Abstract
Introduction
Growth hormone is secreted by the pituitary gland, which forms part of the craniofacial midline. IRF6 encodes a transcription factor involved in the development of the craniofacial midline and mutations in IRF6 are known to disturb craniofacial development. Craniofacial and pituitary development are closely related. After whole exome sequencing revealed a new mutation in IRF6 in a family with Idiopathic Growth Hormone Deficiency (IGHD), we screened the remainder of our IGHD cohort for mutations in this gene and related their genotypes to pituitary and craniofacial morphology.
Materials and methods
We sequenced all coding exons and exon–intron boundaries of IRF6 in 81 patients with IGHD. We performed a multiplex ligation-dependent probe amplification (MLPA) in order to exclude copy number variations in IRF6. We analyzed facial measurements taken from standardized digital pictures of 48 patients.
Results
We found two new variants and eleven polymorphisms. Apart from the new mutation found in the index family (p.Arg233Cys), we found one other new heterozygous missense mutation in IRF6 (Pro456Ser). p.Arg233Cys was reported as extremely rare in exome databases (1 allele out of 120.852 alleles sequenced), strictly conserved among species and was predicted deleterious by all variant predictor programs. Pro456Ser was predicted to be benign. MLPA did not reveal any exon deletions or duplications in any of the patients.
Conclusion
This is the first report of IRF6 analysis in an IGHD cohort. We found one new mutation which, based on in silico analysis, could be of functional relevance. However, we did not find any mutations in the other patients. Therefore, we conclude that IRF6 defects are rare in IGHD patients and further research should focus on new candidate genes.
Electronic supplementary material
The online version of this article (doi:10.1007/s11102-017-0808-8) contains supplementary material, which is available to authorized users.
Keywords: Pituitary, Craniofacial, Midline, Genetics, Growth hormone deficiency, IRF6
Introduction
Growth hormone (GH) is secreted by the pituitary gland, which forms part of the craniofacial midline. Growth Hormone Deficiency (GHD) is the result of insufficient GH secretion, resulting in decreased production of GH-dependent growth factors. The incidence of Idiopathic GH Deficiency (IGHD) is estimated to vary between 1 in 3500 to 1 in 10,000 live births [1–5]. Estimates indicate that between 5 and 30% of idiopathic IGHD case have first-degree relatives with short stature, suggesting a genetic etiology [6].
Although mutations in GH1 and GHRHR can cause IGHD [7–10], the vast majority of patients with IGHD do not carry mutations in these two genes. This suggests that other genes are involved. The identification of these genes is needed to understand the pathogenesis of this complex condition.
Pituitary hormone deficiencies have been associated with defects in the craniofacial midline. Several authors have reported associations between pituitary problems and craniofacial defects [11–17]. Studies investigating hormone deficiencies in larger cohorts of patients with craniofacial clefting show remarkable results. Traggiai et al. studied 19 patients with cleft lip and palate and found GH deficiency in seven of them (37%) [17]. Akin et al. [18] studied 33 patients with median orofacial clefts and found endocrine abnormalities in 22 (70.9%), of which 13 had single and nine multiple hormone deficiencies. Growth hormone deficiency was detected in four of them (12%). There was no relationship between the types of orofacial cleft and endocrine abnormalities. Slavotinek et al. reviewed 31 patients with pituitary duplication. In 19 of them (61%), pituitary duplication was accompanied by cleft palate [19]. Another well-known association between pituitary problems and craniofacial midline defects is called septo-optic dysplasia, defined as the absence of the septum pellucidum combined with hypoplasia of the optic nerve and pituitary dysfunction [20, 21].
Also in patients with Isolated GH Deficiency (IGHD), a broad range of craniofacial midline abnormalities have been described, like hypertelorism, cleft lip and palate and single median maxillary central incisor [22, 23]. The mechanism underlying this association between isolated GH deficiency and craniofacial midline defects is still poorly understood.
Apart from the association between pituitary function and craniofacial features, we also found an association between pituitary morphology and craniofacial features [24].
All these findings are suggestive of an association between craniofacial midline and pituitary development. The mechanism underlying this association has not yet been elucidated.
An important gene involved in craniofacial midline formation is IRF6 on chromosome 1q32.2. IRF6 encodes Interferon Regulatory Factor 6, a transcription factor expressed in oral and nasal epithelia. IRF6 regulates the proliferation and differentiation of epithelial cells during the formation of the craniofacial midline during embryonic development [25]. Mutations in IRF6 are associated with Van der Woude Syndrome (VWS, OMIM #119300) and Popliteal Pterygium Syndrome (PPS, OMIM #119500). Both syndromes are characterized by a cleft lip and/or cleft palate, which are due to defective craniofacial development. Whereas mutations in IRF6 cause syndromic orofacial clefting, IRF6 polymorphisms are associated with non-syndromic cleft lip and/or palate (NSCL/P, OMIM #119530) [26].
Whole exome sequencing (WES) revealed an IRF6 mutation in one of the families of our IGHD cohort. Since IRF6 mutations are known to disturb craniofacial development, which is related to pituitary development, we considered IRF6 a possible candidate gene for IGHD. Furthermore, we hypothesized that variations in IRF6 might be related to craniofacial morphology. The relation between IRF6 and pituitary function, pituitary morphology and craniofacial features has not been studied before.
