Abstract
Extracellular vesicles (EVs) are proposed to play important roles in intercellular communication. Two classes of EVs can be distinguished based on their intracellular origin. Exosomes are generated within endosomes and released when these fuse with the plasma membrane, whereas ectosomes bud directly from the plasma membrane. Studies of EV function have been hindered by limited understanding of their biogenesis. Components of the endosomal sorting complex required for transport (ESCRT) machinery play essential roles in topologically equivalent processes at both the endosome and the plasma membrane and are consistently recovered in EVs, but whether they are generally required to produce EVs is still debated. Here, we study the effects of inhibiting the ESCRT-associated AAA+ ATPase VPS4 on EV release from cultured cells using two methods for EV recovery, differential centrifugation and polyethylene glycol precipitation followed by lectin affinity chromatography. We find that inhibiting VPS4 in HEK293 cells decreases release of EV-associated proteins and miRNA as well as the overall number of EV particles. The tetraspanins CD63 and CD9 are among the most frequently monitored EV proteins, but they differ in their subcellular localization, with CD63 primarily in endosomes and CD9 on the plasma membrane. We find that CD63 and CD9 are enriched in separable populations of EVs that are both sensitive to VPS4 inhibition. Serum stimulation increases release of both types of EVs and is also reduced by inhibiting VPS4. Taken together, our data indicate that VPS4 activity is important for generating exosomes and ectosomes, thereby generally implicating the ESCRT machinery in EV biogenesis.
Introduction
Extracellular vesicles (EVs) are released by virtually all cells and are present at varying concentration in most or all body fluids (1, 2, 3). Early studies described roles for EVs in discarding membrane proteins during reticulocyte maturation (4, 5) and in eliciting immune responses (6, 7). Since then, EVs have emerged as candidates for broad roles in intercellular communication (3, 8). EVs can be classified based on their cell of origin, e.g., prostasomes derive from prostate cells, oncosomes from tumor cells, etc. Each cell can also generate different types of EVs. Exosomes are uniformly sized vesicles released when endosomal multivesicular bodies (MVBs) fuse with the plasma membrane. Ectosomes or shedding microvesicles (also referred to simply as microvesicles) are released directly from the plasma membrane and can be heterogeneous in size (9, 10). Typical EV preparations are likely to contain a mixture of vesicles (11, 12, 13), and an important question for the future is whether EVs that originate from different membranes have overlapping or unique functions.
EV content has been extensively characterized using contemporary -omics methodology, leading to a long list of EV-associated proteins, RNA, and lipids (14). If EVs have specific roles in intercellular communication, mechanisms are needed to selectively incorporate relevant cargo. Insight into these has been slow to emerge. Among molecules overrepresented in EVs are widely expressed tetraspanins, including CD63 and CD9, that share a conserved four-transmembrane-domain structure. Tetraspanins are well known for their role in regulating trafficking of interacting proteins (15, 16), potentially in conjunction with cholesterol (17), and are candidates for organizing EV content. Other proteins implicated in selecting EV protein cargo include ARRDC1, syntenin-1, Alix, and Tsg101 (18, 19). Proteins suggested to play a role in RNA packaging include sumoylated heterogeneous nuclear ribonuclear protein A2B1 (20), annexin a2 (21), and YBX1 (22). However, much remains to be understood about the role of these and other factors in defining EV content.
Creating an EV requires membrane deformation to form a vesicle and membrane fission to release it. Filamentous polymers of endosomal sorting complex required for transport III (ESCRT-III) are thought to drive membrane fission to generate intralumenal vesicles (ILVs) in endosomes and release viruses from the plasma membrane, acting as spiral coils or springs to constrict and sever membranous connections (23, 24, 25). The AAA+ ATPase VPS4 remodels and disassembles ESCRT-III polymers and is required for sustained ESCRT pathway function. ESCRT-III and VPS4 have therefore long been considered likely participants in EV biogenesis, and recent models increasingly connect ESCRTs to EVs (26, 27). Despite this, specific understanding of whether and how ESCRTs contribute to generating EVs is based on variable and sometimes contradictory data (18, 19, 28, 29, 30, 31). Some inconsistencies may be due to difficulties in separating EVs from contaminating membranes including dead cells. Others, however, are likely to be specific to the ESCRT machinery, which is modular and utilizes overlapping but distinct subsets of proteins for different purposes (23, 26). The consistent recovery of ESCRT proteins including Alix, Tsg101, and others in association with EVs (14) strengthens their connection to EV biogenesis, although current thinking about ESCRT function does not account for the presence of ESCRT proteins in released vesicles. Importantly, there are other protein- and lipid-based mechanisms that may operate together with or instead of ESCRTs to generate and release EVs (32). Further insight into how ESCRTs contribute to EV biogenesis is clearly needed and will help to solidify our understanding of EV biology.
In this study, we reasoned that if ESCRTs play a general role in EV biogenesis, interfering with VPS4 activity should have an inhibitory effect regardless of which subset of ESCRT proteins is involved and whether the EVs in question originate in endosomes or at the plasma membrane. We also characterize an efficient and scalable method for collecting EVs using polymer-based precipitation followed by lectin affinity chromatography. Our results demonstrate that inhibiting VPS4 impairs both exosome and ectosome release, thereby generally implicating VPS4 and its ESCRT-III substrates in EV biogenesis.
Materials and Methods
Antibodies, plasmids, and reagents
Antibodies used include mouse monoclonal antibodies against CD63 (Developmental Studies Hybridoma Bank: H5C6; no. 215-820, Ancell Corp., Bayport, MN), CD9 (no. MCA496GA, Serotec/Bio-Rad, Hercules, CA), mCherry (no. 96752, Novus Biologicals, Littleton, CO) and rabbit polyclonal antibodies against CHMP4A (33), calnexin (no. 2433, Cell Signaling Technologies, Danvers, MA), and syntenin-1 (no. 22399, Proteintech, Rosemont, IL). Secondary antibodies used were conjugated to horseradish peroxidase for immunoblotting (Thermo Fisher Scientific, Waltham, MA), IRdye800CW for infrared-based cell quantitation (LiCor, Lincoln, NE), and Alexa Fluor 488 for immunofluorescence (Thermo Fisher Scientific).
pEGFP-CD63 (from Paul Roche, National Institutes of Health, Bethesda, MD) was used to generate pmCherry-CD63 using BamHI and XhoI sites. pmGFP-CD9 was generated by transferring CD9 from pcDNA3.1 FLAG-CD9 (from Michael Caplan, Yale University School of Medicine) into pmEGFP-c2 using the BamHI and XhoI sites. mCherry-GPI was from Gerald Baron (Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health).
HEK293TRex cells with tetracycline-inducible VPS4A(E228Q) or VPS4B(E235Q) (33, 34) were maintained in 5 μg/mL blasticidin and 100 μg/mL zeocin. PANC-1 and HT-29 cells were from ATCC (Manassas, VA). U87 cells were from Joshua Rubin (Washington University School of Medicine, St. Louis, MO). All cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% tetracycline-free fetal bovine serum and 1 mM L-glutamine. HEK293TRex cell lines additionally expressing the indicated fluorescent proteins were generated by transfecting with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) followed by selection with 500 μg/mL G418.
Chemicals were from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. Fetal bovine serum was from Atlanta Biologicals (Flowery Branch, GA). Protein concentrations were determined using a Bio-Rad protein assay reagent with bovine serum albumin as a standard.
