Abstract
Prior genome-wide association studies for oral clefts have focused on clinic-based samples with unclear generalizability. Prior samples were also small for investigating effects by cleft type and exclusively studied isolated clefts (those occurring without other birth defects). We estimated the effects of 17 top loci on cleft types in both isolated and nonisolated cases in the largest consortium to date of European-descent population-based studies. Our analytic approach focused on a mother-child dyad case-control design, but it also allowed analyzing mother-only or child-only genotypes to maximize power. Our total sample included 1,875 cases with isolated clefts, 459 cases with nonisolated clefts, and 3,749 controls. After correcting for multiple testing, we observed significant associations between fetal single-nucleotide polymorphisms (SNPs) at IRF6, PAX7, 8q21.3, 8q24, KIAA1598-VAX1, and MAFB and isolated cleft lip only (CLO) and cleft lip and palate (CLP). Significant associations were observed between isolated CLO and fetal SNPs near TPM1 and NOG1 and between CLP and fetal SNPs at ABCA4-ARHGAP29, THADA, FOXE1, and SPRY2. Overall, effects were similar for isolated CLO and CLP, except for ABCA4-ARHGAP29. A protective effect was observed for the fetal NOG1 SNP on cleft palate only, opposite in direction to the effect on CLO. For most fetal SNPs, a dose-response allelic effect was observed. No evidence of parent-of-origin or maternal genome effects was observed. Overall, effect direction and magnitude were similar between isolated and nonisolated clefts, suggesting that several loci are modifiers of cleft risk in both isolated and nonisolated forms. Our results provide reliable estimates of the effects of top loci on risks of oral clefts in a population of European descent.
Keywords: cleft lip, cleft palate, genetics, genetic epidemiology, molecular genetics, meta-analysis
Introduction
Genome-wide association studies (GWASs) of isolated cleft lip with or without cleft palate (CL/P) occurring without other birth defects have identified multiple loci with evidence of etiologic mutations (Birnbaum et al. 2009; Grant et al. 2009; Beaty et al. 2010; Beaty et al. 2013; Mangold et al. 2010; Camargo et al. 2012; Ludwig et al., 2012; Sun et al. 2015; Wolf et al. 2015). Key loci include (in genomic order) PAX7 (1p36), ABCA4-ARHGAP29 (1p22.1), IRF6 (1q32), THADA (2p21), DCAF4L2 (8q21.3), 8q24, GADD45G (9q22.1), KIAA1598-VAX1 (10q25.3), SPRY2 (13q31.3), TPM1 (15q22.2), NTN1 (17p13.1), NOG1 (17q22), and MAFB (20q12). More recent and larger GWASs have found evidence for these loci and additional ones such as FOXE1 (9q22.3; Leslie, Carlson, et al. 2016; Leslie et al. 2017; Yu et al. 2017). Candidate gene studies first suggested some of these genes/loci for CL/P, such as IRF6 and FOXE1, but also others, such as MSX1, FGFR2, and CRISPLD2 (Chiquet et al. 2007; Leslie and Marazita 2013). Interestingly, except perhaps for FOXE1 (Moreno et al. 2009; Leslie et al. 2017), there is little evidence that the loci for CL/P are related to cleft palate only (CPO) especially at GWAS significance levels.
Despite the convincing evidence for many loci, there are several limitations in prior assessments of their effects. Previous studies have almost exclusively focused on clinic-based rather than population-based samples. Genetic risk estimates from clinic-based samples may have limited generalizability because of potential ascertainment bias (Kukull and Ganguli 2012). In contrast, population-based samples provide more reliable and generalizable estimates for quantifying the epidemiologic and clinical relevance of these top loci. Prior studies have had modest sample sizes that may lack power to accurately estimate separate effects on cleft lip only (CLO) versus cleft lip with palate (CLP) and meaningfully examine potential differences between these 2 phenotypes, and most examined CL/P as 1 phenotype. Small sample sizes have been a challenge for examining genetic effects on CPO, a less common trait with a higher proportion of syndromic cases. Another limitation is focusing on fetal gene effects with little attention to additional underlying genetic mechanisms, such as maternal gene (MG) and parent-of-origin (PoO) effects. MG effects influence fetal intrauterine environment conditional on fetal genes. PoO effects indicate differential effects in fetal genes depending on whether they were inherited from the mother or the father. Two studies of MG and PoO effects based on GWAS data (Shi et al. 2012; Garg et al. 2014) found nonsignificant effects. However, their study samples were relatively modest for a meaningful assessment of such effects. Finally, GWASs have focused on isolated orofacial clefting (OFC), and little is known about whether these loci are relevant for nonisolated OFC occurring with other birth defects or syndromes.
In this study, we estimated the effects of the main OFC loci in GWASs and top candidate gene studies published before February 2016 on risks of isolated CLO, CLP, and CPO examined as separate phenotypes in the largest consortium to date of population-based samples of European descent. We examined fetal, MG, and PoO effects, and we estimated effects on nonisolated OFC separately for CL/P and CPO. We used individual-level data pooled across 5 population-based samples, accounting for study differences through stratified estimation, to study a total of 31 single-nucleotide polymorphisms (SNPs) within 17 genes/loci (Appendix Table 1). Our work provides the most definitive assessment to date of the importance of these loci for each of the 3 isolated OFC phenotypes in populations of European descent, including risk estimates that may be meaningful for clinical counseling and etiologic research. Furthermore, we provide novel evidence on the relevance of these loci for nonisolated OFC.
