Abstract
Tetratricopeptide repeat (TPR) domains are ubiquitous protein interaction domains that adopt a modular antiparallel array of α‐helices. The TPR fold typically adopts a monomeric state, and consensus TPRs sequences successfully fold into the expected monomeric topology. The versatility of the TPR fold also supports different quaternary structures, which may function as regulatory switches. One example is yeast mitochondrial fission 1 (Fis1) that appears to interconvert between monomer and dimer states in regulating division of peroxisomes and mitochondria. Whether human Fis1 can also interconvert like the yeast molecule is unknown. A TPR consensus proline residue present in human Fis1 is absent in the yeast molecule and, when added, prevents yeast Fis1 dimerization suggesting that the TPR consensus proline might have persisted to prevent TPR oligomerization. Here, we address this question with human Fis1 and the consensus TPR protein CTPR3. We demonstrate that human Fis1 does not form a noncovalent dimer via its TPR domain, despite conditions that favor dimerization of the yeast protein. We also show that the presence of the consensus proline is not sufficient to forbid TPR dimerization. Lastly, an analysis of all available TPR protein structures (22 nonredundant structures, totaling 64 TPRs—42 with the consensus proline and 22 without) revealed that the consensus proline is not necessary for turn formation, but does favor shorter turns. This work suggests the TPR consensus proline is not to prevent oligomerization, but to favor tight turns between repeats.
Keywords: TPR protein, tetratricopeptide repeat, protein dimerization, CTPR3, Fis1, protein folding, size exclusion chromatography
Introduction
Repeat proteins are an arrangement of a single structural module repeated multiple times, and one such repeat found in all kingdoms of life is the tetratricopeptide repeat (TPR). The TPR is a 34‐residue helix‐turn‐helix fold that is commonly repeated 2–20 times, which creates an antiparallel array of α‐helices with a superhelical twist.1 This architecture presents two large surface areas—concave and convex faces—well suited for mediating protein interactions. Indeed, both surfaces of the TPR fold are known to mediate protein interactions, however, the concave surface more commonly does so.2 A well‐studied example is heat shocking organizing protein (Hop), which is critical for chaperone activity by acting as a scaffold for Hsp90 to receive substrate from Hsp70.3 Its function is dependent on two distinct TPR domains in Hop (TPR1: 3 TPR motifs; TPR2: 6 TPR motifs).3 TPR1 domain mediates Hsp70 binding, whereas Hop's TPR2 domain mediates Hsp90 binding.3 Both interactions are mediated by TPR concave surfaces, which are absolutely required to bring the two master chaperones together.3 The modularity of the TPR fold has led to the successful design of consensus TPR proteins (CTPRn), where n denotes the number of identical TPRs and ranges from 2 to 20.4 Consensus TPR proteins are stable, monomeric, adopt the canonical TPR helix‐turn‐helix fold, and support binding to native ligands including a peptide derived from Hsp90.2 These findings illustrate a deep understanding of how TPR sequence specifies its fold, at least for monomeric TPRs.5
TPR domains can also mediate self‐association, which in many cases is essential for function.6 The TPR protein rapsyn is responsible for clustering neuronal acetylcholine receptors at the postsynaptic membrane for efficient signaling, which is dependent on TPR‐mediated dimerization.7 Another TPR containing protein YbgF from E. coli, a scaffolding protein important for bacterial cell division,8 adopts TPR‐mediated dimers and trimers in solution. In the YbgF system, three tyrosine residues that are conserved in the YbgF family, but not part of the TPR consensus motif, were found to specify its oligomeric state. Indeed, substitution of these residues to aspartate impedes oligomerization. These results suggest an intrinsic ability of TPRs to form multimers. To test the ability of TPRs to form oligomers, the critical tyrosine residues from YbgF were introduced into the monomeric consensus TPR construct CTPR3, and were found to induce oligomerization, similar to the native YbgF protein.9 These results point to the potential for any TPR protein to oligomerize; a property that might be advantageous as a switch that presents an even larger surface area for interactions that a monomeric TPR fold.
TPR domain mediated oligomers can exist in several topologies.10 One topology is concave surface mediated, a two‐fold symmetry along the concave surface where both convex surfaces are exposed, as seen in the Cut9 TPR dimer,11 Cdc27,11 and human Mps112; in every case dimerization was shown to be critical for function.13, 14 A second topology is terminus mediated as seen in the CTPR3Y3 (trimer), described above.9 This topology involves an interaction between the terminal helices (N or C‐terminal) of the two subunits. The terminus‐mediated topology is also seen in the native Apc7 (N‐term to N‐term dimer)15 and TOM70p (C‐term to C‐term dimer).16 The third topology is convex surface mediated, involving a two‐fold symmetry along the convex surface where both concave surfaces are exposed, as seen in O‐linked GlcNAc Transferase17 and Sgt1.18 The different topologies seen in various TPR containing proteins illustrate the versatility of the fold where oligomerization could potentially allow or prevent TPR binding surfaces.
