Abstract
The accumulation associated protein (Aap) of Staphylococcus epidermidis mediates intercellular adhesion events necessary for biofilm growth. This process depends upon Zn2+‐induced self‐assembly of G5 domains within the B‐repeat region of the protein, forming anti‐parallel, intertwined protein “ropes” between cells. Pleomorphism in the Zn2+‐coordinating residues was observed in previously solved crystal structures, suggesting that the metal binding site might accommodate other transition metals and thereby support dimerization. By use of carefully selected buffer systems and a specialized approach to analyze sedimentation velocity analytical ultracentrifugation data, we were able to analyze low‐affinity metal binding events in solution. Our data show that both Zn2+ and Cu2+ support B‐repeat assembly, whereas Mn2+, Co2+, and Ni2+ bind to Aap but do not support self‐association. As the number of G5 domains are increased in longer B‐repeat constructs, the total concentration of metal required for dimerization decreases and the transition between monomer and dimer becomes more abrupt. These characteristics allow Aap to function as an environmental sensor that regulates biofilm formation in response to local concentrations of Zn2+ and Cu2+, both of which are implicated in immune cell activity.
Keywords: accumulation associated protein, Staphylococcus epidermidis, biofilm, intercellular adhesion, self‐assembly, transition metal, analytical ultracentrifugation
Short abstract
Introduction
Staphylococci are responsible for a large percentage of healthcare‐associated infections; in particular, Staphylococcus epidermidis is the primary causative agent of device‐related infections.1, 2 A major complicating factor in these infections is the propensity of staphylococci to form biofilms, specialized colonies of bacteria that adhere to a surface. Bacteria growing in a biofilm are highly resistant to antibiotic action and immune cell responses.1 The accumulation associated protein (Aap) of S. epidermidis is a key cell wall‐anchored (CWA) protein necessary for intercellular adhesion throughout biofilm growth and maturation.3 Aap‐deficient strains of S. epidermidis fail to form biofilms in culture,3 and Aap was shown to be essential for S. epidermidis infection in a rat catheter model in vivo.4
Aap is a remarkable multi‐functional protein that mediates host cell attachment but can also use environmental cues to switch into the primary intercellular adhesion protein during biofilm formation. Aap forms a highly elongated, thread‐like filament that projects outward from the staphylococcal cell wall via an extended stalk,5, 6 with multiple distinct domains that mediate various functions [Fig. 1(A)]. The N‐terminal region of Aap includes short unstructured A‐repeats followed by a lectin domain; this region has been implicated in mediating host cell attachment.9 In contrast, the downstream B‐repeat superdomain, comprising 5–17 tandem B‐repeats, is responsible for intercellular adhesion in the biofilm.10 The B‐repeat region becomes unmasked and can mediate intercellular adhesion after the secreted S. epidemidis protease SepA cleaves Aap between the lectin and B‐repeat regions.11 Each 128‐aa B‐repeat consists of a 78‐residue G5 subdomain and a 50‐residue spacer subdomain.12 Crystal structures of Aap B‐repeats revealed a highly elongated fibrillar structure primarily composed of long 3‐stranded β‐sheets separated by triple‐helix‐like coils [Fig. 1(B)].7, 8
Figure 1.
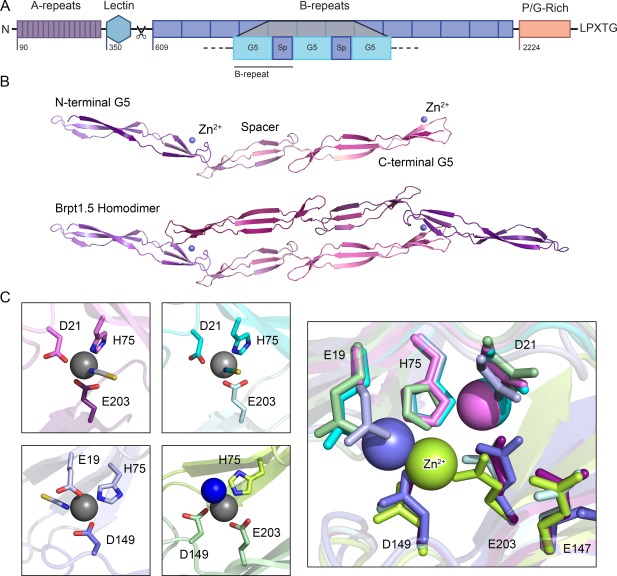
An overview of the structural characteristics of Aap. (A) The distinct regions of Aap include 16 short A‐repeats, a globular lectin domain, between 5–17 tandem B‐repeats (each of which contains a metal‐binding G5 domain followed by a spacer domain, denoted as “Sp”), a proline/glycine‐rich stalk region and a LPXTG cell wall anchoring motif. The A‐repeats and lectin domains are proteolytically processed to expose the B‐repeat superdomain, which can then assemble with B‐repeat regions on other staphylococcal cells to mediate intercellular adhesion in the biofilm. (B) Previously solved crystal structures of the Brpt1.5 construct showing the monomer alone and the Zn2+‐induced dimer.7, 8 Each G5 domain consists of two free‐standing, 3‐stranded β‐sheets separated by a triple‐helix‐like twist; the spacer domain adopts a similar but shorter version of the same fold. The metal‐binding sites are found on the face of the second β‐sheet in each G5 domain (identified by a blue metal ion, labeled Zn2+). (C) Dimerization occurs upon coordination of the Zn2+ ion in trans, creating a tetrahedral binding site with two ligands from one G5 domain and a third ligand from the opposite protomer. The fourth site is occupied by a solvent molecule. Comparing four crystal structures of Brpt1.5 bound to Zn2+, we observed three distinct coordination schemes, each of which was validated in solution by mutagenesis and analytical ultracentrifugation.7 This pleomorphism in metal ion coordination is likely the underlying basis for the ability of the B‐repeat region to self‐assemble with either Zn2+ or Cu2+.
