 85 | 12 | |||
| Mannose + 10 mM acetate | 140 | 8 | 10 | 73 | 2 | 76 | 78 | 8 | |
| Galactose | 99 | 38 | 26 | 22 | 88 | 10 | |||
| Galactose + 10 mM acetate | 144 | 11 | 11 | 75 | 2 | 59 | 77 | 12 | |
| GlcNAc | 162 | 7 | 26 | 98 | 27 | 98 | 2 | ||
| GlcNAc + 10 mM acetate | 192 | 5 | 31 | 84 | 3 | 34 | 81 | 11 | |
TABLE 2 .
Carbon balance of E. hallii on mucin-derived sugars with or without acetate
| Sugar | No. of carbons (mM) |
Carbon recovery (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Substrates |
Products |
||||||||
| Sugar | Acetate | Lactate | Acetate | Butyrate | Formate | CO2 | Avg | SD | |
| Glucose | 122 | 14 | 55 | 7 | 27 | 87 | 30 | ||
| Glucose + 10 mM acetate | 133 | 8 | 56 | 12 | 29 | 79 | 18 | ||
| Mannose | 106 | 19 | 66 | 12 | 24 | 117 | 24 | ||
| Mannose + 10 mM acetate | 115 | 9 | 49 | 12 | 26 | 85 | 21 | ||
| Galactose | 74 | 0,1 | 50 | 5 | 16 | 96 | 29 | ||
| Galactose + 10 mM acetate | 106 | 0,1 | 64 | 13 | 24 | 93 | 14 | ||
| GlcNAc | 147 | 57 | 76 | 11 | 25 | 116 | 40 | ||
| GlcNAc + 10 mM acetate | 160 | 44 | 61 | 11 | 27 | 90 | 19 | ||
Mucus-induced trophic chains of A. muciniphila and butyrate producers A. caccae, E. hallii, and F. prausnitzii results in butyrate production
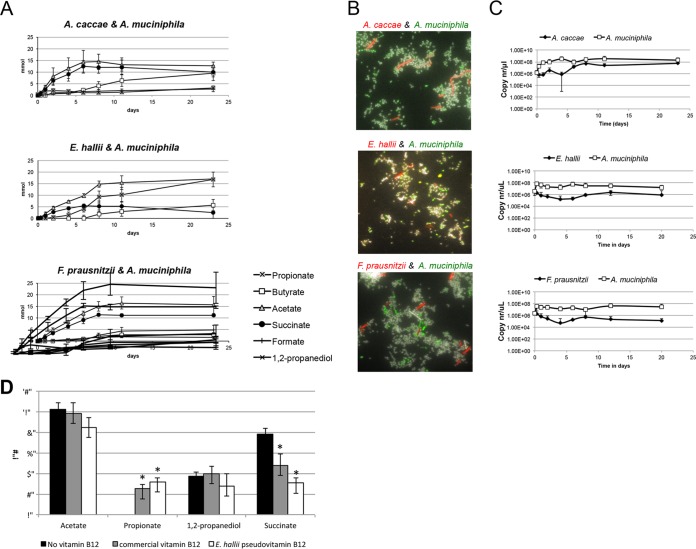
After the monoculture experiments, a series of cocultures of approximately equal amounts of A. muciniphila and butyrate producers were set up to test whether sugars and acetate produced as a result of mucin degradation by A. muciniphila would enable butyrate production of the chosen isolates. Remarkably, this coculturing on mucin-containing media supported growth and butyrate production for all three tested species (Fig. 3). A. caccae produced butyrate in levels comparable to those found in the monoculture conditions that were supplemented with acetate. Similarly, F. prausnitzii also produced butyrate in coculture with A. muciniphila and also produced 5 mM formate indicative of acetate consumption. Butyrate levels produced by E. hallii were in the range of what was seen in the monocultures growing on single sugars. The pH was monitored in all experiments and stayed around pH 6.5 throughout the experiments. Determination by quantitative PCR (Q-PCR) and qualitative presence (fluorescent in situ hybridization [FISH]) of the butyrate-producing species within the cocultures indicated a difference in abundance of the butyrate producers of several log units compared to the abundance of A. muciniphila (Fig. 3 and Table S1). The abundance of A. caccae increased 100-fold over the first 8 days of incubation based on the increase in its 16S rRNA gene copy number. Maximum butyrate levels were reached after 11 days of incubation. In contrast to the results for cultures, no lactate was measured during the cross-feeding experiments with A. caccae. Both Q-PCR and FISH results indicated a ratio of A. muciniphila to A. caccae of approximately 100:1.
FIG 3 .
A. muciniphila degradation and fermentation of mucus enables cross-feeding by the butyrate-producing gut isolates. (A to C) Cocultures of A. muciniphila with butyrate-producing isolates were performed and measurements of product formation and consumption (A), FISH staining (B), and Q-PCR (C) were performed. (D) Measurement of A. muciniphila metabolites on mucus-containing media without the addition of vitamin B12 or with vitamin B12 from E. hallii or pseudovitamin B12 from E. hallii. The graph shows the mean values for the experiment performed a minimum of three times in duplicate. Asterisks indicate a significant difference (P < 0.05) compared to the condition without vitamin B12 added.
