Abstract
In a search for plant homologues of dipeptidyl peptidase III (DPP III) family, we found a predicted protein from the moss Physcomitrella patens (UniProt entry: A9TLP4), which shared 61% sequence identity with the Arabidopsis thaliana uncharacterized protein, designated Nudix hydrolase 3. Both proteins contained all conserved regions of the DPP III family, but instead of the characteristic hexapeptide HEXXGH zinc-binding motif, they possessed a pentapeptide HEXXH, and at the N-terminus, a Nudix box, a hallmark of Nudix hydrolases, known to act upon a variety of nucleoside diphosphate derivatives.
To investigate their biochemical properties, we expressed heterologously and purified Physcomitrella (PpND) and Arabidopsis (AtND) protein. Both hydrolyzed, with comparable catalytic efficiency, the isopentenyl diphosphate (IPP), a universal precursor for the biosynthesis of isoprenoid compounds. In addition, PpND dephosphorylated four purine nucleotides (ADP, dGDP, dGTP, and 8-oxo-dATP) with strong preference for oxidized dATP. Furthermore, PpND and AtND showed DPP III activity against dipeptidyl-2-arylamide substrates, which they cleaved with different specificity.
This is the first report of a dual activity enzyme, highly conserved in land plants, which catalyses the hydrolysis of a peptide bond and of a phosphate bond, acting both as a dipeptidyl peptidase III and an atypical Nudix hydrolase.
Keywords: Arabidopsis thaliana, enzyme kinetics, metalloprotease, plant biochemistry, Physcomitrella patens, substrate specificity
Introduction
The peptidase family M49 (dipeptidyl peptidase III family) was first distinguished among metallopeptidases based on the unique zinc binding motif, the hexapeptide HEXXGH (Fukasawa et al. 1998; Chen and Barrett 2004), and by the presence of additional four conserved regions (consensus sequences) in the primary structures of its members (Abramić et al. 2004). All five evolutionary conserved regions are situated in the C-terminal half of the dipeptidyl peptidase III (DPP III) molecule (Abramić et al. 2004). Determination of the first three dimensional structure, from the yeast DPP III, revealed a two-domain protein with a novel fold (Baral et al. 2008), as an additional feature of the family M49.
The biochemical properties of this zinc-peptidase isolated from various (mostly mammalian) organisms, have been extensively characterized, as have been structure-activity studies of heterologously (in Escherichia coli) expressed enzymes from human, rat, bacterial and yeast origin (Fukasawa et al. 1998; Fukasawa et al. 1999; Abramić et al. 2000; Chen and Barrett 2004; Salopek-Sondi et al. 2008; Špoljarić et al. 2009; Jajčanin-Jozić et al. 2010; Karačić et al. 2012; Vukelić et al. 2012; Abramić et al. 2015). In contrast to biochemistry, molecular enzymology, and structural biology (Baral et al. 2008; Bezerra et al. 2012) knowledge of peptidases belonging to the DPP III family, their physiological roles have been much less elucidated. As DPP III is a cytosolic enzyme, broadly distributed in cells and tissues, it is assumed to participate in the final stages of intracellular protein catabolism (Abramić et al. 1988). In addition, several studies indicated a role for DPP III in the mammalian pain-modulatory system (Chiba et al. 2003; Baršun et al. 2007), and its participation in the endogenous defense against oxidative stress is well supported (Liu et al. 2007). It has been shown that DPP III is an activator in the Keap1-Nrf2 signaling pathway (Hast et al. 2013), which is the major regulator of cytoprotective responses to oxidative and electrophilic stress (Kansanen et al. 2013).
In search for plant homologues of the M49 family, we have found a predicted protein of 770 amino acids from the moss Physcomitrella patens (UniProtKB code: A9TLP4), and an Arabidopsis thaliana uncharacterized protein of 772 amino acids, designated Nudix hydrolase 3 in UniProtKB (entry code Q8L831; gene: NUDT3). A multiple sequence alignment revealed that Physcomitrella protein A9TLP4, and A. thaliana protein Q8L831 contain all conserved sequence regions of the M49 family with one exception: instead of the characteristic hexapeptide zinc-binding motif HEXXGH, they possess the pentapeptide HEXXH. In addition, near their N-termini both plant proteins contained a variant of the «Nudix box» motif (Figure 1), a characteristics of the Nudix hydrolase superfamily.
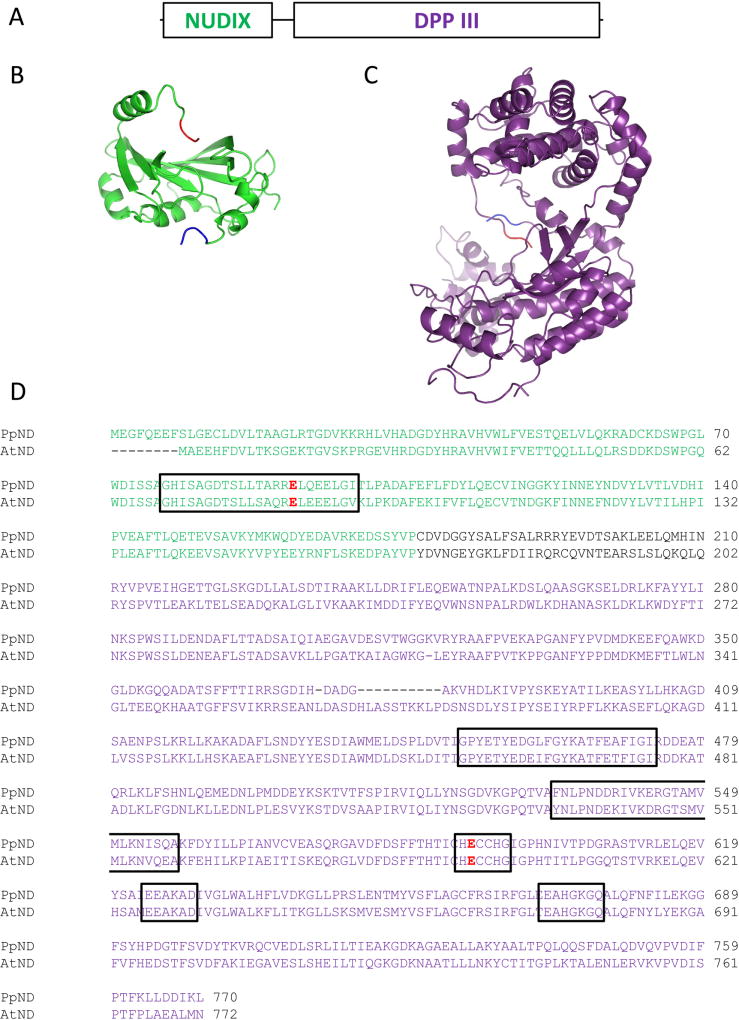
Figure 1.
A) a schematic representation of plant Nudix-DPP III proteins, where the first 200 amino acids form a Nudix fold domain, and the last 550 amino acids form a DPP III fold; B) PpND model (residues 5–177) based on human IDI1 structure (PDB: 2i6k; Zhang et al. 2007); C) PpND model (residues 210–760) based on human DPP III structure (PDB: 3fvy; Bezerra et al. 2012). First three amino acids in the models are colored blue to indicate N-terminus, last three amino acids are colored red to indicate C-terminus. Models were made using Phyre2 server (www.sbg.bio.ic.ac.uk/phyre2) and visualized using PyMOL (www.pymol.org); D) alignment of protein sequences of Physcomitrella patens and Arabidopsis thaliana Nudix-DPP III, with framed variant Nudix box and five conserved regions of the DPP III family. Catalytic glutamates – E92 for Nudix hydrolase and E592 for peptidase activity - are given in red. In all panels, Nudix domain is shown in green and DPP III domain in purple. Putative linker sequence is shown in black.
The Nudix superfamily is widespread among eukaryotes, bacteria, archaea, and viruses and consists mainly of pyrophosphohydrolases that act upon substrates of general structure nucleoside diphosphates linked to some moiety X to yield nucleoside monophosphate plus phosphate-X (Bessman et al. 1996; McLennan 2006; Xu et al. 2006). These enzymes are characterized by a conserved sequence motif GX5EX7REUXEEXGU (Nudix box), where U represents a bulky hydrophobic amino acid such as Ile, Leu, or Val and X is any amino acid. Their substrates include (deoxy)ribonucleoside diphosphates and intact and oxidatively damaged nucleoside triphosphates, nucleotide sugars, dinucleotide coenzymes, dinucleoside polyphosphates, and capped RNA (Mildvan et al. 2005). The Nudix box forms a loop-α helix-loop structural motif which functions as catalytic site (Mildvan et al. 2005). Additional sequences associated with this motif influence the substrate specificity of the enzyme (Yoshimura and Shigeoka 2015).
