Abstract
Proteins anchored to membranes through covalently linked fatty acids and/or isoprenoid groups play crucial roles in all forms of life. Sorting and trafficking of lipidated proteins has traditionally been discussed in the context of partitioning to membrane domains of different lipid composition. We recently showed that membrane shape/curvature can in itself mediate the recruitment of lipidated proteins. However, exactly how membrane curvature and composition synergize remains largely unexplored. Here we investigated how three critical structural parameters of lipids, namely acyl chain saturation, headgroup size, and acyl chain length, modulate the capacity of membrane curvature to recruit lipidated proteins. As a model system we used the lipidated minimal membrane anchor of the GTPase, N-Ras (tN-Ras). Our data revealed complex synergistic effects, whereby tN-Ras binding was higher on planar DOPC than POPC membranes, but inversely higher on curved POPC than DOPC membranes. This variation in the binding to both planar and curved membranes leads to a net increase in the recruitment by membrane curvature of tN-Ras when reducing the acyl chain saturation state. Additionally, we found increased recruitment by membrane curvature of tN-Ras when substituting PC for PE, and when decreasing acyl chain length from 14 to 12 carbons (DMPC versus DLPC). However, these variations in recruitment ability had different origins, with the headgroup size primarily influencing tN-Ras binding to planar membranes whereas the change in acyl chain length primarily affected binding to curved membranes. Molecular field theory calculations recapitulated these findings and revealed lateral pressure as an underlying biophysical mechanism dictating how curvature and composition synergize to modulate recruitment of lipidated proteins. Our findings suggest that the different compositions of cellular compartments could modulate the potency of membrane curvature to recruit lipidated proteins and thereby synergistically regulate the trafficking and sorting of lipidated proteins.
Introduction
Proteins anchored to the membrane through covalently linked fatty acids and/or isoprenoid groups are found in all forms of life, with >500 in humans alone (1). Importantly, lipidation of proteins like the HIV Gag proteins, Src Family kinases, and Ras GTPases is tightly linked to protein function by facilitating membrane binding and sorting (2, 3), and misregulation of lipidation is associated with severe diseases (4).
The lipid composition of organelles has traditionally been considered extremely important for regulating the cellular organization of lipidated proteins through the preferential partitioning between membrane domains with distinct compositional properties (5, 6, 7). The functional significance of the organelle lipid composition is further supported by the active preservation of the compartment-specific localization of selective lipid species (8, 9). One example of this is the systematic variation in the gradient of some lipid types along the ER-Golgi-Plasma membrane secretory pathway, a pathway believed to facilitate lipidated protein trafficking (10). However, recently we introduced an additional organizational cue, when we showed lipidated proteins to be selectively recruited by membrane curvature (11, 12).
Indeed, close examination reveals that the shape of membrane organelles is a highly conserved and regulated cellular phenotype (13, 14). The fact that both composition and morphology is highly conserved suggests that lipidated protein trafficking and sorting may depend on the compartment-specific combinations of membrane composition and morphology; nevertheless, how these might synergize to facilitate selective recruitment of lipidated proteins to membranes remains largely unexplored.
Here we used a lipidated protein model system comprising the membrane anchoring C-tail motif of the GTPase N-Ras (tN-Ras), which controls eukaryotic cell proliferation and survival and constitutes one of the most frequently mutated genes in cancer tumors (15, 16). We employed the dual lipidated (palmitoyl and farnesyl) model peptide in combination with a single liposome-based assay to study how biologically relevant variations in bilayer lipid shape properties (headgroup size, degree of acyl chain saturation, and acyl chain length) affected the membrane curvature-selective recruitment of the bilayer. We show that modulating lipid shape by either reducing PE concentration, increasing the degree of saturated acyl chains, or reducing lipid length all increased the potency by which tN-Ras was recruited by membrane curvature. Molecular field theory calculations revealed lipid shape-dependent variations in the curvature-mediated relief in the lipid packing density of the outer leaflet as the molecular mechanism underlying the differences in the recruitment ability between membrane systems. Our results demonstrated how bilayer lipid shape and membrane curvature synergize in regulating the selective recruitment of lipidated proteins, and we propose that cells might use their inherent compositional heterogeneity between cellular compartments to modulate the curvature-selective recruitment of lipidated proteins.
