Abstract
Trimethylation of lysine 27 on histone 3 (H3K27me3) by the Polycomb repressive complex 2 (PRC2) contributes to localized and inherited transcriptional repression. Kdm6b (Jmjd3) is a H3K27me3 demethylase that can relieve repression-associated H3K27me3 marks, thereby supporting activation of previously silenced genes. Kdm6b is proposed to contribute to early developmental cell fate specification, cardiovascular differentiation, and/or later steps of organogenesis, including endochondral bone formation and lung development. We pursued loss-of-function studies in zebrafish to define the conserved developmental roles of Kdm6b. kdm6ba and kdm6bb homozygous deficient zebrafish are each viable and fertile. However, loss of both kdm6ba and kdm6bb shows Kdm6b proteins share redundant and pleiotropic roles in organogenesis without impacting initial cell fate specification. In the developing heart, co-expressed Kdm6b proteins promote cardiomyocyte proliferation coupled with the initial stages of cardiac trabeculation. While newly formed trabecular cardiomyocytes display a striking transient decrease in bulk cellular H3K27me3 levels, this demethylation is independent of collective Kdm6b. Our results indicate a restricted and likely locus-specific role for Kdm6b demethylases during heart ventricle maturation rather than initial cardiogenesis.
Keywords: H3K27me3, histone demethylase, Kdm6b, cardiogenesis, ventricle, trabeculation
INTRODUCTION
Covalent histone modifications establish chromatin landscapes that influence gene expression programs by promoting active or silenced transcriptional states. For instance, localized trimethylation of lysine 27 on histone 3 (H3K27me3) in vertebrates is deposited by Enhancer of zeste homolog 1 and 2 (Ezh1 and 2), the catalytic subunits of the Polycomb repressive complex 2 (PRC2), and is associated with gene silencing (Cao et al., 2002; Margueron and Reinberg, 2011; Müller et al., 2002). H3K27me3 marks can be removed by the lysine-specific demethylases Kdm6a (Utx) and Kdm6b (Jmjd3) (Cloos et al., 2008; Klose and Y. Zhang, 2007; Mosammaparast and Shi, 2010). Therefore, H3K27me3 modifications likely provide an epigenetically inherited but reversible layer of transcriptional control. In vertebrates, H3K27me3 dynamics are implicated in cell fate specification (Alder et al., 2010; Dahl et al., 2010; Rugg-Gunn et al., 2010) and cell reprogramming associated with acquired pluripotency (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007). Further, vertebrate H3K27me3 demethylases directly support cell signal or transcription factor-induced gene expression programs (Jiang et al., 2013; Kartikasari et al., 2013; Ramadoss et al., 2012) and de-differentiation during organ regeneration (Stewart et al., 2009).
Studies using embryonic stem cells (ES cells) suggest Kdm6b enables cell specification of all three germ layers (Burgold et al., 2008; Kartikasari et al., 2013; Ohtani et al., 2011; 2013). These results predict that Kdm6b is responsible for relieving H3K27me3 marks at fate specifying genes as pluripotent cells adopt distinct cell lineage identities (Bernstein et al., 2006; Mikkelsen et al., 2007). However, in vivo tests of this model using mouse Kdm6b reverse genetics have yielded conflicting results. Mice homozygous for a Kdm6b allele that deletes exons encoding N-terminal regions of the protein are peri-implantation lethal at embryonic day 6.5 (E6.5) (Ohtani et al., 2013). In contrast, mice homozygous for several independently generated Kdm6b mutant alleles that delete the catalytic JmjC domain die perinatally with lung defects (Q. Li et al., 2014; Satoh et al., 2010; Shpargel et al., 2014; F. Zhang et al., 2015). These latter studies suggest that Kdm6b-driven H3K27me3 demethylation does not instructively derepress key cell fate regulatory genes during early embryonic development.
Kdm6b-deficient mouse embryos are smaller and edematous (Q. Li et al., 2014), implicating Kdm6b in cardiovascular development. In further support, H3K27me3 marks are conspicuously lost from key cardiogenic genes during in vitro cardiac differentiation of ES cells (Paige et al., 2012; Wamstad et al., 2012) and PRC2 is required to maintain cardiomyocyte identity (Delgado-Olguín et al., 2012; He et al., 2012; San et al., 2016). Additionally, Kdm6b−/− ESCs induced to undergo cardiomyocyte differentiation misexpress many genes implicated in early heart development, including decreased levels of the cardiac progenitor factors Mesp1, Pdgfra, and Mef2c (Ohtani et al., 2011). Correspondingly, Kdm6b-deficient ES cells exhibit reduced ability to form differentiated cardiomyocytes and contractile embryoid bodies (EBs). Although in vivo mouse and zebrafish studies demonstrate Kdm6a contributes to the activation of core cardiac transcription factors (Lee et al., 2012; Van Laarhoven et al., 2015; Welstead et al., 2012), the in vivo contributions of Kdm6b to cardiovascular development have not been examined.
We pursued in vivo loss-of-function studies in zebrafish to define the evolutionarily conserved contributions of Kdm6b to vertebrate development. Due to an ancestral whole genome duplication in the teleost lineage (Amores, 1998; Jaillon et al., 2004; Meyer and Schartl, 1999), the zebrafish genome contains two Kdm6b orthologs, kdm6ba and kdm6bb. In such instances of duplicated genes, the “ohnologs” frequently acquire distinct roles or one ohnolog becomes nonfunctional due to relaxed selective pressure (Force et al., 1999; Lynch and Force, 2000; Postlethwait, 2007; Steinke et al., 2006). However, redundant functions, with or without compensatory networks, are also possible. While kdm6bb may support the reactivation of embryonic developmental gene programs during fin regeneration (Stewart et al., 2009), kdm6ba has not been studied in any context.
We generated kdm6ba and kdm6bb loss-of-function alleles using CRISPR/Cas9 mutagenesis. Zebrafish homozygous for either individual allele are viable and fertile. However, kdm6ba−/−; kdm6bb−/− zebrafish larvae have pleiotropic yet specific organogenesis defects, including a small heart ventricle, consistent with coinciding heart expression. The small ventricle originates from insufficient proliferation of both endocardial and myocardial cells associated with the initiation of cardiac trabeculation at larval hatching. This burst of trabecular cardiomyocyte proliferation correlates with transient bulk cellular H3K27me3 demethylation, implying a global relaxing of gene repression enables the cell cycle entry transition. However, collective Kdm6b does not appear to promote this observed cell-wide demethylation and therefore likely has locus-specific roles. Our data provide an example of straightforward non-compensatory redundancy between duplicated genes in zebrafish. Further, we show that Kdm6b’s unique functions (those not shared with Kdm6a) are largely restricted to later stages of organ growth and morphogenesis rather than early cell fate specification or progenitor cell activity.
MATERIALS AND METHODS
Zebrafish
The University of Oregon Institutional Animal Care and Use Committee (IACUC) approved and monitored all zebrafish procedures following the guidelines and recommendations outlined by the Guide for the Care and Use of Laboratory Animals (National Academic Press). Wildtype AB, Tg(myl7:dsRedExpress-nuc) (Takeuchi et al., 2011), and Tg(kdrl:EGFP) (Jin et al., 2005) lines were used in this study.
CRISPR-Cas9 generation of mutant alleles
CRISPR-Cas9-mediated mutagenesis was used to generate targeted deletions within the JmjC domains of both kdm6ba and kdm6bb. Guide RNAs (gRNAs) were generated following methods adapted from (Bassett et al., 2013). Guide oligonucleotides were obtained from Integrated DNA Technologies and had the following core sequence: 5′-AATTAATACGACTCACTATA-NNNNNNNNNNNNNNNNNNNN-GTTTTAGAGCTAGAAATAGC-3′, with a T7 promoter sequence followed by a series of Ns indicating the targeting sequence. The gRNA targeting sequences for kdm6ba and kdm6bb were 5′-GCTTAGCATATTCCCACTGG-3′ and 5′-CAGGTGGAAGGC-GCAGTTGC-3′, respectively. Guide oligonucleotides were annealed to a generic gRNA scaffold (5′-GATCCGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACT-AGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′) and extended using Phusion polymerase (New England Biolabs) followed by column-based purification.
In vitro transcription of gRNAs was performed using the Megascript T7 Transcription Kit (Thermo Fisher) and purified via RNA Clean & Concentrator columns (Zymo Research). Cas9 mRNA was in vitro transcribed from Addgene plasmid 46757:pT3TS-nCas9n (Jao et al., 2013) using the T3 mMessage mMachine kit (Thermo Fisher). Cas9 mRNA and gRNA were combined and diluted to a final concentration of 100 ng/μl in Danieau’s buffer containing phenol red. We injected 1–3 nl of the Cas9/gRNA mix into the developing embryo at the one-cell stage. Fish raised from injected embryos (see below) were screened for mutations by amplicon sequencing. Founders carrying isolated mutant alleles were outcrossed to wildtype fish to isolate stable germline transmitting lines used for subsequent crosses and phenotypic analyses.