Assay of ESCRT polymerization
Tetracycline was added at 0.5 μg/mL to inducible HEK293TRex cells to initiate GFP-VPS4(EQ) expression. After 2 h, media were replaced to remove tetracycline. At the indicated time, cells were washed with phosphate-buffered saline (PBS) and solubilized in lysis buffer (1% Triton X-100, 10% sucrose, 1 mM EDTA, 10 mM Tris-Cl (pH 8.0), 1 mM PMSF, and protease inhibitors). Lysates were centrifuged at 10,000 × g for 15 min; pellets were resuspended in the same volume as the soluble fraction and both were boiled in sample buffer for sodium dodecyl sulfate polyacrylamide-gel electrophoresis (SDS-PAGE).
EV collection by differential centrifugation
All EVs were collected into culture medium containing EV-depleted fetal bovine serum prepared by overnight centrifugation at 100,000 × g. Standard protocols were adapted to concentrate EVs (35, 36). Briefly, EV-containing medium collected from the indicated cells was cleared of large membranes by spinning at 1500 × g for 10 min followed by 10,000 × g for 30 min or passage through a PVDF syringe filter (0.2 μm, low protein binding; Millipore, Billerica, MA). EVs were pelleted from this supernatant by centrifugation in a Ti70 rotor (Beckman Coulter, Brea, CA) at 100,000 × g for 2 h, resuspended in 1 mL PBS, and pelleted again in a TL100.3 rotor at 100,000 × g for 1 h. Final EV pellets were resuspended in PBS in a volume normalized to the total protein content of the cells from which they were derived. For reproducible handling of 100k pellets, EVs were typically collected over 16–24 h from at least one 15 cm dish of cells.
EV collection by PEG precipitation and concanavalin A binding
Culture media cleared of larger membranes as above were combined with an equal volume of 40% polyethylene glycol (PEG; 3350 average molecular weight) in PBS. After 1 h on ice, precipitated material was pelleted by spinning at 17,000 × g for 20 min and resuspended in ConA binding buffer (20 mM Tris-Cl (pH 7.4), 150 mM NaCl, 2 mM MnCl2, and 2 mM CaCl2). Samples were incubated for the indicated time with concanavalinA sepharose (GE Healthcare, Chicago, IL) or amylose resin (New England Biolabs, Ipswich, MA). Beads were washed in ConA binding buffer and either directly boiled in SDS-PAGE sample buffer or eluted with 1 M α-D-methylmannoside in binding buffer. Volumes of all EV preparations were normalized to the total protein content of the cells from which they were derived. This method was scalable and used to concentrate EVs from as little as one 35 mm dish of cells.
Sucrose gradients
EV pellets were suspended in 2 mL of 20 mM HEPES (pH 7.4) and 2.5 M sucrose, placed in the bottom of a 14 × 89 mm centrifuge tube, overlaid with a 10 mL linear sucrose gradient (2–0.25 M sucrose in 20 mM HEPES (pH 7.4)), and centrifuged in a Beckman SW41 rotor at 210,000 × g for 16 h. Fractions of 1 mL were collected, diluted with 3 mL buffer, and bound to nitrocellulose using a vacuum-based slot blot apparatus (Midwest Scientific, Valley Park, MO). After drying, nitrocellulose was subjected to immunoblotting for EV proteins as below. The density of each gradient fraction (before dilution) was determined using a hand refractometer (Atago USA, Bellevue, WA). The measured sucrose content (in Brix%) was converted to solution density using a standard room-temperature-based conversion chart.
SDS-PAGE and immunoblotting
Samples were boiled in SDS-PAGE sample buffer (2% SDS, 60 mM Tris (pH 6.8), 0.01% bromophenol blue, and 10% glycerol) with or without 1% β-mercaptoethanol for 5 min and spun for 2 min at 21,000 × g. For immunoblotting CD63 and CD9, β-mercaptoethanol was omitted because the antibodies used recognize nonreduced forms of these proteins. Samples were separated on 10% gels followed by transfer to nitrocellulose. Membranes were blocked and probed in 20 mM Tris (pH 7.5), 100 mM NaCl, 0.05% Tween20, and 5% nonfat dry milk. Blots were imaged on a ChemiDoc MP imaging system (Bio-Rad) and quantitated with Bio-Rad Image Studio software. CD63 typically appeared in multiple poorly resolved bands; boxes for quantitation were drawn to be inclusive of all.
Quantitative RT-PCR analysis of miRNA
EVs collected by differential centrifugation were suspended in PBS, and RNA was isolated using TRIzol LS (Invitrogen, Hercules, CA). Samples were spiked with 100 nM Caenorhabditis elegans synthetic miRNA as a control (cel-miR-39, 5′-UCACCGGGUGUAAACAGCUUG-3′). cDNA synthesis was primed with hairpin stem-loop oligonucleotides as previously described (37) (miR-92a, 5′-GCGTGGTCCCGACCACCACAGCCGCCACGACCACGCACAGGC-3′; miR-150, 5′-GCGTGGTCCCGACCACCACAGCCGCCACGACCACGCCACTGG-3′; cel-miR-39, 5′-GCGTGGTCCCGACCACCACAGCCGCCACGACCACGCCAAGCT-3′). cDNA was amplified in an ABI Prism 7500 fast real-time polymerase chain reaction (PCR) machine for 40 PCR cycles using SYBR green PCR master mixture (Applied Biosystems, Foster City, CA), 100 nM template-specific (miR-92a, 5′-GTGACGATCTATTGCACTTGTCCCG-3′; miR-150, 5′-GTGACGATCTCTCCCAACCCTTGTA-3′; cel-miR-39, 5′-CAGTGACGATCTCACCGGGTGTAAATC-3′), and a 100 nM universal reverse primer (5′-TCCCGACCACCACAGCC-3′). Relative quantification of miRNA expression was performed using the comparative threshold method with normalization to cel-miR-39.
Fluorescent EV particle counts
EVs from cells expressing the indicated fluorescently tagged proteins were collected by differential centrifugation and resuspended in PBS as above. One microliter of an appropriately diluted sample was placed on a glass slide, overlaid with a 12-mm-diameter poly-L-lysine coated glass coverslip, and imaged by widefield fluorescence microscopy. Thresholded images were subjected to particle counting using standard ImageJ plug-ins (38).
Immunofluorescence
Cells on poly-L-lysine coated no. 1.5 glass were fixed in 2% paraformaldehyde in PBS, permeabilized with 0.1% saponin, and blocked and stained with antibodies diluted in PBS containing 5% goat serum and 0.01% saponin. Coverslips were mounted on slides in 100 mM Tris (pH 8.5), 10.5% polyvinyl alcohol, and 21% v/v glycerol.
Microscopy
Imaging was performed on an Olympus IX81 microscope equipped with a 60×/1.42 NA oil objective and a Yokogawa CSU-10 spinning-disc confocal scanner, 488 and 561 nm lasers, and a Photometrics (Tucson, AZ) Cascade 512B camera operating with MicroManager (39). Wide-field epifluorescence images for EV particle counting were acquired with a Hamamatsu (Hamamatsu City, Japan) Flash 2.8 camera.
Infrared immune labeling and quantitation
Cells plated in poly-L-lysine-coated 24-well plates at varying densities were fixed and stained as for immunofluorescence in the absence or presence of saponin using infrared (IR)-dye-conjugated secondary antibodies. Staining was quantitated using an Odyssey (Danbury, CT) IR imager and Li-Cor Image Studio Software Cells labeled with only secondary antibodies were used for background subtraction. Nonpermeabilized and permeabilized samples processed in parallel were used to define the distribution of fluorescence between surface and internal membranes (internal = total – surface).