A large number of Nudix hydrolases from various organisms such as bacteria, yeast, algae, nematodes, plants and vertebrates have been identified and characterized (Dunn et al. 1999; McLennan 2006; Ogawa et al. 2008; Yoshimura and Shigeoka 2015). The prototype member of the Nudix superfamily is the MutT pyrophosphohydrolase from E. coli, a monomeric protein of 129 amino acids. Its three dimensional structure has been determined, and its catalytic mechanism studied in detail (Mildvan et al. 2005). It has been proposed that the biological role of MutT is to remove mutagenic nucleotides, since its best substrate in vitro is 8-oxo-dGTP (Mildvan et al. 2005).
Not only Nudix hydrolases, but also the isopentenyl diphosphate isomerases (IDIs) share the Nudix motif (Yoshumura and Shigeoka 2015). IDI is an metalloenzyme that catalyzes interconversion of isopentenyl diphosphate and dimethylallyl diphosphate (Bonanno et al. 2001). Phylogenetic analysis revealed two distinct families of IDI, IDI1 and IDI2 (Berthelot et al 2012). IDI1s, found from bacteria to human, are members of the Nudix superfamily (Berthelot et al. 2012). E. coli IDI1 is a single α/β domain protein that resembles E. coli MutT (Bonanno et al. 2001).
The specific functions of many Nudix hydrolases have not been elucidated, due to their broad substrate specificities. However, increasing evidence is emerging showing that Nudix enzymes may play protective, regulatory, and signaling roles in metabolism (Mildvan et al. 2005; Yoshimura and Shigeoka 2015).
This is the first report of proteins with both Nudix and DPP III domains. Intrigued by potential for roles in plant stress response and cytoprotection, we undertook the heterologous expression and biochemical characterization of the P. patens A9TLP4 and A. thaliana Q8L831 proteins. We hypothesized that both proteins have dipeptidyl peptidase III activity, due to the presence of evolutionary conserved regions of the M49 family in their primary structure (Figure 1). In addition, we investigated whether they could act as a Nudix enzyme as well.
Results
Search for plant homologues of DPP III family
Plant dipeptidyl peptidases III have been recognized only recently as data from plant genomes sequencing projects have become publicly available. Among the first such hits by BLAST similarity search, we found the homologue of DPP III family in the moss Physcomitrella patens subsp. patens, which is used as a model organism for studies on plant physiology, development and evolution (Prigge and Bezanilla 2010, Müller et al. 2015). Mosses share fundamental physiological processes with higher plants, although they diverged early in evolution. Sequencing of the P. patens genome enabled comparisons with other species, which revealed genomic changes accompanying the evolutionary movement to land, including acquisition of genes for tolerating terrestrial stresses (temperature variation and water availability) (Rensing et al. 2008).
The Physcomitrella patens homologue - a predicted protein A9TLP4 - showed 30.1% identity (E-value: 4e-22) with the Bacteroides thetaiotaomicron DPP III (UniProt entry Q8A6N1), and 22.3% identity (E-value: 4.7e-7) with the human orthologue of M49 family (UniProt Q9NY33).
We tentatively named the P. patens uncharacterized protein A9TLP4, because it contained the Nudix box and DPP III (M49) family consensus sequences, as «P. patens Nudix-DPP III» (PpND) and searched online databases of plant genomes for its homologues. Using Conserved Domain Architecture Retrieval Tool (Geer et al. 2002), we obtained 76 non-redundant sequences containing both Nudix and M49 domain. This type of domain architecture was found only in land plants (not shown).
Among the PpND homologues found, an Arabidopsis thaliana protein (UniProt: Q8L831) annotated as «Nudix hydrolase 3», product of NUDT3 gene, attracted our interest, since its enzyme activity as a Nudix hydrolase was not experimentally confirmed. It had 61.6% identity to PpND (BLAST E-value: 0.0). Ogawa et al. (2005), who extensively investigated cytosolic Nudix hydrolases in A. thaliana, suggested that the product of AtNUDT3 is not likely a typical Nudix hydrolase enzyme, because its predicted amino acid sequence shows an extremely low similarity to those of other AtNUDTs, and its molecular mass (86.9 kDa) is much higher than those of the enzymes in other organisms.
Heterologous production, purification and biochemical characterization of the recombinant P. patens PpND and the recombinant A. thaliana Nudix hydrolase 3 (AtND)
The cDNA cloning, expression and purification of the recombinant PpND and AtND was performed as described in Materials and Methods.
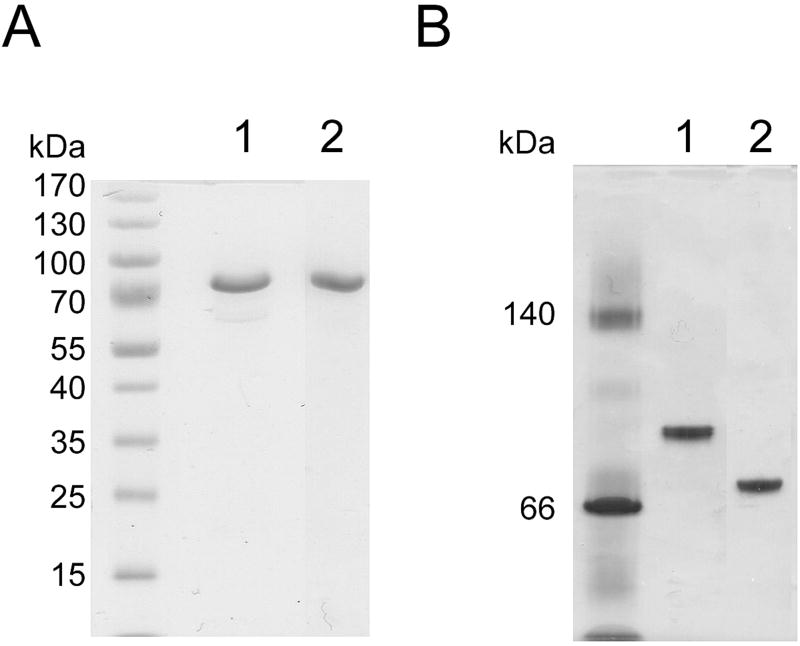
The purified His-tagged proteins were analyzed by SDS-PAGE (Figure 2A) and both exhibited a single protein band, whose molecular weight was estimated to be about 88 000, similar to the theoretical molecular mass. Native PAGE analysis of PpND and AtND also showed one protein band (Figure 2B), indicating that each protein exists in solution as a monomer. The isoelectric point as determined by isoelectric focusing was about pH 5.1 for PpND and about pH 5.5 for AtND. Circular dichroism (CD) spectra were obtained for both proteins indicating similar secondary structures (Supplementary Figure 1).
Figure 2.
Polyacrylamide gel electrophoresis (PAGE) of affinity and size-exclusion purified AtND (lane 1) and PpND (lane 2). A: SDS-PAGE performed under reducing conditions on PhastGel Homogeneous (12.5 %) plate; B: native-PAGE performed on PhastGel Gradient (8–25%) plate. Proteins were visualized by Coomassie Blue staining.
Peptidase activity of purified PpND and AtND was investigated by assaying the hydrolysis of chromogenic substrates of dipeptidyl peptidases, aminopeptidases and endopeptidase. Among them, both plant proteins cleaved only dipeptidyl-2-naphthylamides (Table 1). The best substrate of PpND was Arg-Arg-2-naphthylamide (Arg2-2NA), which is known as the preferred synthetic substrate of dipeptidyl peptidase III type of enzyme (Ellis and Nuenke 1967). In addition, five other dipeptidyl-2-naphthylamides were hydrolyzed by both plant enzymes: Ala-Arg-, Pro-Arg-, Phe-Arg-, Ala-Ala- and Gly-Arg-2NA (Table 1).
Table 1.
