Abstract
Volatile organic compounds (VOCs) are major pollutants that are found in contaminated sites, particularly in developed countries such as Japan. Various microorganisms that degrade individual VOCs have been reported, and genomic information related to their phylogenetic classification and VOC-degrading enzymes is available. However, the biodegradation of multiple VOCs remains a challenging issue. Practical sites, such as chemical factories, research facilities, and illegal dumping sites, are often contaminated with multiple VOCs. In order to investigate the potential of biodegrading multiple VOCs, we initially reviewed the biodegradation of individual VOCs. VOCs include chlorinated ethenes (tetrachloroethene, trichloroethene, dichloroethene, and vinyl chloride), BTEX (benzene, toluene, ethylbenzene, and xylene), and chlorinated methanes (carbon tetrachloride, chloroform, and dichloromethane). We also summarized essential information on the biodegradation of each kind of VOC under aerobic and anaerobic conditions, together with the microorganisms that are involved in VOC-degrading pathways. Interactions among multiple VOCs were then discussed based on concrete examples. Under conditions in which multiple VOCs co-exist, the biodegradation of a VOC may be constrained, enhanced, and/or unaffected by other compounds. Co-metabolism may enhance the degradation of other VOCs. In contrast, constraints are imposed by the toxicity of co-existing VOCs and their by-products, catabolite repression, or competition between VOC-degrading enzymes. This review provides fundamental, but systematic information for designing strategies for the bioremediation of multiple VOCs, as well as information on the role of key microorganisms that degrade VOCs.
Keywords: biodegradation, chlorinated ethene, BTEX, chlorinated methane, multiple VOCs
Volatile organic compounds (VOCs) are major pollutants that are found in soil and groundwater in developed countries. Contamination by tetrachloroethene (PCE), trichloroethene (TCE), benzene, and cis-dichloroethene (cis-DCE) accounts for approximately 11%, 10%, 9%, and 8%, respectively, in areas in which contamination exceeds environmental standards in Japan (121). In the United States, contamination by TCE, vinyl chloride (VC), benzene, and PCE accounts for 22%, 9%, 8%, and 7%, respectively, in the operable units of superfund sites (182). The International Agency for Research on Cancer reported the carcinogenic properties of VOCs, and, among them, TCE, VC, and benzene are associated with high cancer risks to humans (http://monographs.iarc.fr/). Thus, soil and groundwater that are contaminated with VOCs require remediation.
Regarding remediation technologies, bioremediation, which uses the degradation abilities of microorganisms, has received much attention because it is inexpensive, environmentally friendly, and applicable in situ (77, 210). According to a report by the United States Environmental Protection Agency (181), bioremediation accounted for 24% of the remediation technologies for contaminated groundwater. Various environmental microorganisms that are capable of degrading individual VOCs have been reported, and genomic information related to their phylogenetic classification and VOC-degrading enzymes are also available.
However, actual soil and groundwater, e.g., those of chemical factories (139, 149), research facilities (176, 177, 179), military bases (178, 183), landfills (35, 184, 185), and illegal dumping sites (180), are frequently contaminated with multiple pollutants rather than a single type of VOC. Difficulties are associated with the biodegradation of multiple VOCs, which has remained a challenging issue in practice for decades (193). Alexander (4) reported that the effects of one VOC on other co-existing VOCs are largely unknown, and these effects have rarely been examined. Yoshikawa et al. (208) recently described a successful case study on the complete biodegradation of multiple VOCs including chlorinated ethenes, benzene, toluene, and dichloromethane through integrated anaerobic-aerobic biodegradation.
In order to systematically review the biodegradation of VOCs, and further investigate the potential of bioremediating multiple VOCs, we initially reviewed studies on the biodegradation of individual VOCs (Table 1), with an emphasis on information about useful microorganisms and mechanisms for the degradation of different VOCs. We investigated the biodegradation of chlorinated ethenes, BTEX (benzene, toluene, ethylbenzene, and xylene), and chlorinated methanes under aerobic and anaerobic conditions in detail. The effects of microorganisms on the biodegradation of a certain VOC with the co-existence of other VOCs were then evaluated in order to discuss the potential of bioremediation for multiple VOCs.
Table 1.
Mechanisms associated with the initial step in the biodegradation of each type of VOC.
| Category | Compounds | Aerobic degradation | Anaerobic degradation |
|---|---|---|---|
| Chlorinated ethenes | Tetrachloroethene (PCE) | Oxidation*1 | Reductive dechlorination116,164) |
| Trichloroethene (TCE) | Oxidation115,201) | ||
| Dichloroethene (DCE) cis-dichloroethene (cis-DCE) trans-dichloroethene (trans-DCE) 1,1-dichloroethene (1,1-DCE) | |||
| Vinyl chloride (VC) | |||
| BTEX | Benzene | Oxidation57,58,186) | *2 |
| Toluene | Fumarate addition199) | ||
| Ethylbenzene | Oxidation/fumarate addition16,199) | ||
| Xylene o-xylene m-xylene p-xylene |
Fumarate addition96,199) | ||
| Chlorinated methanes | Carbon tetrachloride (CT) | *3 | Reductive dechlorination61,146) |
| Chloroform (CF) | Oxidation28) | ||
| Dichloromethane (DCM) | Dechlorination (glutathione substitution)127) | Fermentation105) |
The aerobic degradation of PCE is limited, except as described by Ryoo et al. (155).
The mechanisms underlying the anaerobic degradation of benzene are unclear, although hydroxylation to phenol, methylation to toluene, and carboxylation to benzoate were proposed by Weelink et al. (199).
The aerobic degradation of CT remains ambiguous.
Biodegradation of chlorinated ethenes
Aerobic biodegradation of chlorinated ethenes
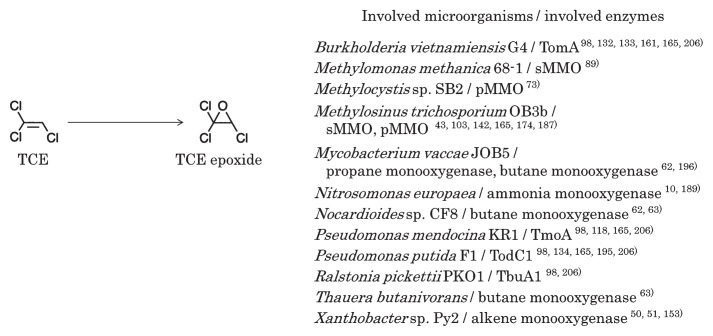
The aerobic biodegradation of chlorinated ethenes with natural gas containing methane, which acts as a co-substrate, was first discovered in the 1980s (201). Besides methane, aromatic compounds (133, 152), alkanes (63, 191, 196), alkenes (50, 64, 192), and ammonia (10) have been confirmed as co-substrates for the degradation of chlorinated ethenes. In addition, phytochemicals from poplar (Populus) leaves also function as co-substrates, resulting in the degradation of TCE (78). Oxygenases that degrade co-substrates lead to the degradation of chlorinated ethenes to epoxide compounds (Fig. 1). The growth-linked oxidation of chlorinated ethenes has only been reported for cis-DCE and VC. Limited information is currently available on the aerobic degradation of PCE (155), and, thus, further studies are required.
Fig. 1.
Possible initial step in the aerobic biodegradation of trichloroethene. TCE denotes trichloroethene. Abbreviations of involved enzymes indicate the following: TomA, toluene 2-monooxygenase; sMMO, soluble methane monooxygenase; pMMO, particulate methane monooxygenase; TmoA, toluene-4-monooxygenase; TodC1, toluene 2,3-dioxygenase; TbuA1, toluene 3-monooxygenase.
Aerobic microorganisms that degrade chlorinated ethenes with oxygenases have been isolated. Methanotrophs such as Methylomonas methanica 68-1 (89), Methylocystis sp. SB2 (73), and Methylosinus trichosporium OB3b (142) use methane monooxygenases to degrade chlorinated ethenes. Aromatic compound degraders, such as Burkholderia vietnamiensis G4 (132) and Pseudomonas putida F1 (134), use toluene monooxygenases and dioxygenases to degrade TCE. Nocardioides sp. CF8 and Thauera butanivorans use butane monooxygenases to degrade TCE, cis-DCE, and VC (63). Mycobacterium ethylenense NBB4, which was isolated on ethene, degrades VC (113). In contrast to the microorganisms described above, Mycobacterium aurum L1 oxidizes VC with growth, and uses an alkene monooxygenase to degrade cis-DCE, trans-DCE, and 1,1-DCE without growth (64, 65). Two microbes, Polaromonas sp. JS666 (38) and Rhodococcus jostii RHA1 (8), are known to oxidize cis-DCE with growth.
Anaerobic biodegradation of chlorinated ethenes
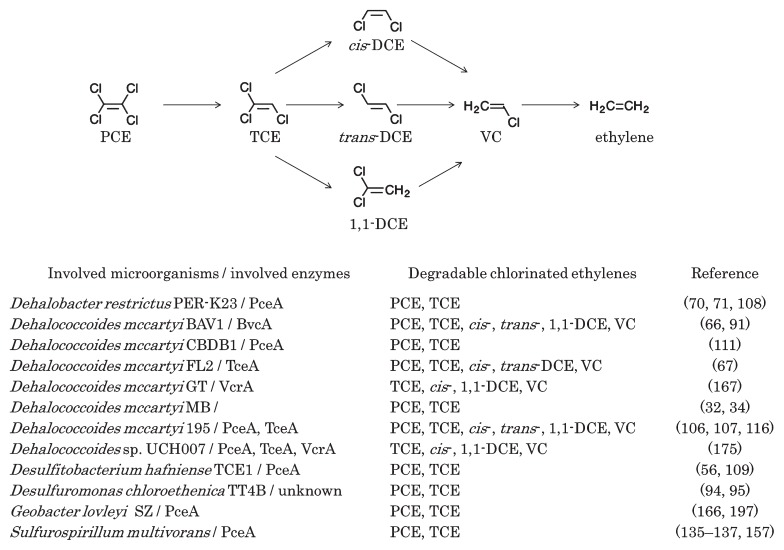
The anaerobic biodegradation of chlorinated ethenes is caused by dechlorination, in which hydrogen sequentially displaces chlorine (186) (Fig. 2). PCE is mainly degraded to TCE, DCEs, VC, and harmless ethene, and among DCEs, cis-DCE predominates over trans-DCE and 1,1-DCE (164, 186). Dechlorination produces energy for degrading microbes; however, they cannot use chlorinated ethenes as a carbon source (86). Besides the main sequential dechlorination pathway described above, the anaerobic oxidization of cis-DCE, VC, and ethene have also been observed under sulfate-reducing and methanogenic conditions (22, 49, 115).
Fig. 2.