Genotyping kdm6ba and kdm6bb mutant alleles
The kdm6ba allele was assessed by PCR using primers 5′-TGGCACAAACATTGACCTGT-3′ and 5′-CATCATGGACAGCAAACCAC-3′ followed by restriction digest with NcoI (New England Biolabs). The kdm6bb mutant allele was assessed by PCR using primers 5′-CTCAAAGCAGAAAGCTGTTGG-3′ and 5′-GTGTGGCCAACATGACTCAG-3′.
In situ hybridization
In situ hybridization probes were synthesized from PCRII-based plasmids (Life Technologies) containing cloned cDNA amplicons. PCR products were obtained from pooled samples of embryonic zebrafish cDNA using the following primers: kdm6bb 5′-CCACTTGACCAACTGCCTTGCAAAAC-3′, 5′-CTGAAAACACACTCCGAGAGGTATCGC-3′; kdm6ba 5′-TGGCACAAACATTGACCTGT-3′, 5′-GTGTGAGGGAAAGGGATGAG-3′. Plasmids were linearized by digestion with NotI and BamHI, respectively. Digoxigenin-11-dUTP (Roche) labeled RNA synthesis was performed using Sp6 (for kdm6bb probe) or T7 (for kdm6ba probe). DIG-labeled probes were DNase treated prior to LiCl precipitation and then resuspended in RNase/DNase free water. Whole-mount in situ hybridizations were performed as described (C. Thisse and B. Thisse, 2008). Hybridized embryos were blocked with 5% normal goat serum in phosphate-buffered saline (PBS) and then incubated overnight with alkaline phosphatase-conjugated anti-DIG antibody (Roche) diluted 1:1000 in blocking buffer. Embryos were developed at room temperature using NBT/BCIP (Promega) and then dehydrated into 100% methanol and stored at −20°C. Embryos were then rehydrated and allowed to equilibrate in 100% glycerol at 4°C prior to imaging on a Leica M165 FC stereomicroscope.
Whole mount immunofluorescence and imaging
Anti-GFP (AVES) immunostaining was performed as described (A. A. Akerberg et al., 2014). Staining for cardiac troponin (CT3, Developmental Studies Hybridoma Bank), DsRed (Clontech) and S46 (Developmental Studies Hybridoma Bank) was performed using a protocol adapted from (Zhou et al., 2011). Embryonic hearts were arrested in diastole with 0.5M KCl and fixed overnight at 4°C in phosphate-buffered saline (PBS) containing 4% paraformaldehyde. The following day, embryos were washed in PBS and dehydrated through a methanol series. Embryos in 100% methanol were stored at −20°C for at least 24 hours prior to rehydration into PBS containing 0.1% Tween-20 (PBST). Embryos were then blocked with PBS containing 1% BSA, 1% Triton X-100, 0.1% DMSO for four hours. Embryos were incubated with primary antibodies diluted in block solution for 4–5 hours at room temperature with gentle agitation followed by three 20′ washes in PBS containing 0.5% Triton X-100 (PB0.5X). Alexa-conjugated secondary antibodies (Thermo Fisher) were diluted 1:1000 in PB0.5X and incubated with embryos for 2–3 hours at room temperature with gentle agitation. Lastly, embryos were washed and stored in TBST (20 mM Tris pH 7.5, 150 mM NaCl, 0.5% Triton X-100) prior to imaging. Primary antibodies CT3, DsRed, and S46 were used at dilutions of 1:500, 1:500, and 1:250, respectively. Immunostained embryos as well as transgenic embryos with native fluorescence were mounted in low melt agarose and imaged with a Leica M165 FC stereomicroscope, Nikon Eclipse Ti inverted fluorescence microscope, or Leica SD6000 spinning disk confocal microscope. 10x DIC images were acquired using a Nikon Eclipse Ti inverted microscope and stitched together using NIS Elements Advanced Research software.
Live embryo imaging
Embryos of the desired stage were anesthetized in Tricaine (Western Chemical) and immobilized in 0.75% low melt agarose dissolved in embryo media (EM). Embryos were imaged on a glass bottom FluoroDish (World Precision Instruments) using epifluorescence or DIC optics with a Nikon Eclipse Ti inverted microscope.
Cell counting and morphometrics
Cardiomyocyte quantification was performed on Tg(myl7:dsRed-nuc) transgenic embryos that first were fixed overnight in PBS containing 4% paraformaldehyde. Embryos were then immediately antibody stained for dsRed and CT3. Stained embryos were mounted in low melt agarose after their heads were removed to provide an unobstructed view of the heart. Mounted embryos were imaged with a Leica SD6000 spinning disk confocal microscope. Image stacks were processed and nuclei counted using ImageJ (National Institutes of Health). Area measurements of trabeculated muscle located between OFT and the atrioventricular canal (AVC) used ImageJ measurement tools. Trabeculation index values were calculated as the ratio between the length of the interior (trabecular) and exterior of the ventricle. Individual data points were normalized to the mean of respective clutch mate control samples.
Skeletal staining
Alcian Blue staining of zebrafish larvae was performed as described (Walker and Kimmel, 2007) with subsequent imaging using a Leica M165 FC stereomicroscope (whole mount) or Leica DM4000B upright microscope (flat mount).
Histological sectioning and immunostaining
Paraffin sections were processed following procedures used for dissected mouse hearts (B. N. Akerberg et al., 2015). Hematoxylin and eosin staining on paraffin sections was performed using conventional reagents and methodology (Ricca Chemical Company). For immunostaining paraffin sections, slides were de-paraffinized using Xylenes and rehydrated into distilled water using a series of graded ethanol washes. Antigens were retrieved by either 1) pressure-cooking for 10′ in 1 mM EDTA and 0.1% Tween-20 (CT3, PCNA (Sigma), myosin heavy chain (MF20, Developmental Studies Hybridoma Bank), GFP, and DsRed antibodies) or 2) 10′ incubation in 0.25% trypsin solution (H3K27me3 antibody, Millipore). Slides were blocked for 1 hour with 10% dried milk, 0.1% Tween-20 or 10% normal goat serum (for anti-H3K27me3) in PBS. Primary antibodies were diluted 1:500 (or 1:50 for MF20) in their respective block solutions and incubated overnight at 4°C. Slides were then washed 5 × 10′ in PBST (1x PBS with 0.1% Tween-20) and incubated with Alexa-conjugated secondary antibodies diluted 1:1000 in block solution for 1 hr. After 3 × 10′ PBST washes, the slides were submerged for 5′ in Hoechst diluted 1:5000 in PBST. Slides were washed in PBST and mounted with Fluorogel (Electron Microscopy Sciences). Imaging was performed with a Leica upright fluorescent microscope or either an Olympus Fluoview FV1000 or Zeiss LSM 880 laser scanning confocal microscope.
