Abstract
Anthropogenic origin pollutants including pesticides, heavy metals, pharmaceuticals and industry chemicals impose many risks to human health and environment and bioremediation has been considered the strategy of choice to reduce the risk of hazardous chemicals. In the current study, we aimed to screen and characterize mycobacteria from the diverse range of Iranian aquatic and terrestrial ecosystems with harsh and unfavorable environmental conditions that can be utilized for biodegradation of target pollutants. Mycobacteria were isolated from a collection of 90 environmental samples and identified to the species level using conventional microbiological and molecular methods including the PCR amplification of hsp65 and sequence analysis of, 16S rRNA genetic markers. The growth rate of the isolates in presence of pollutants, chromatography, Gibbs and turbidometric methods were used to assess their biodegradation activity. A total of 39 mycobacterial isolates (43.3%) were recovered from 90 samples that belonged to 21 various species consisting of M. fortuitum; 6 isolates, M. flavescens and M. paragordonae; 4 isolates each, M. monacense, M. fredriksbergense and M. aurum; 2 isolates each, 7 single isolates of M. conceptionense, M. porcinum, M. simiae, M. celeriflavum, M. novocastrense, M. neoaurum, M. obuense and 12 isolates that belonged to 8 unknown potentially novel mycobacterial species. The isolates were categorized in three groups based on their bioremediation activity, i.e., 5 (12.8%) organisms without biodegradation activity, 20 (51.2%) organisms with previously reported biodegradation activity, and 14 (35.9%) organisms that showed biodegradation activity but not previously reported. Our results showed that the Iranian ecosystems harbor a good reservoir of diverse mycobacterial species with biodegrading potentiality for neutralizing environmental chemical pollutants.
Keywords: Mycobacterium, Bioremediation, 16S rRNA sequencing, Phylogeny
Introduction
During past decades and with the advances in science and technology various chemicals and synthetic products such as petroleum derivatives like petrochemicals and plastic, insecticides and herbicides, radioactive substances and many other similar materials were increasingly introduced into the environment (Eisenbud and Gesell 1997; El-Shahawi et al. 2010; Petry et al. 1996; Samanta et al. 2002; White et al. 2010). Most of these contaminants are of anthropogenic origin and derived from industrial effluent discharges, point contaminant spills, and diffuse agricultural sources (Ritter et al. 2002; Schwarzenbach et al. 2010). Once in the environment, contaminants can be considered as a source for toxicity and pose adverse effect on the health of living organisms (Mastrangelo et al. 1996; Oliva et al. 2001; Reigart 2009; Thompson et al. 2009). In recent years, numerous chemical and physical methods such as dredging and incineration, evaporation, light oxidation, chemical oxidation, adsorption and leaching of soil particles have been applied to decompose and recycle these materials (Arias-Estévez et al. 2008; Perelo 2010; Schaer et al. 2001). However, these approaches are not cost effective and ecofriendly and may cause further damage to the environment by producing and emitting secondary pollutants and in some other cases dispersing pollutant agents make them much more bioavailable and increases their toxicity risk (Khan et al. 2004; Mulligan et al. 2001; Virkutyte et al. 2002). Finding an ecofriendly and rapid degradation approach for these hazardous materials could make a change in promoting public and environmental health. Bioremediation is one of these methods that has attracted much interest amongst scientists and environmentalists for degradation and neutralization of pollutants. Bioremediation is a process that uses organisms, mostly microorganisms and plants, to degrade, reduce toxicity or detoxify waste products and pollutants (Kumar et al. 2011). Therefore, the most important step in bioremediation process is the isolation and characterization of microorganisms capable of decontamination of a particular or a group of pollutants (Adams et al. 2015).
Numerous soil and aquatic bacterial species can breakdown and consume pollutants as a sole carbon and/or energy source, such as Pseudomonas, Burkholderia, Bacillus, Polaromonas, Sphingomonas genera and members of actinomycetes such as Mycobacterium, Rhodococcus and Nocardia (Leja and Lewandowicz 2010; Megharaj et al. 2011; Ward and Singh 2014). Actinomycetes which are among the most abundant groups of microorganisms in soil and water have been considered as a suitable candidate for implementation in bioremediation process due to their high catabolic capacity as well as survival and persistence in unfavorable environmental conditions. Actinomycetes are one of the dominant bacterial phyla on earth, a group of terrestrial or aquatic gram-positive bacteria with very complex to very simple cell structure, capable of decomposing a high variety of organic materials, fixing nitrogen, and producing secondary metabolites (Sharma 2014; Watanabe 2001). Thus, in the current study, we aimed to screen, identify and characterize environmental nontuberculous mycobacteria (NTM) from Iranian ecosystems and to assess their bioremediation activity that can contribute in development and enhancement of bioremediation technology.
Materials and methods
Sampling
In a period of over 2 years between July 2014 and August 2016, a total number of 90 environmental samples, from ecologically diverse sources in Iran; i.e., drinking and non-drinking water, sea and river sediments, and effluents from municipal wastewater, hospitals, private households, industries, animal husbandries, were collected aseptically in sterile bottles and containers (Fig. 1; Table 1).
Fig. 1.
Geographic distribution of sampling site from Iran’s ecosystems
Table 1.
Samples profile, phenotypic and molecular features and bioremediation analysis of mycobacterial isolates from Iranian ecosystems
| Sample profile | Phenotypic features | 16S rRNA analysis | Biodegradation activity | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | Location (city) Province | Source | pH | Temperature | Opt. Tm | Growth rate | Semi quantitative catalase | Tween 80 hydrolyses | Tolerance of NaCl5% | Pyrazinamidase | Reduction of tellurite potassium | Urease | Runyon group | Similarity (%) | Base pair differences | Identification | |
| A (2, 12, 13) | Khorramabad (Lorestan) | River sediment/forest soil | 7 | 8–14 | 35 | R | + | + | + | − | − | + | IV | 99.69 | 4/1285 | M. sacrum like | PAHs/sodium sulfate |
| A28 | Ahwaz (Khuzestan) | River sediment | 7.2 | 18 | 35 | R | + | + | + | − | − | + | IV | 99.69 | 4/1285 | M. sacrum like | PAHs/sodium sulfate |
| A53 | Fulad Shahr (Isfahan) | Forest soil | 7.4 | 30 | 30 | R | − | + | + | − | − | + | IV | 97.74 | 22/974 | M. flavescens | PAHs/crude oil |
| A82 | Dorud (Lorestan) | River sediment | 7.2 | 25 | 30 | R | − | + | + | − | − | + | IV | 97.74 | 22/974 | M. flavescens | PAHs/crude oil |