We demonstrated that the critical environmental factor required for B‐repeat self‐assembly and biofilm formation is the divalent transition metal cation Zn2+. In vitro, Zn2+ induces dimerization of B‐repeat proteins. Furthermore, Zn2+ chelation inhibits staphylococcal biofilm growth, but adding back Zn2+ at physiological levels restores the biofilm.12 Several crystal structures of the terminal intact B‐repeat and the capping G5 domain of Aap (called Brpt1.5) revealed the basis for Zn2+‐mediated self‐assembly [Fig. 1(B)].7, 8 One face of each G5 domain uses residues (Glu, Asp, or His) to tetrahedrally coordinate Zn2+ in trans, which brings two Brpt1.5 protomers together to form an anti‐parallel dimer. Structural modeling of longer B‐repeat constructs indicated that Zn2+‐induced assembly would lead to the formation of a twisted, rope‐like structure between staphylococcal cells.7 This mechanism resembles metal‐induced self‐assembly of multidentate ligands such as porphyrin ladders that coordinate metal ions in trans.13 A notable feature of these systems is that the macroscopic affinity of the ligand for the metal ion increases dramatically with increasing number of metal binding sites. This behavior is related to the chelate effect, in which the initial intermolecular assembly event is followed by one or more intramolecular ligation events with minimal entropic cost due to the tethered nature of the reacting species.14 We observe this phenomenon with Aap B‐repeat constructs; short repeats such as Brpt1.5 require millimolar Zn2+ concentrations to assemble, but longer repeats require lower metal concentrations, culminating in the full‐length native B‐repeat region (with up to 17 repeats) requiring only low micromolar Zn2+ for assembly during biofilm formation.12, 15
An interesting feature of the dimeric Brpt1.5 structures was that different crystal forms showed similar overall conformations but distinct coordination schemes, suggesting pleomorphic coordination of Zn2+ by several nearby acidic residues and a histidine side chain [Fig. 1(C)]. This observed variability in both the ligating residues and coordinating bond lengths, combined with flexibility of the B‐repeat fold,7, 8 suggests that the observed metal‐ binding site may not be highly specific and that other bioavailable metal ions might induce self‐assembly.
When defining the search space for other metal ions that might induce Aap self‐assembly, both the chemistry and geometry of the binding pocket must be considered. The highly electronegative residues that form the bulk of the Zn2+ binding site are surrounded by a secondary shell of hydrophobic and uncharged hydrophilic residues. These help to direct each coordinating residue's orientation and tune its pK a values for favorable interactions with the high charge density of the Zn2+ ion.7 The most realistic metal ion candidates need to have similar electrostatic properties, particularly a high positive charge density. In addition, a coordination number of 4 and the potential for adopting tetrahedral geometry appear to be important based on the structural data. Considering these factors, we selected the first transition series of metals: Mn2+, Co2+, Ni2+, and Cu2+ for comparison to Zn2+ to delineate the fundamental mechanism of metal binding and specificity in Aap.
Results
Sedimentation velocity analytical ultracentrifugation (AUC) experiments were designed to test the ability of these transition metal cations to induce Brpt1.5 dimerization. Under typical buffer conditions used previously for these experiments (20 mM Tris, pH 7.4 with 150 mM NaCl), transition metal cations other than Zn2+ were unable to induce Brpt1.5 dimerization.12 The minimal Brpt1.5 construct used to quantitatively define the metal‐induced dimerization event required high Zn2+ concentrations (in the mM range) to induce assembly.12 However, there are significant technical challenges in measuring weak transition metal ion binding events, including strong absorbance of the metal ions across a wide spectral range, limited solubility of metal ions at high pH, and loss of Zn2+‐binding affinity by Brpt1.5 at acidic pH due to protonation of Zn2+‐ligating residues. These issues necessitated optimization of the approach to definitively address metal specificity. Experimentally, sedimentation velocity data were collected using interference optics rather than absorbance optics and data were analyzed using a modified c(s) model with additional floated parameters in the program Sedfit.16 Second, an optimized buffer system (100 mM Tris pH 7.8, 50 mM NaCl) maintained Brpt1.5 in an assembly‐competent pH range (above pH 6.812), while promoting metal ion solubility by a combination of pH buffering capacity and the ability of Tris to act as a weak chelator of divalent cations.
The optimized buffer included only 50 mM NaCl to minimize electrostatic screening, allowing complete dimerization of Brpt1.5 at 2 mM Zn2+ [Fig. 2(A)] compared to 10 mM Zn2+ under the previous conditions.12 However, concentrations of up to 20 mM total Mn2+, Co2+, and Ni2+ were still incapable of inducing any detectable Brpt1.5 assembly [Fig. 2(D–F)]. In contrast, 10 and 20 mM Cu2+ induced self‐assembly of Brpt1.5 [Fig. 2(B,C)]. The reaction boundaries formed in the presence of Cu2+ shifted to the right, indicating a concentration‐dependent increase in Brpt1.5 self‐assembly with rapid exchange. The sedimentation coefficient at 20 mM Cu2+ was still lower than that observed for the Brpt1.5 dimer induced by Zn2+, indicating incomplete dimer assembly in the presence of Cu2+.
Figure 2.
