Abstract
Methanol-consuming culturable bacteria were isolated from the plant surface, rhizosphere, and inside the stem of Neobuxbaumia macrocephala. All 38 isolates were facultative methylotrophic microorganisms. Their classification included the Classes Actinobacteria, Sphingobacteriia, Alpha-, Beta-, and Gammaproteobacteria. The deduced amino acid sequences of methanol dehydrogenase obtained by PCR belonging to Actinobacteria, Alpha-, Beta-, and Gammaproteobacteria showed high similarity to rare-earth element (REE)-dependent XoxF methanol dehydrogenases, particularly the group XoxF5. The sequences included Asp301, the REE-coordinating amino acid, present in all known XoxF dehydrogenases and absent in MxaF methanol dehydrogenases. The quantity of the isolates showed positive hybridization with a xoxF probe, but not with a mxaF probe. Isolates of all taxonomic groups showed methylotrophic growth in the presence of Ce3+ or Ca2+. The presence of xoxF-like sequences in methylotrophic bacteria from N. macrocephala and its potential relationship with their adaptability to xerophytic plants are discussed.
Keywords: rare-earth elements, lanthanides, pectin metabolism, Tehuacan, xoxF5
Methanol, one of the most common C1 compounds delivered by plants, is released through the stomata. This compound is also produced with the decay of pectin and lignin from dead plant tissue (1, 19, 47). Methanol and organic molecules without C-C bonds are utilized as carbon and energy sources by methylotrophic organisms. These organisms are classified as facultative or obligate methylotrophs depending on their capability to use compounds with multiple C and C-C bonds. Methylotrophic microorganisms are ubiquitous and include organisms of the Classes Actinobacteria, Spirochaetes, Alpha-, Beta-, Gamma-, and Deltaproteobacteria, of the Phyla Firmicutes, Bacteroidetes, Chloroflexi, Acidobacteria, Nitrospirae, Verrucomicrobia, Cyanobacteria, and Planctomycetes, and even of the domain Archaea (5, 8, 15, 22, 25, 29, 30, 35, 38, 43).
Many methylotrophic bacteria are commonly associated with plants. Nevertheless, there have not yet been reports in Cactaceae. Several methylotrophs exert positive effects when inoculated in plants (37–39, 54). These responses have been attributed to different mechanisms such as nitrogen fixation, decreased metal toxicity, the contribution of pyrrolo-quinoline quinone (PQQ), elicitation of plant defenses, decreased plant levels of ethylene, and the synthesis of molecules including phytohormones, vitamin B12, polysaccharides, and osmoprotectants (11, 39–41, 49, 57, 60). Methanol and methane-catabolizing microorganisms oxidize methanol through different dehydrogenases, and the methanol dehydrogenase, MxaFI-MDH has been examined in the most detail. It is a heterotetramer that is encoded by the genes mxaF and mxaI, and its activity depends on PQQ and Ca2+ as co-factors (10). MxaFI-MDH is typically carried by Alphaproteobacteria, Gammaproteobacteria, and a few Betaproteobacteria. Some Betaproteobacteria also possess the PQQ methanol dehydrogenase MDH2, which shows sequence similarity to MxaFI-MDH (24, Fig. 1). Low GC Gram-positive methylotrophs typically have a NADPH-dependent methanol dehydrogenase (6), and a methanol:NDMA (N,N′-dimethyl-4-nitrosoaniline) oxidoreductase has been reported in the Class Actinobacteria (23, 48). Other dehydrogenases phylogenetically related to MxaFI-MDH include a diverse but related group of enzymes called XoxF. Recent studies demonstrated that XoxF dehydrogenases oxidize methanol and depend on rare-earth elements instead of Ca2+ as co-factors (18, 27, 46, 50). A sequence analysis revealed that XoxF enzymes are grouped in at least five classes (55).
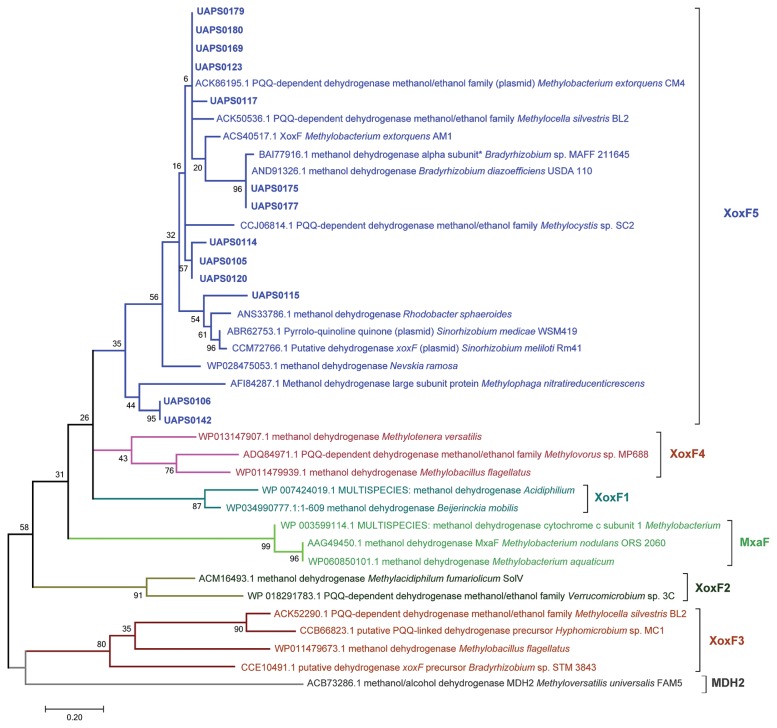
Fig. 1.
Phylogeny of putative methanol dehydrogenase amplicons of N. macrocephala isolates. Sequences of N. macrocephala isolates are shown in bold blue letters. Sequences were aligned by Muscle. Phylogeny was constructed with maximum-likelihood in MEGA 6.0 using deduced amino acid sequences. A total of 500 iterations were used for bootstrapping.
Neobuxbaumia macrocephala is a xerophytic branching columnar Cactaceae with a height from 3 to 15 m. This plant is endemic to the Tehuacán-Cuicatlán Biosphere Reserve and its distribution is confined to a few patches with calcareous soils (44, 51, 58). N. macrocephala has smaller populations than other Neobuxbaumia species that reside in other semi-arid habitats (16).