Methods
Our study employs data from a unique international consortium of 5 OFC population-based case-control studies: the Norway Facial Clefts Study, the Norwegian Mother and Child Cohort, the Utah Child and Family Health Study, the Iowa Case-Control Study, and the US National Birth Defects Prevention Study. Cases and controls for each study were identified through national or state-based registries that provide population-based surveillance of OFC. Mothers and children were enrolled and provided DNA specimens. The controls across all studies included children without oral clefts or other major birth defects who were randomly selected from the same birth year and regions as the cases (Reefhuis et al. 2015; Kummet et al. 2016; Kutbi 2017). Subject enrollment and data collection in each study were approved by the appropriate institutional review boards. Descriptions of these studies and genotyping methods are in the online Appendix.
In the main analysis, we limited our sample to children born with isolated oral clefts (excluding cases with syndromes or other major birth defects). Because of differences in minor allele frequencies and possibly genetic effects by ancestry, we limited the sample from the US studies to self-reported non-Hispanic whites. Most participants in the Norway studies were Caucasians (studies required eligible participants to be native speakers of Norwegian). In additional analyses, we examined effects of SNPs significantly related to isolated clefts on risk of nonisolated clefts.
The total analytic sample included 1,875 cases with isolated clefts, including 1,311 mother-child dyads with genetic data on both mothers and children. The sample included 459 cases with nonisolated clefts, of whom 267 had mother-child dyad data, and 3,749 controls, including 2,481 mother-child dyads with genetic data. Appendix Table 2 shows the sample distribution by case-control group, cleft type, and genetic data availability for each study.
Statistical Analysis
Analyses were performed with the HAPLIN R package version 3.5. HAPLIN is based on a full maximum-likelihood approach and can estimate a variety of genetic parameters for case-triad, case-dyad, case-control, and hybrid designs (Weinberg and Umbach 2005; Gjessing and Lie 2006; Skare et al. 2012). Because we had DNA on children and mothers for the majority of our sample, our estimation was primarily based on a hybrid case-control dyad design, in which fathers’ genotypes were probabilistically inferred from the child-mother dyad data. This design allows for estimating the most relevant genetic parameters. We included the genetic data of the smaller proportion of the sample with DNA only on mothers or children (Appendix Table 2) to optimize statistical power. Appendix Figure 1 shows that we had good power for testing fetal SNP effects even for moderate effect sizes and the power gain based on the total sample versus complete mother-child dyads. When mother-only data were included, a probability distribution for possible child genotypes for a given SNP is estimated for each mother and used in deriving the fetal SNP effect. To ensure that our main estimates combining data from complete dyads with mother-only and child-only data are not biased, we performed sensitivity analyses that included only mother-child dyad data. Results from these analyses were virtually the same as those based on the full sample.
Our hybrid design allowed for complete separation of direct effects of maternal and fetal genes (Gjessing and Lie 2006; Buyske 2008) and estimation of PoO effects for fetal genes (Shi et al. 2012). All analyses were stratified by study to account for differences in allelic frequency and loci effects across studies and then combined in a meta-analysis after testing for effect heterogeneity between sites.
Our analysis proceeded through the following steps. Focusing first on cases with isolated OFC, we performed a screening analysis of each SNP using a model with a multiplicative dose-response effect of fetal alleles for each of the 3 case groups and Bonferroni adjustment for 31 SNPs (P < 0.0016). Our subsequent analyses for fetal effects focused on SNPs that had significant effects after our initial screening. For each SNP, we then estimated associations with each OFC case group for the single dose (heterozygotes) and double dose (homozygotes) of the minor allele. Next, we examined MG effects for each SNP controlling for the fetal genotype. Furthermore, we evaluated PoO effects by estimating the effect of a single dose of a minor allele inherited from the mother and that inherited from the father and tested the difference between these effects in heterozygotes. Finally, we estimated the fetal effects of SNPs with significant effects on isolated OFC from the first screen on nonisolated OFC, combining nonisolated CLO and CLP (because of the smaller sample size of nonisolated cases).
Results
Fetal Allele Effects on Isolated OFC
Of the 31 fetal SNPs in the 17 examined loci, 14 SNPs in 12 loci had significant associations with isolated CLO or CLP or both after Bonferroni adjustment (Tables 1 and 2). SNPs within PAX7, IRF6, 8q21.3, 8q24, VAX1, KIAA1598, and MAFB had significant associations with both CLO and CLP. SNPs near TPM1 and NOG1 had significant associations only with CLO after Bonferroni adjustment, although these were weakly associated with CLP at nominal significance levels (P = 0.005 and 0.03, respectively). In contrast, SNPs at ABCA4-ARHGAP29, THADA, FOXE1, and SPRY2 had significant associations with CLP only. FOXE1 and SPRY2 SNPs were associated with CLO at nominal significant levels (P ≤ 0.01), but there was no evidence for associations of ABCA4-ARHGAP29 or THADA with CLO. We did not observe significant associations at the other 5 loci for either CLO or CLP, including MSX1, FGFR2, CRISPLD2, NTN1, and MYH9. There was little evidence of an association with these loci even at nominal significance levels (only NTN1 was weakly associated with CLO and CLP at P ≤ 0.05). The sensitivity analyses excluding mother-only and child-only genetic data showed similar results, with an overall slight decline in significance as expected; however, the relative risk estimates were very close, indicating no bias when these data were combined with mother-child dyad data (Appendix Tables 3 and 4).
Table 1.