Hydrolytic activitya of P. patens A9TLP4 (PpND)and A. thaliana NUDT3 (AtND) towards peptidase substrates.
| Substrate | Relative hydrolysis rate (%) | |
|---|---|---|
|
|
||
| PpND | AtND | |
| Arg-Arg-2NA | 100 | 100 |
| Ala-Arg-2NA | 62 | 684 |
| Ala-Ala-2NA | 40 | 366 |
| Gly-Arg-2NA | 16 | 197 |
| Pro-Arg-2NA | 14 | 622 |
| Phe-Arg-2NA | 12 | 411 |
| Asp-Arg-2NA | 0 | 0 |
| Gly-Pro-2NA | 0 | 0 |
| Gly-Phe-2NA | 0 | 0 |
| Arg-2NA | 0 | 0 |
| Leu-2NA | 0 | 0 |
| Ala-2NA | 0 | 0 |
| BANA | 0 | 0 |
The enzymes were incubated at 37 °C in Tris-HCl buffer (ionic strength = 0.01) pH 7.5 (PpND), respectively 7.9 (AtND) containing 75 µM CoCl2, with the substrates at a final concentration of 0.04 mM (100 % = 131.4 nmol, or 17.2 nmol of Arg2-2NA hydrolyzed per min and milligram of PpND or AtND, respectively). 2NA: 2-naphthylamide; BANA: Nα-benzoyl-L-Arg-2-naphthylamide
The biochemical properties of plant enzymes were further characterized using their hydrolytic action on Arg2-2NA. The optimal pH for the hydrolysis of this substrate was determined at pH 7.4, for PpND, and at pH 7.9 for AtND. Only Co2+ ions (among examined Co2+, Mg2+, Mn2+and Zn2+ ions) activated the PpND and AtND significantly. Both enzymes showed patterns of inhibition typical for DPPs III: strong inhibition with chelating agents (EDTA, o-phenanthroline) and the sulfhydryl reagent p-hydroxymercuri-benzoate.
Kinetic analyses of PpND and AtND with peptidase substrates
Screening for peptidase activity indicated the difference in specificity between the moss and Arabidopsis enzyme (Table 1). To further investigate the catalytic properties of PpND and AtND hydrolysis of dipeptidyl-2-naphthylamides, which were selected as a good substrates according to the initial results (Table 1), kinetic analyses were performed as described in Materials and methods. The two best substrates of the PpND were Arg2-2NA and Ala-Arg-2NA. The kinetic parameters for Arg2-2NA hydrolysis catalyzed by P. patens enzyme were determined to be: Km = 10.63 ± 1.72 µM, kcat = 0.822 ± 0.180 min−1,, kcat/ Km = 1289 M−1 s−1, and for hydrolysis of Ala-Arg-2NA: Km = 17.21 ± 0.75 µM, kcat = 0.633 ± 0.157 min−1, kcat/ Km = 613 M−1 s−1, revealing a slight preference for diarginyl derivative (two-fold higher kcat/ Km). To investigate the AtND specificity, kinetic parameters were determined for four dipeptidyl-2-naphthylamides, selected as a good substrates. As shown in Table 2, AtND preferred Ala-Arg-2NA. The catalytic efficiency (the kcat/ Km value) for this substrate was 19-fold and 9-fold higher compared to that for Arg2-2NA and Pro-Arg-2NA.
Table 2.
Kinetic parametersa for the hydrolysis of dipeptidyl-2-naphthylamide substrates catalyzed by the Arabidopsis thaliana Nudix-DPP III (AtND).
| Substrate | Km (µM) | kcat (min−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Ala-Arg-2NA | 5.0 ± 0.3 | 2.49 ± 0.21 | 8300.0 |
| Pro-Arg-2NA | 40.85 ± 12.09 | 2.23 ± 0.3 | 908.2 |
| Arg-Arg-2NA | 3.25 ± 1.12 | 0.120 ± 0.001 | 615.4 |
| Ala-Ala-2NA | 240.2 ± 69.4 | 2.28 ± 0.41 | 158.2 |
The kinetic parameters were determined from the initial reaction rates at 25 °C, as described in Materials and methodss. Km and kcat values are presented as mean ± SD
Investigation of the predicted isopentenyl diphosphate isomerase (IDI) activity
In addition to the Nudix hydrolases, Nudix superfamily encompasses other non-hydrolytic enzymes. including the isopentenyl diphosphate (IPP) isomerases (Yoshimura and Shigeoka 2015). Interestingly, Gunawardana et al. (2009) predicted that AtNUDT3 (AtND) is a member of isopentenyl diphosphate isomerase (IDI) family, based on a comprehensive bioinformatic analysis of 25 Nudix hydrolases encoded in the Arabidopsis genome. When we aligned the primary structure of PpND and AtND to proteins with confirmed IDI activity (human IDI1, E. coli IDI, Halobacterium IDI) (Supplementary Figure 2), we noticed that most of the residues necessary for IDI function, inferred from structural studies on human and E. coli IDI (Wouters et al. 2003; Zhang et al. 2007), are conserved in plant proteins. However, in PpND and AtND, the signature active-site motif of IDIs, NTCCSH, is replaced by ISSAGH, in which the catalytic (second) cysteine is substituted by an alanine residue (Ala76 in PpND sequence). Interestingly, IDI activity has been reported for an enzyme from the archaea Halobacterium sr. NRC1 (UniProt: Q9HDP40), although the catalytic Cys is replaced by Ala (Supplementary Figure 2) (Hoshino and Egushi 2007). Therefore, both PpND and AtND were tested for IDI activity by a color conplementation assay (Wang et al. 2009), as described in Materials and methods. Colonies expressing PpND and AtND were yellow, indicating the absence of isomerase activity (Supplementary Figure 3).
Nudix hydrolase activity
The presence of a Nudix box motif at the N-termini of these two proteins motivated further investigation of their potential Nudix hydrolase activity. For that purpose, we used a continuous fluorescence assay (Xu et al 2013), that employs fluorescently tagged E. coli phosphate binding protein (PBP) as a phosphate sensor. We screened PpND and AtND with substrates of Nudix hydrolases, with the common structure nucleoside–diphosphate-X. In addition, we examined isopentenyl diphosphate (IPP) as a substrate for hydrolysis by PpND or AtND. Although these plant enzymes did not have IDI activity, based on their structural similarity (Supplementary Figure 2) we hypothesized that they could bind IPP, in order to hydrolyze it.
Substrate Screening
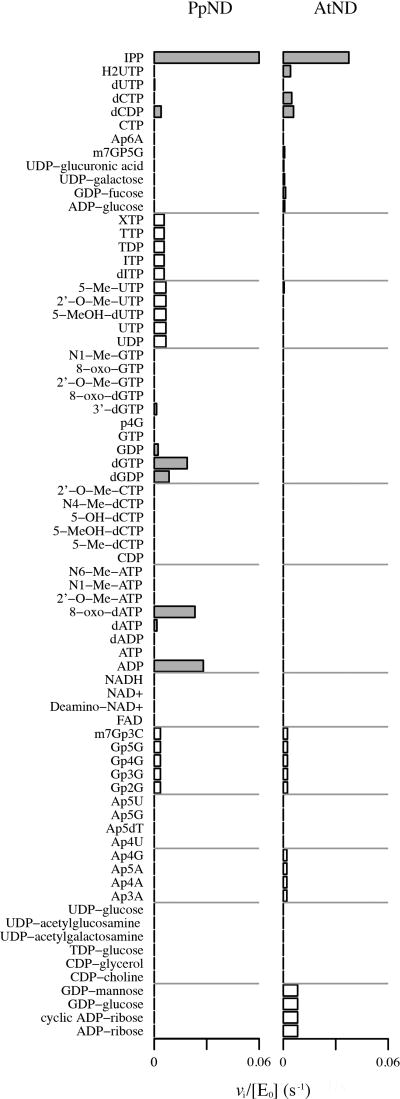
Figure 3 illustrates the screening results of both enzymes against the library of 73 substrates (full list of substrates is given in Supplementary Table 1), along with their vi/[E0] values (s−1). White bars indicate the cases in which the activity against a substrate was measured as part of group mixture. Grey bars indicate the screened activity for a substrate that was assayed individually against the enzyme. More details are given in Supplementary Table 2.
Figure 3.
Substrate specificity screening of PpND and AtND against a 73-compound library by phosphate sensor assay. The vi/[E0] values (s−1) are graphed for every substrate in the library that was found to have detectable activity against the enzymes. These reactions were measured at pH 7.6 and 37 °C, with 5 µM substrate and 50 nM enzyme. Estimated activities for substrates that were screened individually are recorded in gray, while the activities of substrates within a group are recorded in white.