Rhizospheric and non-rhizospheric bacteria associated with cacti mostly include Actinobacteria, Firmicutes, Alphaproteobacteria, Cyanobacteria, Planctomycetes, Bacteroidetes, Chloroflexi, and Acidobacteria (2, 3, 34, 56). Limited information is currently available on the ecological interactions among cacti and microorganisms, including those of N. macrocephala. In order to design any future restoration strategy for endangered plant species, it is desirable to retrieve a broad knowledge of its biology. The diversity of cultured methylotrophic bacteria associated with this plant was investigated as the first step with the aim of gaining insights into the ecology of N. macrocephala with microorganisms, and as a prerequisite for future inoculation experiments using this plant.
Materials and Methods
Sampling
Rhizospheric soil, surface, and endophytic samples were obtained from six plant specimens from the Tehuacán-Cuicatlán Biosphere Reserve. Approximately 10 g of rhizospheric soil (profundity 15–25 cm) was retrieved from a distance within 1 m of the sampled specimen. Approximately 5 cm2 of the stem surface was sampled with sterile swabs soaked in sterile 10 mM MgSO4 solution. The swabs were deposited in 1 mL of the same solution. Regarding endophytic samples, ca. 5 cm2 of the stem surface was disinfected with 70% ethanol, and ca. 1 cm3 of tissue was extracted with a sterile scalpel. All samples were kept in sterile plastic sealed bags and transported under chilled conditions to the lab.
Isolation and DNA extraction
In order to isolate endophytes, approximately 2 mm of surface plant tissues including the cuticle were discarded under sterile conditions. The remaining plant material was macerated in a sterile mortar and resuspended in 10 mM MgSO4 (1:10 w:v). Epiphytic suspensions and soil dilutions in 10 mM MgSO4 were inoculated on plates (1.6% agar) of methanol mineral salts medium (MMSM; 21) containing 0.5% methanol; 6.89 mM K2HPO4; 4.56 mM KH2PO4; 0.228 mM CaCl2; 0.811 mM MgSO4; 1.71 mM NaCl; 3.7 μM FeCl3; 3.8 mM (NH4)2SO4; 20 nM CuSO4; 41.5 nM MnSO4; 38 nM Na2MoO4; 0.163 μM H3BO3; 0.243 μM ZnSO4; and 21 nM CoCl2, and incubated at 30°C for 8–10 d. Isolated bacterial colonies were streaked in the same medium and incubated at 30°C until growth was observed. Isolated colonies were grown in the same medium and also in GP containing (L−1): Casein peptone 10 g, glycerol 10 g, and agar 15 g. DNA was extracted from cells growing in MMSM medium with the DNA Isolation Kit for Cells and Tissues (Roche Diagnostics, Indianapolis, IN, USA) following the recommended instructions of the supplier.
Ca2+ and Ce3+-methanol dependent growth
Isolates were grown in GP plates at 30°C for 4 d. One loopful of bacterial cells was washed twice in 10 mM MgSO4, resuspended in 10 mL of the same solution, and 5 μL of the suspension was inoculated in 5 mL of modified MMSM with 30 μM CaCl2 or lacking Ca2+ but with 30 μM CeCl3. Cells were incubated at 30°C under shaking for 5 d. Bacterial growth was assessed by absorbance at 600 nm 72, 96, 120, and 144 h after the inoculation. The cultures of three independent replicates grown in either Ca2+ or Ce3+-MMSM broths were statistically compared by the unpaired t-test, P<0.05.
Dot blot hybridization
Genomic DNAs were transferred to nylon filters by dot blots, with 1 μg of DNA per dot, except for M. extorquens JCM2802, which had 100 ng. One microgram of U. maydis 207 was used as a negative control. One hundred nanograms of DNA 32P-labeled probes specific for mxaF and xoxF5 were used for hybridizations. These probes were obtained by the PCR amplification of Methylobacterium extorquens JCM2802 genomic DNA with the primers mxa f1003 and mxa r1561 (42); and xoxFf361 and xoxFr603 (Table 1), for mxaF and xoxF5, respectively. The sizes of the probes were ca. 560 bases for mxaF and ca. 240 bases for xoxF5. The probes were labeled with [α-32P]dCTP by polymerase extension using random primers (Amersham Rediprime II DNA Labeling System, GE Healthcare, Pittsburgh, PA, USA). Prehybridization and hybridization were performed at 65°C for 12 h using Rapid Hyb buffer (GE Healthcare). The membranes were washed under high stringency conditions (2×SSC [1×SSC is 0.15 M NaCl plus 0.015 M sodium citrate] plus 0.1% SDS for 10 min, 1×SSC plus 0.1% SDS for 15 min, 0.5×SSC plus 0.1% SDS for 15 min, 0.1×SSC plus 0.1% SDS for 15 min, 0.1×SSC plus 0.1% SDS at 65°C for 30 min, and SDS was then removed with 0.1×SSC) (52).
Table 1.
Methanol dehydrogenase primers.
| Primer* | Sequence (5′-3′) | Target | Reference |
|---|---|---|---|
| mxa f1003 | GCG GCA CCA ACT GGG GCT GGT | mxaF | (42) |
| mxa r1561 | GGG CAG CAT GAA GGG CTC CC | ||
| xoxF361f | CAG GAT CCG TCC GTG AT | M. extorquens xoxF | This work |
| xoxF603r | SGA GAT GCC GAC GAT GA | ||
| mxaFxoxF916f | GGC GAC AAC AAG TGG WCG ATG | mxaF, xoxF4, xoxF5 | This work |
| mxaFxoxF1360r | AGT CCA TGC AGA CRT GGT T |
Numbers indicate approximate position in the gene.
DNA amplification and sequencing
16S rRNA genes were amplified with the primers B27F (5′-TAG AGT TTG ATC CTG GCT CAG-3′) and B1392R (5′-CAG GGG CGG TGT GTA-3′) using the following conditions: one initial denaturation at 95°C for 3 min, 26 cycles at 94°C for 30 s, 57°C for 45 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. Methanol dehydrogenase genes were amplified with the primers mxaFxoxFf916 and mxaFxoxFr1360 (Table 1) designed to preferentially amplify mxaF, xoxF4, and xoxF5, using the following conditions: one initial denaturation at 95°C for 3 min, 35 cycles at 94°C for 20 s, 55°C for 45 s, and 72°C for 1 min, and one final extension at 72°C for 10 min. The design of the primers mxaFxoxFf916 and mxaFxoxFr1360 was based on the alignments of the mxaF, xoxF4, and xoxF5 public sequences. The alignments of other xoxF subfamilies did not show sufficiently long conserved regions for designing potentially acceptable primers. Sanger DNA sequencing were performed at the Instituto de Biotecnología (UNAM, www.ibt.unam.mx) with the primers used for PCR amplification.
