Abstract
Babesia spp. are tick-transmitted haemoparasites causing tick fever in cattle. In Australia, economic losses to the cattle industry from tick fever are estimated at AUD$26 Million per annum. If animals recover from these infections, they become immune carriers. Here we describe a novel multiplex TaqMan qPCR targeting cytochrome b genes for the identification of Babesia spp. The assay shows high sensitivity, specificity and reproducibility, and allows quantification of parasite DNA from Babesia bovis and B. bigemina compared to standard PCR assays. A previously published cytochrome b SYBR Green qPCR was also tested in this study, showing slightly higher sensitivity than the Taqman qPCRs but requires melting curve analysis post-PCR to confirm specificity. The SYBR Green assays were further evaluated using both diagnostic submissions and vaccinated cattle (at 7, 9, 11 and 14 days post-inoculation) showed that B. bigemina can be detected more frequently than B. bovis. Due to fewer circulating parasites, B. bovis detection in carrier animals requires higher DNA input. Preliminary data for a novel fluorescent PCR genotyping based on the Internal Transcribed Spacer 1 region to detect vaccine and field alleles of B. bovis are described. This assay is capable of detecting vaccine and novel field isolate alleles in a single sample.
Keywords: babesiosis, carrier, diagnosis, qPCR, cytochrome b, genotyping, ITS1, vaccines
1. Introduction
Babesia species are tick-borne intra-erythrocytic apicomplexan parasites found in a variety of domestic and wild animals, and in humans. Mixed infections are responsible for widespread morbidity and mortality in livestock of tropical and subtropical regions of the world [1]. Economic losses in cattle due to tick fever (babesiosis and anaplasmosis) are estimated at AUD$26 Million and >US$10 Billion in Australia [2] and worldwide [3] per annum, respectively. The standard method for diagnosis of clinical babesiosis is the microscopic examination of blood smears. Although this method is rapid, inexpensive, and reasonably sensitive in clinical cases, the sensitivity is poor for immune carrier animals because of the low parasitaemia in these animals [4,5]. Serological assays are useful for epidemiological studies and to confirm responses to vaccination, but do not indicate if the animal is persistently infected. Quantitative PCR may be useful for the detection of these latent carrier animals.
Live vaccines for cattle to protect against babesiosis and anaplasmosis have been produced in several countries including South Africa, Israel, Argentina and Australia [6,7]. Following vaccination, the parasite numbers decrease to carrier state levels and are usually no longer detectable in blood smears. In Australia, Tick Fever Centre (TFC) in addition to the production of live tick fever vaccines, also provides diagnostic support and investigations into outbreaks of tick fever in cattle, using microscopy and serology as the principal diagnostic tools. However, quantitative PCR (qPCR) is also a useful tool in this context to discriminate between Babesia bovis and Babesia bigemina infections at very low parasitaemias detected with light microscopy; and for genotype discrimination of the vaccine strain B. bovis from field isolates.
PCR assays are more sensitive than microscopic detection which cannot detect Babesia parasites at early stages of infection and also from immune carrier animals with low levels of parasitaemia [5,8,9]. Several conventional PCR assays have been employed in diagnostic applications and epidemiological studies to detect these parasites with a high degree of sensitivity [10,11,12], but these do not permit the estimation of the initial concentration of target DNA and are at best semi-quantitative [13]. Quantitative PCR has been used to assess disease severity and treatment outcome in various protozoan diseases [14,15]. SYBR Green qPCR detection of cytochrome b genes [16] and TaqMan-based qPCRs detecting 18S ribosomal genes [17] were reported for the detection and quantification of both B. bovis and B. bigemina from Spain and Argentina, and Brazil, respectively.
PCR methods have been invaluable for the differentiation of attenuated vaccine and field isolates supporting tick fever live vaccine production facilities in Australia [18,19], South Africa [20], and Israel [21]. The most significant Babesia spp. associated with these vaccine studies has been B. bovis. In the Australian context, B. bovis causes a much higher incidence of field outbreaks compared with B. bigemina; clinical disease associated with B. bigemina is diagnosed only occasionally. Previous typing studies have focused on single copy genes which may vary due to immune pressure such as the Bv80 and BvVA1 genes [13,20]. A tandem repeat-based multilocus typing system based on 14 B. bovis mini- and micro-satellites was developed demonstrating geographic strain differences, but vaccine and field isolate differentiation was not reported [22].
This paper describes the optimization and development of qPCRs for the identification and differentiation of Babesia spp. (B. bovis and B. bigemina) using Australian isolates. The new multiplex TaqMan qPCR assay was designed to amplify the mitochondrial DNA cytochrome b genes known to have a higher copy number compared with ribosomal RNA genes [23]. Preliminary data for a new genotyping method for B. bovis comparing vaccine and field isolates by amplifying the Internal Transcribed Spacer Sequence 1 region using a novel fluorescence PCR method are also reported [24] analyzed by capillary electrophoresis [25]. This novel data are compared to the single copy BvVA1 PCR assay which is one of the methods routinely used by TFC for the differentiation of vaccine and field isolates.
2. Materials and Methods
2.1. Parasite and Parasite DNA Samples
One isolate of both B. bovis Dixie strain and B. bigemina strain G previously identified by blood smears, standard PCR and sequencing techniques [13], were used as reference strains in the evaluation of all PCR methods described here. Babesia bovis and B. bigemina field strains were either extracted from diagnostic submissions or from previous diagnostic submissions which had been cryopreserved as stabilates at Tick Fever Centre (TFC) as previously described [19]. Babesia bovis diagnostic submissions were also presented to TFC as blood smears (venous blood or capillary blood from organs) on glass slides, or 10 mL blood vacutainers (BD Australia). Vacutainers were also collected from 17 calves vaccinated with Trivalent Tick Fever Vaccine (a live vaccine made with B. bovis Dixie strain, B. bigemina G strain and Anaplasma marginale subsp. centrale) at Days 7, 9, 11 and 14 post-inoculation to compare standard PCR with qPCRs in detecting Babesia parasites (68 samples).
2.2. DNA Isolation and Quantification from Purified Parasites
Babesia bovis (Dixie) and B. bigemina (G strain) positive control parasites were extracted from 75 mL of blood of known parasitaemia. Babesia bigemina and B. bovis infected blood samples were leucocyte filtered using Imugard III blood filters (Terumo BCT, Lakewood, CO, USA) according to the manufacturer’s instructions. Parasites were purified following the OIE method for B. bovis parasite antigen preparation [26]. Parasite DNA was extracted from purified B. bovis and B. bigemina parasites using the QIAamp DNA Blood Mini Kit (QIAGEN Pty Ltd., Victoria, Australia) according to the manufacturer’s instructions and then quantified by NanoDrop 2000 (Thermofisher Scientific Australia Pty Ltd., Victoria, Australia). DNA was subjected to 10-fold dilutions from 10 ng/µL to 1 ag/µL (B. bovis, approximately 3.5 × 106 parasites/10 ng/µL; B. bigemina, approximately 2 × 106 parasites/10 ng/µL). All dilutions of infected red blood cells were prepared in duplicate for the TaqMan qPCR assay in parallel to the SYBR Green qPCR assay and the standard PCR assays. Negative control DNA was isolated from the blood of uninfected cattle.
