Abstract
Adaptor proteins are a class of cytoplasmic proteins that bind to phosphorylated residues in receptor tyrosine kinases and trigger signaling cascades that control critically important cellular processes, such as cell survival, growth, differentiation, and motility. Here, we seek to characterize the interaction between epidermal growth factor receptor (EGFR) and the cytoplasmic adaptor protein growth factor receptor-bound protein 2 (Grb2) in a cellular context. To do so, we explore the utility of a highly biologically relevant model system, mammalian cells under reversible osmotic stress, and a recently introduced Förster resonance energy transfer microscopy method, fully quantified spectral imaging. We present a method that allows us to quantify the stoichiometry and the association constant of the EGFR-Grb2 binding interaction in the plasma membrane, in the presence and absence of activating ligand. The method that we introduce can have broad utility in membrane protein research, as it can be applied to different membrane protein-cytoplasmic protein pairs.
Introduction
Receptor tyrosine kinase (RTK) activity is critically important for proper human growth and development, and has been implicated in many developmental disorders and cancers (1, 2, 3, 4, 5, 6, 7, 8, 9, 10). RTKs are single-pass membrane receptors that consist of a ligand-binding extracellular (EC) domain, a transmembrane (TM) helix, and a tyrosine kinase intracellular (IC) domain. RTK activation occurs as a result of lateral RTK dimerization and is regulated through ligand binding. Dimerization brings the kinase domains into close proximity, allowing for kinase cross-activation and subsequent phosphorylation of tyrosine residues (1, 2, 3, 4). Ligand binding stabilizes these RTK dimers and induces structural changes in the dimer that increase kinase activity (11, 12, 13, 14). Phosphorylated tyrosines in RTK IC domains serve as docking sites for cytoplasmic proteins, which in turn trigger intracellular signaling cascades that control cell survival, growth, differentiation, and motility (1, 2, 3, 4, 15, 16).
Adaptor proteins are a class of cytoplasmic proteins that bind to phosphorylated residues in RTK IC domains (1, 2, 3, 4). These small proteins are composed of highly specific, modular binding domains, such as the Src homology 2 (SH2) and phosphotyrosine-binding domains. Although they have no inherent activity, they are responsible for mediating many protein-protein interactions (15, 16, 17, 18, 19, 20, 21, 22). Adaptor proteins such as Grb2, Shc, and Nck initiate the recruitment and activation of other cytoplasmic proteins, thereby triggering IC signaling networks like the MAPK, PI3K, PKC, and STAT pathways (1, 2, 3, 4, 16, 23).
The interactions between RTKs and adaptor proteins have been studied extensively (20, 21, 24, 25, 26). These experiments can generally be sorted into two broad categories based on the method of investigation: those that rely on traditional biophysical methods and those that use emerging cell-based techniques. In the first category of experiments (17, 18, 27, 28, 29, 30), fluorescence spectroscopy, Förster resonance energy transfer (FRET), surface plasmon resonance, and isothermal titration calorimetry have all been used to characterize RTK-adaptor protein interactions, yielding thermodynamic and/or kinetic binding parameters. However, these techniques require reduction of the experimental system to short peptides representing the RTKs and the adaptor proteins. Furthermore, synthetic lipids or surface displays of RTK peptides are used to model the plasma membrane, or the experiments are conducted in solution rather than on a two-dimensional surface. In the second category of experiments (31, 32, 33, 34, 35, 36), live cells are used to examine the RTK-adaptor protein interaction. These experiments allow for the study of full-length RTKs and adaptor proteins. They also capture the native physicochemical complexities of both the plasma membrane and the cytoplasm. However, the results typically consist of qualitative observations, rather than quantitative measurements, of adaptor protein recruitment to receptors.
Here, we seek to combine the quantitative power of biophysical techniques with the biological relevance of live cells, with the goal of measuring the thermodynamic association constant between RTKs and adaptor proteins in a biologically relevant context. For this, we explore the utility of a recently introduced FRET microscopy method, fully quantified spectral imaging (FSI) (37), and a highly biologically relevant model system, cells under reversible osmotic stress (38).
Recent experiments have overturned the notion that the membrane is a smooth surface, instead showing that cells have an abundance of extra membrane stored in folds and ruffles (38, 39). This geometry complicates the measurement of two-dimensional protein concentrations in the membrane (37), thus preventing calculations of association constants in the membrane. Reversible osmotic stress “unwrinkles” the plasma membrane of cells (38), yielding a simpler, yet still highly biologically relevant, experimental system that makes it possible to directly measure and quantify protein concentrations and protein-protein interactions. Previously, the thermodynamics of lateral association of RTKs have been examined in cells under reversible osmotic stress using the FSI technique (37, 40, 41, 42, 43). Here, we expand the methodology to quantify adaptor protein binding to RTKs, and we compare our results to previous work.
Additionally, we demonstrate that such quantitative biophysical measurements can further our mechanistic knowledge of RTK signaling. Recent work has suggested that RTKs form dimers in the absence of ligand (1, 11, 12, 13, 44, 45), with Western blots showing that these unliganded dimers are often phosphorylated, but to a lower degree than their liganded counterparts (11, 13, 22, 41, 46, 47). It has been hypothesized that RTKs can transition between inactive and active conformations in the absence of ligand (9, 10, 12, 47, 48), but quantitative biophysical parameters describing such transitions are lacking due to experimental limitations (49, 50). We show that we can measure such parameters in cells under reversible osmotic stress by monitoring the recruitment of adaptor proteins to RTKs in the absence of ligand.
For method development, we focus on the interaction between the RTK epidermal growth factor receptor (EGFR) and the adaptor protein growth factor receptor-bound protein 2 (Grb2). After EGFR dimerization and cross-phosphorylation, Grb2 monomers are recruited to the tail of EGFR and mediate the receptor’s connection to the MAPK signaling pathway. Grb2 is known to exist in a monomer-dimer equilibrium in the cytoplasm, but only Grb2 monomers are believed to bind EGFR (18, 27, 51). This interaction takes place through Grb2’s SH2 domain, which binds to the phosphorylated Y1068 and Y1086 residues of EGFR (2, 9, 10, 18, 20, 27, 29, 32, 51). Of the many possible pairs of receptors and adaptor proteins, we selected EGFR and Grb2 because the recruitment of Grb2 to EGFR after phosphorylation has been well documented (17, 18, 27, 28, 29, 31, 32, 33, 34, 35, 36, 52) and because the interaction can be observed using FRET (32, 34, 53, 54). Here, we measure the thermodynamic association constant for Grb2 monomers binding to EGFR dimers in the presence of ligand, and we gain insight into Grb2 recruitment to the EGFR dimer in the absence of ligand.
Materials and Methods
Plasmids
The full-length EGFR gene in the pSSX vector (55) and the empty pSSX vector were kind gifts from Dr. Daniel Leahy (University of Texas, Austin). The Grb2 gene was a gift from Dr. Jin Zhang (University of California, San Diego). Established molecular cloning techniques were used to generate plasmids coding for each protein labeled with a fluorescent protein at the C-terminus. The fluorescent proteins mTurquoise and yellow fluorescent protein (YFP) were selected because they form a FRET pair. EGFR was labeled with mTurquoise by attaching the fluorophore to the C-terminus of the receptor via a flexible GGS linker. To generate this EGFR plasmid, the genes encoding the linker and the fluorescent protein were inserted between the KpnI and NotI restriction sites of the pSSX vector.