Sequence analysis
Sequence analyses were performed with MEGA 7.0 (32). The sequences were aligned with the database sequences of related microorganisms by ClustalW. Pairwise distances and neighbor joining trees were used to elucidate the genus identity of the 16S rRNA sequences. The phylogeny of methanol dehydrogenases was inferred with the maximum likelihood method with the deduced amino acid sequences. Initial trees were assessed by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances, and then selecting the topology with the greatest log likelihood value. Confidence was evaluated by bootstrapping with 500 iterations.
Nucleotide sequences
16S rRNA sequences have been deposited in GenBank under the accession numbers KT936080–KT936091, KT936093, KT936095, KT936096, KT936105, KT936109–KT936114, KT936119, KT936125–KT936127, KT936134, KT936135, KT936140, KT936141, KT936144, KT936145, and KY00648–KY00653; and xoxF sequences under the accession numbers KT932117–KT932121, KT932123, KT932124, KT932126–KT932128 and KY884986–KY884988 (Table 2).
Table 2.
Methylotrophic culturable isolates from N. macrocephala.
| Isolate | Genus | Taxonomic Class | 16S rRNA Acc. Num. | Origin | Hybridization with | Amplicons with mxaF-xoxF primers Acc. Num. | Subjected to the methanol-Ca2+/Ce3+ experiment | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| mxaF | xoxF | |||||||
| UAPS0102 | Arthrobacter | Actinobacteria | KT936093 | Rhizospheric | ND | P | NA | Yes |
| UAPS0104 | Arthrobacter | KT936095 | Epiphytic | ND | S | NA | No | |
| UAPS0105 | Arthrobacter | KT936096 | Rhizospheric | ND | P | KT932119D | Yes | |
| UAPS0126 | Pedobacter | Sphingobacteriia | KT936125 | Rhizospheric | ND | N | NA | Yes |
| UAPS0120 | Microvirga | Alphaproteobacteria | KT936105 | Epiphytic | ND | N | KY884987 | Yes |
| UAPS0121 | Microvirga | KT936119 | Epiphytic | S | S | NA | Yes | |
| UAPS0136 | Microvirga | KT936112 | Epiphytic | ND | S | NA | Yes | |
| UAPS0137 | Microvirga | KT936113 | Epiphytic | ND | P | NA | Yes | |
| UAPS0106 | Inquilinus | KT936134 | Rhizospheric | N | P | KY884986 | Yes | |
| UAPS0142 | Inquilinus | KT936135 | Rhizospheric | P | P | KT932126D | Yes | |
| UAPS0122 | Methylobacterium | KT936114 | Endophytic | P | P | NA | Yes | |
| UAPS0123 | Methylobacterium | KT936111 | Rhizospheric | S | P | KY884988 | Yes | |
| UAPS0160 | Rhizobium | KT936127 | Rhizospheric | P | P | NA | No | |
| UAPS0110 | Sphingomonas | KT936140 | Endophytic | N | P | NA | No | |
| UAPS0115 | Subaequorebacter/ | KT936141 | Endophytic | N | P | KT932127D | Yes | |
| Geminicoccus | ||||||||
| UAPS0114 | Massilia | Betaproteobacteria | KT936109 | Epiphytic | P | P | KT932123D | No |
| UAPS0174 | Massilia | KT936144 | Epiphytic | P | P | NA | No | |
| UAPS0175 | Massilia | KT936145 | Epiphytic | S | P | KT932128D | No | |
| UAPS0177 | Massilia | KT936110 | Epiphytic | N | P | KT932124D | Yes | |
| UAPS0117 | Acinetobacter | Gammaproteobacteria | KT936080 | Epiphytic | S | P | KT932117 | Yes |
| UAPS0118 | Acinetobacter | KT936081 | Epiphytic | P | S | NA | No | |
| UAPS0127 | Acinetobacter | KT936082 | Epiphytic | ND | N | NA | No | |
| UAPS0145 | Acinetobacter | KT936083 | Epiphytic | P | N | NA | No | |
| UAPS0149 | Acinetobacter | KT936084 | Epiphytic | P | N | NA | No | |
| UAPS0156 | Acinetobacter | KT936085 | Epiphytic | P | N | NA | No | |
| UAPS0158 | Acinetobacter | KT936086 | Rhizospheric | P | N | NA | No | |
| UAPS0163 | Acinetobacter | KT936087 | Epiphytic | P | N | NA | Yes | |
| UAPS0165 | Acinetobacter | KT936088 | Epiphytic | ND | N | NA | No | |
| UPAS0168 | Acinetobacter | KT936089 | Epiphytic | P | S | NA | No | |
| UAPS0169 | Acinetobacter | KT936090 | Epiphytic | S | S | KT932118D | No | |
| UAPS0172 | Acinetobacter | KT936091 | Epiphytic | P | S | NA | No | |
| UAPS0179 | Acinetobacter | KY400648 | Rhizospheric | ND | S | KT932120 | Yes | |
| UAPS0180 | Acinetobacter | KY400649 | Endophytic | ND | P | KT932121 | Yes | |
| UAPS0181 | Acinetobacter | KY400650 | Epiphytic | ND | P | NA | No | |
| UAPS0182 | Acinetobacter | KY400651 | Endophytic | ND | P | NA | Yes | |
| UAPS0183 | Acinetobacter | KY400652 | Endophytic | ND | P | NA | No | |
| UAPS0184 | Acinetobacter | KY400653 | Rhizospheric | ND | P | NA | No | |
| UAPS0155 | Pseudomonas | KT936126 | Epiphytic | P | P | NA | Yes | |
N, negative hybridization; P, positive hybridization; S, slight hybridization; ND, not determined; NA, not amplificated with the primersmxaf916 and mxar1360 D, XoxF sequences long enough to cover Asp301.