Purified DNA from B. microti, B. duncani and an unknown Babesia spp. were provided by the Centers for Disease Control and Prevention (Atlanta, GA, USA) to our laboratory previously and utilized as negative controls to evaluate the new PCR assays. DNA from Anaplasma marginale (Dawn strain) and Anaplasma marginale subsp. centrale (anaplasmosis vaccine strain [27]) was prepared from 10 mL EDTA vacutainers. Briefly, the blood samples were centrifuged at 2500× g for 10 min at room temperature, and the plasma and buffy coat layers removed by pipetting. Subsequently 200 µL of the red blood cell pellet was used to extract DNA using the QIAamp DNA Blood Mini Kit (QIAGEN Pty Ltd., Victoria, Australia) according to the manufacturer’s instructions.
2.3. DNA Isolation from Clinical Samples and Vaccinated Cattle
Preparation of samples containing parasites differed depending on the source of the sample which included fresh blood (10 mL EDTA or lithium heparin vacutainers), blood smears on glass slides (for B. bovis field isolates) and cryopreserved stabilates. For the vacutainers, the bloods were centrifuged to allow removal of the plasma and buffy coat, and processed through the QIAamp DNA Blood Mini Kits as described above for Anaplasma spp. For blood smears, the cells were scraped from glass slides using a scalpel blade and re-suspended in 200 µL of phosphate buffered saline (PBS) which was subsequently also processed using the QIAamp DNA Blood Mini Kits. For stabilate DNA extractions using cryopreserved blood samples (2.5 mL parasite infected blood mixed with 2.5 mL 20% polyvinyl-pyrrolidone-40 in PBS stored in liquid nitrogen), the 5 mL stabilate was first centrifuged at 4000× g for 10 min prior to removal of the supernatant, with the pellet re-suspended in 1 mL PBS and re-centrifuged at 10,000× g for 5 min prior to suspension in 200 µL PBS and extracted using the QIAamp DNA Blood Mini Kit (QIAGEN). For the evaluation of the B. bovis genotyping assay, DNA from seven characterized TFC B. bovis stabilates (Table 1) were extracted as described above. Stabilates each have an identifier within TFC’s database described as a capital letter followed by 2 digits e.g., C48, F95, and H03, as shown in Table 1. Table 1 (and Table S1) summarizes the sources and the DNA extraction methods used for all reference and field isolates to confirm assay specificities.
Table 1.
Sources of reference isolates, Babesia bigemina and Babesia bovis vaccine and field samples used to determine specificity of the assays described in this study (details in Table S1).
| Species Strain/No. | Sample Source 1 |
|---|---|
| Reference Isolates: | |
| B. microti-1610 | CDC |
| B. microti-1737 | CDC |
| B. microti-1750 | CDC |
| B. microti-1743 | CDC |
| B. microti-1716 | CDC |
| B. duncani-1671 | CDC |
| Babesia spp. 1749 CDC | CDC |
| A. marginale subsp. central 2 | TFC vaccine |
| A. marginale Dawn strain | TFC stabilate |
| B. bovis-Dixie vaccine strain | TFC vaccine calf |
| B. bigemina-G vaccine strain | TFC vaccine calf |
| B. bovis and B. bigemina field isolates (Species specific qPCR assays): | |
| B. bovis—1–31 | TFC field samples |
| B. bigemina—1–14 | TFC stabilates of field strains |
| B. bovis isolates for genotyping evaluation 3: | |
| B. bovis Dixie vaccine passaged through ticks-H03 4, H10 4 | TFC stabilates |
| B. bovis H92 5 | TFC stabilates of field strains |
| B. bovis H97 6 | |
| B. bovis J40 6 | |
| B. bovis J50 6 | |
| B. bovis—32 6 | TFC field samples |
| B. bovis—33 5 | |
| B. bovis—34 6 | |
1 All samples were from Tick Fever Centre (TFC) except for these control species from the CDC: Centers for Disease Control and Prevention, USA; DNA provided from CDC to our laboratory; 2 Originally from the Onderstepoort Veterinary Institute, South Africa; collected from TFC calf animal number 9710; 3 Compared with positive control B. bovis Dixie vaccine strain; 4 B. bovis Dixie vaccine strain passaged through ticks; 5 Field isolate with no known Babesia spp. vaccination history; 6 Field isolate with known Babesia spp. vaccination history.
For the extraction of DNA from the 17 vaccinated cattle at 4 time points (n = 68), the same method for 10 mL EDTA vacutainers and the QIAGEN DNA Blood Mini Kits was undertaken, as described above. DNA concentrations were determined using the NanoDrop 2000 (ThermoFisher Scientific Australia Pty Ltd., Victoria, Australia). DNA from blood smears and stabilates were not measured due to the presence of bovine DNA (white blood cells) in the samples and thus would not be an accurate measure of parasite DNA.
2.4. Standard PCR
Standard PCRs for the detection of B. bovis and B. bigemina were performed using a Corbett Palm Cycler (QIAGEN Pty Ltd., Victoria, Australia). For B. bovis, a 25 μL reaction included 2.5 μL 10× Taq Buffer (5 PRIME GmbH, Hilden, Germany), 10 pmol of each primer (Bo Fwd and Bo Rev) [10], 200 μM dNTPs, 0.2 μL Taq DNA Polymerase (5 PRIME GmbH, Hilden, Germany) and 1–5 μL DNA template. Temperature cycling conditions were: 60 s at 95 °C, followed by 35 cycles of 60 s at 95 °C, 60 s at 55 °C and 90 s at 73 °C, with the final cycle with an additional 5 min at 73 °C.
For B. bigemina standard detection PCR, a 25 μL reaction included 2.5 μL 10× Taq Buffer (5 PRIME), 25 pmol of each primer (BiIA and BiIB) [28], 200 μM dNTPs, 0.5 μL Taq DNA Polymerase (5 PRIME) and 1–5 μL DNA template. Temperature cycling conditions were: 5 min at 95 °C prior to the addition of Taq polymerase, followed by 35 cycles of 60 s at 95 °C, 60 s at 65 °C and 90 s at 73 °C, with the final cycle with an additional 15 min at 73 °C.
Both B. bigemina and B. bovis standard PCR products were separated on 1% agarose and 1× TBE (45 mM Tris-borate and 1 mM EDTA pH 8) gels at 80 volts for 45 min, and visualized under UV following 5 μg/mL ethidium bromide staining. The molecular weight marker was the HyperLadder II (Bioline Australia Pty Ltd., Narellan, NSW, Australia).
