Abstract
The insertion of newly synthesized membrane proteins is a well-regulated and fascinating process occurring in every living cell. Several translocases and insertases have been found in prokaryotic and eukaryotic cells, the Sec61 complex and the Get complex in the endoplasmic reticulum and the SecYEG complex and YidC in bacteria and archaea. In mitochondria, TOM and TIM complexes transport nuclear-encoded proteins, whereas the Oxa1 is required for the insertion of mitochondria-encoded membrane proteins. Related to the bacterial YidC and the mitochondrial Oxa1 are the Alb3 and Alb4 proteins in chloroplasts. These membrane insertases are comparably simple and can be studied in vitro, after their biochemical purification and reconstitution in artificial lipid bilayers such as liposomes or nanodiscs. Here, we describe the recent progress to study the molecular mechanism of YidC-dependent and unassisted membrane insertion at the single molecule level.
Main Text
Integral membrane proteins display single or multiple α-helical transmembrane segments within the lipid bilayer, while multiple β-sheets form a cylindrical pore structure in the case of β-barrel proteins present in the outer membrane of Gram-negative bacteria and mitochondria. The multispanning α-helical membrane proteins fold into compact structures in the bilayer that generally exclude lipid molecules. Therefore, the globular structure of proteins in the membrane can be compared to the basic structure of soluble globular proteins where water is excluded.
Because membrane proteins are located in a hydrophobic lipid environment, the amino acids (aa) of their transmembrane helices harbor mainly hydrophobic side chains. The outward-facing hydrophobic residues allow lipid-protein interactions while the internal amino acid side chains mediate the protein-protein interaction between the individual helices with the exception of membrane proteins forming hydrophilic pores. This implies that most membrane-spanning 15–20 aa residues are purely hydrophobic, which poses a problem for the biosynthesis of membrane proteins. When the nascent protein chain leaves the ribosome, it releases the hydrophobic segments into the cytosol that would quickly form aggregates if left unattended in the polar solvent. To address this problem, the cellular machinery employs proteins that transiently bind to the hydrophobic segments, as in the case of the bacterial signal recognition particle that binds to hydrophobic segments and directs the ribosome nascent chain complex to the membrane surface during its synthesis (1), or to chaperones such as DnaK or SecB that bind the translated polypeptide chain in a mostly unfolded state (2).
In prokaryotic and eukaryotic cells, proteins are inserted by proteinaceous molecular devices, termed “insertases” and “translocases”. The Sec translocases are present in the endoplasmic reticulum as a heterotrimeric Sec61 complex (3, 4) and in the bacterial inner membrane as multimeric SecYEG-YidC complex (5). In the mitochondrial inner membrane, an insertase, Oxa1, is found that shows homology to the bacterial YidC protein (6, 7). In Escherichia coli, YidC has six transmembrane helices and a large periplasmic loop called “P1” that is not essential for the function of the protein (8). In contrast to the Sec translocon that contains a central pore, YidC does not exhibit a polar channel but rather a hydrophilic groove. This groove is positioned in the inner leaflet of the membrane facing the cytoplasm, facilitating substrate translocation due to decreasing the thermodynamic barrier for this process. The limited size of this groove also explains why the insertase is able to only allow the translocation of protein segments that are short and hydrophilic. The purified YidC protein can be reconstituted into liposomes and is able to efficiently insert YidC-dependent membrane proteins, added to the outside medium (9).
To study the insertion of proteins into the membrane in a reconstituted system, it is possible to couple the insertion with a purified protein translation system (10). This can be achieved by using radiolabeled amino acids and by following the membrane insertion of the newly translated labeled protein by protease protection (11). However, these bulk systems have the limitation that membrane insertion shows efficiencies below 10% (12). Another attractive method is to use a noncoupled membrane insertion system. Here, the membrane protein is purified and kept at high concentrations in a loosely folded state, either with chaotropic agents or in organic solvents. Then, the protein is added in a small volume to the liposomes or proteoliposomes and membrane insertion can be followed at a milli- to nanosecond timescale using high resolution spectroscopy (13). The organic solvents or the chaotropic agents are diluted at least a 100-fold to not interfere with the integrity of the lipid bilayer. This creates a time window for membrane proteins to bind to the liposome (or the insertase/translocase) in an insertion/translocation competent state. Aggregation and misfolding are competing events that can occur in addition to insertion and in-membrane folding. Nevertheless, efficiencies up to 95% were observed using this experimental approach (10, 11). The reconstituted system also allows the analysis of membrane insertion at a single molecule level (see below).