Results
Thirty-eight bacterial isolates (Classes Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Actinobacteria, and Sphingobacteriia) were obtained using methanol as the sole carbon and energy sources (Table 2). All isolates showed facultative growth using other carbon and energy sources. No obligate methylotrophic bacteria were found. Twenty-two strains were isolated from the plant surface (one Actinobacteria, four Alphaproteobacteria, four Betaproteobacteria, and thirteen Gammaproteobacteria); six isolates were endophytic (three Alphaproteobacteria and three Gammaproteobacteria); and ten were rhizospheric (two Actinobacteria, one Sphingobacteriia (Phylum Bacteroidetes), four Alphaproteobacteria, and three Gammaproteobacteria). The identity of methylotrophic bacteria from the plant surface, from inside the plant, or the rhizosphere were as follows: Arthrobacter, one epiphyte and two rhizospheric; Pedobacter, one rhizospheric, Microvirga, four epiphytes; Inquilinus, two rhizospheric; Methylobacterium, one epiphyte, and one rhizospheric; Rhizobium, one rhizospheric; Sphingomonas, one endophyte; Subaequorebacter/Geminicoccus, one endophyte; Massilia, four epiphytes; Acinetobacter, twelve epiphytes, three rhizospheric, and three endophytes; and Pseudomonas, one epiphyte (Table 2, Fig. S1).
All methylotrophic isolates tested showed growth with methanol as the carbon and energy sources and Ca2+ or REE, Ce3+, as co-factors (Table 3). Different isolates showed distinct methylotrophic growth rates. Hence, the time of their maximum growth in the presence of Ce3+ ranged between a 72- and 144-h incubation. Most of the isolates did not show any preference for either co-factor, whereas it was apparent for some that one of the co-factors improved methylotrophic growth. In this assay, 22 isolates were selected to include all taxonomical groups. These strains included two Actinobacteria, one Sphingobacteriia, ten Alphaproteobacteria, one Betaproteobacteria, and eight Gammaproteobacteria.
Table 3.
Methylotrophic growth with Ca2+ or Ce3+ as co-factor for methanol dehydrogenase.
| Time | Genus | Strain | Growth with | |
|---|---|---|---|---|
|
| ||||
| Ca2+ | Ce3+ | |||
| 72 h | Sphingomonas | UAPS0110 | 0.7883 | 0.7637 |
| Methylobacterium | UAPS0123 | 1.0710* | 0.7660 | |
| Rhizobium | UAPS0160 | 0.8717 | 0.9367 | |
|
| ||||
| 96 h | Methylobacterium | UAPS0122 | 0.4123 | 0.3007 |
|
| ||||
| 120 h | Arthrobacter | UAPS0102 | 0.8563 | 1.3483* |
| Arthrobacter | UAPS0105 | 0.7910 | 1.1037 | |
| Subaequorebacter/ | ||||
| Geminicoccus | UAPS0115 | 0.2057 | 0.7513* | |
| Acinetobacter | UAPS0117 | 1.0703* | 0.7873 | |
| Microvirga | UAPS0120 | 0.9390* | 0.4777 | |
| Microvirga | UAPS0121 | 0.9967* | 0.7640 | |
| Pedobacter | UAPS0126 | 0.9957* | 0.7133 | |
| Microvirga | UAPS0137 | 1.1033 | 0.8533 | |
| Inquilinus | UAPS0142 | 0.6683 | 0.9637 | |
| Pseudomonas | UAPS0155 | 0.6140 | 0.6230 | |
| Acinetobacter | UAPS0163 | 1.2073 | 1.2163 | |
| Acinetobacter | UAPS0169 | 0.9680 | 1.4060* | |
| Massilia | UAPS0177 | 0.6817 | 1.0697* | |
| Acinetobacter | UAPS0180 | 1.3707 | 1.1673 | |
| Acinetobacter | UAPS0182 | 0.9920 | 1.0623 | |
| Acinetobacter | UAPS0183 | 0.8610 | 1.1247 | |
|
| ||||
| 144 h | Microvirga | UAPS0136 | 0.6087 | 0.2263 |
| Acinetobacter | UAPS0179 | 0.3757 | 0.8020 | |
Data correspond to absorbance at 600 nm, the media of three replicates. Cells were incubated under shaking at 30°C. The registers correspond to their time of maximum growth in the presence of Ce3+. The growth of each strain in the presence of Ca2+/Ce3+ wascomparedandthesignificance of differences between two values was assessed by the unpaired t-test, P>0.05. Values marked with an asterisk are significantly higher than their counterparts.
Amplicons (approximately 550 bp in length) with mxaFxoxFtargeted primers were obtained in 34.2% (13) of the isolates. All sequences were more similar to XoxF-like methanol dehydrogenases than to MDH-like methanol dehydrogenases (Fig. 1). After the sequence analysis, five Alphaproteobacteria, three Betaproteobacteria, four Gammaproteobacteria, and one Actinobacteria isolates were found to possess xoxF5-like sequences. Furthermore, Asp301 characteristic of XoxF dehydrogenases was detected in all of the amplicons that covered that region (Fig. 2, Table 2).
Fig. 2.
Partial alignment of sequences of methanol dehydrogenases that cover the region encoding Asp301. Asp301 (D) has been detected in all XoxF dehydrogenases and it is necessary for REE coordination. MxaF dehydrogenases do not possess Asp301.
Among the twenty-five isolates from which amplicons were not obtainable with the mxaf and xoxF-targeted primers, eleven clearly hybridized with a xoxF5 probe from M. extorquens (Table 2; Fig. 3): one Actinobacteria, four Alphaproteobacteria, one Betaproteobacteria, and five Gammaproteobacteria. The remaining fourteen isolates did not hybridize to the xoxF5 probe or were not amplified with the mxaFxoxF primers, including one Actinobacteria, one Sphingobacteriia, two Alphaproteobacteria, and ten Gammaproteobacteria. Hybridization with the mxaF probe was very faint; however, some dots indicated that the organism possessed mxaF loci (Fig. S2).
Fig. 3.