2.5. Optimization of SYBR Green Based Quantitative PCR Assay
Primers used for the SYBR Green qPCR assays were as described by Buling and co-workers [16] to specifically amplify the mitochondrial DNA cytochrome b genes of B. bovis and B. bigemina. The pairs of forward and reverse primers are shown in Table 2. PCR cycling conditions were modified and optimized using the reagents and equipment commonly used at the TFC laboratory. Briefly, SYBR Green based qPCR was conducted in a Rotor-Gene 6000 (QIAGEN Pty Ltd., Victoria, Australia). Reactions (20 µL) included 10 µL of SensiMix 2× SYBR no-ROX kit (Bioline Australia Pty Ltd., Sydney, NSW, Australia), 500 nM of each oligonucleotide primer and 1 µL of extracted genomic DNA template. Temperature cycling conditions were: 5 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 56 °C, and 30 s at 72 °C, acquiring SYBR Green fluorescence at the end of each extension step. After preliminary testing, the threshold was set to 0.2 for all assays. Cycle threshold (Ct) scores, corresponding to the cycle number at which the amplification curve intersects the threshold line, were recorded for each sample. Negative (no template or DNA extraction kit buffer) controls were included in each PCR run. Melting curve analysis was performed using the manufacturer’s standard protocol (Bioline Australia Pty Ltd., Alexandria, NSW, Australia). A bovine mitochondrial 16S rRNA as a housekeeping gene was used to optimize the SYBR Green qPCR conditions [29].
Table 2.
Primers and probes for the PCR assays for B. bovis and B. bigemina used in this study.
| Species | Primer/Probe | Sequence 5′-3′ | Product Length (bp) | Target Sequence (Genbank No.) | Reference |
|---|---|---|---|---|---|
| B. bovis | Standard PCR | 356 | (M38218.1) | [10] | |
| BoF | CACGAGGAAGGAACTACCGATGTTGA | 656–681 | |||
| BoR | CCAAGGAGCTTCAACGTACGAGGTCA | 986–1011 | |||
| SYBR Green qPCR | 88 | (GQ214235.1) | [16] | ||
| cbosg-1 (F) | TGTTCCTGGAAGCGTTGATTC | 135–155 | |||
| cbosg-2 (R) | AGCGTGAAAATAACGCATTGC | 202–222 | |||
| TaqMan qPCR | 90 | (AB499088.1) | This study | ||
| bovisF160 | ATATGTTTGCATTTGCTG | 160–178 | |||
| bovisR249 | CTCCAAACCAATATGAAAG | 230–249 | |||
| bovisPb | VIC-CAAACCATAAAGTCATCGGTATATCCTAC-MGB | 196–225 | |||
| Genotyping PCR | 245 | This study (B. bovis) M13 [24] |
|||
| M13BbovITSF | 1 GAGCGGATAACAATTTCACACAGGAAGGAGAAGTCGTAACAAGG | (EF458299.1) | |||
| BbovITSR | GGTCGTGGCAGTCACGGC | 9–28 | |||
| M13-FAM | 6FAM-GAGCGGATAACAATTTCACACAGG | 236–253 | |||
| B. bigemina | Standard PCR | 278 | (S45366.1) | [28] | |
| BiIA | CATCTAATTTCTCTCCATACCCCTCC | 6–31 | |||
| BiIB | CCTCGGCTTCAACTCTGATGCCAAAG | 258–283 | |||
| SYBR Green qPCR | 88 | (GQ214234.1) | [16] | ||
| cbisg-1 (F) | TGTTCCAGGAGATGTTGATTC | 182–202 | |||
| cbisg-2 (R) | AGCATGGAAATAACGAAGTGC | 249–269 | |||
| TaqMan qPCR | 146 | (AB499085.1) | This study | ||
| bigemF295 | GGTCTATTTGGTGGAGTT | 295–313 | |||
| bigemR413 | ACAAGACCAAATGCAATT | 395-413 | |||
| bigemPb | 6FAM-CAATTGTTCTTGGAGCAGCT-TAMRA | 329–349 | |||
| Bovine | SYBR Green qPCR 2 | 108 | (V00654) | [29] | |
| MTFB | GCGATTTTAAAGACTAGACCC | 2642–2662 | |||
| MTRBO | TGAATAGGATTGCGCTGT | 2732–2749 |
2.6. TaqMan qPCR Assay
Species-specific primers (forward and reverse) and fluorescence-labelled probes for the multiplex qPCR assay were designed using Primer 3 software (http://primer3.sourceforge.net; Howard Hughes Medical Institute, Chevy Chase, MD, USA). The VIC and FAM labelled probes specifically target the mitochondrial cytochrome b genes of B. bovis and B. bigemina, respectively. All primers utilized in this study were synthesized through Integrated DNA Technologies (Skokie, IL, USA). TaqMan probes (VIC and FAM labelled) were synthesized through Applied Biosystems (ThermoFisher Scientific Pty Ltd., Victoria, Australia).
TaqMan qPCR was conducted in a Rotor Gene Q (QIAGEN Pty Ltd., Victoria, Australia). Reactions (20 µL) included 10 µL of SensiFAST 2× Probe Mix (Bioline Australia Pty Ltd., Alexandria, NSW, Australia), 500 nM of each oligonucleotide primer, 150 nM of the VIC- (B. bovis) or 6FAM- (B. bigemina) labelled probe, and 1 µL of extracted genomic DNA template. Temperature cycling conditions were: 5 min at 95 °C, followed by 40 cycles of 95 °C for 30 s, 54 °C for 30 s and 72 °C for 30 s, acquiring fluorescence on both the green (FAM) and yellow (VIC) channels at the end of each extension step. Ct scores were recorded for each sample. Negative controls (no template or DNA extraction kit buffer) were included in each PCR run.
2.7. Sensitivity and Specificity of Species Specific Assays
Sensitivity of each assay was determined by using the serial 10 fold dilutions of DNA prepared from the control B. bovis and B. bigemina strains (Dixie and G strains, respectively) described above. To evaluate the analytical specificity, DNA from Babesia microti, Babesia duncani, Anaplasma marginale, and Anaplasma marginale subsp. centrale (vaccine strain for bovine anaplasmosis used in Australia, originally isolated in South Africa [30,31]) and DNA from non-infected bovine blood, were subjected to the qPCR assays for bovine Babesia parasites. The reference isolates are listed in Table 1.
2.8. Clinical Evaluation of Assay Sensitivity and Specificity of Species Specific Assays
Clinical sensitivity and specificity of the standard PCRs, TaqMan and SYBR Green qPCRs were evaluated using diagnostic samples submitted to TFC. Thirty-two B. bovis diagnostic samples and 14 B. bigemina stabilates (stored field samples) were screened (Table 1). In addition, the standard PCR and SYBR Green qPCR were used to screen 17 animals inoculated with both Australian vaccine strains (B. bovis Dixie strain and B. bigemina G strain) at 4 different time points (Days 7, 9, 11 and 14) post-inoculation (n = 68 samples). DNA prepared from the B. bovis Dixie and B. bigemina G strains described in 2.2 above were included as positive controls in these experiments.
2.9. Statistical Analysis
The Student’s t-test (tails 1, type 2) was used to compare the melting temperatures of the different amplification products from the species specific SYBR Green qPCR. Significance was set at p < 0.05.