Possible pathways of anaerobic biodegradation for chlorinated ethenes. Abbreviations of involved enzymes indicate the following: PceA, dehalogenase dechlorinating PCE and TCE to cis-DCE; BvcA, dehalogenase dechlorinating VC; TceA, dehalogenase dechlorinating TCE to VC; VcrA, dehalogenase dechlorinating all DCE isomers to ethene. Abbreviations of VOCs indicate the following: PCE, tetrachloroethene; TCE, trichloroethene; DCE, dichloroethene; VC, vinyl chloride
Anaerobic microbes that degrade chlorinated ethenes are diverse (Fig. 2). However, only the genus Dehalococcoides is known to degrade DCEs and VC. The isolation of anaerobic degraders of DCEs and VC has been a significant issue for a long time, and Dehalococcoides mccartyi 195 was first isolated in 1997 (102, 116). Strain 195 degrades PCE, TCE, cis-DCE, and 1,1-DCE as growth-linked substrates, and degrades trans-DCE and VC as non-growth substrates. Unlike other Dehalococcoides species, D. mccartyi strains MB and CBDB1 dechlorinate TCE and generate trans-DCE, rather than cis-DCE (32, 111). Dehalococcoides has key reductive dehalogenases, such as TceA, which dechlorinate TCE and all DCE isomers to VC, as well as VC to ethene at low dechlorinating rates (107), VcrA, which dechlorinates all DCE isomers to ethene, as well as TCE to cis-DCE at low dechlorinating rates. (129), and BvcA, which dechlorinates all DCE isomers to VC, and dechlorinates TCE without growth (91, 171). A gene expression analysis suggested that the reductive dehalogenase gene mbrA is involved in the production of trans-DCE in the dechlorinating pathway (34). Desulfitobacterium strains as well as Dehalococcoides, have the dehalogenase, PceA, which dechlorinates PCE and TCE to cis-DCE (60, 168). Strains of Dehalococcoides, such as BTF08 highly enriched from groundwater and UCH007 isolated from groundwater in Japan, contain the genes of three well-known reductive dehalogenases, pceA, tceA, and vcrA (148, 175). Accompanied by advances in genome sequencing techniques, putative reductive dehalogenases in Dehalococcoides have been reported (148, 158, 198). Multiple reductive dehalogenase genes may be induced by a single chlorinated ethene in a microbial enrichment culture containing Dehalococcoides, as demonstrated by Futamata et al. (55). The X-ray crystal structure of PceA from Sulfurospirillum multivorans has been reported by Bommer et al. (20), and revealed that cobalamin supports reductive dechlorination.
In engineering practices associated with the bioremediation of chlorinated ethenes, electron donors (e.g. lactate, methanol, molasses, hydrogen release compounds, and vegetable oils) and vitamin B12 are commonly injected into contaminated sites in order to stimulate reductive dechlorination (144, 182). Yeast extract also stimulates reductive dechlorination (122). As for bioaugmentation, microbial consortia containing Dehalococcoides, such as KB-1 (45), have been introduced into contaminated sites. Successful case studies on bioaugmentation have been reported (48, 110). The density of useful microorganisms is used as a criterion for selecting biostimulation or bioaugmentation, and genetic biomarkers such as the Dehalococcoides 16S rRNA gene and reductive dehalogenase genes including tceA, vcrA, and bvcA are used as indicators (75, 182).
Biodegradation of BTEX
Aerobic biodegradation of BTEX
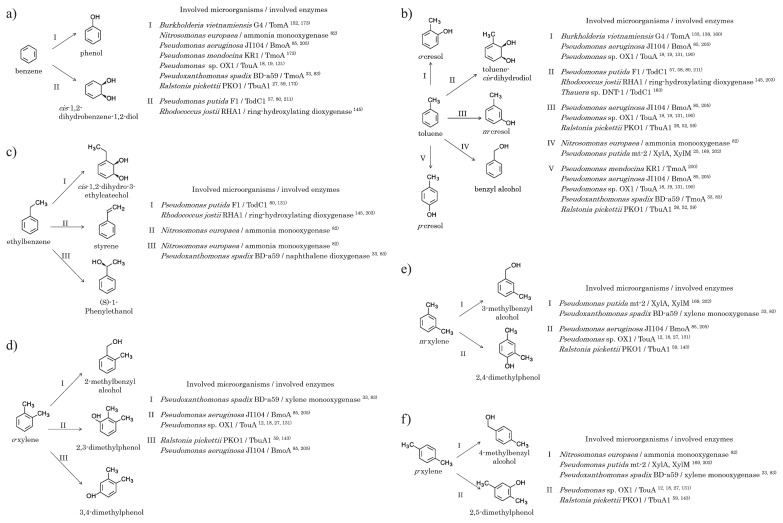
The aerobic biodegradation of BTEX has a long history, and BTEX-degrading pathways may be traced back to the 1960s (57, 58). BTEX are oxidized by oxygenases (Fig. 3). The intermediates, catechol compounds, are produced by these pathways: catechol during benzene and toluene degradation, 3-methylcatechol during toluene, o-xylene, and m-xylene degradation, and 4-methylcatechol during p-xylene degradation.
Fig. 3.
Possible initial steps in the aerobic biodegradation of benzene, toluene, ethylbenzene, and xylene. Each figure shows initial steps for a particular VOC: a), benzene; b), toluene; c), ethylbenzene; d), o-xylene; e), m-xylene; f), p-xylene. Abbreviations of involved enzymes indicate the following: TomA, toluene 2-monooxygenase; TmoA, toluene-4-monooxygenase; BmoA, benzene monooxygenase; TouA, toluene/o-xylene monooxygenase; TbuA1, toluene 3-monooxygenase; TodC1, toluene 2,3-dioxygenase; XylA, xylene monooxygenase; XylM, xylene monooxygenase.
The degradability of BTEX and the degrading pathway used by microorganisms depend on the types of degrading enzymes. Pseudomonas mendocina KR1, Ralstonia pickettii PKO1, and B. vietnamiensis G4 degrade benzene as well as toluene using toluene-4-monooxygenase (TmoA), toluene 3-monooxygenase (TbuA1), and toluene 2-monooxygenase (TomA), respectively (26, 52, 59, 138, 160, 161, 173, 200). Pseudomonas sp. OX1 degrades benzene, toluene, and o-xylene using the toluene/o-xylene monooxygenase TouA (12, 18, 19, 131, 190). P. putida mt-2 degrades toluene and xylenes using the xylene monooxygenases XylA and XylM (25, 159, 169, 202). Pseudomonas aeruginosa JI104 degrades benzene with the benzene monooxygenase BmoA (84, 85, 205). BmoA has low substrate specificity, and attacks toluene, xylene, and ethylbenzene, as well as benzene. Pseudoxanthomonas spadix BD-a59 degrades all BTEX (83), and has genes encoding TmoA, a xylene monooxygenase, and naphthalene monooxygenase (33). Nitrosomonas europaea degrades benzene, toluene, ethylbenzene, and p-xylene with an ammonia monooxygenase (82). In addition to monooxygenases, dioxygenases degrade BTEX. Toluene 2,3-dioxygenase (TodC1) from P. putida F1 degrades benzene, toluene, and ethylbenzene (57, 80, 131, 211, 212). R. jostii RHA1 degrades benzene, toluene, ethylbenzene, and o-xylene with a biphenyl dioxygenase and/or an ethylbenzene dioxygenase (145, 203). Thauera sp. DNT-1 degrades toluene with a dioxygenase under aerobic conditions (163). Strain DNT-1 also degrades toluene under anaerobic conditions via a pathway that produces benzyl succinate.
In the biostimulation of BTEX, an injection of oxygen release compounds (30) and an air sparging technique are commonly used in practical sites (79, 204). Various primer sets for PCR to detect genes coding BTEX-degrading enzymes have been developed (14, 68) and reverse-transcriptase (RT)-quantitative PCR for these genes is used in order to judge the effectiveness of oxygen injections (15).
Anaerobic biodegradation of BTEX
The anaerobic biodegradation of BTEX was regarded as difficult for a long time, and the microbial transformation of xylenes under anoxic conditions was first confirmed in the mid-1980s (96). In addition to xylenes, the biodegradation of aromatic compounds such as benzene, toluene, and ethylbenzene, in the absence of oxygen has been reported since the 1990s (e.g. 44, 97, 151). During the anaerobic biodegradation of BTEX, aromatic compounds supply electrons to various electron acceptors such as NO3−, Fe3+, SO42−, and HCO3− (194, 199). The anaerobic degradation of toluene, as well as xylenes and ethylbenzene, starts with fumarate addition. In addition to fumarate addition, ethylbenzene is oxidized by a dehydrogenase that is produced by nitrate-reducing bacteria (16). Regarding the anaerobic degradation of benzene, the degradation pathway remains unclear; however, possible pathways have been proposed in previous reviews (37, 53, 194, 199).
Various anaerobic BTEX degraders have been isolated (e.g. 199). Among them, those using nitrate as an electron acceptor, such as Aromatoleum aromaticum EbN1 (151), Azoarcus sp. T (44), and Thauera aromatica K172 (5), have been isolated most frequently. In addition, microorganisms that use ferric iron and sulfate as electron accepters, such as Geobacter grbiciae TACP-2T (36) and Desulfobacula toluolica Tol2 (150), have also been isolated. Under methanogenic conditions, members of Desulfobacterales and Coriobacteriaceae are involved in the anaerobic degradation of benzene, which has been confirmed by stable isotope probing (140). Microorganisms that degrade p-xylene were only recently isolated; Desulfosarcina sp. PP31 was isolated as a degrader under sulfate-reducing conditions by Higashioka et al. (69). In the anaerobic toluene degradation pathway, the initial step, fumarate addition to toluene, is catalyzed by a benzyl succinate synthase (BssA) (100). BssA may also catalyze fumarate addition to m-xylene (1, 17).
Biodegradation of chlorinated methanes
Aerobic biodegradation of chlorinated methanes
Although the aerobic biodegradation of carbon tetrachloride (CT) remains uncertain, chloroform (CF) and dichloromethane (DCM) may be degraded under aerobic conditions. Methane, toluene, and butane monooxygenases oxidize CF to phosgene through trichloromethanol (28). Aerobic growth-linked DCM degradation mainly relies on glutathione, and DCM is dechlorinated and transformed to formaldehyde (127). The aerobic oxidation of DCM also occurs when methane and ammonia co-exist, although the degrading microorganisms do not assimilate DCM (142, 189).
Aerobic CF degraders have been obtained, as reported by Cappelletti et al. (28). Microorganisms, such as M. trichosporium OB3b (142), Nocardioides sp. CF8 (63), and P. mendocina KR1 (200), which degrade CF, use methane, butane, and toluene, respectively, as carbon and energy sources. An aerobic DCM-dechlorinating bacterium, Methylopila helvetica DM1, was first reported by Brunner et al. (23). A wide variety of methylotrophic bacteria, such as Ancylobacter, Bacillus, Chryseobacterium, Hyphomicrobium, and Methylobacterium (127) species, have been shown to degrade DCM with growth. Rhodococcus sp. EH831 degrades DCM and BTEX (99), suggesting that it has potential as a degrader of multiple VOCs. Most of these degrading microorganisms have been assessed for the presence of the DCM dehalogenase DcmA, which catalyzes the dechlorination of DCM. Methylobacterium extorquens DM4 is considered to have acquired the dcmA gene through horizontal gene transfer (156). In M. extorquens DM4, the acquired dcmA gene has been shown to participate in enzymatic or metabolic pathways, such as stress responses, metabolic tuning, regulation, cell structure adjustments to the solvent properties of DCM, DNA repair following damage with mutagenic agents, and chloride export (81, 120, 128). In addition, microbes that degrade DCM as non-growth substrates have also been isolated. M. trichosporium OB3b and N. europaea degrade DCM using a methane monooxygenase and ammonia monooxygenase, respectively (142, 189).
Anaerobic biodegradation of chlorinated methanes
CT is dechlorinated under anaerobic conditions, and this process is mediated by cofactors such as corrinoid (93), coenzyme F430 (92), iron compounds (147), cytochromes (29), and humic substances (114). Under sulfate-reducing conditions, CT is mainly degraded to CS2 with the cofactor vitamin B12, a type of corrinoid, while it is degraded to CF in the absence of vitamin B12 (87). The dechlorination of CF to DCM occurs with or without growth. The growth-linked dechlorination of CF was first reported by Grostern et al. (61), and, in their study, Dehalobacter appeared to dechlorinate CF to DCM. The pathway of anaerobic DCM biodegradation remains unknown. Rather than being dechlorinated, DCM is considered to be fermented into formate and acetate (105).