| A (113, 88) | Yazd (Yazd) | Hospital water | 7.4 | 14 | 30 | R | − | + | + | − | − | + | IV | 97.74 | 22/974 | M. flavescens | PAHs/crude oil |
| A38 | Shazand (Markazi) | Oil refinery soil | 8.5 | 6 | 35 | R | + | − | + | + | − | − | IV | 100 | 0/1074 | M. conceptionense | Dibenzothiophene |
| A55 | Abadan (Khuzestan) | Oil refinery soil | 7.6 | 24 | 25 | R | + | + | + | − | − | + | IV | 99.9 | 1/1074 | M. monacense | PAHs/crude oil/pristan |
| A75 | Isfahan (Isfahan) | Porcelain wastewater | 6.5 | 20 | 25 | R | + | + | + | − | − | + | IV | 99.9 | 1/1074 | M. monacense | PAHs/crude oil/pristan |
| A6 | Dezful (Khuzestan) | Dam water | 7.6 | 13 | 30 | R | − | + | + | − | + | + | IV | 99.56 | 6/1378 | M. vanbaalenii like | PAHs |
| A16 | Arak (Markazi) | Salt Lake sediment | 8 | 4 | 25 | R | − | − | − | + | − | + | IV | 98.22 | 24/1352 | M. gadium like | Sodium sulfate |
| A4 | Khorram abad (Lorestan) | Land farm soil | 7 | 14 | 25 | R | + | + | − | − | + | + | IV | 99.48 | 7/1356 | M. tusciae like | PAHs |
| A23 | Kish (Hormozgan) | Sea sediment | 7.9 | 20 | 30 | R | + | − | + | − | − | + | IV | 98.1 | 18/916 | M. pallens like | PAHs |
| A7 | Isfahan (Isfahan) | Hospital water | 7.6 | 12 | 30 | S | − | − | + | + | − | − | II | 99 | 9/960 | M. cookii like | – |
| A15 | Omidie (Khuzestan) | Oil contaminated soil | 6.4 | 25 | 25 | R | + | + | + | − | − | + | IV | 99.9 | 1/1023 | M. porcinum | PAHs/crude oil/phenol |
| A18 | Khorramabad (Lorestan) | Hospital water | 7.6 | 10 | 35 | S | − | − | + | + | + | − | I | 100 | 0/999 | M simiae | – |
| A5 | Bushehr (Bushehr) | Sea sediment | 7.2 | 24 | 30 | R | + | + | + | − | − | + | IV | 100 | 0/872 | M. fredriksbergense | PAHs/crude oil |
| A9 | Isfahan (Isfahan) | River sediment | 7.8 | 8 | 30 | R | + | + | + | − | − | + | IV | 100 | 0/872 | M. .fredriksbergense | PAHs/crude oil |
| A8 | Shahin Shahr (Isfahan) | Oil refinery soil | 8.3 | 18 | 30 | R | + | − | + | − | − | − | IV | 99.4 | 4/663 | M. celeriflavum | PAHs |
| A14 | Omidie (Khuzestan) | Oil well sediments | 6.9 | 20 | 25 | R | + | + | − | − | + | + | IV | 99.2 | 11/1378 | M. vaccae like | PAHs/crude oil |
| A33 | Bandar anzali (Gilan) | River sediments | 6.8 | 20 | 35 | R | + | + | + | + | + | − | IV | 99.7 | 3/983 | M. novocastrense | Sulfate sodium |
| A (24, A25, A52) | Isfahan (Isfahan) | Hospital water and soil | 7–8.8 | 12–28 | 35 | S | + | − | − | − | + | − | II | 99.6–99.9 | 1–4/995 | M. paragordonae | – |
| A90 | Shahr Kurd (kohkiloye) | Hospital water | 7.6 | 14 | 35 | S | + | − | − | − | + | − | II | 99.9 | 1/995 | M. paragordonae | – |
| A11 | Dorud (Lorestan) | Cement factory soil | 6.8 | 14 | 35 | R | + | − | + | + | + | + | IV | 100 | 0/1044 | M. aurum | PAHs/PVC/morpholine/piperidin |
| A89 | Kermanshah (Kermanshah) | Petrochemical factory soil | 7 | 16 | 35 | R | + | − | + | + | + | + | IV | 100 | 0/1044 | M. aurum | PAHs/PVC/morpholine/piperidin |
| A43 | Hamadan (Hamadan) | Power plant soil | 8.5 | 6 | 35 | R | + | − | − | + | + | + | IV | 99.12 | 11/1253 | M. neoaurum | PAHs/crude oil/pristan |
| A27 | Kerman (Kerman) | Sarcheshmeh copper mine soil | 8.4 | 16 | 25 | R | − | + | + | − | + | − | IV | 99.63 | 4/1067 | M. obuense | PAHs/methoxy chloro ethane |
| A85 | Chabahar (Sistan and balochestan) | Sea sediments | 7.4 | 14 | 35 | R | − | + | − | + | + | − | IV | 99.64 | 5/1385 | M. phocaicum like | PAHs/crude oil/fluoro-glycofen ethyl |
| A3 | Ahwaz (Khuzestan) | River sediments | 7.8 | 16 | 35 | R | − | + | − | + | + | − | IV | 99.64 | 5/1385 | M. phocaicum like | PAHs/crude oil/fluoro-glycofen ethyl |
| A (44, 49) | Khorramabad (Lorestan) | Forest soil | 7 –7.6 | 14–26 | 35 | R | − | − | − | + | − | − | IV | 98.56–100 | 0–12/1247 | M. fortuitum | Natural rubber/phenols/squalene |
| A63 | Isfahan (Isfahan) | Hospital water | 7.4 | 16 | 35 | R | − | − | − | + | − | − | IV | 100 | 0/1247 | M. fortuitum | Natural rubber/phenols/squalene |
| A (69, 91) | Najaf abad (Isfahan) | Tile factory sewage | 7.6 | 22 | 35 | R | − | − | − | + | − | − | IV | 98.56–100 | 0–12/1247 | M. fortuitum | Natural rubber/phenols/squalene |
| A115 | Ramsar (Mazandaran) | Forest soil | 7 | 18 | 35 | R | − | − | − | + | − | − | IV | 98.88 | 6/1247 | M. fortuitum | Natural rubber/phenols/squalene |
Opt. Tm optimum temperature, Similarity % similarity to the nearest validated species, Base pair differences the number of nucleotide differences between the isolates and the nearest validated species, R resistance
The samples were processed based on standard procedures. In summary, the aquatic samples were transported at 4 °C to the laboratory and processed within a maximum period of 24 h. The collected water samples were treated with 0.005% cetylpyridinium chloride (CPC) for 15 min to reduce the number of not-desirable microbial contaminants such as fungi, Protista and other bacteria. Afterwards, the pretreated samples were subjected to vacuum filtration (cellulose nitrate 0.45 µm, Sartorius AG, Gottingen, Germany). The filters were rinsed and mashed in tubes containing 15 ml of distilled water. Almost 100 µl aliquots of dissolved samples were transferred into the tubes of Löwenstein–Jensen (LJ) and Sauton’s media (Allen 1998) and incubated at constant temperatures of 25, 30 and 35 °C in an atmosphere of 5% CO2.
For soil and dust, 15–30 g of samples were taken from 3 to 5 cm depth of the sampling points and transferred to the laboratory. Five grams of samples were transferred to 50 ml sterile centrifuge tubes containing 20 ml sterile distilled water and centrifuged at 4300 × g for 20 min at room temperature. The pellets and supernatants were decontaminated in separate tubes by addition of 3% sodium lauryl sulfate and 1% NaOH (Kamala et al. 1994). Afterwards, 100 µl of the decontaminated samples were used to inoculate into Löwenstein–Jensen (LJ) and Sauton’s media and incubated at temperatures of 25, 30 and 35 °C in an atmosphere of 5% CO2 for 12 weeks (Kamala et al. 1994; Lahiri et al. 2014).
For sediment samples, up to 3 g of samples were stirred for 30 min in 100 ml of sterile Ringer’s solution (5% v/v). The suspension was homogenized, tenfold serial dilutions were prepared in sterile water and 200 μl of each pretreated 10−2, 10−3, and 10−4 dilutions was inoculated into the Sauton’s media supplemented with antifungal and antibacterial antibiotics including kanamycin, nystatin and nalidixic acid (each at 50 µg/ml). The samples were incubated for 3 weeks at temperatures of 25, 30 and 35 °C in an atmosphere of 5% CO2 (Baskaran et al. 2011).
The details of environmental samples tested in the current study are given in Table 1.
Conventional identification of the isolates
The isolates were characterized phenotypically by the use of conventional phenotypic and biochemical tests (Wayne 1984). The tests included acid-fast staining, colony characterization, the growth rate at temperatures of 25, 30 and 35 °C, and the standard biochemical assays, i.e., semi quantitative and heat-stable (68 °C) catalase production, tween opacity, pigment production, urease and pyrazinamidase activity, nitrate and tellurite reduction, and niacin accumulation.