5-ethynyl-2-deoxyuridine labeling
Fish were incubated with 5-ethynyl-2-deoxyuridine (EdU) (Thermo Fisher) at a concentration of 0.1 mg/ml in embryo media for 12–15 hours prior to harvest at 5 dpf. Embryos were processed into paraffin sections as described above. EdU Click-It reactions (Thermo Fisher) were performed prior to immunostaining with the samples kept in the dark throughout subsequent staining procedures.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
cDNA was synthesized using SuperScript III (Thermo Fisher) with Trizol-isolated RNA harvested from pooled embryos (5–10 embryos per pool) or dissected embryonic hearts. qRT-PCR was performed using KAPA SYBR FAST qPCR master mix reagents (Kapa Biosystems). Relative mRNA expression was normalized using rpl18 or rps11 to calculate ΔCTs (threshold cycles). Transcript levels were compared using a ΔΔCT approach. ΔCT values were used for two-tailed Student’s t-tests to determine significance with a Bonferroni correction for multiple comparisons. qPCR primer sequences: kdm6bb 5′-GCATGGCGTGGACTATCTG-3′, 5′-CGTGAATCATAGGGACGATTG-3′; kdm6ba 5′-GCCCAAACCTCTCCAATGTA-3′, 5′-GTGTGAGGGAAAGGGATGAG-3′; kdm6a 5′-ACATAAACATTGGCCCTGGA-3′, 5′-GCCACCAAGATCCCATTAGA-3′; kdm6al 5′-CACAACAACTTCTGCGCTGT-3′, 5′-CCTCGTACAGGTCCTCCAGA-3′; rpl18 5′-CCGAGACCAAGAAATCCAGA-3′, 5′-GAGGCCAGCAGTTTCTCTTG-3′; ′rps11 ′5′-GATGGCGGACACTAGAAC-3′, ′5′-CCAATCCAACGTTTCTGTGA -3′; vmhcl 5′-GATGGAACTGAGGATGCTGAC -3′, 5′-TTGACTCTTGGATGGCACAG -3′
RESULTS
Histone demethylases kdm6ba and kdm6bb have non-compensatory redundant functions during zebrafish development
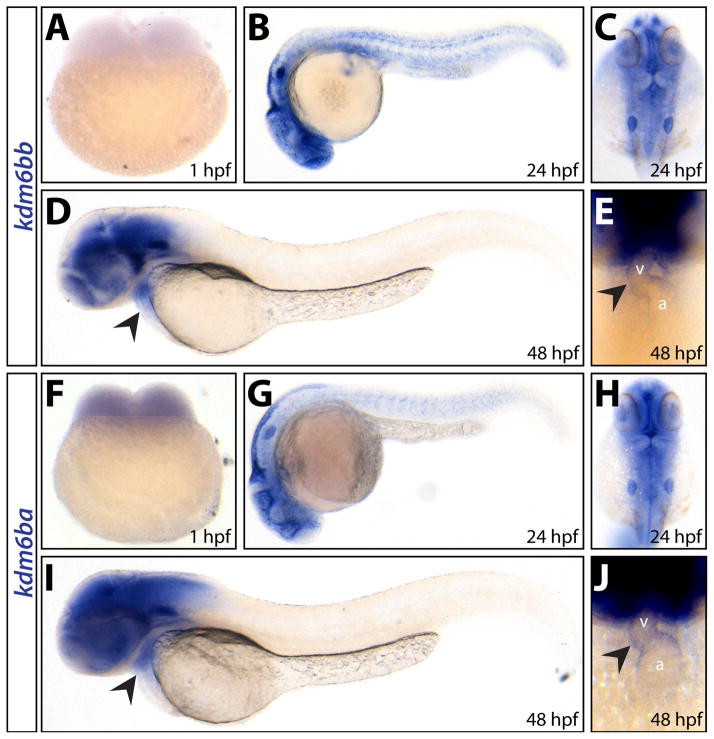
We monitored embryonic expression of the kdm6b H3K27me3 histone demethylase ohnologs by RNA in situ hybridization (Fig. 1A–J). kdm6ba transcripts were uniquely maternally contributed (Fig. 1F) whereas both kdm6ba and kdm6bb transcripts were robustly expressed in the developing brain (Fig. 1B–C and G–H), otic vesicles (Fig. 1C and H), and heart (Fig. 1D–E and I–J). The grossly similar expression patterns of kdm6ba and kdm6bb during early development suggested redundancy. Therefore, we generated mutant alleles of both genes using CRISPR/Cas9 targeted mutagenesis to investigate their unique and shared roles during zebrafish development.
Figure 1. kdm6ba and kdm6bb have broad and largely overlapping developmental expression patterns.
(A–J) Whole mount in situ hybridization for kdm6bb (A–E) and kdm6ba (F–J) transcripts on 1 hpf (A and F), 24 hpf (B, C, G, H) and 48 hpf (D, E, I, J) embryos. Arrowheads denote cardiac expression. Abbreviations: a, atrium; v, ventricle.
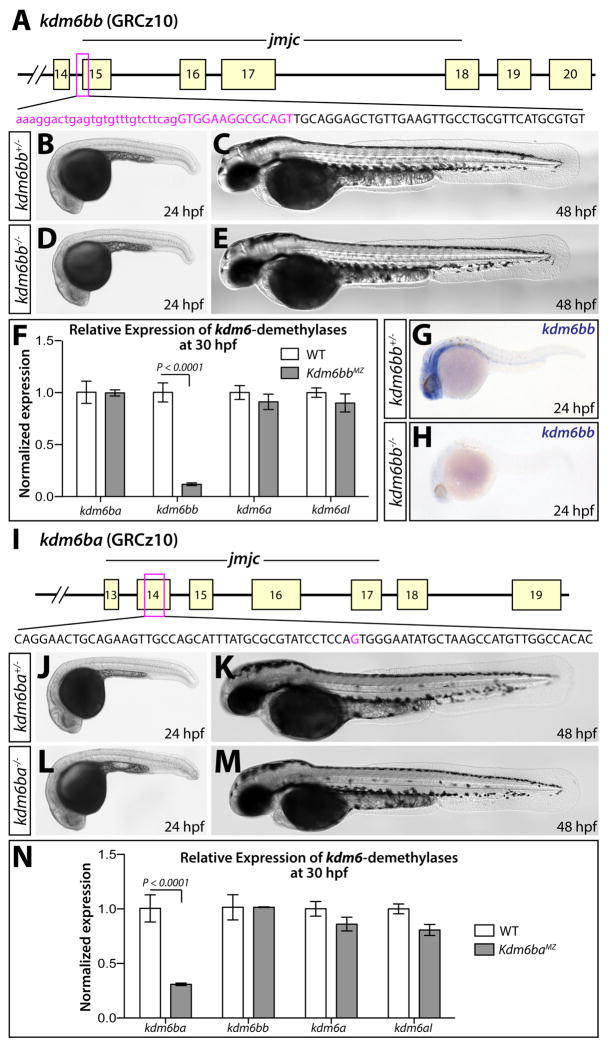
We first targeted insertion-deletion (indel) mutations to the 5′ end of the essential catalytic JmjC domain of kdm6bb (De Santa et al., 2007; Lan et al., 2007; Hong et al., 2007; Xiang et al., 2007), isolating a germline-transmitted allele (kdm6bbb1307, abbreviated kdm6bb− from here forth) harboring a 40 bp deletion spanning the first 14 bp of exon 15 (Fig. 2A). This deletion introduces an early stop codon predicted to result in a truncated Kdm6bb protein lacking the entire JmjC domain. kdm6bb−/− fish did not exhibit any overt developmental defects and were both viable and fertile as adults (Fig. 2B–E). Vascular organization was also unchanged in kdm6bb−/− embryos at 3 dpf as visualized by the Tg(kdrl:EGFP) reporter line (Fig. S1A, B). Notably, kdm6bb transcript levels were decreased nearly nine fold in homozygous kdm6bb−/− animals at 30 hours post fertilization (hpf) presumably due to nonsense mediated decay, as shown by qRT-PCR and in situ hybridization (Fig. 2F–H). Expression levels of the other three Kdm6-family demethylases kdm6ba, kdm6a, and kdm6al were not significantly different in kdm6bb−/− animals. Therefore, genetic deficiency of kdm6bb does not trigger transcriptional compensation (Fig. 2F).
Figure 2. Generation and characterization of kdm6b loss of function alleles.
(A) Schematic showing the deleted sequence in the kdm6bb mutant allele recovered from CRISPR/Cas9 targeted mutagenesis (magenta bases). (B–E) Whole mount differential interference contrast (DIC) microscopy images of 24 (B and D) and 48 hpf (C and E) embryos. (F) Bar graph showing relative kdm6bb transcript levels between wildtype and kdm6bbMZ embryos at 30 hpf determined by qRT-PCR on cDNA from three separate pools of ten embryos each. (G, H) In situ hybridization for kdm6bb on control and kdm6bbMZ 24 hpf zebrafish embryos. (I) Schematic showing the CRISPR/Cas9-generated kdm6ba mutant allele. The deleted base is shown in magenta. (J–M) DIC images of control and kdm6baMZ embryos at 24 hpf (J and L) and 48 hpf (K and M). (N) Bar graphs comparing qRT-PCR-determined normalized expression levels of each H3K27me3 demethylase-encoding transcripts between 30 hpf wildtype and kdm6bbMZ embryos (three independent pools of ten embryos each). Error bars in the bar graphs (F, N) show one standard deviation. P-values indicate a significant difference determined by Student’s two-tailed t-tests after applying a Bonferroni correction for multiple comparisons.
We subsequently isolated a kdm6ba mutant allele (kdm6bab1308, henceforth denoted kdm6ba−) containing a single base deletion within exon 14 of kdm6ba (Fig. 2I). This frameshift mutation is predicted to result in an early stop codon yielding a truncated Kdm6ba protein missing the JmjC domain. kdm6ba−/− embryos lacking the notable maternal contribution (kdm6baMZ) showed no overt embryonic defects at 24 and 48 hpf and were both viable and fertile (Fig. 2J–M). kdm6ba−/− embryos also exhibited normal vascular development at 3 dpf (Fig S1C, D). kdm6baMZ animals expressed three fold less kdm6ba mRNA than their wildtype clutchmates (Fig. 2N), likely reflecting nonsense mediated decay and further signifying kdm6ba loss of function. qRT-PCR showed that kdm6ba-deficiency did not elicit compensatory expression of kdm6bb, kdm6a, or kdm6al at 30 hpf (Fig. 2N).