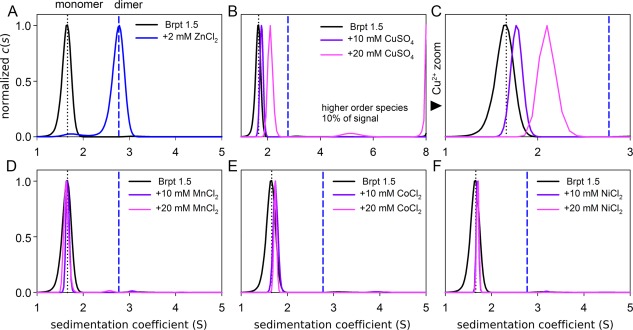
Zn2+ and Cu2+ support Brpt1.5 dimerization; other transition metal ions are not assembly competent. (A) Sedimentation velocity analysis of 8 µM Brpt 1.5 with Zn2+ and (B‐C) Cu2+ reveals dimerization. The dashed black line marks the monomer peak while the heavier dashed blue lines marks the Zn2+‐induced dimer's peak. Increasing concentrations of Cu2+ drive the population toward increasing dimer but are unable to reach the terminal boundary observed with Zn2+. Under identical conditions, (D) Mn2+, (E) Co2+ and (F) Ni2+ induced no appreciable Brpt1.5 assembly, even at high concentrations.
Furthermore, with 20 mM total Cu2+, an additional higher‐order species was observed near 5.2 S comprising about 10% of the total signal [Fig. 2(B)]. Significant higher‐order aggregation was also observed at 20 mM Cu2+, precluding analysis of higher Cu2+ concentrations. These data demonstrate that although Mn2+, Co2+, and Ni2+ do not induce Brpt1.5 assembly, Cu2+ supports oligomerization, although it differs from Zn2+ both in its efficacy and the oligomerization states accessible.
The Zn2+ binding site of each G5 domain features a strongly electronegative patch due to a cluster of acidic Glu and Asp side chains surrounding a central metal‐binding residue (varying between His and Glu in different repeats).7, 8 In general, Glu, Asp, and His side chains frequently act as ligands for transition metal ions17; thus, although neither Mn2+, Ni2+, nor Co2+ support Aap self‐assembly, they may still bind to the G5 domain metal‐binding site. Indeed, we observed that each of these metal ions was able to competitively inhibit Zn2+‐mediated Brpt1.5 self‐assembly [Fig. 3(A–C)]. This demonstrates that the Brpt1.5 protein features a relatively nonspecific metal binding site, but that only Zn2+ and Cu2+ are competent to induce self‐assembly. Based on the concentrations of inhibitor metal ions required to inhibit Zn2+‐mediated assembly of Brpt1.5, the apparent binding affinity of the transition metal ions followed the pattern Mn2+ < Co2+ < Ni2+ < Cu2+ < Zn2+, in accordance with that predicted by the Irving‐Williams series.18
Figure 3.
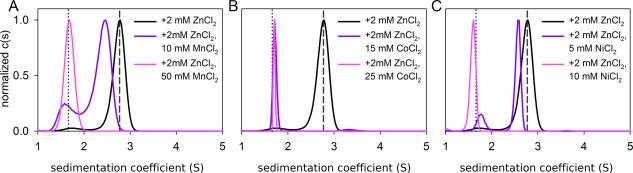
Competition experiments reveal that the assembly‐incompetent transition metal ions bind to Brpt1.5 and competitively inhibit Zn2+‐induced assembly. In each panel, Brpt1.5 formed a dimer in the presence of 2 mM Zn2+ (black trace) that could be inhibited competitively by increasing concentrations (purple to pink traces) of A) Mn2+; B) Co2+; or C) Ni2+.
Both Zn2+ and Cu2+ were capable of inducing Brpt1.5 oligomerization, but the differences in the efficacy and extent of oligomerization raise the question of whether both metal ions act at the same site on Brpt1.5. Thus, we leveraged the previously described E203A mutant of Brpt1.5 that is assembly‐incompetent with Zn2+ 7 to compare assembly induced by both metal ions. The E203A mutant did not assemble in the presence of either Zn2+ [Fig. 4(A)] or Cu2+ [Fig. 4(B)], suggesting that both metals use the same coordination site to induce Brpt1.5 dimerization. The slight rightward shift in the sedimentation coefficient distribution at high Cu2+ concentration [Fig. 4(B)] is similar to that observed with Co2+ and Ni2+ [Fig. 2(E,F)], suggesting that this is likely due to slight changes in sedimentation rate of monomeric Brpt1.5 upon metal binding.
Figure 4.
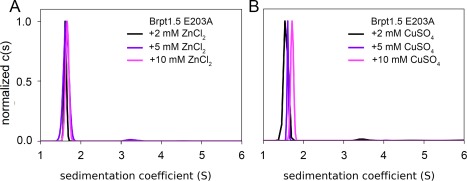
The E203A mutant of Brpt1.5 fails to assemble with either Zn2+ or Cu2+. (A) The Brpt1.5 E203A mutant7 is assembly‐incompetent even at high concentrations of Zn2+. (B) Likewise, the E203A mutant fails to assemble in the presence of Cu2+, indicating that Cu2+ and Zn2+‐induced assembly events occur via the same metal binding site targeted by the E203A mutation.