Molecular identification of the isolates
DNA extraction and purification
Chromosomal DNA was extracted using simple boiling method or the modified Pitcher method (Pitcher et al. 1989; Rahdar et al. 2017; Shojaei et al. 2011). In brief, the simple boiling method was performed as follows: a few colonies of bacteria added into 2 ml of TE buffer (tris EDTA), and boiled for 30 min, centrifuged at 11,900×g for 10 min. The supernatant was transferred to a sterile microtube and centrifuged at 20,000×g for 10 min. The modified Pitcher method included the lysis of actinomycetes cells obtained with the pretreatment with lipase (2 mg/ml) followed by the cell wall disruption using lysozyme (200 mg/ml final concentration) and proteinase K (300 μg/ml final concentration) in the presence of 3% sodium dodecyl sulfate (SDS). The DNA-containing aqueous phase was purified with phenol–chloroform–isoamyl alcohol (25:24:1, vol/vol/vol) and chloroform-isoamyl alcohol (24:1, vol/vol). The precipitation of DNA by either of two methods, was carried out using ammonium acetate (7.5 mol/l) and ethanol at −20 °C. The Precipitated DNA was washed with 70% ethanol and re-suspended in 100 µl of Milli-Q water.
Molecular identification of mycobacterial isolates
The environmental isolates that were identified phenotypically as Mycobacterium were further verified to the genus level using a specific PCR protocol based on a 228-bp fragment of the 65-kDa heat shock protein (hsp65). The primers included hsp65F: 5′-CTGGTCAAGGAAGGTCTGCG-3′, and hsp65R: 5′-GATGACACCCTCGTTGCCAAC-3′) as recommended by Khan and Yadav (2004). For identification of the isolates at the species level the amplification and direct sequence analysis of 16S rRNA gene was used as described previously (Shojaei et al. 2011) using the primers 27F: 5′-AGA GTT TGA TCM TGG CTC AG-3′and 1492R: 5′-CG GTT ACC TTG TTA CGA CTT-3′) (Wilson et al. 1990). The sequencing was performed by ABI 3100 genetic analyzer in Bioneer Company (South Korea). The obtained sequences were aligned manually with all existing sequences of the closely related mycobacteria retrieved from the GenBankTM database, compared with the relevant sequences and analyzed using the jPhydit program (Jeon et al. 2005).
Nucleotide sequence accession numbers
The GenBank accession numbers for the 16S rRNA sequencing of the mycobacteria isolated in this study are; M. coockii like (JX566888), M. simiae (KF028776), M. frederiksbergense (KF019696), M. sacrum like (KU564076), M. vanbaalenii like (KU564074), M. gadium like (KU564078), M. aurum (KU564075), M. tusciae like (KU564073), M. novocastrense (NR_029208), M. obuense (KF028777) and M. phocaicum like (KF019699).
Strain culture collection numbers
The culture collection numbers for two potentially novel strains of mycobacteria in this study are listed below: M. cookii like (A7) JCM 30922 and CCUG 67561 and M. phocaicum like (A3 and A85) JCM 30989 and CCUG 67787.
Bioremediation analysis
The biodegradation activity of the strains isolated from environmental samples in the current study was evaluated according to Kanaly and Harayama (2000). The details are as follows:
Chemicals and media
PAHs mix solution (1-1) [Acenaphthene, Acenaphthylene, Anthracene, Benzo(b)fluoranthene, 1,2-Benz anthracene, Benzo(a)pyrene, Benzo(k)fluoranthene, Benzo(g,h,i)perylene, Chrysene, Dibenz(a,h)anthracene, Fluoranthene, Fluorene, Indeno(1,2,3-cd)pyrene, Phenanthrene, Naphthalene, Pyrene] were purchased from AccuStandard. PAHs stock solution was 0.2 mg/ml in dichloromethane and methanol. Phenol was purchased from Merck, Germany. Phenol stock solution was 10 mg/ml in deionized water. Sodium sulfate was purchased from Merck, Germany. Sodium sulfate stock solution was 10 mg/ml dissolved in deionized water. All other solvents and chemicals used were of reagent grade. Mineral Salt Medium (MSM) contained (g/l), g/l were KH2PO4 (0.42), K2HPO4 (0.375), (NH4)2SO4 (0.244), NaCl (0.015), CaCl2·2H2O (0.015), MgSO4·7H2O (0.05), and FeCl3·6H2O (0.054).
Evaluation of biodegradation activity of isolates was carried out by adding 1 ml of direct colony suspension is made in normal saline and turbidity adjusted to 0.5 McFarland standard (1.5 × 108 CFU/ml) into 100 ml aliquots of % 1 PAHs, % 1 phenol and % 1 sodium sulfate enriched MSM were prepared in 250 ml flask and incubated for 6 days at 30 °C in an orbital shaker (90 rpm). To evaluate the bacterial growth, samples were collected at 24-h intervals and the absorbance at 560 nm was measured by Spectrophotometer.
Showing sign of growth in the media indicates consumption and/or decomposition of material by studied isolates. Standard procedure was used to final confirmation of degradation of the studied material, (Chesnin and Yien 1951; Gibbs 1927; Manoli and Samara 1996) explained in the following method.
Determination of PAHs degradation
An amount 5 ml of the MSM medium containing PAHs that showed bacterial growth was transferred to a screw cap glass tube and supplemented with 0.6 ml of a mixture of tetrachlorethylene and methanol (1:100) as the extraction solvent, vortexed for 10 s, then centrifuged at 3000×g for 10 min. The organic phase was then collected and transferred to a clean tube to be further analyzed by HPLC. PAHs were analyzed by Manager 5000 HPLC systems (Knauer, Germany). A reversed phase column C18 (60 × 2 mm, particle size 2.2 µm) using mobile phase with acetonitrile/water (gradient elution 20 min at a constant flow rate 5:95, and 0.3 ml/min 50 °C) was used to separate PAHs (Manoli and Samara 1996). The UV absorbance at 254 nm was measured and the PAHs content of the sample was calculated using the standard curve and function calculated using sterile PAHs standards.
Determination of phenol degradation
An amount of 5 ml of MSM medium containing phenol which showed bacterial growth was transferred to a sterile tube and the pH was adjusted to 8.0, then subjected to centrifugation at 2700g for 20 min. An amount of 150 µl of the collected supernatant was mixed with 30 µl NaHCO3, and 20 µl of Gibbs reagent (2,6-dichloroquinone 4-chloroimide) was added to the mixture and shacked for 30 min at room temperature (Gibbs 1927). The UV absorbance at 620 nm was recorded and the phenol content determined using a standard curve and function calculated using sterile phenol standards.
Determination of sodium sulfate degradation
An amount of 5 ml of MSM medium containing sodium sulfate was transferred to a sterile tube. One ml 1% acetic acid and 1 ml acetate buffer was added and mixed for 3 min. Afterwards, 1 ml of barium chloride was added and mixed for another 3 min. The turbidity was calculated using Spectrophotometry at a wavelength of 420 nm (Chesnin and Yien 1951). The amount of consumed sodium sulfate was measured by using the standard curve calculated using sterile sodium sulfate standards.
Results
Isolation and characterization of strains
The recorded temperature and pH of the soil samples were in the range of 4–30 °C and 6.0–8.0 respectively. For wastewater and sediment samples, these measures were in the range of 7–24 °C and 6.8–7.8, respectively. The corresponding figures for the water samples were 5–23 °C and 7.0–8.2, respectively, and the total dissolved solids (TDS) for the water samples ranged between 600 and 1200 mg/l (Table 1).
The details of water, soil, wastewater and sediment samples and the isolates key properties are presented in Table 1.
From 90 water, soil, wastewater and sediment samples, a total number of 39 isolates were recovered and identified as non-tuberculosis mycobacteria based on culture, morphological and biochemical features, and by molecular analysis.
The 16S rRNA gene sequencing of the isolates revealed that all isolates had nucleotide signatures of mycobacteria at positions 70–98 (A–T), 293–304 (G–T), 307 (C), 328 (T), 614–626), 661–744 (G–C), (A–T), 631 (G), 824–876 (T–A), 825–875 (A–T), 843 (C), and 1122–1151 (A–T) (Stackebrandt et al. 1997). Of these 39 isolates, 15 were slowly growing mycobacteria and 24 were rapidly growing mycobacteria. All rapidly growing isolates contained the short helix 18 (position 451–482) that is characteristic of the rapidly growing mycobacteria. In contrast, all slowly growing isolates, except for the isolate A18, characterized by an extended helix, which is present in slowly growing mycobacteria. The isolate A18 was an exception in this scheme since it belonged to M. simiae complex that contrary to other slow growing mycobacteria present the signature of rapid growers (short helix 18) in the 16S rRNA (Tortoli 2003).