2.10. Babesia bovis ITS Genotyping Method
New primers were designed flanking the ITS1 variable length region using a conserved forward primer and a B. bovis specific reverse primer to amplify fragments of ~200–250 bp depending on the isolate (Table 1). The forward primer, M13BbovITSF, is anchored in the 3’ end of the 18S rRNA gene. The forward primer was given an M13 extension so that an additional 6-FAM-labeled M13 forward primer could be included in the reaction for product detection [24]. The reverse primer, BbovITSR, was anchored in the conserved ITS1 3’ end flanking the variable regions of the ITS1. A Multiplex PCR kit (QIAGEN Pty Ltd., Victoria, Australia) was used to amplify 2 µL of DNA purified from previously cryopreserved stabilate samples as described above. PCRs were conducted in 12 µL volumes using 0.2 pmol M13Bbov ITSF, 2 pmol M13FAM, 2 pmol BbovITSR, 1.2 µL 5× Q solution and 6 µL QIAGEN Master Mix 2.5× (QIAGEN Pty Ltd., Victoria, Australia). The thermo-cycling conditions were as previously described: 95 °C 15 min, 94 °C 30 s, 50 °C 45 s, 72 °C 90 s for 35 cycles followed by 72 °C for 45 min in DNA Engine Peltier thermocycler (BioRad, Gladesville, NSW, Australia). The subsequent analyses of these PCR products were undertaken through the Animal Genetics Laboratory, the University of Queensland as an external service provider (http://animal-genetics.lab.uq.edu.au/). Briefly, the PCR products were diluted 1 in 10 with water and 3 µL of each diluted product mixed with 10 µL of formamide plus LIZ 500 size standard (1:0.004 ratio) (Applied Biosystems, Foster City, CA, USA). Samples were denatured at 95 °C for 3 min and then chilled on ice before being electrokinetically injected into a POP-7 polymer matrix using a 50 cm, 16 capillary, 3130XL DNA Genetic Analyzer (Applied Biosystems, Foster City, USA). Allele sizes were determined using Genemapper (v3.7, Applied Biosystems, Foster City, USA) as described previously [25].
3. Results
3.1. Sensitivity and Specificity
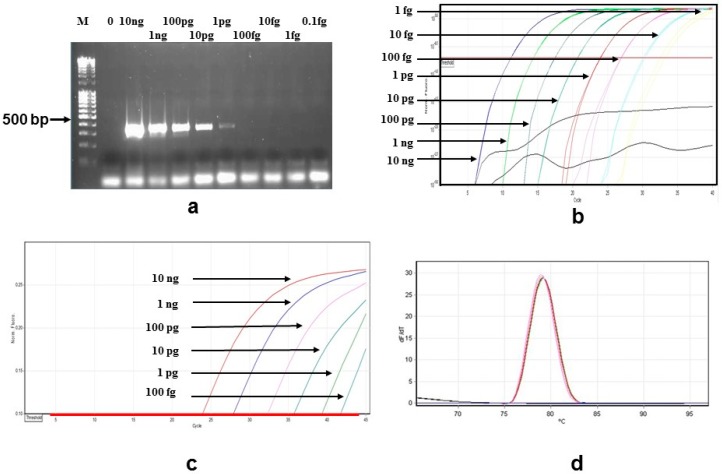
To evaluate the sensitivity of both SYBR Green and the TaqMan qPCRs (this study), serial dilutions of DNA extracted from B. bovis and B. bigemina-infected bovine blood were used. Sensitivity of B. bovis amplifications was 1 pg with standard PCR (Figure 1a), 1 fg with SYBR Green qPCR (Figure 1b) and 100 fg with TaqMan qPCR (Figure 1c). This corresponded to 35 B. bovis parasites/µL and 0.35 B. bovis parasites/µL for TaqMan and SYBR Green detection limits, respectively. Melting curve analysis for the SYBR Green qPCRs is shown in Figure 1d, at 79.06 ± 0.05 °C. No positive signal was observed in qPCR assays with DNA from other species listed in Table 1 (See Table S1).
Figure 1.
Results obtained from all B. bovis PCR assays examined in this study: (a) Sensitivity of the standard PCR assay determined following agarose gel electrophoresis. M is the Bioline HyperLadder II and the concentrations of B. bovis DNA are shown above each lane with PCR products estimated at 356 bp; (b) SYBR Green qPCR plots of B. bovis dilutions with DNA concentrations indicated; (c) TaqMan qPCR plots of B. bovis dilutions with DNA concentrations indicated; (d) Melting curves from the SYBR Green qPCRs in Figure 1b.
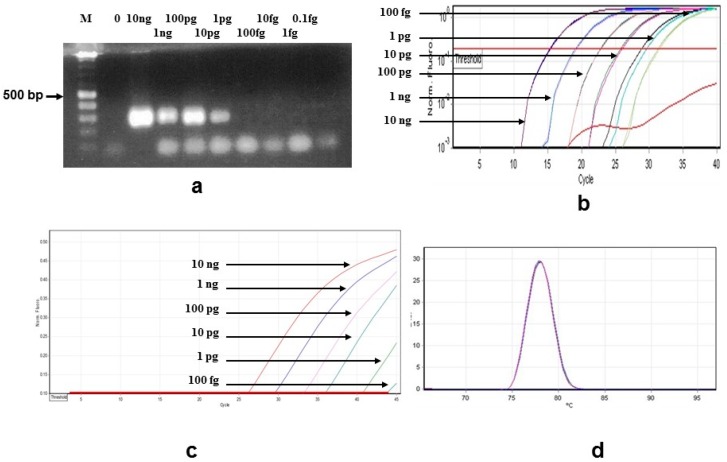
Sensitivity of B. bigemina amplifications was 10 pg with standard PCR (Figure 2a), 100 fg for both the SYBR Green (Figure 2b) and the TaqMan qPCRs (Figure 2c). For the qPCRs, this corresponds to the detection of 2 B. bigemina parasites/µL. Melting curve analysis for the SYBR Green qPCRs is shown in Figure 2d, at 77.83 ± 0.09 °C. Although Ct values were observed with B. microti (5 isolates at Ct values higher than 35) and B. duncani (1 isolate, Ct = 34.81) in the B. bigemina SYBR Green qPCR, the melting curve analysis showed that these other non-specific species have melting temperatures of 81.7 ± 0.95 °C (data not shown). TaqMan qPCR results were negative for all species listed in Table 1 including B. microti and B. duncani (See Table S1).
Figure 2.
Results obtained from all B. bigemina PCR assays examined in this study: (a) Sensitivity of the standard PCR assay determined following agarose gel electrophoresis. M is the Bioline HyperLadder II and the concentrations of B. bigemina DNA are shown above each lane with PCR products estimated at 278 bp; (b) SYBR Green qPCR plots of B. bigemina dilutions with DNA concentrations indicated; (c) TaqMan qPCR plots of B. bigemina dilutions with DNA concentrations indicated; (d) Melting curves from the SYBR Green qPCRs in Figure 2b.
Melting curve analysis of the SYBR Green qPCR discriminated between B. bigemina- and B. bovis-infected samples (Figure 1d and Figure 2d). A student’s t-test found a significant difference between the melt temperatures of the two species. The melting temperature for B. bigemina was 77.83 ± 0.09 °C (n = 9), and for B. bovis was 79.06 ± 0.05 °C (n = 9, p < 0.001; Student’s t-test).
A summary of assay sensitivities is presented in Table 3 which compares published methods with that obtained here including our new Multiplex TaqMan qPCR targeting the cytochrome b genes of B. bigemina and B. bovis. The Buling et al. [16] SYBR Green qPCR was not as sensitive when applied to our samples in this study with 10-fold and 100-fold less detection for B. bovis and B. bigemina respectively. Buling et al. claim the detection of 1000 target copies using PCR products and not genomic DNA for this calculation. The multiplex TaqMan qPCR assay targeting the cytochrome b genes was also less sensitive compared to Kim et al. [17] targeting 18S rRNA genes however our parasite count would not be accurate as it was estimated from initial parasitaemias from large volumes of blood processed to isolate pure parasites. Overall specificities are comparable at 100%.