Fetal SNP Effects on Isolated Cleft Lip Only.
| Genes/Locus | SNP | RRa | 95% CI | P Valueb | P Value–Sitec |
|---|---|---|---|---|---|
| PAX7 | rs742071 | 1.52 | 1.31 to 1.75 | 3.74E-08 | 0.90 |
| ABCA4-ARHGAP29 | rs560426 | 1.10 | 0.95 to 1.27 | 0.20 | 0.52 |
| IRF6 | rs2235371 | 0.80 | 0.45 to 1.43 | 0.45 | 0.95 |
| IRF6 | rs642961 | 1.60 | 1.36 to 1.87 | 1.42E-08 | 0.69 |
| THADA | rs7590268 | 1.16 | 0.98 to 1.36 | 0.08 | 0.83 |
| MSX1 | rs3111689 | 1.15 | 0.98 to 1.36 | 0.09 | 0.11 |
| 8q21.3 | rs12543318 | 1.51 | 1.31 to 1.75 | 4.46E-08 | 0.87 |
| 8q24 | rs987525 | 1.93 | 1.65 to 2.25 | 5.88E-16 | 0.65 |
| FOXE1 | rs7864322 | 0.86 | 0.74 to 1.01 | 0.07 | 0.78 |
| FOXE1 | rs10818094 | 0.99 | 0.84 to 1.18 | 0.95 | 0.88 |
| FOXE1 | rs1443433 | 0.91 | 0.74 to 1.12 | 0.40 | 0.75 |
| FOXE1 | rs74934500 | 0.73 | 0.45 to 1.18 | 0.19 | 0.97 |
| FOXE1 | rs3758249 | 0.80 | 0.69 to 0.93 | 0.004 | 0.95 |
| FOXE1 | rs10984103 | 0.82 | 0.70 to 0.96 | 0.01 | 0.89 |
| KIAA1598-VAX1 | rs7078160 | 1.32 | 1.11 to 1.57 | 0.002 | 0.16 |
| KIAA1598-VAX1 | rs4752028 | 1.34 | 1.13 to 1.60 | 8.34E-04 | 0.20 |
| FGFR2 | rs4752566 | 0.90 | 0.78 to 1.05 | 0.18 | 0.64 |
| FGFR2 | rs2912760 | 1.12 | 0.96 to 1.32 | 0.16 | 0.02 |
| FGFR2 | rs3135761 | 1.10 | 0.92 to 1.32 | 0.30 | 0.43 |
| FGFR2 | rs2912771 | 0.83 | 0.70 to 0.99 | 0.04 | 0.04 |
| FGFR2 | rs2981428 | 1.09 | 0.94 to 1.26 | 0.26 | 0.77 |
| FGFR2 | rs3750817 | 1.00 | 0.86 to 1.16 | 1.00 | 0.50 |
| SPRY2 | rs8001641 | 0.82 | 0.71 to 0.95 | 0.008 | 0.81 |
| TPM1 | rs1873147 | 1.31 | 1.13 to 1.53 | 5.82E-04 | 0.31 |
| CRISPLD2 | rs1546124 | 1.00 | 0.86 to 1.17 | 0.96 | 0.57 |
| NTN1 | rs4791331 | 1.16 | 0.95 to 1.42 | 0.14 | 0.20 |
| NTN1 | rs8069536 | 1.18 | 1.02 to 1.36 | 0.03 | 0.49 |
| NOG1 | rs227731 | 1.36 | 1.18 to 1.58 | 2.79E-05 | 0.45 |
| MAFB | rs13041247 | 0.71 | 0.61 to 0.82 | 9.85E-06 | 0.81 |
| MYH9 | rs3752462 | 1.10 | 0.94 to 1.28 | 0.24 | 0.11 |
| MYH9 | rs1002246 | 1.12 | 0.96 to 1.30 | 0.14 | 0.99 |
95% CI, 95% confidence interval; RR, relative risk; SNP, single-nucleotide polymorphism.
RR for heterozygotes. RR for homozygotes is estimated as the square.
Effect of child’s allele in multiplicative model. Significant with Bonferroni adjustment if P < 0.0016.
Test of heterogeneity of estimates across sites.
Table 2.
Fetal SNP Effects on Isolated Cleft Lip with Palate.