Although anaerobic CT degraders have been isolated (146), the microorganisms that use CT as a carbon source have not. Acetogens, iron reducers, and methanogens degrade CT with cofactors. An acetogenic microorganism, Acetobacterium woodii DSM1030, anaerobically degrades CT and CF with vitamin B12 (46). Iron-reducing microorganisms, such as Geobacter metallireducens and G. sulfurreducens, degrade CT with iron compounds (109). Chloroform-reductive dehalogenases that are involved in CF degradation with growth have recently been revealed from Dehalobacter sp. CF50 (170, 172) and Desulfitobacterium sp. PR (42). As anaerobic DCM degraders, Dehalobacterium formicoaceticum DMC (104) and Dehalobacter strains (76) have been successfully isolated; however, the enzymes involved in the fermentative degradation of DCM have yet to be identified. In addition to degrading DCM under aerobic conditions, Hyphomicrobium sp. DM2 also degrades DCM using DcmA under anaerobic conditions (90).
Interactions among co-existing VOCs
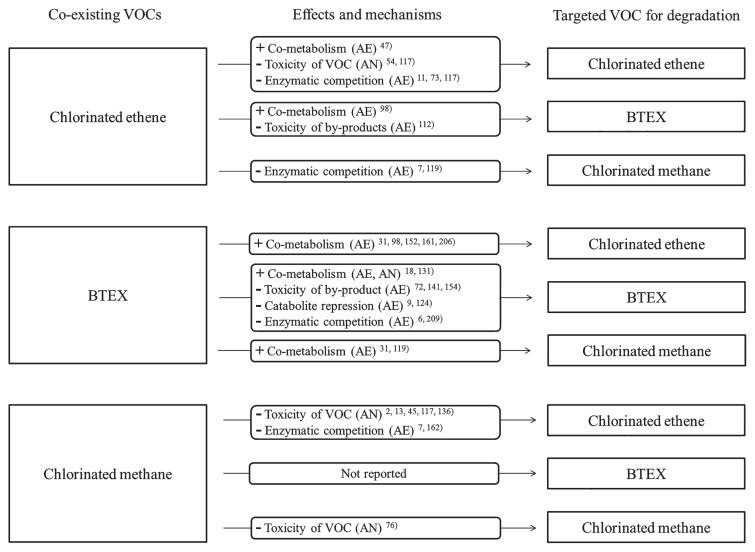
VOC biodegradation may be enhanced (207), constrained (141), and/or unaffected (24) by co-existing VOCs. In most cases, the enhancement of VOC degradation occurs because of the co-metabolism of VOC-degrading enzymes. Conversely, constraints of VOC degradation occur because of the toxicity of co-existing VOCs and their degradation products, catabolite repression, and competition with VOC-degrading enzymes (Fig. 4). We analyze the interactions among co-existing chlorinated ethenes, BTEX, and chlorinated methanes below.
Fig. 4.
Possible interaction among the targeted VOC for degradation and co-existing VOCs. BTEX means benzene, toluene, ethylbenzene, and xylene. + and − indicate enhancement and constraint, respectively. AE and AN in brackets mean the effects occurring under aerobic and anaerobic conditions, respectively.
Enhancement
Co-metabolism of multiple VOCs
Co-metabolism is defined as the transformation of an organic compound by a microorganism that is unable to use the compound as a source of energy or one of its constituent elements (3, 4). The co-existence of multiple VOCs may lead to co-metabolism in which one VOC is degraded as a growth-linked substrate and the other is co-metabolically degraded as a non-growth substrate. In co-metabolism, VOCs may be degraded by the same enzymes or one VOC functions as an inducer for the degradation of the other VOCs. Other VOC-degrading enzymes may be gratuitously induced by growth-linked substrates or their metabolites.
As described earlier, chlorinated ethenes are known to be degraded under aerobic conditions while degrading microorganisms utilize another chlorinated ethene, benzene, toluene, or xylene as the growth-linked substrate (Table 2). Degrading enzymes for BTEX, such as TouA, work on multiple BTEX in some cases, while BTEX are utilized as a growth-linked or non-growth substrate (18, 131).
Table 2.
Enhancement of VOC degradation by co-metabolism.
| Microorganism | Targeted VOC for degradation | Growth-linked VOCs | Possible degrading enzyme | Reference |
|---|---|---|---|---|
| Burkholderia vietnamiensis G4 | TCE | benzene, toluene | TomA | (152, 161, 173) |
| Pseudomonas mendocina KR1 | CF | toluene | TmoA | (119) |
| Pseudomonas putida F1 | o-xylene | ethylbenzene | *1 | (131) |
| Pseudomonas sp. ENVBF1 | CF | toluene | *1 | (119) |
| Pseudomonas sp. ENVCP5 | CF | toluene | *1 | (119) |
| Pseudomonas sp. OX1 | TCE*2 | toluene, o-xylene | TouA | (31, 131) |
| 1,1-DCE*2 | (31, 131) | |||
| CF*2 | (31, 131) | |||
| ethylbenzene*2 | (18, 131) | |||
| m-xylene*2 | (18, 131) | |||
| p-xylene*2 | (18, 131) | |||
| Ralstonia pickettii PKO1 | TCE | toluene | TbuA1 | (98, 206) |
| Ralstonia sp. TRW-1 | cis-DCE | VC | *1 | (47) |
| trans-DCE | *1 | (47) |
Unidentified enzymes degrading growth-linked VOCs and/or enzymes induced by growth-linked VOCs or their metabolites may be related to degradation.
The degradation of VOCs was confirmed withEscherichia coli JM109 (pBZ1260) expressing touA.
Abbreviations of VOCs indicate the following: TCE, trichloroethene; DCE, dichloroethene; VC, vinyl chloride; CF, chloroform. Abbreviations of degrading enzymes denote the following: TomA, toluene 2-monooxygenase; TmoA, toluene-4-monooxygenase; TouA, toluene/o-xylene monooxygenase; TbuA1, toluene 3-monooxygenase.
The chlorinated methane, CF, is degraded as a non-growth substrate under aerobic conditions with a growth-linked substrate such as toluene and o-xylene (31, 119).
Constraints
Toxicity of co-existing VOCs
The toxicity of VOCs to microorganisms is caused by their inability to detoxify VOCs. The toxicity of VOCs influences microbial growth (88) and the degradability of VOCs (54). These effects are generally greater at high VOC concentrations (13, 45, 88). Tolerance to the toxicity of VOCs differs among microorganisms. Koenig et al. (88) reported that fast-growing microorganisms in VOC-free cultures, such as Klebsiella spp., have a higher tolerance to VOCs than Desulfovibrio vulgaris.
The constraints caused by the toxicity of co-existing VOCs occur in the anaerobic degradation of chlorinated ethenes (Table 3). In addition, the co-existence of chlorinated methanes inhibits the anaerobic degradation of chlorinated ethenes. During the anaerobic degradation of DCM, CF-mediated inhibition occurs, and this is attributed to its toxicity (76).
Table 3.
Constraints of VOC degradation caused by the toxicity of co-existing VOCs to microorganisms.
| Microorganism | Targeted VOC for degradation | Co-existing toxic VOCs | Concentration of co-existing toxic VOCs | Reference |
|---|---|---|---|---|
| Dehalobacter sp. | DCM | CF | 42 μM | (76) |
| Desulfitobacterium hafniense Y51 | PCE | cis-DCE | 5 mM*1 | (54) |
| TCE | 5 mM*1 | (54) | ||
| Sulfurospirillum multivorans | PCE | cis-DCE | 14 mM*2 | (136) |
| CT | 100 μM*2 | (136) | ||
| CF | 25 μM*2 | (136) | ||
| DCM | 50 μM*2 | (136) | ||
| Microcosm | PCE | CT | 10 15 μM | (2) |
| VC | 10 15 μM | (2) | ||
| Microcosm | PCE | CT | 19 μM | (13) |
| CF | 4 μM | (13) | ||
| Microcosm | VC | CF | 2.5 μM | (45) |
| Microcosm | TCE | CF | 1.6 μM | (117) |
Desulfitobacterium hafniense Y51 lost the pceA gene.
The concentration indicates the inhibition of PCE dehalogenase activity by 50%.
Abbreviations of VOCs indicate the following: PCE, tetrachloroethene; TCE, trichloroethene; DCE, dichloroethene; VC, vinyl chloride; CT, carbon tetrachloride; CF, chloroform; DCM, dichloromethane.
Toxicity of by-products following the degradation of co-existing VOCs
When multiple VOCs co-exist, the toxicity of their by-products may affect the degradation of other VOCs. The by-products of VOC degradation, such as epoxide compounds and catechol compounds, are toxic. Epoxide compounds, which may be toxic to microorganisms and inhibit VOC degradation, are produced from the aerobic degradation of chlorinated ethenes (188). Of a mixture of four toluene-degrading bacteria, P. putida mt-2, P. putida F1, P. putida GJ31, and B. vietnamiensis G4, only P. putida mt-2 survived exposure to TCE and subsequent TCE degradation (112). This was because the other three microorganisms degraded TCE and then died because of the toxicity of the TCE by-product. In order to avoid the toxicity of epoxide compounds, a system, such as the epoxyalkane:coenzyme M transferase (EaCoMT) of Mycobacterium sp. JS60 (39, 40), is required to metabolize and/or detoxify by-products. The etnE gene, which encodes EaCoMT, is distributed in various environments, and has been detected in Mycobacterium, Nocardioides-like microorganisms, and Haliea-like microorganisms (101).
Catechol compounds are the main by-products of BTEX degradation, and concerns have been expressed regarding their toxicity (130). P. putida PPO1 produces toxic by-products, such as catechol compounds, during the degradation of p-xylene in the presence of benzene (141). The by-products from p-xylene inhibit benzene degradation, and the accumulation of these by-products increases the inhibition of VOC degradation. 3-Methylcatechol is produced in the degradation pathway of toluene, o-xylene, and m-xylene. Microbial growth ceases with the accumulation of 3-methylcatechol and toluene degradation is limited by P. putida strains (72, 154). In order to avoid constraints, microorganisms need enzymes, such as catechol 2,3-dioxygenase encoded by xylE of P. putida mt-2 (74, 202) and 3-methylcatechol 2,3-dioxygenase encoded by todE of P. putida F1 (21, 211), which degrade 3-methylcatechol.
Catabolite repression
Catabolite repression occurs when microbes are exposed to multiple carbon sources. This leads the microorganisms to use a rapidly metabolizable carbon source first. Catabolite repression has been extensively studied in Escherichia coli, which uses glucose and other carbon sources (41), and, thus, catabolite repression may occur in the presence of multiple VOCs.
The degradation of toluene and xylene is inhibited by catabolite repression, which is induced by a rapidly metabolizable carbon source, such as succinate (8), a by-product of benzene and toluene degradation. The phosphotransferase enzyme IIA component encoded by the pstN gene, as well as the catabolite repression control (Crc) protein, is involved in this repression (9, 123). The Crc protein produced by P. putida has been studied in detail, and regulates toluene and xylene degradation by binding the translation initiation sites of mRNAs that are in the toluene/xylene degradation pathway (125). The mRNA levels of toluene/xylene degradation pathway genes, such as xylA and xylM, are more than 50% lower in a wild-type P. putida strain than in a crc mutant. Two small RNAs, corresponding to the crcY and crcZ genes, control Crc protein levels (126). Crc also inhibits the degradation of the by-product of toluene, benzoate, to catechol (124). These findings suggest that the presence of multiple VOCs leads to an excess of easily metabolizable carbon sources, as well as VOC by-products, which may cause catabolite repression and inhibit VOC degradation.