Materials and methods
Study subjects
We studied 81 patients with IGHD participating in the Dutch HYPOPIT study, which investigates the genetic causes of idiopathic GH deficiency. These patients had been recruited from the Endocrinology Departments at six university and two non-university hospitals, and had been registered in the Dutch National Registry of Growth Hormone Treatment between 1992 and 2003. IGHD was defined as a peak GH response <6.7 µg/L during Growth Hormone test (mostly arginine tests), or <10 µg/L combined with serum IGF-I < −2 SDS (Standard Deviation Score, according to age- and gender-specific values) and normal serum levels of other pituitary hormones. Exclusion criteria were: GH deficiency of known cause, such as a brain tumor, brain surgery, brain radiation or known syndromes. We obtained approval from the medical ethics committees of all participating hospitals. Informed consent was obtained from all participants and their parents if they were less than 18 years old. 81 IGHD patients who were treated in the participating hospitals and who fulfilled the criteria for IGHD, agreed to participate in the study. All patients received GH treatment. Clinical data of the patients had previously been collected from the Dutch National Registry of Growth Hormone Treatment. GH1 and GHRHR mutations had been ruled out in all patients. MRI reports were available for 67 patients.
DNA analysis
DNA of 81 patients with IGHD was isolated from whole blood collected in EDTA tubes using standard procedures. We made dilutions of 25 µl/ng of all the collected DNA samples and stored them in a 96-wells plate at −20 °C.
Whole exome sequencing
DNA samples of the index patient and his first-degree relatives was analyzed by Whole Exome Sequencing: Nimblegen SeqCap EZ Exome v2.0 44 Mb in combination with Illumina Paired-End Library Preparation and 2 × 100 bp Sequencing at 4 Gb per sample. Additional analysis was done with Illumina Human CytoSNP850K SNP arrays.
IRF6 Sequencing
After WES revealed the IRF6 mutation in the index family, we screened the remaining 80 IGHD patients for IRF6 defects as well. For all patients, we amplified coding exons and exon–intron boundaries by PCR, using primers described by Wang et al. [36]. For exons 3, 4, 8 and 9 we designed new primers using the program Primer3 (reference sequence NM_006147.3). Primers sequences are shown in supplementary Table S1. For PCR, we used the following Qiagen reagents: 10x PCR buffer, 0.2 mM dNTP mixture and 5 units/μl Taq DNA Polymerase. For the primers used to amplify exon 4, we also added 0.5 μl 25 mM MgCl. For primer pairs 1, 2, 5, 6, 7a, 7b, 9a and 9c, we used the standard PCR program. For primer pairs 3, 4, 8 and 9b, we used a Touch Down PCR, lowering the annealing temperature one degree per cycle, during the first ten cycles. PCR reagents and detailed amplification programs are shown in supplementary Tables S2 and S3. After gel electrophoresis, we purified the PCR products using the High Pure PCR product Purification kit (Qiagen) or the Illustra GFX 96 PCR Purification Kit (GE Healthcare) according to the supplied protocols. Sequencing was outsourced to Baseclear Sequencing Services (www.baseclear.com) and carried out using the ABI3730XL sequencer (Life Technologies). The results were analyzed for mutations using Sequencher4.1 (Genecards). For each new variant, the Variant Effect Predictor (www.Ensembl.org) was consulted to predict functional impact. The predictions of this tool are based on the structure and function of the protein and the degree of conservation.
Multiplex ligation-dependent probe amplification
Copy number analysis was performed using Multiplex Ligation-dependent Probe Amplification (MLPA). We used SALSA MLPA EK1 reagent and P304 IRF6 probemix according to the protocol of MRC-Holland. We analyzed the results with the program Genemarker (SOFTGENETICS).
Craniofacial measurements
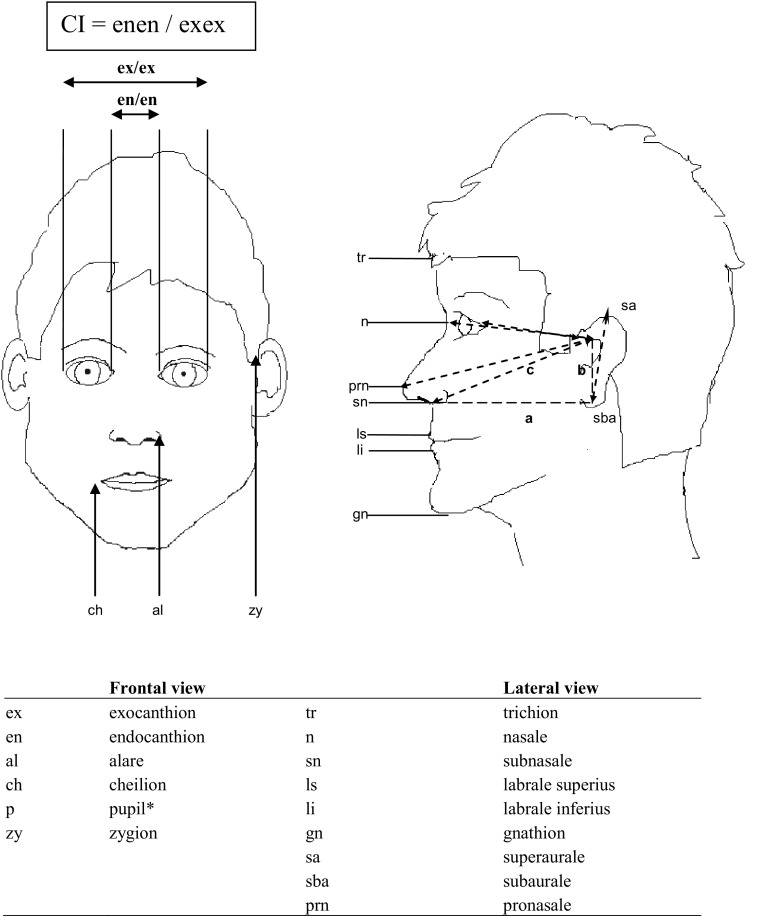
For the current analysis, we measured Canthal Index (CI) from digital photographs taken previously [24]. Frontal and lateral digital photographs were taken under standardized conditions in order to obtain comparable images of reliable quality. Pictures were taken using a Canon® 4.0 Megapixel digital camera. A squared background plate was used to position pictures parallel to the lower extreme of the computer screen, according to the method used by Bishara [27]. Pictures were taken in 48 patients, 4 patients were excluded because of suboptimal quality due to movement artefacts. Adobe Photoshop was used for morphometric analysis. Facial measurements of all pictures were performed by an independent observer (JB). The landmarks we used were those described by Farkas [28] (Fig. 1). Horizontal and vertical lines were drawn and distance between the lines were measured.