Table 3.
Summary of sensitivity and specificity of published qPCR assays for the detection of Babesia bovis and Babesia bigemina.
| Reference | Assay Type | Species | Target Gene | Specificity | Sensitivity | Source of Isolates |
|---|---|---|---|---|---|---|
| [16] | SYBR Green |
B. bovis B. bigemina |
cytochrome b | 100% | 0.1 fg 1 (1000 target copies) | Spain, Argentina |
| B. bovis | 100% | 1 fg or 0.35 parasites/µL | Australia 2 | |||
| B. bigemina | 100% | 100 fg or 20 parasites/µL | ||||
| This study | TaqMan probes | B. bovis | cytochrome b | 100% | 100 fg or 35 parasites/µL | Australia |
| B. bigemina | 100% | 100 fg or 20 parasites/µL | ||||
| [32] | SYBR Green | B. bovis | msa2c | 100% | 1000 copies/mL | Brazil |
| [17] | TaqMan probes |
B. bovis B. bigemina |
18S rRNA genes | 100% | 2.5 parasites/µL | Brazil |
1 Parasite numbers per µL sensitivity was not determined; 2 Buling et al. method [16] using Australian samples from this study.
3.2. Clinical Sensitivity of SYBR Green qPCR Assays
Clinical sensitivity was determined by screening field samples from babesiosis diagnostic submissions as well as monitoring cattle vaccinated with B. bovis Dixie strain and B. bigemina G strain at four different time points post-inoculation. Table 4 summarizes the results showing that the SYBR Green qPCRs were able to detect each species from all clinical samples. The B. bovis standard PCR had negative results for 9/31 samples whereas the B. bigemina standard PCR assay amplified all 14 samples successfully. In addition, some of these diagnostic samples demonstrated mixed infections of B. bovis and B. bigemina as determined by qPCR. Supplementary Table S1 details results across reference and field isolates (31 B. bovis and 14 B. bigemina) for standard PCR, TaqMan and SYBR Green qPCRs. Results show mixed infections in four and six of the B. bigemina and B. bovis field samples, respectively. For the B. bigemina samples, two samples with a known history of tick fever vaccination (Table 1) were not positive in B. bovis qPCR (Table S1), indicating the lack of detection of carrier level parasites.
Table 4.
Summary of clinical sensitivities for Babesia bovis and Babesia bigemina standard PCRs and SYBR Green qPCRs.
| Clinical Samples 1 | Standard PCR B. bovis |
Standard PCR B. bigemina |
SYBR qPCR B. bovis |
SYBR qPCR B. bigemina |
|---|---|---|---|---|
| 31 B. bovis | 22/31 | ND | 31/31 | 6/31 |
| 14 B. bigemina | ND | 14/14 | 4/14 | 14/14 |
The standard PCR and SYBR qPCR assays were also used to detect B. bovis and B. bigemina in 17 vaccinated cattle sampled at four different time points post-inoculation (Days 7, 9, 11, 14) (see Table S2). All 17 animals were positive in B. bigemina qPCR at three or four time points. Only three animals were negative at Day 7, with the remaining 14 positive for qPCR at each time point. The B. bigemina standard PCR had 33 negative results out of the total 68 samples from the 17 cattle. For B. bovis SYBR Green qPCR, one animal was negative at all time points, two animals positive at one time point only, five positive at two time points, and nine with three or four positive time points per animal. In most cases, the negative time points were Day 7 and Day 9. The total number of positive B. bovis qPCR results was 25/68, which is much less than for B. bigemina with 65/68 positive. When the B. bovis standard PCR was correlated with the SYBR Green qPCR results, a further eight time points were negative where qPCR was positive, with only 17 positive samples overall by standard PCR. The results from this vaccination study indicate that the B. bovis qPCR is less sensitive “clinically” for the detection of B. bovis in comparison with B. bigemina detection.
3.3. Babesia Bovis ITS Genotyping
Allele sizes are summarized in Table 5. The current Australian B. bovis Dixie vaccine strain produces two alleles at 224 and 234 bp, which correspond to BvVA1 alleles at 3.5 and 6.8 kb, respectively. Two stabilates derived from this vaccine following tick transmission shows that one of these has a further two new alleles at 233 and 243 bp with extra alleles previously not detected using BvVA1 PCR (Table 5). B. bovis H97, J40, 32 and 34 are field isolates which show that these isolates also contain Dixie vaccine strain alleles detected using the ITS assay. Using the BvVA1 PCR, one B. bovis Dixie allele was also detected in strain H97. H92 is a field isolate from an animal with no vaccination history; hence the presence of one of the “Dixie through ticks” ITS1 allele is likely to be a coincidence. Note no BvVA1 Dixie alleles are detected in H92. Field isolate J50 appears to contain alleles commensurate with Dixie vaccine as well as an additional allele seen following tick transmission of Dixie vaccine (see H10). Isolate 33 did not yield positive BvVA1 results however using the ITS assay several alleles were detected with one matching a Dixie vaccine allele. Babesia bigemina and A. marginale did not produce PCR products in this assay (data not shown). Supplementary Figure S1 shows an example of GeneMapper plots for Dixie vaccine and H97 field isolate. Note that the molecular weights for Dixie vaccine were recorded at 225 and 235 bp for this particular analysis. Each analysis can vary by 1–2 bp, thus the incorporation of appropriate sample controls is recommended for each run to enable within run comparisons.
Table 5.
Summary of Babesia bovis genotyping showing BvVAI PCR results compared with ITS alleles determined following capillary electrophoresis and GeneMapper analysis.
| B. bovis Isolates 1 | Isolate Description | BvVA1 1,2 kb |
ITS Alleles 1,2 bp |
|---|---|---|---|
| Dixie | Vaccine isolate | 3.5, 6.8 | 224, 234 |
| H03 | Dixie vaccine following tick passage | 6.8 | 224, 234 |
| H10 | Dixie vaccine following tick passage | 3.5, 6.8 | 224, 233, 234, 243 |
| H92 3 | Field isolate 3 | 5.5, 6.1 | 218, 233, 235, 239, 242, 247 |
| H97 | Field isolate | 6.4, 6.8 | 224, 234, 242 |
| J40 | Field isolate | 6.1, (6.2) | 224, 226, 227, 229, 231, 234, 236, 238, 239, 242 |
| J50 | Field isolate | 6.1 | 224, 233, 234 |
| 32 | Field isolate | 6.2, 8.0 | 224, 234, 237, (309), (337) |
| 33 | Field isolate | Nil | (142), (189), 220, 224, 250, 256, 315, 325, 337, 342 |
| 34 | Field isolate | 6.5 | 218, (224), 234, 244, (302), (337) |
1 Dixie vaccine strain alleles are shown in bold font; 2 Faint bands are indicated by a bracket; 3 No history of Dixie vaccine strain vaccination.