Grb2 was labeled with YFP via a flexible GGS linker. To generate the Grb2 plasmid, genes for the GGS linker and YFP were inserted into the empty pSSX vector, between the XbaI and NotI restriction sites. Next, a Kozak sequence and the gene encoding Grb2 were added between the BamHI and XbaI restriction sites. All plasmids were sequenced to confirm their identity (Genewiz, Fredrick, MD).
Cell culture
Chinese hamster ovary (CHO) cells were used in these experiments, because they do not exhibit endogenous EGFR expression (56). The CHO cells were passed every other day using standard tissue culture techniques. The full culture media consisted of Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Sugar Land, TX) supplemented with 1.8 g/L glucose, 1.5 g/L sodium bicarbonate, 1 mM nonessential amino acids (Thermo Fisher Scientific), and 10% fetal bovine serum (HyClone, Logan, UT). The cells were kept at 37°C in a 5% carbon dioxide incubator.
CHO cells were transfected using FugeneHD (Promega Corporation, Madison, WI), at a ratio of 3 μL of transfection reagent per 1 μg of plasmid. To calibrate the microscope for FSI measurements (37), cells were singly transfected with 8 μg of the pSSX-EGFR-GGS-mTurquoise plasmid or 3 μg of the pSSX-Grb2-GGS-YFP plasmid. For FRET experiments, cells were co-transfected with 6 μg of pSSX-EGFR-GGS-mTurquoise and 2–4 μg of pSSX-Grb2-GGS-YFP. Cells were incubated for 24 h post-transfection.
Twelve hours before the start of an experiment, the full culture media was replaced with starvation culture media (identical to the full culture media, but lacking fetal bovine serum and phenol red). This process removes any factors that might stimulate EGFR.
Preparation of cells under reversible osmotic stress
After 12 h, the starvation culture media was replaced with a hypotonic buffer, thus subjecting the cells to reversible osmotic stress (38). The hypotonic buffer is composed of one part starvation culture media to nine parts deionized water, plus 25 mM HEPES buffer and 1 mg/mL bovine serum albumin (Sigma-Aldrich, St. Louis, MO). In the presence of the hypotonic buffer, CHO cells swell, which unwrinkles the folds of the plasma membrane (37, 38). This is a reversible process (37, 38, 40, 41, 42, 43), and so the thermodynamic reactions under investigation occur in live cells.
For experiments performed in the presence of ligand, the hypotonic buffer was supplemented with 300 nM of the ligand EGF, a saturating quantity of ligand (14, 57). The EGF used in these experiments was a kind gift from Daniel Leahy (University of Texas, Austin).
Before initiating FRET measurements, cells were incubated in the hypotonic buffer for 20 min. This time allowed cells to reach osmotic equilibrium with the hypotonic buffer and allowed the ligand to fully bind to the receptors.
FRET measurements
Here, we utilized the FSI method, a quantitative fluorescence microscopy technique that measures FRET efficiency, donor concentration, and acceptor concentration in selected regions of a cell. The FSI method has been described in detail in previous work (37). Briefly, cells under reversible osmotic stress were imaged using a two-photon microscope (58) equipped with the Aurora Spectral Technologies (Milwaukee, WI) OptiMiS system (59). The OptiMiS system allows us to acquire a full fluorescence spectrum for every pixel of the image. We imaged the cross section of each co-transfected cell twice, once upon excitation of the donor fluorophore and again upon excitation of the acceptor fluorophore. The fluorescence intensity to fluorophore concentration calibration curves were generated by imaging solutions of the purified fluorescent proteins (37). The donor and acceptor fluorophores’ emission spectra were acquired by imaging cells transfected with only the donor or only the acceptor. These calibration measurements established the relationship between fluorophore concentration and fluorescence intensity and allowed for pixel-level deconvolution of fluorescence spectra into donor concentration, acceptor concentration, and FRET efficiency, as described (37).
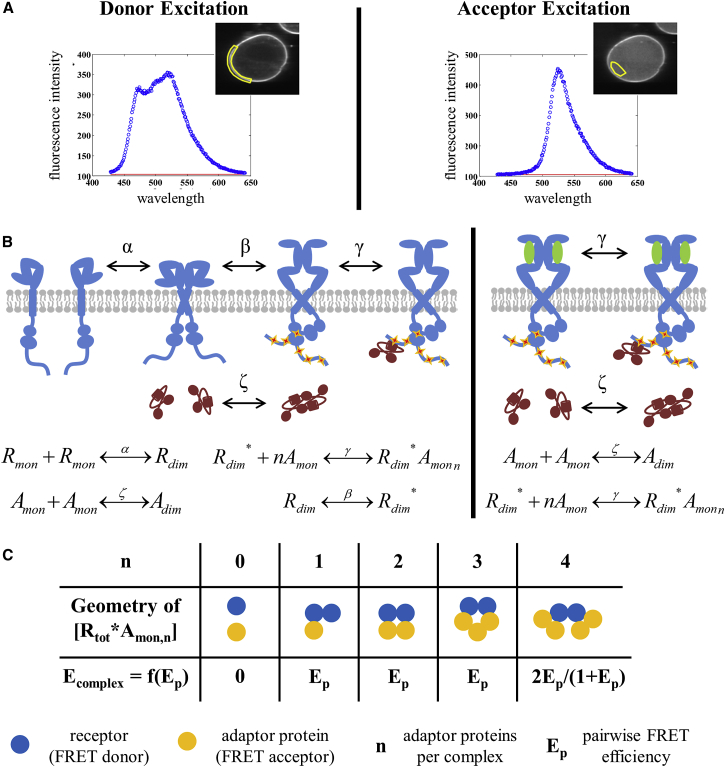
Fig. 1 A shows FSI images and fluorescence spectra for a CHO cell under reversible osmotic stress. In this work, we were interested in the interaction between a membrane protein and a cytoplasmic protein, so two regions in a cell were selected for FSI analysis: one region along the membrane (Fig. 1 A, left) and another region in the cytoplasm (Fig. 1 A, right).
Figure 1.
An overview of the methodology used to quantify the interactions between membrane proteins and cytoplasmic proteins. (A) Representative images (insets) of a CHO cell under reversible osmotic stress and fluorescence emission spectra collected using the FSI technique. FSI yields approximation-free measurement of donor (EGFR-mTurquoise) concentration, acceptor (Grb2-YFP) concentration, and FRET efficiency in selected regions of a cell under reversible osmotic stress (see (37) for details). (B) Cartoon representation of the thermodynamic equilibrium reactions in the presence of saturating concentrations of the EGF ligand (right) and in the absence of ligand (left). The receptors are shown in blue, the ligand in green, the phosphorylation sites in orange, and the adaptor proteins in red. The adaptor protein Grb2 exists in a monomer-dimer equilibrium in the cytoplasm, where the dimerization constant ζ is known (23, 51). EGFR kinase domains are believed to adopt either an active asymmetric dimer configuration that is phosphorylated and capable of interacting with Grb2, or an inactive symmetric configuration (64, 65, 66). In the presence of saturating amounts of ligand, we assume that all receptors exist as active dimers that are either free or bound to Grb2, and we determine the association constant, γ. In the absence of ligand, we assume that the receptors are found as monomers, inactive dimers, active dimers, and RTK dimer-adaptor monomer complexes. We determine the conformational parameter, β, describing the transition from the inactive dimer to the active dimer configuration. (C) Possible oligomer sizes and geometries for the assembly of an RTK dimer with monomeric adaptor proteins. The FRET efficiency, Ecomplex, in each complex of fluorophores is calculated as a function of the pairwise FRET efficiency, Ep, as described previously (74). To see this figure in color, go online.