Quality Control of Insertion-competent Liposomes and Proteoliposomes
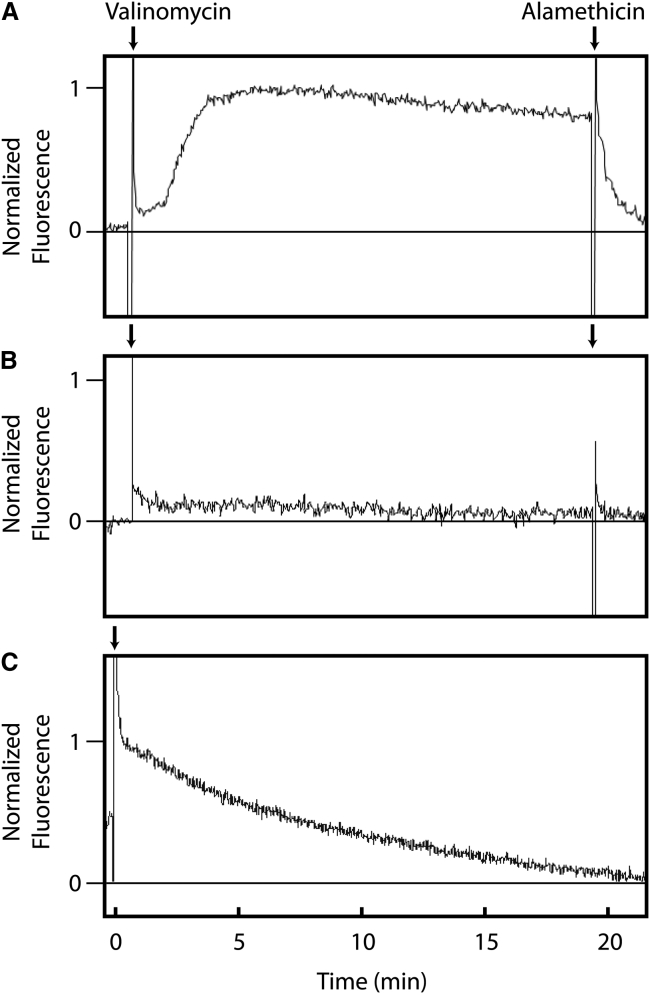
To generate liposomes from a solvent-free lipid film, the use of an extruder is a recommended method producing unilamellar liposomes of a defined size (14). Critical for the quality of the bilayer membranes is the lipid source. Pure lipids are crucial for single-molecule experiments, as contaminants contribute to the noise in a measurement system, making analysis difficult. In addition, contaminations in the lipid preparation might result in unsealed membranes and disordered bilayers. Therefore, the testing of the liposome membranes addressing their ability to maintain an electrochemical ion potential is recommended (14). The membrane potential can be measured by using an asymmetric K+/Na+ buffer system, e.g., by generating liposomes in a buffer containing 200 mM Na+, which are then washed and resuspended in a buffer containing 200 mM K+ (Fig. 1). Then oxonol VI, a voltage-sensitive dye, is added (15). To generate the potential, the peptide valinomycin is added, which makes the membrane permeant to potassium ions. When following the fluorescence signal of oxonol, a sealed membrane shows a constant potential for at least 20 min (Fig. 1 A). A defective membrane shows a rapid loss of the potential (Fig. 1 C).
Figure 1.
Membrane potential across liposomes. An electrochemical potential was generated by adding 0.25 μM valinomycin (left arrow) to liposomes with 200 mM Na+ inside and 200 mM K+ outside measured by oxonol VI fluorescence (A). Alamethicin was added as a control, which forms a nonselective cation peptide pore and therefore leads to the collapse of the membrane potential (right arrow). No potential was measured when the liposomes had inside and outside 200 mM Na+ (B). When the liposomes were generated from insufficiently purified lipids and treated as in (A), the initial potential steadily decreased (C), indicating leaky bilayers (9, 29).