Qualitative measurements in coculture experiments by FISH. Download TABLE S1, PDF file, 0.04 MB (40.8KB, pdf) .
Copyright © 2017 Belzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In the F. prausnitzii-A. muciniphila cocultures, F. prausnitzii 16S rRNA gene copy numbers decreased, and a small amount of butyrate appeared after 8 days of incubation. FISH staining revealed the presence of F. prausnitzii cells within the cocultures but confirmed its slow growth. Finally, within the E. hallii-A. muciniphila cocultures, low levels of butyrate started to build up after 8 days. This was associated with an increase in 16S rRNA gene copy numbers of E. hallii on day 8. Q-PCR and FISH staining showed an A. muciniphila-to-E. hallii ratio of 100:1 after 8 to 24 days (Fig. 3 and Table S1).
Vitamin B12-dependent syntrophy between E. hallii and A. muciniphila.
Analyses of the metabolites produced in cocultures showed that in the A. muciniphila-E. hallii coculture, the proportion of succinate to propionate had shifted compared to the proportion in monocultures of A. muciniphila (Fig. 3). This was not observed in the other cocultures. Notably, 1,2-propanediol, found as a result of fucose degradation by A. muciniphila in monocultures, was not detected in the coculture with E. hallii.
Conversion of propionate to succinate involves vitamin B12-dependent methylmalonyl-CoA mutase.
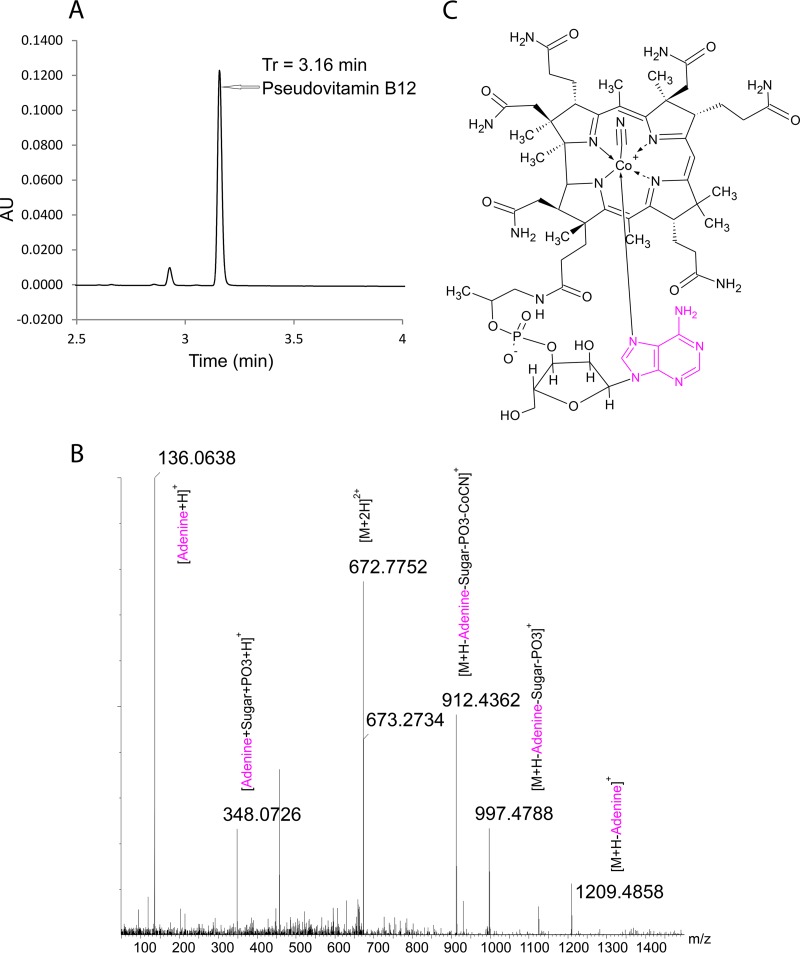
Detailed mass spectroscopy analysis confirmed that E. hallii is capable of synthesizing a B12 vitamer in monocultures as described previously (24). Our analyses show that the structure of this vitamer (Fig. 4) is pseudovitamin B12, as the lower ligand contained adenine instead of 5,6-dimethyl benzimidazole (DMBI). No effect of DMBI addition was observed on the structure of the produced B12 vitamer.
FIG 4 .
UHPLC-UV chromatogram of E. hallii vitamin B12. (A) Immunoaffinity-purified cell extract of E. hallii (in arbitrary units [AU]) is shown on the y axis, and time (in minutes) is shown on the x axis. Tr, retention time. (B) LC-MS/MS identified a peak at 3.16 min. (C) Chemical structure of pseudovitamin B12 from E. hallii.
To test the hypothesis that A. muciniphila can use the pseudovitamin B12 produced by E. hallii for the conversion of succinate to propionate, the effects of both purified E. hallii and commercially available vitamin B12 on A. muciniphila growth were tested. Indeed, the addition of pseudovitamin B12 and vitamin B12 resulted in significant lower succinate levels and significant higher propionate production. The addition of either vitamin B12 resulted in a profile identical to the profile observed for A. muciniphila-E. hallii coculture (Fig. 3).