The screening results against individual substrates initially found that PpND exhibited the highest activity against IPP (vi/[E0] value: 0.06 s−1), 8-oxo-dATP (0.02 s−1), ADP (0.03 s−1), and dGTP (0.03 s−1). Notable activity for AtND was only found against IPP (0.04 s−1).
Michaelis-Menten kinetics
For all substrates that were found to have notable activity in the screening, kinetic parameters were measured carefully. Supplementary Figure 4 shows the results of PpND and AtND against these substrates. A summary of the kcat/Km ratios with their standard errors is given in Table 3.
Table 3.
Kinetic parameters for Nudix hydrolase catalyzed reactions.
| Enzyme | Substrate | kcat /Km (M−1 s−1) |
|---|---|---|
| PpND | IPP | (1.9 ± 0.16) × 104 |
| 8-oxo-dATP | (1.1 ± 0.64) × 104 | |
| ADP | 280 ± 26 | |
| dGDP | 170 ± 18 | |
| dGTP | 185 ± 15 | |
| AtND | IPP | (1.8 ± 0.13) × 104 |
The most reactive substrate of PpND is IPP, with a kcat/Km = (1.9 ± 0.16) × 104 M−1 s−1, followed by 8-oxo-dATP with a kcat/Km = (1.1 ± 0.64) × 104 M−1 s−1. Despite showing high activity readings in the screening process, ADP was found to have activity closer to that of dGTP and the less reactive dGDP, on the order of magnitude of 102 M−1 s−1. For IPP, the comparison of vi/[E0] values with and without inorganic pyrophosphatase (PPase) found no appreciable change in activity. AtND was recorded to have a kcat/Km of (1.8 ± 0.13) × 104 M−1 s−1 against IPP. The addition of PPase also did not have an affect in the recorded activity measured.
Additionally, because IPP was a new substrate for this screen panel, IPP was also screened against MutT enzyme, to determine whether it could lead to any artifactual enzyme activity. No such activity was seen (not shown).
Identification of isopentenyl monophosphate (IP) as a product of Nudix hydrolase activity of PpND towards IPP
The activity of PpND toward IPP was additionally examined by LC-MS/MS as described in Materials and methods. Reactions with two concentrations of PpND (60 nM and 600 nM) were performed at 37 °C for 10 min and their products were analyzed. Negative ESI tandem mass detection was run in multiple reaction monitoring (MRM) mode. The values of m/z of IPP and IP standard parent ions were 244.70 and 164.90, respectively. Collision-induced dissociation of parental ions yielded characteristic fragments of IPP (m/z=226.7, 158.5, 79.0) and IP (m/z= 148.6, 96.9, 79.0). As shown in Supplementary Figure 5, IP was identified as the product of PpND enzyme reaction toward IPP as a substrate. In addition, the progress of enzyme reaction (Supplementary Figure 5, B and C) was shown in comparison to the control (Supplementary Figure 5A). Based on calculation, the ratio of characteristic ion m/z=79.00, generated from IPP and those from IP was 4.5:1 (IPP:IP) in enzyme reaction with 60 nM PpND (Supplementary Figure 5B) and 1:4 (IPP:IP) in reaction with 10 times higher concentration of PpND (Supplementary Figure 5C).
Structure – activity relationships
We observed a significantly lower kcat value for hydrolysis of the diarginyl-2NA substrate catalyzed by the PpND and AtND, in comparison with human and bacterial orthologues of the DPP III family (Jajčanin-Jozić and Abramić 2013; Vukelić et al. 2012). This was not surprising, since the plant enzymes contain a pentapeptide HECCH, instead of the characteristic active-site hexapeptide HEXXGH. The importance of this hexapeptide motif for full activity of DPP III had been shown previously for the rat orthologue, by mutational analysis (Fukasawa et al. 1999). Substitution of E592 in Physcomitrella DPP III, corresponding to the catalytic glutamate in the canonical DPP III motif, HEXXXH, resulted in a complete loss of activity towards Arg2-2NA, while the IPP hydrolase activity remained (Figure 4, PpND_E592A). Two insertion variants were produced to investigate if introduction of the consensus active-site motif into the P. patens protein would enhance peptidase activity. A variant with the hexapeptide HECCGH, created by inserting a glycine into the pentapeptide H591ECCH595 motif of wild-type PpND, showed a 29-fold decreased hydrolytic activity towards Arg2-2NA compared to the wild-type enzyme. A second mutant with the hexapeptide HECLGH, designed to mimic the active-site motif of the Bacteroides thetaiotaomicron DPP III (Vukelić et al. 2012), again resulted in a much less active (57-fold) enzyme. CD measurements showed that the reduced activity is not due to changes in the secondary structure (not shown). Separate expression of the isolated DPP III domain resulted in substantial loss in activity, while expression of the Nudix domain failed to yield soluble Nudix domain protein.
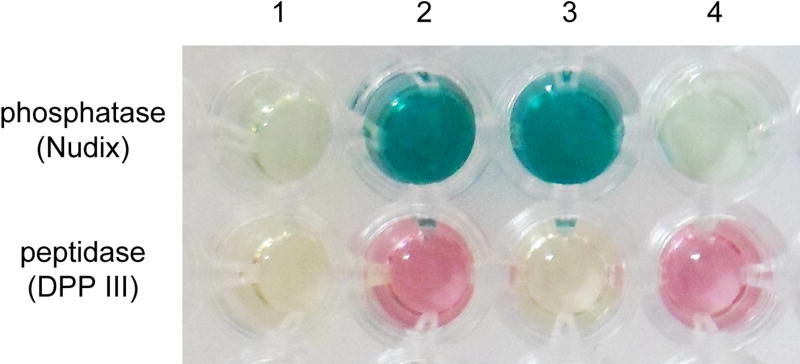
Figure 4.
Activity of the wild-type and PpND mutants towards IPP (phosphatase) and Arg2-2NA (peptidase) determined using colorimetric assays described in Materials and methods. 1 - negative control (reaction mixture without the enzyme); 2 – wild-type PpND; 3 – PpND E592A, disrupting DPP III motif; 4 – PpND E92A, disrupting variant Nudix box
As the first glutamate of the Nudix box motif is essential for catalysis in different Nudix hydrolases (Gunawardana et al. 2009), E92 of the PpND RE92LQEE motif was replaced by alanine. The variant PpND_E92A showed no IPP hydrolase activity, but retained DPP III activity towards Arg2-2NA (Figure 4). As A76 from the PpND motif ISSA76GH aligns with the catalytic cysteine of the active-site motif of IDIs (Supplementary Figure 2), variant PpND_A76C was generated to investigate whether a potential isomerase activity could be found. As with wild-type PpND, this was not detected in the color complementation assay (not shown). Finally, the double mutant PpND_A76C_E92A was prepared, based on the possibility that IPP isomerase activity might only be detected when IPP hydrolase activity is not present due to the design of the in vivo assay. However, this double mutant (PpND_A76C_E92A) also did not have IPP isomerase activity (not shown).
Discussion
Proteolytic enzymes (peptidases) are involved in many aspects of plant physiology, growth and development (Schaller 2004). Considering their numerous and diverse biological functions, it is not surprising that a large number of peptidases has been recognized in plant genomes. According to the MEROPS database (http://merops.sanger.ac.uk/) (Rawlings et al. 2014), the count of known and putative peptidases in plants P. patens and A. thaliana is 343 and 745, respectively. However, only a limited number of proteolytic enzymes from these two plants have been characterized by biochemical approaches, none of them being reported as a dipeptidyl peptidase.
In this study, recombinant A9TLP4 protein originating from the moss Physcomitrella patens and recombinant NUDT3 protein from the flowering plant Arabidopsis thaliana were obtained by heterologous expression in E. coli, and their enzymatic properties were examined. Using bioinformatic approaches we have recognized these two proteins as atypical members of the metallopeptidase family M49 (DPP III family), which instead of the hexapeptide HEXXGH active-site zinc-binding motif contain the pentapeptide HECCH. In addition they harbour a variant Nudix box motif, a characteristics of Nudix hydrolases, near their N-termini. Therefore, we called them «Nudix-DPP III» (ND), PpND and AtND.