The Cu2+‐mediated assembly of Brpt1.5 is quite interesting and potentially relevant biologically, given that the bioavailability of Cu2+ in serum is comparable to that of Zn2+.19 To further evaluate Aap assembly with a physiologically relevant construct, we compared the assembly properties of a longer Brpt5.5 construct in the presence of Zn2+ and Cu2+. Brpt5.5 contains five intact B‐repeats and the C‐terminal capping G5 (i.e., six total G5 domains instead of two); this is the minimal number of repeats that can support biofilm formation at the bacterial cell surface with the orthologous protein SasG.20 Use of the longer construct increased apparent metal affinity, as expected based on the chelate effect.12 The lower metal concentrations required for assembly allowed the use of an even more optimal buffer system containing 100 mM MOPS (pH 7.2) with 50 mM NaCl to minimize metal interaction with the buffer and increase metal solubility, but 20 mM Tris (pH 8.6) was still required to solubilize the metal ions in the Cu2+ sample. The buffer mismatch model (BMM) in Sedfit was needed to successfully fit the sedimentation data due to the mismatch between sample and buffer and co‐sedimentation of metal ions under these conditions [Fig. 5(A,B)].21
Figure 5.
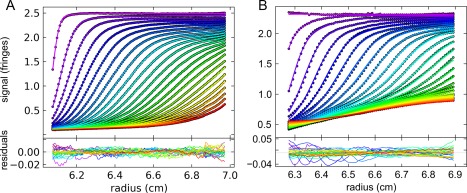
Use of the Buffer Mismatch Model (BMM) in Sedfit allows for correction of nonoptimal sedimentation velocity conditions. (A) A typical c(s) interference data set from a Brpt1.5 experiment in metal‐free MOPS buffer. Circles show every 10th data point of every 3rd scan from the first half of the loaded data set, superimposed by lines showing the modeled fit. (B) The same selection of data from the Brpt5.5 sample with 4 mM CuSO4 is an example of a case where the BMM rescues a condition that would not be accessible using traditional c(s) methods. Note the distinct differences in the shape of the boundaries and the abnormal baseline caused by co‐sedimentation of the Cu2+ ions and mismatch with the buffer blank in the reference sector.
Using the MOPS buffer system, Brpt5.5 in the presence of 1 mM Zn2+ sedimented with a reaction boundary consistent with a dimer species [Fig. 6(A), dashed blue line], based on 2D size‐and‐shape analysis of the 3.9 S peak.22, 23 In contrast to Brpt1.5 assembly, which terminates at the dimer, increasing concentrations of Zn2+ drive Brpt5.5 to assemble into even larger oligomers (6.2 S). Cu2+ also induced Brpt5.5 oligomerization, although higher Cu2+ concentrations (4 mM Cu2+ vs. 1 mM Zn2+) were required to produce a reaction boundary approaching the 3.9 S peak observed with Zn2+ [Fig. 6(B)]. Furthermore, the transition from monomer to oligomer occurred over an extremely narrow range of Cu2+ concentrations: an increase of less than 0.5 mM Cu2+ was sufficient to shift Brpt5.5 from mostly monomer to mostly putative dimer (from 3.65 to 4.00 mM Cu2+). Metal‐induced self‐assembly is a form of linked equilibrium between ligand binding and protein assembly.15, 24 A general feature of such equilibria is that the greater the number of ligands linked to assembly, the steeper the transition between monomer and oligomer.25 We previously demonstrated with Brpt1.5 and Brpt2.5 constructs that binding of 1–2 Zn2+ ions per G5 domain is linked to dimerization.12 The steep transition observed for Cu2+ suggests that a greater number of Cu2+ ions are bound during Brpt5.5 assembly compared to Zn2+.
Figure 6.
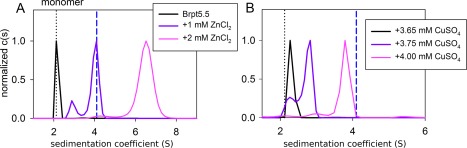
The longer Brpt5.5 construct has a higher affinity for both Zn2+ and Cu2+ and is more sensitive to changes in metal concentration. Sedimentation velocity analysis of 12 µM Brpt5.5 in the presence of both Zn2+ and Cu2+ reveals more extensive assembly than observed with Brpt1.5. In each panel, the dotted black line shows the S value of the Brpt5.5 monomer, while the blue dashed line marks the location of the primary reaction boundary induced by 1 mM Zn2+. (A) A Zn2+ concentration of 1 mM results in Brpt5.5 assembly consistent with a putative dimer, whereas addition of 2 mM Zn2+ to Brpt5.5 drives the reaction boundary into a higher‐order assembly state. (B) Brpt5.5 shows a stark transition from nearly complete monomer toward a putative dimer over a very narrow concentration range (from 3.65 mM to 4.00 mM total Cu2+).
Comparison of the zinc and copper AUC data yields hints about the conformation of these B‐repeat:metal complexes. In addition to the sedimentation coefficient (S), sedimentation velocity data yields information on the weight‐average frictional ratio (f/f 0) for the sample. This ratio is a shape parameter that describes the deviation of the sedimenting particle from an ideal sphere (which would have an f/f 0 value of 1.0). Nearly spherical globular proteins have frictional ratios near 1.2, while elongated species have values of 1.6 or higher.26 In previous analyses, we observed that the f/f 0 value for free Brpt1.5 was near 1.75 (highly elongated); the ratio decreased to around 1.5 upon Zn2+‐induced dimerization. This is consistent with the highly elongated crystal structures of free B‐repeat constructs and the structure of the Zn2+‐induced Brpt1.5 dimer, which forms a side‐by‐side antiparallel arrangement that is wider than the extended monomer.7, 8 In the current study we observed a similar trend in the frictional ratios as Brpt1.5 shifted from monomer toward dimer, suggesting a similar conformation in both Zn2+‐ and Cu2+‐induced Brpt1.5 dimers. This would be expected if the Cu2+ is bound in the same electronegative pocket to which Zn2+ binds (as demonstrated with the E203A mutant in Fig. 4), with a similar overall dimer configuration.