The last topology identified to date is a 3D domain swapped dimer described for S. cerevisiae protein Fis1 [Fig. 1(B)]. Fis1 is a mitochondrial outer membrane anchored protein with a cytoplasmic TPR domain thought to act as a receptor for proteins involved in yeast mitochondrial fission, which is the process by which one mitochondrion divides into two daughter mitochondria.26, 27 Mutations in the TPR domain of Fis1 were identified from different genetic screens as nonfunctional alleles.28, 29 When introduced into the cytoplasmic domain and isolated, these mutants were found to populate a dimeric state that is barely populated by the wild type sequence.20 The ability of Fis1 mutants to reveal a nascent dimer state was surprising since the NMR and x‐ray structures of wild type Fis1 were monomeric.19, 30 These seemingly disparate observations were reconciled by subsequent realization that Fis1 is kinetically trapped as a monomer, and can be driven into dimer equilibrium by heat or chemical denaturation. Mutations that enhance either the monomer or dimer states are nonfunctional suggesting that the interconversion might be important for function.20
Figure 1.
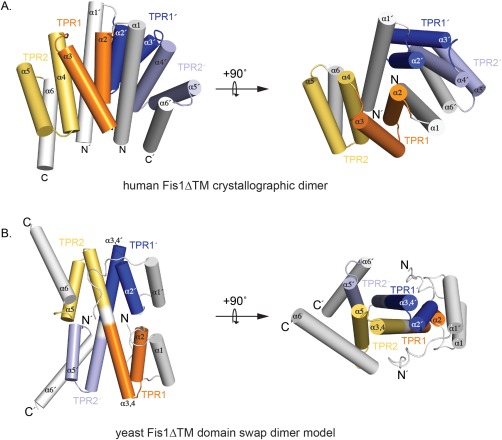
Structural models for Fis1 dimerization. (A) A cartoon representation shows the dimer model of human Fis1 that requires reciprocating interactions between helix 1 and the TPR concave surface. TPR1 is orange/blue, TPR2 is yellow/light blue and the N‐terminal helices 1 and C‐terminal helices 6 are colored white. The transmembrane domain is not shown. PDB code: 1NZN.pdb.19 (B) A cartoon representation shows the 3D domain swap dimer model of yeast Fis1 cytoplasmic domain which requires a loop to helix transition between TPR1 and TPR2.20 The N‐terminal arm (white) binds into the concave surface, which is comprised of 2 TPR motifs from two protomers (TPR1 orange/blue; TPR2 yellow/light blue). This model was docked using ClusPro.21, 22, 23, 24, 25
Yeast Fis1 dimerization is governed by the so‐called Fis1 arm, an N‐terminal region of 16 amino acids that binds into the TPR concave surface, possibly regulating access to the binding surface.31 A variant of yFis1 lacking the arm (ΔN16) appears as an obligate dimer and cannot support fission. The arm of Fis1 is not the only structural aspect that appears important for dimerization. A structural model of the yeast Fis1 dimer shows intervening residues between TPRs 1 and 2 form an extended helix that allows for a 3D domain swapped orientation of the dimer [Fig. 1(B)]. In the yeast Fis1 monomer, these intervening residues form an unstructured loop linking TPRs 1 and 2, as is observed in other TPR proteins. The observation that a coil‐to‐helix transition can allow for dimerization in yeast Fis1 raises the possibility that domain swapping dimers could occur in other TPRs as they allow for the same interhelical interactions that stabilize the fold.32, 33 However, TPRs contain a highly conserved proline residue at the end of the repeat that is a TPR consensus residue. Proline has poor helical propensity and would be expected to prevent a coil to helix transition that supports dimerization of yeast Fis1. Curiously, the consensus proline is missing in yeast Fis1 where it is an alanine. Substitution of a proline at this position (A72P) prevents dimerization and cannot support fission in a growth assay.20 Thus, it appears that the absence of the consensus proline may be sufficient to allow for dimerization, which in the case of yeast Fis1 is normally prevented by the presence of the N‐terminal Fis1 arm.
The consensus proline and the Fis1 arm are both present in mammalian Fis1, consistent with monomeric structures of both human and mouse Fis1.31 However, Fis1 is conserved from yeast to human, with a sequence similarity of approximately 66% and sequence identity of 27%, raising the possibility that the TPR domain of human Fis1 might also mediate dimerization. Indeed, Fis1 isolated from rat cell mitochondria migrates as a ∼200 kDa complex by BN‐PAGE that differential tagging experiments show contain more than one Fis1 molecule.34 The complex may be mediated by human Fis1 dimerization and curiously, human Fis1 crystallized as a dimer [Fig. 1(A)], adopting a topology resembling the Cut9 concave surface mediated‐dimer, and differs from the 3D domain‐swapped structural model of the yeast dimer [Fig. 1(B)]. Human Fis1 was also found to crosslink as multimers that were dependent on the removal of the N‐terminal arm.35 These findings together with the precedent of yeast Fis1 being autoinhibited by its N‐terminal arm for dimerization, raise the question of whether human Fis1 also forms a dimer that is governed by its arm. Here we address these questions and find that, unlike the yeast molecule, human Fis1 is unable to noncovalently self‐associate via its cytoplasmic TPR domain despite removal of N‐terminal arm. We also find that upon removal of the TPR consensus proline dimerization remains unaffected in either human Fis1 or a consensus TPR protein. Our work supports the conclusion that the TPR fold intrinsically specifies a monomeric state, and removal of the consensus proline is not sufficient to allow dimerization of this ubiquitous fold.