Fig. 1.
Anthropometric landmarks for frontal facial photographs carried out in our patients
Statistical analysis
We used SPSS 22.0 to analyze craniofacial measurements in relation to growth-related parameters (GH levels during arginine testing, IGF-I SDS and height SDS at start GH). We used one way ANOVA and Chi square analysis to assess differences in clinical parameters according to genotype.
Results
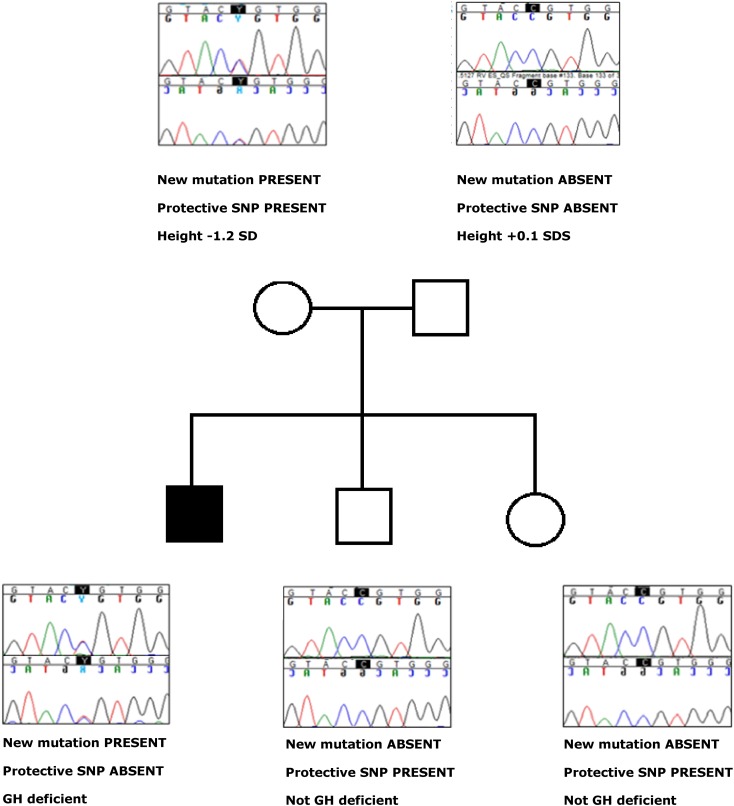
The IRF6 mutation found in the index family is a heterozygous missense mutation in exon 7, encoding part of the protein binding domain of IRF6. The substitution of the C by a T at this position (c.697C>T) results in an amino acid change of Arginine to Cysteine (p.Arg233Cys). The mutation was found in a GH deficient boy with non-consanguineous parents. His father’s height was 182 cm (0.1SD) and his mother’s height was 160 cm (−1.2 SD). His younger brother and sister had growth retardation as well, but they were not GH deficient. The boy was born after 39 weeks of pregnancy and was small for gestational age (2400 and 47 cm). He suffered neonatal jaundice for which he received phototherapy. His arginine test showed a peak GH of 1.7 µg/L, glucagon test showed a peak GH of 2.5 µg/L and his IGF-I SDS before start of GH was −1.4. He did not have any other pituitary hormone deficiencies. At start of GH treatment his height SDS was −2.4. At that time, his bone age was 2 years delayed. He did not have any midline defect and his pituitary MRI was normal. He responded very well to GH therapy; his height SDS increased with 2 SDS and his IGF-I increased from −1.4 to 0.4 SDS. p.Arg233Cys is reported as extremely rare in exome databases (1 allele out of 120.852 alleles sequenced). Multiple species amino acid sequence alignment showed that arginine at AA position 233 is conserved in many different species (overview of species available upon request). The Variant Effect Predictor showed this mutation is deleterious. After finding p.Arg233Cys in the index case, we sequenced both parents, the brother and the sister and found that the mother also carried the mutation (Fig. 2). The mother’s phenotype was normal. However, apart from the new mutation p.Arg233Cys, the mother carried the minor allele of rs2235371. Rs2235371 is a polymorphism known to protect against orofacial cleft formation. Figure 2 shows the genotypes for the new mutation p.Arg233Cys and the protective SNP rs2235371, as well as phenotypic data for the index patient and his first-degree relatives. As seen in this figure, the only person with IGHD is the only person with the new mutation p.Arg233Cys without the protective SNP rs2235371 (see also "Discussion" section).