Results
CD63 and CD9 are enriched in separable populations of EVs
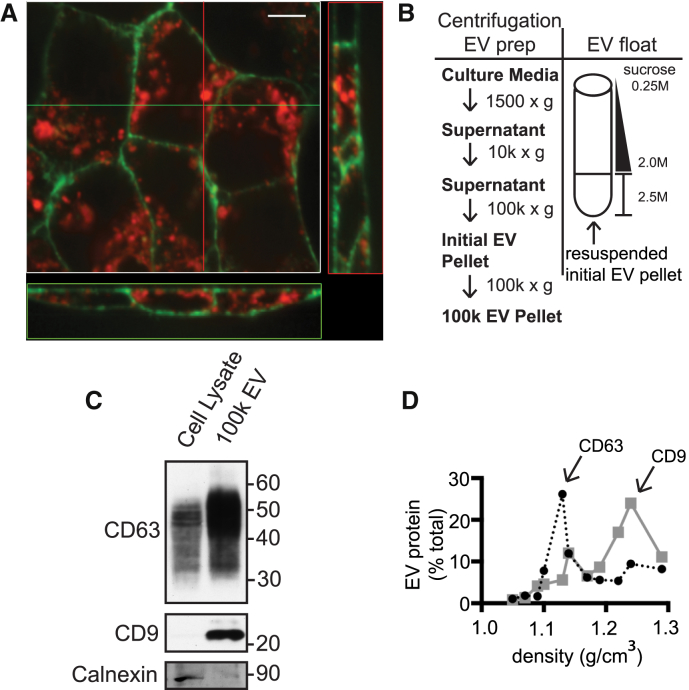
The tetraspanins CD63 and CD9 are among the proteins most commonly recovered in EVs (14). Interestingly, these two proteins do not share the same overall subcellular distribution: CD63 is enriched on ILVs within MVBs, whereas CD9 localizes primarily to the plasma membrane (11, 15, 40). Imaging mCherry-CD63 and mGFP-CD9 in stably transfected HEK293 cells demonstrated that these proteins segregate even when overexpressed (Fig. 1 A).
Figure 1.
CD63 and CD9 are released on distinct EVs. (A) Confocal z-stacks in orthogonal view of HEK293 cells stably expressing mGFP-CD9 on the plasma membrane and mCherry-CD63 on intracellular vesicles. Scale bars, 10 μm. (B) Flow chart of EV isolation and sucrose density gradient flotation protocols. (C) For the indicated proteins, 8 μg of total cellular protein and 1 μg of 100k EVs prepared from HT-29 cells were separated by SDS-PAGE and immunoblotted. 100k EVs from HEK293 cells showed similar protein profile (not shown). (D) Distribution of tetraspanin proteins after flotation of 100k EVs from HEK293 cells into linear sucrose density gradients. The x axis indicates the density of each fraction as determined by refractometry. Fractions were immunoblotted for CD63 (black line) and CD9 (gray line) and quantitated, and the values were plotted as the percent of total signal across all fractions. The experiment shown is representative of four biological replicates. To see this figure in color, go online.
Based on their differential localization, we wondered if CD63 and CD9 might be enriched in distinct EVs that separately originate from the MVB or the plasma membrane. We collected EVs using a conventional differential centrifugation protocol, and we refer to these pelleted EVs as “100k EVs” (Fig. 1 B; (35, 36). Immunoblotting whole cell lysates and 100k EVs confirmed that both CD63 and CD9 are enriched in 100k EVs, whereas the cell-associated endoplasmic reticulum protein calnexin was absent (Fig. 1 C). To determine whether CD63 and CD9 are on equivalent membranes, we resuspended 100k EVs in 2.5 M sucrose and floated them into a sucrose density gradient (Fig. 1 D). Immunoblotting demonstrated that CD63-containing membranes peak at a density characteristic of exosomes (∼1.15 g/cm3) (6, 13, 40, 41). In contrast, CD9 was primarily found on membranes with density 1.20–1.24 g/cm3. Others have also noted differences in the density of CD63 versus CD9-enriched extracellular membranes (11, 42). Together with their localization, we suggest that these tetraspanins preferentially mark distinct extracellular membranes that correspond, at least in part, to exosomes and ectosomes. Experiments described below further explore and support this proposal.
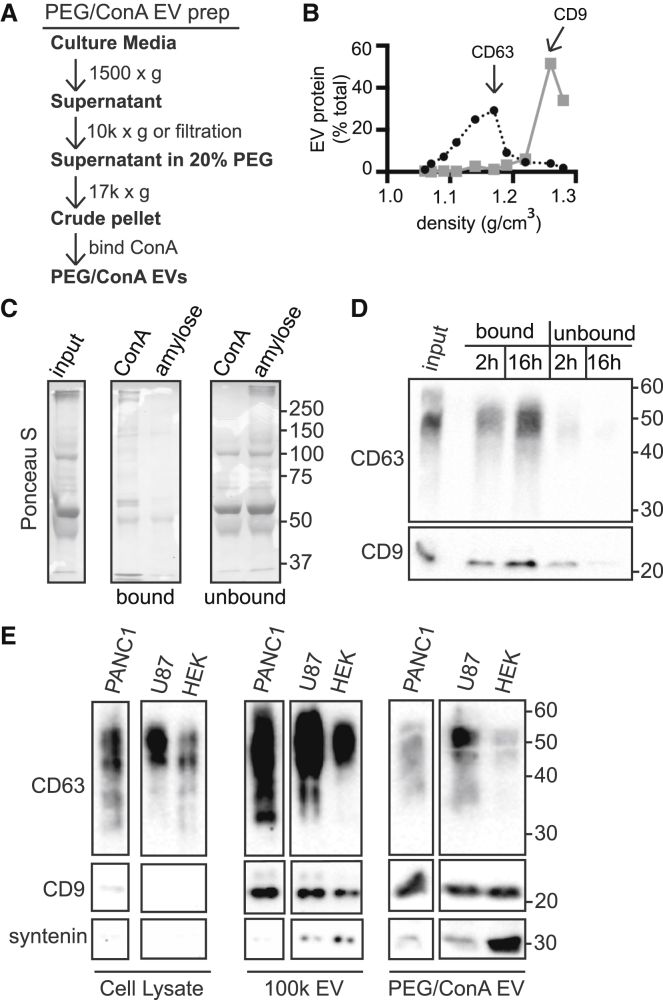
Collecting EVs by PEG precipitation and binding to concanavalin A
Recovering EVs by ultracentrifugation is inherently inefficient (43), and below a threshold sample size, 100k EV pellets become too small for reproducible handling. As a result, most preparations of EVs from cultured cells use large numbers of cells and long collection periods, typically 24 h. This creates challenges for studies of factors involved in EV release. To decrease sample requirements, we turned to PEG precipitation (Fig. 2 A). Our protocol was adapted from procedures used to concentrate low-titer viruses (44) and is similar to those described by others over the past few years to concentrate EVs (45, 46). To compare PEG-precipitated membranes with those in the 100k EV pellet, we resuspended PEG pellets in 2.5 M sucrose and floated them into sucrose density gradients. Similar to 100k EVs, PEG-precipitated membranes containing CD63 peaked at a density of ∼1.15 g/cm3, whereas CD9 containing membranes peaked at ∼1.25 g/cm3 (Fig. 2 B).
Figure 2.