Experimental characterization of purified proteins revealed biochemical properties (hydrolysis of DPP III specific substrate, Arg2-2NA and activation by Co2+ ions) which classify them as members of the DPP III family. The catalytic efficiency (kcat/Km) of both plant enzymes for Arg2-2NA was similar to the yeast DPP III, that has 12-fold higher kcat/Km for Arg2-2NA hydrolysis (Jajčanin-Jozić et al. 2010), but much lower (3 orders of magnitude), compared to the values determined for human and bacterial DPPs III (Vukelić et al. 2012; Jajčanin-Jozić and Abramić 2013). At present, the structural basis for this low efficiency of the plant enzymes is not known, and our data on insertion mutants do not support the explanation that this is due to the shorter active-site motif (HECCH).
The biochemical characterization of DPP III activity revealed significant differences between the two plant enzymes: PpND showed lower pH optimum than AtND (at pH 7.4 vs. pH 7.9). In addition, they differed in their substrate specificity. Among the substrates of X-Arg-2NA series, the Physcomitrella enzyme moderately preferred Arg2-2NA. In contrast, the Arabidopsis enzyme showed low activity for Arg2-2NA, and pronounced preference for Ala-Arg-2NA (Table 1 and Table 2).
Screening of PpND and AtND against a library of potential Nudix hydrolase substrates by phosphate sensor assay (Figure 3, Supplementary Table 2) revealed that these plant proteins possess Nudix hydrolase activity as well. Thus, experimental finding of dual activity of PpND and AtND confirmed our starting hypothesis, based on their primary structures. These two plant enzymes show both Nudix hydrolase and dipeptidyl peptidase III activity, catalysing the hydrolysis of phosphate bonds, and of peptide bonds. To our knowledge, this is the first report on Nudix and DPP III activity contained in a monomeric enzyme. Nevertheless, we were unable to express the individual domains of PpND separately, leading us to conclude that both domains are needed for proper folding and activity of this protein.
The existence of two distinct active sites (Nudix and DPP III) is supported by site-directed mutagenesis. Glu592 from the active-site motif H591ECCH595 (Figure 1) is essential for DPP III activity of PpND. The importance of the glutamic acid residue from the characteristic zinc-binding motif HELLGH has been already established for the catalytic activity of mammalian and yeast DPP III (Fukasawa et al. 1999; Baral et al. 2008; Bezerra et al. 2012). Mutational analysis further revealed that Glu92 from the Nudix box motif is crucial for the Nudix hydrolase (IPP hydrolase) activity of PpND. This residue aligns with Glu53 in E. coli MutT (multiple sequence alignment, MSA, not shown). The major role of Glu53 as a general base in MutT catalysis has been confirmed (Mildvan et al. 2005).
Results of site-directed mutagenesis on essential glutamic acid residues (E92 and E592) in PpND indicate two distinct active sites (Nudix and DPP III).
Our results provide clear evidence that both Nudix-DPP III (PpND and AtND) have phosphohydrolase activity against IPP though its physiological relevance is unproven. Since the addition of inorganic pyrophosphatase in the assay did not have an effect on this hydrolytic activity, we inferred that both plant enzymes catalyze the hydrolysis of the terminal phosphate bond. This was confirmed by LC-MS/MS analysis of the reaction mixture in which the formation of isopentenyl monophosphate was identified. Isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) are elemental (C5 building blocks) for the biosynthesis of isoprenoids, naturally occurring compounds, produced by all living organisms. They are considered to be the largest and most diverse group of natural products (George et al. 2015), composed of over 50,000 compounds which include primary metabolites (sterols, carotenoids) and secondary metabolites. Isoprenoid compounds in vivo perform a large variety of functions (Berthelot et al. 2012). In plants they are broadly represented and play essential roles acting as pigments, hormones, pollinator attractors, insecticides, pesticides. IPP and DMAPP are synthesized by two independent pathways, the mevalonic acid (MVA) and methylerythritol phosphate (MEP) pathways. Most recently, it was reported that, in a modified MVA pathway in the cytosol of plant cells, isopentenyl phosphate kinase (IPK) catalyses the formation of IPP from IP (Henry et al. 2015). An IPP phosphatase was postulated. The authors suggest that IP/DMAP formation is likely an adaptive trait, and not just the consequence of general phophatase activity. Both AtND and PpND are cytosolic proteins. Their ability to hydrolyze IPP distinguishes them from the majority of known Nudix hydrolases, though few others have been tested for such activity. Moreover, no IPP hydrolase has been characterized which has a level of activity that could be physiologically relevant. Our results support that PpND and AtND could participate in the plant isoprenoid metabolic network. These enzymes may have uses in biotechnology where IPP phosphohydrolase activity is useful.
Most recently, Magnard et al. (2015) have shown that the cytosolic Nudix hydrolase RhNUDX1 from rose possesses geranyl diphosphate diphosphohydrolase activity in vitro and contributes to the biosynthesis of monoterpene alcohol geraniol in plants. Our findings on PpND and AtND hydrolytic activity towards IPP thus corroborates the idea that isoprenoid diphosphates may be natural substrates of Nudix hydrolases.
In summary, for the first time, two atypical members of the DPP III (M49) family, containing a Nudix box motif, were identified, heterologously expressed and biochemically characterized: Physcomitrella patens protein (PpND) and Arabidopsis thaliana protein (AtNUDT3, AtND). Our results revealed that both plant proteins possess dual enzymatic activity, acting as a dipeptidyl peptidase III and Nudix hydrolase, in agreement with our starting hypothesis based on the bioinformatic analysis. Two glutamic acid residues, E592 and E92, catalytically important for DPP III and Nudix hydrolase activity, respectively, were confirmed by site-directed mutagenesis. AtND and PpND cleaved DPP III substrates with different specificity. Both enzymes hydrolyzed isopentenyl diphosphate (IPP), a universal precursor for the biosynthesis of isoprenoid compounds, with similar catalytic efficiency. The activity was significant but insufficient to be certain of its physiological role. Future studies are needed to prove whether PpND and AtND participate in plant isoprenoid metabolism, and whether PpND is involved in protection against oxidative stress in the moss Physcomitrella patens, both suggested by the biochemical rationale of our present results.
Materials and methods
Cloning and mutagenesis
The full-length cDNA clone for the P. patens A9TLP4 (PpND) was generated by GeneArt AG (Regensburg, Germany). The codon usage was adapted to the codon bias of Escherichia coli genes. The cDNA was cloned into the pET21a_A028 bacterial expression vector between NdeI and XhoI restriction sites adding His-tag codons at the 3' end. The full-length cDNA clone for the Arabidopsis thaliana protein Q8L831, which we designated as «AtND», was prepared from A. thaliana cDNA clone U16680 obtained from the Arabidopsis Biological Resource Center (Yamada et al. 2003) in the pLATE31 plasmid (aLICator kit, Thermo Fisher Scientific, Waltham, MA, USA).
Mutants of PpND were constructed using the QuikChange® XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The template used for mutagenesis was the pET21a-A9TLP4-His6 construct (GeneArt AG). Domain fragments of PpND were cloned into pLATE31 with a C-terminal His-tag. The Nudix domain fragment contained residues 1 to 206, and the DPP III domain fragment contained residues 207–770. The primers, listed in Supplementary Table 3 were custom synthesized by Sigma-Aldrich (St. Louis, MO, USA) and Macrogen (Seoul, South Korea).
Production, purification and characterization of recombinant PpND and AtND proteins
Cells of Escherichia coli BL21- Codon Plus (DE3) - RIL were transformed with the appropriate plasmid (constructs containing PpND wild-type, mutant or domain fragment of PpND, or AtND), grown in Luria Bertani broth with ampicillin to OD600 0.6, and were then cooled to 18 °C. The culture was supplemented with magnesium and zinc sulfate to final concentrations of 1mM MgSO4 and 10µM ZnSO4. Expression was induced with 0.25 mM IPTG (isopropyl β-D-1-thiogalactopyranoside), and cells were harvested after 20 h at 18 °C and 130 rpm. Cell harvesting and protein purification from bacterial pellet was performed according to Jajčanin-Jozić et al. (2010). In brief, cell pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 10 mM imidazole) and lysed enzymatically by lysozyme as well as mechanically with sonication. Proteins were purified using Ni-NTA affinity chromatography and gel filtration on the column of HiLoad 16/60 Superdex 200 prep grade. Protein purity was confirmed by SDS-PAGE according to Laemmli (1970) on a BioRad Mini-Protean Tetra Cell, and by SDS-PAGE with PhastGel Homogeneous 12.5 plates on PhastSystem (Pharmacia, Uppsala, Sweden). Molecular mass of purified proteins was determined by SDS-PAGE using PhastGel 8–25% gradient plates and low molecular weight standard mixture from Pharmacia for reference proteins. Isoelectric focusing for pI determination, and native-PAGE with PhastGel Gradient plates were performed on PhastSystem following manufacturer’s procedures. Protein bands were stained with Coomassie Brilliant Blue R-250.