Competition for degrading enzymes
Degrading enzymes work on different co-existing VOCs in some cases (Table 4). Methane monooxygenases degrade chlorinated ethenes and chlorinated methanes (43, 73, 142), and toluene monooxygenases also degrade chlorinated ethene and chlorinated methane compounds such as TCE and CF (119, 161). The oxygenases of BTEX react with multiple compounds of BTEX (57, 80). Thus, these enzymes compete for substrates.
Table 4.
Constraints of VOC degradation caused by competition for degrading enzymes.
| Microorganism | Degrading enzyme | VOCs causing competitive inhibition | Reference |
|---|---|---|---|
| Methylocystis sp. SB2 | pMMO | TCE, cis-DCE, and VC | (73) |
| Methylosinus trichosporium OB3b | sMMO | TCE and trans-DCE | (11) |
| cis-DCE and trans-DCE | |||
| Pseudomonas mendocina KR1 | toluene monooxygenase | TCE and CF*1 | (119) |
| Pseudomonas putida F1 | toluene dioxygenase | benzene and toluene | (209) |
| toluene and p-xylene | |||
| Pseudomonas sp. CFS-215 | toluene dioxygenase | benzene and toluene | (6) |
| Pseudomonas sp. ENVBF1 | toluene monooxygenase | TCE and CF*1 | (119) |
| Pseudomonas sp. OX1 | TouA | TCE and CF*2 | (162) |
| Methanotrophic microcosm | methane monooxygenase | TCE and CF | (7) |
The co-existence of TCE inhibited the degradation of CF, while TCE degradation was not affected. Abbreviations of VOCs indicate the following: TCE, trichloroethene; DCE, dichloroethene; VC, vinyl chloride; CF, chloroform. Abbreviations of degrading enzymes denote the following: pMMO, particulate methane monooxygenase; sMMO, soluble methane monooxygenase; TouA, toluene/o-xylene monooxygenase; MMO, methane monooxygenase.
The TCE degradation rate decreased from 82% to 57% because of the co-existence of CF, while CF degradation did not change.
Future perspectives
Previous studies on VOC biodegradation mostly examined the degradation of a single VOC, even though contaminated sites are often polluted with multiple VOCs. In this review, a systematic survey associated with the biodegradation of chlorinated ethenes, BTEX, and chlorinated methanes was performed. The enhancement and constraint of VOC degradation were discussed with an emphasis on the effects of co-existing VOCs on useful microorganisms for a certain VOC. This review may provide fundamental, but useful knowledge for developing novel approaches to the biodegradation of multiple VOCs. There are diverse interactions among co-existing VOCs, depending on the kinds of degrading microorganisms and types of VOCs. In order to achieve effective designs and operations associated with the bioremediation of multiple VOCs in practice, the use of combined multiple microorganisms that degrade VOC and/or the introduction of microorganisms that degrade multiple VOCs may be a feasible strategy. Further studies on the interactions among VOCs are required, particularly on stimulatory interactions for increasing the efficiency of bioremediation. The use of new tools, such as isotopic and enzymatic analyses, will increase our understanding of the detailed mechanisms associated with interactions among co-existing VOCs.
References
- 1.Achong G.R., Rodriguez A.M., Spormann A.M. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J Bacteriol. 2001;183:6763–6770. doi: 10.1128/JB.183.23.6763-6770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson D.T., Parkin G.F. Impact of mixtures of chlorinated aliphatic hydrocarbons on a high-rate, tetrachloroethenedechlorinating enrichment culture. Environ Sci Technol. 2000;34:1959–1965. [Google Scholar]
- 3.Alexander M. The breakdown of pesticides in soils. In: Brady N.C., editor. Agriculture and the Quality of Our Environment. American Association for the Advancement of Science; Washington, D.C: 1967. pp. 331–342. [Google Scholar]
- 4.Alexander M. Biodegradation and Bioremediation. Academic Press; San Diego: 1994. [Google Scholar]
- 5.Altenschmidt U., Fuchs G. Anaerobic degradation of toluene in denitrifying Pseudomonas sp.: indication for toluene methylhydroxylation and benzoyl-CoA as central aromatic intermediate. Arch Microbiol. 1991;156:152–158. doi: 10.1007/BF00290990. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez P.J., Vogel T.M. Substrate interactions of benzene, toluene, and para-xylene during microbial degradation by pure cultures and mixed culture aquifer slurries. Appl Environ Microbiol. 1991;57:2981–2985. doi: 10.1128/aem.57.10.2981-2985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez-Cohen L., McCarty P.L. Product toxicity and cometabolic competitive inhibition modeling of chloroform and trichloroethylene transformation by methanotrophic resting cells. Appl Environ Microbiol. 1991;57:1031–1037. doi: 10.1128/aem.57.4.1031-1037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araki N., Chino K., Kasai D., Masai E., Fukuda M. Degradation of cis-1,2-dichloroethylene by Rhodococcus jostii RHA1. The 66th Annual Meeting of the Society of Biotechnology of Japan 3P-091; 2014. (in Japanese) [Google Scholar]
- 9.Aranda-Olmedo I., Marín P., Ramos J.L., Marqués S. Role of the ptsN gene product in catabolite repression of the Pseudomonas putida TOL toluene degradation pathway in chemostat cultures. Appl Environ Microbiol. 2006;72:7418–7421. doi: 10.1128/AEM.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arciero D., Vannelli T., Logan M., Hooper A.B. Degradation of trichloroethylene by the ammonia-oxidizing bacterium Nitrosomonas europaea. Biochem Biophys Res Commun. 1989;159:640–643. doi: 10.1016/0006-291x(89)90042-9. [DOI] [PubMed] [Google Scholar]
- 11.Aziz C.E., Georgiou G., Speitel G.E., Jr Cometabolism of chlorinated solvents and binary chlorinated solvent mixtures using M. trichosporium OB3b PP358. Biotechnol Bioeng. 1999;65:100–107. doi: 10.1002/(sici)1097-0290(19991005)65:1<100::aid-bit12>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 12.Baggi G., Barbieri P., Galli E., Tollari S. Isolation of a Pseudomonas stutzeri strain that degrades o-xylene. Appl Environ Microbiol. 1987;53:2129–2132. doi: 10.1128/aem.53.9.2129-2132.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagley D.M., Lalonde M., Kaseros V., Stasiuk K.E., Sleep B.E. Acclimation of anaerobic systems to biodegrade tetrachloroethene in the presence of carbon tetrachloride and chloroform. Water Res. 2000;34:171–178. [Google Scholar]
- 14.Baldwin B.R., Nakatsu C.H., Nies L. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl Environ Microbiol. 2003;69:3350–3358. doi: 10.1128/AEM.69.6.3350-3358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldwin B.R., Biernacki A., Blair J., Purchase M.P., Baker J.M., Sublette K., Davis G., Ogles D. Monitoring gene expression to evaluate oxygen infusion at a gasoline-contaminated site. Environ Sci Technol. 2010;44:6829–6834. doi: 10.1021/es101356t. [DOI] [PubMed] [Google Scholar]
- 16.Ball H.A., Johnson H.A., Reinhard M., Spormann A.M. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J Bacteriol. 1996;178:5755–5761. doi: 10.1128/jb.178.19.5755-5761.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beller H.R., Spormann A.M. Substrate range of benzylsuccinate synthase from Azoarcus sp. strain T. FEMS Microbiol Lett. 1999;178:147–153. doi: 10.1111/j.1574-6968.1999.tb13771.x. [DOI] [PubMed] [Google Scholar]
- 18.Bertoni G., Bolognese F., Galli E., Barbieri P. Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1996;62:3704–3711. doi: 10.1128/aem.62.10.3704-3711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertoni G., Martino M., Galli E., Barbieri P. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1998;64:3626–3632. doi: 10.1128/aem.64.10.3626-3632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bommer M., Kunze C., Fesseler J., Schubert T., Diekert G., Dobbek H. Structural basis for organohalide respiration. Science. 2014;346:455–458. doi: 10.1126/science.1258118. [DOI] [PubMed] [Google Scholar]
- 21.Bordel S., Muñoz R., Díaz L.F., Villaverde S. New insights on toluene biodegradation by Pseudomonas putida F1: influence of pollutant concentration and excreted metabolites. Appl Microbiol Biotechnol. 2007;74:857–866. doi: 10.1007/s00253-006-0724-8. [DOI] [PubMed] [Google Scholar]
- 22.Bradley P.M., Chapelle F.H. Microbial mineralization of ethene under sulfate-reducing conditions. Bioremediat J. 2002;6:1–8. [Google Scholar]
- 23.Brunner W., Staub D., Leisinger T. Bacterial degradation of dichloromethane. Appl Environ Microbiol. 1980;40:950–958. doi: 10.1128/aem.40.5.950-958.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bucheli-Witschel M., Hafner T., Rüegg I., Egli T. Benzene degradation by Ralstonia pickettii PKO1 in the presence of the alternative substrate succinate. Biodegradation. 2009;20:419–431. doi: 10.1007/s10532-008-9233-z. [DOI] [PubMed] [Google Scholar]
- 25.Bühler B., Schmid A., Hauer B., Witholt B. Xylene monooxygenase catalyzes the multistep oxygenation of toluene and pseudocumene to corresponding alcohols, aldehydes, and acids in Escherichia coli JM101. J Biol Chem. 2000;275:10085–10092. doi: 10.1074/jbc.275.14.10085. [DOI] [PubMed] [Google Scholar]
- 26.Byrne A.M., Kukor J.J., Olsen R.H. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PKO1. Gene. 1995;154:65–70. doi: 10.1016/0378-1119(94)00844-i. [DOI] [PubMed] [Google Scholar]
- 27.Cafaro V., Notomista E., Capasso P., Di Donato A. Regiospecificity of two multicomponent monooxygenases from Pseudomonas stutzeri OX1: molecular basis for catabolic adaptation of this microorganism to methylated aromatic compounds. Appl Environ Microbiol. 2005;71:4736–4743. doi: 10.1128/AEM.71.8.4736-4743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cappelletti M., Frascari D., Zannoni D., Fedi S. Microbial degradation of chloroform. Appl Microbiol Biotechnol. 2012;96:1395–1409. doi: 10.1007/s00253-012-4494-1. [DOI] [PubMed] [Google Scholar]
- 29.Castro C.E., Wade R.S., Belser N.O. Biodehalogenation: reactions of cytochrome P-450 with polyhalomethanes. Biochemistry. 1985;24:204–210. doi: 10.1021/bi00322a029. [DOI] [PubMed] [Google Scholar]
- 30.Chapman S.W., Byerley B.T., Smyth D.J.A., Mackay D.M. A pilot test of passive oxygen release for enhancement of in situ bioremediation of BTEX-contaminated ground water. Ground Water Monit Remed. 1997;17:93–105. [Google Scholar]
- 31.Chauhan S., Barbieri P., Wood T.K. Oxidation of trichloroethylene, 1,1-dichloroethylene, and chloroform by toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1998;64:3023–3024. doi: 10.1128/aem.64.8.3023-3024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng D., He J. Isolation and characterization of “Dehalococcoides” sp. strain MB, which dechlorinates tetrachloroethene to trans-1,2-dichloroethene. Appl Environ Microbiol. 2009;75:5910–5918. doi: 10.1128/AEM.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi E.J., Jin H.M., Lee S.H., Math R.K., Madsen E.L., Jeon C.O. Comparative genomic analysis and benzene, toluene, ethylbenzene, and o-, m-, and p-xylene (BTEX) degradation pathways of Pseudoxanthomonas spadix BD-a59. Appl Environ Microbiol. 2013;79:663–671. doi: 10.1128/AEM.02809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow W.L., Cheng D., Wang S., He J. Identification and transcriptional analysis of trans-DCE-producing reductive dehalogenases in Dehalococcoides species. ISME J. 2010;4:1020–1030. doi: 10.1038/ismej.2010.27. [DOI] [PubMed] [Google Scholar]