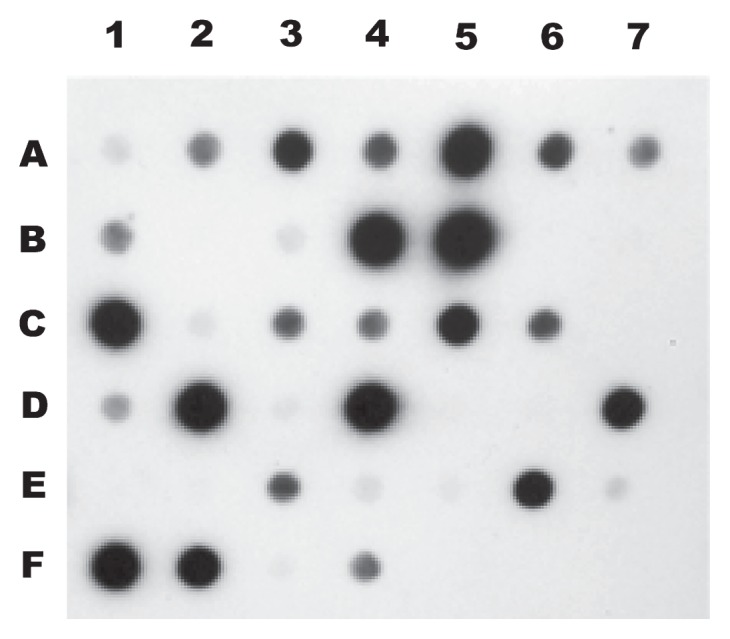
Dot-blot hybridization with xoxF. Lines A1, UAPS0104; A2, UAPS0105; A3, UAPS0106; A4, UAPS0181; A5, UAPS0110; A6, UAPS0102; A7, UAPS0184; B1, UAPS0182; B2, UAPS0149; B3, UAPS0121; B4, UAPS0122; B5, UAPS0123; B6, UAPS0126; B7, UAPS0127; C1, UAPS0114; C2, UAPS0136; C3, UAPS0137; C4, UAPS0180; C5, UAPS0115; C6, UAPS0142; C7, UAPS0145; D1, UAPS0179; D2, UAPS0174; D3, UAPS0118; D4, UAPS0155; D5, UAPS0156; D6, UAPS0158; D7, UAPS0177; E1, UAPS0165; E2, UAPS0120; E3, UAPS0160; E4, UAPS0168; E5, UAPS0169; E6, UAPS0175; E7, UAPS0172; F1, UAPS0117; F2, UAPS0183; F3, UAPS0163; F4, M. extorquens JCM2802 (100 ng); F5, Ustilago maydis 207; F6 and F7, void. One microgram of total DNA of the bacterial strains evaluated was transferred to nylon membranes. PCR probes (100 ng) were obtained by the PCR amplification of Methylobacterium extorquens JCM2802 with the primers xoxF5f361 5′-CAG GAT CCG TCC GTG AT-3′ and xoxF5r603 5′-SGA GAT GCC GAC GAT GA-3′.
Discussion
Methanol and methane are very common carbon compounds produced by plants (19, 28). Methylotrophy is distributed in many different taxa (31). In this study, bacteria of the Classes Actinobacteria, Sphingobacteria, Alpha-, Beta-, and Gammaproteobacteria were isolated in a methanol-based medium. Since this mostly plant-originated compound is a very common C-source in nature, numerous plant-associated microorganisms have the capability to use it.
Among the methylotrophs cultivated from N. macrocephala and its rhizosphere, most were isolated from the stem surface. We hypothesize that this relates to the presence of stomata and consequently to the main source of methanol from inner plant tissues (19). All the dehydrogenase sequences obtained were similar to xoxF5, genes that are phylogenetically related to other xoxF subfamilies and to mxaF. These xoxF5-like sequences were obtained from isolates belonging to the Classes Actinobacteria, and Alpha-, Beta-, and Gammaproteobacteria. mxaF-like sequences were previously identified in these classes and the phyla Bacteroidetes and Verrucomicrobia (4, 9, 29). Aspartic acid 301, the amino acid responsible for REE coordination (27), was detected in all of the sequences that covered that region. In contrast, none of the sequences showed different amino acids to Asp in that position. Additionally, none of the amplicons with mxaFxoxFtargeted primers were mxaF; they were xoxF5. Therefore, the sequenced amplicons coded for XoxF dehydrogenases. Nevertheless, we cannot rule out that some of the isolates possessed mxaF due to faint dot-blot hybridization with a mxaF probe. Positive hybridization with the xoxF probe indicated that these strains may possess xoxF5. Although we cannot exclude sequences of other xoxF subfamilies cross-hybridizing with the probe, hybridization and washing stringency conditions reduce that possibility. Some of the isolates that did not hybridize with the xoxF5 and mxaF probes or were not amplified with mxaF-xoxF primers may possess other sequences of the xoxF subfamilies or other methanol dehydrogenases such as MDH2 or NAD-dependent methanol dehydrogenase. Although we also designed primers and unsuccessfully attempted the amplification of methanol:NDMA oxidoreductase (Table S1, Results not shown), its presence cannot be excluded. In some of the cases in which we detected hybridization to mxaF or xoxF5, we did not obtain amplicons of methanol dehydrogenase genes. This inconsistency may be related to the design of the primers. All isolates tested in the methylotrophy assay grew using Ce3+, as expected, but also used Ca2+ as a co-factor. Therefore, it currently remains unclear whether XoxF enzymes accept Ca2+ besides REE, as suggested by Keltjens et al. 2014 (27).
The ubiquities of xoxF, of their peptides, and of the bacteria carrying them in nature have been demonstrated in different studies, including the N. macrocephala-related ecosystem. XoxF has been detected in the phyllospheres of rice, clover, soybean, and Arabidopsis (15, 30). A previous study in a particular marine environment also showed the high abun-dance of XoxF (53). In an autecological approach, a semi in situ SIP assay detected the strong expression of a xoxF-like locus in Methylotenera mobilis (59). Furthermore, methanol oxidation in Methylomicrobium buryatense, possessing xoxF and mxaFI functional loci appeared to be mainly accomplished by XoxF (12).
It has not yet been established whether there is a biogeography of subfamilies of xoxF. New studies on methylotrophy with non-culture and culture approaches in different environments are needed. A pioneer ecological study of the different xoxF subfamilies in coastal marine water only detected sequences of the clusters xoxF4 and xoxF5 (55). In a different environment, the methanol dehydrogenase peptides XoxF and MxaF of Methylobacterium, a microorganism that only possesses xoxF5 and mxaF sequences, were abundantly detected in the phyllosphere of soybean, clover, rice, and A. thaliana (15, 30). The present culture-dependent study demonstrated the presence of microorganisms possessing sequences of the subfamily xoxF5 in the semi-arid environment of N. macrocephala.
A previous study with some XoxF enzymes reported high affinity for methanol (27, 50). If the enzymes of more diverse microorganisms exhibit similar behaviors, XoxF may be crucial for methylotrophic bacteria that thrive in plants showing slow metabolic properties and producing methanol at low rates, such as cacti. The presence of XoxF may be favored in environments in which sand, and, thus, REEs, are abundant, such as arid lands (50).