Comparison of EV collection by PEG/ConA and ultracentrifugation methods. (A) Flow chart of the PEG/ConA EV collection procedure. (B) Sucrose density gradient analysis of HEK293 cell EVs precipitated with PEG. As in Fig. 1D, the x axis indicates the density of each fraction as determined by refractometry. Fractions were immunoblotted for CD63 (black line) and CD9 (gray line) and quantitated, and the values were plotted as the percent of total signal across all fractions. (C) SDS-PAGE of PEG precipitated protein binding to ConA sepharose versus control amylose resin. Ponceau S stain shows the total protein in input, bound, and unbound fractions. (D) PEG precipitated EVs from HEK293 cells were incubated with ConA sepharose for the indicated time, released from beads by boiling in SDS-PAGE sample buffer, and immunoblotted for CD63 and CD9. (E) Comparison of 100k and PEG/ConA EVs. EVs released by the indicated cell lines over 24 h were harvested from culture media by ultracentrifugation (100k EV) or PEG/ConA precipitation (PEG/ConA EV). 100k EVs from 20% and PEG/ConA EVs from 10% of the cell culture media were separated by SDS-PAGE and immunoblotted for CD63, CD9, and syntenin. Whole-cell lysate blots were loaded with 10 μg of total protein. The images shown are from a single exposure and were spliced to remove an irrelevant lane. Lighter exposure (not shown) of 100k EV fractions confirmed that U87 cells released the most CD63.
One difficulty with PEG and similar precipitants is that they are nonselective, concentrating EVs along with other material present in serum-containing media. This material frequently interfered with immunoblotting, particularly when the amount of EV-associated protein was low. We therefore added an enrichment step, taking advantage of the fact that EV surface glycoproteins readily bind to lectins (47, 48). We found that concanavalin A (ConA) sepharose did not efficiently bind the major serum-derived proteins present in PEG precipitates (Fig. 2 C) but did recruit CD63 and CD9 (Fig. 2 D), which could be eluted with SDS-sample buffer or methyl-α-D-mannopyranoside, a high-affinity ConA ligand (49). Using this PEG/ConA enrichment protocol we were able to reliably collect EVs from as little as 1 mL conditioned cell media for subsequent analysis by immunoblotting.
To compare PEG/ConA EVs to those isolated by differential centrifugation, we prepared EVs from two cancer cell lines (U87 and PANC1) that release abundant EVs (50), as well as from the HEK293 cells used in the rest of this study. The relative recovery and enrichment of EV-associated proteins was comparable using the two protocols (Fig. 2 E). Both methods also recovered syntenin-1, a cytoplasmic adaptor protein that binds CD63 and is typically enriched in exosomes (13, 18) (Fig. 2 E). Interestingly, there was more variability between cell lines in CD63 than in CD9 release. This has been previously noted (51) and suggests greater variation among cultured cells in exosome than ectosome release.
Inhibiting VPS4 function in cells
Conflicting reports concerning the role of ESCRTs in EV biogenesis raise questions about whether ESCRTs are generally required to create EVs or instead operate in parallel with other, ESCRT-independent machinery (2). A potent way to inhibit AAA+ ATPases such as VPS4 is to express ATPase-deficient subunits that coassemble with endogenous enzyme to create nonfunctional holoenzymes (52). In cells, VPS4 mutants block ESCRT-III disassembly, thereby globally interfering with ESCRT dynamics and ESCRT-dependent processes (33, 34, 53). To date, however, mutant VPS4 has been found to have varying effects on EV release (18, 19, 28, 29, 30), possibly because of heterogeneities associated with transient protein expression. Experiments using siRNA-mediated depletion of VSP4 and other ESCRT components have also come to differing conclusions (31). To revisit the role of VPS4 activity in EV release, we turned to well-characterized cell lines that inducibly and uniformly express inhibitory Walker B mutants of VPS4 (VPS4A E228Q or VPS4B E235Q) (33, 34). These cell lines have previously been used to address and resolve controversies surrounding the role of VPS4 in viral budding events (34, 54).
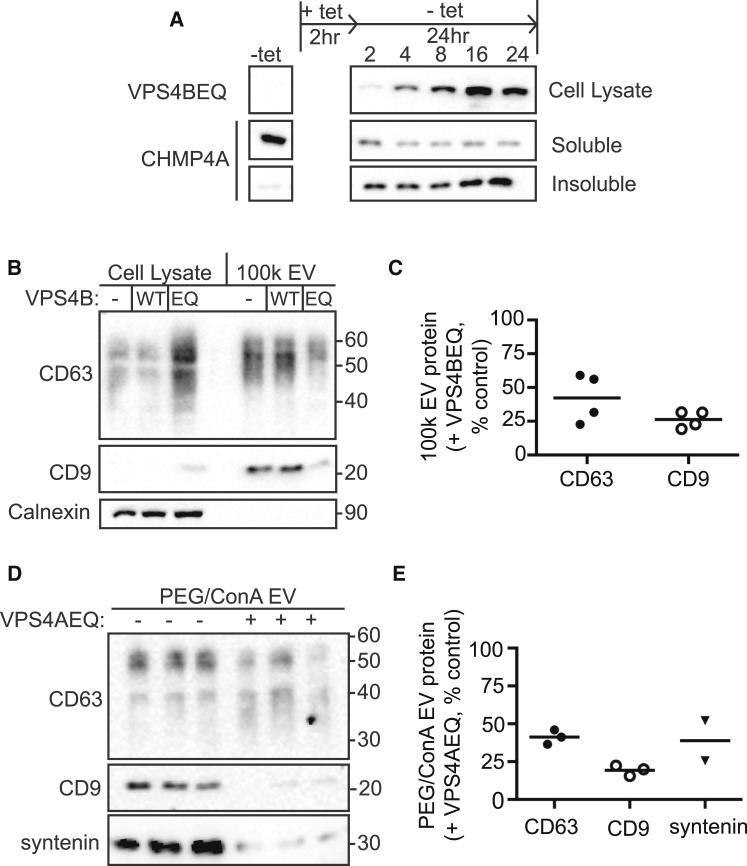
To set conditions for inhibiting VPS4 function while minimizing expression of mutant enzyme over the extended time course used to collect EVs, we monitored effects of VPS4EQ expression on the status of endogenous ESCRT-III proteins. CHMP4A, an essential ESCRT-III subunit, shifted into an insoluble fraction within 2 h of tetracycline-induced mutant VPS4 expression, confirming disruption of ESCRT dynamics (Fig. 3 A). As in earlier studies of viral budding (34, 54), effects of mutant VPS4A or VPS4B were indistinguishable; individual figures therefore indicate the particular mutant used, whereas the text designation “VPS4EQ” describes results seen with both inhibitory mutants.
Figure 3.
Release of CD63 and CD9 in EVs requires VPS4 activity. (A) Top: Immunoblot of whole-cell lysate showing the GFP-VPS4BEQ expression induced by a 2 hr pulse of tetracycline. Middle and Bottom: Immunoblot of ESCRT-III protein CHMP4A in soluble and insoluble fractions separated by centrifugation at 17,000 × g. Pellets were resuspended in the same volume as supernatants, and equal volumes of each were loaded. (B) Cell lysate from 0.05 × 106 cells and 100k EVs from 10 × 106 cells as indicated were immunoblotted for CD63, CD9, and calnexin. (C) Quantitation of CD63 and CD9 on immunoblots from four independent experiments comparing 100k EVs from control uninduced or VPS4EQ-expressing HEK293 cells. The lines indicate mean. (D) CD63, CD9, and syntenin in EVs were collected by PEG/ConA precipitation. Each lane represents a technical replicate prepared from one-third of the total culture supernatant; for quantitation, these were averaged to represent a single experiment. (E) Quantitation of CD63, CD9, and syntenin on immunoblots from two (syntenin) or three (CD63 and CD9) independent experiments comparing PEG/ConA EVs from control uninduced or VPS4AEQ-expressing HEK293 cells. The lines indicate mean.