| Genes/Locus | SNP | RRa | 95% CI | P Valueb | P Value–Sitec |
|---|---|---|---|---|---|
| PAX7 | rs742071 | 1.33 | 1.18 to 1.50 | 3.84E-06 | 0.61 |
| ABCA4-ARHGAP29 | rs560426 | 1.25 | 1.10 to 1.41 | 3.91E-04 | 0.47 |
| IRF6 | rs2235371 | 0.73 | 0.43 to 1.25 | 0.25 | 0.86 |
| IRF6 | rs642961 | 1.33 | 1.16 to 1.53 | 6.66E-05 | 0.55 |
| THADA | rs7590268 | 1.27 | 1.12 to 1.46 | 3.96E-04 | 0.74 |
| MSX1 | rs3111689 | 1.05 | 0.91 to 1.21 | 0.50 | 0.50 |
| 8q21.3 | rs12543318 | 1.21 | 1.07 to 1.37 | 0.002 | 0.30 |
| 8q24 | rs987525 | 1.85 | 1.63 to 2.10 | 1.47E-19 | 0.12 |
| FOXE1 | rs7864322 | 0.84 | 0.74 to 0.96 | 0.01 | 0.25 |
| FOXE1 | rs10818094 | 1.08 | 0.94 to 1.24 | 0.29 | 0.53 |
| FOXE1 | rs1443433 | 1.03 | 0.87 to 1.21 | 0.74 | 0.51 |
| FOXE1 | rs74934500 | 0.69 | 0.45 to 1.05 | 0.08 | 0.52 |
| FOXE1 | rs3758249 | 0.78 | 0.69 to 0.88 | 8.18E-05 | 0.95 |
| FOXE1 | rs10984103 | 0.81 | 0.71 to 0.92 | 0.0011 | 0.79 |
| KIAA1598-VAX1 | rs7078160 | 1.33 | 1.15 to 1.53 | 1.04E-04 | 0.80 |
| KIAA1598-VAX1 | rs4752028 | 1.36 | 1.18 to 1.56 | 3.20E-05 | 0.67 |
| FGFR2 | rs4752566 | 1.01 | 0.89 to 1.14 | 0.92 | 0.67 |
| FGFR2 | rs2912760 | 0.98 | 0.86 to 1.12 | 0.81 | 0.50 |
| FGFR2 | rs3135761 | 0.99 | 0.85 to 1.16 | 0.90 | 0.64 |
| FGFR2 | rs2912771 | 0.96 | 0.84 to 1.11 | 0.62 | 0.23 |
| FGFR2 | rs2981428 | 1.00 | 0.88 to 1.12 | 0.96 | 1.00 |
| FGFR2 | rs3750817 | 1.01 | 0.89 to 1.14 | 0.90 | 0.59 |
| SPRY2 | rs8001641 | 0.79 | 0.70 to 0.90 | 2.05E-04 | 0.84 |
| TPM1 | rs1873147 | 1.20 | 1.06 to 1.37 | 0.005 | 0.49 |
| CRISPLD2 | rs1546124 | 1.01 | 0.89 to 1.15 | 0.86 | 0.98 |
| NTN1 | rs4791331 | 1.21 | 1.03 to 1.42 | 0.02 | 0.17 |
| NTN1 | rs8069536 | 1.12 | 0.99 to 1.27 | 0.06 | 0.26 |
| NOG1 | rs227731 | 1.14 | 1.01 to 1.29 | 0.03 | 0.11 |
| MAFB | rs13041247 | 0.67 | 0.59 to 0.76 | 2.32E-09 | 0.63 |
| MYH9 | rs3752462 | 1.01 | 0.88 to 1.14 | 0.94 | 0.30 |
| MYH9 | rs1002246 | 1.10 | 0.97 to 1.24 | 0.15 | 0.21 |
95% CI, 95% confidence interval; RR, relative risk; SNP, single-nucleotide polymorphism.
RR for heterozygotes. RR for homozygotes is estimated as the square.
Effect of child’s allele in multiplicative model. Significant with Bonferroni adjustment if P < 0.0016.
Test of heterogeneity of estimates across sites.
Of all the examined loci, only the SNP at NOG1 had a significant association with isolated CPO (Table 3) in the opposite direction (minor allele protective) from that with CLO (or CLP). All other SNPs had insignificant associations even at P < 0.05, and most P values were large, ruling out any role for these loci in isolated CPO.
Table 3.
Fetal SNP Effects on Isolated Cleft Palate Only.
| Genes/Locus | SNP | RRa | 95% CI | P Valueb | P Value–Sitec |
|---|---|---|---|---|---|
| PAX7 | rs742071 | 1.03 | 0.90 to 1.19 | 0.68 | 1.00 |
| ABCA4-ARHGAP29 | rs560426 | 0.94 | 0.82 to 1.08 | 0.39 | 0.46 |
| IRF6 | rs2235371 | 1.25 | 0.79 to 1.98 | 0.34 | 0.62 |
| IRF6 | rs642961 | 0.92 | 0.77 to 1.09 | 0.33 | 0.37 |
| THADA | rs7590268 | 1.03 | 0.87 to 1.21 | 0.74 | 0.66 |
| MSX1 | rs3111689 | 1.01 | 0.86 to 1.19 | 0.87 | 0.98 |
| 8q21.3 | rs12543318 | 0.98 | 0.85 to 1.13 | 0.80 | 0.86 |
| 8q24 | rs987525 | 0.87 | 0.73 to 1.04 | 0.12 | 0.11 |
| FOXE1 | rs7864322 | 0.88 | 0.75 to 1.02 | 0.10 | 0.22 |
| FOXE1 | rs10818094 | 1.08 | 0.92 to 1.27 | 0.34 | 0.47 |
| FOXE1 | rs1443433 | 0.83 | 0.68 to 1.02 | 0.07 | 0.78 |
| FOXE1 | rs74934500 | 0.80 | 0.46 to 1.37 | 0.42 | 0.02 |
| FOXE1 | rs3758249 | 0.87 | 0.75 to 1.00 | 0.05 | 0.53 |
| FOXE1 | rs10984103 | 0.91 | 0.79 to 1.05 | 0.22 | 0.54 |
| KIAA1598-VAX1 | rs7078160 | 1.04 | 0.87 to 1.24 | 0.68 | 0.55 |
| KIAA1598-VAX1 | rs4752028 | 1.06 | 0.88 to 1.26 | 0.56 | 0.38 |
| FGFR2 | rs4752566 | 0.98 | 0.85 to 1.13 | 0.81 | 0.91 |
| FGFR2 | rs2912760 | 0.94 | 0.80 to 1.10 | 0.45 | 0.31 |
| FGFR2 | rs3135761 | 0.98 | 0.82 to 1.18 | 0.87 | 0.10 |
| FGFR2 | rs2912771 | 0.98 | 0.84 to 1.15 | 0.86 | 0.74 |
| FGFR2 | rs2981428 | 1.02 | 0.88 to 1.17 | 0.82 | 0.21 |
| FGFR2 | rs3750817 | 0.96 | 0.83 to 1.11 | 0.61 | 0.08 |
| SPRY2 | rs8001641 | 1.08 | 0.94 to 1.24 | 0.30 | 0.90 |
| TPM1 | rs1873147 | 0.90 | 0.76 to 1.05 | 0.18 | 0.11 |
| CRISPLD2 | rs1546124 | 0.96 | 0.83 to 1.11 | 0.58 | 0.16 |
| NTN1 | rs4791331 | 0.98 | 0.80 to 1.20 | 0.84 | 0.96 |
| NTN1 | rs8069536 | 0.94 | 0.82 to 1.08 | 0.40 | 0.77 |
| NOG1 | rs227731 | 0.74 | 0.64 to 0.85 | 3.83E-05 | 0.14 |
| MAFB | rs13041247 | 0.94 | 0.81 to 1.08 | 0.38 | 0.41 |
| MYH9 | rs3752462 | 1.10 | 0.95 to 1.28 | 0.20 | 0.35 |
| MYH9 | rs1002246 | 0.97 | 0.84 to 1.12 | 0.68 | 0.66 |
95% CI, 95% confidence interval; RR, relative risk; SNP, single-nucleotide polymorphism.