Besides its participation in methylotrophic metabolism, XoxF may be involved in the regulation of stress responses and in denitrification metabolism (17, 45). Its putative role in stress responses may be particularly important in semi-arid areas and in plant surfaces.
Although the typical methanol dehydrogenase from Actinobacteria is methanol:NDMA oxidoreductase, they do not exclusively carry it. The synthesis of PQQ by Actinobacteria in the presence of methanol suggested the presence of a PQQ-dependent methanol dehydrogenase (22). In another study, a Brevibacterium casei strain, an actinobacterial methylotrophic human mouth microorganism, carried a mxaF methanol dehydrogenase sequence (4; see Fig. 1), and more recently, metagenomic studies in the desert of Atacama detected Pseudonocardia PQQ methanol dehydrogenase genes (36). The presence of xoxF genes in Actinobacteria isolated in this study may have originated from lateral transfer events, as has been detected in the locus mxaF of methanotrophic bacteria (7, 33) and in methylotrophic Alphaproteobacteria (7).
The methylotrophic isolates from the environment of N. macrocephala belonged to Proteobacteria, Actinobacteria, and Sphingobacteriia. Among them, Acinetobacter spp. (Gammaproteobacteria) were the most frequently isolated organisms. It has been reported that Acinetobacter uses methanol as a carbon source (20, 61) and a methanol dehydrogenase sequence coding Asp301 has previously been detected in this genus (20). Similar to these findings, other studies identified Proteobacteria and Actinobacteria as some of the most common taxa in the rhizosphere and soil from cacti and other plants from arid lands (2, 11, 13, 26).
Methylotrophic bacteria are ubiquitous and have meaningful roles in ecosystems. Since water is mostly limited in arid environments, perennial plants from these environments show restrained growth, particularly throughout the dry season. The community of methylotrophic culturable bacteria associated with the semi-arid thriving cactus N. macrocephala include xoxF-like dehydrogenases-possessing microorganisms. Their ecological role in xerophytic plants warrants further study. Since the cultivation procedures employed in the present study do not necessarily produce a real picture of bacterial diversity, the future application of non-culture approaches will enrich knowledge on methylotrophic diversity in this environment. In future inoculation experiments, we intend to detect the isolates of methylotrophic bacteria that may stimulate the growth of N. macrocephala, particularly in the vulnerable juvenile stage.
Supplementary Material
Acknowledgements
This work was partially funded by the projects CONACYT CB-2009-128235-Z and BUAP-VIEP. We thank Antonino Báez (Instituto de Ciencias, BUAP) for editing the manuscript. We are grateful to Ángeles Domínguez, Liliana López (Instituto de Ciencias, BUAP), and Aracely Jacinto for their statistical and technical assistance. We are grateful to Rebeca Martínez and Mónica Martínez (Instituto de Ciencias, BUAP) for providing the DNA of U. maydis 207. We acknowledge the suggestions of anonymous referees. In memoriam of Jesús Caballero, an exemplary scientist and excellent friend.
References
- 1.Abanda-Nkpwatt D., Müsch M., Tschiersch J., Boettner M., Schwab W. Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J Exp Bot. 2006;57:4025–4032. doi: 10.1093/jxb/erl173. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre-Garrido J.F., Montiel-Lugo D., Hernández-Rodríguez C., Torres-Cortes G., Millán V., Toro N., Martínez-Abarca F., Ramírez-Saad H.C. Bacterial community structure in the rhizosphere of three cactus species from semi-arid highlands in central Mexico. Anton Leeuw Int J Gen Mol Microbiol. 2012;101:891–904. doi: 10.1007/s10482-012-9705-3. [DOI] [PubMed] [Google Scholar]
- 3.Andrew D.R., Fitak R.R., Munguia-Vega A., Racolta A., Martinson V.G., Dontsov K. Abiotic factors shape microbial diversity in Sonoran desert soils. Appl Environ Microbiol. 2012;78:7527–7537. doi: 10.1128/AEM.01459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anesti V., McDonald I.R., Ramaswamy M., Wade W.G., Kelly D.P., Wood A.P. Isolation and molecular detection of methylotrophic bacteria occurring in the human mouth. Environ Microbiol. 2005;7:1227–1238. doi: 10.1111/j.1462-2920.2005.00805.x. [DOI] [PubMed] [Google Scholar]
- 5.Antony C.P., Kumaresan D., Ferrando L., Boden R., Moussard H., Fernández-Scavino A., Shouche Y.S., Murrell J.C. Active methylotrophs in the sediments of Lonar Lake, a saline and alkaline ecosystem formed by meteor impact. ISME J. 2010;4:1470–1480. doi: 10.1038/ismej.2010.70. [DOI] [PubMed] [Google Scholar]
- 6.Arfman N., Hektor H.J., Bystrykh L.V., Govorukhina N.I., Dijkhuizen L., Frank J. Properties of an NAD(H)-containing methanol dehydrogenase and its activator protein from Bacillus methanolicus. Eur J Biochem. 1997;244:426–433. doi: 10.1111/j.1432-1033.1997.00426.x. [DOI] [PubMed] [Google Scholar]
- 7.Beck D.A.C., McTaggart T.L., Setboonsarng U., Vorobev A., Goodwin L., Shapiro N., Woyke T., Kalyuzhnaya M.G., Lidstrom M.E., Chistoserdova L. Multiphyletic origins of methylotrophy in Alphaproteobacteria, exemplified by comparative genomics of Lake Washington isolates. Environ Microbiol. 2015;17:547–554. doi: 10.1111/1462-2920.12736. [DOI] [PubMed] [Google Scholar]
- 8.Borodina E., Kelly D.P., Rainey F.A., Ward-Rainey N.L., Wood A.P. Dimethylsulfone as a growth substrate for novel methylotrophic species of Hyphomicrobium and Arthrobacter. Arch Microbiol. 2000;173:425–437. doi: 10.1007/s002030000165. [DOI] [PubMed] [Google Scholar]
- 9.Chistoserdova L., Lidstrom M.E. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology. 1997;143:1729–1736. doi: 10.1099/00221287-143-5-1729. [DOI] [PubMed] [Google Scholar]
- 10.Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13:2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury S.P., Schmid M., Hartmann A., Tripathi A.K. Diversity of 16S-rRNA and nifH genes derived from rhizosphere soil and roots of an endemic drought tolerant grass, Lasiurus sindicus. Eur J Soil Biol. 2009;45:114–122. [Google Scholar]
