Abstract
A new study clarifies a relationship between growth, gene expression, and cell size in cyanobacteria. Quite unexpectedly, cyanobacteria and Escherichia coli appear to share an invariance principle to coordinate growth and chromosome replication. This principle allows quantitative predictions of cell size across a wide range of growth conditions in both organisms.
Physics has a long history of discovering invariance principles that are intimately connected to conservation laws. In classical physics, examples of such laws include conservation of energy, momentum, electric charge, and mass. These laws are important because they help us understand the inner workings of physical systems so that we can predict their behavior. One may say that our ability to predict directly reflects our understanding of the system.
At first glance, biology seems quite different. The power of mathematical representations of physical laws seems to stem from the fundamental simplicity of physical interactions. However, every measurement in biology involves a huge underlying complexity of molecular detail. And yet, the search for mathematical regularities in biological data has been surprisingly fruitful, because in part reducing a large data set to a simple mathematical rule sharpens our thinking. It compels us to ask for an explanation of the formula, and it draws our attention to anomalous mutants or conditions that break the mathematical rule.
In a recent study, Zheng and O’Shea have taken an elegant, minimalist approach to understand the relationship between gene expression, chromosome copy number, and cell size in cyanobacteria [1]. They expressed yellow fluorescent proteins from a constitutive promoter as a readout of global regulation of protein levels, simultaneously measuring the chromosome copy number and cell size using microscopy.
They noticed that the protein concentration is constant from cell to cell despite variation in chromosome copy number and therefore in gene dosage. For a stable protein, the average rate at which the number of protein copies in the cell increases should be proportional to the product of the average transcription rate, the average translation rate, and the gene dosage. Therefore, for the concentration of the protein to remain constant during growth, this total protein synthesis rate should be the same as the rate growth.
Zheng and O’Shea saw a gratifying resolution when they realized that the number of genome copies increases linearly with cell volume in individual cells. Thus, increased gene dosage supports the higher rate of protein production in a longer cell. This is an elegant way to keep protein concentration independent of size, because it means that the cytoplasm of long cells and short cells has approximately the same capacity for transcription and translation and that all copies of the genome are transcriptionally active. This linear relationship is in agreement with previous findings [2–3].
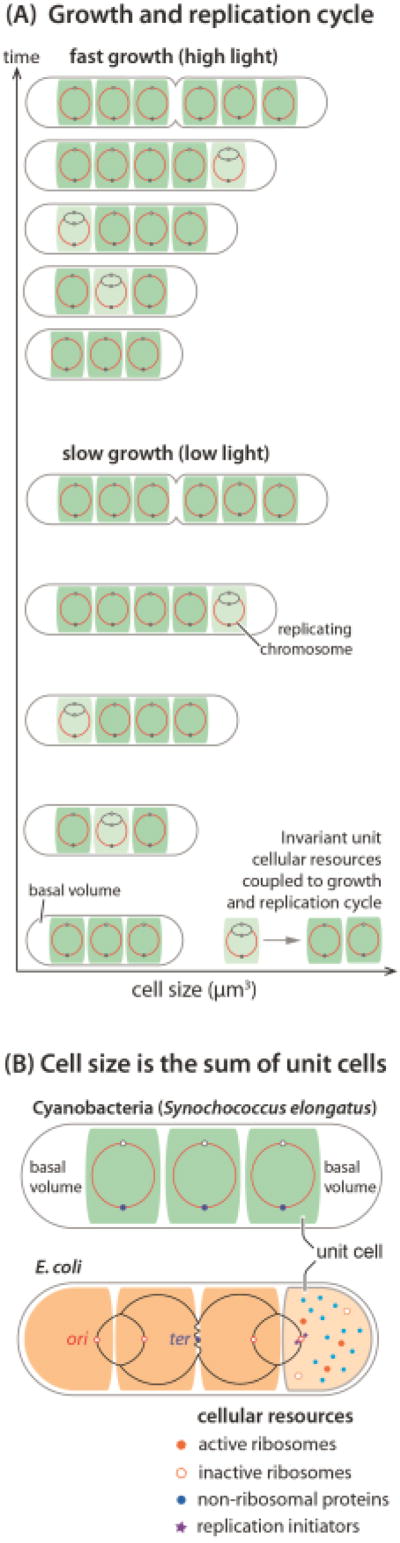
But there is more to the story. Cyanobacteria are photosynthetic prokaryotes and their growth rate depends on the intensity of illumination. Surprisingly, close examination of Zheng and O’Shea’s data reveals that neither the cell size distributions nor the chromosome copy number distributions are affected by the illumination-imposed growth rate in their experimental conditions. Most newborn cells contain on average three chromosomes, and double their number by the time they divide, consistent with previous results [2–3]. The average newborn size is independent of the growth rate. Furthermore, previous work suggests that replication initiation is asynchronous and, at any given time, only one of the chromosome copies undergoes DNA replication [2–4]. Taken together, current findings show that growth and the chromosome replication cycle are coupled such that the amount of protein produced during the replication cycle of one chromosome is invariant [Figure 1(A)].
Figure 1.