Materials and Methods
Materials
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphatidylserine, 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine-N-(cap biotinyl), 1-palmitoyl-2-oleoyl-sn-glycero-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-phosphatidylserine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC), 1,2-dioleoyl-sn-glycero-3-phosphatidylserine, 1,2-myristoyl-sn-glycero-3-phosphocholine (DMPC), and 1,2-dimyristoyl-sn-glycero-3-phosphatidylserine were all acquired from Avanti Polar Lipids (Alabaster, AL). 1,2-Dioleoyl-sn-glycero-3-phosphatidylethanolamine-Atto655 (DOPE-Atto655) was acquired from Atto-Tec (Siegen, Germany). For specific lipid compositions, please see the Supporting Material. The tN-Ras-Alexa488 (tN-Ras) peptide was synthesized and handled as described in Larsen et al. (11).
Liposome preparation
Liposomes were prepared using a previously described lipid hydration method (12, 17). In brief, lipids dissolved in chloroform were thoroughly mixed in a glass vial, at the molar ratios described in the previous section. The solution was dried under nitrogen flow and incubated in vacuum for 4 h. Liposomes were rehydrated by carefully adding a 200 mM D-Sorbitol solution to the lipid film, for a final lipid concentration of 1 g/L. The mixture was incubated overnight at 50°C, which is above the melting temperature of all lipid components, ensuring that lipid film rehydration occurs from a fluid state. The liposomes were then subjected to 10 freeze-thaw cycles to minimize multilamellarity, by immersion in liquid nitrogen followed by thawing in a water bath. After freeze thawing, liposomes were extruded once through a single Isopore polycarbonate membrane with a pore size of 800 nm from Millipore (Billerica, MA) in a Mini-Extruder (Avanti Polar Lipids, Alabaster, AL). The liposomes were flash frozen in liquid nitrogen and stored at −21°C. We have previously shown, using electron microscopy imaging, that the multilamellarity of our liposome preparations is negligible (<5%) (12) and that variation of membrane curvature within the ensemble does not skew the average liposome composition (11, 18, 19).
Single liposome curvature assay
We used the single liposome curvature (SLiC) assay along with the tN-Ras peptide, as described in Larsen et al. (11). We rely on the intrinsic size polydispersity of unilamellar liposomes that are formed during freeze thawing, thus the final single extrusion step through an 800-nm pore filter merely eliminates large aggregates (20). Therefore we are imaging a wide range of curvatures with access to the individual diameters at the single liposome level. In brief, vesicles were labeled by including 1 mol % of DOPE-Atto655 and immobilized on a passivated glass surface. tN-Ras kept in DMSO was added to the solution and allowed to bind at saturating conditions of 1 μM. The final DMSO concentration was kept <0.5%. After a 10 min equilibration period, we sequentially acquired confocal fluorescence micrographs for both membrane (Atto-655) and tN-Ras (Alexa488) channels.
As previously published (12, 20, 21), we can extract the integrated intensity of the membrane dye and convert liposome intensity to diameter, because the integrated intensity of the membrane label is proportional to the number of membrane dyes, which scales with the surface area (12, 20, 21). The liposome diameter is consequently proportional to the square root of the integrated intensity, scaled by a calibration factor. The calibration factor is obtained from a reference sample, produced by extruding liposomes 20 times through a 50-nm filter to ensure as narrow a size distribution as possible. The mean diameter of this population is now measured using dynamic light scattering and correlated to the mean integrated intensity calculated from an intensity histogram obtained by imaging the control sample at the microscope (20). The accuracy of this calibration approach has been validated using cryo-electron microscopy, and from this independent technique we estimated a ±5 nm uncertainty on the liposome diameter determined by the fluorescence approach (20).