These observations provide evidence for bidirectional metabolic cross-feeding between A. muciniphila and E. hallii. A. muciniphila liberates sugars from mucus and produces 1,2-propanediol for growth support of E. hallii. In return, A. muciniphila is provided with a vitamin B12 analogue used as a cofactor for the conversion of succinate to propionate via methylmalonyl-CoA synthase (Fig. 5). Apparently both vitamin B12 and pseudovitamin B12 can be used as a cofactor by A. muciniphila to activate the methylmalonyl-CoA synthase. Hence, the B12 vitamer produced by E. hallii is in the pseudovitamin B12 form and can be used by other intestinal microorganisms, but it has lower affinity than vitamin B12 for the human intrinsic factor (25).
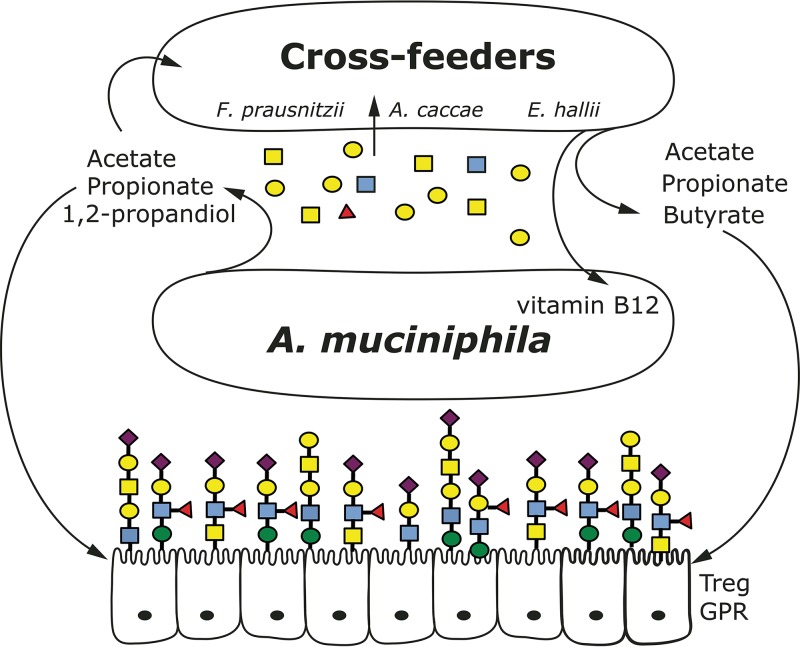
FIG 5 .
Schematic overview of mucus-dependent cross-feeding network. Keystone mucolytic bacteria, such as A. muciniphila, degrade mucin glycans resulting in oligosaccharides (mainly galactose, fucose, mannose, and GlucNac) and SCFAs (acetate, propionate, and 1,2-propanediol) that can be used for growth, as well as propionates, butyrate, and vitamin B12 production by cross-feeding partners. Treg GPR, regulatory T cell G-protein-coupled protein receptor.
DISCUSSION
In spite of the great interest in metabolic conversions in the human gut, there is limited information on actual product sharing mechanisms and trophic dependencies of individual members of the intestinal microbiota. One such syntrophic relationship has been described for the species Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii (26). F. prausnitzii can metabolize acetate produced by B. thetaiotaomicron to produce butyrate. This butyrate is then utilized by host epithelial cells and regulates host immunity via epithelial cell signaling, colonic T regulatory cells, and macrophages (19, 27). In addition, a few studies demonstrated the use of lactate and acetate produced by Bifidobacterium spp. by colonic butyrate producers (28–30). Specifically, this form of cross-feeding has been described for Bifidobacterium adolescentis and F. prausnitzii (30).
Moreover, cocultivation of amylolytic bacteria from the human colon, such as Eubacterium rectale, B. thetaiotaomicron, or Bifidobacterium adolescentis, with Ruminococcus bromii L2-63 can lead to increased starch utilization (31). In addition, coculturing of the non-starch-degrading species Anaerostipes hadrus with R. bromii has been shown to result in the removal of the reducing sugars that accumulate in R. bromii monocultures (32). Similarly, by stable isotope probing with 13C-labeled resistant starch has revealed a butyrogenic trophic chain between R. bromii and E. rectale in an in vitro human colon model (33, 34).
Various studies have coupled cooccurrence networks of bacteria to their genome content to model possible metabolic cross-feeding (22, 35). It should be noted that the studies discussed above all focus on cross-feeding that relies on diet-derived colonic sugars. However, mucin-derived sugars are the main source of energy for a group of microbiota members that can directly impact host cross talk at the mucosa (26). Mucus-dependent microbial networks at the mucosal layer would yield butyrate and other components with health benefits to the host (26). Our study supports the hypothesis that cross-feeding between microbiota members can take place when mucus is the only carbon source to support growth. Such mucosal trophic networks could determine host microbial cross talk in immune and metabolic regulation.
The mucosa-colonizing bacterium A. muciniphila is strongly correlated with a lean phenotype and increased barrier function (36–38). The correlation between A. muciniphila and host might depend on an additional microbial player. Indeed, we have shown that the mucus-degrading capacity of A. muciniphila may provide substrates to butyrate producers tested.