Effect of inhibiting VPS4 activity on EVs
To monitor the effect of inhibiting VPS4 on EV biogenesis, we examined release of EV-associated proteins from cells with or without inducing VPS4EQ expression. Immunoblots of CD63, CD9, and syntenin showed that expressing VPS4EQ decreased the amount of each in EVs by 50% or more (Fig. 3, B–E). Importantly, VPS4EQ decreased CD63 release in EVs despite the significant increase in cellular CD63 (Fig. 3 B, Cell Lysate) caused by this mutant’s known inhibitory effects on degradative trafficking to the lysosome (55). The decrease in both CD63 and CD9 in EVs suggests that ESCRTs are involved in vesicle biogenesis at both the MVB and the plasma membrane. Residual release of EV-associated proteins from VPS4EQ-expressing cells may result from incomplete pathway inhibition over the extended EV collection period, although we cannot exclude a minor role for VPS4 and ESCRT-independent machinery.
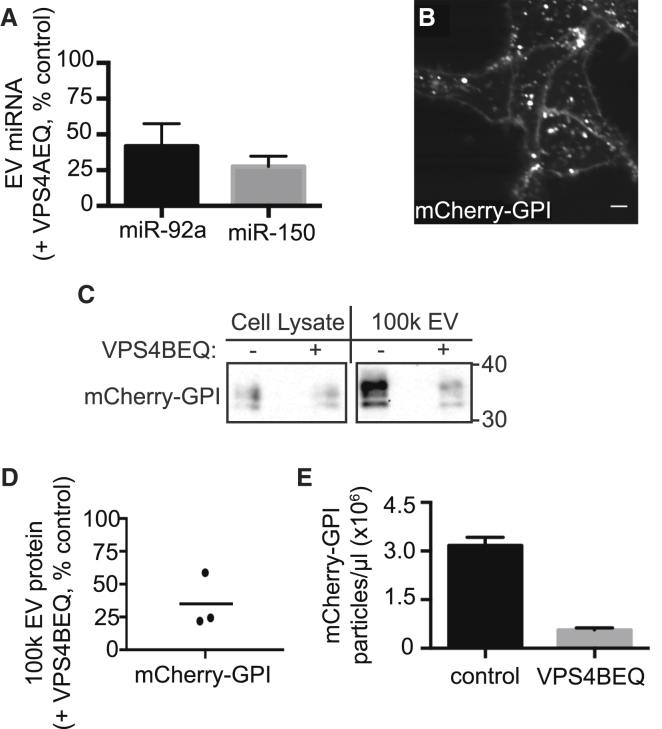
To look at the effect of VPS4EQ on other potential EV cargo, we examined two representative EV miRNAs. We selected miR-150, which is known to be released by HEK293 cells (56), and miR-92a, which is broadly expressed and implicated in regulating cell proliferation (57). Both miRNAs have been identified extracellularly, likely in EVs (58). qRT-PCR analysis of 100k EVs showed that both are present in EVs released by HEK293 cells and that inducing expression of VPS4EQ reduced the amount of both recovered in EVs (Fig. 4 A).
Figure 4.
VPS4-dependent release of miRNA and GPI-labeled vesicles. (A) qRT-PCR analysis of 100k EVs from cells expressing VPS4EQ. Values were normalized to spike-in cel-miR-39 RNA and compare mi-RNA in EVs released by control uninduced or VPS4EQ-expressing HEK293 cells. Data are expressed as the mean ± SE of three independent experiments. (B) Confocal image of VPS4EQ-inducible HEK293 cells stably expressing mCherry-GPI. Scale bar, 10 μm. (C) Immunoblot of cell lysate and 100k EVs from HEK293 cells expressing mCherry-GPI with or without VPS4EQ. (D) Quantitation of mCherry-GPI in EVs from three independent experiments, comparing mCherry-GPI release from cells with or without induction of VPS4EQ. The line indicates mean. (E) Quantification of mCherry-GPI particles released from cells comparing control uninduced to VPS4BEQ-expressing HEK293 cells. Values are average particle counts from five fields. Error bars indicate the standard deviation. Data are from one of two biological replicates.
Decreased amounts of EV-associated proteins or miRNA typically correlate with release of fewer EVs, but could instead reflect a change in EV composition. We therefore counted EVs using fluorescence microscopy (59). To label EVs originating from different membranes with a single fluorescent protein, we generated a VPS4BEQ-inducible HEK293 cell line constitutively expressing glycosylphosphatidylinositol (GPI)-anchored mCherry (60). In these cells, mCherry-GPI was present both on the cell surface and on internal organelles (Fig. 4 B). Expressing VPS4BEQ reduced the amount of mCherry-GPI detected on EV immunoblots (Fig. 4, C and D). Parallel counting of fluorescent puncta immobilized on polylysine-coated glass showed that expressing VPS4EQ reduced the number of released particles by ∼5-fold (Fig. 4 E), confirming that the observed decreases in EV-associated cargo correspond to fewer released EVs.
Serum triggers VPS4-dependent EV release
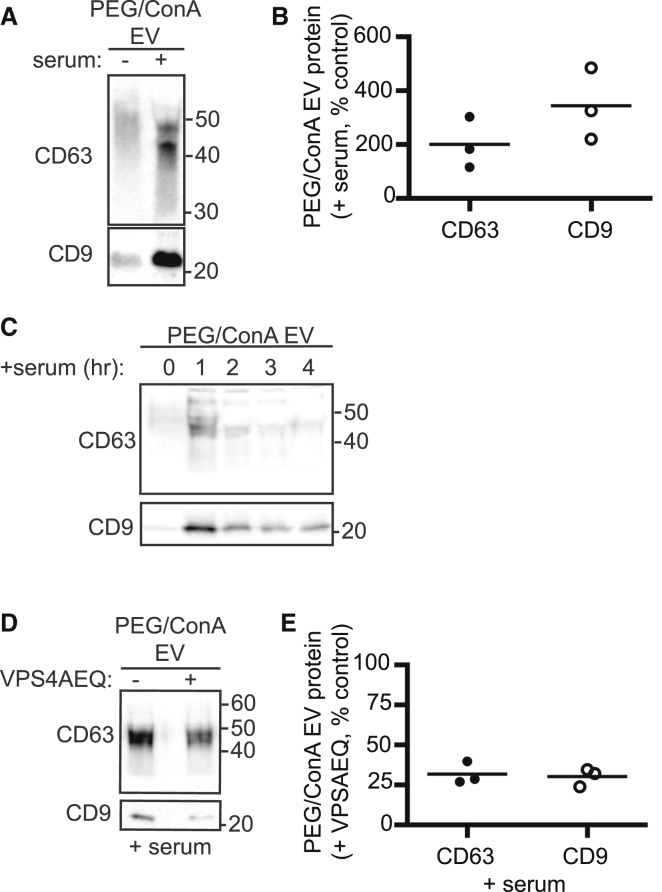
EV production is increasingly thought to be a physiological response enabling intercellular communication and can be triggered by a wide variety of stimuli (61, 62, 63, 64). To further explore the role of ESCRTs in EV release, we collected EVs from serum-stimulated cells and observed a robust increase in CD63, and especially CD9, release (Fig. 5, A and B). Taking advantage of the efficient EV recovery enabled by PEG/ConA enrichment, we collected EVs at 1 h intervals from serum-stimulated cells. Interestingly, most tetraspanins were released during the first hour of stimulation, demonstrating that the response to serum is both rapid and transient (Fig. 5 C). Importantly, inhibiting VPS4 by expressing VPS4EQ before serum stimulation decreased CD63 and CD9 in EVs by ∼75% (Fig. 5, D and E).