Fig. 2.
Genotypic and phenotypic data of the family with the new mutation p.Arg233Cys. For each individual, phenotype and presence or absence of the new mutation (p.Arg233Cys) and the protective SNP (p.Val274Ile,rs2235371) is shown
After finding this new mutation in IRF6 in the index family, we screened another 80 patients with IGHD and we found one more new mutation and 11 known polymorphisms in IRF6 (Table 1). MLPA did not show any copy number variations. The genotype distributions of the known IRF6 SNPs in our cohort were consistent with Hardy–Weinberg equilibrium (data not shown) and not significantly different from those in European control cohorts (www.ensembl.org).
Table 1.
Known and new variants found in the IRF6 gene in our IGHD cohort
| Exon | SNP ID | Position | Impact | Base change | AA change |
|---|---|---|---|---|---|
| 1 | rs34743335 | 5′upstream | c.-313T>A | ||
| rs12403006 | 5′UTR | c.-302A>T | |||
| 2 | rs2235377 | Intronic | c.-75-4A>G | ||
| rs861019 | 5′UTR | c.-73T>C | |||
| 3 | No variants found | ||||
| 4 | rs7552506 | Intronic | c.175-5C>G | ||
| 5 | rs2013162 | Exonic | Silent | c.459G>T | p.Ser58= |
| 6 | No variants found | ||||
| 7 | New mutation | Exonic | Missense | c.697C>T | p.Arg233Cys |
| rs2235371 | Exonic | Missense | c.820G>A | p.Val274Ile | |
| rs41303263 | Exonic | Silent | c.759T>C | p.Tyr158= | |
| rs2235373 | c.1060+37C>T | ||||
| 8 | No variants found | ||||
| 9 | New mutation | Exonic | Missense | c.1366C>T | p.Pro456Ser |
| rs17317411 | 3′UTR | c.*451A>G | |||
| rs75012801 | 3′UTR | c.*479T>G |
Variants printed in bold are discussed in the “Discussion” section
The other new mutation, p.Pro456Ser was found in a boy diagnosed as GH deficient at the age of 9 years and started GH treatment when his height SDS was −3.8. He did not have any other pituitary hormone deficiencies. Exome databases showed that Pro456Ser was also rare, but the mutation is not strictly conserved among species and predicted to be benign (not damaging). His unaffected father also carried this mutation (data not shown).
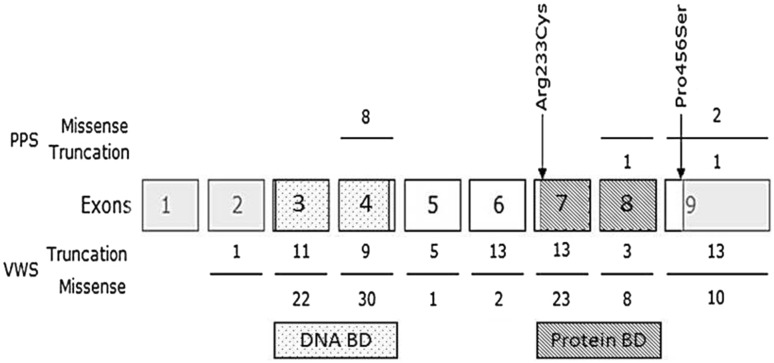
Figure 3 shows IRF6 variants previously described in the literature, as well as the new IRF6 mutations found in the current study. Apart from the two new mutations, we found 11 known polymorphisms. We focused on rs2235371 (V274I), which has been reported to be protective against orofacial cleft formation (see also “Discussion” section). The clinical characteristics of our patient cohort according to rs2235371 genotype are shown in Table 2. Patients carrying the protective minor (A) allele showed a trend towards a less severe phenotype, which was consistent for all clinical parameters measured: they had a higher GH peak during arginine testing, a larger height SDS and a higher IGF-1 SDS at start of GH treatment. Mid-facial hypoplasia, a facial characteristic often seen in patients with severe GHD, was also less often seen in patients with the protective minor allele. None of the patients with the protective allele had orofacial clefts, whereas of the 76 patients without the protective allele, two had orofacial clefts (one female patient had a cleft lip and palate and one male patient had a cleft lip and palate and bifid tongue). Pituitary anomalies on MRI were absent among the patients with the protective allele, whereas they were present in 51% of the patients without the protective allele (0% vs. 51% p = 0.017) (Table 3). Although the milder phenotype was clear for all clinical parameters measured, only the difference in MRI results reached statistical significance (p = 0.017).
Fig. 3.
Overview of IRF6 variants reported in the literature. Numbers above the exons indicate mutations found in patients with PPS, number underneath the exons indicate mutations found in patient with VWS. The grey boxes represent non-coding parts of IRF6. Exons 3 and 4 (dotted) encode the DNA-binding domain whereas exons 7 and 8 (striped) encode the protein-binding domain. The two arrows indicate the new mutations found in the current cohort
Table 2.