4. Discussion
We investigated the development of a novel multiplex TaqMan qPCR for the detection of B. bovis and B. bigemina simultaneously. The SYBR Green qPCR (also targeting the cytochrome b gene) appeared to be slightly more sensitive than our new multiplex assay. We also report preliminary data for a novel fluorescent PCR to genotype B. bovis targeting the ITS1, moreover detecting both vaccine and field alleles in single samples for the first time in Australia.
The detection of Babesia spp. in vaccinated cattle and disease outbreaks, and in immune carrier animals, can be problematic when low numbers of parasites are present. In comparison with conventional PCR assay, SYBR Green and Taqman qPCR assays have several advantages for detection and quantification of parasite DNA. Detection and quantification of a qPCR product can take place in a single tube, obviating the need for post-PCR manipulation and thus reducing the risk of contamination [33]. It is quick, taking less than three hours to set up, perform the assay and analyse the data; and primers for both B. bovis and B. bigemina can be added in one multiplex qPCR reaction. Therefore, qPCR has a faster turnaround time than the conventional PCR assay.
Our study confirmed that qPCRs are more sensitive than standard PCRs, being able to detect Babesia infection in bovine blood at 1000-fold lower concentrations than standard PCR. The cytochrome b gene was targeted due its reported higher copy number than ribosomal genes [23]. The same detection levels reported by Buling et al. [16] were not achieved for the SYBR Green qPCR, particularly for B. bigemina. Some of these differences could be attributed to the use of different reagents, thermocyclers, DNA extraction protocols and the samples used to calculate assay sensitivities. Specifically, Buling et al. [16] utilized DNA from species specific PCR products to calculate DNA concentrations for their assay sensitivity calculations. In comparison, we determined the DNA concentration using genomic parasite DNA extracts which would explain the decrease in sensitivity for our samples. A 10-fold decrease in the number of Babesia parasites was detected in our TaqMan assays compared with the published protocol [17]. Kim et al. [17] processed small volumes of parasites from in vitro culture using minimal steps compared to our extraction procedure and targeted 18S rRNA genes. Our parasite numbers would have been over-inflated as we first purified parasites from large volumes of infected bovine blood (~400 mL) taken directly from our vaccine donor animals. Subsequently a subset of these purified parasites (equating to ~75 mL of infected blood) were used to purify DNA and the number of parasites screened in the cytochrome b TaqMan PCRs was “extrapolated”. It is known that the purification of parasites from whole blood leads to loss of parasites through the centrifugation steps thus our extrapolated parasite count is far from accurate. Overall, the SYBR Green assays appeared to be more sensitive than the TaqMan assays for the same target, melting temperature analysis is needed for SYBR Green specificity which could be problematic for some diagnostic laboratories. It thus may be more beneficial to use the multiplex TaqMan assay (described here) with higher template DNA in order to obtain clearer results and better robust diagnostic specificity. Standardization of assays between laboratories would be best achieved by sharing samples.
This study also demonstrated the successful detection of B. bovis using DNA preparations from blood smears. This was undertaken using Giemsa’s stained smears where microscopy could not differentiate Babesia species; or the presence of Babesia species was suspected, but was not certain, due to smear quality. qPCR detection of B. bovis parasites was successful from smear material which provides another option for diagnosticians when blood smears cannot be interpreted. This is the first time that DNA and PCR technology has been applied for the diagnosis of bovine babesiosis using stained blood smears, to our knowledge.
Recently, a nested PCR targeting the cytochrome b genes from both B. bovis and B. bigemina detected as low as 0.1 fg DNA [34]. Nested PCRs thus can have similar sensitivity to real time or qPCRs and have been applied in the investigation of bovine babesiosis pathogens in the Philippines, Syria, Vietnam, Thailand and Kenya [35,36,37,38]. However, nested PCR assays are at high risk of introducing PCR product contamination, hence our evaluation using TaqMan and SYBR Green qPCR methods.
This study also showed that in clinical samples, the detection of B. bovis can be compromised due to lower parasitaemia compared with B. bigemina. It is known that B. bovis parasitaemia during acute infection can be lower than 1% compared with B. bigemina at >10% [26]. We thus recommend for qPCR of B. bovis, that a higher concentration of DNA be used as template. As we routinely purified 200 µL from a RBC pellet prepared from 10 mL blood tubes, we recommend that for B. bovis qPCR that the complete RBC pellet be processed and concentrated prior to addition as PCR template. This would increase the ability to detect B. bovis in immune carriers, as demonstrated in the vaccine qPCR study and in field isolates known to have been vaccinated where B. bovis could not be detected using the methods described.
In this study we employed a novel fluorescence technology for the discrimination of PCR fragments ranging from 142–337 bp using capillary electrophoresis. We verified the presence of allele variants for three of the B. bovis isolates (Dixie vaccine, H97 and J40) through sequencing (data not shown). This method was originally described by Schuelke (2000) reporting the usefulness of this approach as an alternative to agarose gel electrophoresis which usually cannot discriminate similarly sized fragments [24]. Godwin and Morgan (2014) further demonstrated the ability of this technology to recognize 10 species of poultry Eimeria parasites targeting a non-coding mitochondrial genome region between the cytochrome c oxidase subunit 3 (Cox3) and the Large Subunit 1 ribosomal RNA genes [25]. Previous methods using PCR to target single copy genes were useful as they detected different populations of B. bovis [13]. The B. bovis internal transcribed spacer gene 1 (between the 18S and the 5.8S rRNA coding regions) has not been exploited for genotyping to our knowledge, however, as more than one rDNA copy is present per Babesia genome, it is feasible that the sensitivity will increase over single copy genes previously used (BvVA1, Bv80) [13].
This is thus the first time that B. bovis vaccine genotypes have been demonstrated in field isolates confirming the presence of the Australian B. bovis Dixie vaccine strain. This has been previously shown with the South African B. bovis vaccine using Bv80 gene genotyping and a cartridge gel system capable of separating fragments differing by 6–10 bp [39]. We also demonstrated the presence of two extra alleles in one sample of the B. bovis Dixie vaccine strain which had been passaged through ticks. The other “Dixie through ticks” sample did not demonstrate non-Dixie vaccine alleles which may be due to the concentration of DNA used in the assay. The concentration of DNA in all B. bovis PCRs appears to be a contentious issue and must be addressed by using higher template DNA concentrations in all B. bovis PCR assays. In our previous studies using Bv80 and BvVA1 genotyping PCRs, vaccine genotypes were not detected in vaccinated field samples possible due to the low numbers of parasites at these carrier levels [18]. The new fluorescent ITS PCR using capillary electrophoresis was able to confirm the presence of different alleles including vaccine genotypes, and thus is more sensitive than the previous gel electrophoresis methods reported [18]. Further research is needed to conclude whether the Australian vaccine strain is co-transmitted with field isolates by ticks as described recently for the South African live B. bovis vaccine [39].
Acknowledgments
This research was supported by the Tick Fever Centre. The authors acknowledge Wayne Jorgensen for suggesting the qPCR research program. The authors also acknowledge Amielle Kerudin (UQ Honours Student) for her preliminary research associated with the fluorescent ITS genotyping for comparing B. bovis isolates. We also thank Susan Anderson for her technical support with DNA extractions and qPCRs. The authors thank Phil Carter (TFC) for a critical review of the manuscript. We acknowledge the University of Queensland’s Animal Genetics Laboratory for analysing the B. bovis fluorescent ITS PCR products and for providing a report of allele sizes following GeneMapper analysis.