- 12.Chu F., Lidstrom M.E. XoxF acts as the predominant methanol dehydrogenase in the Type I methanotroph Methylomicrobium buryatense. J Bacteriol. 2016;198:1317–1325. doi: 10.1128/JB.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connon S.A., Lester E.D., Shafaat H.S., Obenhuber D.C., Ponce A. Bacterial diversity in hyperarid Atacama Desert soils. J Geophys Res Biogeosciences. 2007;112:G04S17. [Google Scholar]
- 14.de Vries G.E., Arfman N., Terpstra P., Dijkhuizen L. Cloning, expression, and sequence analysis of the Bacillus methanolicus Cl methanol dehydrogenase gene. J Bacteriol. 1992;74:5346–5353. doi: 10.1128/jb.174.16.5346-5353.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delmotte N., Knief C., Chaffron S., Innerebner G., Roschitzki B., Schlapbach R., von Mering C., Vorholt J. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci USA. 2009;106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esparza-Olguin L., Valverde T., Mandujano M.C. Comparative demographic analysis of three Neobuxbaumia species (Cactaceae) with differing degree of rarity. Popul Ecol. 2005;47:229–245. [Google Scholar]
- 17.Firsova Y.E., Torgonskaya M.L., Trotsenko Y.A. Functionality of the xoxF gene in Methylobacterium dichloromethanicum DM4. Microbiology (Russia) 2015;84:796–803. [PubMed] [Google Scholar]
- 18.Fitriyanto N.A., Fushimi M., Matsunaga M., Pertiwiningrum A., Iwama T., Kawai K. Molecular structure and gene analysis of Ce3+-induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J Biosci Bioeng. 2011;111:613–617. doi: 10.1016/j.jbiosc.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Galbally L.E., Kirstine W. The production of methanol by flowering plants and the global cycle of methanol. J Atmos Chem. 2002;43:195–229. [Google Scholar]
- 20.Ghosh A., Goyal A., Rakesh K.J. Study of methanol-induced phenotypic changes in a novel strain of Acinetobacter lwoffi. Arch Microbiol. 2007;188:533–539. doi: 10.1007/s00203-007-0268-z. [DOI] [PubMed] [Google Scholar]
- 21.Green P.N. Methylobacterium. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., editors. The Prokaryotes. Vol. 5. Springer; New York: 2006. pp. 257–265. [Google Scholar]
- 22.Hazeu W., de Bruijn J.C., van Dijken J.P. Nocardia sp. 239, a facultative methanol utilizer with the ribulose monophosphate pathway of formaldehyde fixation. Arch Microbiol. 1983;135:205–210. [Google Scholar]
- 23.Hektor H.J., Kloosterman H., Dijkhuizen L. Nicotinoprotein methanol dehydrogenase enzymes in Gram-positive methylotrophic bacteria. J Mol Catal B-Enzym. 2000;8:103–109. [Google Scholar]
- 24.Kalyuzhnaya M.G., Hristova K.R., Lidstrom M.E., Chistoserdova L. Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: implications for environmental detection of methylotrophy and evidence for convergent evolution. J Bacteriol. 2008;190:3817–3823. doi: 10.1128/JB.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalyuzhnaya M.G., Lapidus A., Ivanova N., et al. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol. 2008;26:1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan D., Maymon M., Agapakis C.M., Lee A., Wang A., Prigge B.A., Volkogon M., Hirsh A.M. A survey of the microbial community in the rhizosphere of two dominant shrubs of the Negev desert highlands, Zygophyllum dumosum (Zygophyllaceae) and Atriplex halimus (Amaranthaceae), using cultivation-dependent and cultivation-independent methods. Am J Bot. 2013;100:1–13. doi: 10.3732/ajb.1200615. [DOI] [PubMed] [Google Scholar]
- 27.Keltjens J.T., Pol A., Reimann J., Op den Camp H.J.M. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98:6163–6183. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 28.Keppler F., Hamilton J.T.G., Brass M., Röckmann T. Methane emissions from terrestrial plants under aerobic conditions. Nature. 2006;439:187–191. doi: 10.1038/nature04420. [DOI] [PubMed] [Google Scholar]
- 29.Kist J., Tate R.L., III Phylogeny of bacterial methylotrophy genes reveals robustness in Methylobacterium mxaF sequences and mxa operon construction. Soil Biol Biochem. 2013;59:49–57. [Google Scholar]
- 30.Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R., von Mering C., Vorholt J. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6:1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolb S., Stacheter A. Prerrequisites for amplicon pyrosequencing of microbial methanol utilizers in the environment. Frontiers Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00268. article 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S., Stecher G., Tamura K. MEGA: Molecular Evolutionary Genetic Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau E., Fisher M.C., Steudler P.A., Cavanaugh C.M. The methanol dehydrogenase gene, mxaF, as a functional and phylogenetic marker for proteobacterial methanotrophs in natural environments. PLoS ONE. 2013;8:e56993. doi: 10.1371/journal.pone.0056993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.D., Seong C.N. Nocardioides opuntiae sp nov., isolated from soil of a cactus. Int J Syst Evol Microbiol. 2014;64:2094–2099. doi: 10.1099/ijs.0.060400-0. [DOI] [PubMed] [Google Scholar]
- 35.Lidstrom M.E. Aerobic methylotrophic Prokaryotes. In: Balows A., Truper H.G., Dworkin M., Harder W., Schleifer K.-H., editors. The Prokaryotes. Vol. 2. Springer; New York: 2006. pp. 618–634. [Google Scholar]
- 36.Lynch R.C., Darcy J.L., Kane N.C., Nemergut D.R., Schmidt S.C. Metagenomic evidence for metabolism of trace atmospheric gases by high-elevation desert Actinobacteria. Front Biol. 2014;5:698. doi: 10.3389/fmicb.2014.00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madhaiyan M., Poonguzhali S., Lee H.S., Hari K., Sundaram S.P., Sa T.M. Pink-pigmented facultative methylotrophic bacteria accelerate germination, growth and yield of sugarcane clone Co86032 (Saccharum officinarum L.) Biol Fertil Soils. 2005;41:350–358. [Google Scholar]
- 38.Madhaiyan M., Reddy S., Anandham R., Senthilkumar M., Poonguzhali S., Sundaram S.P., Sa T.M. Plant growth–promoting Methylobacterium induces defense responses in groundnut (Arachis hypogaea L.) compared with rot pathogens. Curr Microbiol. 2006;53:270–276. doi: 10.1007/s00284-005-0452-9. [DOI] [PubMed] [Google Scholar]