A model describing RTK-adaptor protein interactions
Here, we present a model to describe the interaction of RTKs with adaptor proteins. This model accounts for RTK monomers and dimers, adaptor protein monomers and dimers, ligand binding, phosphorylation, and adaptor protein binding.
The formation of RTK dimers can be described by a two-state thermodynamic model, as discussed in previous work (44, 45, 60, 61, 62):
| (1) |
where Rmon and Rdim denote RTK monomers and dimers, respectively. The association constant for RTK dimerization, α, can be written as
| (2) |
where [Rmon] and [Rdim] are in two-dimensional units of proteins/μm2 and α has units of μm2/protein.
Some adaptor proteins can form homodimers in the cytoplasm (23, 51, 52, 63). This process can also be described by a two-state thermodynamic model:
| (3) |
where Amon and Adim indicate adaptor protein monomers and dimers, respectively. The association constant for adaptor protein dimerization, ζ, can be written as
| (4) |
where [Amon] and [Adim] are in three-dimensional units of proteins/μm3 and ζ has units of μm3/protein.
The kinase domains in active EGFR dimers are known to adopt an asymmetric configuration (see Fig. 1 B), which allows for tyrosine phosphorylation and subsequent interaction with adaptor proteins and other soluble proteins (2, 20, 64, 65). This active configuration is believed to be adopted in the plasma membrane upon ligand binding (12, 64, 65, 66). It is also believed to be adopted in the absence of ligand, with a finite probability that is unknown (9, 20, 35, 65). We model this process as a conformational change from an inactive dimer (Rdim) to an active dimer :
| (5) |
The equilibrium constant describing this conformational change, β, can be written as
| (6) |
where [Rdim] and are in two-dimensional units of proteins/μm2 and β is dimensionless.
We consider the equilibrium binding reaction of adaptor protein monomers (Amon) to active RTK dimers , following the lead of previous work (18, 24, 67, 68, 69). For the EGFR-Grb2 interactions examined here, we assume that adaptor protein monomers bind to active RTK dimers (23, 51, 52, 63). In the general case of n identical monomeric adaptor proteins binding to one active RTK dimer, we can write
| (7) |
The association constant, γ, can be written as
| (8) |
where and are, respectively, the two-dimensional concentrations of active RTK dimers and RTK dimer-adaptor monomer complexes in the plasma membrane, in units of proteins/μm2. [Amon] is the three-dimensional concentration of adaptor protein monomers in the cell cytoplasm, in units of proteins/μm3. The association constant γ is in units of (μm3/protein)n.
Cells under reversible osmotic stress are known to not undergo endocytosis (70). Hence, we can apply the principle of mass conservation to receptors in the plasma membrane:
| (9) |
where [Rtot] denotes the total concentration of receptors in the membrane. We also apply the principle of mass conservation to adaptor proteins in the cytoplasm:
| (10) |
where [Atot] is the total concentration of adaptor proteins in the cytoplasm.
The biological activity of RTKs is expected to depend on the fractions of receptors that exist as monomers, inactive dimers, active dimers, and RTK dimer-adaptor monomer complexes (see Fig. 1 B). These fractions, denoted as fRmon, fRdim, fRdim∗, and fRdim∗Amon,n, respectively, can be written as
| (11) |
Interpretation of FRET data
If the receptor is labeled with a FRET donor and the adaptor is labeled with a FRET acceptor, then the binding of adaptor protein monomers to a phosphorylated RTK dimer in the active configuration brings the two fluorophores into close proximity and the reaction can be observed via FRET. The concentration of the donor (equivalently, the receptor), the acceptor (equivalently, the adaptor protein), and the FRET efficiency are quantities that can be measured experimentally using FSI (37). The principles of FSI imaging and FSI data analysis are briefly explained in FRET measurements and are illustrated in Fig. 1 A. For a detailed description of the FSI methodology, please see (37).
The FRET efficiency (E), which is defined in terms of the change in donor fluorescence, is directly related to the formation of the RTK dimer-adaptor monomer complexes (71, 72, 73) according to
| (12) |
where Ecomplex is the FRET efficiency in the complex, and [donor] is the concentration of the donor. In our experiments, the receptor is labeled with the donor fluorophore, so we substitute [donor] with [Rtot] in all subsequent equations.
The value of Ecomplex depends on both the geometric configuration and the oligomeric size of the RTK dimer-adaptor monomer complex. The geometric configuration of the RTK dimer-adaptor monomer complex determines d, the inter-fluorophore separation. The dependence of Ecomplex on d can be formalized through consideration of the parameter Ep, which is the pairwise FRET efficiency between a donor and an acceptor in the complex. The oligomeric size of the complex dictates the number of donors and acceptors available to undergo FRET. The dependence of Ecomplex on Ep for different sizes of oligomers has been reported by Raicu and Singh (74) in previous work. In Fig. 1 C, we depict possible configurations of the RTK dimer-adaptor monomer complex (varying n, the number of adaptor monomers per complex) alongside the expected value of Ecomplex as a function of Ep (calculated according to (74)). The pairwise FRET efficiency can be written as
| (13) |
where d is the distance between the donor and acceptor and R0 is the Förster radius of the FRET pair (in this case, 54.5 Å). Since the exact stoichiometry of the RTK dimer-adaptor monomer complex considered here (EGFR-Grb2) is under debate (20, 28, 31, 32, 34, 36), all cases need to be considered to properly interpret experimental data.
The model under saturating ligand conditions
In the presence of ligand, we assume that all receptors exist as ligand-bound, active dimers. Fig. 1 B, right, shows a cartoon representation of the species present and the relationships between them. In this case, the mass conservation of receptors statement (Eq. 9) is reduced to
| (14) |
We then use Eq. 6 to rewrite Eq. 14 in terms of only:
| (15) |
To calculate the concentration of adaptor proteins found as monomers, we first substitute Eq. 4 into Eq. 10 to obtain
| (16) |
Then, we use the quadratic formula (75) to solve Eq. 16 for the concentration of monomeric adaptor proteins:
| (17) |
where [Atot] is measured and ζ must be known from prior work.
Combining Eqs. 15 and 17, we arrive at a single equation to describe the interaction between receptors and adaptor proteins in the presence of ligand:
| (18) |
To facilitate comparison of this model with FRET data, we rearrange Eq. 12 to obtain as a function of FRET efficiency:
| (19) |
and substitute into Eq. 18 to find
| (20) |
The quantities [Rtot], [Atot], and E are experimentally measurable. Nonlinear least squares (76, 77) (we use MATLAB’s “nlinfit”) can be used to fit Eq. 20 to experimental data, yielding the association constant, γ, and the geometrical parameter, Ecomplex, and their 95% confidence intervals. This fitting process can be repeated for the different RTK dimer-adaptor monomer complexes shown in Fig. 1 C. For each possible stoichiometry, the mean-square error (MSE) between the best-fit model and the experimental data can be calculated according to
| (21) |
where k is the number of experimental data points, FRETex,i is the experimentally measured FRET efficiency, and FRETth,i is the theoretical FRET efficiency predicted by solving Eq. 20 for a given [Rtot] and [Atot]. The model that minimizes the MSE provides the best fit to the experimental data, thus yielding the best-fit stoichiometry of the RTK dimer-adaptor monomer complex.