Based on morphological, culture and biochemical properties and the genus specific marker, i.e., the presence of a 228-bp fragment of the hsp65, all 39 isolates were identified as Mycobacterium of which, 2 isolates fit into Runyon group I, 11 isolates in Runyon group II, 2 isolates in Runyon group III and 24 isolates, i.e., the rapidly growing mycobacteria were classified in Runyon group IV (Table 1). The isolates belonged to 14 validated species and 7 unknown or potentially novel species. The most prevalent mycobacteria were M. fortuitum (6 isolates, 15.38%). This followed by M. flavescens and M. paragordonae (4 isolates each; 10.25%) and a potentially novel species closely related to uncharacterized species M. sacrum (4 isolates, 10.25%). The other thirteen isolates comprised a single isolates each belonged to seven established species that were M. conceptionense, M. porcinum, M simiae, M. celeriflavum, M. obuense, M. novocastrense and M. neoaurum, as well as six unknown isolates potentially novel species of mycobacteria that were closely related to M. tusciae (isolate A4), M. vanbaalenii (isolate A6), M. gadium (isolate A16), M. pallens (isolate A23), M. cookii (isolate A7), and M. vaccae (isolate A14).
The relationship between our isolates and the standard established species of mycobacteria was supported by a high bootstrap value in phylogenetic tree based on 16S rRNA gene depicted by MEGA 6. The corresponding value for the isolates that showed substantial nucleotide differences with the known mycobacterial species and named as novel species requiring further investigation found to be rather low (Fig. 2).
Fig. 2.
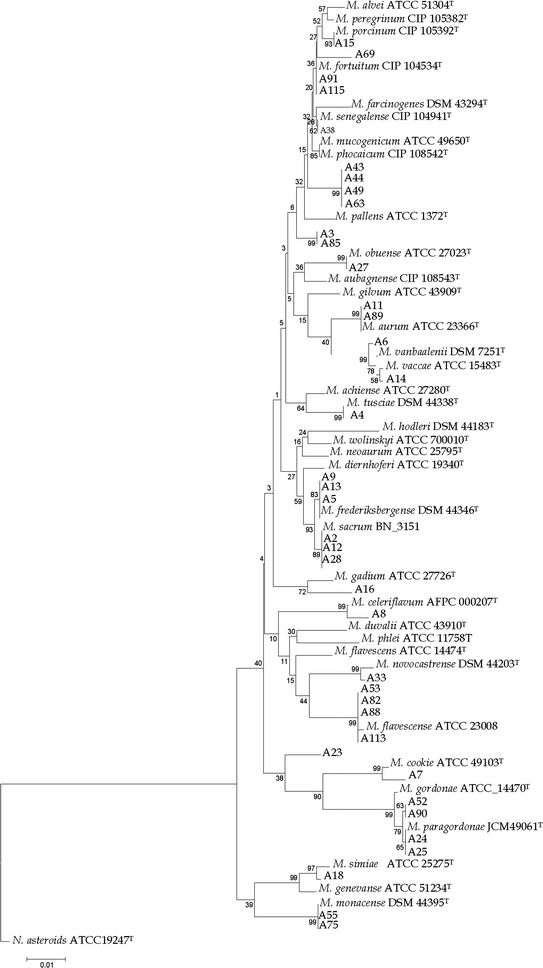
16S rRNA sequence based phylogenetic tree for Iranian biodegrading NTM isolates and the nearest validated species of mycobacteria by using the neighbor-joining method. The figures at each node represent bootstrapping values. The tree was rooted with N. asteroides
Bioremediation analysis
Based on biodegradation activity, our isolates can be classified in three categories:
The isolates without biodegradation activity that included, four isolates of M. paragordonae, i.e., the isolates A24, A25, A52 and A90 and one isolate of M. simiae that is, the isolate A18 (Table 1).
The isolates with biodegradation activity based on the previous reports by other investigators, that included two isolates of M. frederiksbergense, i.e., A5 and A9, with capability of PAHs degradation (Willumsen et al. 2001), two isolates of M. phocaicum, i.e., A3 and A85 with capability of PAHs and fluoroglycofen ethyl degradation (Chen et al. 2011), one isolate M. obuense, i.e., A27 with capability of Methoxychlor ethane degradation (Satsuma and Masuda 2012), Two isolates of M. aurum, i.e., A11 and A89 with capability of PAHs, polyvinyl chloride (PVC), morpholine, thiomorpholine and piperidin degradation (Combourieu et al. 1998; Hartmans and De Bont 1992), One isolate of M. conceptionense, i.e., A38 with capability of dibenzothiophene degradation (Akhtar et al. 2016), One isolate of M. neoaurum, i.e., A43 with capability of pristan, soybean phytosterols, crude oil and PAHs degradation (Bastiaens et al. 2000; Mikolasch et al. 2009; Wei et al. 2010), Six isolates of M. fortuitum, i.e., A44, A49, A63, A69, 91 and A115 with capability of poly chlorinated and brominated biphenyls (PCB and PBBs), natural rubber and squalene degradation (Berekaa and Steinbüchel 2000; Rose and Steinbüchel 2005; Uotila et al. 1992), four isolates of M. flavescens, i.e., A53, A83, A88 and A113 with capability of crude oil and PAHs degradation (Miller et al. 2004), and two isolates of M. monacense, i.e., A55 and A75 with capability of crude oil and PAH degradation (Miller et al. 2004).
The isolates that were either the previously established mycobacteria without known biodegradation activity or the novel species that were first isolated and reported in the current study as capable of biodegradation of environmental chemical pollutants.
A growth curve was plotted with O.D at 560 nm on Y axis and time in hours on axis respectively. On the basis of growth curve plotted, it was found that isolates A3 and A85 were identified as M. phocaicum like had maximum growth against a concentration of 0.2 µg/ml for PAHs mix solution, followed by the isolates A2, A12, A13 and A28 were identified as M. sacrum-like, the isolate A4 was identified as M. tusciae like, the isolate A6 was identified as M. vanbaalenii like, the isolate A8 was identified as M. celeriflavum, the isolate A14 was identified as M. vaccae like, the isolate A15 was identified as M. porcinum and the isolate A23 was identified as M. pallens like. The growth curves are shown in Fig. 3. A3 and A85 was found to be highest growth rate in presence of PAHs followed by the growth of isolate A23, A2, A6 and A14 isolate and the isolate A8 was found to lowest growth rate. The growth response indicates that, organisms were able to utilize PAHs mix solution as their sole carbon source. As the growth proceeded, the metabolites produced during the degradation of PAHs were toxic at a higher level which reduced the growth rate (Table 1; Fig. 3).
Fig. 3.
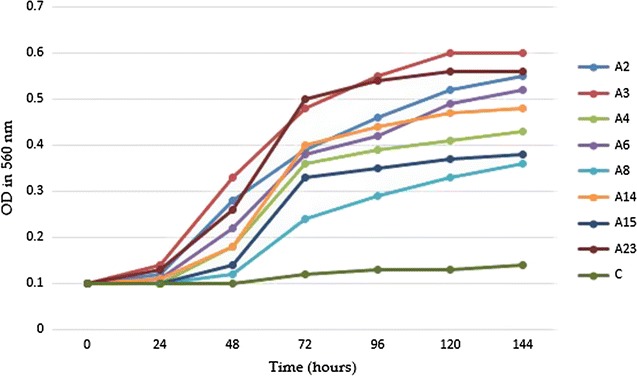
Growth curves of Iranian isolates of mycobacteria over a 24 h. Incubation period at 30 °C in the presence of PAHs
According to the chromatogram obtained from the analysis, the results are as follows. Isolate A3 and A83 showed a higher rate of mixed PAHs degradation with 90% degradation of it (Fig. 4).
Fig. 4.