The case is different for the Brpt5.5 construct. In the absence of metal, monomeric Brpt5.5 sedimented with a frictional ratio of 3.2 (extremely elongated). As seen for Brpt1.5, the Zn2+‐induced transition from monomer to apparent dimer at 3.9 S leads to a decrease in f/f 0, yielding a value of 2.2. However, the transition from putative dimer to higher‐order oligomers then reverses the trend; the weight‐average f/f 0 increases from 2.2 up to 2.9 in samples with the highest metal concentrations. This trend in the f/f 0 suggests that successive Brpt5.5 protomers might form larger assemblies (beyond dimer) by staggering their register to create even longer complexes, rather than assembling side‐by‐side, which would increase the width relative to the length and thereby reduce the frictional ratio even further than that seen in the dimer. The 3.9 S reaction boundary may not exclusively reflect a dimer population but instead might represent a reaction boundary consisting, for example, of monomers, dimers, and higher‐order oligomers in rapid exchange. Regardless, the increasing trend of weight‐averaged f/f 0 values as Zn2+ concentrations increase would suggest that higher‐order oligomers are increasingly more elongated.
Hydrodynamic modeling [Fig. 7(A)] using the four Brpt1.5 crystal structures previously solved in our lab7 predicts the sedimentation coefficient of monomeric Brpt1.5 to range from 1.63–1.72 S and the f/f 0 to be 1.73–1.75, values that correspond closely to the experimentally determined parameters (s = 1.69 S; f/f 0 = 1.75). Modeling of the Brpt1.5 Zn2+‐induced dimer [Fig. 7(B)] predicted a sedimentation coefficient of 2.66–2.72 S and a decrease in f/f 0 to 1.57–1.68, also similar to experimental values (s = 2.75 S; f/f 0=1.55). While no crystal structures of Brpt5.5 have been reported, using the suite of existing Brpt1.5 variant structures7, 8 it was possible to construct homology models [Fig. 7(C)]. Atomic shell modelling, a more computationally intensive approach than that used for Brpt1.5, predicts a sedimentation coefficient of 2.26 S, which is fairly close to the experimental value of 2.15 S. Although quasi‐rigid modeling of flexible proteins such as Brpt1.5 are only estimations, the simulation results help correlate monomer and dimer to both observed reaction boundaries of Brpt1.5 and verify that an elongated configuration of monomeric Brpt5.5 is consistent with its sedimentation behavior.
Figure 7.
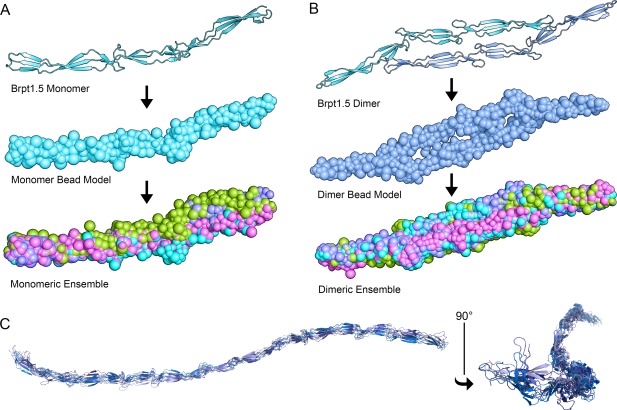
Hydrodynamic modeling of Brpt1.5 and Brpt5.5. Brpt1.5 crystal structures7 were used for hydrodynamic modeling of monomer and dimer species. (A) A low‐resolution bead model of the Brpt1.5 monomer is created from the coordinates of each crystal structure and combined into an ensemble used to predict the average sedimentation coefficient and frictional ratio. (B) Structures of the Zn2+‐induced Brpt1.5 dimer7 were used to create a bead model to estimate the sedimentation parameters of a homogenous Brpt1.5 dimer population. (C) Since no structure of Brpt5.5 has currently been solved, various Brpt1.5 structures7, 8 were combined to create an ensemble of homology models for the longer construct, yielding predicted sedimentation parameters that were similar to experimental values.
Discussion
The coordination number (CN) of a metal ion describes the preferred number of coordinating ligands (e.g., protein side chains). In metal‐protein complexes, Mn2+ can be found with a CN of 5 or 6, while both Co2+ and Ni2+ prefer a CN of 6, typically octahedral geometry.27 The only two metals that induced Aap B‐repeat assembly, even under optimized conditions, were Zn2+ and Cu2+. Zn2+ is almost exclusively found with a CN of 4 (tetrahedral coordination), whereas Cu2+ can adopt a CN of 3 or 4 and shows a preference for square planar geometry in metal‐protein complexes.27 However, protein‐imposed geometry can create a tetrahedral Cu2+ coordination site.28 Plastocyanins,29, 30 stellacyanin,31 and nitrite reductase32 are a few examples that demonstrate tetrahedral geometry in a Cu2+ binding site. Energetically, tetrahedral coordination by Cu2+ is less favorable, which may explain the lower relative affinity both Brpt1.5 and Brpt5.5 show for Cu2+ versus Zn2+ despite their nearly identical residue coordination distances,17, 27 similar atomic radii,33 and high charge densities. This also suggests that the selectivity of the G5 domains for transition metal ions comes mostly from their ability to adopt tetrahedral geometry rather than other properties of the cation.