Abbreviations
The following abbreviations are used in this manuscript:
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TFC | Tick Fever Centre |
| PCR | Polymerase Chain Reaction |
| qPCR | Quantitative PCR |
| M13 | M13 bacteriophage |
| EDTA | Ethylenediaminetetraacetic acid |
| PBS | Phosphate buffered saline |
| dNTPs | Deoxynucleotides |
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/2306-7381/3/3/23/s1. Figure S1: Example Genemapper plots showing peaks for Dixie vaccine and one field isolate H97. Note fragment sizes 225 and 235 bp for Dixie vaccine strain for this electrophoresis run, Table S1: Comparison of standard PCR, TaqMan PCR and SYBR Green qPCRs for the detection of B. bovis and B. bigemina in reference isolates and field samples, Table S2: Use of standard PCR and SYBR Green qPCR to detect both B. bovis and B. bigemina in 17 vaccinated cattle at 4 time points post-inoculation (Days 7, 9, 11 and 14), total 68 samples.
Author Contributions
Bing Zhang developed the TaqMan assays and supervised the activities of Jacqueline L. Sambono who undertook all evaluations associated with PCR testing for B. bovis and B. bigemina testing. Ala E. Lew-Tabor provided further interpretation of the data as described in this manuscript. Peter Rolls sourced and assisted with background diagnostic and infection information for the samples tested. Jess A. T. Morgan was responsible for the development of the ITS B. bovis typing PCR which was subsequently screened by Bronwyn Venus and Ala E. Lew-Tabor to demonstrate “proof of concept” of this assay. The manuscript was written equally by Bing Zhang and Ala E. Lew-Tabor with contributions from Peter Rolls and Jacqueline L. Sambono.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bock R.E., Jackson L., DeVos A.J., Jorgensen K. Babesiosis of cattle. Parasitology. 2004;129:S247–S269. doi: 10.1017/S0031182004005190. [DOI] [PubMed] [Google Scholar]
- 2.Sackett D., Holmes P., Abbott K., Jephcott S., Barber M. Assessing the Economic Cost of Endemic Disease on the Profitability of Australian Beef Cattle and Sheep Producers. Meat & Livestock Australia Limited; North Sydney, NSW, Australia: 2006. [Google Scholar]
- 3.Lew-Tabor A.E., Rodriguez Valle M. A review of reverse vaccinology approaches for the development of vaccines against ticks and tick borne diseases. Ticks Tick Borne Dis. 2016;7:573–585. doi: 10.1016/j.ttbdis.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Fahrimal Y., Goff W.L., Jasmer D.P. Detection of Babesia bovis carrier cattle by using polymerase chain reaction amplification of parasite DNA. J. Clin. Microbiol. 1992;30:1373–1379. doi: 10.1128/jcm.30.6.1374-1379.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oura C.A., Bishop R.P., Wampande E.M., Lubega G.W., Tait A. Application of a reverse line blot assay to the study of haemoparasites in cattle in Uganda. Int. J. Parasitol. 2004;34:603–613. doi: 10.1016/j.ijpara.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Bock R.E., De Vos A.J. Immunity following use of the Australian tick fever vaccine: A review of the evidence. Aust. Vet. J. 2001;79:832–839. doi: 10.1111/j.1751-0813.2001.tb10931.x. [DOI] [PubMed] [Google Scholar]
- 7.De Castro J.J. Sustainable tick and tickborne disease control in livestock improvement in developing countries. Vet. Parasitol. 1997;71:77–97. doi: 10.1016/S0304-4017(97)00033-2. [DOI] [PubMed] [Google Scholar]
- 8.Almeria S., Castella J., Ferrer D., Ortuno A., Estrada-Pena A., Gutierrez J.F. Bovine piroplasms in Minorca (Balearic Islands, Spain): A comparison of PCR-based and light microscopy detection. Vet. Parasitol. 2001;99:249–259. doi: 10.1016/S0304-4017(01)00464-2. [DOI] [PubMed] [Google Scholar]
- 9.Calder J.A.M., Reddy G.R., Chieves L., Courtney C.H., Littell R., Livengood J.R., Norval R.A.I., Smith C., Dame J.B. Monitoring Babesia bovis infections in cattle by using PCR-based tests. J. Clin. Microbiol. 1996;34:2748–2755. doi: 10.1128/jcm.34.11.2748-2755.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa J.V., Chieves L.P., Johnson G.S., Buening G.M. Multiplex polymerase chain reaction based assay for the detection of Babesia bigemina, Babesia bovis and Anaplasma marginale DNA in bovine blood. Vet. Parasitol. 1993;50:69–81. doi: 10.1016/0304-4017(93)90008-B. [DOI] [PubMed] [Google Scholar]
- 11.Luo J., Yin H., Liu Z., Yang D., Guan G., Liu A., Ma M., Dang S., Lu B., Sun C., et al. Molecular phylogenetic studies on an unnamed bovine Babesia sp. based on small subunit ribosomal RNA gene sequences. Vet. Parasitol. 2005;133:1–6. doi: 10.1016/j.vetpar.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira-Sequeira T.C.G., Oliveira M.C.S., Araujo J.P., Amarante A.F.T. PCR-based detection of Babesia bovis and Babesia bigemina in their natural host Boophilus microplus and cattle. Int. J. Parasitol. 2005;35:105–111. doi: 10.1016/j.ijpara.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Lew A.E., Dalrymple B.P., Jeston P.J., Bock R.E. PCR methods for the discrimination of Babesia bovis isolates. Vet. Parasitol. 1997;71:223–237. doi: 10.1016/S0304-4017(97)00025-3. [DOI] [PubMed] [Google Scholar]
- 14.Francino O., Altet L., Sanchez-Robert E., Rodriguez A., Solano-Gallego L., Alberola J., Ferrer L., Sanchez A., Roura X. Advantages of real-time PCR assay for diagnosis and monitoring of canine leishmaniosis. Vet. Parasitol. 2006;137:214–221. doi: 10.1016/j.vetpar.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Rama Iniguez S., Dea-Ayuela M.A., Sanchez-Brunete J.A., Torrado J.J., Alunda J.M., Bolas-Fernandez F. Real-time reverse transcription-PCR quantification of cytokine mRNA expression in golden Syrian hamster infected with Leishmania infantum and treated with a new amphotericin B formulation. Antimicrob. Agents Chemother. 2006;50:1195–1201. doi: 10.1128/AAC.50.4.1195-1201.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buling A., Criado-Fornelio A., Asenzo G., Benitez D., Barba-Carretero J.C., Florin-Christensen M. A quantitative PCR assay for the detection and quantification of Babesia bovis and B. bigemina. Vet. Parasitol. 2007;147:16–25. doi: 10.1016/j.vetpar.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 17.Kim C., Iseki H., Herbas M.S., Yokoyama N., Suzuki H., Xuan X., Fujisaki K., Igarashi I. Development of TaqMan-based real-time PCR assays for diagnostic detection of Babesia bovis and Babesia bigemina. Am. J. Trop. Med. Hyg. 2007;77:837–841. [PubMed] [Google Scholar]