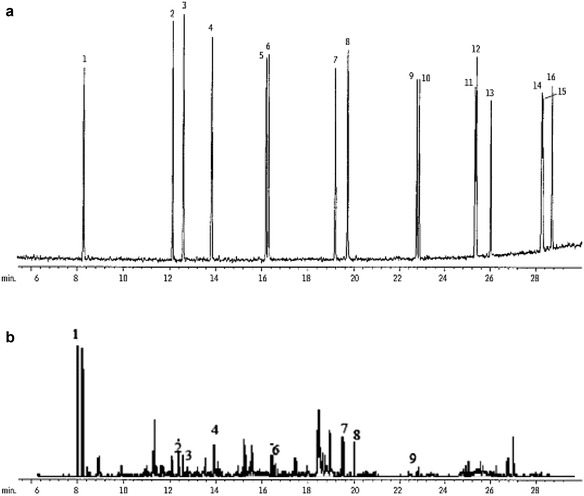
HPLC chromatograms of PAHs mix solution by selected mycobacterial isolates, a control samples, b after 144 incubation at 30 °C. (1) Naphthalene, (2) acenaphthylene, (3) acenaphthene, (4) fluorene, (5) phenanthrene, (6) anthracene, (7) fluoranthene, (8) pyrene, (9) benzo[a] anthracene, (10) chrysene, (11) benzo[b]fluoranthene, (12) benzo[k]fluoranthene, (13) benzo[a]pyrene, (14) indeno[1,2,3-cd]pyrene, (15) dibenzo[a,h]anthracene
On the basis of growth curve plotted, it was found that isolate A15 was identified as M. porcinum had maximum growth against a concentration of 1 mg/ml for phenol. The growth response indicates that, organisms were able to utilize phenol as their sole carbon source (Table 1; Fig. 5). The results of Gibbs method showed that isolate A15 was able to degrade the 87% of phenol after 144 h.
Fig. 5.
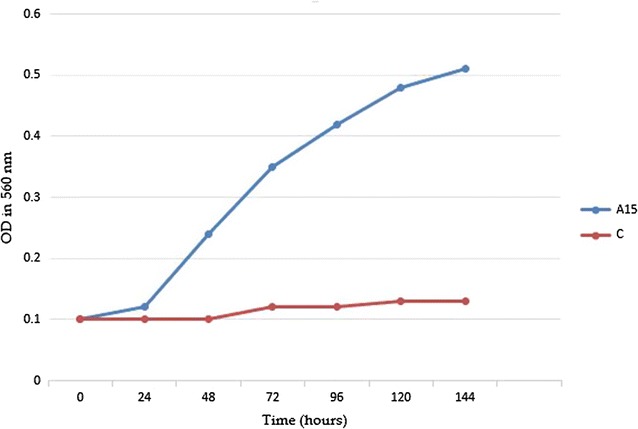
Growth curves of Iranian mycobacterium isolates over a 24 h. Incubation period at 30 °C in the presence of phenol
On the basis of growth curve plotted, it was found that isolate A16 had maximum growth against a concentration of 1 mg/ml for sodium sulfate, followed by the isolates A2, A12, A13, A28 and the isolate A33 was identified as M. novocastrense. The growth curves are shown in Fig. 6 isolate A16 was found to be highest growth rate in presence of %1 sodium sulfate, followed by the growth of isolate A2 and isolate A33 and the other tested isolate was not able to grow significantly (Table 1; Fig. 6). The growth response indicates that, organisms were able to utilize sodium sulfate as their sole carbon source.
Fig. 6.
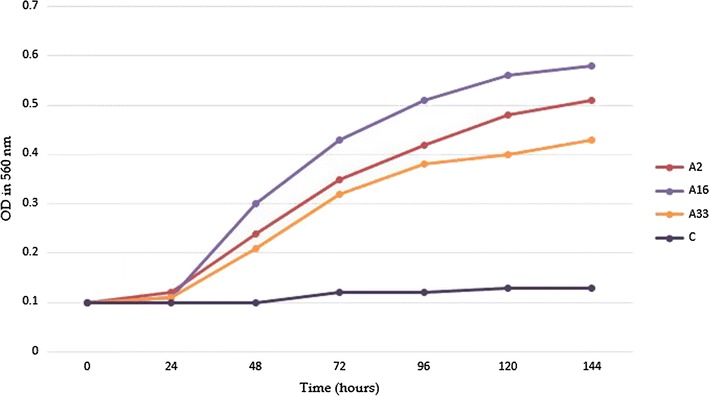
Growth curves of Iranian isolates of mycobacteria over a 24 h. Incubation period at 30 °C in the presence of sodium sulfate
The results of turbidimetry method, indicates that isolate A16 showed highest sodium sulfate biodegradation activity and was able to degrade 100% of the sodium sulfate followed by A2 and A33 that was able to degrade 80 and 65% of sodium sulfate in medium after 144 h respectively.
Discussion
Since the exploration of oil in 1908, Iran has experienced an economic transition, i.e. transformed from traditional agriculture led to industry led. Development, and industrialization in particular, have made immense positive contributions to health, including greater personal and social wealth, as well as vastly improved health, transportation and communication. Iranian people are living longer and are healthier than they were centuries and even decades ago. However, industrialization has also had adverse health consequences not only for workforces, but for the general population as well. These effects have been caused either directly by exposure to safety hazards and harmful agents, or indirectly through environmental degradation locally and regionally. Iranian people are being exposed to these environmental health hazards through a range of ways that include traditional hazards of industrial contamination of air, water, food and land, as well as new pollution phenomena such as dust and sand storms.
Destroyed ecosystems and health risks have emphasized on the necessity for cleanup and remedies for soil and water, which have prompted many researchers to develop environment clean up strategies. These approaches include the physical, chemical and thermal processes (Arias-Estévez et al. 2008; Schaer et al. 2001). These techniques however have some adverse effects on the environment and are also expensive (Mulligan et al. 2001; Virkutyte et al. 2002). Recently, biological techniques like phytoremediation and bioremediation are being evaluated for the remediation of environment contaminated with chemical pollutants (Adams et al. 2015; Brown et al. 2011; Susarla et al. 2002).
Bioremediation is a waste management technique that involves the use of naturally occurring organisms to neutralize pollutants from a contaminated site. Up to now, many bacterial strains, such as Pseudomonas, Alcanivorax, Acinetobacter, Rhodococcus, and Bacillus have been isolated from soil and water, and were effectively used for the bioremediation of petroleum contaminants (Das and Mukherjee 2007; Girma 2015; Ron and Rosenberg 2002). Depending on the site and its contaminants, bioremediation may be safer and less expensive than alternative solutions such as incineration or landfilling of the contaminated materials. It also has the advantage of treating the contamination in place so that large quantities of soil, sediment or water do not have to be dug up or pumped out of the ground for treatment (Adams et al. 2015).
Till date several investigators have described the ability of sludge-derived mycobacteria to degrade chemical pollutants (Guo et al. 2010; Miserez et al. 1999). Mycobacteria have shown ability to degrade polychlorophenols, heavy metals and diverse PAHs (Girma 2015; Kumar et al. 2011; Xun 2012).
This study was undertaken to screen various Iranian ecosystems in search of mycobacteria with capability to degrade environmental contamination and in particular the oil contaminating pollutants.
A total of 39 biodegrading mycobacteria were isolated from 90 samples collected from the diverse ecosystems of Iran. The isolates A5 and A9 were identified as M. frederiksbergense. This organism is a rapidly growing scotochromogenic Mycobacterium which was first isolated and characterized in 2001 from soil in Denmark (Willumsen et al. 2001). M. frederiksbergense is able to degrade light and heavy chain PAHs (Willumsen et al. 2001).
The isolates A3 and A85 were identified as M. phocaicum. This organism was first isolated and characterized in 2006 from clinical samples (Adékambi et al. 2006a). Afterwards, Chen and his colleagues found out that M. phocaicum was able to degrade fluoroglycofen ethyl and PAHs (Chen et al. 2011).