Two distinct types of trophic chains between A. muciniphila and butyrate-producing species were observed in this study. In the case of A. caccae, liberated sugars from mucus could sustain growth but A. muciniphila-derived acetate increased growth and metabolic production of butyrate even further, indicative of metabolic syntrophic interactions. In the case of E. hallii, a specific metabolic and cofactor syntrophic interaction was observed; pseudovitamin B12 affected the carbon flux within A. muciniphila, resulting in propionate production.
It is known from human studies that propionate delivered to the colon has various beneficial effects, including the regulation of satiety (39). Remarkably, E. hallii was able to utilize mucus sugars, in agreement with an earlier report (40). However, E. hallii had no clear advantage when acetate was present, possibly due to its own production of acetate when grown on mucus-derived sugars that already reached levels comparable to that of A. muciniphila monoculture.
Recently, it was reported that E. hallii is also able to use 1,2-propanediol for the production of propionate. Our data show the lack of 1,2-propanediol in the A. muciniphila-E. hallii coculture and supports the previous suggested syntrophic possibilities between intestinal microbes (24). 1,2-Propanediol is produced by A. muciniphila from fucose. As such, the presence or absence of fucose in the intestinal mucosa (FUT2 polymorphism) may help explain microbial networks at the mucosal layer (41). Furthermore, in coculture experiments with A. muciniphila and F. prausnitzii, low levels of butyrate were measured accompanied by the presence of cells and 16S rRNA copies of this butyrate producer as opposed to monocultures of the organism on the same medium (Table S2A). These results further indicate that the association of butyrate Clostridium cluster XIVa and IV species could indeed yield the production of butyrate as a result of a microbial metabolic network in the mucosal layer, which is poor in usable carbon sources.
Growth and metabolic measurements of butyrogens. Download TABLE S2, PDF file, 0.1 MB (55.7KB, pdf) .
Copyright © 2017 Belzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The fact that a changed metabolic profile for A. muciniphila in the presence of E. hallii was found is further evidence supporting a mutualistic syntrophic interaction. The availability of pseudovitamin B12 in vivo can be of importance for the microbial ecosystem as well as the host. Microorganisms are the only natural sources of the pseudovitamin B12 derivatives, and several intestinal microbes have been reported to contribute to the pseudovitamin B12 levels in the intestine (42). The approximate concentration of the cobalamin analogue adenine (as produced by E. hallii) is 164 ng/g (wet weight) of feces (43), and this is also in the range of what we found to be needed for A. muciniphila propionate induction (100 ng/ml). It is not clear whether pseudovitamin B12 can be used by intestinal cells. While the affinity of human intrinsic factor for pseudovitamin B12 is lower than that for vitamin B12, it is equally bound by transcobalamin and haptocorrin human corroid factors (25) and is not antagonistic to vitamin B12 (44), and it may be transported without intrinsic factor (45). Moreover, it has been shown that pseudovitamin B12 produced by Lactobacillus reuteri, also an abundant mouse intestinal bacterium, can alleviate vitamin B12 deficiency in mice (46, 47).
In summary, the present data indicate that pseudovitamin B12 is biologically active in A. muciniphila propionate metabolism that involves methylmalonyl-CoA mutase (48). Hence, the synthropic partners together produce a higher propionate-to-succinate ratio, and this in turn is beneficial for host cell metabolism. It also implies that stimulating or diminishing a keystone species, such as A. muciniphila, from the microbiota can have dramatic effect on a complete microbial network and associated host-microbe homeostasis. In this case, stimulating or administrating A. muciniphila within the intestine might benefit from addition of another organism or solely pseudovitamin B12 to stimulate the organism’s production of propionate and a healthy mucosal environment (Fig. 5).
Many gastrointestinal disorders have been associated with mucosal damage and lower gut barrier function. The fact that intestinal bacteria may have an impact on both these factors, either directly or via specific immune and metabolic stimulation, further emphasizes the importance of having the right bacteria at the right place. Loss of mucosal integrity and the associated mucobiome could be indicative of disease states and its development. A. muciniphila has been positively associated with a lean phenotype and beneficial metabolic gene regulation in human cell types (36, 49). Its presence might be essential for a mucosal adherent network of beneficial microorganisms that together prompt these effects of the host. As a matter of fact, weight loss studies usually report increased abundance of Verrucomicrobia (mainly A. muciniphila) as well as several other microbial species (50–52). Taken together, these results further indicate the possible importance of mucosa-associated microbial networks and their metabolic cross-feeding for regulation of host health-related parameters and prevention of disease.
MATERIALS AND METHODS
Bacterial growth conditions.