- 39.Madhaiyan M., Poonguzhali S., Sa T. Metal tolerating methylotrophic bacteria reduces nickel and cadmium toxicity and promotes plant growth on tomato (Lycopersicum sculentum L.) Chemosphere. 2007;69:220–228. doi: 10.1016/j.chemosphere.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 40.Madhaiyan M., Poonguzhali S., Senthilkumar M., Sundaram S.P., Sa T. Nodulation and plant-growth promotion by methylotrophic bacteria isolated from tropical legumes. Microbiol Res. 2009;164:114–120. doi: 10.1016/j.micres.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Madhaiyan M., Poonguzhali S., Lee J.-S., Lee K.C., Sundaram S. Flavobacterium glycines sp. nov., a facultative methylotroph isolated from the rhizosphere of soybean. Int J Syst Evol Microbiol. 2010;60:2187–2192. doi: 10.1099/ijs.0.014019-0. [DOI] [PubMed] [Google Scholar]
- 42.McDonald I.R., Murrell J.C. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol. 1997;638:3218–3224. doi: 10.1128/aem.63.8.3218-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McTaggart T.L., Beck D.A.C., Setboonsarng U., Shapiro N., Woyke T., Lidstrom M.E., Kalyuzhnaya M.G., Chistoserdova L. Genomics of methylotrophy in Gram-positive methylamine-utilizing bacteria. Microorganisms. 2015;3:94–112. doi: 10.3390/microorganisms3010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miquelajauregui Y., Valverde T. Survival and early growth of two congeneric cacti that differ in their level of rarity. J Arid Environ. 2010;74:1624–1631. [Google Scholar]
- 45.Mustakhimov I., Kalyuzhnaya M.G., Lidstrom M.E., Chistoserdova L. Insights into denitrification in Methylotenera mobilis from denitrification pathway and methanol metabolism mutants. J Bacteriol. 2013;195:2207–2211. doi: 10.1128/JB.00069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagawa T., Mitsui R., Tani A., Sasa K., Tashiro S., Iwama T., Hayakawa T., Kawa K. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS ONE. 2012;7:e50480. doi: 10.1371/journal.pone.0050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nemecek-Marshall M., MacDonald R.C., Franzen J.J., Wojciechowski C., Fall R. Methanol emission from leaves: enzymatic detection of gas phase methanol and relation of methanol fluxes to stomata conductance and leaf development. Plant Physiol. 1995;108:1359–1368. doi: 10.1104/pp.108.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park H., Lee H., Ro Y.T., Kim Y.M. Identification and functional characterization of a gene for the methanol: N,N′-dimethyl-4-nitrosoaniline oxidoreductase from Mycobacterium sp. strain JC1 (DSM 3803) Microbiology. 2010;156:463–471. doi: 10.1099/mic.0.034124-0. [DOI] [PubMed] [Google Scholar]
- 49.Pigoleva S.V., Zakharchenko N.S., Pigolev A.V., Trotsenko Y.A., Buryanov Y.I. Influence of colonizing methylobacteria on morphogenesis and resistance of sugar beet and white cabbage plants to Erwinia carotovora. Microbiology (Russia) 2009;45:604–609. [PubMed] [Google Scholar]
- 50.Pol A., Barends T.R.M., Dietl A., Khadem A.F., Eygensteyn J., Jetten M.S.M., Op den Camp H.J.M. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol. 2014;16:255–264. doi: 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- 51.Ruedas M., Valverde T., Zavala-Hurtado J.A. Analysis of the factors that affect the distribution and abundance of three Neobuxbaumia species (Cactaceae) that differ in their degree of rarity. Acta Oecol-Int J Ecol. 2006;29:155–164. [Google Scholar]
- 52.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- 53.Sowell S.M., Abraham P.E., Shah M., Verberkmoes N.C., Smith D.P., Barofsky D.F., Giovannoni S.J. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J. 2011;5:856–865. doi: 10.1038/ismej.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tani A., Takai Y., Suzukawa I., Akita M., Murase H., Kimbara K. Practical application of methanol-mediated mutualistic symbiosis between Methylobacterium species and a roof greening moss, Racomitrium japonicum. PLoS ONE. 2012;7:e33800. doi: 10.1371/journal.pone.0033800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taubert M., Grob C., Howat A.M., Burns O.J., Dixon J.L., Chen Y., Murrell J.C. XoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ Microbiol. 2015;17:3937–3948. doi: 10.1111/1462-2920.12896. [DOI] [PubMed] [Google Scholar]
- 56.Torres-Cortés G., Millán V., Fernández-González A.J., Aguirre-Garrido J.F., Ramírez-Saad H.C., Fernández-López M., Toro N., Martínez-Abarca F. Bacterial community in the rhizosphere of the cactus species Mammillaria carnea during dry and rainy seasons assessed by deep sequencing. Plant Soil. 2012;357:275–288. [Google Scholar]
- 57.Trotsenko Y.A., Ivanova E.G., Doronina N.V. Aerobic methylotrophic bacteria as phytosymbionts. Microbiology (Russia) 2001;70:725–736. [PubMed] [Google Scholar]
- 58.Valiente-Banuet A., Rojas-Martínez A., del Coro-Arizmendi M., Davila P. Pollination biology of two columnar cacti (Neobuxbaumia mezcalaensis and Neobuxbaumia macrocephala) in Tehuacan Valley, Central Mexico. Am J Bot. 1997;84:452–455. [Google Scholar]
- 59.Vorobev A., Beck D.A.C., Kalyuzhnaya M.G., Lidstrom M.E., Chistoserdova L. Comparative transcriptomics in three Methylophilaceae species uncover different strategies for environmental adaptation. PeerJ. 2013;1:e115. doi: 10.7717/peerj.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yim W.J., Kim K.Y., Lee Y.W., Sundaram S.P., Lee Y., Sa T.M. Real time expression of ACC oxidase and PR-protein genes mediated by Methylobacterium spp. in tomato plants challenged with Xanthomonas campestris pv. vesicatoria. J Plant Physiol. 2014;171:1064–1075. doi: 10.1016/j.jplph.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 61.Yuan H., Yao J., Masakorala K., Wang F., Cai M., Yu C. Isolation and characterization of a newly isolated pyrene-degrading Acinetobacter strain USTB-X. Environ Sci Pollut Res. 2014;21:2724–2732. doi: 10.1007/s11356-013-2221-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.