The isolate A27 was identified as M. obuense. This organism was first isolated and characterized in 2006 from clinical samples (Tsukamura and Mizuno 1971). In subsequent studies it was reported that this Mycobacterium has the capacity to degrade PAHs and Methoxychlor ethane which is an organochlorine insecticide (Satsuma and Masuda 2012).
The isolates A11 and A89 were identified as M. aurum that was first isolated and characterized in 1993 from environmental samples (Yokota et al. 1993). Based on the results of the current study and the studies by other investigators, M. aurum has the ability to degrade polyvinyl chloride, PAHs, morpholine, thiomorpholine and piperidin, which is a common additive for pH adjustment in both fossil fuel and nuclear power plant steam systems (Combourieu et al. 1998; Hartmans and De Bont 1992).
The isolate A33 was identified as M. novocastrense which was first isolated and characterized from clinical samples (Shojaei et al. 1997), However there has been no reports on its biodegradation activity. In the present study, we showed that M. novocastrense has the capacity to degrade sodium sulfate.
The isolate A38 was identified as M. conceptionense. This organism is a rapidly growing scotochromogenic Mycobacterium which was first isolated and characterized in 2006 from clinical samples (Adékambi et al. 2006b). It has been shown that M. conceptionense is able to desulfurize dibenzothiophene which is an organosulfur compound used in oil refinery (Akhtar et al. 2016).
The isolate A43 was identified as M. neoaurum which was first isolated and characterized in 1972 from clinical samples (Tsukamura and Mizuno 1972). In several studies the capability of M. neoaurum to degrade PAHs, crude oil and pristan (used as a lubricant, a transformer oil, an immunologic adjuvant, and an anti-corrosion agent) was reported (Bastiaens et al. 2000; Mikolasch et al. 2009; Wei et al. 2010).
The isolates A44, A49, A63, A69, A91 and A115 were identified as M. fortuitum. This species has shown an extended capacity for degradation of hexamethyltetracosane, halogenated phenol derivatives such as polychlorinated biphenyl (PCB) and polybrominated biphenyl (PBB) as well as natural and synthetic rubber (Berekaa and Steinbüchel 2000; Rose and Steinbüchel 2005; Uotila et al. 1992).
The isolates A53, A83, A88 and A113 were identified as M. flavescens. The organism was first isolated and characterized from a drug treated tuberculous guinea pig (Bojalil et al. 1962). Subsequently, it was reported that this species has ability to use and degrade PAHs and crude oil as sole carbon and energy sources (Miller et al. 2004).
The isolates A55 and A75 were identified as M. monacense which was first isolated from clinical samples (Reischl et al. 2006). Miller and his colleagues reported that M. monacense is capable of crude oil and various PAHs degradation (Miller et al. 2004).
The isolate A8 was identified as M. celeriflavum. This species is a rapid growing scotochromogenic Mycobacterium that was first isolated and characterized from clinical sample in Iran in 2015 (Shahraki et al. 2015). We isolated this organism from the river sediments and showed that it has the ability of PAHs degradation.
The isolate A15 was identified as M. porcinum which was first isolated from the lymph node of porcine (Tsukamura et al. 1983). We showed that this organism has the capacity of PAHs and phenol degradation.
In our study, ten isolates including A2, A4, A6, A7, A12, A13, A14, A16, A23 and A28, were found to have phenotypic and molecular characteristics of novel Mycobacterium species. The isolates were evaluated for biodegradation activity against PAHs, phenol and sodium sulfate. All isolates, except for the isolate A7, showed biodegradation activity. The isolates A2, A12, A13 and A28 were found to degrade PAHs and sodium sulfate. The isolates A4, A6, A14 and A23 were found to degrade the PAHs and the isolate A16 was able to degrade sodium sulfate.
The characterization of these unknown mycobacteria remains to be completed using a thorough phenotypic and molecular assays including cell wall composition analysis, sequence analysis of key genetic markers such as gyrB, secA1, and DNA-DNA relatedness.
In conclusion, the results of our study showed that the diversity of mycobacteria, particularly prokaryotes, offers the potential for adaptation to various habitats, including environments severely contaminated with hydrocarbons and heavy metals. This broad adaptability has great value for the bioremediation of damaged ecosystems. Our study also confirms the idea that despite being abundant in environment, mycobacteria have been simply ignored for such significant usage. Indeed, there is an untapped potential with regard to bioremedial actinomycetes particularly mycobacteria that has yet to be discovered and administered in bioremediation of hazardous chemicals.
Authors’ contributions
DA and HS carried out the design of the study and performed the statistical analysis, and drafted the manuscript DA, HS, and ADN carried out the experimental studies and participated in the experiments. HS, SM revised the molecular analysis. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to office of vice-chancellor for Research of Isfahan University of Medical Sciences for the support of the current study.
Competing interests
The authors declare that they have no competing interests. The authors alone are responsible for the content and writing of the manuscript.
Availability of data and materials
All data in this manuscript were deposited in publicly available repositories in the Department of Microbiology, School of Medicine, Isfahan University of Medical Sciences, and Isfahan, Iran.
Consent for publication
Not applicable.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This study was funded by Isfahan University of Medical Sciences (Grant Number: 393405).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- NTM
nontuberculous mycobacteria
- CPC
cetylpyridinium chloride
- RCF
relative centrifugal force
- LJ
Löwenstein–Jensen
- SDS
sodium dodecyl sulfate
- TE
tris EDTA
- hsp
heat shock protein
- PAHs
polycyclic aromatic hydrocarbon
- MSM
mineral salt medium
- TDS
total dissolved solids
- PVC
polyvinyl chloride
- PCB
polychlorinated biphenyl
- PBB
polybrominated biphenyl
Contributor Information
Davood Azadi, Email: Davood.azadi@gmail.com.
Hasan Shojaei, Phone: +98-31-33359359, Email: hasanshojaei@msn.com, Email: h_shojaei@idrc.mui.ac.ir.
Sina Mobasherizadeh, Email: mobasherizadeh@gmail.com.
Abass Daei Naser, Email: s_daeinasrei@yahoo.com.