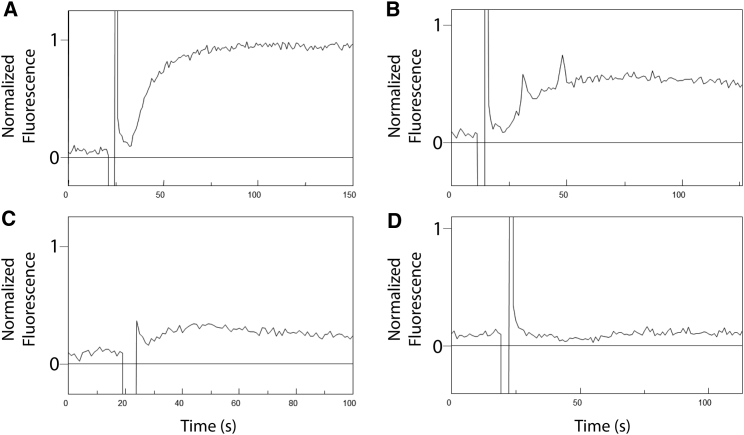
In this system, the membrane potential across the liposomal membrane is determined by the concentration gradient between the liposomal lumen and the outside medium. As visualized by the oxonol VI fluorescence, defined concentration gradients of potassium sulfate between 1:100 and 1:104 lead to potentials of −10 to −210 mV, respectively (Fig. 2). In this concentration range the fluorescence showed a linear relation to the calculated Nernst potential (15). The data show that the transmembrane potential can be easily adjusted by the applied ion gradients.
Figure 2.
Dosing the electrochemical membrane potential. Inside/outside K+ concentrations were set to 20 μM/200 mM K+ (A), 200 μM/200 mM K+ (B), 2 mM/200 mM K+μM (C), and 40 mM/200 mM K+ (D). From the apparent oxonol VI fluorescence and the calculated Nernst potential, Δψ was determined as −240 mV (A), −177 mV (B), and 74−50 mV (C).
Reconstitution of YidC into Proteoliposomes
To generate proteoliposomes for the subsequent insertion experiments, detergent-purified YidC protein was mixed with a lipid/protein ratio of at least 5000:1 (mol/mol). Several cycles with the extruder through a 0.4 μm pore size were required to generate the proteoliposomes with a diameter of ∼200 nm. Proteoliposomes can be produced from a variety of lipids and mixtures thereof as the insertion and folding can be lipid dependent. Here, we used 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), a common bilayer lipid, absent in bacteria. Previously, we also used a lipid mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) with a molar ratio of 3:1, mimicking the membrane composition of bacteria. Both the PC and the PE/PG YidC proteoliposomes showed equivalent properties regarding reconstitution and membrane insertion of YidC (13). DOPC can be useful to investigate the influence on the membrane charge toward the insertion and folding of membrane proteins, as this lipid exhibits a net-neutral surface charge in contrast to the cationic composition of POPE/POPG liposomes.
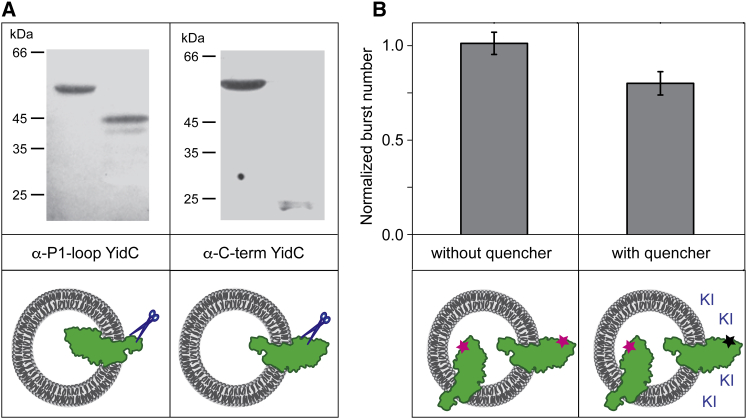
The topology of the reconstituted YidC protein can be determined by protease accessibility (9). Externally added trypsin digests the YidC protein and generates protease-protected fragments depending on its orientation. If the large P1 domain is in the lumen of the proteoliposome, a 42-kDa fragment is found, whereas in the inverted orientation a 20-kDa fragment is generated. Depending on the intensity of either band, the topology distribution can be calculated (Fig. 3 A). This approach can be used for many membrane proteins; if the protein has both a cytoplasmic and a periplasmic domain, a domain-specific antibody is ideally suited to distinguish the protected from the exposed domain.