Results
Human Fis1 cytoplasmic domain does not form a noncovalent dimer, despite global denaturation and refolding
Recombinant yeast Fis1ΔTM expressed in E. coli is monomeric.31 However, upon heating or chemical denaturation a dimeric state was populated indicating that yeast Fis1ΔTM molecule was isolated as a kinetically trapped monomer.20 We asked whether the “kinetic trap” property was shared by human Fis1. To address that question, the cytoplasmic domain (residues 1–125, Fis1ΔTM) was purified using nickel affinity chromatography and size exclusion chromatography (SEC) [Fig. 2(A)]. Fis1ΔTM eluted as two species: species A at 75 mL and species B at 82 mL. Based on molecular weight standards (Supporting Information Fig. S2), species A and B eluted at molecular weights consistent with that for a dimer and monomer of Fis1ΔTM (28 and 14 kDa), respectively. However, using SDS‐PAGE and NMR spectroscopy, we determined that species A was a fusion artifact from purification (also 28 kDa) while species B was Fis1ΔTM. Chromatographic fractions corresponding to the Fis1ΔTM monomer were pooled and concentrated to 150 µM, unfolded for 15 hours in 6 M guanidine HCl, and subsequently dialyzed into a refolding buffer containing no chemical denaturant. For the yeast molecule, this protocol removes the kinetic barrier for monomer‐dimer interconversion (app Kd ∼15 µM). The refolded human sample was analyzed by SEC and eluted as two species: species A at 75 mL and species B at 82 mL [Fig. 2(B)]. By reducing SDS‐PAGE of the peak fractions, Species A migrated as a single band at the same molecular weight as species B [Fig. 2(B), inset], which indicated that the species was not fusion construct contaminant, but rather a disulfide‐linked Fis1 dimer. A control sample not unfolded/refolded eluted as a single species [Fig. 2(C)] suggesting that unfolding promoted disulfide formation. Both unfolded/refolded and control monomer samples were well folded by NMR spectroscopy (Supporting Information Fig. S3). Since Fis1ΔTM contains a single, solvent accessible cysteine, C41, we conducted refolding experiments in the absence and presence of the reductant dithiothreitol (DTT). Fis1ΔTM refolded from 6 M GdHCl in the absence of DTT eluted as two species with ∼70% found as species A [Fig. 2(D)]. This result is in stark contrast to the observation of <5% species A with DTT (2 mM) in the refolding buffer [Fig. 2(C)]. Species A from the sizing column [Fig. 2(D)] was collected and treated with 10 mM DTT and reapplied to the sizing column and eluted as primarily (∼80%) species B (Supporting Information Fig. S4). Size exclusion chromatography and multi‐angle laser light scattering (SEC‐MALLS) of Fis1ΔTM in the absence of DTT shows that the molecular weights of the two eluting species correspond to the molecular weights of monomer (average calculated molecular weight ∼15 kDa) and dimer (average calculated molecular weight ∼30 kDa) (Supporting Information Fig. S5). Taken together, these data indicate that species A was formed by a disulfide bridge between two Fis1ΔTM molecules. To confirm the covalent nature of species A, we purified a variant of Fis1ΔTM in which the native Cys was replaced with Ala (C41A). After unfolding and refolding in a similar manner as above, Fis1ΔTM C41A variant eluted as predominantly species B [Fig. 2(E)]. Refolding Fis1ΔTM C41A resulted in very few changes in conformation as assessed by 2D NMR spectroscopy (Supporting Information Fig. S6). We conclude that the human Fis1 cytoplasmic domain readily forms a covalent dimer mediated by a C41 cystine linkage that is enhanced upon protein unfolding. Unlike the yeast ortholog, we find little evidence to support a noncovalent dimeric state.
Figure 2.
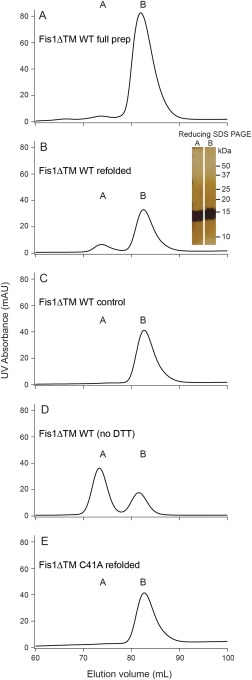
Recombinant Fis1 appears to form a disulfide bridged dimer. (A) Purified recombinant Fis1ΔTM elutes as two species from a Superdex 75 column: A (75 mL elution volume) and B (82 mL elution volume). B) Species B from (A) was concentrated to 150 µM protein, unfolded, refolded, and reapplied to a Superdex 75 column and again elutes as two species. Both species A and B are the same molecular weight on a silver stained reducing SDS‐PAGE gel (inset). (C) Species B from (A) was concentrated to 150 µM protein and applied to a Superdex 75 column and did not populate species A. D) Species B from (A) was concentrated to 150 µM protein, refolded in the absence of reductant, and applied to a Superdex 75 column and predominantly populated species A. (E) Fis1ΔTM C41A species B was isolated from a Superdex 75 column, concentrated to 150 µM, refolded, and reapplied to a Superdex 75 column and mainly populated species B
Removal of the human Fis1 N‐terminal arm is predominantly monomeric, unlike the yeast Fis1 cytoplasmic domain
In the yeast molecule, dimer formation is prevented by the first 16 amino acids of the protein, which has been dubbed the Fis1 arm.20 Removal of the N‐terminal arm enhances yeast Fis1 dimerization to the extent that it is essentially an obligate dimer.20 In human Fis1, the N‐terminal arm is 8 residues shorter, and may act in an autoinhibitory manner similar to yeast Fis1.30 To test whether the human Fis1 arm might prevent dimerization, a variant of human Fis1 lacking the arm (residues 9–125, ΔNFis1ΔTM) was unfolded and refolded as described above. Application to a size exclusion column, resulted in elution of two populations: a small population (<10%) eluting at 75 mL (species A) and the main species (∼90%) at 82 mL (species B) [Supporting Information Fig. S7(A)]. Species B was found to be well folded by 2D NMR spectroscopy of 15N‐labeled protein despite being unfolded and refolded [Supporting Information Fig. S7(B)]. Both species A and B were assayed by reducing SDS‐PAGE and migrated as a single band at the expected molecular weight for ΔNFis1ΔTM (data not shown), thus verifying that species A was a ΔNFis1ΔTM dimer and not a contaminant. These studies revealed that removal of the human Fis1 arm appears to have little influence on disulfide bridge formation of the cytoplasmic domain. Unlike the yeast domain, the N‐terminal arm of human Fis1 does not inhibit dimerization.