For each liposome, the integrated Alexa488 intensity scales with the number of bound tN-Ras peptides and the integrated DOPE-Atto655 intensity scales with the surface area, thus the intensity ratio gives us the absolute density in arbitrary units (11, 12, 21). We then convert the absolute density to normalized densities by dividing all data points with the fit value found at 400 nm in diameter (11, 12, 21). All experiments were performed in a 10 mM HEPES, 95 mM NaCl, pH 7.4 buffer. Neither the immobilization (22) nor the binding of tN-Ras influenced liposome shape (11). Furthermore, tN-Ras binding was investigated under equilibrium conditions, as previously demonstrated by fluorescence recovery after photobleaching experiments (11). It is known that Alexa488 essentially does not interact with membranes (23), and we have previously demonstrated that the attachment of Alexa488 or other fluorophores does not impede the recruitment by membrane curvature of lipid anchors (11). In principle, curvature could impose sorting of the membrane lipids of the liposomes, which would tend to negate any curvature-dependent distortion of lipid packing. Some theoretical calculations suggest that lipids with very high intrinsic spontaneous curvature (like lyso-lipids) could be significantly sorted by high membrane curvatures, although this has not been experimentally validated. On the contrary, for the majority of lipids with smaller intrinsic spontaneous curvatures, both experimental and theoretical studies have concluded that lateral sorting of lipids by membrane curvatures found in vivo is energetically unfavorable, due to the entropic cost of lipid demixing (24, 25, 26, 27, 28, 29, 30). The only way to overcome this limitation is to bestow the individual lipids with collective properties. Thus, the examples where lipids have been shown to be laterally sorted by curvature are limited to membrane systems close to a phase separation (29, 31) or if individual lipids are cross-linked (28). In this work, we are taking extra care not to work with systems close to a phase separation or lipid dyes shown to cluster. Finally, we used previously published data sets to validate that lipid lateral sorting based on lipid spontaneous curvature does not occur in the SLiC assay through control experiments with premixed pairs of lipid fluorophores displaying a constant average intensity ratio value, as a function of liposome diameter (18, 19) (Fig. S1).
Equipment and settings
For imaging liposomes and tN-Ras, we used a TCS SP5 inverted confocal microscope and an oil immersion objective HCX PL APO CS × 100 (NA 1.4); Leica Microsystems, Wetzlar, Germany. Detection of Alexa488 labeled tN-Ras was performed at 495–580 nm (exc. 488 nm); detection of Atto655-labeled vesicles was performed at 640–750 nm (exc. 633 nm) using photomultipliers. In all cases, sequential imaging was used to avoid cross excitation. Images had a resolution of 2048 × 2048 pixels, with a pixel size of 25.2 nm and a bit depth of 16. Temperature control (±0.5°C) in the microscope chamber was achieved by enclosing the entire microscope within a box heated by stable air flow (The Cube 2 Temperature Controller; Life Imaging Services, Basel, Switzerland). All experiments were performed at 22°C, except for the DLPC and DMPC experiments, which were performed at 30°C. Image analysis and data treatment were performed using custom-made routines in the softwares Igor Pro (WaveMetrics, Lake Oswego, OR) and Fiji (ImageJ; National Institutes of Health, Bethesda, MD).