RR for heterozygotes. RR for homozygotes is estimated as the square.
Effect of child’s allele in multiplicative model. Significant with Bonferroni adjustment if P < 0.0016.
Test of heterogeneity of estimates across sites.
For all examined SNPs, there was no evidence of heterogeneity in effects across the pooled studies for each cleft type (Tables 1–3). The hypothesis of homogenous effects across studies could not be rejected, even at nominal significance levels for most SNPs, which supports the pooled results from stratified estimation across studies.
Dosage Effects of Fetal Alleles on Isolated OFC
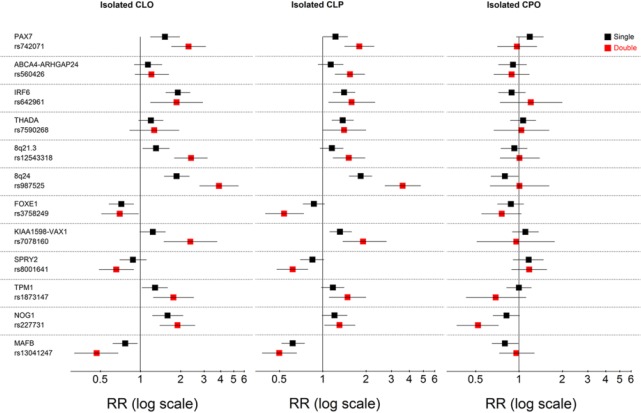
Figure 1 shows the associations of single and double doses of minor fetal alleles for the SNPs that had significant associations with any cleft type. The overall patterns of associations were remarkably similar between isolated CLO and CLP, suggesting that CLO and CLP share similar etiology across most examined loci. Dose-response associations were consistent for most SNPs. For both CLO and CLP, the double doses of PAX7, 8q24, 8q21.3, KIAA1598-VAX1, SPRY2, TPM1, NOG1, and MAFB had larger associations than those for the single-dose alleles. This indicates that the simpler multiplicative model for the screening analyses fit the data well, with the advantage of estimating fewer parameters.
Figure 1.
Single- and double-dose effects measured by relative risks (RRs) for minor alleles of the 15 fetal single-nucleotide polymorphisms that had significant effects for 1 of the case categories: isolated cleft lip only (CLO), isolated cleft lip and palate (CLP), and isolated cleft palate only (CPO). Black squares represent single-dose effects; red squares represent double-dose effects; and lines are 95% confidence intervals. We include only rs3758249 in FOXE1 in this figure because haplotype analysis indicated that the effect of rs10984103 is explained by the minor allele of the first single-nucleotide polymorphism (Appendix Fig. 2).
The largest association among all the loci was for SNP rs987525 of 8q24; the risk for both CLO and CLP increased 4-fold with the double dose, compared with a 2-fold increase for the single dose. The next-largest positive associations for CLO risk were nearly 2-fold increased risks with the double minor allele doses of PAX7, IRF6, 8q21.3, KIAA1598-VAX1, TPM1, and NOG1. For CLP, the next-largest positive associations were nearly 2-fold higher for the double minor allele doses of PAX7 and KIAA1598-VAX1, followed by smaller associations for ABCA4-ARHGAP29, IRF6, and 8q21.3. A few negative associations were also observed. The double minor allele dose of MAFB rs13041247 was associated with 50% reduction in risk of CLO or CLP. Similarly, the double minor allele dose of FOXE1 rs3758249 was associated with 25% reduction in CLO risk and 50% reduction in CLP risk, while the double minor allele dose of SPRY2 rs8001641 was associated with 30% reduction in risk of CLO or CLP. Appendix Table 5 reports the estimates for risks associated with the major alleles of these 3 loci (MAFB, FOXE1, and SPRY2) when the minor allele is used as reference.
A very different pattern was observed for isolated CPO, with most single and double minor allele dose effects being close to the null and nonsignificant. Only NOG1 rs227731 had significant single- and double-dose associations with 50% reduction in risk with the double-dose effect.
MG Effects
We observed no evidence of MG effects after adjusting for multiple comparisons, and most effects were close to the null (Appendix Tables 6–8). It is worth noting, however, that IRF6 rs2235371 (P = 0.02) and CRISPLD2 rs1546124 (P = 0.003) were significant without such adjustment for isolated CLP (Appendix Table 7) and that FOXE1 rs74934500 (P = 0.02), KIAA1598-VAX1 rs4752028 (P = 0.03), and FGFR2 rs3750817 (P = 0.01) were significant for isolated CPO without adjustment (Appendix Table 8).