The model in the absence of ligand
In the absence of ligand, we assume the presence of RTK monomers, inactive RTK dimers, active RTK dimers, and RTK dimer-adaptor monomer complexes in the plasma membrane. Fig. 1 B, left, shows a cartoon representation of the species present and the relationships between them. We can rearrange the thermodynamic relations in Eqs. 2, 6, and 8 to relate the various concentrations:
| (22) |
If we substitute these statements into Eq. 9, we can write the equation for mass conservation in terms of RTK dimer-adaptor monomer complex concentration only:
| (23) |
Finally, if we substitute Eqs. 17 and 19 into Eq. 23, we obtain a single equation describing the RTK-adaptor protein interaction in the absence of ligand:
| (24) |
Nonlinear least squares (for instance, MATLAB’s “nlinfit”) (76, 77) is used to fit Eq. 24 to the experimental data to find the conformational parameter β and its 95% confidence interval.
Results
A method to examine the interaction between membrane proteins and cytoplasmic proteins
In this work, we develop and implement a method to quantify the stoichiometry and thermodynamics of membrane protein-cytoplasmic protein interactions. This method includes both a theoretical and an experimental component. We focus on the interaction between RTKs, the second largest class of membrane proteins, and adaptor proteins, a class of cytoplasmic proteins that link membrane proteins to various signaling cascades. To validate the method, we study the interaction between the RTK EGFR and the adaptor protein Grb2.
The theoretical component of the method is a quantitative model describing RTK-adaptor protein interactions. This thermodynamic model considers RTK monomers and dimers, adaptor protein monomers and dimers, ligand binding, phosphorylation, and adaptor protein binding. RTKs form dimers in the membrane, and this process is described using a two-state thermodynamic model (44, 45, 60, 61, 62). EGFR dimers exist in both inactive symmetrical and active asymmetrical configurations (9, 12, 20, 35, 64, 65, 66). The transition between these two dimer configurations is modeled using a single conformational parameter. Finally, the equilibrium between adaptor protein monomers and dimers is also described using a two-state model.
Phosphorylated RTK dimers are capable of interacting with adaptor proteins, which in turn triggers various signaling cascades (2, 20, 64, 65). Grb2 is known to bind EGFR as a monomer (18, 27, 51), so our model examines the equilibrium binding reaction of active RTK dimers and adaptor protein monomers to form complexes. The stoichiometry of the EGFR-Grb2 complex is unknown, so we consider the formation of complexes with different numbers of adaptor protein monomers. A cartoon representation of this thermodynamic model can be found in Fig. 1 B. Please see Materials and Methods for full equations.
The experimental component of our method is a FRET microscopy technique, FSI, that was established recently (37). Specifically, we measure the binding of the adaptor protein Grb2 to the RTK EGFR using FRET. Full-length EGFR is labeled at the C-terminus with mTurquoise (a FRET donor) via a flexible GGS linker. Full-length Grb2 is labeled at the C-terminus with YFP (a FRET acceptor) via a GGS linker. Earlier studies have verified that the interaction between EGFR and Grb2 is observable in this experimental design (32, 53).
CHO cells are co-transfected with the EGFR and Grb2 plasmids. After 24 h, the full culture media is replaced with a limited culture media (called starvation media) that lacks growth factors, to eliminate the presence of any EGFR ligands. Twelve hours later, the medium is replaced again, this time with a hypotonic buffer that consists of one part starvation medium and nine parts water (see Materials and Methods for additional details). This buffer places CHO cells under osmotic stress, causing the cells to swell in a reversible process that unwrinkles the plasma membrane. This allows for accurate measurements of donor concentration, acceptor concentration, and FRET efficiency in selected regions of the membrane (37, 40, 41, 42, 43). To conduct experiments in the presence of ligand, swelling medium is supplemented with 300 nM EGF and 1 mg/mL bovine serum albumin.
After the application of osmotic stress, cells are incubated in the hypotonic buffer for 20 min. Then, the FSI method (37) is used to image and then analyze the fluorescence of individual cells. The cross section of each cell is imaged using a two-photon microscope (58) equipped with the Aurora Spectral Technologies (Milwaukee, WI) OptiMiS system (59). Each cell is imaged twice, once at the excitation of the donor fluorophore and once at the excitation of the acceptor fluorophore (see Fig. 1 A). To calibrate the microscope, the same two images are collected for solutions of known concentrations of fluorescent proteins and for cells transfected with only the donor-labeled protein or only the acceptor-labeled protein. The calibration directly correlates fluorescence intensity to fluorophore concentration and specifies the emission spectra of the donor and acceptor fluorophores, thereby enabling deconvolution of fluorescence spectra into donor, acceptor, and FRET contributions. In each cell, we make measurements in two regions: the plasma membrane and the cytoplasm. We select a region of homogeneous plasma membrane and we measure 1) the concentration of the donor (equivalently, the receptor) and 2) the FRET efficiency. We also select a region of homogeneous cytoplasm and measure 3) the concentration of the acceptor (equivalently, the adaptor protein). Together, these three measurements correspond to one data point, as shown in both Figs. 2 B and 3 B. On average, two data points are acquired per cell. See Fig. 1 A for a representation of plasma membrane and cytoplasm regions, as well as the fluorescence spectra.
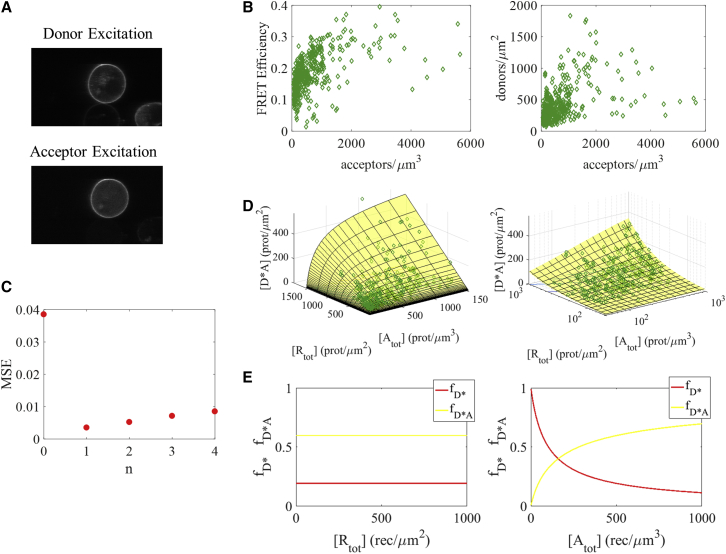
Figure 2.
FRET data for the interaction between EGFR and Grb2, under saturating ligand conditions. (A) A representative image of a CHO cell under reversible osmotic stress, in the presence of EGF. Fluorescence in the donor excitation image indicates the location of EGFR; fluorescence in the acceptor excitation image denotes Grb2. (B) In the left-hand plot, FRET efficiency in the plasma membrane is shown as a function of Grb2 (acceptor) concentration in the cytoplasm. The right-hand plot shows EGFR (donor) concentration in the membrane versus Grb2 (acceptor) concentration in the cytoplasm. (C) The MSE for fits of Eq. 20 to the experimental data when the number of monomeric adaptor proteins, n, is varied between 0 and 4. The model with one Grb2 monomer per EGFR dimer gives the best fit. (D) The theoretical and the experimental concentrations of the RTK dimer-adaptor monomer complex, [Rtot∗Amon,n], are calculated using Eqs. 18 and 19, respectively. The best-fit model (yellow surface) and experimental data (green diamonds) are plotted as a function of both total receptor concentration, [Rtot], and adaptor protein concentration, [Atot], on linear (left) and semi-log (right) scales. (E) The fraction of receptors found in active dimers with no adaptors bound (red line) and in RTK dimer-adaptor monomer complexes (yellow line) for the best-fit model as a function of total receptor concentration (left) and adaptor protein concentration (right). To see this figure in color, go online.