Fis1 P63A mutation does not alter cytoplasmic domain oligomeric state
Yeast Fis1 is thought to form a domain swapped dimer via a coil‐to‐helix transition of the loop between TPR repeats 1 and 2 [Fig. 1(B)]. Consistent with this interpretation, substitution of a proline for an alanine in the loop (A72P) abrogated dimerization in all conditions that allowed dimerization of the wild‐type sequence including introduction of dimer‐promoting variants.20 Notably, the A72 in yeast Fis1 sequence aligns to position #32 (out of 34), which is most frequently a proline.4 Yeast Fis1 does not contain the TPR consensus proline yet still folds into a TPR fold, which illustrates that the TPR consensus proline at position #32 is not necessary to specify the TPR fold. The finding that the yeast molecule can adopt a 3D domain‐swapped dimer that is prevented by introduction of a proline, raises the possibility that the consensus proline prevents dimerization in other TPR proteins. We tested this idea by replacing the TPR proline with alanine in the human Fis1 cytoplasmic domain (P63A), and the resulting protein product was assayed for oligomeric state by SEC. Fis1ΔTM C41A/P63A was unfolded and refolded as described above and eluted as predominantly (>95%) monomer (Fig. 3). Similarly, ΔNFis1ΔTM C41A/P63A was unfolded and refolded and eluted as a monomer (data not shown). These mutations resulted in modest structural changes as assessed by heteronuclear 2D NMR spectroscopy (Supporting Information Fig. S8). These data and similar experiments conducted at a range of concentrations (data not shown) support the conclusion that removal of the consensus proline is not sufficient for dimerization of human Fis1.
Figure 3.
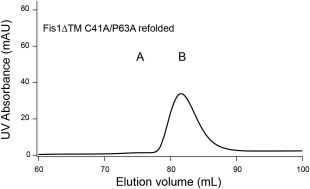
Mutation of a TPR consensus proline residue to alanine (P63A) does not change oligomeric state. Fis1ΔTM C41A/P63A species B was isolated from a Superdex 75 column, concentrated to 150 µM, refolded, and reapplied to a Superdex 75 column and mainly populated species B
Substitution of the consensus proline in a consensus TPR protein is not sufficient for dimerization
To test whether this finding was generalizable to other TPRs, we utilized a consensus TPR protein, CTPR3, whose sequence was designed based on a statistical analysis of predicted TPR proteins and consists of 3 identical TPRs.4 We tested whether the consensus proline is a TPR‐specific mechanism for preventing dimerization by substituting an Ala for Pro66, which is TPR position #32 in the second TPR motif of CTPR3. The P66A substitution was assayed for oligomeric state in the same way as with the human Fis1 molecule. Isolated CTPR3 has been shown by SEC to be monomeric at high concentration,9 which was reproduced in our study, where CTPR3 eluted as a single peak at 80 mL [Fig. 4(A)]. Refolding either the CTPR3 [Fig. 4(B)] or the P66A [Fig.4(C)] mutant were unable to access other oligomeric states at protein concentrations as high as 350 µM. As observed by 1D proton NMR spectroscopy, refolding of the CTPR3 proteins did not change the overall fold of the protein (Supporting Information Fig. S9). We conclude that removal of the consensus proline at position #32 is not sufficient to induce dimerization of the CTPR3 protein.
Figure 4.