Clinical data of our patients according to IRF6 rs2235371 genotype
| IRF6 rs2235371 | ||||
|---|---|---|---|---|
| G/G | G/A | Total | p | |
| Sex | ||||
| F/M | 24/52 | 1/4 | 81 | 0.63 |
| Height at start GH | ||||
| Mean ± SD | −3.2 ± 0.8 | −2.9 ± 0.7 | 81 | 0.45 |
| Range | (−5.8 to −0.7) | (−3.7 to −2.1) | ||
| Arginin test peak GH | ||||
| Mean ± SD | 9.5 ± 6.3 | 11.5 ± 2.7 | 65 | 0.53 |
| Range | (0–24) | (8.8–15) | ||
| IGF-I SDS | ||||
| Mean ± SD | −3.2 ± 2.4 | −2.6 ± 1.5 | 73 | 0.55 |
| Range | (−9.0 to 0.9) | (−5.0 to −1.1) | ||
| Birth weight (kg) | ||||
| Mean ± SD | 3.1 ± 0.6 | 2.8 ± 0.5 | 78 | 0.25 |
| Range | (1.2–4.3) | (2.1–3.2) | ||
| Birth length (cm) | ||||
| Mean ± SD | 48 ± 3.5 | 45.5 ± 4.2 | 60 | 0.18 |
| Range | (35–52) | (41–50) | ||
| Gestational age at birth (w) | ||||
| Mean ± SD | 38.9 ± 2.8 | 38.9 ± 1.3 | 76 | 0.94 |
| Range | (32–43) | (37–40) | ||
| Pituitary anomalies on MRI | ||||
| No | 24 (31%) | 4 (80%) | 81 | 0.017 |
| Yes | 39 (51%) | 0 (0%) | ||
| Unknown | 13 (17%) | 1 (20%) | ||
| Midfacial hypoplasiaa | ||||
| Yes | 9 (12%) | 0 (0%) | 81 | 0.414 |
| No | 67 (88%) | 5 (100%) | ||
| Midline defectb | ||||
| Yes | 2 (3%) | 0 (0%) | 81 | 0.713 |
| No | 74 (97%) | 5 (100%) | ||
GHD growth hormone deficiency
Significant differences (between people with and without the protective SNP) are shown in bold
aMidfacial hypoplasia reported by treating physician, before start of GH treatment
bOne female patient had a cleft lip and palate, one male patient had a cleft lip and palate and a bifid tongue
Table 3.
Pituitary morphology in our patients, according to IRF6 SNP rs2235371 genotype
| rs2235371 genotype | ||
|---|---|---|
| G/G | G/A | |
| Normal pituitary | 24 (31%)a | 4 (100%)a |
| MRI | 22 | 4 |
| CTb | 2 | 0 |
| Abnormal pituitary (MRI) | 39 (51%)a | 0 (0%)a |
| Classic triad | 2 | 0 |
| Other pituitary anomalies on MRIc | 2 | 0 |
| Aplastic AP | 1 | 0 |
| (Partial) empty sella | 2 | 0 |
| EPP, invisible AP | 1 | 0 |
| EPP, AP not described | 1 | 0 |
| EPP, normal AP | 8 | 0 |
| EPP, small AP | 8 | 0 |
| AP small, PP small | 6 | 0 |
| AP small, PP invisible | 3 | 0 |
| AP small, PP and stalk normal | 5 | 0 |
| No pituitary imaging available | 13 (17%) | 1 (20%) |
| Total | 76 | 5 |
EPP ectopic posterior pituitary, AP anterior pituitary
aG/G versus G/A: p = 0.017
bMRI not available
cOne patient had an ‘abnormal form’ of the pituitary (not otherwise specified), the other had a hypodense area within the pituitary
Craniofacial measurements were not related to rs2235371 genotype.
As expected, there were slight differences between males and female craniofacial features. Craniofacial measurements according to sex are shown in Table 4.
Table 4.