PoO Effects
There was no evidence of PoO effects, and most associations were similar between maternal and paternal alleles (Appendix Tables 9 and 10). However, 8q21.3 had a marginally significant larger effect on isolated CLO risk when originating from the mother (relative risk = 1.54 vs. 1.08, P value of difference = 0.06), while 8q24 had a marginally significant larger effect on isolated CLP risk when originating from the father (relative risk = 2.11 vs. 1.66, P value of difference = 0.08). We examined PoO effects for the 14 SNPs for isolated CPO for comparison and observed no significant PoO differences, including for NOG1 (Appendix Table 11). There was, however, increased risk with paternal allele of KIAA1598-VAX1 but reduced risk with maternal allele (P value of difference = 0.06).
Fetal Effects on Isolated versus Nonisolated OFC
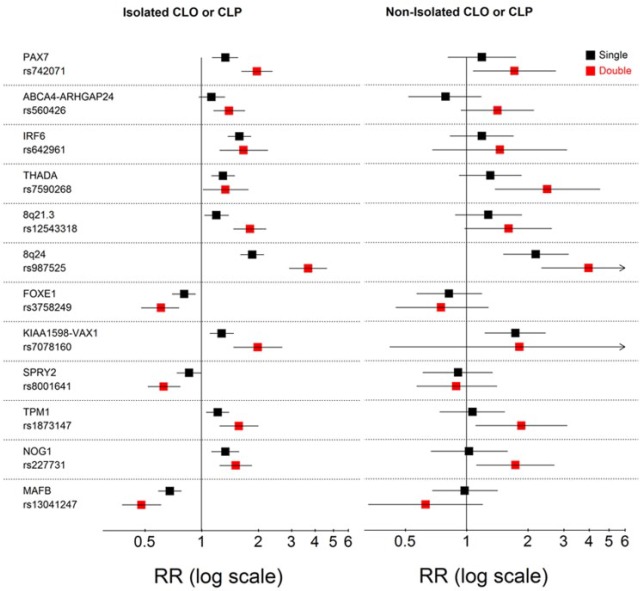
Since we found associations that were generally similar for isolated CLO and CLP, we performed an analysis combining these 2 groups of cases to examine whether associations for isolated CL/P were different from those of nonisolated CL/P (Fig. 2). Overall, the associations with most loci had similar directions and close magnitudes between isolated and nonisolated CL/P, although many effects for the smaller sample of nonisolated CL/P were nonsignificant. However, the associations with the single and double minor allele doses of 8q24, the single allele dose of KIAA1598-VAX1, and double allele doses of PAX7, THADA, TPM1, and NOG1 on nonisolated CL/P were significant at nominal levels (P < 0.05). Overall, these results indicate that these loci play a similar role in both isolated and nonisolated CL/P.
Figure 2.
Single- and double-dose effects measured by relative risks (RRs) for minor alleles of 15 fetal single-nucleotide polymorphisms for the poled category of isolated cleft lip only (CLO) or isolated cleft lip and palate (CLP; first panel) and for nonisolated cleft lip only or nonisolated cleft lip and palate (second panel). Black squares represent single-dose effects; red squares represent double-dose effects; and lines are 95% confidence intervals.
We performed a similar analysis comparing isolated CPO with nonisolated CPO. The only SNP with a significant association for isolated CPO (NOG1 rs227731) had similar risks for nonisolated CPO (Appendix Fig. 3). The double minor allele for PAX7 was associated with a significant increase in nonisolated but not isolated CPO risk. No other significant associations were observed for nonisolated CPO.
Discussion
Using the largest sample to date of individuals of European descent, we investigated 31 SNPs within 17 top loci for OFC from previous GWASs and candidate gene studies. Our initial screening of fetal alleles with Bonferroni adjustment for isolated CLO, CLP, and CPO found significant associations for 14 SNPs in 12 loci; most associations were consistent with a dose-response allelic effect. Most SNPs had remarkably similar effects between isolated CLO and CLP. Potential exceptions were ABCA4-ARHGAP29 and THADA, which were significantly associated with isolated CLP but not CLO. In contrast, TPM1 and NOG1 had stronger associations with isolated CLO than CLP. There was no evidence of significant associations with isolated CPO except for NOG1 (rs227731), which had opposite effects on CPO and CLO—reduction (increase) in CPO (CLO) risk with the minor allele. Our study is the first to report differential effects of ABCA4-ARHGAP29 on CLP and CLO and opposite NOG1 effects on CLO and CPO, including the significant reduction in CPO risk. The lack of significant associations of other loci with isolated CPO is consistent with prior studies (Beaty et al. 2011; Böhmer et al. 2013; Leslie et al. 2017; Ludwig et al. 2017) and studies of familial recurrence suggesting a distinct etiology for CPO in many, if not most, cases (Sivertsen et al. 2008; Grosen et al. 2010). Among all examined loci, 8q24 (rs987525) had the strongest effects on isolated CLO and CLP. There was little evidence of prominent MG or PoO effects consistent with previous studies (Shi et al. 2012; Garg et al. 2014) and findings of similar mother-offspring and father-offspring OFC recurrences (Sivertsen et al. 2008; Grosen et al. 2010), suggesting that such effects do not play a major role in the etiologic mechanisms of almost all the examined loci. A detailed discussion of key loci findings is in the online Appendix. We did not find significant associations for CLO or CLP with MSX1, FGFR2, CRISPLD2, NTN1, and MYH9, suggesting that prior findings for these loci are perhaps not generalizable to population-based samples and are possibly more specific to selective clinic-based samples.