Akkermansia muciniphila MucT (ATTC BAA-835) was grown as described previously (17, 53). Purified mucin was prepared as follows. Ten grams of hog gastric mucin (type III; Sigma-Aldrich) was dissolved in 500 ml of 0.1 M NaCl (pH 7.8) containing 0.02 M phosphate buffer (0.02 M NaH2PO4 and Na2HPO4) (pH 7.8), stirring for 24 h at 4°C. After 1 h, the pH was adjusted to pH 7.2 using 1 M NaOH. After centrifugation, the supernatant was cooled on ice and precipitated with 60% (vol/vol) prechilled ethanol. After centrifugation, the pellet was dissolved in 0.1 M NaCl. These last two steps were repeated twice. After the last centrifugation step, the pellet was washed once with 100% ethanol, dissolved in 100 ml Milli-Q, and dialyzed using Spectra/Por 6 8,000-Da MWCO (molecular weight cutoff) protein dialysis with four changes. Last, the dialyzed mucin was freeze dried and dissolved in Milli-Q at a concentration of 5% (wt/vol). Mucin was added to the medium after autoclaving. The resulting purified mucin was tested for the absence of oligosaccharides. Incubations were performed in serum bottles sealed with butyl rubber stoppers at 37°C under anaerobic conditions provided by a gas phase of 182 kPa (1.5 atm) N2/CO2 (80/20 ratio). Growth was measured by a spectrophotometer as the optical density at 600 nm (OD600).
Faecalibacterium prausnitzii A2-165 was grown anaerobically at 37°C in YCFA medium supplemented with 33 mM acetate and 25 mM glucose (53). Anaerostipes caccae L1-92 (54) was grown anaerobically at 37°C in either PYG medium (DSMZ) or minimal medium (55) containing 25 mM glucose. Eubacterium hallii L2-7 was grown anaerobically at 37°C in YCFA medium without the addition of fatty acids (propionate, isovaleric acid, valeric acid, isobutyrate, and butyrate). Mucin sugar utilization was performed in minimal medium with or without the addition of 10 mM acetate. In some cases, the experiments were performed with mucin-derived single sugars (mannose [Sigma-Aldrich]), fucose (Sigma-Aldrich), galactose (Biochemika), N-acetylgalactosamine (Sigma-Aldrich), or N-acetylglucosamine (Sigma-Aldrich); these were used at a concentration of 25 mM. Growth was monitored for 24 h, and samples were collected regularly for OD600 and high-performance liquid chromatography (HPLC) analysis.
Coculture experiments were performed in minimal medium supplemented with mucus (17), and the medium was buffered to reduce pH changes due to fermentation products. Optimal coculture conditions were established as follows. A. muciniphila was added to media containing mucin, and the media containing bacteria were incubated for 8 h to reach measurable concentrations of acetate and liberate sugars. Subsequently, 108 cells of A. caccae, E. hallii, or F. prausnitzii were added to the A. muciniphila-containing cultures. All cells had been washed twice with phosphate-buffered saline (PBS) before being added to the coculture to prevent carryover of products from the preculture. During the coculture, 0.15% mucin was added to the medium every 48 h to maintain sufficient substrate availability for A. muciniphila. All growth experiments were repeated a minimum of three times in duplicate.
High-performance liquid chromatography.
For fermentation product analysis, 1 ml of bacterial culture was centrifuged, and the supernatant was stored at −20°C for high-performance liquid chromatography (HPLC) analysis. Substrate conversion and product formation were measured with a Thermo Scientific Spectrasystem high-performance liquid chromatography (HPLC) system equipped with a Varian Metacarb 67H column (300 by 6.5 mm) kept at 45°C and with 0.005 mM sulfuric acid as the eluent. The eluent had a flow rate of 0.8 ml/min, and metabolites were detected by determining the refractive index. Carbon balances were calculated by the amount of carbon of the products/amount of carbon of the substrate × 100%, using sugars and short-chain fatty acids (SCFAs) as measured by HPLC with biological triplicate samples and technical duplicate samples. We used theoretical CO2 calculations: 6 mol glucose yields 8 mol CO2, and 1 mol lactate yields 1 mol CO2.
Ultrahigh performance liquid chromatography-mass spectrometry (UHPLC-MS).
For vitamin B12 analysis, E. hallii cells (0.2 g) were mixed with 10 ml of extraction buffer (8.3 mM sodium hydroxide and 20.7 mM acetic acid [pH 4. 5]) containing 100 µl of 1% NaCN. The vitamin was extracted in its cyano form by subjecting the mixture to a boiling water bath for 30 min. After cooling, the extract was recovered by centrifugation (6,900 × g for 10 min; Hermle, Wehingen, Germany) and finally purified by immunoaffinity column chromatography (Easy-Extract; R-Biopharma, Glasgow, Scotland). The reconstituted extract was analyzed for vitamin content using an HSS T3 C18 column (2. 1 by 100 mm; 1.8 µm) on a Waters Acquity UPLC (ultraperformance liquid chromatography) system (Milford, MA) equipped with a photodiode array detector (PDA) (210 to 600 nm) and interfaced to a high-resolution quadrupole time of flight mass spectrometer (QTOF; Synapt G2-Si, Waters). The eluent was a gradient flow (0.32 ml/min) of water (solvent A) and acetonitrile (solvent B), both acidified with 0.1% formic acid: 0 to 0.5 min (95 parts solvent A to 5 parts of solvent B [95:5]), 0.5 to 5 min (60:40), 5 to 6 min (60:40), and 6 to 10 min (95:5). The column was maintained at 30°C, and the UV detection was recorded at 361 nm. The MS analysis was done in positive ion mode with electrospray ionization, using a scanning range set for m/z of 50 to 1,500. The parent ions corresponding to the vitamin peak were further fragmented (tandem mass spectrometry [MS/MS]) and analyzed.