Facial measurements of our patients according to sex #=mean of left and right lateral photographs
| N | Mean | SD | Minimum | Maximum | p | |
|---|---|---|---|---|---|---|
| Canthal index (enen/exex) | ||||||
| Male | 36 | 0.38 | 0.04 | .31 | .55 | 0.051 |
| Female | 15 | 0.36 | 0.02 | .32 | .39 | |
| Total | 51 | 0.37 | 0.03 | .31 | .55 | |
| Eye width (exen/exex #) | ||||||
| Male | 36 | 0.31 | 0.02 | .28 | .37 | 0.047 |
| Female | 15 | 0.32 | 0.01 | .30 | .33 | |
| Total | 51 | 0.31 | 0.02 | .28 | .37 | |
| Nose width (alal/exex) | ||||||
| Male | 36 | 0.42 | 0.04 | .36 | .55 | 0.493 |
| Female | 15 | 0.42 | 0.03 | .35 | .46 | |
| Total | 51 | 0.42 | 0.04 | .35 | .55 | |
| Mouth width (chch/exex) | ||||||
| Male | 34 | 0.56 | 0.07 | .39 | .73 | 0.240 |
| Female | 12 | 0.59 | 0.05 | .48 | .66 | |
| Total | 46 | 0.57 | 0.07 | .39 | .73 | |
| Forehead height (trn/trls #) | ||||||
| Male | 23 | 0.51 | 0.05 | .41 | .59 | 0.054 |
| Female | 7 | 0.55 | 0.03 | .50 | .60 | |
| Total | 30 | 0.52 | 0.05 | .41 | .60 | |
| Nose length (nsn/trls #) | ||||||
| Male | 23 | 0.37 | 0.05 | .31 | .48 | 0.225 |
| Female | 7 | 0.35 | 0.03 | .28 | .38 | |
| Total | 30 | 0.37 | 0.04 | .28 | .48 | |
| Philtrum length (snls/trls #) | ||||||
| Male | 23 | 0.11 | 0.02 | .09 | .15 | 0.074 |
| Female | 7 | 0.10 | 0.02 | .08 | .12 | |
| Total | 30 | 0.11 | 0.02 | .08 | .15 | |
| Ear length (sasba/trls #) | ||||||
| Male | 21 | 0.48 | 0.04 | .42 | .56 | 0.120 |
| Female | 6 | 0.51 | 0.02 | .48 | .53 | |
| Total | 27 | 0.48 | 0.04 | .42 | .56 | |
Discussion
We studied IRF6 as a new candidate gene for Idiopathic Growth Hormone Deficiency, based on literature data and on Exome Sequencing results of one family of our IGHD cohort. In 81 IGHD patients, we found two new mutations, of which one has a possible deleterious effect.
IRF6 is a transcriptional activator, which belongs to a family of nine transcription factors that share a highly conserved DNA binding domain and a less conserved protein-binding domain [29]. Defects in IRF6 can prevent the expression of a large number of genes directly controlled by IRF6, listed by Bottia et al. [30].
The new mutation we found in the index family, p.Arg233Cys, is located in the protein-binding domain of IRF6. The Arginine residue at position is conserved in many different species (overview of species available upon request), suggesting functional importance. The introduction of a cysteine residue in the protein could cause formation of a disulfide bond, which is likely to affect protein stability. This supports the prediction of Ensembl’s Variant Effect Predictor that the new mutation p.Arg233Cys is deleterious. Apart from the index case, his mother also carried the p.Arg233Cys variant. Interestingly, the mother had a normal phenotype. However, the mother also carried the protective allele of rs2235371 (V274I), whereas the index case did not. Rs2235371 (V274I) affects a highly conserved Valine in the protein-binding domain of IRF6, replacing it with Isoleucine [31]. Although some authors conclude otherwise [32], the vast majority of studies conclude that rs2235371 has an independent protective effect on orofacial cleft formation [33–37]. The only person in this family with GH deficiency was the boy with the new mutation p.Arg233Cys without the protective SNP rs2235371. Our hypothesis is that rs2235371 might also have a protective effect on the IGHD phenotype in this family. Although the functional impact of p.Arg233Cys remains to be confirmed, its effect might be counteracted by the protective rs2235371 allele. Further research should include functional analyses to characterize the effect of the new mutation, also in combination with the protective SNP.
Defects in the craniofacial midline are associated with deficiencies of pituitary hormones, like GH [11–17]. GH deficiency was found in 37% of patients with cleft lip and palate [17]. Although one might expect to find combined pituitary hormone deficiencies in patients with midline defects, Akin et al. [18] found isolated pituitary hormone deficiency in 13 of 33 patients with median orofacial clefts. GH deficiency was detected in four of the patients (12%). The mechanism underlying the association between craniofacial midline defects and GH deficiency has not yet been elucidated.
In our study, we found associations which, prudently, might be interpreted as a protective effect of the rs2235371 minor allele on IGHD phenotype. This is in line with literature findings showing an independent protective effect of rs2235371 on orofacial cleft formation. In our cohort, patients carrying the minor allele for rs2235371 had a slightly less severe phenotype. This was consistent for all clinical parameters measured: they had a larger height SDS at start of GH treatment, higher GH peak during arginine testing, higher IGF-1 SDS at start of GH treatment, no pituitary anomalies on MRI and less mid-facial hypoplasia (a facial characteristic often seen in patients with severe GHD). However, since patient numbers are small, p values were not below 0.05 and therefore we cannot draw any firm conclusions from this.
Miller et al. [38] reported that IRF6 SNP rs2235371 was related to craniofacial measurements. However, we did not find any association between rs2235371 genotypes and craniofacial measurements.
To our knowledge, this is the first study investigating IRF6, a gene involved in craniofacial midline formation, in relation to facial and pituitary morphology of patients with idiopathic growth hormone deficiency. Although the mutation p.Arg233Cys might have functional impact and thus be a rare cause of IGHD, the majority of patients in our cohort did not have any defects in IRF6. We therefore conclude that IRF6 defects are rare in IGHD patients and further research should focus on other genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully acknowledge Pfizer for their financial support to this research project. We like to acknowledge Professor Annelies de Klein and research technician Hannie Douben of the clinical genetics laboratory for their help with the MLPA analysis.