In contrast to the specificity observed with metal‐induced B‐repeat assembly, all tested transition metal ions were capable of binding to the G5 domain. This is perhaps not surprising, given that Asp, Glu, and His are preferred ligands for each of these metals, and these residues are highly enriched in the pleomorphic G5 binding site.17 Although Mn2+, Co2+, and Ni2+ at high concentrations act as competitive inhibitors for Zn2+‐induced B‐repeat assembly in vitro, none of these is likely to inhibit assembly in vivo. S. epidermidis can form Aap‐mediated biofilms at the low micromolar Zn2+ concentrations present in serum.12 Considering that the bioavailability of Zn2+ (reported as 11.2–15.1 µM) and Cu2+ (15.1 µM) in blood serum is much higher than that of Mn2+ (28.4 nM), Co2+ (2.44 nM), and Ni2+ (44.6 nM),19 none of the latter metal ions is likely to inhibit assembly in vivo.
In summary, the linked equilibrium between metal binding and self‐assembly, combined with the strong chelate effect observed for tandem B‐repeats, allows Aap to act as a sensor molecule to regulate biofilm formation based on local Zn2+ or Cu2+ levels. Zn2+ plays a critical role in the function of a wide range of leukocytes34 and granulocytes store high levels of Zn2+,35 released upon degranulation. Likewise, serum Cu2+ levels increase during infection due to ceruloplasmin activity; macrophages use Cu2+ to enhance killing of pathogens in the phagolysosome.36 Thus, Zn2+ and Cu2+ regulate the host immune response and represent a danger signal to staphylococci. Metal‐induced Aap assembly is a mechanism to sense local immune activation and respond by initiating biofilm formation. At the same time, the high surface density of cell‐surface Aap may act as a local sink for metals critical to immune cell function, adding an additional hurdle to clearance of the infection.
Materials and Methods
Molecular cloning
Brpt1.5 and Brpt1.5 E203A constructs were generated from previously described Invitrogen Gateway System entry vectors.7, 12 Brpt5.5 is an elongated B‐repeat construct from Aap containing the six most C‐terminal G5 domains (residues 1505–2223) from Staphylococcus epidermidis RP62A (AAW53239.1) with an N‐terminal tobacco etch virus (TEV) protease cleavage site. The final construct in the destination vector pHisMBP‐DEST encoded an N‐terminal hexahistidine‐tagged maltose binding protein (His‐MBP) followed by the TEV protease cut site and terminating with the B‐repeats.
Protein expression and purification
Novagen BLR DE3 Escherichia coli competent cells were transformed with destination B‐repeat plasmids and grown to an OD600 of 0.9–1.2 at 37°C. Cultures were cooled to 10°C using an ice bath before the addition of 200 µM Isopropyl β‐D‐thiogalactopyranoside (IPTG) and 2% v/v ethanol. Cultures were shaken overnight at 20°C. Centrifugation at 4200 rpm yielded pellets that were then sonicated. Protein was purified from filtered lysate by passage over a Ni‐affinity column containing His‐Trap Chelating resin from GE Healthcare Life Sciences. Addition of TEV protease and dithiothreitol (DTT) at a final concentration of 0.2 µM released the His‐MBP. After cleavage, free MBP was removed by passage over the same HisTrap column followed by a polishing passage over a Superdex 75 or 200 gel filtration column for the Brpt1.5 and Brpt5.5 constructs, respectively.
Analytical ultracentrifugation
Sedimentation velocity (SV) experiments with Brpt1.5 (at 8 µM) and Brpt5.5 (at 12 µM) were conducted on the Beckman XL‐I analytical ultracentrifuge using ProteomeLab 6.2 for scan collection with or without various transition metal cations. The An‐60 Ti rotor was run at 48,000 rpm at 20°C until complete sedimentation of sample occurred. The Rayleigh interference optical detection system was used for samples with metal ions that absorbed at 235 nm: Co2+, Ni2+, and Cu2+. Samples were dialyzed into 50 mM tris(hydroxymethyl)aminomethane (Tris) pH 7.6 with 50 mM NaCl in the case of the Brpt1.5 construct or either 50 mM 3‐(N‐morpholino)propanesulfonic acid (MOPS) pH 7.2 with 50 mM NaCl or 4‐(N‐Morpholino)butanesulfonic acid (MOBS) pH 7.2 with 50 mM NaCl and 10 mM Tris pH 8.5 for the Brpt5.5 construct. The respective metals were added to the samples and the pH was verified just before loading into the cell. The high metal concentrations required were found to be incompatible with overnight dialysis; during the process of dialysis we observed significant metal hydroxide precipitate. Simply adjusting to a lower pH was not a possibility due to the range in which Brpt1.5 is capable of assembly.12 By adding the metals after dialysis and eliminating the temperature change, metals stayed fully solubilized throughout each AUC run and maintained the protein in its functional range. Our previous quantitative studies of Zn2+‐induced Brpt1.5 assembly were conducted with defined free Zn2+ concentrations after dialysis.7, 8, 12 Even though that approach was not possible in the current study, the metal ions used here (1–50 mM) were in large excess over protein (8–12 μM), so the total M2+ concentrations approximate the free M2+ concentrations.
Buffer optimization
Tris is known to weakly chelate metals,37 which can help to solubilize certain metals but also complicates determination of the true free metal concentration and the exact concentration required to reach the fully assembled reaction boundary. Chelation can be largely removed by the use of alternate buffering agents, MOPS or its derivative MOBS.38 Repeating the sedimentation velocity experiment with Brpt1.5 in the presence of Zn2+ using MOPS showed only minor differences in the protein's sensitivity to metal. No difference was seen with MOPS versus MOBS. Taken together, these experiments suggested that weak chelation by Tris was not inhibiting assembly of Brpt1.5 in the presence of Mn2+, Co2+, and Ni2+; rather, these metals are not competent to support Brpt1.5 self‐assembly.