Fluorescent in situ hybridization (FISH).
The following rRNA-targeted oligonucleotide probes were used: (i) Cy3-labeled universal EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′), which is complementary to a conserved region of the bacterial 16S rRNA molecule specific to most eubacteria except phyla of Plantomycetales and Verrucomicrobia (17); and (ii) Cy5-labeled EUB338 III (5′-GCTGCCACCCGTAGGTGT-3′), the supplementary probes for eubacteria to target Verrucomicrobia (56).
Cell fixation, in situ hybridization, DAPI staining, and microscopy.
Bacterial cultures (0.5 ml) were fixed overnight with 1.5 ml of 4% paraformaldehyde (PFA) at 4°C. Working stocks were prepared by harvesting bacterial cells by 5 min centrifugation at 8,000 × g, followed by resuspension in ice-cold phosphate-buffered saline (PBS) and 96% ethanol at a 1:1 (vol/vol) ratio. Three microliters of the PBS-ethanol working stocks were spotted into 18 wells (round wells with a 6-mm diameter) on gelatin-coated microscope slides. The slides were hybridized with the DNA probes by applying 10 μl of hybridization mixture per well, which contained 1 volume of probe mixture (probe concentration of 20 μM) and 9 volumes of hybridization buffer (20 mM Tris-HCl, 0.9 M NaCl, 0.1% SDS [pH 7.2]). The slides were hybridized for at least 3 h in a moist chamber at 50°C; this was followed by 30-min incubation in washing buffer (20 mM Tris-HCl, 0.9 M NaCl [pH 7. 2]) at 50°C for washing. The slides were rinsed briefly with Milli-Q and air dried. The slides were stained with a 4,6-diamine-2-phenylindole dihydrochloride (DAPI) mixture containing 200 μl PBS and 1 μl DAPI dye (100 ng/μl) for 5 min in the dark at room temperature, followed by Milli-Q rinsing and air drying. The slides were then covered with Citifluor AF1 and a coverslip. The bacteria on the slides were enumerated using an Olympus MT ARC/HG epifluorescence microscope. A total of 25 positions per well were automatically analyzed in three-color channels (DAPI, Cy3, and Cy5) using a quadruple band filter.
Quantitative real-time PCR.
The abundances of A. muciniphila and butyrate producers in coculture were determined by quantitative real-time PCR. Bacterial cultures were harvested at 16,100 × g for 10 min. DNA extractions were performed using MasterPure Gram-positive DNA purification kit. The DNA concentrations were determined fluorometrically (Qubit dsDNA HS [double-stranded DNA high-sensitivity] assay; Invitrogen) and adjusted to 1 ng/μl prior to use as the template in quantitative PCR (Q-PCR). Primers targeting A. muciniphila, A. caccae, and E. hallii based on specific variable regions of the 16S rRNA gene (Table 3) were used for quantification. Standard template DNA was prepared from the 16S rRNA gene of each bacterium by amplification with primers 27F (F stands for forward) and 1492R (R stands for reverse). Standard curves were prepared with nine standard concentrations of 100 to 108 gene copies μl−1. PCRs were performed in triplicate with iQ SYBR green supermix (Bio-Rad) in a total volume of 10 μl with primers at 500 nM in the wells on 384-well plates with the wells sealed with optical sealing tape. Amplification was performed with an iCycler (Bio-Rad) and the following protocol: one cycle of 95°C for 10 min; 35 cycles of 95°C for 15 s, 60°C for 20 s, and 72°C for 30 s each; one cycle of 95°C for 1 min; one cycle of 60°C for 1 min; and a stepwise increase of the temperature from 60 to 95°C (at 0.5°C per 5 s) to obtain melt curve data. Data were analyzed using the Bio-Rad CFX Manager 3.0.
TABLE 3 .
PCR primers used in this study and their amplification products
| Bacterium | Primer | Primer sequence | Product size (bp) | Reference |
|---|---|---|---|---|
| Akkermansia muciniphila | AM1 | 5′ CAGCACGTGAAGGTGGGGAC 3′ | 327 | 57 |
| AM2 | 5′ CCTTGCGGTTGGCTTCAGAT 3′ | |||
| Anaerostipes caccae subgroup | OFF2555 | 5′ GCGTAGGTGGCATGGTAAGT 3′ | 83 | 58 |
| OFF2556 | 5′ CTGCACTCCAGCATGACAGT 3′ | |||
| Eubacterium hallii L2-7 | EhalF | 5′ GCGTAGGTGGCAGTGCAA 3′ | 278 | 59 |
| EhalR | 5′ GCACCGRAGCCTATACGG 3′ | |||
| Faecalibacterium prausnitzii | FPR2F | 5′ GGAGGAAGAAGGTCTTCGG 3′ | 248 | 59 |
| Fprau645R | 5′ AATTCCGCCTACCTCTGCACT 3′ | |||
Statistics.