Figure 3.
Topology analysis of YidC in the reconstituted proteoliposomes. Purified YidC protein was mixed with DOPC lipids and unilamellar proteoliposomes were generated with an extruder. The topology of YidC can be analyzed by protease accessibility by adding trypsin (A) (9). The proteolytic fragments were detected using immunoblotting by either an antiserum directed to the large periplasmic domain (left panel) or to the cytoplasmic tail (right panel). The topology of YidC modified with a fluorophore can be determined by the relative amount that is protected from the quencher added to the outside of the proteoliposome (B). The periplasmic domain of YidC was labeled with Atto520. For both cases (A and B), >80% of the YidC periplasmic domain was found in the liposomal lumen in the case of DOPC-reconstituted YidC.
Alternatively, fluorescently labeled YidC protein can be used to determine its membrane topology. Depending where the fluorophore is located on the protein, accessibility to a fluorescence quencher, e.g., KI, can be determined. This approach allows us to determine the orientation of the protein within the liposome. In case of a protein with a fluorescent label in its periplasmic part, YidC insertion should lead to the protection of the fluorophore from externally added quenchers, and is therefore a measure of its topology. Interestingly, in case of YidC we observed directional insertion during reconstitution, favoring the periplasmic domain to be inside the lumen of the liposome (Fig. 3 B).
Model Proteins for Fluorescence-based Insertion Studies
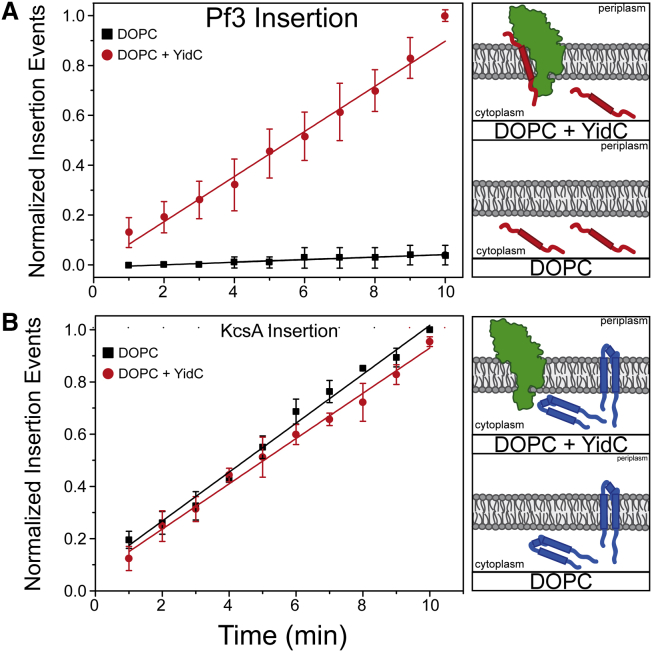
The single-spanning Pf3 coat protein is the best-studied YidC substrate protein. The protein is inserted into the inner membrane of Pseudomonas aeruginosa before it is assembled into the coat of the extruding filamentous phage by a membrane-embedded molecular motor (16, 30). Also when expressed from a plasmid in E. coli, the protein is inserted into the bacterial membrane with an C-in and N-out topology, having no requirement for SecYEG (17) but being strictly dependent on YidC (9), which was shown with the respective E. coli depletion strains. For fluorescence labeling, a fluorophore can be introduced into the protein by mutating a functionally unimportant residue to a cysteine at the desired position using site-directed mutagenesis (18). Then, the protein was purified and modified by maleimide-sulfhydryl chemistry. The labeled protein was then separated from the unbound dye by size-exclusion chromatography. When the Pf3 coat protein was modified close to the N-terminus (residue 3) and added to YidC proteoliposomes, it was inserted with the fluorophore protected from the quencher KI, demonstrating the translocation of the N-terminus into the lumen of the liposome (Fig. 4 A). When the protein was added to liposomes that did not contain the insertase YidC, no protection of the fluorophore was observed, which shows the absolute requirement of the insertase for the membrane insertion of Pf3. In the presence of YidC, the Pf3 protein was found in the orientation with the fluorophore inside the liposome, which is identical to the native conformation of Pf3 in the inner membrane, as the liposome lumen corresponds to the periplasm of a bacterial cell. When the fluorophore was positioned at the C terminus, it was quenched by KI regardless of whether YidC was present. We concluded that the protein is inserted with the C-terminus positioned outside of the liposome in the in vitro approach, corresponding to the cytoplasm of the cell (C-inside topology) (18).