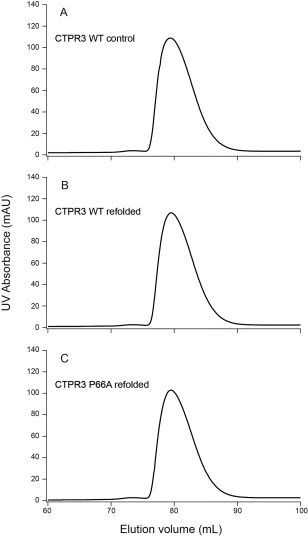
The TPR consensus proline does not appear to govern the oligomeric state of the CTPR3 protein. (A) CTPR3 was isolated from a Superdex 75 column, concentrated to 350 µM and reapplied to a Superdex 75 column and did not extensively populate higher order species. (B) CTPR3 was isolated from a Superdex 75 column, concentrated to 350 µM, refolded, and reapplied to a Superdex 75 column and did not extensively populate higher order species. (C) CTPR3 P66A variant (removal of the second TPR consensus proline) species B was isolated from a Superdex 75 column, concentrated to 350 µM, refolded, and reapplied to a Superdex 75 column and did not extensively populate higher order species
Discussion
Repeat proteins form major classes of protein interaction domains necessary to sustain life. The modularity of repeat protein folds is well described by a 1D Ising model,32 which intrinsically lends itself to 3D domain swapping as found for the yeast Fis1 molecule. For the human molecule, four lines of evidence suggested this TPR domain might also form high order species. First, human Fis1 crosslinked as dimers and trimers on mitochondria and oligomers were mediated by helix 1, which includes the N‐terminal arm.35 Second, Fis1 migrated as a ∼200 kDa complex by BN‐PAGE that was dependent on the N‐terminal arm, and pull‐down experiments showed the complex contains at least two Fis1 molecules.34 The present study was well motivated by these lines of evidence to answer the question whether human Fis1 cytoplasmic domain can dimerize. Third, recombinant human Fis1 cytoplasmic domain crystallized as a dimer.19 Lastly, human Fis1 shares 66% sequence similarity to the yeast Fis1 cytoplasmic domain that forms a dimer, which appears to be important in mitochondrial fission.20 Surprisingly, our study shows that the human Fis1 cytoplasmic domain cannot form a noncovalent dimer. Thus, the Fis1 multimer observed by BN‐PAGE likely could be transmembrane domain mediated or possibly disulfide‐linked. Though disulfide bonds are not typically seen in the reducing environment of the cytoplasm, some proteins use reversible disulfide linkages for redox sensing36 and whether Fis1 acts to sense cellular redox status via disulfide formation is unknown.
This study shows that substitution of a TPR consensus proline to alanine is not sufficient to allow for dimerization of human Fis1 or CTPR3. As noted previously, CTPR3 self‐associates upon replacing three consensus TPR aspartates to tyrosines.9 This finding might raise the question of whether tyrosines at similar positions in yeast Fis1 contribute to its dimerization. A sequence alignment of yeast Fis1 with the CTPR3Y3 mutant showed that Fis1 shared only one of three critical tyrosines (D73Y in CTPR3 mutant is equivalent to Y69 in yeast Fis1); the position that contributed least to dimerization propensity.9 These considerations suggest that the mechanism of CTPR3Y3 oligomerization is fundamentally different from that of yeast Fis1, which is borne out from structural studies in which CTPR3 multimers associate via the engineered tyrosines in an antiparallel orientation. By contrast, yeast Fis1 dimer derives from a 3D domain‐swap between TPRs 1 and 2. TPRs can also form multimers in other ways illustrating the versatility of the TPR sequence/fold.9, 10
TPR protein oligomerization could potentially be another form of protein activity regulation. In the case of YbgF, TolA is responsible for binding and dissociating the TPR oligomer, which is thought to change its function.37 Another scenario is the opposite; it might be that small proteins must oligomerize to enlarge their functional surface area. In the case of TPRs, since they are typically scaffolding proteins, it could be that small (∼2–3 repeats) might oligomerize to enlarge their contact area with binding partners. Oligomerization could serve as another regulation step. Yeast Fis1 appears to have an oligomerization ability when a critical TPR proline is absent, which appears important for its function, given that yeast Fis1 A72P does not fully support fission in a growth assay.20 What the work of others has demonstrated is the importance of other positions in the TPR consensus sequence that might indicate an ability to oligomerized.9
The present study shows that the presence of the consensus proline does not unilaterally govern the oligomeric state of a TPR containing protein. Persistence of the TPR consensus proline as part of the turn between helices was suggested previously,4 because the consensus proline likely forces a tight turn, which has been suggested to fold independently of the rest of the protein.38 An analysis of all available TPR protein structures in the Protein Data Bank included 22 nonredundant TPR protein structures, totaling 64 nonredundant TPRs. Each TPR was followed by a turn, typically starting at residue #31 of the TPR and, of these turns, 42 contained the consensus proline and 22 did not. This structural analysis revealed that the TPR consensus proline was not necessary for turn formation between TPRs, but does strongly favor shorter turns (Fig. 5) by adopting αR space torsion angle that is common in tight turns between helices.38 In summary, we find that the TPR consensus proline is neither necessary nor sufficient for turn formation, and its removal is not sufficient to allow for dimerization.
Figure 5.
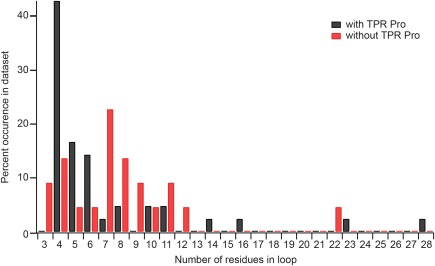
Structural analysis of 64 unique TPR motifs suggests that the TPR consensus proline favors shorter turns between motifs. Turns at the end of predicted TPR motifs were separated by the number of amino acid residues in the turn and whether they contained the TPR consensus proline. Each TPR was followed by a turn, typically starting at residue #31 of the TPR and, of these turns, 42 contained the consensus proline and 22 did not. These turns were tallied and are shown as a histogram normalized to the number of turns with (black bars) and without (red bars) the TPR consensus proline
Methods
Cloning
The human Fis1 gene (1–125) was expressed as a 6xHis‐Smt3‐fusion using a modified pQE30 plasmid as described previously.39, 40 The construct contained at the N‐terminus a 6xHis‐Smt3 tag that was cleavable by ULP1 protease, leaving a native N‐terminus. The ΔNFis1ΔTM was cloned in the same way, where the gene contained amino acids 9–125. Point mutations C41A and P63A of human Fis1 and P66A of CTPR3 were made by site‐directed mutagenesis. The double mutations were made by combining the 5′ amplification of C41A and the 3′ amplification of P63A. All constructs were sequence verified by Retrogen.