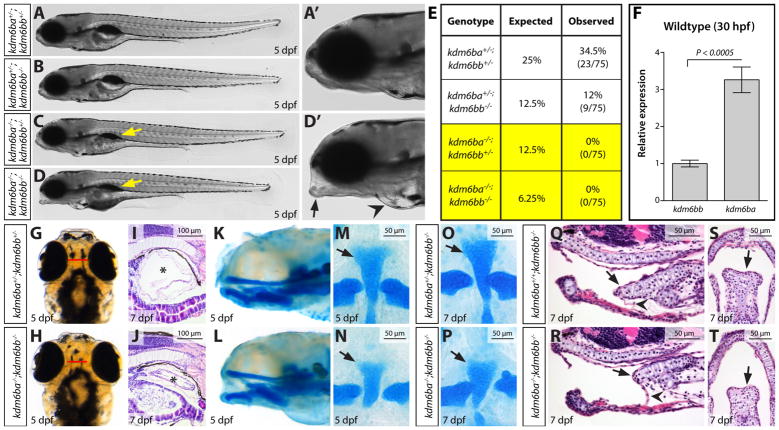
To determine if the H3K27me3 demethylases kdm6ba and kdm6bb have redundant functions, we generated kdm6ba−/−; kdm6bb−/− double mutant (or kdm6b-deficient) animals. Although kdm6ba−/−; kdm6bb−/− fish developed normally for their first three days (Fig. S2A–D), they failed to inflate their swim bladder upon hatching and were not viable past 10–12 dpf (Fig. 3A–E). kdm6b-deficient larvae also consistently exhibited cardiac edema (Fig. 3D–D′) and a craniofacial “slack jaw” abnormality in which the jaw was unable to close (Fig. 3D–D′). kdm6ba−/−; kdm6bb+/− animals also failed to develop a swim bladder and did not survive to adulthood (Fig. 3C and E). Conversely, kdm6ba+/−; kdm6bb−/− animals were completely viable with no obvious embryonic defects (Fig. 3B and E). This weighted dose-dependency suggests that Kdm6ba contributes relatively more to its and Kdm6bb’s shared developmental roles, possibly explained by threefold higher transcript levels of kdm6ba over kdm6bb at 30 hpf (Fig. 3F).
Figure 3. Combined genetic deficiency of kdm6ba and kdm6bb causes pleiotropic larval development defects.
(A–D) Whole mount stitched DIC microscopy images of clutch mate 5 dpf larvae of the indicated genotypes. (A′ & D′) Zoomed anterior region of the control and kdm6ba−/−; kdm6bb−/− fish shown in A and D, respectively. Yellow arrows mark the absence of an inflated swim bladder. The black arrow and black arrowhead highlight the slack jaw and cardiac edema phenotypes, respectively, of a kdm6ba−/−; kdm6bb−/− larvae. (E) Viability table showing the expected versus observed adult genotype frequencies from kdm6ba+/−; kdm6bb+/− in-crosses. (F) Bar graph displaying relative kdm6ba and kdm6bb transcript levels in 30 hpf wildtype embryos. Data is normalized to rpl18 expression. P-value is from a Student’s two-tailed t-test with error bars showing one standard deviation. (G, H) Whole mount dorsal view of 5 dpf control and clutch mate kdm6ba/kdm6bb-deficient larvae. (I, J) H&E-stained sagittal sections through 7 dpf larvae centered on the swim bladder (asterisk). (K, L) Sagittal whole mount view of alcian blue stained 5 dpf larvae. (M–P) Flat mounts of alcian blue-stained 5 dpf (M, N) and 7 dpf (O, P) larvae of the indicated genotypes. Black arrows mark the basihyal cartilage. (Q–T) H&E-stained sagittal (Q, R) and coronal (S, T) midline sections through 7 dpf kdm6bb-null and kdm6ba/kdm6bb-double deficient larvae. Arrows indicate the basihyal cartilage and arrowheads point at the cartilage’s associated ligament. 50 or 100 μm scale bars are shown.
A further analysis of 5 dpf kdm6ba−/−; kdm6bb−/− larvae revealed several additional defects. kdm6b-deficient animals exhibited cranial swelling arising between 4 and 5 dpf (Fig. 3G–H). Hematoxylin and eosin (H&E) staining of paraffin sections confirmed an intact but un-inflated swim bladder in kdm6ba−/−; kdm6bb−/− larvae (Fig. 3I–J). As Kdm6b promotes endochondral bone formation in mice (F. Zhang et al., 2015), we performed Alcian Blue staining to determine if kdm6b-deficient larvae exhibited abnormal cartilaginous bone. Whereas the majority of craniofacial skeletal development was unaffected, kdm6ba−/−; kdm6bb−/− larvae displayed a truncated basihyal cartilage at 5 dpf and 7 dpf that likely accounts for the “slack-jaw” phenotype (Fig. 3K–P). By H&E staining of 7 dpf larvae, we found that the reduced basihyal in kdm6b-deficient larvae contained subjectively fewer but morphologically normal chondrocytes and retained associated ligaments (Fig. 3Q–T). These phenotypes and the larval stage lethality of kdm6ba−/−; kdm6bb−/− fish are congruent with those mouse studies that proposed Kdm6b largely functions during later stages of organogenesis, rather than during early cell lineage specification (Burgold et al., 2012; Q. Li et al., 2014; Satoh et al., 2010; Shpargel et al., 2014; F. Zhang et al., 2015).
Kdm6b proteins promote larval stage cell proliferation associated with heart ventricle trabeculation
The absence of embryonic phenotypes in kdm6ba−/−; kdm6bb−/− embryos does not eliminate the possibility that maternally contributed mRNA or protein contribute to early lineage specification (Burgold et al., 2008; Kartikasari et al., 2013; Ohtani et al., 2013; 2011). Although the lethality of kdm6ba−/−; kdm6bb−/− animals precluded us from eliminating maternal contributions of both transcripts, only kdm6ba transcripts were present prior to zygotic genome activation (Fig. 1A and F). To achieve maximum kdm6b depletion in the early embryo with respect to both maternal and zygotic contributions, we crossed kdm6ba−/−; kdm6bb+/+ females to kdm6ba+/−; kdm6bb−/− males. kdm6baMZ; kdm6bb+/− progeny from this cross were phenotypically indistinguishable from kdm6ba−/−; kdm6bb+/− animals, lacking a swim bladder and dying as larvae. Further, kdm6baMZ; kdm6bb+/− larvae did not develop the severe edema and slack-jaw phenotype seen in kdm6ba−/−; kdm6bb−/− fish with intact maternal but not zygotic kdm6ba (Fig. S3A–C). Therefore, kdm6b demethylases likely do not have unique early embryogenesis roles necessitating maternally contributed mRNA or protein.
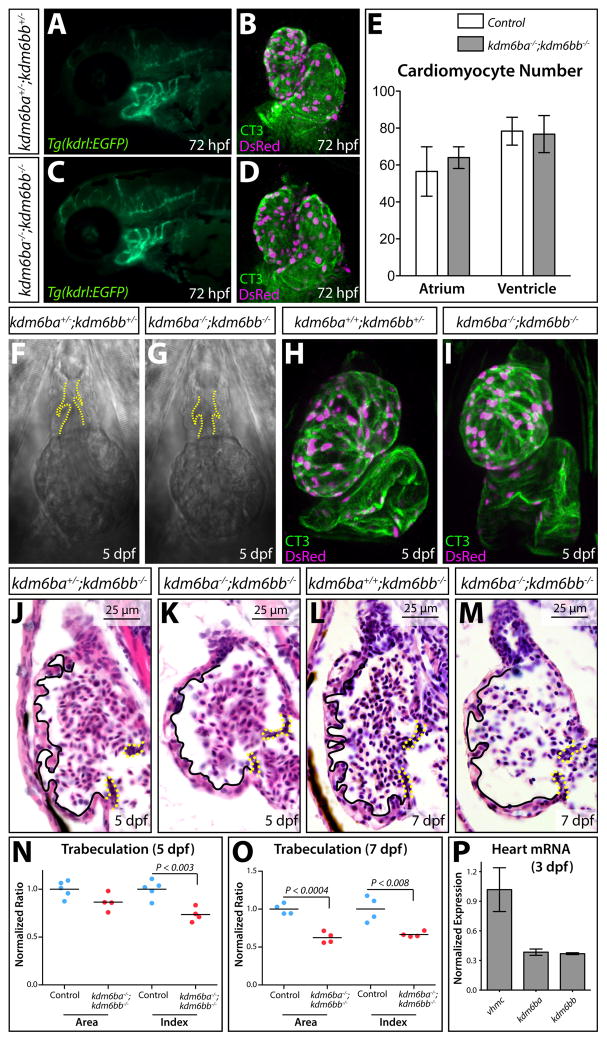
Despite the lack of embryonic cardiovascular phenotypes, the presence of cardiac edema in 5 dpf kdm6ba−/−; kdm6bb−/− larvae implied Kdm6b demethylases are involved in cardiovascular development. The Tg(kdrl:EGFP) vascular-marking transgene showed that 3 dpf kdm6ba−/−; kdm6bb−/− larvae have grossly normal aortic arch artery development (Fig. 4A and C). Likewise, cardiogenesis through 3 dpf, including the generation of Tg(myl7:DsRed-nuc)-marked cardiomyoctes, was unaffected by kdm6b deficiency (Fig. 4B and D–E). Live imaging revealed that hearts of kdm6b-deficient 5 dpf larvae produced unidirectional and steady blood flow, consistent with their normal OFTs and OFT valves (Movie S1, Fig. 4F–G). However, kdm6ba−/−; kdm6bb−/− larvae had smaller and more spherically-shaped ventricles, which was also apparent by whole mount staining for cardiac troponin (CT3) and DsRed in a Tg(myl7:DsRed-nuc) background (Fig. 4H–I). To further characterize ventricle morphology in kdm6b-deficient animals, we analyzed H&E stained sagittal sections of 5 and 7 dpf larvae. 5 dpf kdm6ba−/−; kdm6bb−/− larvae had a smaller and more rounded ventricle with greatly reduced number and size of trabecular projections that normally form on the outer curvature of the zebrafish ventricle between 3 and 4 dpf (Liu et al., 2010) (Fig. 4J–K). The OFT, atria, and atrioventricular canal (AVC) valves were notably unaffected. The specific trabeculation defect was even more apparent at 7 dpf (Fig. 4L–M). Quantitatively, kdm6b-deficient larvae first showed diminished trabeculation at 5 dpf followed by reduced ventricular muscle at 7 dpf (Fig. 4N, O, Fig. S4). Both kdm6ba and kdm6bb were robustly expressed in isolated 3.5 dpf hearts as determined by qRT-PCR (Fig. 4P) We conclude that kdm6ba and kdm6bb share a specific and heart autonomous role promoting early stages of zebrafish cardiac trabeculation.