Figure 4.
Insertion of Pf3 (A) or KcsA (B) in DOPC liposomes in the absence or presence of YidC, measured by FCS. The proteins were purified and labeled with Atto520, then added to DOPC liposomes (black) or to YidC proteoliposomes (red). The fluorescence was measured for 10 min in the presence of 100 mM KI. Whereas the insertion of KcsA was independent of the YidC insertase, the Pf3 coat protein strictly requires YidC.
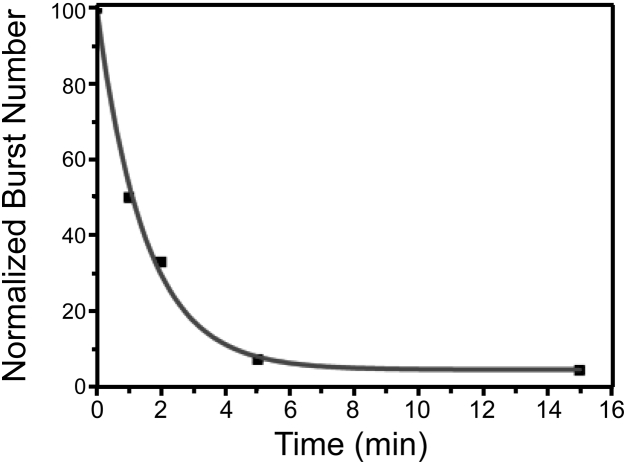
Proteins rapidly lose their ability for YidC-mediated insertion, as can be shown by the following experiment: The Pf3 coat protein was added to an aqueous solution and incubated for defined periods of time before YidC-proteoliposomes were added. The rate of insertion decreased dramatically with only about half of the YidC substrate Pf3 coat being able to insert after incubation without proteoliposomes for 60 s (Fig. 5). This observation indicated that in addition to insertion events, side reactions occur that render the protein unable to insert, most likely due to misfolding and/or aggregation.
Figure 5.
Loss of insertion competence of Pf3 coat protein after incubation in aqueous media in the absence of YidC-proteoliposomes. Low nanomolar concentrations of Pf3 were added to the buffer, followed by the addition of proteoliposomes after defined time points (1, 2, 5, or 15 min) or simultaneously (0 min), displayed on the x axis. Immediately after the proteoliposomes have been added, relative rates of insertion were tested by determining the number of bursts, according to Fig. 4, during 6.5 min measurements (y axis).
KcsA is a tetrameric potassium channel protein from the soil bacterium Streptomyces lividans. Recently, we studied the mechanism of the unassisted and directional insertion of this two-transmembrane protein (19). We analyzed individual steps of protein insertion on a single molecule level. Interestingly, no proteinaceous factors such as the translocase SecYEG or the insertase YidC are required for the directional insertion of KcsA. This was investigated in E. coli cells where YidC or SecE were depleted (19, 20). Intriguingly, the charged amino acid residues in the N-terminal region of KcsA are essential for the insertion of the unfolded protein into the phospholipid bilayer. We could demonstrate that electrostatic forces mediate the first step toward membrane insertion.
KcsA is an ideal model protein to investigate spontaneous insertion and the subsequent processes that occur before the formation of a functional homotetrameric channel: folding and assembly. In all processes, the lipid composition seems to play a major role. It has been reported that the lipid composition drastically affects the function of KcsA (21), and we could show that the structure and thermal stability of KcsA is strongly dependent on the chemistry of the lipid bilayer. The insertion also depends on the composition of the membrane. Because the membrane insertion process cannot be synchronized, we employed single molecule methods. For the insertion into DOPC, a suitable bilayer lipid, we could demonstrate that the process is independent of YidC, in contrast to the Pf3 protein. Thus, in the presence of YidC, KcsA was not inserted with a higher efficiency or kinetics into the YidC-proteoliposomes compared to liposomes (Fig. 4 B). In a parallel experiment, we verified that YidC is absolutely required for the membrane insertion of the single-transmembrane protein Pf3 (Fig. 4 A).