The His‐TEV‐CTPR3 pPROEX‐HTam plasmid was a generous gift from the Regan lab.6, 41
Protein expression and purification
DNA plasmids encoding various Fis1ΔTM constructs were transformed into the BL21(pREP4) strain of E. coli. A 25 mL LB starter culture was inoculated and grown overnight at 37°C before being diluted into 1L of 15N‐labeled minimal media containing 3 g/L 15N ammonium chloride (Cambridge Isotope Laboratories, Tewksbury, MA), 8 g/L unlabeled glucose, 2 mM magnesium sulfate, 30 µM calcium chloride, 30 mg/L of kanamycin, and 50 mg/L of ampicillin. Expression was induced at an OD600 of ∼1 using 1 mM isopropyl β‐D‐1‐thiogalactopyranoside and the cultures were shaken overnight at 18°C. Cells were harvested by centrifugation at 5000g for 30 minutes and resuspended in a 1:40 dilution volume (2.5 mL/g wet cell paste) of Buffer A (25 mM Tris, 500 mM NaCl, 30 mM imidazole, 0.02% sodium azide, pH 7.4). Cells were lysed by 10 passes at 15,000 psi using an EmulsiFlex‐C3 cell homogenizer (Avestin, Ottawa, ON, Canada) and lysate was clarified by centrifugation. The lysate supernatant was applied to a column with His60 resin (Clontech, Mountain View, CA), equilibrated in Buffer A, and batch eluted with Buffer A supplemented with 500 mM imidazole. Imidazole was removed by dialysis into Buffer A without imidazole and with 0.1% BME. During dialysis, His‐tagged Smt3 tags were removed from the Fis1 constructs by 12 hour incubation with the yeast SUMO protease, ULP1, at 4°C. Dialysate was reapplied to the nickel resin and the cleaved protein was collected and concentrated using 3 kD MWCO VivaSpin concentrators (GE Healthcare, Marlborough, MA). Fis1ΔTM constructs were further purified by size exclusion chromatography using a HiLoad 16/60 Superdex S‐75 prep grade column (GE Healthcare, Marlborough, MA) in Buffer B (50 mM sodium phosphate, 184 mM NaCl, 0.02% azide, and 2 mM DTT, pH 7.4). Protein concentrations were determined by absorbance at 280 nm using the molar extinction coefficient estimated from its primary sequence: 15,930 M−1•cm−1 for all Fis1ΔTM constructs. CTPR3 protein samples were prepared as described previously.6, 41
Chemical denaturation and size exclusion chromatography
Fis1ΔTM (150 µM) and CTPR3 (350 µM) samples were dialyzed into 500‐fold excess of Buffer B supplemented with 6 M Guanidine HCl (GdHCl) to a final GdHCl concentration of 5.99 M at room temperature overnight before being dialyzed against 1000‐fold excess of Buffer B to remove GdHCl. As a control experiment, identical samples were dialyzed in Buffer B without GdHCl. Sample dilution upon dialysis was estimated to be <10% total protein volume. Samples were then applied to a HiLoad 16/60 Superdex S‐75 prep grade column (GE Healthcare, Marlborough, MA), equilibrated in Buffer B at a flow rate of 0.75 mL/min. Relative populations of monomer and dimer species in size exclusion chromatography experiments were estimated by fitting chromatographic peaks to a Gaussian function in IGOR Pro (Wavemetrics, Inc., Portland, OR): . The fitted values were used to calculate the area under the curve (AUC) using the equation: . Multiangle laser light scattering was measured by DAWN HELEOS using 18 light scattering angles and Optilab for refractive index (Wyatt Technologies, Santa Barbara, CA). Molecular weights were determined using the Zimm formalism using the ASTRA software.
NMR spectroscopy
Labeled (15N) samples of Fis1 constructs contained Buffer B supplemented with 0.02% azide and 10% 2H2O. HSQC spectra were recorded on a Bruker Avance 500 spectrometer (Bruker, Billerica, MA) at 25°C with 4 scans, 1024 (t2) × 300 (t1) complex points with acquisition times of 51 ms (1H) and 90 ms (15N). Unlabeled samples of CTPR3 constructs were used for 1D 1H NMR spectrum collection. Spectra were collected on a Bruker Avance 500 spectrometer at 25°C with 128 scans and 2048 t1 points in 1H, with an acquisition time of 82 ms. NMR data processing was carried out using NMRPipe.42 Proton 1D spectra were subsequently analyzed using NMRDraw42 while 2D HSQC spectra were analyzed using XEASY.43
TPR structural analysis
The Protein Data Bank was searched for TPR containing proteins on April 4, 2017 and returned 32 results; of these, 22 were analyzed as nonredundant, meaning that each protein sequence and structure was counted and analyzed once: 1NZN, 1NA0, 2WQH, 1C0M, 2FBN, 1ZU2, 2C2L, 1WM5, 1YA0, 1WAO, 1XNF, 1TJC, 1W3B, 1P5Q, 1IYG, 1KT0, 1IHG, 1HXI, 1QQE, 2PQN, 1EL2, 1ELR. The 22 nonredundant protein sequences were analyzed by TPRPred44 to uniformly define TPR boundaries. WHATIF45 was used to extract torsion angles of each turn separating TPRs. From the 22 structures analyzed, 64 turns were identified and included turns between TPRs and capping helices. The backbone torsion angles of the TPR motif were classified into Ramachandran space proposed by Efimov38 (αR: ψ < −30°, −120°≤ ϕ ≤ −60°; : −30°≤ ψ ≤ 30°, −120°≤ ϕ ≤ −60°; β: ψ > 80°, −120°≤ ϕ ≤ −60°; δ: 40°≤ ψ ≤ 80°, −110°≤ ϕ ≤ −70°; αL: −40°≤ ψ ≤ −20°, 40°≤ ϕ ≤ 100°; ɛ: −180°≤ ψ ≤ −140°, 70°≤ ϕ ≤ 130°). Turns were identified as separating the end of one TPR motif and the start of another TPR (or capping helix). Helical boundaries were defined by TPRPred where >4 consecutive backbone torsion angles adopted αR.