- 18.Lew A.E., Bock R.E., Croft J.M., Minchin C.M., Kingston T.G., Dalgliesh R.J. Genotypic diversity in field isolates of Babesia bovis from cattle with babesiosis after vaccination. Aust. Vet. J. 1997;75:575–578. doi: 10.1111/j.1751-0813.1997.tb14197.x. [DOI] [PubMed] [Google Scholar]
- 19.Bock R.E., DeVos A.J., Lew A., Kingston T.G., Fraser I.R. Studies on failure of T strain live Babesia bovis vaccine. Aust. Vet. J. 1995;72:296–300. doi: 10.1111/j.1751-0813.1995.tb03558.x. [DOI] [PubMed] [Google Scholar]
- 20.Combrink M.P., Troskie P.C., Pienaar R., Latif A.A., Mans B.J. Genotypic diversity in Babesia bovis field isolates and vaccine strains from South Africa. Vet. Parasitol. 2014;199:144–152. doi: 10.1016/j.vetpar.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Molad T., Fleiderovich L., Leibovitz B., Wolkomirsky R., Behar A., Markovics A. Differentiation between Israeli B. bovis vaccine strain and field isolates. Vet. Parasitol. 2015;208:159–168. doi: 10.1016/j.vetpar.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Llaneza A., Caballero M., Baravalle E., Mesplet M., Mosqueda J., Suarez C.E., Echaide I., Katzer F., Pacheco G.M., Florin-Christensen M., et al. Development of a tandem repeat-based multilocus typing system distinguishing Babesia bovis geographic isolates. Vet. Parasitol. 2010;167:196–204. doi: 10.1016/j.vetpar.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 23.Salem G.H., Liu X., Johnsrude J.D., Dame J.B., Roman Reddy G. Development and evaluation of an extra chromosomal DNA-based PCR test for diagnosing bovine babesiosis. Mol. Cell. Probes. 1999;13:107–113. doi: 10.1006/mcpr.1998.0223. [DOI] [PubMed] [Google Scholar]
- 24.Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- 25.Godwin R.M., Morgan J.A. A simple, one-tube assay for the simultaneous detection and diagnosis of ten Australian poultry Eimeria. Electrophoresis. 2014;35:494–502. doi: 10.1002/elps.201300286. [DOI] [PubMed] [Google Scholar]
- 26.Rolls P.J., Bock R.E., de Vos A.J., Waldron S.J. OIE Manual of Diagnostic Tests and Vaccine for Terrestrial Animals. 7th ed. Volume 1. Work Organisation for Animal Health (OIE); Paris, France: 2012. Bovine Babesiosis; pp. 601–615. [Google Scholar]
- 27.Herndon D.R., Palmer G.H., Shkap V., Knowles D.P., Jr., Brayton K.A. Complete genome sequence of Anaplasma marginale subsp. centrale. J. Bacteriol. 2010;192:379–380. doi: 10.1128/JB.01330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figueroa J.V., Chieves L.P., Johnson G.S., Buening G.M. Detection of Babesia bigemina-infected carriers by polymerase chain reaction amplification. J. Clin. Microbiol. 1992;30:2576–2582. doi: 10.1128/jcm.30.10.2576-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cawthraw S., Saunders G.C., Martin T.C., Sawyer J., Windl O., Reaney S.D. Real-time PCR detection and identification of prohibited mammalian and avian material in animal feeds. J. Food Prot. 2009;72:1055–1062. doi: 10.4315/0362-028x-72.5.1055. [DOI] [PubMed] [Google Scholar]
- 30.Theiler A. Further Investigation into Anaplasmosis of South African Cattle. Government Printer and Stationery Office; Pretoria, South Africa: 1911. pp. 7–46. [Google Scholar]
- 31.Rogers R.J., Shiels I.A. Epidemiology and control of anaplasmosis in Australia. J. S. Afr. Vet. Assoc. 1979;50:363–366. [PubMed] [Google Scholar]
- 32.Ramos C.A., Araujo F.R., Souza I.I.F., Bacanelli G., Luiz H.L., Russi L.S., Oliveira R.H., Soares C.O., Rosinha G.M., Alves L.C. Real-time polymerase chain reaction based on msa2c gene for detection of Babesia bovis. Vet. Parasitol. 2011;176:79–83. doi: 10.1016/j.vetpar.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 33.Monis P.T., Giglio S., Keegan A.R., Andrew Thompson R.C. Emerging technologies for the detection and genetic characterization of protozoan parasites. Trends Parasitol. 2005;21:340–346. doi: 10.1016/j.pt.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Romero-Salas D., Mira A., Mosqueda J., Garcia-Vazquez Z., Hidalgo-Ruiz M., Vela N.A., de Leon A.A., Florin-Christensen M., Schnittger L. Molecular and serological detection of Babesia bovis- and Babesia bigemina-infection in bovines and water buffaloes raised jointly in an endemic field. Vet. Parasitol. 2016;217:101–107. doi: 10.1016/j.vetpar.2015.12.030. [DOI] [PubMed] [Google Scholar]
- 35.Adjou Moumouni P.F., Aboge G.O., Terkawi M.A., Masatani T., Cao S., Kamyingkird K., Jirapattharasate C., Zhou M., Wang G., Liu M., et al. Molecular detection and characterization of Babesia bovis, Babesia bigemina, Theileria species and Anaplasma marginale isolated from cattle in Kenya. Parasit. Vectors. 2015;8:496. doi: 10.1186/s13071-015-1106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Luo Y., Cao S., Terkawi M.A., Lan D.T., Long P.T., Yu L., Zhou M., Gong H., Zhang H., et al. Molecular and seroepidemiological survey of Babesia bovis and Babesia bigemina infections in cattle and water buffaloes in the central region of Vietnam. Trop. Biomed. 2014;31:406–413. [PubMed] [Google Scholar]
- 37.Terkawi M.A., Alhasan H., Huyen N.X., Sabagh A., Awier K., Cao S., Goo Y.K., Aboge G., Yokoyama N., Nishikawa Y., et al. Molecular and serological prevalence of Babesia bovis and Babesia bigemina in cattle from central region of Syria. Vet. Parasitol. 2012;187:307–311. doi: 10.1016/j.vetpar.2011.12.038. [DOI] [PubMed] [Google Scholar]
- 38.Yu L., Terkawi M.A., Cruz-Flores M.J., Claveria F.G., Aboge G.O., Yamagishi J., Goo Y.K., Cao S., Masatani T., Nishikawa Y., et al. Epidemiological survey of Babesia bovis and Babesia bigemina infections of cattle in Philippines. J. Vet. Med. Sci. 2013;75:995–998. doi: 10.1292/jvms.12-0425. [DOI] [PubMed] [Google Scholar]
- 39.Combrink M.P., Troskie P.C., de Klerk D.G., Pienaar R., Latif A.A., Mans B.J. Co-transmission of the non-transmissible South African Babesia bovis S24 vaccine strain during mixed infection with a field isolate. Ticks Tick Borne Dis. 2015;6:158–163. doi: 10.1016/j.ttbdis.2014.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.