Figure 4. Kdm6ba and kdm6bb redundantly promote cardiac trabeculation.
(A, C) Fluorescent imaged whole mount 72 hpf embryos showing the aortic arch arteries of control and kdm6b-deficient Tg(kdrl:EGFP) animals. (B, D) Confocal heart images of whole mount 72 hpf Tg(myl7:DsRed-nuc) larvae antibody stained for DsRed (red, cardiomyocyte nuclei) and cardiac troponin (CT3, heart muscle). Control and combined kdm6ba/kdm6bb-deficient larvae are shown. (E) Bar graphs comparing the absolute number of atrial and ventricular cardiomyocytes between 72 hpf control (kdm6ba+/−; kdm6bb+/− and kdm6ba+/+; kdm6bb+/−) and kdm6ba−/−; kdm6bb−/− larvae. Error bars represent one standard deviation (n=6 control and 7 kdm6ba/bb-deficient fish from two clutches). (F, G) DIC microscopy images showing the OFT and ventricle of control and kdm6b-deficient embryos at 5 dpf. Dashed yellow lines outline the OFT valves. (H, I) Whole mount confocal imaged hearts of immunostained 5 dpf control and kdm6ba−/−; kdm6bb−/− larvae. These Tg(myl7:DsRed-nuc) fish are stained with anti-DsRed (red, cardiomyocyte nuclei) and anti-cardiac troponin (CT3, green) antibodies. (J–M) H&E stained sagittal sections through the heart of 5 and 7 dpf control and kdm6ba−/−; kdm6bb−/− larvae. Solid lines outline the trabeculae and dashed yellow lines indicate the AVC valves. (N, O) Scatterplot graphs showing the normalized outer curvature ventricular sectional area and trabeculation extent (“index”) of 5 and 7 dpf control compared to clutch mate kdm6ba−/−; kdm6bb−/− larvae. Each data point represents one animal. P-values are from Student’s two-tailed t-tests. (P) Bar graph of qRT-PCR data comparing kdm6ba and kdm6bb to vmhcl transcript levels using three pools of isolated 3 dpf zebrafish embryonic hearts (normalized to vmhcl levels). Error bars are one standard deviation.
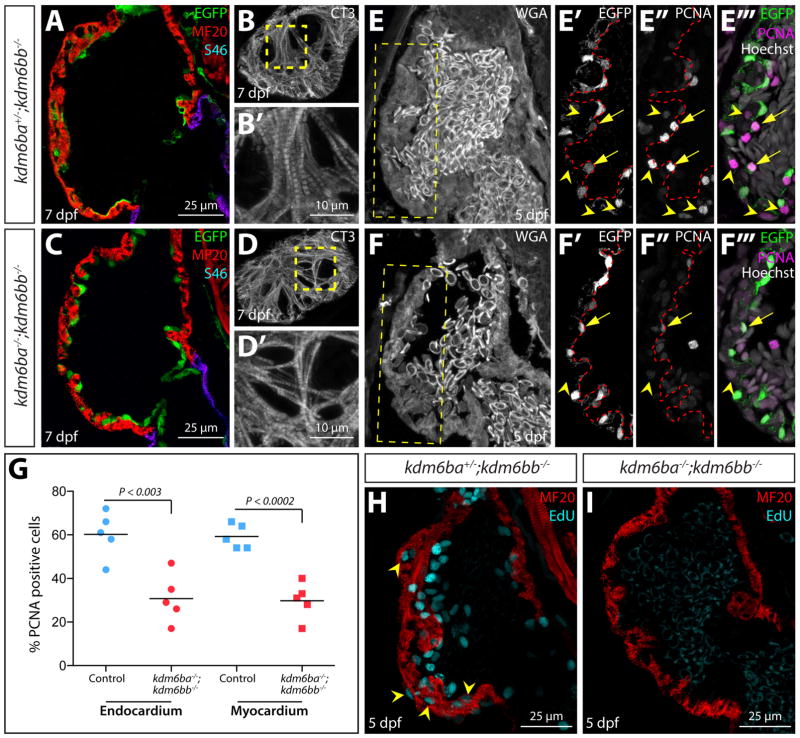
By immunostaining sectioned larvae, we found that myosin heavy chain (recognized using the pan-myocardial MF20 antibody) and S46 (atrial cardiomyocyte-specific) were expressed normally in 5 dpf kdm6ba−/−; kdm6bb−/− larval hearts (Fig. 5A–D′). Therefore, the diminished cardiac trabeculation in kdm6b-deficient larvae did not reflect an altered chamber identity. CT3 antibody staining further confirmed that ventricular cardiomyocytes of kdm6-deficient larvae were differentiated and formed myofibrils (Fig. 5B–B′ and D–D′). Finally, we used the Tg(kdrl:EGFP) reporter line and anti-GFP staining to determine that the endocardium of kdm6b-deficient embryos was present and maintained an epithelial organization (Fig. 5E′–F′). Collectively, these results show that Kdm6b is not required for the specification or differentiation of endocardial and myocardial cells and therefore that the trabeculation defects in kdm6b-deficient larvae likely represents a direct role of Kdm6b in trabecular growth and/or morphogenesis.
Figure 5. Kdm6b proteins enable coordinated endocardial and myocardial proliferation associated with early stage ventricle maturation.
(A–C) Confocal immunofluorescence images of sagittal heart sections from 5 dpf kdm6b- and kdm6a/kdm6b-deficient Tg(kdrl:EGFP) larvae stained with anti-GFP (green, endocardium), anti-myosin heavy chain (MF20, red, myocardium), and anti-S46 (magenta, atrial muscle) antibodies. (B, D) Cardiac troponin (CT3) antibody staining of superficial sections through the heart of 5 dpf kdm6bb−/− and kdm6ba−/−; kdm6bb−/− larvae. B′ and D′ are zoomed images of the dashed yellow box regions. (E–F) Confocal imaged sagittal sections of 5 dpf control and combined kdm6b-deficient Tg(kdrl:EGFP) embryos stained with wheat germ agglutinin (WGA, grey) as well as anti-GFP (green in overlay panels, endocardium) and anti-PCNA (magenta in overlay panels) antibodies. Yellow boxes in E and F indicate zoomed regions shown in E′–F‴. Arrows and arrowheads mark PCNA-positive endocardial and myocardial cells, respectively. (G) Scatterplot graphs showing the number of PCNA positive endocardial and myocardial cells within the outer curvature ventricular wall scored from matched sagittal heart sections of 5 dpf control and kdm6ba−/−; kdm6bb−/− larvae. Each point represents a distinct fish. P-values are from two-tailed Student’s t-tests. (H, I) Confocal heart images of sagittal sectioned 5 dpf control and kdm6a/kdm6b-deficient larvae stained for EdU incorporation (cyan, proliferating cells) and with anti-myosin heavy chain antibody (red, MF20, cardiomyocytes).