Supplementary Materials
Supplementary material includes figures that show the native fold of the Fis1 and CTPR3 mutants as well as additional chromatograms to support the conclusion that Fis1 does not form a noncovalent dimer. Also included are the protein standard curve for the Superdex S‐75 column and multiangle light scattering data for Fis1ΔTM monomer and dimer. TPRManuscript_SuppFigures‐resub.pdf
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Supporting Information Figures.
Acknowledgments
We thank R.C. Wells for the yeast Fis1 domain swap dimer model as well as Davin Jensen from the Volkman laboratory at MCW for the pQE30‐Smt3 plasmid and ULP1 protease plasmid.
References
- 1. D'Andrea LD, Regan L (2003) TPR proteins: The versatile helix. Trends Biochem Sci 28:655–662. [DOI] [PubMed] [Google Scholar]
- 2. Cortajarena AL, Wang J, Regan L (2010) Crystal structure of a designed tetratricopeptide repeat module in complex with its peptide ligand. FEBS J 277:1058–1066. [DOI] [PubMed] [Google Scholar]
- 3. Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I (2000) Structure of TPR domain‐peptide complexes: Critical elements in the assembly of the Hsp70‐Hsp90 multichaperone machine. Cell 101:199–210. [DOI] [PubMed] [Google Scholar]
- 4. Main ERG, Xiong Y, Cocco MJ, D'Andrea L, Regan L (2003) Design of stable alpha‐helical arrays from an idealized TPR motif. Structure 11:497–508. [DOI] [PubMed] [Google Scholar]
- 5. Phillips JJ, Javadi Y, Millership C, Main ERG (2012) Modulation of the multistate folding of designed TPR proteins through intrinsic and extrinsic factors. Protein Sci 21:327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Main ERG, Jackson SE, Regan L (2003) The folding and design of repeat proteins: Reaching a consensus. Curr Opin Struct Biol 13:482–489. [DOI] [PubMed] [Google Scholar]
- 7. Ramarao MK, Bianchetta MJ, Lanken J, Cohen JB (2001) Role of rapsyn tetratricopeptide repeat and coiled‐coil domains in self‐association and nicotinic acetylcholine receptor clustering. J Biol Chem 276:7475–7483. [DOI] [PubMed] [Google Scholar]
- 8. Gray AN, Egan AJF, Van't Veer IL, Verheul J, Colavin A, Koumoutsi A, Biboy J, Altelaar AFM, Damen MJ, Huang KC, Simorre JP, Breukink E, Den Blaauwen T, Typas A, Gross CA, Vollmer W (2015) Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Elife 4:e07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krachler AM, Sharma A, Kleanthous C (2010) Self‐association of TPR domains: Lessons learned from a designed, consensus‐based TPR oligomer. Proteins 78:2131–2143. [DOI] [PubMed] [Google Scholar]
- 10. Zeytuni N, Zarivach R (2012) Structural and functional discussion of the tetra‐trico‐peptide repeat, a protein interaction module. Structure 20:397–405. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D (2010) The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo‐dimer interface similar to Cdc27. EMBO J 29:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thebault P, Chirgadze DY, Dou Z, Blundell TL, Elowe S, Bolanos‐Garcia VM (2012) Structural and functional insights into the role of the N‐terminal Mps1 TPR domain in the SAC (spindle assembly checkpoint). Biochem J 448:321–328. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Z, Roe SM, Diogon M, Kong E, El Alaoui H, Barford D (2010) Molecular structure of the N‐terminal domain of the APC/C subunit Cdc27 reveals a homo‐dimeric tetratricopeptide repeat architecture. J Mol Biol 397:1316–1328. [DOI] [PubMed] [Google Scholar]
- 14. Lee S, Thebault P, Freschi L, Beaufils S, Blundell TL, Landry CR, Bolanos‐Garcia VM, Elowe S (2012) Characterization of spindle checkpoint kinase Mps1 reveals domain with functional and structural similarities to tetratricopeptide repeat motifs of Bub1 and BubR1 checkpoint kinases. J Biol Chem 287:5988–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han D, Kim K, Kim Y, Kang Y, Lee JY, Kim Y (2009) Crystal structure of the N‐terminal domain of anaphase‐promoting complex subunit 7. J Biol Chem 284:15137–15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu Y, Sha B (2006) Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol 13:589–593. [DOI] [PubMed] [Google Scholar]
- 17. Jínek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E (2004) The superhelical TPR‐repeat domain of O‐linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol 11:1001–1007. [DOI] [PubMed] [Google Scholar]