Statistics were performed using t test and corrected for multiple testing using false-discovery rate (FDR) correction for multiple comparisons. P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
We thank Harm Wopereis, Nam Bui, Caroline Plugge, and Fons Stams for their constructive comments.
This work was supported by Advanced research grant 250172 (MicrobesInside) of the European Research Council to W.M.D.V., the Gravity 024.002.002 (SIAM) and Spinoza grants of the Netherlands Organization for Scientific Research (NWO) to W.M.D.V.
Jan Knol and Loo Wee Chia are financially supported by Nutricia Research, but there was no involvement of the company in the content of this work.
Footnotes
Citation Belzer C, Chia LW, Aalvink S, Chamlagain B, Piironen V, Knol J, de Vos WM. 2017. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. mBio 8:e00770-17. https://doi.org/10.1128/mBio.00770-17.
REFERENCES
- 1.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoetendal EG, de Vos WM. 2014. Effect of diet on the intestinal microbiota and its activity. Curr Opin Gastroenterol 30:189–195. doi: 10.1097/MOG.0000000000000048. [DOI] [PubMed] [Google Scholar]
- 3.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, Lieber A, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8:343ra382. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, Vipperla K, Naidoo V, Mtshali L, Tims S, Puylaert PG, DeLany J, Krasinskas A, Benefiel AC, Kaseb HO, Newton K, Nicholson JK, de Vos WM, Gaskins HR, Zoetendal EG. 2015. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouwehand AC, Derrien M, de Vos W, Tiihonen K, Rautonen N. 2005. Prebiotics and other microbial substrates for gut functionality. Curr Opin Biotechnol 16:212–217. doi: 10.1016/j.copbio.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Pacheco AR, Barile D, Underwood MA, Mills DA. 2015. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci 3:419–445. doi: 10.1146/annurev-animal-022114-111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tailford LE, Crost EH, Kavanaugh D, Juge N. 2015. Mucin glycan foraging in the human gut microbiome. Front Genet 6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouwerkerk JP, de Vos WM, Belzer C. 2013. Glycobiome: bacteria and mucus at the epithelial interface. Best Pract Res Clin Gastroenterol 27:25–38. doi: 10.1016/j.bpg.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Wang W, Zhou R, Ng SC, Li J, Huang M, Zhou F, Wang X, Shen B, Kamm MA, Wu K, Xia B. 2014. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore) 93:e51. doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong PY, Croix JA, Greenberg E, Gaskins HR, Mackie RI. 2011. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS One 6:e25042. doi: 10.1371/journal.pone.0025042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. 2014. The intestinal microbiome in early life: health and disease. Front Immunol 5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, Sommer F, Bäckhed F, Hansson GC, Johansson ME. 2015. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 16:164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belzer C, de Vos WM. 2012. Microbes inside—from diversity to function: the case of Akkermansia. ISME J 6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 18.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 19.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, De Weirdt R, Kerckhof FM, Van de Wiele T. 2013. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J 7:949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison DJ, Mackay WG, Edwards CA, Preston T, Dodson B, Weaver LT. 2006. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br J Nutr 96:570–577. [PubMed] [Google Scholar]
- 22.Muñoz-Tamayo R, Laroche B, Walter E, Doré J, Duncan SH, Flint HJ, Leclerc M. 2011. Kinetic modelling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbiol Ecol 76:615–624. doi: 10.1111/j.1574-6941.2011.01085.x. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. 2012. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol 78:420–428. doi: 10.1128/AEM.06858-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engels C, Ruscheweyh HJ, Beerenwinkel N, Lacroix C, Schwab C. 2016. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol 7:713. doi: 10.3389/fmicb.2016.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stupperich E, Nexø E. 1991. Effect of the cobalt-N coordination on the cobamide recognition by the human vitamin B12 binding proteins intrinsic factor, transcobalamin and haptocorrin. Eur J Biochem 199:299–303. doi: 10.1111/j.1432-1033.1991.tb16124.x. [DOI] [PubMed] [Google Scholar]
- 26.Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, Langella P, Thomas M. 2013. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang PV, Hao L, Offermanns S, Medzhitov R. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falony G, Vlachou A, Verbrugghe K, De Vuyst L. 2006. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol 72:7835–7841. doi: 10.1128/AEM.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. 2015. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett 362:fnv176. doi: 10.1093/femsle/fnv176. [DOI] [PubMed] [Google Scholar]
- 31.Ze X, Duncan SH, Louis P, Flint HJ. 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ze X, Le Mougen F, Duncan SH, Louis P, Flint HJ. 2013. Some are more equal than others: the role of “keystone” species in the degradation of recalcitrant substrates. Gut Microbes 4:236–240. doi: 10.4161/gmic.23998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovatcheva-Datchary P, Egert M, Maathuis A, Rajilić-Stojanović M, de Graaf AA, Smidt H, de Vos WM, Venema K. 2009. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol 11:914–926. doi: 10.1111/j.1462-2920.2008.01815.x. [DOI] [PubMed] [Google Scholar]