(A) In cyanobacteria, growth and chromosome replication are coupled so that the amount of protein produced (thus added cell size) during the replication cycle of one chromosome is invariant regardless of the growth rate. The average cell volume (V) increases linearly with respect to the number (N) of unit volume (V0) from the basal volume (Vbasal), i.e., V = N* V0 + Vbasal. (B) The general growth law states that cell size is the sum of all invariant unit cells, where the unit cell is the average cell size per replication origins at initiation [8]. Both cyanobacteria and E. coli appear to follow this principle, with additional basal volume (Vbasal) for cyanobacteria (white space in the illustrated cyanobacteria cell). This basal term may reflect specialized structures associated with the cell poles that do not scale with the number of chromosomes.
These results suggest a common principle of cell size control between cyanobacteria and Escherichia coli, which was once thought unlikely. One of the major lessons from the studies of E. coli physiology can be summarized as the “(nutrient) growth law”, which relates cell size to growth rate [5]. Based on this foundational work, later studies showed that the increase in the average cell size is directly proportional to the average number of replication origins present during multifork replication [6–7] [Figure 1(B)]. In fact, the average cell size per replication origin is invariant even when the biosynthetic capacity of the cell is severely perturbed [8].
Therefore, both cyanobacteria and E. coli appear to follow the “general growth law” that cell size is the sum of all invariant “unit cells,” where the number of unit cells is determined by the number of replication origins simultaneously present in the cells [Figure 1(B)] [8].
An obvious and important biological question is what mechanism underlies the observed invariance of the unit cell in both cyanobacteria and E. coli. In E. coli, a long-standing idea is based on the accumulation of a fixed critical amount of replication initiators (e.g. DnaA) at the origin. Following initiation, these initiators are thought to be titrated away by binding sites in the newly replicated DNA [8]. Initiator expression is known to be autoregulated so that their concentration is maintained constant independent of cell size and growth rate [9]. In principle, the same mechanism could apply to cyanobacteria so that a fixed amount of initiators accumulate per chromosome cycle, implying a constant increase in cell volume. Highly cooperative binding of initiators [10] might also provide a clue to the mechanism that selects only a single cyanobacterial chromosome copy for replication at a time—once a particular origin is selected stochastically by the binding of a pioneer initiator protein, cooperative interactions might ensure that initiators continue to accumulate predominantly at that site. The invariance of the unit cell under growth inhibition is consistent with the “initiator threshold” idea [8], and would be a straightforward hypothesis to test in cyanobacteria.
Cyanobacteria like S. elongatus have a very different lifestyle from well-studied bacteria like E. coli. S. elongatus has a rhythmic growth environment controlled by the light-dark cycle, it maintains multiple copies of its chromosomes which replicate asynchronously, and the relationship between chromosome number and the initiation of cytokinesis is flexible, depending on both illumination and time of day. There are many unanswered questions about the molecular mechanisms in cyanobacteria that underlie these phenomena. Despite these differences, the simple mathematical rules that both E. coli and S. elongatus appear to follow—the invariance of the unit cell—allow us to predict the cell size of either organism by simply counting the average number of chromosomes in a given condition. This is reminiscent of how physicists can make predictions based on a conservation law without knowing all of the details of a system, made even more remarkable that it applies to bacteria from widely divergent phyla. We believe that it likely points to a fundamental coordination principle of the bacterial cell.
Acknowledgments
We thank David Savage for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zheng X, O’Shea E. Cell Rep. Cyanobacteria Maintain Constant Protein Concentration despite Genome Copy-Number Variation. 2017 doi: 10.1016/j.celrep.2017.03.067. [DOI] [PubMed] [Google Scholar]
- 2.Jain I, Vijayan V, O’Shea E. Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc. Nat. Acad. Sci. USA. 2012;109(34):13638–13643. doi: 10.1073/pnas.1211144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen A, Afonso B, Silver P, Savage D. Spatial and Temporal Organization of Chromosome Duplication and Segregation in the Cyanobacterium Synechococcus elongatus PCC 7942. PLoS ONE. 2012;7(10):e47837. doi: 10.1371/journal.pone.0047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe S, Ohbayashi R, Shiwa Y, Noda A, Kanesaki Y, Chibazakura T, Yoshikawa H. Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Molecular microbiology. 2012;83(4):856–65. doi: 10.1111/j.1365-2958.2012.07971.x. [DOI] [PubMed] [Google Scholar]
- 5.Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on Medium and Temperature of Cell Size and Chemical Composition during Balanced Growth of Salmonella typhimurium. J. Gen. Microbiol. 1958;19(3):592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 6.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 7.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 8.Si F, Li D, Cox S, et al. Invariance of Initiation Mass and Predictability of Cell Size in Escherichia coli. Curr Biol. 2017 doi: 10.1016/j.cub.2017.03.022. http://dx.doi.org/10.1016/j.cub.2017.03.022. [DOI] [PMC free article] [PubMed]
- 9.Skarstad K, Katayama T. Regulating DNA replication in bacteria. Cold Spring Harbor Perspectives in Biology. 2013;5(4):a012922–a012922. doi: 10.1101/cshperspect.a012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walldén M, Fange D, Lundius EG, Baltekin Ö, Elf J. The Synchronization of replication and division cycles in individual E. coli Cells. Cell. 2016;166(3):729–739. doi: 10.1016/j.cell.2016.06.052. [DOI] [PubMed] [Google Scholar]