- 18. Taube M, Pieńkowska JR, Jarmołowski A, Kozak M (2014) Low‐resolution structure of the full‐length barley (Hordeum vulgare) SGT1 protein in solution, obtained using small‐angle X‐ray scattering. PLoS One 9:e93313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dohm JA, Lee SJ, Hardwick JM, Hill RB, Gittis AG (2004) Cytosolic domain of the human mitochondrial fission protein Fis1 adopts a TPR fold. Proteins 54:153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lees JPB, Manlandro CM, Picton LK, Tan AZE, Casares S, Flanagan JM, Fleming KG, Hill RB (2012) A designed point mutant in fis1 disrupts dimerization and mitochondrial fission. J Mol Biol 423:143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S (2017) The ClusPro web server for protein‐protein docking. Nat Protoc 12:255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, Vajda S (2013) How good is automated protein docking? Proteins 81:2159–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozakov D, Brenke R, Comeau SR, Vajda S (2006) PIPER: an FFT‐based protein docking program with pairwise potentials. Proteins 65:392–406. [DOI] [PubMed] [Google Scholar]
- 24. Comeau SR, Gatchell DW, Vajda S, Camacho CJ (2004) ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20:45–50. [DOI] [PubMed] [Google Scholar]
- 25. Comeau SR, Gatchell DW, Vajda S, Camacho CJ (2004) ClusPro: a fully automated algorithm for protein‐protein docking. Nucleic Acids Res 32:W96–W99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM (1999) The dynamin‐related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol 1:298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mozdy AD, McCaffery JM, Shaw JM (2000) Dnm1p GTPase‐mediated mitochondrial fission is a multi‐step process requiring the novel integral membrane component Fis1p. J Cell Biol 151:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karren MA, Coonrod EM, Anderson TK, Shaw JM (2005) The role of Fis1p‐Mdv1p interactions in mitochondrial fission complex assembly. J Cell Biol 171:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tieu Q, Nunnari J (2000) Mdv1p is a WD repeat protein that interacts with the dynamin‐related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol 151:353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N (2003) The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR‐like helix bundle. J Mol Biol 334:445–458. [DOI] [PubMed] [Google Scholar]
- 31. Suzuki M, Neutzner A, Tjandra N, Youle RJ (2005) Novel structure of the N terminus in yeast Fis1 correlates with a specialized function in mitochondrial fission. J Biol Chem 280:21444–21452. [DOI] [PubMed] [Google Scholar]
- 32. Aksel T, Barrick D (2009) Chapter 4 Analysis of repeat‐protein folding using nearest‐neighbor statistical mechanical models, 1st ed. Elsevier Inc, pp. 95–125. [DOI] [PMC free article] [PubMed]
- 33. Kajander T, Cortajarena AL, Main ERG, Mochrie SGJ, Regan L (2005) A new folding paradigm for repeat proteins. J Am Chem Soc 127:10188–10190. [DOI] [PubMed] [Google Scholar]
- 34. Jofuku A, Ishihara N, Mihara K (2005) Analysis of functional domains of rat mitochondrial Fis1, the mitochondrial fission‐stimulating protein. Biochem Biophys Res Commun 333:650–659. [DOI] [PubMed] [Google Scholar]
- 35. Serasinghe MN, Yoon Y (2008) The mitochondrial outer membrane protein hFis1 regulates mitochondrial morphology and fission through self‐interaction. Exp Cell Res 314:3494–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D (2004) Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem 279:21749–21758. [DOI] [PubMed] [Google Scholar]
- 37. Krachler AM, Sharma A, Cauldwell A, Papadakos G, Kleanthous C (2010) TolA modulates the oligomeric status of YbgF in the bacterial periplasm. J Mol Biol 403:270–285. [DOI] [PubMed] [Google Scholar]
- 38. Efimov AV (1991) Structure of α‐α‐hairpins with short connections. Protein Eng 4:245–250. [DOI] [PubMed] [Google Scholar]
- 39. Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, Butt TR (2004) SUMO fusions and SUMO‐specific protease for efficient expression and purification of proteins. J Struct Funct Genomics 5:75–86. [DOI] [PubMed] [Google Scholar]
- 40. Takekoshi T, Ziarek JJ, Volkman BF, Hwang ST (2012) A locked, dimeric CXCL12 variant effectively inhibits pulmonary metastasis of CXCR4‐expressing melanoma cells due to enhanced serum stability. Mol Cancer Ther 11:2516–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cortajarena AL, Mochrie SGJ, Regan L (2011) Modulating repeat protein stability: the effect of individual helix stability on the collective behavior of the ensemble. Protein Sci 20:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 43. Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K (1995) The program XEASY for computer‐supported NMR spectral analysis of biological macromolecules. J Biomol NMR 6:1–10. [DOI] [PubMed] [Google Scholar]
- 44. Karpenahalli MR, Lupas AN, Söding J (2007) TPRpred: a tool for prediction of TPR‐, PPR‐ and SEL1‐like repeats from protein sequences. BMC Bioinformatics 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hekkelman ML, Te Beek TAH, Pettifer SR, Thorne D, Attwood TK, Vriend G (2010) WIWS: a protein structure bioinformatics Web service collection. Nucleic Acids Res 38:W719–W723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figures.