In order to check the secondary structure similarity between the PpND and AtND proteins, circular dichroism (CD) spectra were recorded on a Jasco J-815 spectropolarimetar (JASCO, Easton, MD, USA) with automatic temperature control using a cylindrical quartz cuvette of 0.1 mm or 1 mm path length. Enzyme samples prepared for CD measurements were exchanged into 10 mM Tris-HCl buffer, pH 7.4. The protein concentrations used were from 0.25 to 0.40 mg ml−1.
Enzyme activity determination and kinetic analysis
Dipeptidyl peptidase activity
DPP III activity was determined by a standard assay with Arg-Arg-2-naphthylamide (Arg2-2NA) as substrate, at 37 °C and at pH 7.4 (for PpND), or at pH 7.9 (for AtND) in Tris-HCl buffer (I=0.01) containing 75 µM CoCl2, using the colorimetric method (Špoljarić et al. 2009).
The influence of metal cations on PpND hydrolytic activity towards Arg2-2NA was examined by adding metal salts (CoCl2, MnCl2, Mg-acetate, or Zn-acetate) at concentrations between 2 µM to 100 µM in the preincubation mixture with the enzyme, and after 3 min at 37 °C the reaction was started with substrate. The effect of peptidase inhibitors was investigated by preincubation of the enzyme in 50 mM buffer with selected effectors for 30 min at 25 °C, followed by the determination of the activity with the standard assay (15 min at 37 °C, in the presence of CoCl2).
Kinetic parameters (Km and kcat) for hydrolysis of Arg2-2NA and of other dipeptidyl-2-naphthylamides were determined with initial rate measurements (Jajčanin-Jozić et al. 2010) at 25 °C and at pH 7.4 (PpND), or at pH 7,9 (AtND) in Tris-HCl buffer (ionic strength 0.01) in the presence of the activator, 75 µM CoCl2.
Protein concentrations were determined by the Bradford method (1976).
Nudix hydrolase activity
Nudix hydrolase activity was determined by a continuous fluorescence assay (Xu et al. 2013), using fluorescently tagged E. coli phosphate binding protein (PBP) as a phosphate sensor. The phosphate sensor was produced and purified using the methods described by Xu et al. (2013). The assay contained 10 mM Tris HCl pH 7.6, 1 mM MgCl2, and 5–10 µM phosphate sensor. In the case where pyrophosphate was a possible product of the reaction, 0.05 U/mL inorganic pyrophosphatase (PPase) was added and where a nucleoside monophosphate was a possible product, 1 U/mL of alkaline phosphatase (APase) was added.
The enzymes were screened using a GENios microplate reader (λex 425 nm, λem 465 nm, gain 50, 100 cycles). The reactions were recorded over 25 minutes at 37 °C to determine substrates with significant kinetic activity. The substrate concentrations were kept at 5 µM and screened against enzyme concentration of 50 nM. Kinetic parameters were found via FluoroMax-4 spectrofluorometer (λex 430 nm, slit width 1–2.5 nm, λem 465 nm, slit width 1–5 nm). The concentration of Nudix enzyme ranged from 20 – 344 nM. These reactions were measured over 5 minutes at 37 °C over varying concentrations of substrates. The slope of the standard curve of the sensor was measured to be 1.8 × 106 RFU/µM phosphate by Fluoromax and 2.5 × 104 RFU/µM phosphate by GENios.
The enzyme was initially screened against a library of 73 possible substrates. See Supplementary Table 1 for the full list of substrates and their groupings. Initial screening used 11 individual substrates and 11 substrates in pooled groups according to the structural similarity. Further screening of individual substrates within groups was conducted as needed. Specifically, PpND was additionally screened against ADP, ATP, dADP, dATP, 8-oxo-dATP, 2’-O-Me-ATP, N1-Me-ATP, N6-Me-ATP, dGDP, dGTP, GDP, GTP, p4G, 3’-dGDP, 8-oxo-dGTP, 2’-O-Me-GTP, 8-oxo-GTP, and N1-Me-GTP. The substrates with notable activity were then measured by careful kinetic analysis. The plots of vi/[E0] over different substrate concentrations were measured up to 20 µM, in which saturation of the enzyme was not achieved for any of the substrates for either PpND or AtND. Therefore, this substrate concentration range was deemed low enough (s<Km), to justify simplifying the Michaelis Menten equation to the linear form at a fixed intercept at 0:
The ratio kcat/Km was obtained via linear regression for all reactions.
In order to identify isopentenyl monophosphate (IP) as a product of phosphatase activity of PpND towards IPP, liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS-MRM) was performed. Standard chemicals IPP and IP were purchased from Sigma Aldrich. IPP (50 µM) was incubated at 37 °C for 10 min with PpND, 60 nM or 600 nM, in reaction mixture (total volume 100 µl) containing 10 mM Tris-HCl buffer pH 7.6 and 1 mM MgCl2. Enzyme reaction was stopped by addition of 5mM EDTA. As a control, IPP was incubated under the identical conditions, without the enzyme. All reaction mixtures were subjected to custom service analysis at BIOCenter, Zagreb, Croatia. In brief, reaction products were analyzed by liquid chromatography coupled to tandem MS/MS (Agilent Technologies 1290 Infinity LC System and Agilent Technologies 6460 Triple Quadropole mass spectrometer, Agilent, San Jose, CA, USA) on an Elipse Plus C18 RRHD column (50 mm length, 2.1 mm internal diameter, and 1.8 µm particle size). Products of enzyme reactions were eluted isocratically with a mix of phase A (20 mM NH4HCO3 containing 0.1% triethylamine) and phase B (80% acetonitrile, v/v, containing 0.1% triethylamine) in ratio 70:30, with a flow-rate of 0.2 ml min−1. The effluent (1 µl) was introduced into the ion source of a 6460 Triple Quadropole mass spectrometer. The capillary voltage was set to – 3500 V, desolvation gas flow was 5 l min−1, and desolvation temperature was 300 °C. Negative ESI tandem mass detection was run in the multiple reaction monitoring (MRM) mode.
The Malachite Green Phosphate Detection Kit (R&D Systems, Minneapolis, MN, USA) was used to determine the liberated inorganic phosphate when Nudix hydrolase (phosphatase) activity of the PpND mutants was investigated towards IPP (Figure 4). Enzyme (60–70 nM) was incubated with IPP (50 µM) for 10 min at 37 °C in 500-µl reaction mixture containing 10 mM TrisHCl (pH 7.6) and 1 mM MgCl2,, and 125 µl of 5 mM EDTA was added to stop the reaction.
Isopentenyl diphosphate isomerase (IDI) assay
In order to test for potential IDI activity, a color complementation assay (Wang et al. 2009) was performed. In this assay, E. coli XL1Blue cells are transformed with pAC-BETA plasmid carrying four genes necessary for production of β-carotene in E. coli. With addition of a plasmid carrying a gene for an IDI, the production of β-carotene in E.coli is significantly increased, resulting in visible change of colony color from yellow (without IDI) to orange. We obtained pAC-BETA and pTrcAtipi plasmids (containing IDI gene from A. thaliana) as a generous gift from Dr. F. X. Cunningham. The pTrc plasmid was produced from pTrcAtipi by cutting out the AtIDI gene with PstI enzyme and recircularization of the empty plasmid. Empty pTrc plasmid represented a negative control. The AtND and PpND sequences were cloned into the pTrc plasmid using specific primers (Supplementary Table 3), and restriction enzymes BamHI and XbaI, so that the ORF began with MSRSAAGGS and continued with full length PpND and AtND. The variant PpND_A76C was prepared by site-directed mutagenesis using pTrcPpND as the template. The double mutant PpND_A76C_E92A was generated using plasmid pTrcPpND_E92A as template for Ala76Cys mutagenesis. To test Physcomitrella and Arabidopsis NDs for IDI activity, E. coli cells carrying pAC-BETA plasmid (CmR) were transformed with one of the prepared pTrc plasmids (empty pTrc as a negative control, pTrcAtND, pTrcPpND, pTrcPpND_A76C and pTrcPpND_A76C_E92A to be tested, and pTrcAtIDI as a positive control, all AmpR). Cells were grown on LB plates supplemented with chloramphenicol and ampicillin overnight at 37 °C and 5 days at room temperature for the development of color.