Abbreviations
- AP
Anterior pituitary
- CLP
Cleft lip and palate
- CP
Cleft palate alone
- EPP
Ectopic posterior pituitary
- GH
Growth hormone
- GHD
Growth hormone deficiency
- HAP
Hypoplastic anterior pituitary
- HSDS
Height SDS
- IGF-I
Insulin-like growth factor-I
- IGHD
Idiopathic GHD
- IRF6
Interferon regulatory factor 6
- MLPA
Multiplex ligation-dependent probe amplification
- NSCL/P
Non-syndromic cleft lip and/or palate
- PP
Posterior pituitary
- PPS
Popliteal pterygium syndrome
- SDS
Standard deviation score
- SNP
Single nucleotide polymorphism
- VWS
Van der Woude syndrome
Compliance with ethical standards
Conflict of interest
ES, AHK, TV, JB, RP and LdG declare that they have no conflict of interest.
Ethical approval
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. We obtained approval from the medical ethics committees of all participating hospitals.
Funding
For this project, we received financial support from Pfizer.
Informed consent
Informed consent was obtained from all individuals participating in this study and their parents if they were less than 18 years old.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s11102-017-0808-8) contains supplementary material, which is available to authorized users.
References
- 1.Lacey KA, Parkin JM. Causes of short stature: a community study of children in Newcastle upon Tyne. Lancet. 1974;1(7846):42–45. doi: 10.1016/S0140-6736(74)93041-4. [DOI] [PubMed] [Google Scholar]
- 2.Lindsay R, Feldkamp M, Harris D, Robertson J, Rallison M. Utah growth study: growth standards and the prevalence of growth hormone deficiency. J Pediatr. 1994;125(1):29–35. doi: 10.1016/S0022-3476(94)70117-2. [DOI] [PubMed] [Google Scholar]
- 3.Thomas M, Massa G, de Craen M, Bourguignon JP, et al. Prevalence and demographic features of childhood growth hormone deficiency in Belgium during the period 1986–2001. Eur J Endocrinol. 2004;151(1):67–72. doi: 10.1530/eje.0.1510067. [DOI] [PubMed] [Google Scholar]
- 4.Vimpani GV, Vimpani AF, Lidgard GP, Cameron EH, Farquhar JW. Prevalence of severe growth hormone deficiency. Br Med J. 1977;2(6084):427–430. doi: 10.1136/bmj.2.6084.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez Jurado LA, Argente J. Molecular basis of familial growth hormone deficiency. Horm Res. 1994;42(4–5):189–197. doi: 10.1159/000184192. [DOI] [PubMed] [Google Scholar]
- 6.Pfaeffle RW, Blum WF. Understanding the genetics of growth hormone deficiency: a reference guide. Oxfordshire: TMG Healthcare Communications; 2000. [Google Scholar]
- 7.Alatzoglou KS, Webb EA, Le Tissier P, Dattani MT. Isolated growth hormone deficiency (GHD) in childhood and adolescence: recent advances. Endocr Rev. 2014;5(3):376–432. doi: 10.1210/er.2013-1067. [DOI] [PubMed] [Google Scholar]
- 8.Corazzini V, Salvatori R. Molecular and clinical aspects of GHRH receptor mutations. Endocr Dev. 2013;24:106–117. doi: 10.1159/000342575. [DOI] [PubMed] [Google Scholar]
- 9.Alatzoglou KS, Dattani MT. Phenotype-genotype correlations in congenital isolated growth hormone deficiency (IGHD) Indian J Pediatr. 2012;79(1):99–106. doi: 10.1007/s12098-011-0614-7. [DOI] [PubMed] [Google Scholar]
- 10.Pfäffle R, Kies W, Klammt J. From GHRH to IGF-1 and downstream: clinical phenotypes and biological mechanisms. Pediatr Endocrinol Rev. 2011;9(Suppl 1):529–534. [PubMed] [Google Scholar]
- 11.Roitman A, Laron Z. Hypothalamo-pituitary hormone insufficiency associated with cleft lip and palate. Arch Dis Child. 1978;53:952–955. doi: 10.1136/adc.53.12.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luiz MC, Barbieri MA, Bettiol H, et al. Growth impairment of children with different types of lip and palate clefts in the first 2years of life: a cross-sectional study. J Pediatr. 2005;81(6):461–465. doi: 10.2223/JPED.1420. [DOI] [PubMed] [Google Scholar]
- 13.Bowers EJ, Mayro RF, Whitaker LA, et al. General body growth in children with cleft palate and related disorders: age differences. Am J Phys Anthropol. 1988;75(4):503–515. doi: 10.1002/ajpa.1330750408. [DOI] [PubMed] [Google Scholar]
- 14.Rudman D, Davis T, Priest JH, et al. Prevalence of growth hormone deficiency in children with cleft lip or palate. J Pediatr. 1978;93(3):378–382. doi: 10.1016/S0022-3476(78)81141-X. [DOI] [PubMed] [Google Scholar]
- 15.Zuppinger KA, Sutter M, Zurbrügg RP. Cleft lip and chorioideal coloboma associated with multiple hypothalamo-pituitary dysfunctions. J Clin Endocrinol Metab. 1971;33(6):934–939. doi: 10.1210/jcem-33-6-934. [DOI] [PubMed] [Google Scholar]
- 16.Tweedie AR, Keith A. Ectopia of the pituitary, with other congenital anomalies of the nose, palate, and upper lip. Proc R Soc Med. 1911;4:47–54. [PMC free article] [PubMed] [Google Scholar]
- 17.Traggiai C, Stanhope R. Endocrinopathies associated with midline cerebral and cranial malformations. J Pediatr. 2002;140(2):252–255. doi: 10.1067/mpd.2002.121822. [DOI] [PubMed] [Google Scholar]