Our comparison of locus effects between isolated and nonisolated CL/P revealed remarkable similarity in direction and magnitude between the 2 cleft groups for most loci. The strongest evidence was observed for 8q24 (rs987525), which had similar significant effects (at P < 0.05) on nonisolated CL/P in both single and double minor allele doses, despite the much smaller sample size. Other loci had significant associations (at P < 0.05) in double minor allele forms (PAX7, THADA, TPM1, and NOG) or single minor allele dose (KIAA1598-VAX1). This finding suggests that these loci are modifiers of cleft risk in both isolated and nonisolated forms and that combining isolated and nonisolated CLO and CLP may be considered in initial GWAS analyses to increase power for identifying such variants. Such analyses would complement analyses specific to isolated forms. Furthermore, it indicates that genetic pathways and possibly even the same etiologic variants converge to contribute risk for CL/P under syndromic and nonsyndromic genetic backgrounds. This reduction in heterogeneity facilitates the design of studies to disentangle the contribution of such variants to OFC etiology. However, it is possible that loci effects on OFC risk within the nonisolated group may vary by etiology of the noncleft malformations, for example, between cases with known syndromes and those of unknown etiologies. Examining this potential heterogeneity in future work is important for understanding the contribution of these loci to nonisolated OFC risk. In conclusion, this study represents the largest population-based examination of top loci suggested for isolated OFC to date. We provide further evidence of the notion of different genetic etiologies between isolated CL/P and CPO and similar genetic etiologies between CLO and CLP for most top loci identified to date. The similarity in effects of most loci between CLO and CLP supports pooling these phenotypes in preliminary GWAS analyses to maximize power. We provide novel evidence of remarkable similarity in effects of top loci between isolated and nonisolated forms of CL/P, suggesting that nonisolated forms may be included in initial GWAS analyses to maximize power and that studying syndromic forms can continue to inform our understanding of molecular mechanisms underlying the nonsyndromic forms. We consistently observed dose-response allelic effects for most loci, allowing for more accurate estimation of genetic effects and providing additional evidence on the likelihood that the effects have a biological basis.
The strengths of our study include a pooled analysis of individual-level data from large population-based samples allowing for precise estimation of loci effects on risk and reducing ascertainment bias. One potential caveat is incomplete accounting for population stratification since we do not have GWAS data to fully capture ancestry. However, this is unlikely to meaningfully affect our results since the majority of our sample is of Caucasian ancestry. While it is possible that some variants may vary in allelic distribution or risk associations across different ancestries among self-reported whites, we did not find significant heterogeneity in association across the 5 study populations we examined. Furthermore, some of the dyad-case samples with isolated OFC from the Utah and Norway studies were included in a previous GWAS (Beaty et al. 2010). However, these cases were previously analyzed with a model of allelic transmission within triads with OFC and not with the combined dyad and case-control design as we did here. Therefore, there is little overlap with previous work that included these samples.
Our results can be considered the most reliable estimates to date of the effects of these top loci on risks of isolated OFC in a population of European descent. These estimates can be used to generate polygenic risk scores to predict risks of CLO or CLP in other smaller data sets with data on the top significant SNPs. Furthermore, they may be informative for predicting risk of oral clefts in cases for whom screening is being considered such as individuals with a family history of clefts. Our analysis focused on the effects of each SNP on its own. Obviously, individuals can have combinations of risk alleles across these SNPs that are associated with different cleft risks, and investigating these combinations in future studies is meaningful to better understand and quantify risk heterogeneity. Also, estimating the effects of causal variants (not only GWAS lead SNPs) and new loci as they are identified, such as GRHL3 for CPO (Leslie, Liu, et al. 2016), is needed for more accurate risk prediction.
Author Contributions
L.M. Moreno Uribe, G.L. Wehby, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; T. Fomina, H.K. Gjessing, M. Gjerdevik, contributed to data analysis, critically revised the manuscript; R.G. Munger, P.A. Romitti, M.M. Jenkins, K. Christensen, contributed to data acquisition, critically revised the manuscript; A.J. Wilcox, J.C. Murray, contributed to data acquisition and interpretation, critically revised the manuscript; R.T. Lie, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Footnotes
A supplemental appendix to this article is available online.