Observing Substrate Interaction with YidC during the Insertion of Single Proteins
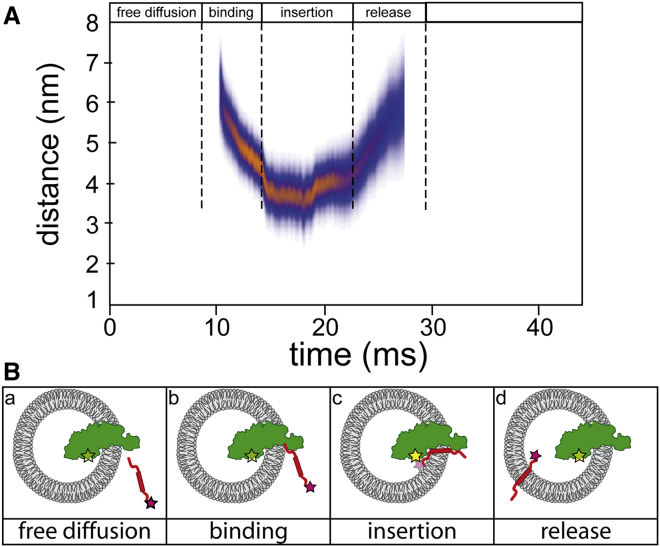
The membrane insertion of the fluorescently labeled substrate proteins can be followed by single-molecule techniques such as fluorescence correlation spectroscopy (FCS) in real time. To investigate protein interactions during translocation/insertion, single-molecule fluorescence resonance energy transfer (FRET) is a suitable method. To this end, the membrane insertase YidC is labeled with a fluorescent acceptor dye and used to generate YidC proteoliposomes (Fig. 6). The proximity between YidC and the substrate labeled with a donor dye is then measured in a confocal volume by FCS-based FRET. Here, the sample is prepared in such a way that only one YidC protein is present in each liposome, which is then diluted to a concentration that only one proteoliposome diffuses through the confocal volume at each time point. Single fluorescent bursts are analyzed for FRET efficiencies and the changes over time are recorded. Fig. 6 B illustrates how the substrate protein Pf3 slowly approaches YidC, stays close together with the insertase before it eventually leaves YidC. While the entire event takes ∼20 ms, the interaction with YidC lasts for ∼4 ms, from binding to release into the membrane (13).
Figure 6.
Insertion of single protein molecules into YidC proteoliposomes observed by FCS-based FRET. Atto520-labeled Pf3 protein was added to a solution containing Atto647N-labeled YidC, reconstituted into proteoliposomes. (A) The high temporal resolution of distance measurements between fluorophores, determined by FRET, allows resolving the individual events taking place on a molecular level, as illustrated in (B). The protein Pf3 (red) freely diffuses in solution (a), until it encounters a YidC-containing proteoliposome and binds to the insertase (green, b). The distance between the fluorophores decreases, which demonstrates a close interaction between YidC and Pf3 during the membrane insertion process (c). The increasing distance between donor and acceptor fluorophor and finally the loss of a FRET signal indicate the release of Pf3 from the insertase YidC (d, adapted from (13)). To see this figure in color, go online.
Using this approach, mutants of YidC can now be studied that show alterations in substrate binding, in translocation of the periplasmic domain, or in the release of the inserted protein from YidC. This last step can be inhibited, e.g., by introducing single cysteines into the transmembrane region of the Pf3 substrate and also into the TM3 of YidC. Coexpression of both proteins leads to disulfides and an accumulation of the cross-linked YidC-Pf3 complex in vivo (22). The detailed analysis of the process of membrane insertion will provide an understanding of the molecular mechanisms of each step taking place during this fundamental process.
A fascinating question is what determines a protein to be a YidC substrate. YidC-dependent proteins are usually small, single or double transmembrane proteins with short (2–30 aa long) periplasmic regions. The Pf3 coat protein can become a YidC-independent protein by extending the hydrophobic region with three leucines (23). Here, the efficiency of the unassisted membrane insertion directly correlates with the hydrophobicity of the membrane anchor sequence (18). Therefore, increasing the hydrophobicity of the membrane anchor region allows spontaneous membrane insertion of the Pf3 coat protein. On the other extreme, when the periplasmic domain is extended with polar residues or its charge is increased, a previously YidC-dependent protein might now require the Sec-system (24, 25, 26). A possible explanation is the hydrophilic groove found in YidC which can accommodate only short hydrophilic peptides (27, 28), and once this length is exceeded, alternative pathways for membrane insertion are required. In the case of the spontaneously inserting KcsA, two hydrophobic sequences flank the periplasmic region and probably contribute to its translocation. This process might be driven by the insertion of these two hydrophobic sequences into the membrane bilayer before the translocation of the periplasmic loop. The combined hydrophobicity of these sequences provides sufficient energy to allow membrane insertion to be independent of YidC.