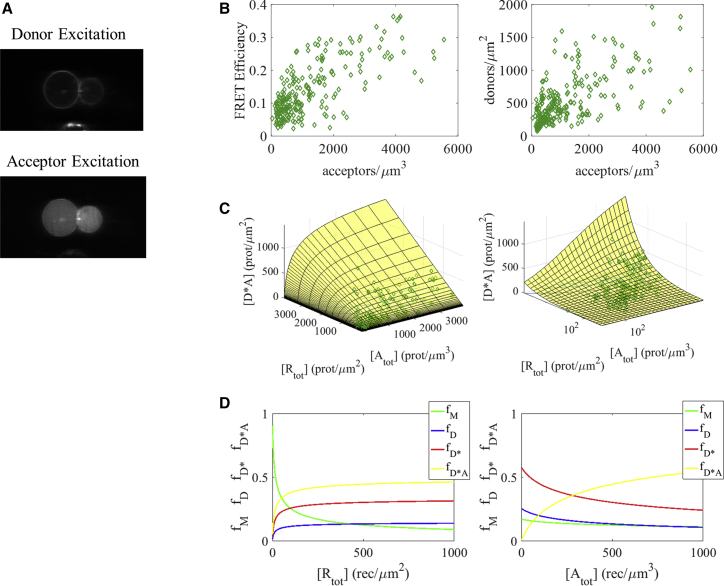
Figure 3.
FRET data for the interaction between EGFR and Grb2 in the absence of ligand. (A) A representative image of a CHO cell under reversible osmotic stress, in the absence of EGF. Fluorescence in the donor excitation image indicates the location of EGFR; fluorescence in the acceptor excitation image shows Grb2. (B) In the left-hand plot, FRET efficiency in a plasma membrane region is shown as a function of Grb2 (acceptor) concentration in the cytoplasm. The right-hand plot shows EGFR (donor) concentration in the membrane versus Grb2 (acceptor) concentration in the cytoplasm. (C) We fit Eq. 23 to the experimental data to determine the parameter, β, describing the conformational switch from inactive symmetric RTK dimers to active asymmetric RTK dimers (see Fig. 1B). The theoretical and experimental RTK dimer-adaptor monomer complex concentrations are calculated using Eqs. 23 and 19, respectively. The best fit model (yellow surface) is plotted alongside experimental data (green diamonds) as a function of both total receptor concentration, [Rtot], and adaptor protein concentration, [Atot], on both a linear (left) and semi-log (right) scale. (D) The fractions of receptors found as monomers, in dimers in an inactive configuration, dimers in an active configuration, and active RTK dimer-adaptor monomer complexes are graphed as functions of total receptor concentration (left) and adaptor protein concentration (right). To see this figure in color, go online.
The thermodynamics of the binding reaction between EGFR and Grb2 can be quantified
In Fig. 2, we show the results for the binding of Grb2 to EGFR in the presence of ligand. Data are collected in the presence of saturating ligand concentration, 300 nM EGF. This concentration exceeds the EGF-EGFR binding coefficient (reported as ∼2 nM (14, 57)) by more than two orders of magnitude. Under these conditions, all EGFR molecules are expected to exist as ligand-bound dimers. Indeed, as demonstrated by Needham et al. (78), all EGFR molecules are found as ligand-bound dimers in CHO cells at this high EGF concentration. The high ligand concentration therefore allows us to control the association state of EGFR in the plasma membrane, allowing quantitative data interpretation.
Fig. 2 A shows typical images of a cell under reversible osmotic stress in the presence of 300 nM EGF, when the donor fluorophore (top) or the acceptor fluorophore (bottom) is excited. Fig. 2 B displays the experimentally measured receptor concentration, adaptor protein concentration, and FRET efficiency. Each data point (green diamonds) corresponds to a membrane region and nearby cytoplasmic region pair. A total of 475 data points were acquired in the presence of ligand. In the left-hand plot, the FRET efficiency is plotted as a function of acceptor concentration. The steep dependence of the FRET efficiency on acceptor concentration is indicative of specific interactions. In the right-hand plot, we graph donor concentration versus acceptor concentration. The variation in donor concentration explains the spread of measured FRET efficiencies observed for a given acceptor concentration.
We fit Eq. 20 to the experimental data while varying the number of adaptor monomers per RTK dimer-adaptor monomer complex, n. The association constant for Grb2 dimerization was previously measured as (2.4 ± 0.8) × 10−3 μm3/protein (23, 51); we use that value as the parameter ζ. Each fit to Eq. 20 yields the association constant between active EGFR dimers and Grb2 monomers, γ, and the geometrical FRET parameter, Ecomplex, for a particular n. We calculate the MSE for each n according to Eq. 21 and we plot the results in Fig. 2 C. The minimum MSE corresponds to n = 1, which suggests that there is one Grb2 monomer bound to each EGFR dimer. For the case of n = 1, the calculated association constant between EGFR dimers and Grb2 monomers is γ = (6.3 ± 0.8) × 10−3 μm3/protein. The FRET efficiency in the RTK dimer-adaptor monomer complex is Ecomplex = 0.77 ± 0.04.
In Fig. 2 D, we compare the theoretical and experimental concentrations of the RTK dimer-adaptor monomer complexes. We plot the best-fit model (yellow surface, calculated using Eq. 18) and the experimental data (green diamonds, calculated according to Eq. 19) on a linear scale in the left-hand plot and on a semi-log scale in the right-hand plot.
In Fig. 2 E, the best-fit values are used to show how the fraction of active RTK dimers with no Grb2 bound (red) and the fraction of RTK dimer-adaptor monomer complexes (yellow) depend on the total receptor concentration (left-hand plot) and on the adaptor protein concentration (right-hand plot). In the left-hand plot, the adaptor protein concentration, [Atot], is fixed at 500 proteins/μm3 and the total receptor protein concentration, [Rtot], is varied between 0 and 1000 receptors/μm2. Neither fraction depends on total receptor concentration. In the right-hand plot, [Rtot] is fixed at 500 proteins/μm3 and [Atot] is varied between 0 and 1000 receptors/μm2. As expected, the fraction of receptors found in dimer-adaptor monomer complexes increases (accompanied by a decrease in the fraction of active dimers with no Grb2 bound) as the concentration of the adaptor protein increases.
A fraction of unliganded EGFR dimers are in the active configuration and bind Grb2
In Fig. 3, we present data collected for EGFR-Grb2 interactions in the absence of ligand. A total of 249 data points were acquired in the absence of ligand. Fig. 3 A shows typical images of a cell under reversible osmotic stress in the absence of EGF when the donor (top) or the acceptor (bottom) is excited. Fig. 3 B displays the experimentally measured receptor concentration, adaptor protein concentration, and FRET efficiency. The left-hand plot shows the FRET efficiency as a function of acceptor concentration. The FRET efficiency increases with increasing acceptor concentration, but the dependence is less pronounced than that observed in the presence of ligand. The right-hand plot shows the donor concentration versus acceptor concentration. Each green diamond corresponds to a membrane region and nearby cytoplasmic region pair.