- 35.Christensen T.H., Kjeldsen P., Albrechtsen H.-J., Heron G., Nielsen P.H., Bjerg P.L., Holm P.E. Attenuation of landfill leachate pollutants in aquifers. Crit Rev Environ Sci Technol. 1994;24:119–202. [Google Scholar]
- 36.Coates J.D., Bhupathiraju V.K., Achenbach L.A., Mclnerney M.J., Lovley D.R. Geobacter hydrogenophilus, Geobacter chapellei and Geobacter grbiciae, three new, strictly anaerobic, dis-similatory Fe(III)-reducers. Int J Syst Evol Microbiol. 2001;51:581–588. doi: 10.1099/00207713-51-2-581. [DOI] [PubMed] [Google Scholar]
- 37.Coates J.D., Chakraborty R., McInerney M.J. Anaerobic benzene biodegradation—a new era. Res Microbiol. 2002;153:621–628. doi: 10.1016/s0923-2508(02)01378-5. [DOI] [PubMed] [Google Scholar]
- 38.Coleman N.V., Mattes T.E., Gossett J.M., Spain J.C. Biodegradation of cis-dichloroethene as the sole carbon source by a β-proteobacterium. Appl Environ Microbiol. 2002;68:2726–2730. doi: 10.1128/AEM.68.6.2726-2730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman N.V., Spain J.C. Epoxyalkane: coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J Bacteriol. 2003;185:5536–5545. doi: 10.1128/JB.185.18.5536-5545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coleman N.V., Spain J.C. Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl Environ Microbiol. 2003;69:6041–6046. doi: 10.1128/AEM.69.10.6041-6046.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Ding C., Zhao S., He J. A Desulfitobacterium sp. strain PR reductively dechlorinates both 1,1,1-trichloroethane and chloroform. Environ Microbiol. 2014;16:3387–3397. doi: 10.1111/1462-2920.12387. [DOI] [PubMed] [Google Scholar]
- 43.DiSpirito A.A., Gulledge J., Shiemke A.K., Murrell J.C., Lidstrom M.E., Krema C.L. Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II, and type X methanotrophs. Biodegradation. 1992;2:151–164. [Google Scholar]
- 44.Dolfing J., Zeyer J., Binder E.P., Schwarzenbach R.P. Isolation and characterization of a bacterium that mineralizes toluene in the absence of molecular oxygen. Arch Microbiol. 1990;154:336–341. doi: 10.1007/BF00276528. [DOI] [PubMed] [Google Scholar]
- 45.Duhamel M., Wehr S.D., Yu L., Rizvi H., Seepersad D., Dworatzek S., Cox E.E., Edwards E.A. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res. 2002;36:4193–4202. doi: 10.1016/s0043-1354(02)00151-3. [DOI] [PubMed] [Google Scholar]
- 46.Egli C., Tschan T., Scholtz R., Cook A.M., Leisinger T. Transformation of tetrachloromethane to dichloromethane and carbon dioxide by Acetobacterium woodii. Appl Environ Microbiol. 1988;54:2819–2824. doi: 10.1128/aem.54.11.2819-2824.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elango V.K., Liggenstoffer A.S., Fathepure B.Z. Biodegradation of vinyl chloride and cis-dichloroethene by a Ralstonia sp. strain TRW-1. Appl Microbiol Biotechnol. 2006;72:1270–1275. doi: 10.1007/s00253-006-0424-4. [DOI] [PubMed] [Google Scholar]
- 48.Ellis D.E., Lutz E.J., Odom J.M., Buchanan R.J., Bartlett C.L., Lee M.D., Harkness M.R., Deweerd K.A. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ Sci Technol. 2000;34:2254–2260. [Google Scholar]
- 49.Elsgaard L. Reductive transformation and inhibitory effect of ethylene under methanogenic conditions in peat-soil. Soil Biol Biochem. 2013;60:19–22. [Google Scholar]
- 50.Ensign S.A., Hyman M.R., Arp D.J. Cometabolic degradation of chlorinated alkenes by alken monooxygenase in a propylene-grown Xanthobacter strain. Appl Environ Microbiol. 1992;58:3038–3046. doi: 10.1128/aem.58.9.3038-3046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ensign S.A. Aliphatic and chlorinated alkenes and epoxides as inducers of alkene monooxygenase and epoxidase activities in Xanthobacter strain Py2. Appl Environ Microbiol. 1996;62:61–66. doi: 10.1128/aem.62.1.61-66.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fishman A., Tao Y., Wood T.K. Toluene 3-monooxygenase of Ralstonia pickettii PKO1 is a para-hydroxylating enzyme. J Bacteriol. 2004;186:3117–3123. doi: 10.1128/JB.186.10.3117-3123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foght J. Anaerobic biodegradation of aromatic hydrocarbons: pathways and prospects. J Mol Microbiol Biotechnol. 2008;15:93–120. doi: 10.1159/000121324. [DOI] [PubMed] [Google Scholar]
- 54.Furukawa K., Suyama A., Tsuboi Y., Futagami T., Goto M. Biochemical and molecular characterization of a tetrachloroethene dechlorinating Desulfitobacterium sp. strain Y51: a review. J Ind Microbiol Biotechnol. 2005;32:534–541. doi: 10.1007/s10295-005-0252-z. [DOI] [PubMed] [Google Scholar]
- 55.Futamata H., Kaiya S., Sugawara M., Hiraishi A. Phylogenetic and transcriptional analyses of a tetrachloroethenedechlorinating “Dehalococcoides” enrichment culture TUT2264 and its reductive-dehalogenase genes. Microbes Environ. 2009;24:330–337. doi: 10.1264/jsme2.me09133. [DOI] [PubMed] [Google Scholar]
- 56.Gerritse J., Drzyzga O., Kloetstra G., Keijmel M., Wiersum L.P., Hutson R., Collins M.D., Gottschal J.C. Influence of different electron donors and acceptors on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieri TCE1. Appl Environ Microbiol. 1999;65:5212–5221. doi: 10.1128/aem.65.12.5212-5221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibson D.T., Koch J.R., Kallio R.E. Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymatic formation of catechol from benzene. Biochemistry. 1968;7:2653–2662. doi: 10.1021/bi00847a031. [DOI] [PubMed] [Google Scholar]
- 58.Gibson D.T., Hensley M., Yoshioka H., Mabry T.J. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry. 1970;9:1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- 59.Gibson T.L., Abdul A.S., Olsen R.H. Microbial degradation of aromatic hydrocarbons in hydrogeological materials: microcosm studies. Groundwater and Geophysical Methods; Proceedings of the Second National Outdoor Action Conference on Aquifer Restoration; Dublin, Ohio: National Water Well Association; 1988. pp. 53–69. [Google Scholar]
- 60.Goris T., Hornung B., Kruse T., Reinhold A., Westermann M., Schaap P.J., Smidt H., Diekert G. Draft genome sequence and characterization of Desulfitobacterium hafniense PCE-S. Stand Genomic Sci. 2015;10:15. doi: 10.1186/1944-3277-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grostern A., Duhamel M., Dworatzek S., Edwards E.A. Chloroform respiration to dichloromethane by a Dehalobacter population. Environ Microbiol. 2010;12:1053–1060. doi: 10.1111/j.1462-2920.2009.02150.x. [DOI] [PubMed] [Google Scholar]
- 62.Halsey K.H., Sayavedra-Soto L.A., Bottomley P.J., Arp D.J. Trichloroethylene degradation by butane-oxidizing bacteria causes a spectrum of toxic effects. Appl Microbiol Biotechnol. 2005;68:794–801. doi: 10.1007/s00253-005-1944-z. [DOI] [PubMed] [Google Scholar]
- 63.Hamamura N., Page C., Long T., Semprini L., Arp D.J. Chloroform cometabolism by butane-grown CF8, Pseudomonas butanovora, and Mycobacterium vaccae JOB5 and methane-grown Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1997;63:3607–3613. doi: 10.1128/aem.63.9.3607-3613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartmans S., de Bont J.A.M., Tramper J., Luyben K.Ch.A.M. Bacterial degradation of vinyl chloride. Biotechnol Lett. 1985;7:383–386. [Google Scholar]
- 65.Hartmans S., de Bont J.A.M. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl Environ Microbiol. 1992;58:1220–1226. doi: 10.1128/aem.58.4.1220-1226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He J., Ritalahti K.M., Yang K.L., Koenigsberg S.S., Löffler F.E. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424:62–65. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- 67.He J., Sung Y., Krajmalnik-Brown R., Ritalahti K.M., Löffler F.E. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)—and 1,2-dichloroethene-respiring anaerobe. Environ Microbiol. 2005;7:1442–1450. doi: 10.1111/j.1462-2920.2005.00830.x. [DOI] [PubMed] [Google Scholar]
- 68.Hendrickx B., Junca H., Vosahlova J., et al. Alternative primer sets for PCR detection of genotypes involved in bacterial aerobic BTEX degradation: distribution of the genes in BTEX degrading isolates and in subsurface soils of a BTEX contaminated industrial site. J Microbiol Methods. 2006;64:250–265. doi: 10.1016/j.mimet.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 69.Higashioka Y., Kojima H., Fukui M. Isolation and characterization of novel sulfate-reducing bacterium capable of anaerobic degradation of p-xylene. Microbes Environ. 2012;27:273–277. doi: 10.1264/jsme2.ME11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holliger C., Schraa G., Stams A.J.M., Zehnder A.J.B. A highly purified enrichment culture couples the reductive dechlorination of tetrachloroethene to growth. Appl Environ Microbiol. 1993;59:2991–2997. doi: 10.1128/aem.59.9.2991-2997.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holliger C., Hahn D., Harmsen H., Ludwig W., Schumacher W., Tindall B., Vazquez F., Weiss N., Zehnder A.J.B. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch Microbiol. 1998;169:313–321. doi: 10.1007/s002030050577. [DOI] [PubMed] [Google Scholar]
- 72.Hüsken L.E., Beeftink R., de Bont J.A., Wery J. High-rate 3-methylcatechol production in Pseudomonas putida strains by means of a novel expression system. Appl Microbiol Biotechnol. 2001;55:571–577. doi: 10.1007/s002530000566. [DOI] [PubMed] [Google Scholar]
- 73.Im J., Semrau J.D. Pollutant degradation by a Methylocystis strain SB2 grown on ethanol: bioremediation via facultative methanotrophy. FEMS Microbiol Lett. 2011;318:137–142. doi: 10.1111/j.1574-6968.2011.02249.x. [DOI] [PubMed] [Google Scholar]
- 74.Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of TOL genes xylB and xylE in Escherichia coli. J Bacteriol. 1981;145:1137–1143. doi: 10.1128/jb.145.3.1137-1143.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Interstate Technology & Regulatory Council. EMD-2. Interstate Technology & Regulatory Council, Environmental Molecular Diagnostics Team; Washington, DC: 2013. Environmental Molecular Diagnostics, New Site Characterization and Remediation Enhancement Tools. [Google Scholar]
- 76.Justicia-Leon S.D., Ritalahti K.M., Mack E.E., Löffer F.E. Dichloromethane fermentation by a Dehalobacter sp. in an enrichment culture derived from river sediment. Appl Environ Microbiol. 2012;78:1288–1291. doi: 10.1128/AEM.07325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Juwarkar A.A., Singh S.K., Mudhoo A. A comprehensive overview of elements in bioremediation. Rev Environ Sci Biotechnol. 2010;9:215–288. [Google Scholar]
- 78.Kang J.W., Doty S.L. Cometabolic degradation of trichloroethylene by Burkholderia cepacia G4 with poplar leaf homogenate. Can J Microbiol. 2014;60:487–490. doi: 10.1139/cjm-2014-0095. [DOI] [PubMed] [Google Scholar]