Myocardial proliferation around the time of larval hatching is required for the initiation of trabeculation in zebrafish (Liu et al., 2010). To determine if cardiac proliferation was impaired in kdm6ba−/−; kdm6bb−/− larvae, we immunostained sections for proliferating cell nuclear antigen (PCNA), a marker of cycling cells (Fig. 5E–F‴). kdm6b-deficient animals displayed a near 50% reduction of both PCNA positive endocardial and myocardial cells at 5 dpf (Fig. 5G). This decrease in proliferative ventricular cells in kdm6ba−/−; kdm6bb−/− larvae was especially striking using EdU incorporation assays to monitor cells that passed through S-phase between 4 and 5 dpf (Fig. 5H and I). In contrast, the normal number of cardiomyocytes in 72 hpf kdm6ba−/−; kdm6bb−/− fish (Fig. 4B–E) suggests that embryonic cardiac proliferation was not disrupted in the absence of Kdm6b. We conclude that kdm6bb and kdm6ba redundantly promote trabecular outgrowth by enabling the coordinated endo- and myocardial proliferation that accompanies the larval stage initiation of trabeculation.
Transient global H3K27me3 demethylation in cardiomyocytes at the onset of myocardial trabeculation
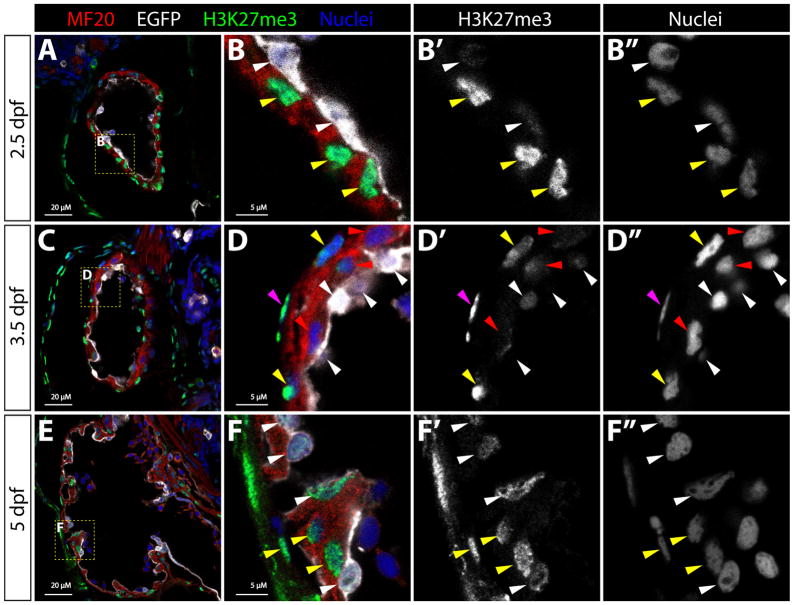
Bulk cellular H3K27me3 levels dramatically increase upon cardiomyocyte differentiation in mice (Delgado-Olguin et al., 2012). Therefore, Kdm6b likewise could regulate widespread rather than locus-specific changes in H3K27me3 levels during heart ventricle maturation. To assess this possibility, we first immunostained sections of 2.5, 3.5, and 5 dpf wildtype zebrafish with anti-H3K27me3 antibodies. Strikingly, H3K27me3 levels varied widely between different cardiac cell types and through the developmental course (Fig. 6). Endocardial cells consistently had very low H3K27me3 levels at 2.5 dpf that progressively increased from 3.5 to 5 dpf. In contrast, the flat nuclei of presumptive epicardial cells (first seen at 3.5 dpf and lacking associated myosin heavy chain immunoreactivity) had high H3K27me3 levels. At 2.5 dpf, prior to trabeculation, all cardiomyocytes displayed robust H3K27me3 levels. However, at 3.5 dpf, newly derived trabecular myocardial cells adjacent to the ventricular cavity frequently showed reduced bulk H3K27me3. In contrast, myocardial cells located on the outer ventricular surface generally retained high H3K27me3 signal. By 5 dpf, trabecular cardiomyocyte H3K27me3 levels were largely normalized with those of the outer cardiomyocyte layer. However, Kdm6b is not uniquely responsible for the bulk transient H3K27me3 decrease associated with trabeculating cardiomyocytes in 3 dpf kdm6b-deficient larvae (Fig. S5). Instead, Kdm6b likely mediates myocardial cell cycle entry by promoting expression of a restricted set of target genes.
Figure 6. Bulk cellular H3K27me3 levels transiently decrease in trabeculating cardiomyocytes.
(A–F) Confocal microscopy immunofluorescence images of sagittal sections through the heart of 2.5, 3.5, and 5 dpf Tg(kdrl:EGFP) embryos stained with anti-myosin heavy chain (red, MF20, myocardium), anti-EGFP (white, endocardium), and anti-H3K27me3 (green) antibodies. Hoechst-stained nuclei are blue. Zoomed images of the yellow boxed areas (in A, C, and E) are shown in (B, D, and F) with adjacent single channel H3K27me3 and nuclei staining to highlight differences in bulk H3K27me3 levels. Magenta and white arrowheads indicate nuclei of epicardial and endocardial cells, respectively. Yellow and red arrowheads mark H3K27me3-high and H3K27me3-low cardiomyocyte nuclei, respectively. 20 μM and 5 μM (zoom panels) scale bars are shown.
DISCUSSION
Our investigation of histone demethylases kdm6ba and kdm6bb during zebrafish development presents several insights. First, we show how duplicated orthologous genes with retained overlapping expression can obscure loss-of-function phenotypes due to simple non-compensatory genetic redundancy. Second, we show that Kdm6b-family demethylases have pleiotropic roles during larval organ development rather than during embryonic cell specification or the establishment of initial organ “frameworks.” Third, Kdm6b demethylases specifically contribute to cardiogenesis by supporting augmented cellular proliferation associated with the initiation of ventricular trabeculation during larval metamorphosis. Fourth, while developing heart ventricle cells display strikingly variable cellular H3K27me3 levels including a transient decrease in trabeculating cardiomyocytes, these bulk dynamics are not driven by Kdm6b proteins alone.
Kdm6b reverse genetic studies provide an example of straightforward non-compensatory redundancy between duplicated genes
Our combined genetic analysis of kdm6bb and kdm6ba illustrates how redundant functions between ohnologous genes can obscure the effects of single gene mutation without involving a compensatory network. As such, our study exemplifies how the lack of phenotypes frequently observed in homozygous mutant zebrafish can be attributed to buffering redundancy originating from the ancestral teleost whole genome duplication event (Amores, 1998; Postlethwait et al., 1998; Lawson, 2016). We suggest that straightforward non-compensatory redundancy be thoroughly considered during reverse genetic studies of ohnologous gene pairs, especially when expression patterns largely overlap.
Histone demethylases kdm6ba and kdm6bb share likely conserved roles in vertebrate organ maturation
Independently generated Kdm6b presumptive null alleles in mice are either pre-implantation (Ohtani et al., 2013) or perinatal (Burgold et al., 2012; Q. Li et al., 2014; Satoh et al., 2010; Shpargel et al., 2014; F. Zhang et al., 2015) lethal. Our zebrafish studies are consistent with the latter publications by suggesting that Kdm6b proteins primarily contribute to organ maturation rather than early cell fate specification, in contrast to some predictions (Kartikasari et al., 2013; Ohtani et al., 2011). These roles are likely evolutionarily conserved as the pleiotropic defects we observe in kdm6b-deficient zebrafish are reminiscent of those reported in mice. For example, kdm6ba−/−; kdm6bb−/− fish display stunted craniofacial cartilage formation and Kdm6b-deficient mice develop skeletal dwarfism during early stages of bone development (Zhang et al., 2015). However, further experiments are necessary to determine if Kdm6b proteins have restricted roles in certain skeletal structures (e.g. the notably truncated basihyal cartilage in kdm6b double mutant zebrafish) or widely contribute to bone formation, as seen in mice. Similarly, the swim bladder defects we observed in our kdm6b-deficient zebrafish may parallel lung structure and respiration defects in Kdm6b-null mice (Q. Li et al., 2014; Satoh et al., 2010) given the evolutionary homology between the teleost swim bladder and lungs of terrestrial vertebrates (Perry et al., 2001; Winata et al., 2009).
kdm6b-deficient zebrafish complete the initial stages of cardiac development, forming discrete heart chambers, an outflow tract, and valves. As such, earlier events including the generation and expansion of cardiac progenitors, specification and differentiation of endocardial and myocardial cells, and initial heart tube extension and looping do not require Kdm6b. This conclusion controverts reported roles of Kdm6b in mesodermal and cardiovascular differentiation derived from embryonic stem cell studies (Ohtani et al., 2011). However, our zebrafish studies are consistent with the preponderance of Kdm6b loss-of-function mouse models that survive to late fetal stages, indicating that overt cardiogenesis is substantially Kdm6b independent.