Figure 5.
Characteristics of serum-triggered EV release. (A and B) HEK293 cells grown for 16 h without serum were incubated in fresh medium without (−) or with (+) 10% serum for 4 h. EVs harvested by PEG/ConA were immunoblotted for CD63 and CD9 and quantitated across three experiments, as shown. (C) HEK293 cells grown for 16 h without serum were incubated in fresh serum-free media for 1 h (0). This was collected and replaced with fresh medium containing 10% serum every hour for 4 h (1–4). EVs were harvested by PEG/ConA precipitation and immunoblotted for CD63 and CD9. (D) Serum-triggered EVs from control and VPS4EQ-expressing cells were harvested by PEG/ConA and immunoblotted for CD63 and CD9. (E) Quantitation of three independent experiments. Values show the percent of protein in PEG/ConA EVs from serum stimulated cells expressing VPS4AEQ compared to EVs from control serum stimulated cells not induced to express VPS4AEQ. The lines indicate mean.
Release of CD63, but not CD9, requires intact microtubules
To differentiate between EVs that originate in MVBs and those that come directly from the plasma membrane, we took advantage of the fact that MVB exocytosis requires transport along microtubules to the plasma membrane (65). We asked how depolymerizing microtubules affects CD63 and CD9 release, reasoning that perturbed MVB trafficking would selectively affect exosomal EVs. CD63 release induced by serum stimulation in cells pretreated with nocodazole was less than half that in untreated cells (Fig. 6). In contrast, CD9 release was largely unchanged (Fig. 6). This reinforces the idea that CD63- and CD9-enriched EVs originate from different membranes that can be independently mobilized in response to serum stimulation.
Figure 6.
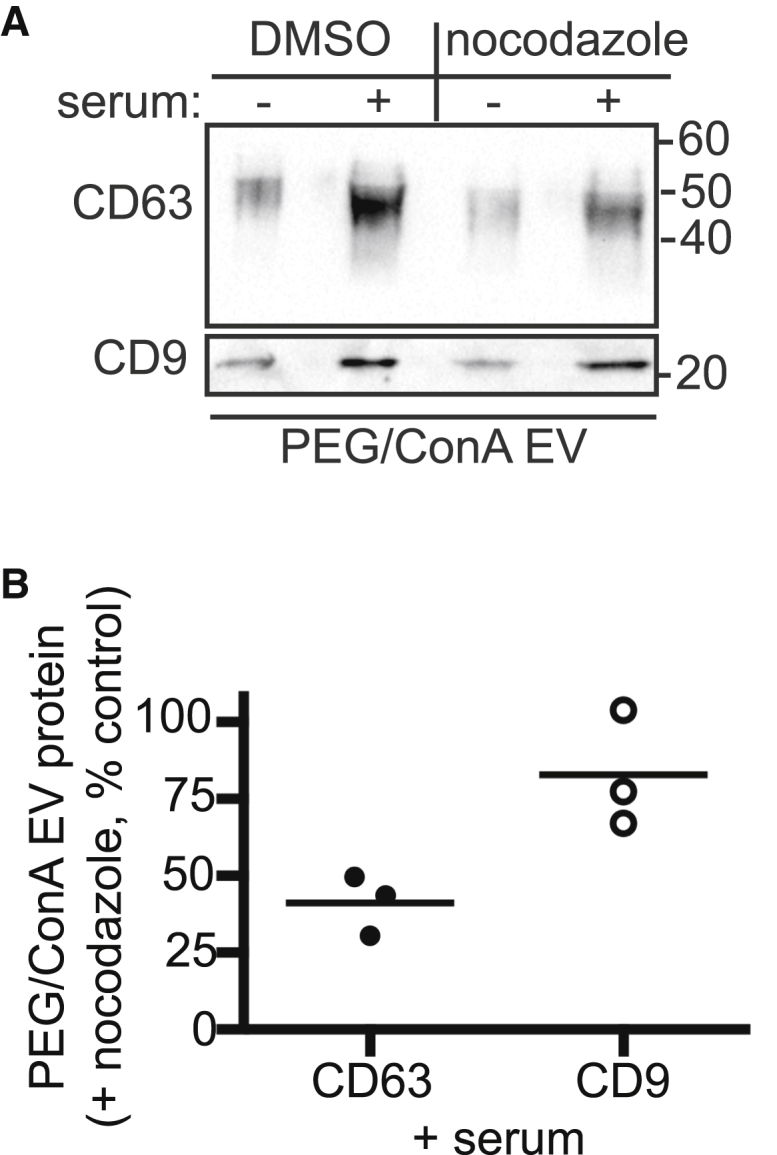
Nocodazole selectively decreases CD63 release. (A) Serum-triggered EVs released from HEK293 cells treated with either 12 μM nocodazole or dimethylsulfoxide (DMSO) were harvested by PEG/ConA precipitation and immunoblotted for CD63 and CD9. (B) Quantitation of three independent experiments, with the line indicating the mean. Values show the percent of protein in PEG/ConA EVs from nocodazole treated cells compared to EVs from control untreated cells. The lines indicate mean.
CD63 retargeted to the plasma membrane is released together with CD9 in EVs
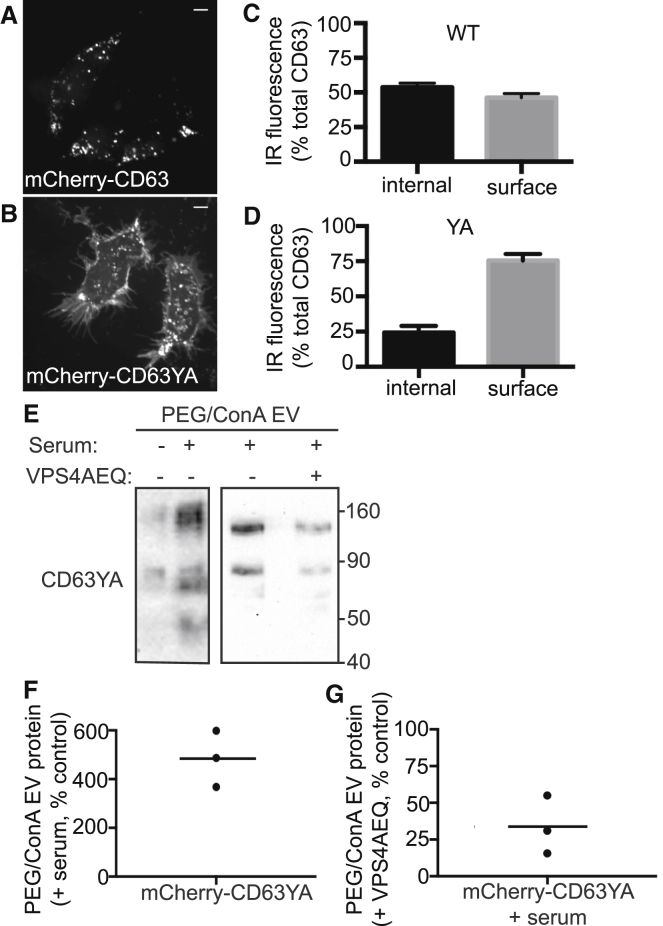
Although the majority of CD63 normally resides on intracellular membranes, a proportion is present on the plasma membrane, where it colocalizes with CD9. Given that the increase in CD9 released in response to serum stimulation was typically greater than that of CD63 (Fig. 5, A and B), we wondered if serum-stimulated EVs preferentially originate at the plasma membrane. To compare release of the same protein from different subcellular locations, we asked how redistributing CD63 from endosomes to the plasma membrane affects its release in EVs. CD63 contains a C-terminal GYXXΦ motif that binds clathrin adaptor proteins and mediates primary targeting to late endosomes and/or lysosomes (16, 66). Mutating this motif to GAXXΦ is reported to shift much of CD63 to the plasma membrane (66), and it did so in HEK293 cells stably expressing mCherry-CD63 with or without this mutation (Fig. 7, A–D). Stimulating cells expressing mCherry-CD63 Y236A with serum increased CD63 release ∼5-fold (Fig. 7 F), similar to endogenous CD9 (Fig. 5 B). Although limited by the fact that we are monitoring changes in a population of vesicles released over time, these data are consistent with the idea that cells respond to serum stimulation by substantially enhancing EV release directly from the plasma membrane. Further supporting a general role for ESCRTs in the release of EVs from the plasma membrane, we found that expressing VPSAEQ also reduced mCherry-CD63 Y236A release (Fig. 7, E and G).