Bioinformatics methods
Sequences of PpND homologues from other species were obtained from UniProt KB (July 2015, www.uniprot.org) (The UniProt Consortium 2011) following BLASTP search (Altschul et al. 1997). Multiple sequence alignment was done using Clustal O 1.2.1 (Sievers at al. 2011) at http://www.ebi.ac.uk/Tools/msa/clustalo/. Online databases of plant genomes were searched for homologues of the Physcomitrella A9TLP4 protein (PpND) using Conserved Domain Architecture Retrieval Tool (July 2015, Conserved Domain Database v. 3.14) (Geer et al. 2002).
Supplementary Material
Supplementary Figure 1. Circular dichroism spectra of wild-type PpND and of AtND proteins.
Supplementary Figure 2. Multiple sequence alignment of human, Escherichia coli and Halobacterium salinarium IDIs with PpND (A9TLP4) and AtND (Q8L831) proteins (made using Clustal O 1.2.1.). Residues important for IDI activity (known from human and E. coli IDI structures) are highlighted yellow, the catalytic cysteine is marked with a black circle, and the IDI active-site motif (NTCCSH) with a line above the sequence.
Supplementary Figure 3. IDI color complementation assay. pTrc is a negative control, pTrcAtIDI is a positive control containing IDI gene from Arabidopsis thaliana, pTrcPpND contains Nudix-DPP III (PpND) from Physcomitrella patens, and pTrcAtND containes Nudix-DPP III (AtND) fom A. thaliana.
Supplementary Figure 4. Kinetic characterization of PpND and AtND against their most reactive substrates. The reactions against the substrates with the highest recorded activity identified in the screening were measured using kinetic analysis. These reactions were measured at pH 7.6 and 37 °C, with enzyme concentrations from 20 to 344 nM. Error bars are shown where justified. In all cases, linear regression was used to determine the ratio of kcat/Km. The reported values of the kinetic parameters for enzyme and substrate are recorded in Table 3.
Supplementary Figure 5. Extracted LC-MS chromatograms of ion product m/z=79.00 of IPP (244.70) and IP (164.90): A) control, IPP incubated at 37 °C for 10 min, no enzyme added; B) IPP incubated at 37 °C for 10 min with PpND (60 nM); C) IPP incubated at 37 °C for 10 min with PpND (600 nM).
Acknowledgments
Support for this study by the Alexander von Humboldt foundation (project name: „Study of plant enzymes from metallopeptidase families M20 and M49"), by the Croatian Science Foundation (project number 7235 “Flexibility, activity and structure correlations in the dipeptidyl peptidase III family”) and NIH R01 GM071749 (to S.E.B.) is gratefully acknowledged. We thank Francis X. Cunningham, Jr., for his generous gift of pAC-BETA and pTrcAtipi plasmids, and Annsea Park for valued guidance.
References
- Abramić M, Zubanović M, Vitale Lj. Dipeptidyl peptidase III from human erythrocytes. Biol. Chem. Hoppe-Seyler. 1988;369:29–38. doi: 10.1515/bchm3.1988.369.1.29. [DOI] [PubMed] [Google Scholar]
- Abramić M, Schleuder D, Dolovčak Lj, Schröder W, Strupat K, Šagi D, Peter-Katalinić J, Vitale Lj. Human and Rat Dipeptidyl Peptidase III: Biochemical and Mass Spectrometric Arguments for Similarities and Differences. Biol. Chem. 2000;381:1233–1243. doi: 10.1515/BC.2000.151. [DOI] [PubMed] [Google Scholar]
- Abramić M, Špoljarić J, Šimaga Š. Prokaryotic homologs help to define consensus sequences in peptidase family M49. Period. biol. 2004;106:161–168. [Google Scholar]
- Abramić M, Karačić Z, Šemanjski M, Vukelić B, Jajčanin-Jozić N. Aspartate 496 from the subsite S2 drives specificity of human dipeptidyl peptidase III. Biol. Chem. 2015;396:359–366. doi: 10.1515/hsz-2014-0247. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral PK, Jajčanin-Jozić N, Deller S, Macheroux P, Abramić M, Gruber K. The first structure of dipeptidyl-peptidase III provides insight into the catalytic mechanism and mode of substrate binding. J. Biol. Chem. 2008;283:22316–22324. doi: 10.1074/jbc.M803522200. [DOI] [PubMed] [Google Scholar]
- Baršun M, Jajčanin N, Vukelić B, Špoljarić J, Abramić M. Human dipeptidyl peptidase III acts as a post-proline-cleaving enzyme on endomorphins. Biol. Chem. 2007;388:343–348. doi: 10.1515/BC.2007.039. [DOI] [PubMed] [Google Scholar]
- Berthelot K, Estevez Y, Deffieux A, Peruch F. Isopentenyl diphosphate isomerase: A checkpoint to isoprenoid biosynthesis. Biochimie. 2012;94:1621–1634. doi: 10.1016/j.biochi.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O'Handley SF. The MutT proteins or "Nudix" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- Bezerra GA, Dobrovetsky E, Viertlmayr R, Dong A, Binter A, Abramić M, Macheroux P, Dhe-Paganon S, Gruber K. Entropy-driven binding of opioid peptides induces a large domain motion in human dipeptidyl peptidase III. Proc. Natl. Acad. Sci. USA. 2012;109:6525–6530. doi: 10.1073/pnas.1118005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno JB, Edo C, Eswar N, Pieper U, Romanowski MJ, Ilyin V, Gerchman SE, Kycia H, Studier FW, Sali A, Burley SK. Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis. Proc. Natl. Acad. Sci. USA. 2001;98:12986–12901. doi: 10.1073/pnas.181466998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chen J-M, Barrett AJ. Dipeptidyl-peptidase III. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. Vol. 1. Amsterdam, The Netherlands: Elsevier Academic Press; 2004. pp. 809–812. [Google Scholar]
- Chiba T, Li Y-H, Yamane T, Ogikubo O, Fukuoka M, Arai R, Takahashi S, Ohtsuka T, Ohkubo I, Matsui N. Inhibition of recombinant dipeptidyl peptidase III by synthetic hemorphin-like peptides. Peptides. 2003;24:773–778. doi: 10.1016/s0196-9781(03)00119-0. [DOI] [PubMed] [Google Scholar]
- Dunn CA, O'Handley SF, Frick DN, Bessman MJ. Studies on the ADP-ribose pyrophosphatase subfamily of the nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J. Biol. Chem. 1999;274:32318–32324. doi: 10.1074/jbc.274.45.32318. [DOI] [PubMed] [Google Scholar]
- Ellis S, Nuenke JM. Dipeptidyl arylamidase III of the pituitary: purification and characterization. J. Biol. Chem. 1967;242:4623–4629. [PubMed] [Google Scholar]
- Fukasawa K, Fukasawa KM, Kanai M, Fujii S, Hirose J, Harada M. Dipeptidyl peptidase III is a zinc metallo-exopeptidase: Molecular cloning and expression. Biochem. J. 1998;329:275–282. doi: 10.1042/bj3290275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K, Fukasawa KM, Iwamoto H, Hirose J, Harada M. The HELLGH motif of rat liver dipeptidyl peptidase III is involved in zinc coordination and the catalytic activity of the enzyme. Biochemistry. 1999;38:8299–8303. doi: 10.1021/bi9904959. [DOI] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George KW, Alonso-Gutierrez J, Keasling JD, Lee TS. Isoprenoid drugs, biofuels, and chemicals – Artemisinin, farnesene, and beyond. Adv. Biochem. Eng. Biotechnol. 2015;148:355–389. doi: 10.1007/10_2014_288. [DOI] [PubMed] [Google Scholar]
- Gunawardana D, Likic VA, Gayler KR. A comprehensive bioinformatics analysis of the nudix superfamily in Arabidopsis thaliana. Comp. Funct. Genom. 2009 doi: 10.1155/2009/820381. Article Number: 820381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hast BE, Goldfarb D, Mulvaney KM, Hast MA, Siesser PF, Yan F, Hayes DN, Major MB. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res. 2013;73:2199–2210. doi: 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry LK, Gutensohn M, Thomas ST, Noel JP, Dudareva N. Orthologs of the archaeal isopentenyl phosphate kinase regulate terpenoid production in plants. Proc. Natl. Acad. Sci. USA. 2015;112:10050–10055. doi: 10.1073/pnas.1504798112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T, Egushi T. Functional analysis of type 1 isopentenyl diphosphate isomerase from Halobacterium sp. NRC-1. Biosci. Biotechnol. Biochem. 2007;71:2588–2591. doi: 10.1271/bbb.70330. [DOI] [PubMed] [Google Scholar]