The situation becomes more complex when considering Cu2+. Its solubility is much lower in MOPS at the target pH. The Ksp of zinc hydroxide is 4.1 × 10−17 while that of copper hydroxide is only 1.6 × 10−19. This insolubility at physiological pH requires copper to be associated with other biological molecules, rather than existing as free copper. An ideal carrier for the Brpt5.5 assembly experiments would be capable of very weakly chelating the Cu2+ ions to maintain solubility but would not outcompete binding to the G5 metal binding site, thus still permitting B‐repeat assembly. Based on its ability to maintain Cu2+ solubility in the Brpt1.5 experiments, we chose Tris as the carrier. Titrations were performed to find the lowest concentration of Tris required to solubilize copper in the desired concentration range of 1–5 mM Cu2+. By separating the buffering and chelating functions, we were able to minimize effects on apparent affinity between Brpt5.5 and the copper while still allowing for enough buffering capacity to keep the experiment within a functional pH range and the Cu2+ fully soluble.
Sedimentation velocity data analysis
SV experiments were analyzed using Sedfit 15.01b.16 Figures were prepared using GUSSI 1.0.8d.39 Ideally, when using interference optical detection, complete subtraction of the reference from the sample signal eliminates any unwanted buffer signal offsets arising from co‐solvent molecules. However, because the system's limitations required addition of the metals post‐dialysis, a buffer mismatch is inevitable. It is possible to computationally correct for this by modeling co‐solute redistribution with specific Lamm Equation solutions implemented in Sedfit.21 The BMM accounts for any concentration difference as a ratio of sample sector to reference sector with a value of 1.0 equating to a perfect concentration match. The value for c 0* (sample)/ (reference) was determined as a floating parameter in a separate fit after radial and time‐invariant noise were subtracted. Similarly, because of small volume differences due to addition of the metal ions, a different meniscus position is expected in the sample and reference sectors. In cases when this mismatch was optically relevant, it was modeled using the Δm parameter, the difference between sample and reference sector menisci, as a fixed value based on the fringe interference. By applying just two conditions of the BBM, it was possible to recover reliable data from a system that initially appeared experimentally intractable.
Hydrodynamic and homology modeling
The Solution Modeler module of UltraScan 9.9 Rev 3087 40 was used to predict the hydrodynamic parameters using the coordinates from four Brpt1.5 crystal structures (PDB codes: 4FUN, 4FUM, 4FUO, 4FUP).7 In the case of 4FUP, both chains in the asymmetric unit were isolated and run separately. The SOMO overlapping bead model was used for all simulations. The Brpt5.5 homology models were constructed using Modeller 9.17 41 with 4FUN, 4FUO, 4FUP, 5TU7, 5TU9, 4FUM, 5TU8 7, 8 as templates in decreasing order of coverage. Of the twenty models created, the five with the best GA341 and lowest zDOPE scores were selected for the ensemble. Atomic shell modelling was performed using Hydropro 10.42
Conflict of Interest
ABH is a co‐inventor on a patent related to inhibition of biofilm formation by Zn2+ chelators.
Acknowledgments
We thank Dr. Deb Conrady, Dr. Catie Shelton, and Alex Yarawsky for helpful discussions.
Broad Audience Statement: Staphylococci resist immune responses by forming biofilms, dense surface‐adherent bacterial colonies. Intercellular adhesion in S. epidermidis biofilms occurs via Zn2+‐dependent self‐assembly of the accumulation associated protein (Aap), forming intertwined filaments between cells. We now show that Cu2+ also supports self‐assembly, whereas other transition metals (Mn2+, Co2+, and Ni2+) bind to Aap but do not support assembly. Thus, Aap acts as a sensor of Zn2+ and Cu2+, critical regulators of the immune response.
References
- 1. Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. [DOI] [PubMed] [Google Scholar]
- 2. Allegranzi B (2011) Report on the burden of endemic Health Care‐Associated Infection Worldwide. World Health Organization, Geneva, Switzerland. ISBN 978 92 4 150150 7. [Google Scholar]
- 3. Hussain M, Herrmann M, von Eiff C, Perdreau‐Remington F, Peters G (1997) A 140‐kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun 65:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schaeffer CR, Woods KM, Longo GM, Kiedrowski MR, Paharik AE, Buttner H, Christner M, Boissy RJ, Horswill AR, Rohde H, Fey PD (2015) Accumulation‐associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect Immun 83:214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banner MA, Cunniffe JG, Macintosh RL, Foster TJ, Rohde H, Mack D, Hoyes E, Derrick J, Upton M, Handley PS (2007) Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation‐associated protein. J Bacteriol 189:2793–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yarawsky AE, English LR, Whitten ST, Herr AB (2017) The proline/glycine‐rich region of the biofilm adhesion protein aap forms an extended stalk that resists compaction. J Mol Biol 429:261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conrady DG, Wilson JJ, Herr AB (2013) Structural basis for Zn2+‐dependent intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA 110:E202–E211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shelton CL, Conrady DG, Herr AB (2017) Functional consequences of B‐repeat sequence variation in the staphylococcal biofilm protein Aap: deciphering the assembly code. Biochem J 474:427–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macintosh RL, Brittan JL, Bhattacharya R, Jenkinson HF, Derrick J, Upton M, Handley PS (2009) The terminal A domain of the fibrillar accumulation‐associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J Bacteriol 191:7007–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, Mack D (2005) Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation‐associated protein by staphylococcal and host proteases. Mol Microbiol 55:1883–1895. [DOI] [PubMed] [Google Scholar]