Figure 7.
Serum-triggered release of plasma-membrane-localized CD63. (A and B) Confocal images of HEK293 cells stably expressing mCherry-CD63 (A) or mCherry-CD63YA (Y236A) (B). Scale bars, 10 μm. (C and D) Cells stably expressing CD63 (C) or CD63YA (D) were immunolabeled for CD63 in the absence or presence of detergent to measure surface and total protein, respectively. Internal fluorescence was derived by subtracting surface from total fluorescence. The values are represented as the mean ± SD of three independent experiments. (E) Anti-mCherry immunoblot of EVs released from HEK293 cells stably expressing mCherry-CD63YA and treated as indicated. (F) Quantitation of immunoblots from three independent experiments showing the effect of serum stimulation on mCherry-CD63YA release. (G) Quantitation of immunoblots from three independent experiments showing the effect of expressing VPS4AEQ on mCherry-CD63YA release. The lines indicate mean.
Discussion
Cells release a large variety of EVs that differ in membrane of origin, cargo composition, and triggers controlling their release. This heterogeneity has complicated investigations of mechanisms responsible for EV biogenesis and definitive studies of their physiologic function. Components of the ESCRT machinery are frequently implicated in EV biogenesis based on vesicle topology and a growing but sometimes inconsistent body of experimental literature. The AAA+ ATPase VPS4 is responsible for ESCRT-III polymer dynamics, and connections between its activity and ESCRT-III function in MVB formation, virus budding, and cytokinesis are well established (26, 67). Previous studies characterizing the role of VPS4 in EV biogenesis came to varying conclusions. Some reported no effect of inhibiting VPS4 on release of EVs marked by CD63 (18, 28) or proteolipid protein (28), whereas others found that inhibiting VPS4 decreased production of a variety of EVs, including T-cell ectosomes (29), ARRDC1-containing microvesicles (19) and hedgehog-containing EVs—likely ectosomes—released from epithelial cells in Drosophila melanogaster wing imaginal discs (68). RNAi-based depletion of VPS4 has also been associated with varying effects on EV release (18, 31, 69). Our findings extend and build on this literature, demonstrating that uniformly and rapidly inhibiting VPS4 decreases both constitutive and serum-stimulated release of EVs from the widely used HEK293 cultured cell line.
To assess VPS4 function in generating different types of EVs, we studied two tetraspanins, CD63 and CD9, which are both widely used EV markers but differ in their steady-state subcellular localization to MVBs (CD63) and the plasma membrane (CD9). Release of these proteins on EVs of differing density (Figs. 1 and 2), differential response to serum stimulation (Figs. 5 and 7), and differential sensitivity to microtubule disruption (Fig. 6) support our proposal that these tetraspanins are enriched in and can serve as useful markers of exosomes (CD63) and ectosomes (CD9). Earlier studies showing that release of CD63, but not CD9, depends on the exocytic GTPase Rab27a/b are consistent with this distinction (70, 71). Recently published comprehensive analyses of size-fractionated EVs from dendritic cells further validate CD63 as a marker of bona fide exosomes and the existence of CD9-only EVs (13), but they also highlight additional levels of heterogeneity among EVs that become apparent after extensive separation and proteomic profiling. Whether such subclasses of EVs are fundamentally different from each other, and from the simple classification we describe here, remains to be determined. Our finding that inhibiting VPS4 decreases release of both CD63 and CD9 implicates ESCRT-III in biogenesis and/or release of most, or even all, of these vesicles. Interestingly, both earlier studies and trends in our data (Fig. 3) are consistent with the possibility that inhibiting VPS4 interferes with the “one step” event needed to release ectosomes from the plasma membrane more acutely than with the “two step” process needed to create and then release endosome-encapsulated exosomes.
Expressing mutant VPS4 efficiently traps ESCRT-III proteins in a polymerized state (Fig. 3; (33, 72)), connecting ESCRT-III dynamics to the changes in EV release seen here. It is, however, likely that not all 12 ESCRT-III proteins in human cells are essential for EV biogenesis, and precisely which are involved may differ depending on where the EVs originate. Future studies will be needed to determine which ESCRT-III proteins are essential for creating EVs and whether selective manipulation of those involved in EV biogenesis is possible.
A compelling connection between the ESCRT machinery and EVs comes from the consistent recovery of ESCRT and ESCRT-associated proteins in vesicles from many different cells, tissues, fluids, and organisms (14). In fact, after tetraspanins such as CD63 and CD9, Alix and Tsg101 are among the most frequently cited markers of EVs. Based on studies in other pathways, these are likely to act in recruiting and activating the ESCRT-III machinery (23, 26). However, why they are recovered within EVs remains unclear. Given the accessibility of EVs to quantitative analyses, further study of EV ESCRT content should provide needed insight into the relationship between ESCRTs and the vesicles they create.
An important aspect of this study was the use of a facile “PEG/ConA” method for concentrating EVs in a form amenable to immunoblot analysis. Comparison to traditional differential centrifugation showed that both methods recover comparable EVs, albeit with somewhat differing efficiency (Fig. 2). The PEG/ConA protocol described here joins a number of recently described strategies for concentrating EVs without ultracentrifugation (including, among others, (45, 46, 70)). Addition of a lectin affinity chromatography step is optimally suited for concentrating low amounts of EVs released into serum-containing media. The ability to scale EV analysis to smaller samples should facilitate further studies of EV biogenesis.
Author Contributions
C.E.J., B.S.S., J.E.S., and P.I.H. designed research. C.E.J. and B.S.S. performed research. C.E.J., B.S.S., J.E.S., and P.I.H. analyzed data. C.E.J. and P.I.H. wrote manuscript.
Acknowledgments
We thank Teresa Naismith for cloning and experimental support, Gregg Fox for early work optimizing the PEG/ConA protocol, and Anil Cashikar, Philip Stahl, and members of the Hanson laboratory for helpful discussions.
This work was supported by R01 GM076686 and GM122434 from the National Institutes of Health to P.I.H.
Editor: Lukas Tamm.