References
- Adams GO, Fufeyin PT, Okoro SE, Ehinomen I. Bioremediation, biostimulation and bioaugmention: a review. Int J Environ Bioremediat Biodegrad. 2015;3(1):28–39. [Google Scholar]
- Adékambi T, Berger P, Raoult D, Drancourt M. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int J Syst Evol Microbiol. 2006;56(1):133–143. doi: 10.1099/ijs.0.63969-0. [DOI] [PubMed] [Google Scholar]
- Adékambi T, Stein A, Carvajal J, Raoult D, Drancourt M. Description of Mycobacterium conceptionense sp. nov., a Mycobacterium fortuitum group organism isolated from a posttraumatic osteitis inflammation. J Clin Microbiol. 2006;44(4):1268–1273. doi: 10.1128/JCM.44.4.1268-1273.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar N, Ghauri MA, Akhtar K. Dibenzothiophene desulfurization capability and evolutionary divergence of newly isolated bacteria. Arch Microbiol. 2016;198(6):509–519. doi: 10.1007/s00203-016-1209-5. [DOI] [PubMed] [Google Scholar]
- Allen BW. Mycobacteria: general culture methodology and safety considerations. Mycobact Protoc. 1998;101:15–30. doi: 10.1385/0-89603-471-2:15. [DOI] [PubMed] [Google Scholar]
- Arias-Estévez M, López-Periago E, Martínez-Carballo E, Simal-Gándara J, Mejuto J-C, García-Río L. The mobility and degradation of pesticides in soils and the pollution of groundwater resources. Agric Ecosyst Environ. 2008;123:247–260. doi: 10.1016/j.agee.2007.07.011. [DOI] [Google Scholar]
- Baskaran R, Vijayakumar R, Mohan P. Enrichment method for the isolation of bioactive actinomycetes from mangrove sediments of Andaman Islands, India. Malays J Microbiol. 2011;7:1–7. [Google Scholar]
- Bastiaens L, Springael D, Wattiau P, Harms H, Verachtert H, Diels L. Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl Environ Microbiol. 2000;66:1834–1843. doi: 10.1128/AEM.66.5.1834-1843.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berekaa MM, Steinbüchel A. Microbial degradation of the multiply branched alkane 2, 6, 10, 15, 19, 23-hexamethyltetracosane (squalane) by Mycobacterium fortuitum and Mycobacterium ratisbonense. Appl Environ Microbiol. 2000;66:4462–4467. doi: 10.1128/AEM.66.10.4462-4467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojalil L, Cerbon J, Trujillo A. Adansonian classification of mycobacteria. Microbiology. 1962;28:333–346. doi: 10.1099/00221287-28-2-333. [DOI] [PubMed] [Google Scholar]
- Brown C, Milke M, Seville E. Disaster waste management: a review article. Waste Manag. 2011;31:1085–1098. doi: 10.1016/j.wasman.2011.01.027. [DOI] [PubMed] [Google Scholar]
- Chen L, Cai T, Wang Q. Characterization of fluoroglycofen ethyl degradation by strain Mycobacterium phocaicum MBWY-1. Curr Microbiol. 2011;62(6):1710. doi: 10.1007/s00284-011-9918-0. [DOI] [PubMed] [Google Scholar]
- Chesnin L, Yien C. Turbidimetric determination of available sulfates. Soil Sci Soc Am J. 1951;15(C):149–151. doi: 10.2136/sssaj1951.036159950015000C0032x. [DOI] [Google Scholar]
- Combourieu B, Poupin P, Besse P, Sancelme M, Veschambre H, Truffaut N, Delort A-M. Thiomorpholine and morpholine oxidation by a cytochrome P450 in Mycobacterium aurum MO1. Evidence of the intermediates by in situ 1H NMR. Biodegradation. 1998;9:433–442. doi: 10.1023/A:1008321610465. [DOI] [PubMed] [Google Scholar]
- Das K, Mukherjee AK. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour Technol. 2007;98(7):1339–1345. doi: 10.1016/j.biortech.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Eisenbud M, Gesell TF. Environmental radioactivity from natural, industrial and military sources: from natural, industrial and military sources. Cambridge: Academic Press; 1997. [Google Scholar]
- El-Shahawi M, Hamza A, Bashammakh A, Al-Saggaf W. An overview on the accumulation, distribution, transformations, toxicity and analytical methods for the monitoring of persistent organic pollutants. Talanta. 2010;80(5):1587–1597. doi: 10.1016/j.talanta.2009.09.055. [DOI] [PubMed] [Google Scholar]
- Gibbs H. Phenol tests III. The indophenol test. J Biol Chem. 1927;72:649–664. [Google Scholar]
- Girma G. Microbial bioremediation of some heavy metals in soils: an updated review. Indian J Sci Res. 2015;6(1):147. [Google Scholar]
- Guo C, Dang Z, Wong Y, Tam NF. Biodegradation ability and dioxgenase genes of PAH-degrading Sphingomonas and Mycobacterium strains isolated from mangrove sediments. Int Biodeterior Biodegrad. 2010;64:419–426. doi: 10.1016/j.ibiod.2010.04.008. [DOI] [Google Scholar]
- Hartmans S, De Bont J. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl Environ Microbiol. 1992;58(4):1220–1226. doi: 10.1128/aem.58.4.1220-1226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y-S, Chung H, Park S, Hur I, Lee J-H, Chun J. jPHYDIT: a JAVA-based integrated environment for molecular phylogeny of ribosomal RNA sequences. Bioinformatics. 2005;21(14):3171–3173. doi: 10.1093/bioinformatics/bti463. [DOI] [PubMed] [Google Scholar]
- Kamala T, Paramasivan C, Herbert D, Venkatesan P, Prabhakar R. Evaluation of procedures for isolation of nontuberculous mycobacteria from soil and water. Appl Environ Microbiol. 1994;60(3):1021–1024. doi: 10.1128/aem.60.3.1021-1024.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaly RA, Harayama S. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J Bacteriol. 2000;182(8):2059–2067. doi: 10.1128/JB.182.8.2059-2067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IU, Yadav JS. Development of a single-tube, cell lysis-based, genus-specific PCR method for rapid identification of mycobacteria: optimization of cell lysis, PCR primers and conditions, and restriction pattern analysis. J Clin Microbiol. 2004;42(1):453–457. doi: 10.1128/JCM.42.1.453-457.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan FI, Husain T, Hejazi R. An overview and analysis of site remediation technologies. J Environ Manag. 2004;71:95–122. doi: 10.1016/j.jenvman.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bisht B, Joshi V, Dhewa T. Review on bioremediation of polluted environment: a management tool. Int J Environ Sci. 2011;1:1079. [Google Scholar]
- Lahiri A, Kneisel J, Kloster I, Kamal E, Lewin A. Abundance of Mycobacterium avium ssp. hominissuis in soil and dust in Germany—implications for the infection route. Lett Appl Microbiol. 2014;59:65–70. doi: 10.1111/lam.12243. [DOI] [PubMed] [Google Scholar]
- Leja K, Lewandowicz G. Polymer biodegradation and biodegradable polymers—a review. Pol J Environ Stud. 2010;19:255–266. [Google Scholar]
- Manoli E, Samara C. Polycyclic aromatic hydrocarbons in waste waters and sewage sludge: extraction and clean-up for HPLC analysis with fluorescence detection. Chromatographia. 1996;43:135–142. doi: 10.1007/BF02292941. [DOI] [Google Scholar]
- Mastrangelo G, Fadda E, Marzia V. Polycyclic aromatic hydrocarbons and cancer in man. Environ Health Perspect. 1996;104:1166. doi: 10.1289/ehp.961041166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R. Bioremediation approaches for organic pollutants: a critical perspective. Environ Int. 2011;37:1362–1375. doi: 10.1016/j.envint.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Mikolasch A, Klenk H-P, Schauer F. Degradation of the multiple branched alkane 2, 6, 10, 14-tetramethyl-pentadecane (pristane) in Rhodococcus ruber and Mycobacterium neoaurum. Int Biodeterior Biodegrad. 2009;63(2):201–207. doi: 10.1016/j.ibiod.2008.09.002. [DOI] [Google Scholar]
- Miller C, Hall K, Liang Y, Nieman K, Sorensen D, Issa B, Sims RC. Isolation and characterization of polycyclic aromatic hydrocarbon-degrading Mycobacterium isolates from soil. Microb Ecol. 2004;48(2):230–238. doi: 10.1007/s00248-003-1044-5. [DOI] [PubMed] [Google Scholar]
- Miserez K, Philips S, Verstraete W. New biology for advanced wastewater treatment. Water Sci Technol. 1999;40:137–144. [Google Scholar]
- Mulligan C, Yong R, Gibbs B. Remediation technologies for metal-contaminated soils and groundwater: an evaluation. Eng Geol. 2001;60(1):193–207. doi: 10.1016/S0013-7952(00)00101-0. [DOI] [Google Scholar]