- 11. Paharik AE, Kotasinska M, Both A, Hoang TN, Buttner H, Roy P, Fey PD, Horswill AR, Rohde H (2017) The metalloprotease SepA governs processing of accumulation‐associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis . Mol Microbiol 103:860–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB (2008) A zinc‐dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA 105:19456–19461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taylor PN, Anderson HL (1999) Cooperative self‐assembly of double‐strand conjugated porphyrin ladders. J Am Chem Soc 12:11538–11545. [Google Scholar]
- 14. Page MI, Jencks WP (1971) Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci USA 68:1678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herr AB, Conrady DG (2011) Thermodynamic analysis of metal ion‐induced protein assembly. Methods Enzymol 488:101–121. [DOI] [PubMed] [Google Scholar]
- 16. Schuck P (2000) Size‐distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78:1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harding MM, Hsin KY (2014) Mespeus–a database of metal interactions with proteins. Methods Mol Biol 1091:333–342. [DOI] [PubMed] [Google Scholar]
- 18. Irving H, Williams RJP (1948) Order of stability of metal complexes. Nature 162:746–747. [Google Scholar]
- 19. Schultze B, Lind PM, Larsson A, Lind L (2014) Whole blood and serum concentrations of metals in a Swedish population‐based sample. Scand J Clin Lab Invest 74:143–148. [DOI] [PubMed] [Google Scholar]
- 20. Corrigan RM, Rigby D, Handley P, Foster TJ (2007) The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153:2435–2446. [DOI] [PubMed] [Google Scholar]
- 21. Zhao H, Brown PH, Balbo A, Fernandez‐Alonso Mdel C, Polishchuck N, Chaudhry C, Mayer ML, Ghirlando R, Schuck P (2010) Accounting for solvent signal offsets in the analysis of interferometric sedimentation velocity data. Macromol Biosci 10:736–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown PH, Schuck P (2006) Macromolecular size‐and‐shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J 90:4651–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaton CT, Herr AB (2015) Elucidating complicated assembling systems in biology using size‐and‐shape analysis of sedimentation velocity data. Methods Enzymol 562:187–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wyman J, Gill SJ (1990) Binding and linkage: functional chemistry of biological macromolecules. Mill Valley, CA: University Science Books. [Google Scholar]
- 25. Murtaugh ML, Fanning SW, Sharma TM, Terry AM, Horn JR (2011) A combinatorial histidine scanning library approach to engineer highly pH‐dependent protein switches. Protein Sci 20:1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schuck P, Zhao H, Brautigam CA, Ghirlando R (2015) Basic principles of analytical ultracentrifugation, CRC Press, New York. [Google Scholar]
- 27. Harding MM (2001) Geometry of metal‐ligand interactions in proteins. Acta Cryst D57:401–411. [DOI] [PubMed] [Google Scholar]
- 28. Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber‐Emmons MT, Kjaergaard CH, Hadt RG, Tian L (2014) Copper active sites in biology. Chem Rev 114:3659–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garrett TP, Clingeleffer DJ, Guss JM, Rogers SJ, Freeman HC (1984) The crystal structure of poplar apoplastocyanin at 1.8‐A resolution. The geometry of the copper‐binding site is created by the polypeptide. J Biol Chem 259:2822–2825. [DOI] [PubMed] [Google Scholar]
- 30. Diaz‐Moreno I, Diaz‐Quintana A, Diaz‐Moreno S, Subias G, De la Rosa MA (2006) Transient binding of plastocyanin to its physiological redox partners modifies the copper site geometry. FEBS Lett 580:6187–6194. [DOI] [PubMed] [Google Scholar]
- 31. Hart PJ, Nersissian AM, Herrmann RG, Nalbandyan RM, Valentine JS, Eisenberg D (1996) A missing link in cupredoxins: crystal structure of cucumber stellacyanin at 1.6 A resolution. Protein Sci 5:2175–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jacobson F, Pistorius A, Farkas D, De Grip W, Hansson O, Sjolin L, Neutze R (2007) pH dependence of copper geometry, reduction potential, and nitrite affinity in nitrite reductase. J Biol Chem 282:6347–6355. [DOI] [PubMed] [Google Scholar]
- 33. Shannon RD (1976) Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Cryst A 32:751–767. [Google Scholar]
- 34. Wellinghausen N, Rink L (1998) The significance of zinc for leukocyte biology. J Leukoc Biol 64:571–577. [DOI] [PubMed] [Google Scholar]
- 35. Szmigielski S, Litwin J (1965) The histochemical study of zinc content in granulocytes in normal adults and in hematologic disorders. Blood 25:56–62. [PubMed] [Google Scholar]
- 36. Besold AN, Culbertson EM, Culotta VC (2016) The Yin and Yang of copper during infection. J Biol Inorg Chem 21:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fischer BE, Haring UK, Tribolet R, Sigel H (1979) Metal ion/buffer interactions. Stability of binary and ternary complexes containing 2‐amino‐2(hydroxymethyl)‐1,3‐propanediol (Tris) and adenosine 5'‐triphosphate (ATP). Eur J Biochem 94:523–530. [DOI] [PubMed] [Google Scholar]
- 38. Ferreira CMH, Pinto ISS, Soares EV, Soares HMVM (2015) (Un)suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions – a review. RSC Adv 5:30989–31003. [Google Scholar]
- 39. Brautigam CA (2015) Calculations and publication‐quality illustrations for analytical ultracentrifugation data. Methods Enzymol 562:109–133. [DOI] [PubMed] [Google Scholar]
- 40. Brookes E, Demeler B, Rosano C, Rocco M (2010) The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur Biophys J 39:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. [DOI] [PubMed] [Google Scholar]
- 42. Ortega A, Amoros D, Garcia de la Torre J (2011) Prediction of hydrodynamic and other solution properties of rigid proteins from atomic‐ and residue‐level models. Biophys J 101:892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]