Although our study shows that combined kdm6ba and kdm6bb are not required for early embryonic development, they may have roles masked by further redundancy with kdm6a (Utx) genes. Mouse genetic studies show that Kdm6a has unique essential developmental functions, including in mesoderm lineage differentiation and cardiogenesis (Lee et al., 2012; Shpargel et al., 2012; Wang et al., 2012; Welstead et al., 2012). In contrast, Kdm6b and Kdm6a are proposed to function redundantly during murine thymocyte differentiation (Manna et al., 2015). Mechanistically, both Kdm6a and Kdm6b interact with the Brg1-associated factor chromatin remodeling complex (Miller et al., 2010). Conflicting reports also indicate that both Kdm6a and Kdm6b proteins may be components of MLL/COMPASS-like H3K4 methyltransferase complexes (De Santa et al., 2007; Issaeva et al., 2007; Williams et al., 2014). Collectively, these studies suggest Kdm6a and Kdm6b share a limited redundancy that is context dependent. The zebrafish genome contains two Kdm6a orthologs, kdm6a and kdm6al. While neither gene has been explored genetically, morpholino studies implicate both kdm6a and kdm6al in Hox-dependent body plan patterning, craniofacial cartilage development, heart looping, and brain morphology (Lan et al., 2007; Van Laarhoven et al., 2015). However, given concerns about morpholino specificity, reverse genetic studies examining kdm6a and kdm6al individually, together, and combined with our kdm6b alleles are necessary to validate and extend these studies.
Dynamic bulk H3K27me3 levels during cardiogenesis support a non-specific mode for H3K27me3 demethylases in enabling cell transitions
Zebrafish embryo genome-wide H3K27me3 and H3K4me3 studies show both marks are established at or prior to zygotic gene activation (3.3 hpf) and can co-localize at key regulatory genes (Lindeman et al., 2011; Vastenhouw et al., 2010). These observations support the ES-cell inspired model that targeted removal of H3K27me3 marks supports activation of epigenetically “poised” lineage-defining regulatory genes. In contrast, our kdm6b genetic studies are consistent with an opposing view that this bivalency model incompletely represents in vivo H3K4me3/H3K27me3 observations and the effects of disrupting either PRC2 or MLL/COMPASS activity (reviewed in Piunti and Shilatifard, 2016). In further support, genetic loss of combined maternal and zygotic ezh2 and, resultantly, all H3K27me3 does not disrupt gastrulation or overall body plan establishment in zebrafish (San et al., 2016). Nevertheless, ezh2-null fish do show pleiotropic embryonic defects, including in heart development, that are consistent with widespread poorly maintained cell identities (San et al., 2016).
Bulk H3K27me3 levels are initially high in all ventricular cardiomyocytes, become notably depleted upon the initiation of cardiac trabeculation, and are then rapidly restored during ventricle maturation. These observed bulk H3K27me3 dynamics may reflect a large scale “reprogramming” of the overall cellular H3K27me3 load that enables signaling effectors to promote a major cell transition at the onset of trabeculation. A conceptually similar but opposite process may occur during initial cardiac differentiation, as bulk H3K27me3 levels rapidly increase as progenitor-like second heart field cells initiate differentiation while appending to the cardiac poles (Delgado-Olguin et al., 2012).
Extending this paradigm, the epigenetic “de-silencing” of repressed genes sometimes may occur by the “bulk” demethylation of H3K27me3-marked nucleosomes rather than by the local action of demethylases on specific subsets of silenced regulatory regions. The specificity of transcriptional responses then is driven by transcription factors acting largely independently from “passively” enabling histone demethylases. This concept supports the re-emerging idea that PRC2-catalyzed H3K27me3 marks maintain rather than specify a transcriptionally repressed state (Comet et al., 2016), as indicated by original Drosophila Polycomb/Trithorax studies. By extension, bulk removal of H3K27me3 marks, as we observed during cardiogenesis, may occur only in specialized developmental, homeostatic, or repair instances. Such roles potentially would enable, but not instruct, major cell state reversals (e.g. de-differentiation, epithelial-to-mesenchymal transitions, or cell cycle re-entry).
Histone demethylases kdm6ba and kdm6bb promote proliferation coupled with the initiation of cardiac trabeculation
Trabeculation of the zebrafish ventricle initiates around 72 hpf with the specification of a small number of trabeculating cardiomyocytes that separate from the otherwise single cell-thick outer myocardial layer (Gupta and Poss, 2012; Han et al., 2016; Liu et al., 2010; Peshkovsky et al., 2011). These specialized trabeculating cardiomyocytes then clonally expand during early larval stages to form extensive ridge-like cardiac trabeculae that protrude into the ventricular cavity. The initial phase of trabeculation driven by neuregulin/ErbB2 signaling (Han et al., 2016; Liu et al., 2010; Peshkovsky et al., 2011) does not depend on Kdm6b as we saw that small groups of multiple cell layer thick myocardium still form in kdm6b-deficient fish. Instead, Kdm6b is required for the subsequent boost of proliferation at larval hatching (around 96 hpf) seen in trabeculating myocardial cells (Gupta and Poss, 2012; Han et al., 2016; Liu et al., 2010) as well as adjacent endocardium that concomitantly expands to enshroud the forming trabeculae.
Kdm6b alone appears unlikely to drive the bulk H3K27me3 demethylation in trabeculating cardiomyocytes. Instead, Kdm6b could provide an “epigenetic” component to transcriptional switches acting on a discrete set of target genes by locally removing H3K27me3 marks or, by a non-catalytic role as part of a MLL/COMPASS H3K4 methylase complex. Of potential relevance, the Kdm6b-cooperating Brg1-associated factor (BAF) chromatin remodeling complex (Miller et al., 2010) enables trabeculation in mice by repressing endocardial transcription of the trabeculation-suppressing Adamts1 matrix protease (Stankunas et al., 2008). Another possibility is that Kdm6b acts in both myocardial and endocardial cells to integrate a systemic signal that widely coordinates organ maturation events coupled to larval metamorphosis. Thyroid hormone is one immediate candidate that is upregulated upon hatching (Chang et al., 2012) and a suspected cardiomyocyte mitogen in mammals (Ledda-Columbano et al., 2006; M. Li et al., 2014; Naqvi et al., 2014). Regardless, the role of Kdm6b in promoting cardiomyocyte proliferation establishes these histone demethylases and downstream target genes as candidate genes for congenital heart defects of the ventricle such as hypoplastic left ventricle syndrome.
The recognition that differentiated cardiomyocytes can re-acquire robust proliferative ability during heart regeneration in both zebrafish and mice (Jopling et al., 2010; Porrello et al., 2011; Poss et al., 2002) and that mammalian cardiomyocytes may retain some proliferative capacity during pre-adolescence (Ali et al., 2014; Haubner et al., 2016; Mollova et al., 2013; Naqvi et al., 2014; Polizzotti et al., 2015; Ye et al., 2016) has focused efforts to recapitulate these mechanisms for injury repair, notably for congestive heart failure. The role of Kdm6b histone demethylases in developmentally timed cardiomyocyte (but not cardiac progenitor) proliferation suggests the reversal of epigenetic H3K27me3 silencing marks may be an essential aspect of innate heart regeneration and a promising path to augment therapeutically delivered cardiomyocytes for heart disease.
Supplementary Material
Highlights.
H3K27me3 demethylases kdm6ba and kdm6bb exhibit simple non-compensatory redundancy
Kdm6b has pleiotropic organogenesis roles without affecting cell specification
Kdm6b supports cardiomyocyte proliferation at the onset of cardiac trabeculation
Bulk H3K27me3 levels transiently decrease in trabeculating cardiomyocytes
Kdm6b alone does not drive global H3K27me3 dynamics during ventricle maturation
Acknowledgments
We thank the University of Oregon zebrafish facility and staff. The Stankunas lab and the University of Oregon zebrafish community provided critical guidance and support. Caroline and Geoffrey Burns shared qRT-PCR primers for rps11 and vmhcl and provided technical advice. Jamie Nichols and Chuck Kimmel shared insights about and protocols for studying craniofacial development. Judith Eisen provided critical feedback on the manuscript.
FUNDING SOURCES
A.A.A. received support from a NIH/NICHD Developmental Biology Training Grant (5T32HD007348). Research funding was provided by the University of Oregon and the National Institutes of Health (1R01HL115294, K.S.).