In the absence of ligand, we assume that the number of adaptor protein monomers per complex (n), the association constant between dimers and adaptor monomers (γ), and the FRET efficiency in the complex (Ecomplex) are the same as in the liganded case (n = 1 protein, γ = 6.3 × 10−3 μm3/protein, and Ecomplex = 0.77), whereas β, the conformational parameter describing the relationship between inactive and active RTK dimers, is unknown. The EGFR association constant in the absence of ligand, α, has been measured previously as (8.8 ± 0.7) × 10−3 μm2/receptor by Macdonald and Pike (79). Using this value, as well as the Grb2 association constant ζ =(2.4 ± 0.8) × 10−3 μm3/protein, we fit Eq. 24 to the experimental data, thus obtaining β = 2.3 ± 0.7. This means that just over two-thirds of the unliganded EGFR dimers that are not bound to Grb2 exist in the active configuration.
In Fig. 3 C, the best-fit model (yellow surface) for the concentration of RTK dimer-adaptor monomer complexes, [Rdim∗Amon,n], is plotted alongside experimental data (green diamonds) on a linear scale (left) and a semi-log scale (right). The theoretical model and experimental data points are calculated using Eqs. 23 and 19, respectively.
Fig. 3 D shows how the fractions of receptors found as monomers (green line), inactive dimers (blue line), dimers in the active configuration with no Grb2 bound (red line), and RTK dimer-adaptor monomer complexes (yellow line) vary as a function of the total receptor concentration, [Rtot], and the adaptor protein concentration, [Atot]. In Fig. 3 D, left, the adaptor protein concentration is fixed at 500 proteins/μm2 while [Rtot] varies, and in Fig. 3 D, right, the total receptor concentration is fixed at 500 receptors/μm2 while [Atot] varies. The fraction of dimer-adaptor monomer complexes increases with both [Atot] and [Rtot] and can reach a significant fraction even in the absence of ligand.
Discussion
In this work, we present a methodology for the study of the interaction between RTKs and adaptor proteins. We perform measurements in live cells under reversible osmotic stress. CHO cells are our cell line of choice because they do not natively express EGFR (56). However, they do express Grb2 (56), and we assume that endogenous Grb2 concentrations are negligible relative to transiently overexpressed YFP-labeled Grb2 concentrations. The plasma membrane of CHO cells under reversible osmotic stress lacks its native folds and ruffles, thereby allowing quantitative measurements of protein concentrations and FRET efficiencies. Earlier work with live cells under reversible osmotic stress has shown that the resultant flattening of the plasma membrane does not involve clathrin-coated pits or the actin cortex (38), and does not alter the lateral interactions between membrane receptors (as assessed by FRET (37)). Hence, we expect receptor interactions to be largely unaltered. Additionally, since endocytosis is inhibited in cells under osmotic stress (70), we expect mass conservation of receptors in the membrane, even upon ligand treatment. Thus, cells under reversible osmotic stress appear to be a highly relevant model system that allows for quantitative biophysical measurements and subsequent comparison of data with thermodynamic models. Cell signaling in these cells should be fully characterized in the future to better understand the relevance and the limitations of quantitative biophysical investigations in this model system.
The thermodynamic model developed and implemented in this work accounts for RTK monomers and dimers, adaptor protein monomers and dimers, ligand binding, phosphorylation, and adaptor protein binding, and it relies on several assumptions. Since we use this model to study the interaction between EGFR and Grb2, we incorporate prior knowledge about both proteins into our theoretical model. In our model, EGFR forms only dimers, not higher-order oligomers. Although recent studies have shown that ligand binding can induce EGFR oligomerization (78, 80), EGFR exists as a dimer for the ligand concentration used in our study (78). Grb2 exists as both monomers and dimers in the cytoplasm, but only Grb2 monomers interact with EGFR. Although both Grb2 monomers and Grb2 dimers have been shown to bind to other active RTK dimers (22, 23, 51), the literature consensus is that Grb2 binds EGFR dimers in the monomeric form (18, 27, 51). The binding of EGF to unliganded EGFR dimers is known to induce a conformational change in the kinase domain, which activates the dimer and initiates interactions with cytoplasmic proteins. We describe this switch from an inactive to an active kinase conformation with a single parameter, β.
Additionally, our thermodynamic model makes several assumptions about the EGFR-Grb2 interaction. First, we assume that only one model of EGFR:Grb2 stoichiometry applies over the broad EGFR and Grb2 concentration ranges covered in these experiments. Second, in the presence of saturating amounts of ligand, we assume that receptors are found only as active RTK dimers and RTK dimer-adaptor monomer complexes. Third, we assume that the binding reaction between active RTK dimers and adaptor protein monomers is independent of ligand (that is, that n, γ, and Ecomplex are identical in the absence and presence of ligand).
Although it is known that the SH2 domain of Grb2 binds to the phosphorylated Y1068 and Y1086 residues of EGFR (2, 9, 10, 18, 20, 27, 29, 32), the stoichiometry of this reaction is debated in the literature (20, 28, 31, 32, 34, 36), with estimates ranging between one and four Grb2 monomers per EGFR dimer. We fit models with one, two, three, and four Grb2 monomers per EGFR dimer (Fig. 1 C) to our experimental data (Fig. 2), and we calculate the MSE to assess how well each model fits the data. The model with one Grb2 monomer per EGFR dimer produces the best fit to the experimental data. This finding is in agreement with recent articles suggesting that EGFR dimers are incompletely phosphorylated due to steric clashes (78, 80). The incomplete phosphorylation of EGFR dimers rationalizes the observation that only one Grb2 monomer binds to the EGFR dimer.
The association constant between an active EGFR dimer and a Grb2 monomer is measured as γ = (6.3 ± 0.8) × 10−3 μm3/protein, which is equivalent to 263 ± 34 nM. Using traditional biophysical techniques and peptide/lipid model systems, this dissociation constant was previously measured as 30–713 nM, with most values on the order of 102 nM (18, 20, 27, 28, 29, 30). Our result is similar to these previous measurements, verifying the methodology that we present and implement here. We also report Ecomplex = 0.77 ± 0.04, which is the FRET efficiency in an RTK dimer-adaptor monomer complex. Using the measured value of Ecomplex and Eqs. 12 and 13, and assuming free fluorophore rotation, the separation of the fluorescent proteins in the EGFR-Grb2 complex can be estimated as 44.6 ± 1.7 Å.
Furthermore, we examine the EGFR-Grb2 binding interaction in the absence of ligand. There is a consensus in the literature that some unliganded EGFR dimers may adopt the active configuration (9, 20, 35), but there are very few quantitative estimates for the relative populations of inactive and active dimers. By following the recruitment of the adaptor protein Grb2 to EGFR via FRET, we calculate the conformational parameter describing the equilibrium between the inactive and the active EGFR dimer configurations as β = 2.3 ± 0.7. This value can be used to predict EGFR activity in the absence of ligand and contributes to the growing mechanistic understanding of the EGFR activation process.
Many RTKs are over-expressed and over-active in tumor tissue, and adaptor proteins are the direct link between receptors and biological activity. Molecules that disrupt the interaction between RTKs and adaptor proteins can decrease aberrant RTK activity (19, 35). In future work, the method presented here can be used to examine the action of such inhibitors, and to screen for new inhibitors. Furthermore, the method can be applied to different receptor-adaptor protein pairs, and the assumptions of the model can be refined as our knowledge of signaling molecules expands. Thus, the method can have broad utility in membrane protein research.