- 79.Kao C.M., Chen C.Y., Chen S.C., Chien H.Y., Chen Y.L. Application of in situ biosparging to remediate a petroleum-hydrocarbon spill site: Field and microbial evaluation. Chemosphere. 2008;70:1492–1499. doi: 10.1016/j.chemosphere.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 80.Kasahara Y., Morimoto H., Kuwano M., Kadoya R. Genome-wide analytical approaches using semi-quantitative expression proteomics for aromatic hydrocarbon metabolism in Pseudomonas putida F1. J Microbiol Methods. 2012;91:434–442. doi: 10.1016/j.mimet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 81.Kayser M.F., Stumpp M.T., Vuilleumier S. DNA polymerase I is essential for growth of Methylobacterium dichloromethanicum DM4 with dichloromethane. J Bacteriol. 2000;182:5433–5439. doi: 10.1128/jb.182.19.5433-5439.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keener W.K., Arp D.J. Transformations of aromatic compounds by Nitrosomonas europaea. Appl Environ Microbiol. 1994;60:1914–1920. doi: 10.1128/aem.60.6.1914-1920.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim J.M., Le N.T., Chung B.S., Park J.H., Bae J.W., Madsen E.L., Jeon C.O. Influence of soil components on the biodegradation of benzene, toluene, ethylbenzene, and o-, m-, and p-xylenes by the newly isolated bacterium Pseudoxanthomonas spadix BD-a59. Appl Environ Microbiol. 2008;74:7313–7320. doi: 10.1128/AEM.01695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kitayama A., Achioku T., Yanagawa T., Kanou K., Kikuchi M., Ueda H., Suzuki E., Nishimura H., Nagamune T., Kawakami Y. Cloning and characterization of extradiol aromatic ring-cleavage dioxygenases of Pseudomonas aeruginosa JI104. J Ferment Bioeng. 1996;82:217–223. [Google Scholar]
- 85.Kitayama A., Suzuki E., Kawakami Y., Nagamune T. Gene organization and low regiospecificity in aromatic-ring hydroxylation of a benzene monooxygenase of Pseudomonas aeruginosa JI104. J Ferment Bioeng. 1996;82:421–425. [Google Scholar]
- 86.Kittelmann S., Friedrich M.W. Identification of novel perchloroethene-respiring microorganisms in anoxic river sediment by RNA-based stable isotope probing. Environ Microbiol. 2008;10:31–46. doi: 10.1111/j.1462-2920.2007.01427.x. [DOI] [PubMed] [Google Scholar]
- 87.Koenig J.C., Lee M.J., Manefield M. Successful microcosm demonstration of a strategy for biodegradation of a mixture of carbon tetrachloride and perchloroethene harnessing sulfate reducing and dehalorespiring bacteria. J Hazard Mater. 2012;219–220:169–175. doi: 10.1016/j.jhazmat.2012.03.076. [DOI] [PubMed] [Google Scholar]
- 88.Koenig J.C., Groissmeier K.D., Manefield M.J. Tolerance of anaerobic bacteria to chlorinated solvents. Microbes Environ. 2014;29:23–30. doi: 10.1264/jsme2.ME13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koh S.C., Bowman J.P., Sayler G.S. Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl Environ Microbiol. 1993;59:960–967. doi: 10.1128/aem.59.4.960-967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kohler-Staub D., Leisinger T. Dichloromethane dehalogenase of Hyphomicrobium sp. strain DM2. J Bacteriol. 1985;162:676–681. doi: 10.1128/jb.162.2.676-681.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krajmalnik-Brown R., Hölscher T., Thomson I.N., Michael Saunders F., Ritalahti K.M., Löffler F.E. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol. 2004;70:6347–6351. doi: 10.1128/AEM.70.10.6347-6351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krone U.E., Laufer K., Thauer R.K., Hogenkamp H.P. Coenzyme F430 as a possible catalyst for the reductive dehalogenation of chlorinated C1 hydrocarbons in methanogenic bacteria. Biochemistry. 1989;28:10061–10065. doi: 10.1021/bi00452a027. [DOI] [PubMed] [Google Scholar]
- 93.Krone U.E., Thauer R.K., Hogenkamp H.P.C. Reductive dehalogenation of chlorinated C1-hydrocarbons mediated by corrinoids. Biochemistry. 1989;28:4908–4914. doi: 10.1021/bi00452a027. [DOI] [PubMed] [Google Scholar]
- 94.Krumholz L.R., Sharp R., Fishbain S. A freshwater anaerobe coupling acetate oxidation to tetrachloroethene dehalogenation. Appl Environ Microbiol. 1996;62:4108–4113. doi: 10.1128/aem.62.11.4108-4113.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krumholz L.R. Desulfuromonas chloroethenica sp. nov. uses tetrachloroethylene and trichloroethylene as electron acceptors. Int J Syst Bacteriol. 1997;47:1262–1263. [Google Scholar]
- 96.Kuhn E.P., Colberg P.J., Schnoor J.L., Wanner O., Zehnder A.J.P., Schwarzenbach R.P. Microbial transformations of substituted benzenes during infiltration of river water to groundwater: laboratory column studies. Environ Sci Technol. 1985;19:961–968. [Google Scholar]
- 97.Kunapuli U., Jahn M.K., Lueders T., Geyer R., Heipieper H.J., Meckenstock R.U. Desulfitobacterium aromaticivorans sp. nov. and Geobacter toluenoxydans sp. nov., iron-reducing bacteria capable of anaerobic degradation of monoaromatic hydrocarbons. Int J Syst Evol Microbiol. 2010;60:686–695. doi: 10.1099/ijs.0.003525-0. [DOI] [PubMed] [Google Scholar]
- 98.Leahy J.G., Byrne A.M., Olsen R.H. Comparison of factors influencing trichloroethylene degradation by toluene-oxidizing bacteria. Appl Environ Microbiol. 1996;62:825–833. doi: 10.1128/aem.62.3.825-833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee E.-H., Kim J., Cho K.-S., Ahn Y.G., Hwang G.-S. Degradation of hexane and other recalcitrant hydrocarbons by a novel isolate, Rhodococcus sp. EH831. Environ Sci Poll Res. 2010;17:64–77. doi: 10.1007/s11356-009-0238-x. [DOI] [PubMed] [Google Scholar]
- 100.Leuthner B., Leutwein C., Schulz H., Hörth P., Haehnel W., Schiltz E., Schägger H., Heider J. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol Microbiol. 1998;28:615–628. doi: 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 101.Liu X., Mattes T.E. Epoxyalkane:coenzyme M transferase gene diversity and distribution in groundwater samples from chlorinated-ethene-contaminated sites. Appl Environ Microbiol. 2016;82:3269–3279. doi: 10.1128/AEM.00673-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Löffler F.E., Yan J., Ritalahti K.M., Adrian L., Edwards E.A., Konstantinidis K.T., Müller J.A., Fullerton H., Zinder S.H., Spormann A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol. 2013;63:625–635. doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- 103.Lontoh S., Semrau J.D. Methane and trichloroethylene degradation by Methylosinus trichosporium OB3b expressing particulate methane monooxygenase. Appl Environ Microbiol. 1998;64:1106–1114. doi: 10.1128/aem.64.3.1106-1114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mägli A., Wendt M., Leisinger T. Isolation and characterization of Dehalobacterium formicoaceticum gen. nov. sp. nov., a strictly anaerobic bacterium utilizing dichloromethane as source of carbon and energy. Arch Microbiol. 1996;166:101–108. [Google Scholar]
- 105.Mägli A., Messmer M., Leisinger T. Metabolism of dichloromethane by strict anaerobic Dehalobacterium formicoaceticum. Appl Environ Microbiol. 1998;64:646–650. doi: 10.1128/aem.64.2.646-650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Magnuson J.K., Stern R.V., Gossett J.M., Zinder S.H., Burris D.R. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl Environ Microbiol. 1998;64:1270–1275. doi: 10.1128/aem.64.4.1270-1275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Magnuson J.K., Romine M.F., Burris D.R., Kingsley M.T. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: Sequence of tceA and substrate range characterization. Appl Environ Microbiol. 2000;66:5141–5147. doi: 10.1128/aem.66.12.5141-5147.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maillard J., Schumacher W., Vazquez F., Regeard C., Hagen W.R., Holliger C. Characterization of the corrinoid iron-sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl Environ Microbiol. 2003;69:4628–4638. doi: 10.1128/AEM.69.8.4628-4638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maithreepala R.A., Doong R.A. Transformation of carbon tetrachloride by biogenic iron species in the presence of Geobacter sulfurreducens and electron shuttles. J Hazard Mater. 2009;164:337–344. doi: 10.1016/j.jhazmat.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 110.Major D.W., McMaster M.L., Cox E.E., Edwards E.A., Dworatzek S.M., Hendrickson E.R., Starr M.G., Payne J.A., Buonamici L.W. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ Sci Technol. 2002;36:5106–5116. doi: 10.1021/es0255711. [DOI] [PubMed] [Google Scholar]
- 111.Marco-Urrea E., Nijenhuis I., Adrian L. Transformation and carbon isotope fractionation of tetra- and trichloroethene to trans-dichloroethene by Dehalococcoides sp. strain CBDB1. Environ Sci Technol. 2011;45:1555–1562. doi: 10.1021/es1023459. [DOI] [PubMed] [Google Scholar]
- 112.Mars A.E., Prins G.T., Wietzes P., de Koning W., Janssen D.B. Effect of trichloroethylene on the competitive behavior of toluene-degrading bacteria. Appl Environ Microbiol. 1998;64:208–215. doi: 10.1128/aem.64.1.208-215.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Martin K.E., Ozsvar J., Coleman N.V. SmoXYB1C1Z of Mycobacterium sp. strain NBB4: a soluble methane monooxygenase (sMMO)-like enzyme, active on C2 to C4 alkanes and alkenes. Appl Environ Microbiol. 2014;80:5801–5806. doi: 10.1128/AEM.01338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martínez C.M., Alvarez L.H., Cervantes F.J. Simultaneous biodegradation of phenol and carbon tetrachloride mediated by humic acids. Biodegradation. 2012;23:635–644. doi: 10.1007/s10532-012-9539-8. [DOI] [PubMed] [Google Scholar]
- 115.Mattes T.E., Alexander A.K., Coleman N.V. Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol Rev. 2010;34:445–475. doi: 10.1111/j.1574-6976.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 116.Maymó-Gatell X., Chien Y.T., Gossett J.M., Zinder S.H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 117.Maymó-Gatell X., Nijenhuis I., Zinder S.H. Reductive dechlorination of cis-1,2-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes”. Environ Sci Technol. 2001;35:516–521. doi: 10.1021/es001285i. [DOI] [PubMed] [Google Scholar]
- 118.McClay K., Streger S.H., Steffan R.J. Induction of toluene oxidation activity in Pseudomonas mendocina KR1 and Pseudomonas sp. strain ENVPC5 by chlorinated solvents and alkanes. Appl Environ Microbiol. 1995;61:3479–3481. doi: 10.1128/aem.61.9.3479-3481.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McClay K., Fox B.G., Steffan R.J. Chloroform mineralization by toluene-oxidizing bacteria. Appl Environ Microbiol. 1996;62:2716–2722. doi: 10.1128/aem.62.8.2716-2722.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Michener J.K., Camargo Neves A.A., Vuilleumier S., Bringel F., Marx C.J. Effective use of a horizontally-transferred pathway for dichloromethane catabolism requires post-transfer refinement. Elife. 2014;3:e04279. doi: 10.7554/eLife.04279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ministry of the Environment, Japan. Results of the survey on implementation of soil contamination countermeasures act and cases of investigation and countermeasures in 2014. Ministry of the Environment; Japan, Tokyo: 2016. (In Japanese) [Google Scholar]