- Oliva A, Spira A, Multigner L. Contribution of environmental factors to the risk of male infertility. Hum Reprod. 2001;16:1768–1776. doi: 10.1093/humrep/16.8.1768. [DOI] [PubMed] [Google Scholar]
- Perelo LW. Review: in situ and bioremediation of organic pollutants in aquatic sediments. J Hazard Mater. 2010;177(1):81–89. doi: 10.1016/j.jhazmat.2009.12.090. [DOI] [PubMed] [Google Scholar]
- Petry T, Schmid P, Schlatter C. The use of toxic equivalency factors in assessing occupational and environmental health risk associated with exposure to airborne mixtures of polycyclic aromatic hydrocarbons (PAHs) Chemosphere. 1996;32:639–648. doi: 10.1016/0045-6535(95)00348-7. [DOI] [PubMed] [Google Scholar]
- Pitcher D, Saunders N, Owen R. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8(4):151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- Rahdar HA, Azadi D, Shojaei H. Molecular analysis and species diversity of Nocardia in hospital environment of a developing country, a potential health hazard. J Med Microbiol. 2017;66(3):334–341. doi: 10.1099/jmm.0.000436. [DOI] [PubMed] [Google Scholar]
- Reigart JR. Recognition and management of pesticide poisonings. Collingdale: DIANE Publishing; 2009. [Google Scholar]
- Reischl U, Melzl H, Kroppenstedt RM, Miethke T, Naumann L, Mariottini A, Mazzarelli G, Tortoli E. Mycobacterium monacense sp. nov. I. Int J Syst Evol Microbiol. 2006;56(11):2575–2578. doi: 10.1099/ijs.0.64527-0. [DOI] [PubMed] [Google Scholar]
- Ritter KS, Sibley P, Hall K, Keen P, Mattu G, Beth Linton L. Sources, pathways, and relative risks of contaminants in surface water and groundwater: a perspective prepared for the Walkerton inquiry. J Toxicol Environ Health Part A. 2002;65:1–142. doi: 10.1080/152873902753338572. [DOI] [PubMed] [Google Scholar]
- Ron EZ, Rosenberg E. Biosurfactants and oil bioremediation. Curr Opin Biotechnol. 2002;13:249–252. doi: 10.1016/S0958-1669(02)00316-6. [DOI] [PubMed] [Google Scholar]
- Rose K, Steinbüchel A. Biodegradation of natural rubber and related compounds: recent insights into a hardly understood catabolic capability of microorganisms. Appl Environ Microbiol. 2005;71(6):2803–2812. doi: 10.1128/AEM.71.6.2803-2812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta SK, Singh OV, Jain RK. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol. 2002;20:243–248. doi: 10.1016/S0167-7799(02)01943-1. [DOI] [PubMed] [Google Scholar]
- Satsuma K, Masuda M. Reductive dechlorination of methoxychlor by bacterial species of environmental origin: evidence for primary biodegradation of methoxychlor in submerged environments. J Agric Food Chem. 2012;60(8):2018–2023. doi: 10.1021/jf2048614. [DOI] [PubMed] [Google Scholar]
- Schaer M, Nüesch F, Berner D, Leo W, Zuppiroli L. Water vapor and oxygen degradation mechanisms in organic light emitting diodes. Adv Funct Mater. 2001;11:116–121. doi: 10.1002/1616-3028(200104)11:2<116::AID-ADFM116>3.0.CO;2-B. [DOI] [Google Scholar]
- Schwarzenbach RP, Egli T, Hofstetter TB, von Gunten U, Wehrli B. Global water pollution and human health. Ann Rev Environ Res. 2010;35:109–136. doi: 10.1146/annurev-environ-100809-125342. [DOI] [Google Scholar]
- Shahraki AH, Çavuşoğlu C, Borroni E, Heidarieh P, Koksalan OK, Cabibbe AM, Hashemzadeh M, Mariottini A, Mostafavi E, Cittaro D, Feizabadi MM, Lazarevic D, Yaghmaei F, Molinari GL, Camaggi A, Tortoli E. Mycobacterium celeriflavum sp. nov., a rapidly growing scotochromogenic bacterium isolated from clinical specimens. Int J Syst Evol Microbiol. 2015;65(2):510–515. doi: 10.1099/ijs.0.064832-0. [DOI] [PubMed] [Google Scholar]
- Sharma M. Actinomycetes: source, identification, and their applications. Int J Curr Microbiol App Sci. 2014;3:801–832. [Google Scholar]
- Shojaei H, Goodfellow M, Magee J, Freeman R, Gould F, Brignall C. Mycobacterium novocastrense sp. nov., a rapidly growing photochromogenic mycobacterium. Int J Syst Evol Microbiol. 1997;47(4):1205–1207. doi: 10.1099/00207713-47-4-1205. [DOI] [PubMed] [Google Scholar]
- Shojaei H, Heidarieh P, Hashemi A, Feizabadi MM, Naser AD. Species identification of neglected nontuberculous mycobacteria in a developing country. Jpn J Infect Dis. 2011;64:265–271. [PubMed] [Google Scholar]
- Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Evol Microbiol. 1997;47(2):479–491. [Google Scholar]
- Susarla S, Medina VF, McCutcheon SC. Phytoremediation: an ecological solution to organic chemical contamination. Ecol Eng. 2002;18:647–658. doi: 10.1016/S0925-8574(02)00026-5. [DOI] [Google Scholar]
- Thompson RC, Moore CJ, Vom Saal FS, Swan SH. Plastics, the environment and human health: current consensus and future trends. Philos Trans R Soc B Biol Sci. 2009;364:2153–2166. doi: 10.1098/rstb.2009.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin Microbiol Rev. 2003;16(2):319–354. doi: 10.1128/CMR.16.2.319-354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamura M, Mizuno S. Mycobacterium obuense, a rapidly growing scotochromogenic mycobacterium capable of forming a black product from p-aminosalicylate and salicylate. Microbiol. 1971;68:129–134. doi: 10.1099/00221287-68-2-129. [DOI] [PubMed] [Google Scholar]
- Tsukamura M, Mizuno S. A new species of rapidly growing scotochromogenic mycobacteria, Mycobacterium neoaurum. Med Biol. 1972;85:229–233. [Google Scholar]
- Tsukamura M, Nemoto H, Yugi H. Mycobacterium porcinum sp. nov., a porcine pathogen. Int J Syst Evol Microbiol. 1983;33(2):162–165. [Google Scholar]
- Uotila J, Kitunen V, Saastamoinen T, Coote T, Häggblom M, Salkinoja-Salonen M. Characterization of aromatic dehalogenases of Mycobacterium fortuitum CG-2. J Bacteriol. 1992;174:5669–5675. doi: 10.1128/jb.174.17.5669-5675.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virkutyte J, Sillanpää M, Latostenmaa P. Electrokinetic soil remediation—critical overview. Sci Total Environ. 2002;289:97–121. doi: 10.1016/S0048-9697(01)01027-0. [DOI] [PubMed] [Google Scholar]
- Ward OP, Singh A. Biodegradation and bioremediation. Encyclopedia of natural resources: land. Boca Raton: Taylor & Francis, CRC Press; 2014. pp. 39–46. [Google Scholar]
- Watanabe K. Microorganisms relevant to bioremediation. Curr Opin Biotechnol. 2001;12:237–241. doi: 10.1016/S0958-1669(00)00205-6. [DOI] [PubMed] [Google Scholar]
- Wayne L. Mycobacterial speciation. The mycobacteria: a sourcebook: 42–43. New York: Marcel Dekker, Inc.; 1984. [Google Scholar]
- Wei W, Wang F-Q, Fan S-Y, Wei D-Z. Inactivation and augmentation of the primary 3-ketosteroid-Δ1-dehydrogenase in Mycobacterium neoaurum NwIB-01: biotransformation of soybean phytosterols to 4-androstene-3, 17-dione or 1,4-androstadiene-3, 17-dione. Appl Environ Microbiol. 2010;76(13):4578–4582. doi: 10.1128/AEM.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PM, Potter TL, Culbreath AK. Fungicide dissipation and impact on metolachlor aerobic soil degradation and soil microbial dynamics. Sci Total Environ. 2010;408:1393–1402. doi: 10.1016/j.scitotenv.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Willumsen P, Karlson U, Stackebrandt E, Kroppenstedt RM. Mycobacterium frederiksbergense sp. nov., a novel polycyclic aromatic hydrocarbon-degrading Mycobacterium species. Int J Syst Evol Microbiol. 2001;51(5):1715–1722. doi: 10.1099/00207713-51-5-1715. [DOI] [PubMed] [Google Scholar]
- Wilson KH, Blitchington R, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun L (2012) Microbial degradation of polychlorophenols. In: Microbial degradation of xenobiotics. Springer, Berlin, pp 1–30
- Yokota A, Takeuchi M, Weiss N. Proposal of two new species in the genus Microbacterium: Microbacterium dextranolyticum sp. nov. and Microbacterium aurum sp. nov. Int J Syst Evol Microbiol. 1993;43(3):549–554. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data in this manuscript were deposited in publicly available repositories in the Department of Microbiology, School of Medicine, Isfahan University of Medical Sciences, and Isfahan, Iran.