Conclusions
The biogenesis of membrane proteins is a highly complex and fascinating process. An estimate of 20–30% of genes code for membrane proteins, and many more proteins have to cross the cell membrane or the membrane of an organelle at some point to reach their final destination. Insertases and translocases provide platforms to facilitate this process. Studying membrane protein insertion in vivo is very tedious and often depends on depletion strains that have to be constructed and are notoriously difficult to handle. Such strains can inhibit the synthesis of chaperones or translocation components that are often required for the growth of cells. Also, when using depletion strains, cells often have residual amounts of the translocation components and thus the experimental results often do not give clear answers. A different approach is to study membrane protein insertion with a reconstituted system in vitro. Here, the protein components of interests have to be purified and can then be studied even on the single-molecule level. Using these methods, a kinetic determination of the process is possible as well as the analysis of thermodynamic factors of proteins inserting, folding, or assembling in the membrane. Only by studying single molecules can individual processes be understood with high temporal resolution. Moreover, mechanistic steps can be determined, e.g., from membrane binding to insertion, and subsequent processes such as folding and assembly to functional protein complexes.
The data provided here clearly show that some proteins are able to insert into the membrane without the assistance of another protein. This sets this group apart from the majority of proteins that require insertases or translocases and have been well characterized.
Author Contributions
A.K. and S.L. conceived and designed the experiments. M.H. performed the experiments. M.H. and S.L. analyzed the data. A.K. and S.L. wrote the manuscript. All authors approved of the final manuscript.
Acknowledgments
This work was supported by the DFG (grant No. LE 3055/3-1).
Editor: Daniel Muller.
References
- 1.Saraogi J., Shan S.O. Co-translational protein targeting to the bacterial membrane. Biochim. Biophys. Acta. 2014;1843:1433–1441. doi: 10.1016/j.bbamcr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castanié-Cornet M.P., Bruel N., Genevaux P. Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim. Biophys. Acta. 2014;1843:1442–1456. doi: 10.1016/j.bbamcr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Dudek J., Pfeffer S., Zimmermann R. Protein transport into the human endoplasmic reticulum. J. Mol. Biol. 2015;427(6 pt. A):1159–1175. doi: 10.1016/j.jmb.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Park E., Rapoport T.A. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn A., Koch H.G., Dalbey R.E. Targeting and insertion of membrane proteins. EcoSal Plus. 2017;7:1–27. doi: 10.1128/ecosalplus.esp-0012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scotti P.A., Urbanus M.L., Luirink J. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnefoy N., Chalvet F., Dujardin G. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- 8.Jiang F., Chen M., Dalbey R.E. Defining the regions of Escherichia coli YidC that contribute to activity. J. Biol. Chem. 2003;278:48965–48972. doi: 10.1074/jbc.M307362200. [DOI] [PubMed] [Google Scholar]
- 9.Serek J., Bauer-Manz G., Kuhn A. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 2004;23:294–301. doi: 10.1038/sj.emboj.7600063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Laan M., Houben E.N., Driessen A.J. Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Rep. 2001;2:519–523. doi: 10.1093/embo-reports/kve106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Laan M., Nouwen N., Driessen A.J. SecYEG proteoliposomes catalyze the Δϕ-dependent membrane insertion of FtsQ. J. Biol. Chem. 2004;279:1659–1664. doi: 10.1074/jbc.M306527200. [DOI] [PubMed] [Google Scholar]