- Jajčanin-Jozić N, Deller S, Pavkov T, Macheroux P, Abramić M. Identification of the reactive cysteine residues in yeast dipeptidyl peptidase III. Biochimie. 2010;92:89–96. doi: 10.1016/j.biochi.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Jajčanin-Jozić N, Abramić M. Hydrolysis of dipeptide derivatives reveals the diversity in the M49 family. Biol. Chem. 2013;394:767–771. doi: 10.1515/hsz-2012-0347. [DOI] [PubMed] [Google Scholar]
- Kansanen E, Kuosmanen SM, Leinonen H, Levonen A-L. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karačić Z, Špoljarić J, Rožman M, Abramić M. Molecular determinants of human dipeptidyl peptidase sensitivity to thiol modifying reagents. Biol. Chem. 2012;393:1523–1532. doi: 10.1515/hsz-2012-0181. [DOI] [PubMed] [Google Scholar]
- Kemmer G, Keller S. Nonlinear least-squares data fitting in Excel spreadsheets. Nat. Protocols. 2010;5:267–281. doi: 10.1038/nprot.2009.182. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kern JT, Walker JR, Johnson JA, Schultz PG, Luesch H. A genomic screen for activators of the antioxidant response element. Proc. Natl. Acad. Sci. USA. 2007;104:5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard J-L, Roccia A, Caissard J-C, Vergne P, Sun P, et al. Biosynthesis of monoterpene scent compounds in roses. Science. 2015;349:81–83. doi: 10.1126/science.aab0696. [DOI] [PubMed] [Google Scholar]
- McLennan AG. Substrate ambiguity among the nudix hydrolases: biologically significant, evolutionary remnant, or both? Cell. Mol. Life Sci. 2006;70:373–385. doi: 10.1007/s00018-012-1210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang L-W, Amzel LM. Arch. Biochem. Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Müller SJ, Gütle DD, Jacquot J-P, Reski R. Can mosses serve as model organisms for forest research? Ann. For. Sci. 2015;468 doi: 10.1007/s13595-015-0468-7. [DOI] [Google Scholar]
- Ogawa T, Ueda Y, Yoshimura K, Shigeoka S. Comprehensive Analysis of Cytosolic Nudix Hydrolases in Arabidopsis thaliana. J. Biol. Chem. 2005;280:25277–25283. doi: 10.1074/jbc.M503536200. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yoshimura K, Miyake H, Ishikawa K, Ito D, Tanabe N, Shigeoka S. Molecular characterization of organelle-type Nudix hydrolases in Arabidopsis. Plant Physiol. 2008;148:1412–1424. doi: 10.1104/pp.108.128413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Bezanilla M. Evolutionary crossroads in developmental biology: Physcomitrella patens. Development. 2010;137:3535–3543. doi: 10.1242/dev.049023. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Salopek-Sondi B, Vukelić B, Špoljarić J, Šimaga Š, Vujaklija D, Makarević J, Jajčanin N, Abramić M. Functional tyrosine residue in the active center of human dipeptidyl peptidase III. Biol. Chem. 2008;389:163–167. doi: 10.1515/BC.2008.021. [DOI] [PubMed] [Google Scholar]
- Schaller A. A cut above the rest: the regulatory function of plant proteases. Planta. 2004;220:183–197. doi: 10.1007/s00425-004-1407-2. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Špoljarić J, Salopek-Sondi B, Makarević J, Vukelić B, Agić D, Šimaga Š, Jajčanin-Jozić N, Abramić M. Absolutely conserved tryptophan in M49 family of peptidases contributes to catalysis and binding of competitive inhibitors. Bioorg. Chem. 2009;37:70–76. doi: 10.1016/j.bioorg.2009.03.002. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukelić B, Salopek-Sondi B, Špoljarić J, Sabljić I, Meštrović N, Agić D, Abramić M. Reactive cysteine in the active-site motif of Bacteroides thetaiotaomicron dipeptidyl peptidase III is a regulatory residue for enzyme activity. Biol. Chem. 2012;393:37–46. doi: 10.1515/BC-2011-193. [DOI] [PubMed] [Google Scholar]
- Wang Y, Qiu C, Zhang F, Guo B, Miao Z, Sun X, Tang K. Molecular cloning, expression profiling and functional analyses of a cDNA encoding isopentenyl diphosphate isomerase from Gossypium barbadense. Biosci. Rep. 2009;29:111–119. doi: 10.1042/BSR20070052. [DOI] [PubMed] [Google Scholar]
- Wouters J, Oudjama Y, Barkley SJ, Tricot C, Stalon V, Droogmans L, Poulter CJ. Catalytic mechanism of Escherichia coli isopentenyl diphosphate isomerase involves Cys-67, Glu-116, and Tyr-104 as suggested by crystal structures of complexes with transition state analogues and irreversible inhibitors. J. Biol. Chem. 2003;278:11903–11908. doi: 10.1074/jbc.M212823200. [DOI] [PubMed] [Google Scholar]
- Xu W, Dunn CA, O'Handley SF, Smith DL, Bessman MJ. Three new nudix hydrolases from Escherichia coli. J. Biol. Chem. 2006;281:22794–22798. doi: 10.1074/jbc.M603407200. [DOI] [PubMed] [Google Scholar]
- Xu A, Desai AM, Brenner SE, Kirsch JF. A continuous fluorescence assay for the characterization of Nudix hydrolases. Anal. Biochem. 2013;437:178–184. doi: 10.1016/j.ab.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, et al. Empirical analysis of transcriptional activity in the Arabidosis Genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Shigeoka S. Versatile physiological functions of the Nudix hydrolase family in Arabidopsis. Biosci. Biotech. Bioch. 2015;79:354–366. doi: 10.1080/09168451.2014.987207. [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu L, Xu H, Wei Z, Wang Y, Lin Y, Gong W. Crystal structures of human IPP isomerase: New insights into the catalytic mechanism. J. Mol. Biol. 2007;366:1437–1446. doi: 10.1016/j.jmb.2006.10.092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Circular dichroism spectra of wild-type PpND and of AtND proteins.
Supplementary Figure 2. Multiple sequence alignment of human, Escherichia coli and Halobacterium salinarium IDIs with PpND (A9TLP4) and AtND (Q8L831) proteins (made using Clustal O 1.2.1.). Residues important for IDI activity (known from human and E. coli IDI structures) are highlighted yellow, the catalytic cysteine is marked with a black circle, and the IDI active-site motif (NTCCSH) with a line above the sequence.
Supplementary Figure 3. IDI color complementation assay. pTrc is a negative control, pTrcAtIDI is a positive control containing IDI gene from Arabidopsis thaliana, pTrcPpND contains Nudix-DPP III (PpND) from Physcomitrella patens, and pTrcAtND containes Nudix-DPP III (AtND) fom A. thaliana.
Supplementary Figure 4. Kinetic characterization of PpND and AtND against their most reactive substrates. The reactions against the substrates with the highest recorded activity identified in the screening were measured using kinetic analysis. These reactions were measured at pH 7.6 and 37 °C, with enzyme concentrations from 20 to 344 nM. Error bars are shown where justified. In all cases, linear regression was used to determine the ratio of kcat/Km. The reported values of the kinetic parameters for enzyme and substrate are recorded in Table 3.
Supplementary Figure 5. Extracted LC-MS chromatograms of ion product m/z=79.00 of IPP (244.70) and IP (164.90): A) control, IPP incubated at 37 °C for 10 min, no enzyme added; B) IPP incubated at 37 °C for 10 min with PpND (60 nM); C) IPP incubated at 37 °C for 10 min with PpND (600 nM).