Author Contributions
N.D.P. designed the study, performed the experiments, analyzed the data, developed the model, and wrote the article. K.H. designed the study, developed the model, and wrote the article.
Acknowledgments
We thank Dr. Daniel Leahy for providing the EGFR gene and EGF, and Dr. Jin Zhang for providing the Grb2 gene. We thank Dr. Patrick Byrne, Dr. Kirill Gorshkov, Michael Paul, and Fozia Ahmed for many useful discussions. We thank Dr. Christopher King for developing and implementing the imaging and analysis tools used in this manuscript.
This work was supported by National Institutes of Health grant GM68619.
Editor: Lukas Tamm.
References
- 1.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemmon M.A., Schlessinger J., Ferguson K.M. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2014;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger J. Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 2014;6:a008912. doi: 10.1101/cshperspect.a008912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eswarakumar V.P., Lax I., Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Cohen M.M., Jr. Some chondrodysplasias with short limbs: molecular perspectives. Am. J. Med. Genet. 2002;112:304–313. doi: 10.1002/ajmg.10780. [DOI] [PubMed] [Google Scholar]
- 7.Cohen M.M., Jr. The new bone biology: pathologic, molecular, and clinical correlates. Am. J. Med. Genet. A. 2006;140:2646–2706. doi: 10.1002/ajmg.a.31368. [DOI] [PubMed] [Google Scholar]
- 8.Gschwind A., Fischer O.M., Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 9.Hynes N.E., Lane H.A. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 10.Olayioye M.A., Neve R.M., Hynes N.E. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarabipour S., Hristova K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016;7:10262. doi: 10.1038/ncomms10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung I., Akita R., Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 13.Sarabipour S., Ballmer-Hofer K., Hristova K. VEGFR-2 conformational switch in response to ligand binding. eLife. 2016;5:e13876. doi: 10.7554/eLife.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macdonald-Obermann J.L., Pike L.J. Different epidermal growth factor (EGF) receptor ligands show distinct kinetics and biased or partial agonism for homodimer and heterodimer formation. J. Biol. Chem. 2014;289:26178–26188. doi: 10.1074/jbc.M114.586826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuriyan J., Cowburn D. Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 16.Schlessinger J., Lemmon M.A. SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE. 2003;2003:RE12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- 17.Rahuel J., Gay B., Grütter M.G. Structural basis for specificity of Grb2-SH2 revealed by a novel ligand binding mode. Nat. Struct. Biol. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- 18.Lemmon M.A., Ladbury J.E., Schlessinger J. Independent binding of peptide ligands to the SH2 and SH3 domains of Grb2. J. Biol. Chem. 1994;269:31653–31658. [PubMed] [Google Scholar]
- 19.Kraskouskaya D., Duodu E., Gunning P.T. Progress towards the development of SH2 domain inhibitors. Chem. Soc. Rev. 2013;42:3337–3370. doi: 10.1039/c3cs35449k. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh M.Y., Yang S., Wilson B.S. Spatio-temporal modeling of signaling protein recruitment to EGFR. BMC Syst. Biol. 2010;4:57. doi: 10.1186/1752-0509-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladbury J.E., Arold S. Searching for specificity in SH domains. Chem. Biol. 2000;7:R3–R8. doi: 10.1016/s1074-5521(00)00067-3. [DOI] [PubMed] [Google Scholar]
- 22.Belov A.A., Mohammadi M. Grb2, a double-edged sword of receptor tyrosine kinase signaling. Sci. Signal. 2012;5:pe49. doi: 10.1126/scisignal.2003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C.C., Melo F.A., Ladbury J.E. Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell. 2012;149:1514–1524. doi: 10.1016/j.cell.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 24.Day Y.S., Baird C.L., Myszka D.G. Direct comparison of binding equilibrium, thermodynamic, and rate constants determined by surface- and solution-based biophysical methods. Protein Sci. 2002;11:1017–1025. doi: 10.1110/ps.4330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Songyang Z., Shoelson S.E., Lechleider R.J. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 26.Waksman G., Kominos D., Kuriyan J. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature. 1992;358:646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 27.Chook Y.M., Gish G.D., Pawson T. The Grb2-mSos1 complex binds phosphopeptides with higher affinity than Grb2. J. Biol. Chem. 1996;271:30472–30478. doi: 10.1074/jbc.271.48.30472. [DOI] [PubMed] [Google Scholar]
- 28.Morimatsu M., Takagi H., Sako Y. Multiple-state reactions between the epidermal growth factor receptor and Grb2 as observed by using single-molecule analysis. Proc. Natl. Acad. Sci. USA. 2007;104:18013–18018. doi: 10.1073/pnas.0701330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batzer A.G., Rotin D., Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol. Cell. Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones R.B., Gordus A., MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 31.Fortian A., Sorkin A. Live-cell fluorescence imaging reveals high stoichiometry of Grb2 binding to the EGF receptor sustained during endocytosis. J. Cell Sci. 2014;127:432–444. doi: 10.1242/jcs.137786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorkin A., McClure M., Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr. Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 33.Ayoub M.A., See H.B., Pfleger K.D. Profiling epidermal growth factor receptor and heregulin receptor 3 heteromerization using receptor tyrosine kinase heteromer investigation technology. PLoS One. 2013;8:e64672. doi: 10.1371/journal.pone.0064672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozer N., Barua D., Clayton A.H. Recruitment of the adaptor protein Grb2 to EGFR tetramers. Biochemistry. 2014;53:2594–2604. doi: 10.1021/bi500182x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanzerstorfer P., Borgmann D., Weghuber J. Quantification and kinetic analysis of Grb2-EGFR interaction on micro-patterned surfaces for the characterization of EGFR-modulating substances. PLoS One. 2014;9:e92151. doi: 10.1371/journal.pone.0092151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh D., Ogiue-Ikeda M., Yu J. Fast rebinding increases dwell time of Src homology 2 (SH2)-containing proteins near the plasma membrane. Proc. Natl. Acad. Sci. USA. 2012;109:14024–14029. doi: 10.1073/pnas.1203397109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King C., Stoneman M., Hristova K. Fully quantified spectral imaging reveals in vivo membrane protein interactions. Integr. Biol. (Camb.) 2016;8:216–229. doi: 10.1039/c5ib00202h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinha B., Köster D., Nassoy P. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 2011;144:402–413. doi: 10.1016/j.cell.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adler J., Shevchuk A.I., Parmryd I. Plasma membrane topography and interpretation of single-particle tracks. Nat. Methods. 2010;7:170–171. doi: 10.1038/nmeth0310-170. [DOI] [PubMed] [Google Scholar]