- 122.Miura T., Yamazoe A., Ito M., Ohji S., Hosoyama A., Takahata Y., Fujita N. The impact of injections of different nutrients on the bacterial community and its dechlorination activity in chloroethene-contaminated groundwater. Microbes Environ. 2015;30:164–171. doi: 10.1264/jsme2.ME14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morales G., Linares J.F., Beloso A., Albar J.P., Martínez J.L., Rojo F. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J Bacteriol. 2004;186:1337–1344. doi: 10.1128/JB.186.5.1337-1344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moreno R., Rojo F. The target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the BenR transcriptional regulator. J Bacteriol. 2008;190:1539–1545. doi: 10.1128/JB.01604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moreno R., Fonseca P., Rojo F. The Crc global regulator inhibits the Pseudomonas putida pWW0 toluene/xylene assimilation pathway by repressing the translation of regulatory and structural genes. J Biol Chem. 2010;285:24412–24419. doi: 10.1074/jbc.M110.126615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moreno R., Fonseca P., Rojo F. Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol Microbiol. 2012;83:24–40. doi: 10.1111/j.1365-2958.2011.07912.x. [DOI] [PubMed] [Google Scholar]
- 127.Muller E.E.L., Bringel F., Vuilleumier S. Dichloromethane-degrading bacteria in the genomic age. Res Microbiol. 2011;162:869–876. doi: 10.1016/j.resmic.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 128.Muller E.E.L., Hourcade E., Louhichi-Jelail Y., Hammann P., Vuilleumier S., Bringel F. Functional genomics of dichloromethane utilization in Methylobacterium extorquens DM4. Environ Microbiol. 2011;13:2518–2535. doi: 10.1111/j.1462-2920.2011.02524.x. [DOI] [PubMed] [Google Scholar]
- 129.Müller J.A., Rosner B.M., Abendroth G., Meshulam-Simon G., McCarty P.L., Spormann A.M. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70:4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Muñoz R., Díaz L.F., Bordel S., Villaverde S. Inhibitory effects of catechol accumulation on benzene biodegradation in Pseudomonas putida F1 cultures. Chemosphere. 2007;68:244–252. doi: 10.1016/j.chemosphere.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 131.Nagarajan K., Loh K.C. Formulation of microbial cocktails for BTEX biodegradation. Biodegradation. 2015;26:51–63. doi: 10.1007/s10532-014-9715-0. [DOI] [PubMed] [Google Scholar]
- 132.Nelson M.J.K., Montgomery S.O., O’Neill E.J., Pritchard P.H. Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl Environ Microbiol. 1986;52:383–384. doi: 10.1128/aem.52.2.383-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nelson M.J.K., Montgomery S.O., Mahaffey W.R., Pritchard P.H. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl Environ Microbiol. 1987;53:949–954. doi: 10.1128/aem.53.5.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nelson M.J.K., Montgomery A.O., Pritchard P.H. Trichloroethylene metabolism by microorganisms that degrade aromatic compounds. Appl Environ Microbiol. 1988;54:604–606. doi: 10.1128/aem.54.2.604-606.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Neumann A., Scholz-Muramatsu H., Diekert G. Tetrachloroethene metabolism of Dehalospirillum multivorans. Arch Microbiol. 1994;162:295–301. doi: 10.1007/BF00301854. [DOI] [PubMed] [Google Scholar]
- 136.Neumann A., Wohlfarth G., Diekert G. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J Biol Chem. 1996;271:16515–16519. doi: 10.1074/jbc.271.28.16515. [DOI] [PubMed] [Google Scholar]
- 137.Neumann A., Wohlfarth G., Diekert G. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J Bacteriol. 1998;180:4140–4145. doi: 10.1128/jb.180.16.4140-4145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Newman L.M., Wackett L.P. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry. 1995;34:14066–14076. doi: 10.1021/bi00043a012. [DOI] [PubMed] [Google Scholar]
- 139.Nijenhuis I., Nikolausz M., Koth A., Felfoldi T., Weiss H., Drangmeister J., Grobmann J., Kastner M., Richnow H.-H. Assessment of the natural attenuation of chlorinated ethenes in an anaerobic contaminated aquifer in the Bitterfeld/Wolfen area using stable isotope techniques, microcosm studies and molecular biomarkers. Chemosphere. 2007;67:300–311. doi: 10.1016/j.chemosphere.2006.09.084. [DOI] [PubMed] [Google Scholar]
- 140.Noguchi M., Kurisu F., Kasuga I., Furumai H. Time-resolved DNA stable isotope probing links Desuldobacterales- and Coriobacteriaceae-related bacteria to anaerobic degradation of benzene under methanogenic conditions. Microbes Environ. 2014;29:191–199. doi: 10.1264/jsme2.ME13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Oh Y.S., Shareefdeen Z., Baltzis B.C., Bartha R. Interactions between benzene, toluene, and p-xylene (BTX) during their biodegradation. Biotechnol Bioeng. 1994;44:533–538. doi: 10.1002/bit.260440417. [DOI] [PubMed] [Google Scholar]
- 142.Oldenhuis R., Vink R.L., Janssen D.B., Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989;55:2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Olsen R.H., Kukor J.J., Kaphammer B. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol. 1994;176:3749–3756. doi: 10.1128/jb.176.12.3749-3756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Parsons . Principles and practices of enhanced anaerobic bioremediation of chlorinated solvents. Air Force Center for Environmental Excellence; Brooks City-Base, Texas: Naval Facilities Engineering Service Center; Port Hueneme, California: Environmental Security Technology Certification Program; Arlington, Virginia: 2004. [Google Scholar]
- 145.Patrauchan M.A., Florizone C., Eapen S., Gómez-Gil L., Sethuraman B., Fukuda M., Davies J., Mohn W.W., Eltis L.D. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1. J Bacteriol. 2008;190:37–47. doi: 10.1128/JB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Penny C., Vuilleumier S., Bringel F. Microbial degradation of tetrachloromethane: mechanisms and perspectives for bioremediation. FEMS Microbiol Ecol. 2010;74:257–275. doi: 10.1111/j.1574-6941.2010.00935.x. [DOI] [PubMed] [Google Scholar]
- 147.Petrovskis E.A., Vogel T.M., Adriaens P. Effects of electron acceptors and donors on transformation of tetrachloromethane by Shewanella putrefaciens MR-1. FEMS Microbiol Lett. 1994;121:357–363. doi: 10.1111/j.1574-6968.1994.tb07126.x. [DOI] [PubMed] [Google Scholar]
- 148.Pöritz M., Goris T., Wubet T., Tarkka M.T., Buscot F., Nijenhuis I., Lechner U., Adrian L. Genome sequences of two dehalogenation specialists—Dehalococcoides mccartyi strains BTF08 and DCMB5 enriched from the highly polluted Bitterfeld region. FEMS Microbiol Lett. 2013;343:101–104. doi: 10.1111/1574-6968.12160. [DOI] [PubMed] [Google Scholar]
- 149.Priya V.S., Philip L. Biodegradation of dichloromethane along with other VOCs from pharmaceutical wastewater. Appl Biochem Biotechnol. 2013;169:1197–1218. doi: 10.1007/s12010-012-0005-1. [DOI] [PubMed] [Google Scholar]
- 150.Rabus R., Nordhaus R., Ludwig W., Widdel F. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol. 1993;59:1444–1451. doi: 10.1128/aem.59.5.1444-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rabus R., Widdel F. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch Microbiol. 1995;163:96–103. doi: 10.1007/BF00381782. [DOI] [PubMed] [Google Scholar]
- 152.Radway J.C., Santo Domingo J.W., Hazen T.C., Wilde E.W. Evaluation of biodegradation potential of foam embedded Burkholderia cepacia G4. Biotechnol Lett. 1998;20:663–666. [Google Scholar]
- 153.Reij M.W., Kieboom J., de Bont J.A.M., Hartmans S. Continuous degradation of trichloroethylene by Xanthobacter sp. strain Py2 during growth on propene. Appl Environ Microbiol. 1995;61:2936–2942. doi: 10.1128/aem.61.8.2936-2942.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Robinson G.K., Stephens G.M., Dalton H., Geary P.J. The production of catechols from benzene and toluene by Pseudomonas putida in glucose fed-batch culture. Biocatalysis. 1992;6:81–100. [Google Scholar]
- 155.Ryoo D., Shim H., Canada K., Barbieri P., Wood T.K. Aerobic degradation of tetrachloroethylene by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1. Nat Biotechnol. 2000;18:775–778. doi: 10.1038/77344. [DOI] [PubMed] [Google Scholar]
- 156.Schmid-Appert M., Zoller K., Traber H., Vuilleumier S., Leisinger T. Association of newly discovered IS elements with the dichloromethane utilization genes of methylotrophic bacteria. Microbiology. 1997;143:2557–2567. doi: 10.1099/00221287-143-8-2557. [DOI] [PubMed] [Google Scholar]
- 157.Scholz-Muramatsu H., Neumann A., Meßmer M., Moore E., Diekert G. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch Microbiol. 1995;163:48–56. [Google Scholar]
- 158.Seshadri R., Adrian L., Fouts D.E., et al. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307:105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- 159.Shaw J.P., Harayama S. Purification and characterisation of the NADH:acceptor reductase component of xylene monooxygenase encoded by the TOL plasmid pWW0 of Pseudomonas putida mt-2. Eur J Biochem. 1992;209:51–61. doi: 10.1111/j.1432-1033.1992.tb17260.x. [DOI] [PubMed] [Google Scholar]
- 160.Shields M.S., Montgomery S.O., Chapman P.J., Cuskey S.M., Pritchard P.H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium G4. Appl Environ Microbiol. 1989;55:1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shields M.S., Montgomery S.O., Cuskey S.M., Chapman P.J., Pritchard P.H. Mutants of Pseudomonas cepacia G4 defective in catabolism of aromatic compounds and trichloroethylene. Appl Environ Microbiol. 1991;57:1935–1941. doi: 10.1128/aem.57.7.1935-1941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shim H., Wood T.K. Aerobic degradation of mixtures of chlorinated aliphatics by cloned toluene-o-xylene monooxygenase and toluene o-monooxygenase in resting cells. Biotechnol Bioeng. 2000;70:693–698. doi: 10.1002/1097-0290(20001220)70:6<693::aid-bit12>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 163.Shinoda Y., Sakai Y., Uenishi H., Uchihashi Y., Hiraishi A., Yukawa H., Yurimoto H., Kato N. Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1. Appl Environ Microbiol. 2004;70:1385–1392. doi: 10.1128/AEM.70.3.1385-1392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Smidt H., de Vos W.M. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- 165.Sun A.K., Wood T.K. Trichloroethylene degradation and mineralization by pseudomonads and Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol. 1996;45:248–256. doi: 10.1007/s002530050679. [DOI] [PubMed] [Google Scholar]