- 12.Schulze R.J., Komar J., Collinson I. Membrane protein insertion and proton-motive-force-dependent secretion through the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Proc. Natl. Acad. Sci. USA. 2014;111:4844–4849. doi: 10.1073/pnas.1315901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winterfeld S., Ernst S., Kuhn A. Real time observation of single membrane protein insertion events by the Escherichia coli insertase YidC. PLoS One. 2013;8:e59023. doi: 10.1371/journal.pone.0059023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn A., Stiegler N., Schubert A.K. Membrane insertion of small proteins. Methods Mol. Biol. 2010;619:39–62. doi: 10.1007/978-1-60327-412-8_3. [DOI] [PubMed] [Google Scholar]
- 15.Apell H.J., Bersch B. Oxonol VI as an optical indicator for membrane potentials in lipid vesicles. Biochim. Biophys. Acta. 1987;903:480–494. doi: 10.1016/0005-2736(87)90055-1. [DOI] [PubMed] [Google Scholar]
- 16.Luiten R.G., Schoenmakers J.G., Konings R.N. The major coat protein gene of the filamentous Pseudomonas aeruginosa phage Pf3: absence of an N-terminal leader signal sequence. Nucleic Acids Res. 1983;11:8073–8085. doi: 10.1093/nar/11.22.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohrer J., Kuhn A. The function of a leader peptide in translocating charged amino acyl residues across a membrane. Science. 1990;250:1418–1421. doi: 10.1126/science.2124001. [DOI] [PubMed] [Google Scholar]
- 18.Ernst S., Schönbauer A.K., Kuhn A. YidC-driven membrane insertion of single fluorescent Pf3 coat proteins. J. Mol. Biol. 2011;412:165–175. doi: 10.1016/j.jmb.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 19.Altrichter S., Haase M., Leptihn S. Mechanism of the spontaneous and directional membrane insertion of a 2-transmembrane ion channel. ACS Chem. Biol. 2017;12:380–388. doi: 10.1021/acschembio.6b01085. [DOI] [PubMed] [Google Scholar]
- 20.van Dalen A., van der Laan M., de Kruijff B. Components required for membrane assembly of newly synthesized K+ channel KcsA. FEBS Lett. 2002;511:51–58. doi: 10.1016/s0014-5793(01)03278-1. [DOI] [PubMed] [Google Scholar]
- 21.Iwamoto M., Oiki S. Amphipathic antenna of an inward rectifier K+ channel responds to changes in the inner membrane leaflet. Proc. Natl. Acad. Sci. USA. 2013;110:749–754. doi: 10.1073/pnas.1217323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klenner C., Kuhn A. Dynamic disulfide scanning of the membrane-inserting Pf3 coat protein reveals multiple YidC substrate contacts. J. Biol. Chem. 2012;287:3769–3776. doi: 10.1074/jbc.M111.307223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiefer D., Kuhn A. Hydrophobic forces drive spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J. 1999;18:6299–6306. doi: 10.1093/emboj/18.22.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pross E., Soussoula L., Kuhn A. Membrane targeting and insertion of the C-tail protein SciP. J. Mol. Biol. 2016;428:4218–4227. doi: 10.1016/j.jmb.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn A. Alterations in the extracellular domain of M13 procoat protein make its membrane insertion dependent on secA and secY. Eur. J. Biochem. 1988;177:267–271. doi: 10.1111/j.1432-1033.1988.tb14372.x. [DOI] [PubMed] [Google Scholar]
- 26.Soman R., Yuan J., Dalbey R.E. Polarity and charge of the periplasmic loop determine the YidC and sec translocase requirement for the M13 procoat lep protein. J. Biol. Chem. 2014;289:1023–1032. doi: 10.1074/jbc.M113.522250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalbey R.E., Kuhn A. How YidC inserts and folds proteins across a membrane. Nat. Struct. Mol. Biol. 2014;21:435–436. doi: 10.1038/nsmb.2823. [DOI] [PubMed] [Google Scholar]
- 28.Dalbey R.E., Kuhn A., Kiefer D. The membrane insertase YidC. Biochim. Biophys. Acta. 2014;1843:1489–1496. doi: 10.1016/j.bbamcr.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Stiegler N., Dalbey R.E., Kuhn A. M13 procoat protein insertion into YidC and SecYEG proteoliposomes and liposomes. J. Mol. Biol. 2011;406:362–370. doi: 10.1016/j.jmb.2010.12.036. [DOI] [PubMed] [Google Scholar]
- 30.Loh B., Haase M., Leptihn S. The transmembrane morphogenesis protein gp1 of filamentous phages contains walker A and walker B motifs essential for phage assembly. Viruses. 2017;9:73. doi: 10.3390/v9040073. [DOI] [PMC free article] [PubMed] [Google Scholar]