This work was supported by the National Institutes of Health, National Institute of Dental and Craniofacial Research (grant 1 R01 DE020895). It was also supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences. This work was further supported by cooperative agreements U01DD000492 and U01DD001035 from the Centers for Disease Control and Prevention. It was additionally supported by Bergen Research Foundation and the Norwegian Research Council through Biobank Norway. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institutes of Health or the Centers for Disease Control and Prevention or other funding sources.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, et al. 2010. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near mafb and abca4. Nat Genet. 42(6):525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, Murray T, Redett RJ, Fallin MD, Liang KY, et al. 2011. Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genet Epidemiol. 35(6):469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, Parker MM, Hetmanski JB, Balakrishnan P, Mansilla MA, et al. 2013. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 132(7):771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de, Assis N, et al. 2009. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 41(4):473–477. [DOI] [PubMed] [Google Scholar]
- Böhmer AC, Mangold E, Tessmann P, Mossey PA, Steegers-Theunissen RP, Lindemans J, Bouwman-Both M, Rubini M, Franceschelli P, Aiello V, et al. 2013. Analysis of susceptibility loci for nonsyndromic orofacial clefting in a European trio sample. Am J Med Genet A. 161(10):2545–2549. [DOI] [PubMed] [Google Scholar]
- Buyske S. 2008. Maternal genotype effects can alias case genotype effects in case-control studies. Eur J Hum Genet. 16(7):783–785. [DOI] [PubMed] [Google Scholar]
- Camargo M, Rivera D, Moreno L, Lidral AC, Harper U, Jones M, Solomon BD, Roessler E, Velez JI, Martinez AF, et al. 2012. GWAS reveals new recessive loci associated with non-syndromic facial clefting. Eur J Med Genet. 55(10):510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Lidral AC, Stal S, Mulliken JB, Moreno LM, Arcos-Burgos M, Valencia-Ramirez C, Blanton SH, Hecht JT. 2007. CRISPLD2: a novel NSCLP candidate gene. Hum Mol Genet. 16(18):2241–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Ludwig KU, Böhmer AC, Rubini M, Steegers-Theunissen R, Mossey PA, Mangold E, Sharp AJ. 2014. Genome-wide analysis of parent-of-origin effects in non-syndromic orofacial clefts. Eur J Hum Genet. 22(6):822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjessing HK, Lie RT. 2006. Case-parent triads: estimating single- and double-dose effects of fetal and maternal disease gene haplotypes. Ann Hum Genet. 70(Pt 3):382–396. [DOI] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, Bradfield JP, Glessner JT, Thomas KA, Garris M, et al. 2009. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr. 155(6):909–913. [DOI] [PubMed] [Google Scholar]
- Grosen D, Chevrier C, Skytthe A, Bille C, Mølsted K, Sivertsen A, Murray JC, Christensen K. 2010. A cohort study of recurrence patterns among more than 54,000 relatives of oral cleft cases in Denmark: support for the multifactorial threshold model of inheritance. J Med Genet. 47(3):162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukull WA, Ganguli M. 2012. Generalizability: the trees, the forest, and the low-hanging fruit. Neurology. 78(23):1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummet CM, Moreno LM, Wilcox AJ, Romitti PA, DeRoo LA, Munger RG, Lie RT, Wehby GL. 2016. Passive smoke exposure as a risk factor for oral clefts—a large international population-based study. Am J Epidemiol. 183(9):834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutbi H, Wehby GL, Moreno Uribe LM, Romitti PA, Carmichael S, Shaw GM, Olshan AF, DeRoo L, Rasmussen SA, Murray JC, et al. 2017. Maternal underweight and obesity and risk of orofacial clefts in a large international consortium of population-based studies. Int J Epidemiol. 46(1):190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Butali A, Buxó CJ, Castilla EE, Christensen K, Deleyiannis FW, Leigh Field L, Hecht JT, et al. 2017. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum Genet. 136(3):275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, McHenry T, et al. 2016. A multi-ethnic genome-wide association study identifies novel loci for nonsyndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q31. Hum Mol Genet. 25(13):2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Liu H, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, et al. 2016. A genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3. Am J Hum Genet. 98(4):744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Marazita ML. 2013. Genetics of cleft lip and cleft palate. Am J Med Genet C Semin Med Genet. 163C(4):246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Böhmer AC, Bowes J, Nikolic M, Ishorst N, Wyatt N, Hammond NL, Gölz L, Thieme F, Barth S, et al. 2017. Imputation of orofacial clefting data identifies novel risk loci and sheds light on the genetic background of cleft lip ± cleft palate and cleft palate only. Hum Mol Genet. 26(4):829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, Becker J, Herberz R, AlChawa T, Nasser E, et al. 2012. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 44(9):968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, Reutter H, de Assis NA, Chawa TA, Mattheisen M, et al. 2010. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 42(1):24–26. [DOI] [PubMed] [Google Scholar]
- Moreno LM, Mansilla MA, Bullard SA, Cooper ME, Busch TD, Machida J, Johnson MK, Brauer D, Krahn K, Daack-Hirsch S, et al. 2009. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum Mol Genet. 18(24):4879–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reefhuis J, Gilboa SM, Anderka M, Browne ML, Feldkamp ML, Hobbs CA, Jenkins MM, Langlois PH, Newsome KB, Olshan AF, et al. 2015. The National Birth Defects Prevention Study: a review of the methods. Birth Defects Res A Clin Mol Teratol. 103(8):656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Wu T, Murray T, Redett RJ, Wilcox AJ, et al. 2012. Genome wide study of maternal and parent-of-origin effects on the etiology of orofacial clefts. Am J Med Genet A. 158(4):784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivertsen A, Wilcox AJ, Skjaerven R, Vindenes HA, Abyholm F, Harville E, Lie RT. 2008. Familial risk of oral clefts by morphological type and severity: population based cohort study of first degree relatives. BMJ. 336(7641):432–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare O, Jugessur A, Lie RT, Wilcox AJ, Murray JC, Lunde A, Nguyen TT, Gjessing HK. 2012. Application of a novel hybrid study design to explore gene-environment interactions in orofacial clefts. Ann Hum Genet. 76(3):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, Du Y, Wang M, Lan F, Hu Z, et al. 2015. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun. 6:6414. [DOI] [PubMed] [Google Scholar]
- Weinberg CR, Umbach DM. 2005. A hybrid design for studying genetic influences on risk of diseases with onset early in life. Am J Hum Genet. 77(4):627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf ZT, Brand HA, Shaffer JR, Leslie EJ, Arzi B, Willet CE, Cox TC, McHenry T, Narayan N, Feingold E, et al. 2015. Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet. 11(3):e1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zuo X, He M, Gao J, Fu Y, Qin C, Meng L, Wang W, Song Y, Cheng Y, et al. 2017. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 8:14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.