- 40.Singh D.R., Ahmed F., Hristova K. EphA2 receptor unliganded dimers suppress EphA2 pro-tumorigenic signaling. J. Biol. Chem. 2015;290:27271–27279. doi: 10.1074/jbc.M115.676866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh D.R., Cao Q., Hristova K. Unliganded EphA3 dimerization promoted by the SAM domain. Biochem. J. 2015;471:101–109. doi: 10.1042/BJ20150433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh D.R., Ahmed F., Hristova K. The SAM domain inhibits EphA2 interactions in the plasma membrane. Biochim. Biophys. Acta. 2017;1864:31–38. doi: 10.1016/j.bbamcr.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh D.R., Pasquale E.B., Hristova K. A small peptide promotes EphA2 kinase-dependent signaling by stabilizing EphA2 dimers. Biochim. Biophys. Acta. 2016;1860:1922–1928. doi: 10.1016/j.bbagen.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Del Piccolo N., Sarabipour S., Hristova K. A new method to study heterodimerization of membrane proteins and its application to fibroblast growth factor receptors. J. Biol. Chem. 2017;292:1288–1301. doi: 10.1074/jbc.M116.755777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del Piccolo N., Placone J., Hristova K. Effect of thanatophoric dysplasia type I mutations on FGFR3 dimerization. Biophys. J. 2015;108:272–278. doi: 10.1016/j.bpj.2014.11.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He L., Shobnam N., Hristova K. FGFR3 heterodimerization in achondroplasia, the most common form of human dwarfism. J. Biol. Chem. 2011;286:13272–13281. doi: 10.1074/jbc.M110.205583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moriki T., Maruyama H., Maruyama I.N. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 48.He L., Hristova K. Physical-chemical principles underlying RTK activation, and their implications for human disease. Biochim. Biophys. Acta. 2012;1818:995–1005. doi: 10.1016/j.bbamem.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Heijne G. A day in the life of Dr K. or how i learned to stop worrying and love lysozyme: a tragedy in six acts. J. Mol. Biol. 1999;293:367–379. doi: 10.1006/jmbi.1999.2998. [DOI] [PubMed] [Google Scholar]
- 50.Hong H., Joh N.H., Tamm L.K. Methods for measuring the thermodynamic stability of membrane proteins. Methods Enzymol. 2009;455:213–236. doi: 10.1016/S0076-6879(08)04208-0. [DOI] [PubMed] [Google Scholar]
- 51.Ahmed Z., Timsah Z., Ladbury J.E. Grb2 monomer-dimer equilibrium determines normal versus oncogenic function. Nat. Commun. 2015;6:7354. doi: 10.1038/ncomms8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maignan S., Guilloteau J.P., Ducruix A. Crystal structure of the mammalian Grb2 adaptor. Science. 1995;268:291–293. doi: 10.1126/science.7716522. [DOI] [PubMed] [Google Scholar]
- 53.Poland S.P., Krstajić N., Ameer-Beg S.M. A high speed multifocal multiphoton fluorescence lifetime imaging microscope for live-cell FRET imaging. Biomed. Opt. Express. 2015;6:277–296. doi: 10.1364/BOE.6.000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turk H.F., Barhoumi R., Chapkin R.S. Alteration of EGFR spatiotemporal dynamics suppresses signal transduction. PLoS One. 2012;7:e39682. doi: 10.1371/journal.pone.0039682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu P., Cleveland T.E., 4th, Leahy D.J. A single ligand is sufficient to activate EGFR dimers. Proc. Natl. Acad. Sci. USA. 2012;109:10861–10866. doi: 10.1073/pnas.1201114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baycin-Hizal D., Tabb D.L., Betenbaugh M. Proteomic analysis of Chinese hamster ovary cells. J. Proteome Res. 2012;11:5265–5276. doi: 10.1021/pr300476w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klein P., Mattoon D., Schlessinger J. A structure-based model for ligand binding and dimerization of EGF receptors. Proc. Natl. Acad. Sci. USA. 2004;101:929–934. doi: 10.1073/pnas.0307285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lichtman J.W., Conchello J.A. Fluorescence microscopy. Nat. Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 59.Biener G., Stoneman M.R., Raicu V. Development and experimental testing of an optical micro-spectroscopic technique incorporating true line-scan excitation. Int. J. Mol. Sci. 2013;15:261–276. doi: 10.3390/ijms15010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L., Novicky L., Hristova K. Measuring the energetics of membrane protein dimerization in mammalian membranes. J. Am. Chem. Soc. 2010;132:3628–3635. doi: 10.1021/ja910692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarabipour S., Del Piccolo N., Hristova K. Characterization of membrane protein interactions in plasma membrane derived vesicles with quantitative imaging Förster resonance energy transfer. Acc. Chem. Res. 2015;48:2262–2269. doi: 10.1021/acs.accounts.5b00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merzlyakov M., Chen L., Hristova K. Studies of receptor tyrosine kinase transmembrane domain interactions: the EmEx-FRET method. J. Membr. Biol. 2007;215:93–103. doi: 10.1007/s00232-007-9009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDonald C.B., Seldeen K.L., Farooq A. Grb2 adaptor undergoes conformational change upon dimerization. Arch. Biochem. Biophys. 2008;475:25–35. doi: 10.1016/j.abb.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 64.Jura N., Endres N.F., Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X., Gureasko J., Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 66.Mi L.Z., Lu C., Springer T.A. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat. Struct. Mol. Biol. 2011;18:984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Devaux P.F., Seigneuret M. Specificity of lipid-protein interactions as determined by spectroscopic techniques. Biochim. Biophys. Acta. 1985;822:63–125. doi: 10.1016/0304-4157(85)90004-8. [DOI] [PubMed] [Google Scholar]
- 68.Smith E.M., Hennen J., Mueller J.D. In situ quantification of protein binding to the plasma membrane. Biophys. J. 2015;108:2648–2657. doi: 10.1016/j.bpj.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dill K.A., Bromberg S. Garland Science; New York: 2003. Molecular Driving Forces: Statistical Thermodynamics in Chemistry and Biology. [Google Scholar]
- 70.Rauch C., Farge E. Endocytosis switch controlled by transmembrane osmotic pressure and phospholipid number asymmetry. Biophys. J. 2000;78:3036–3047. doi: 10.1016/S0006-3495(00)76842-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Merzlyakov M., You M., Hristova K. Transmembrane helix heterodimerization in lipid bilayers: probing the energetics behind autosomal dominant growth disorders. J. Mol. Biol. 2006;358:1–7. doi: 10.1016/j.jmb.2006.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li E., You M., Hristova K. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Förster resonance energy transfer suggest weak interactions between fibroblast growth factor receptor 3 (FGFR3) transmembrane domains in the absence of extracellular domains and ligands. Biochemistry. 2005;44:352–360. doi: 10.1021/bi048480k. [DOI] [PubMed] [Google Scholar]
- 73.You M., Li E., Hristova K. Forster resonance energy transfer in liposomes: measurements of transmembrane helix dimerization in the native bilayer environment. Anal. Biochem. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raicu V., Singh D.R. FRET spectrometry: a new tool for the determination of protein quaternary structure in living cells. Biophys. J. 2013;105:1937–1945. doi: 10.1016/j.bpj.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Press W.H., Teukolsky S.A., Flannery B.P. Cambridge University Press; Cambridge, United Kingdom: 2007. Numerical Recipes, 3rd Edition: The Art of Scientific Computing. [Google Scholar]
- 76.Johnson M.L., Faunt L.M. Parameter estimation by least-squares methods. Methods Enzymol. 1992;210:1–37. doi: 10.1016/0076-6879(92)10003-v. [DOI] [PubMed] [Google Scholar]
- 77.Motulsky H.J., Ransnas L.A. Fitting curves to data using nonlinear regression: a practical and nonmathematical review. FASEB J. 1987;1:365–374. [PubMed] [Google Scholar]
- 78.Needham S.R., Roberts S.K., Martin-Fernandez M.L. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat. Commun. 2016;7:13307. doi: 10.1038/ncomms13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Macdonald J.L., Pike L.J. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc. Natl. Acad. Sci. USA. 2008;105:112–117. doi: 10.1073/pnas.0707080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang Y., Bharill S., Kuriyan J. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife. 2016;5:e14107. doi: 10.7554/eLife.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]