- 34.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy R, Borenstein E. 2013. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci U S A 110:12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, de Vos WM, Satokari R. 2015. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol 81:3655–3662. doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mack I, Cuntz U, Grämer C, Niedermaier S, Pohl C, Schwiertz A, Zimmermann K, Zipfel S, Enck P, Penders J. 2016. Weight gain in anorexia nervosa does not ameliorate the faecal microbiota, branched chain fatty acid profiles, and gastrointestinal complaints. Sci Rep 6:26752. doi: 10.1038/srep26752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SE, MacDougall K, Preston T, Tedford C, Finlayson GS, Blundell JE, Bell JD, Thomas EL, Mt-Isa S, Ashby D, Gibson GR, Kolida S, Dhillo WS, Bloom SR, Morley W, Clegg S, Frost G. 2015. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64:1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duncan SH, Louis P, Flint HJ. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka T, Scheet P, Giusti B, Bandinelli S, Piras MG, Usala G, Lai S, Mulas A, Corsi AM, Vestrini A, Sofi F, Gori AM, Abbate R, Guralnik J, Singleton A, Abecasis GR, Schlessinger D, Uda M, Ferrucci L. 2009. Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am J Hum Genet 84:477–482. doi: 10.1016/j.ajhg.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kräutler B. 2005. Vitamin B12: chemistry and biochemistry. Biochem Soc Trans 33:806–810. doi: 10.1042/BST0330806. [DOI] [PubMed] [Google Scholar]
- 43.Allen RH, Stabler SP. 2008. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr 87:1324–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe F, Katsura H, Takenaka S, Fujita T, Abe K, Tamura Y, Nakatsuka T, Nakano Y. 1999. Pseudovitamin B12 is the predominant cobamide of an algal health food, Spirulina tablets. J Agric Food Chem 47:4736–4741. doi: 10.1021/jf990541b. [DOI] [PubMed] [Google Scholar]
- 45.Doets EL, In’t Veld PH, Szczecińska A, Dhonukshe-Rutten RAM, Cavelaars AEJM, van’t Veer P, Brzozowska A, de Groot LCPGM. 2013. Systematic review on daily vitamin B12 losses and bioavailability for deriving recommendations on vitamin B12 intake with the factorial approach. Ann Nutr Metab 62:311–322. doi: 10.1159/000346968. [DOI] [PubMed] [Google Scholar]
- 46.Molina VC, Médici M, Taranto MP, Font de Valdez G. 2009. Lactobacillus reuteri CRL 1098 prevents side effects produced by a nutritional vitamin B deficiency. J Appl Microbiol 106:467–473. doi: 10.1111/j.1365-2672.2008.04014.x. [DOI] [PubMed] [Google Scholar]
- 47.Santos F, Vera JL, Lamosa P, de Valdez GF, de Vos WM, Santos H, Sesma F, Hugenholtz J. 2007. Pseudovitamin B12 is the corrinoid produced by Lactobacillus reuteri CRL1098 under anaerobic conditions. FEBS Lett 581:4865–4870. doi: 10.1016/j.febslet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 48.van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. 2011. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, Roeselers G. 2014. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio 5:e01438-14. doi: 10.1128/mBio.01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward EK, Schuster DP, Stowers KH, Royse AK, Ir D, Robertson CE, Frank DN, Austin GL. 2014. The effect of PPI use on human gut microbiota and weight loss in patients undergoing laparoscopic Roux-en-Y gastric bypass. Obes Surg 24:1567–1571. doi: 10.1007/s11695-014-1275-1. [DOI] [PubMed] [Google Scholar]
- 51.Liou AP, Paziuk M, Luevano JM Jr, Machineni S, Turnbaugh PJ, Kaplan LM. 2013. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 5:178ra41. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Remely M, Tesar I, Hippe B, Gnauer S, Rust P, Haslberger AG. 2015. Gut microbiota composition correlates with changes in body fat content due to weight loss. Benef Microbes 6:431–439. doi: 10.3920/BM2014.0104. [DOI] [PubMed] [Google Scholar]
- 53.Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 52:2141–2146. doi: 10.1099/00207713-52-6-2141. [DOI] [PubMed] [Google Scholar]
- 54.Schwiertz A, Hold GL, Duncan SH, Gruhl B, Collins MD, Lawson PA, Flint HJ, Blaut M. 2002. Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst Appl Microbiol 25:46–51. doi: 10.1078/0723-2020-00096. [DOI] [PubMed] [Google Scholar]
- 55.Plugge CM. 2005. Anoxic media design, preparation, and considerations. Methods Enzymol 397:3–16. doi: 10.1016/S0076-6879(05)97001-8. [DOI] [PubMed] [Google Scholar]
- 56.Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 57.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. 2007. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, DuBois A, Khlebnikov A, van Hylckama Vlieg JET, Punita S, Glickman JN, Onderdonk A, Glimcher LH, Garrett WS. 2010. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci USA 107:18132–18137. doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. 2009. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Qualitative measurements in coculture experiments by FISH. Download TABLE S1, PDF file, 0.04 MB (40.8KB, pdf) .
Copyright © 2017 Belzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth and metabolic measurements of butyrogens. Download TABLE S2, PDF file, 0.1 MB (55.7KB, pdf) .
Copyright © 2017 Belzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.