- 166.Sung Y., Fletcher K.E., Ritalahti K.M., Apkarian R.P., Ramos-Hernandez N., Sanford R.A., Mesbah N.M., Löffler F.E. Geobacter lovleyi sp. nov. strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl Environ Microbiol. 2006;72:2775–2782. doi: 10.1128/AEM.72.4.2775-2782.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sung Y., Ritalahti K.M., Apkarian R.P., Löffler F.E. Quantitative PCR confirms purity of strain GT, a novel trichloroetheneto-ethene-respiring Dehalococcoides isolate. Appl Environ Microbiol. 2006;72:1980–1987. doi: 10.1128/AEM.72.3.1980-1987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Suyama A., Yamashita M., Yoshino S., Furukawa K. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J Bacteriol. 2002;184:3419–3425. doi: 10.1128/JB.184.13.3419-3428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Suzuki M., Hayakawa T., Shaw J.P., Rekik M., Harayama S. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J Bacteriol. 1991;173:1690–1695. doi: 10.1128/jb.173.5.1690-1695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tang S., Gong Y., Edwards E.A. Semi-automatic in silico gap closure enabled de novo assembly of two Dehalobacter genomes from metagenomic data. PLoS ONE. 2012;7:e52038. doi: 10.1371/journal.pone.0052038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tang S., Chan W.W.M., Fletcher K.E., Seifert J., Liang X., Löffler F.E., Edwards E.A., Adrian L. Functional characterization of reductive dehalogenases by using blue native polyacrylamide gel electrophoresis. Appl Environ Microbiol. 2013;79:974–981. doi: 10.1128/AEM.01873-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tang S., Edwards E.A. Identification of Dehalobacter reductive dehalogenases that catalyse dechlorination of chloroform, 1,1,1-trichloroethane and 1,1-dichloroethane. Philos Trans R Soc Lond, B, Biol Sci. 2013;368:20120318. doi: 10.1098/rstb.2012.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tao Y., Fishman A., Bentley W.E., Wood T.K. Oxidation of benzene to phenol, catechol, and 1,2,3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pickettii PKO1. Appl Environ Microbiol. 2004;70:3814–3820. doi: 10.1128/AEM.70.7.3814-3820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tsien H.C., Brusseau G.A., Hanson R.S., Wackett L.P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989;55:3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Uchino Y., Miura T., Hosoyama A., Ohji S., Yamazoe A., Ito M., Takahata Y., Suzuki K., Fujita N. Complete genome sequencing of Dehalococcoides sp. strain UCH007 using a differential reads picking method. Stand Genomic Sci. 2015;10:102. doi: 10.1186/s40793-015-0095-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.United States Environmental Protection Agency. Final five-year review report for Brookhaven National Laboratory Superfund Site, Town of Brookhaven. United States Environmental Protection Agency; Washington, DC: 2006. [Google Scholar]
- 177.United States Environmental Protection Agency. Final comprehensive five-year ROD review report, Third five-year ROD review for Allegany Ballistics Laboratory. United States Environmental Protection Agency; Washington, DC: 2008. [Google Scholar]
- 178.United States Environmental Protection Agency. First fiveyear review joint base Andrews naval air facility Washington. United States Environmental Protection Agency; Washington, DC: 2011. [Google Scholar]
- 179.United States Environmental Protection Agency. Final first five-year review report. National Aeronautics and Space Administration Jet Propulsion Laboratory, United States Environmental Protection Agency; Washington, DC: 2012. [Google Scholar]
- 180.United States Environmental Protection Agency. Fourth five-year review report, The Rose Township dump site Oakland County, Michigan. United States Environmental Protection Agency; Washington, DC: 2012. [Google Scholar]
- 181.United States Environmental Protection Agency. Superfund remedy report. 14th ed. United States Environmental Protection Agency; Washington, DC: 2013. EPA 542-R-13-016. [Google Scholar]
- 182.United States Environmental Protection Agency. Introduction to in situ bioremediation of groundwater. United States Environmental Protection Agency; Washington, DC: 2013. EPA 542-R-13-018. [Google Scholar]
- 183.United States Environmental Protection Agency. Basewide five-year reviews Dover air force base, Delaware. United States Environmental Protection Agency; Washington, DC: 2014. [Google Scholar]
- 184.United States Environmental Protection Agency. Third fiveyear review report for parker landfill superfund site, SDMS Doc ID 567594. United States Environmental Protection Agency; Washington, DC: 2014. [Google Scholar]
- 185.United States Environmental Protection Agency. Fourth five-year review report for the Colesville municipal landfill site. United States Environmental Protection Agency; Washington, DC: 2015. [Google Scholar]
- 186.van Agteren M.H., Keuning S., Janssen D.B. Handbook on biodegradation and biological treatment of hazardous organic compounds. Kluwer Academic Publishers; Dordrecht, Netherland: 1998. [Google Scholar]
- 187.van Hylckama Vlieg J.E.T., de Koning W., Janssen D.B. Transformation kinetics of chlorinated ethenes by Methylosinus trichosporium OB3b and detection of unstable epoxides by on-line gas chromatography. Appl Environ Microbiol. 1996;62:3304–3312. doi: 10.1128/aem.62.9.3304-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.van Hylckama Vlieg J.E.T., Janssen D.B. Formation and detoxification of reactive intermediates in the metabolism of chlorinated ethenes. J Biotechnol. 2001;85:81–102. doi: 10.1016/s0168-1656(00)00364-3. [DOI] [PubMed] [Google Scholar]
- 189.Vannelli T., Logan M., Arciero D.M., Hooper A.B. Degradation of halogenated aliphatic compounds by the ammoniaoxidizing bacterium Nitrosomonas europaea. Appl Environ Microbiol. 1990;56:1169–1171. doi: 10.1128/aem.56.4.1169-1171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vardar G., Wood T.K. Protein engineering of toluene-oxylene monooxygenase from Pseudomonas stutzeri OX1 for synthesizing 4-methylresorcinol, methylhydroquinone, and pyrogallol. Appl Environ Microbiol. 2004;70:3253–3262. doi: 10.1128/AEM.70.6.3253-3262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Verce M.F., Freedman D.L. Modeling the kinetics of vinyl chloride cometabolism by an ethane-grown Pseudomonas sp. Biotechnol Bioeng. 2000;71:274–285. doi: 10.1002/1097-0290(2000)71:4<274::aid-bit1017>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 192.Verce M.F., Ulrich R.L., Freedman D.L. Transition from cometabolic to growth-linked biodegradation of vinyl chloride by a Pseudomonas sp. isolated on ethene. Environ Sci Technol. 2001;35:4242–4251. doi: 10.1021/es002064f. [DOI] [PubMed] [Google Scholar]
- 193.Vidali M. Bioremediation. An overview. Pure Appl Chem. 2001;73:1163–1172. [Google Scholar]
- 194.Vogt C., Kleinsteuber S., Richnow H.H. Anaerobic benzene degradation by bacteria. Microb Biotechnol. 2011;4:710–724. doi: 10.1111/j.1751-7915.2011.00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wackett L.P., Gibson D.T. Degradation of trichloroethylene by toluene dioxygenase in whole-cell studies with Pseudomonas putida F1. Appl Environ Microbiol. 1988;54:1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wackett L.P., Brusseau G.A., Householder S.R., Hanson R.S. Survey of microbial oxygenases: trichloroethylene degradation by propane-oxidizing bacteria. Appl Environ Microbiol. 1989;55:2960–2964. doi: 10.1128/aem.55.11.2960-2964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wagner D.D., Hug L.A., Hatt J.K., Spitzmiller M.R., Padilla-Crespo E., Ritalahti K.M., Edwards E.A., Konstantinidis K.T., Löffler F.E. Genomic determinants of organohalide-respiration in Geobacter lovleyi, an unusual member of the Geobacteraceae. BMC Genomics. 2012;13:200. doi: 10.1186/1471-2164-13-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang S., Chng K.R., Chen C., Bedard D.L., He J. Genomic characterization of Dehalococcoides mccartyi strain JNA that reductively dechlorinates tetrachloroethene and polychlorinated biphenyls. Environ Sci Technol. 2015;49:14319–14325. doi: 10.1021/acs.est.5b01979. [DOI] [PubMed] [Google Scholar]
- 199.Weelink S.A.B., van Eekert M.H.A., Stams A.J.M. Degradation of BTEX by anaerobic bacteria: physiology and application. Rev Environ Sci Biotechnol. 2010;9:359–385. [Google Scholar]
- 200.Whited G.M., Gibson D.T. Toluene-4-monooxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991;173:3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wilson J.T., Wilson B.H. Biotransformation of trichloroethylene in soil. Appl Environ Microbiol. 1985;49:242–243. doi: 10.1128/aem.49.1.242-243.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Worsey M.J., Williams P.A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yamada A., Kishi H., Sugiyama K., Hatta T., Nakamura K., Masai E., Fukuda M. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl Environ Microbiol. 1998;64:2006–2012. doi: 10.1128/aem.64.6.2006-2012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yang X., Beckmann D., Fiorenza S., Niedermeier C. Field study of pulsed air sparging for remediation of petroleum hydrocarbon contaminated soil and groundwater. Environ Sci Technol. 2005;39:7279–7286. doi: 10.1021/es050084h. [DOI] [PubMed] [Google Scholar]
- 205.Yarmoff J.J., Kawakami Y., Yago T., Maruo H., Nishimura H. cis-Benzeneglycol production using a mutant Pseudomonas strain. J Ferment Technol. 1988;66:305–312. [Google Scholar]
- 206.Yeager C.M., Arthur K.M., Bottomley P.J., Arp D.J. Trichloroethylene degradation by toluene-oxidizing bacteria grown on non-aromatic substrates. Biodegradation. 2004;15:19–28. doi: 10.1023/b:biod.0000009947.09125.35. [DOI] [PubMed] [Google Scholar]
- 207.Yoshikawa M., Zhang M., Toyota K. Enhancement and biological characteristics related to aerobic biodegradation of toluene with co-existence of benzene. Water Air Soil Pollut. 2016;227:340. [Google Scholar]
- 208.Yoshikawa M., Zhang M., Toyota K. Integrated anaerobic-aerobic biodegradation of multiple contaminants including chlorinated ethylenes, benzene, toluene and dichloromethane. Water Air Soil Pollut. 2017;228:25. doi: 10.1007/s11270-016-3216-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yu H., Kim B.J., Rittmann B.E. A two-step model for the kinetics of BTX degradation and intermediate formation by Pseudomonas putida F1. Biodegradation. 2001;12:465–475. doi: 10.1023/a:1015012913426. [DOI] [PubMed] [Google Scholar]
- 210.Zhang M., Yoshikawa M. Proceedings of Geo-Chicago 2016: Sustainability, Energy, and the Geoenvironment. ASCE Geotechnical Special Publication; Reston: 2016. An overview of remediation technologies for sites contaminated with volatile organic compounds; pp. 295–301. [Google Scholar]
- 211.Zylstra G.J., McCombie W.R., Gibson D.T., Finette B.A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988;54:1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zylstra G.J., Gibson D.T. Toluene degradation by Pseudomonas putida F1. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]