References
- 1.Lo Cicero A., Stahl P.D., Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr. Opin. Cell Biol. 2015;35:69–77. doi: 10.1016/j.ceb.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Maas S.L., Breakefield X.O., Weaver A.M. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 4.Pan B.T., Johnstone R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 5.Harding C., Heuser J., Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raposo G., Nijman H.W., Geuze H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zitvogel L., Regnault A., Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 8.Desrochers L.M., Antonyak M.A., Cerione R.A. Extracellular vesicles: satellites of information transfer in cancer and stem cell biology. Dev. Cell. 2016;37:301–309. doi: 10.1016/j.devcel.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocucci E., Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Bobrie A., Colombo M., Théry C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles. 2012 doi: 10.3402/jev.v1i0.18397. Published online April 16, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tauro B.J., Greening D.W., Simpson R.J. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell Proteomics. 2013;12:587–598. doi: 10.1074/mcp.M112.021303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowal J., Arras G., Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keerthikumar S., Chisanga D., Mathivanan S. ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charrin S., Jouannet S., Rubinstein E. Tetraspanins at a glance. J. Cell Sci. 2014;127:3641–3648. doi: 10.1242/jcs.154906. [DOI] [PubMed] [Google Scholar]
- 16.Pols M.S., Klumperman J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009;315:1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman B., Kelly B., Blacklow S.C. Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell. 2016;167:1041–1051.e11. doi: 10.1016/j.cell.2016.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baietti M.F., Zhang Z., David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 19.Nabhan J.F., Hu R., Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagiwara K., Katsuda T., Ochiya T. Commitment of annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 2015;589(24 Pt B):4071–4078. doi: 10.1016/j.febslet.2015.11.036. [DOI] [PubMed] [Google Scholar]
- 22.Shurtleff M.J., Temoche-Diaz M.M., Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson P.I., Cashikar A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 24.Schöneberg J., Lee I.H., Hurley J.H. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiaruttini N., Redondo-Morata L., Roux A. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell. 2015;163:866–879. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurley J.H. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beer K.B., Wehman A.M. Mechanisms and functions of extracellular vesicle release in vivo—what we can learn from flies and worms. Cell Adhes. Migr. 2016;11:135–150. doi: 10.1080/19336918.2016.1236899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trajkovic K., Hsu C., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 29.Choudhuri K., Llodrá J., Dustin M.L. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507:118–123. doi: 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong J., Körner R., Klein R. Exosomes mediate cell contact-independent ephrin-Eph signaling during axon guidance. J. Cell Biol. 2016;214:35–44. doi: 10.1083/jcb.201601085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colombo M., Moita C., Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 32.Babst M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 2011;23:452–457. doi: 10.1016/j.ceb.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y., Kimpler L.A., Hanson P.I. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 34.Taylor G.M., Hanson P.I., Kielian M. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J. Virol. 2007;81:13631–13639. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tauro B.J., Greening D.W., Simpson R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Théry C., Amigorena S., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;Chapter 3:22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 37.Michel C.I., Holley C.L., Schaffer J.E. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14:33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edelstein A., Amodaj N., Stuurman N. Computer control of microscopes using μManager. Curr. Protoc. Mol. Biol. 2010;Unit14.20:1–17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escola J.M., Kleijmeer M.J., Geuze H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 41.Hill A.F., Pegtel D.M., Nolte-’t Hoen E.N. ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J. Extracell. Vesicles. 2013;2:22859. doi: 10.3402/jev.v2i0.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu R., Greening D.W., Simpson R.J. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods. 2015;87:11–25. doi: 10.1016/j.ymeth.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 43.Cvjetkovic A., Lötvall J., Lässer C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles. 2014;3:23111. doi: 10.3402/jev.v3.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohno T., Mohan S., Sano K. A new improved method for the concentration of HIV-1 infective particles. J. Virol. Methods. 2002;106:167–173. doi: 10.1016/s0166-0934(02)00162-3. [DOI] [PubMed] [Google Scholar]
- 45.Rider M.A., Hurwitz S.N., Meckes D.G., Jr. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 2016;6:23978. doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng Y., Sui Z., Zhang Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst. 2016;141:4640–4646. doi: 10.1039/c6an00892e. [DOI] [PubMed] [Google Scholar]
- 47.Krishnamoorthy L., Bess J.W., Jr., Mahal L.K. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat. Chem. Biol. 2009;5:244–250. doi: 10.1038/nchembio.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batista B.S., Eng W.S., Mahal L.K. Identification of a conserved glycan signature for microvesicles. J. Proteome Res. 2011;10:4624–4633. doi: 10.1021/pr200434y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novogrodsky A., Katchalski E. Lymphocyte transformation induced by concanavalin A and its reversion by methyl-α-D-mannopyranoside. Biochim. Biophys. Acta. 1971;228:579–583. doi: 10.1016/0005-2787(71)90064-5. [DOI] [PubMed] [Google Scholar]
- 50.Vader P., Breakefield X.O., Wood M.J. Extracellular vesicles: emerging targets for cancer therapy. Trends Mol. Med. 2014;20:385–393. doi: 10.1016/j.molmed.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshioka Y., Konishi Y., Ochiya T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles. 2013;2:20424. doi: 10.3402/jev.v2i0.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanson P.I., Whiteheart S.W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 53.Yoshimori T., Yamagata F., Ohsumi Y. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell. 2000;11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe R., Lamb R.A. Influenza virus budding does not require a functional AAA+ ATPase, VPS4. Virus Res. 2010;153:58–63. doi: 10.1016/j.virusres.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujita H., Yamanaka M., Himeno M. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J. Cell Sci. 2003;116:401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- 56.Guduric-Fuchs J., O’Connor A., Simpson D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y., Zhang Z. Potential microRNA-mediated oncogenic intercellular communication revealed by pan-cancer analysis. Sci. Rep. 2014;4:7097. doi: 10.1038/srep07097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J., Li S., Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higginbotham J.N., Demory Beckler M., Coffey R.J. Amphiregulin exosomes increase cancer cell invasion. Curr. Biol. 2011;21:779–786. doi: 10.1016/j.cub.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.López-Cobo S., Campos-Silva C., Valés-Gómez M. Glycosyl-Phosphatidyl-Inositol (GPI)-anchors and metalloproteases: their roles in the regulation of exosome composition and NKG2D-mediated immune recognition. Front. Cell Dev. Biol. 2016;4:97. doi: 10.3389/fcell.2016.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aatonen M.T., Ohman T., Siljander P.R. Isolation and characterization of platelet-derived extracellular vesicles. J. Extracell. Vesicles. 2014;3:24692. doi: 10.3402/jev.v3.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soo C.Y., Song Y., Powis S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abache T., Le Naour F., Rubinstein E. The transferrin receptor and the tetraspanin web molecules CD9, CD81, and CD9P-1 are differentially sorted into exosomes after TPA treatment of K562 cells. J. Cell. Biochem. 2007;102:650–664. doi: 10.1002/jcb.21318. [DOI] [PubMed] [Google Scholar]
- 64.Savina A., Furlán M., Colombo M.I. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 65.Akhmanova A., Hammer J.A., 3rd Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rous B.A., Reaves B.J., Luzio J.P. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell. 2002;13:1071–1082. doi: 10.1091/mbc.01-08-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alonso Y Adell M., Migliano S.M., Teis D. ESCRT-III and Vps4: a dynamic multipurpose tool for membrane budding and scission. FEBS J. 2016;283:3288–3302. doi: 10.1111/febs.13688. [DOI] [PubMed] [Google Scholar]
- 68.Matusek T., Wendler F., Thérond P.P. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature. 2014;516:99–103. doi: 10.1038/nature13847. [DOI] [PubMed] [Google Scholar]
- 69.Wehman A.M., Poggioli C., Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 2011;21:1951–1959. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ostrowski M., Carmo N.B., Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 71.Bobrie A., Krumeich S., Théry C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920–4930. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- 72.Hanson P.I., Roth R., Heuser J.E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]