Footnotes
AUTHOR CONTRIBUTIONS
A.A.A. designed and performed most of the experiments and wrote the manuscript. A.H. performed experiments and assisted with figure preparation. S.S. and K.S. designed and performed experiments and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerberg AA, Stewart S, Stankunas K. Spatial and temporal control of transgene expression in zebrafish. PLoS ONE. 2014;9:e92217. doi: 10.1371/journal.pone.0092217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerberg BN, Sarangam ML, Stankunas K. Endocardial Brg1 disruption illustrates the developmental origins of semilunar valve disease. Dev Biol. 2015;407:158–172. doi: 10.1016/j.ydbio.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P, Pombo A, O’Neill L, Azuara V. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development. 2010;137:2483–2492. doi: 10.1242/dev.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SR, Hippenmeyer S, Saadat LV, Luo L, Weissman IL, Ardehali R. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc Natl Acad Sci USA. 2014;111:8850–8855. doi: 10.1073/pnas.1408233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A. Zebrafish hox Clusters and Vertebrate Genome Evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Voituron N, Caganova M, Tripathi PP, Menuet C, Tusi BK, Spreafico F, Bévengut M, Gestreau C, Buontempo S, Simeone A, Kruidenier L, Natoli G, Casola S, Hilaire G, Testa G. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Reports. 2012;2:1244–1258. doi: 10.1016/j.celrep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chang J, Wang M, Gui W, Zhao Y, Yu L, Zhu G. Changes in thyroid hormone levels during zebrafish development. Zoolog Sci. 2012;29:181–184. doi: 10.2108/zsj.29.181. [DOI] [PubMed] [Google Scholar]
- Cloos PAC, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P. Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS ONE. 2010;5:e9150. doi: 10.1371/journal.pone.0009150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Delgado-Olguín P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012;44:343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Bloomekatz J, Ren J, Zhang R, Grinstein JD, Zhao L, Burns CG, Burns CE, Anderson RM, Chi NC. Coordinating cardiomyocyte interactions to direct ventricular chamber morphogenesis. Nature. 2016;534:700–704. doi: 10.1038/nature18310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubner BJ, Schneider J, Schweigmann U. Functional recovery of a human neonatal heart after severe myocardial infarction. Circulation. 2016;118:216–221. doi: 10.1161/CIRCRESAHA.116.307017. [DOI] [PubMed] [Google Scholar]
- He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, Daley GQ, Kong SW, Orkin SH, Seidman CE, Seidman JG, Pu WT. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res. 2012;110:406–415. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biémont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigó R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quétier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang J, Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res. 2013;23:122–130. doi: 10.1038/cr.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SW, Beis D, Mitchell T, Chen JN, Stainier DYR. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Martí M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartikasari AER, Zhou JX, Kanji MS, Chan DN, Sinha A, Grapin-Botton A, Magnuson MA, Lowry WE, Bhushan A. The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 2013;32:1393–1408. doi: 10.1038/emboj.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lawson ND. Reverse Genetics in Zebrafish: Mutants, Morphants, and Moving Forward. Trends in Cell Biology. 2016;26:77–79. doi: 10.1016/j.tcb.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Ledda-Columbano GM, Molotzu F, Pibiri M, Cossu C, Perra A, Columbano A. Thyroid hormone induces cyclin D1 nuclear translocation and DNA synthesis in adult rat cardiomyocytes. FASEB J. 2006;20:87–94. doi: 10.1096/fj.05-4202com. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Iismaa SE, Naqvi N, Nicks A, Husain A, Graham RM. Thyroid hormone action in postnatal heart development. Stem Cell Res. 2014;13:582–591. doi: 10.1016/j.scr.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang HY, Chepelev I, Zhu Q, Wei G, Zhao K, Wang RF. Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development. PLoS Genet. 2014;10:e1004524. doi: 10.1371/journal.pgen.1004524.s011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, Winata C, Mathavan S, Müller F, Aleström P, Collas P. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell. 2011;21:993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Liu J, Bressan M, Hassel D, Huisken J, Staudt D, Kikuchi K, Poss KD, Mikawa T, Stainier DYR. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137:3867–3875. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S, Kim JK, Baugé C, Cam M, Zhao Y, Shetty J, Vacchio MS, Castro E, Tran B, Tessarollo L, Bosselut R. Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat Commun. 2015;6:8152. doi: 10.1038/ncomms9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Current Opinion in Cell Biology. 1999;11:699–704. doi: 10.1016/S0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX Play a Demethylase-Independent Role in Chromatin Remodeling to Regulate T-Box Family Member-Dependent Gene Expression. Molecular Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, Silberstein LE, Dos Remedios CG, Graham D, Colan S, Kühn B. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci USA. 2013;110:1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/S0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Naqvi N, Li M, Calvert JW, Tejada T, Lambert JP, Wu J, Kesteven SH, Holman SR, Matsuda T, Lovelock JD, Howard WW, Iismaa SE, Chan AY, Crawford BH, Wagner MB, Martin DIK, Lefer DJ, Graham RM, Husain A. A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell. 2014;157:795–807. doi: 10.1016/j.cell.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Vlachojannis GJ, Koyanagi M, Boeckel JN, Urbich C, Farcas R, Bonig H, Marquez VE, Zeiher AM, Dimmeler S. Epigenetic Regulation of Endothelial Lineage Committed Genes in Pro-Angiogenic Hematopoietic and Endothelial Progenitor Cells. Circ Res. 2011;109:1219–1229. doi: 10.1161/CIRCRESAHA.111.247304. [DOI] [PubMed] [Google Scholar]
- Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, Rieger MA, Zeiher AM, Dimmeler S. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res. 2013;113:856–862. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, Moon RT, Stamatoyannopoulos J, Murry CE. A Temporal Chromatin Signature in Human Embryonic Stem Cells Identifies Regulators of Cardiac Development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SF, Wilson RJ, Straus C, Harris MB, Remmers JE. Which came first, the lung or the breath? Comp Biochem Physiol, Part A Mol Integr Physiol. 2001;129:37–47. doi: 10.1016/s1095-6433(01)00304-x. [DOI] [PubMed] [Google Scholar]
- Peshkovsky C, Totong R, Yelon D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev Dyn. 2011;240:446–456. doi: 10.1002/dvdy.22526. [DOI] [PubMed] [Google Scholar]
- Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- Polizzotti BD, Ganapathy B, Walsh S, Choudhury S, Ammanamanchi N, Bennett DG, Dos Remedios CG, Haubner BJ, Penninger JM, Kühn B. Neuregulin stimulation of cardiomyocyte regeneration in mice and human myocardium reveals a therapeutic window. Sci Transl Med. 2015;7:281ra45. doi: 10.1126/scitranslmed.aaa5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient Regenerative Potential of the Neonatal Mouse Heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zool B Mol Dev Evol. 2007;308:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. Journal of Biological Chemistry. 2012;287:44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci USA. 2010;107:10783–10790. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San B, Chrispijn ND, Wittkopp N, van Heeringen SJ, Lagendijk AK, Aben M, Bakkers J, Ketting RF, Kamminga LM. Normal formation of a vertebrate body plan and loss of tissue maintenance in the absence of ezh2. Sci Rep. 2016;6:24658. doi: 10.1038/srep24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T. KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development. PLoS Genet. 2014;10:e1004507. doi: 10.1371/journal.pgen.1004507.s012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke D, Salzburger W, Braasch I, Meyer A. Many genes in fish have species-specific asymmetric rates of molecular evolution. BMC Genomics. 2006;7:20. doi: 10.1186/1471-2164-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Tsun ZY, Izpisua Belmonte JC. A histone demethylase is necessary for regeneration in zebrafish. Proc Natl Acad Sci USA. 2009;106:19889–19894. doi: 10.1073/pnas.0904132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguín P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, Zhou YQ, Yeh RF, Henkelman RM, Harvey RP, Metzger D, Chambon P, Stainier DYR, Pollard KS, Scott IC, Bruneau BG. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Van Laarhoven PM, Neitzel LR, Quintana AM, Geiger EA, Zackai EH, Clouthier DE, Artinger KB, Ming JE, Shaikh TH. Kabuki syndrome genes KMT2D and KDM6A: functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum Mol Genet. 2015;24:4443–4453. doi: 10.1093/hmg/ddv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and Coordinated Epigenetic Regulation of Developmental Transitions in the Cardiac Lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, Ge K. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci USA. 2012;109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci USA. 2012;109:13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Rappsilber J, Nielsen AL, Johansen JV, Helin K. The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLoS ONE. 2014;9:e96545. doi: 10.1371/journal.pone.0096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winata CL, Korzh S, Kondrychyn I, Zheng W, Korzh V, Gong Z. Development of zebrafish swimbladder: The requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev Biol. 2009;331:222–236. doi: 10.1016/j.ydbio.2009.04.035. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- Ye L, Qiu L, Zhang H, Chen H, Jiang C, Hong H, Liu J. Cardiomyocytes in Young Infants With Congenital Heart Disease: a Three-Month Window of Proliferation. Sci Rep. 2016;6:23188–23188. doi: 10.1038/srep23188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Xu L, Xu L, Xu Q, Li D, Yang Y, Karsenty G, Chen CD. JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. Journal of Molecular Cell Biology. 2015;7:23–34. doi: 10.1093/jmcb/mjv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, Heideman W, Burns CE, Burns CG. Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature. 2011;474:645–648. doi: 10.1038/nature10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.