- 18.Akin MA, Kurtoğlu S, Sarici D, et al. Endocrine abnormalities of patients with cleft lip and/or cleft palate during the neonatal period. Turk J Med Sci. 2014;44(4):696–702. doi: 10.3906/sag-1303-89. [DOI] [PubMed] [Google Scholar]
- 19.Slavotinek A, Parisi M, Heike C, et al. Craniofacial defects of blastogenesis: duplication of pituitary with cleft palate and orophgaryngeal tumors. Am J Med Genet A. 2005;135(1):13–20. doi: 10.1002/ajmg.a.30694. [DOI] [PubMed] [Google Scholar]
- 20.Zoric L. Septo-optic dysplasia plus: a case report. BMC Res Notes. 2014;7(1):191. doi: 10.1186/1756-0500-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Senawi R, et al. Septo-optic dysplasia complex: clinical and radiological manifestations in Omani children. Oman J Ophthalmol. 2013;6(3):193. doi: 10.4103/0974-620X.122277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yassin OM, El-Tal YM. Solitary maxillary central incisor in the midline associated with systemic disorders. Oral Surg Oral Med Oral Pathol Oral Radiol Endodontology. 1998;85(5):548–551. doi: 10.1016/S1079-2104(98)90289-X. [DOI] [PubMed] [Google Scholar]
- 23.Kjær I. Brain malformation in single median maxillary central incisor. Neuropediatrics. 2009;40(6):280–283. doi: 10.1055/s-0030-1248245. [DOI] [PubMed] [Google Scholar]
- 24.de Graaff LC, Baan J, Govaerts LC, et al. Facial and pituitary morphology are related in Dutch patients with GH deficiency. Clin Endocrinol. 2008;69(1):112–116. doi: 10.1111/j.1365-2265.2007.03167.x. [DOI] [PubMed] [Google Scholar]
- 25.Kwa MQ, et al. Interferon Regulatory Factor 6 differentially regulates toll-like receptor 2-dependent chemokine gene expression in epithelial cells. J Biol Chem. 2014;289(28):19758–19768. doi: 10.1074/jbc.M114.584540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuppia L, Capogreco M, Marzo G, et al. Genetics of syndromic and nonsyndromic cleft lip and palate. J Craniofac Surg. 2011;22(5):1722–1726. doi: 10.1097/SCS.0b013e31822e5e4d. [DOI] [PubMed] [Google Scholar]
- 27.Bishara SE, Jorgensen GJ, Jakobsen JR. Changes in facial dimensions assessed from lateral and frontal photographs: part I–methodology. Am J Orthod Dentofac Orthop. 1995;108:389–393. doi: 10.1016/S0889-5406(95)70036-6. [DOI] [PubMed] [Google Scholar]
- 28.Farkas LG. Anthropometry of the head and face in medicine. New York: Elsevier; 1981. [Google Scholar]
- 29.Little HJ, et al. Missense mutations that cause Van der Woude syndrome and popliteal pterygium syndrome affect the DNA-binding and transcriptional activation functions of IRF6. Hum Mol Genet. 2009;18(3):535–545. doi: 10.1093/hmg/ddn381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bottia E, Spallonea G, Morettia F, et al. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. PNAS. 2011;108(33):13710–13715. doi: 10.1073/pnas.1110931108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zucchero TM, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351(8):769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 32.Rahimov F, Marazita ML, Visel A, et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40(11):1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JW, McIntosh I, Hetmanski JB, et al. Association between IRF6 and nonsyndromic cleft lip with or without cleft palate in four populations. Genet Med. 2007;9:219–227. doi: 10.1097/GIM.0b013e3180423cca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jugessur A, Rahimov F, Lie RT, et al. Genetic variants in IRF6 and the risk of facial clefts: single-marker and haplotype- based analyses in a population-based case-control study of facial clefts in Norway. Genet Epidemiol. 2008;32:413–424. doi: 10.1002/gepi.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birnbaum S, Ludwig KU, Reutter H, et al. IRF6 gene variants in Central European patients with non-syndromic cleft lip with or without cleft palate. Eur J Oral Sci. 2009;117(6):766–769. doi: 10.1111/j.1600-0722.2009.00680.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Pan Y, Zhang Z, et al. Three polymorphisms in IRF6 and 8q24 are associated with nonsyndromic cleft lip with or without cleft palate: evidence from 20 studies. Am J Med Genet Part A. 2012;158A:3080–3086. doi: 10.1002/ajmg.a.35634. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Liu Q, Xu W, Li Z, Jiang M, et al. TGFA and IRF6 contribute to the risk of nonsyndromic cleft lip with or without cleft palate in Northeast China. PLoS ONE. 2013;8(8):e70754. doi: 10.1371/journal.pone.0070754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller SF, Weinberg S, Nidey N, et al. Exploratory genotype–phenotype correlations of facial form and asymmetry in unaffected relatives of children with non-syndromic cleft lip and/or palate. J Anat. 2014;224:688–709. doi: 10.1111/joa.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.