Results
Modulating acyl chain saturation affects tN-Ras binding to both curved and planar membranes
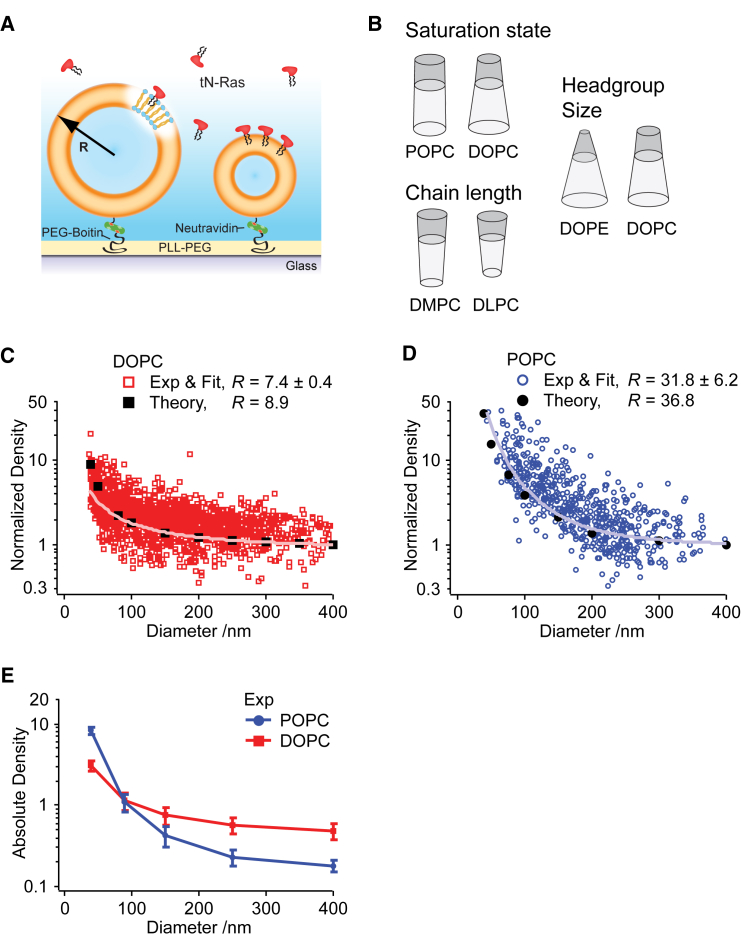
To study how variations in lipid shape affected the selective recruitment by membrane curvature, we used tN-Ras and our previously described SLiC assay (11, 12, 17). We employed fluorescently labeled liposomes, immobilized on a passivated glass surface, and allowed fluorescently labeled tN-Ras to bind from solution (Fig. 1 A). Tuning the density of liposomes on the surface allowed us to image individual liposomes using confocal microscopy and to extract the integrated intensities of both the membrane dye (DOPE-Atto655) and the tN-Ras dye (Alexafluor488) in a parallel manner (>100 liposomes/frame). Because immobilization does not perturb the spherical shape of individual liposomes (22), we could convert their integrated Atto655 membrane dye intensity to liposome diameter in units of nanometers, using a combination of microscopy and dynamic light scattering, as described in Kunding et al. (20). Because we previously demonstrated tN-Ras to display reversible membrane binding and knowing that the integrated intensity arising from fluorescently labeled tN-Ras is proportional to the number of molecules, the ratio of the integrated Alexa488 and Atto655 intensities allowed us to calculate the absolute density of bound tN-Ras on individual liposomes of various curvatures (11, 12).
Figure 1.
Recruitment by membrane curvature of tN-Ras is modulated by lipid shape. (A) Shown here is a single liposome curvature assay for studying the effect of membrane composition on the recruitment by membrane curvature of tN-Ras. Single liposomes of various sizes and thus curvatures are immobilized on a passivated surface and tN-Ras is bound from solution. By imaging fluorescently marked liposomes and tN-Ras with confocal microscopy, liposome diameter and tN-Ras density on individual liposomes was quantified. (B) Given here is an illustration of the lipid shapes compared in the study, highlighting how lipid shape is determined by the relation between the volume of the headgroup (dark gray) and acyl chains (light gray). (C) Shown here is normalized tN-Ras density as a function of liposome diameter for liposomes prepared from DOPC lipids (dark red markers) with corresponding off-set power function fit (light red line) and molecular field theory calculations depicting the equilibrium binding concentrations of tN-Ras on DOPC membranes (black markers). The recruitment ratio (R) represents the increase in tN-Ras density when reducing the diameter by a factor of 10 as quantified either from power function fitting to the experimental data or from the theoretical calculations. (D) Given here is normalized tN-Ras density and experimental fit as a function of liposome diameter for liposomes prepared from POPC lipids (dark-blue markers and light-blue line) and molecular field theory calculations depicting the equilibrium binding concentrations of tN-Ras on POPC membranes (black markers). R is reported as the average mean ± SE of nDOPC = 9 and nPOPC = 8 independent experiments. (E) Given here is average absolute tN-Ras density on either POPC (blue) or DOPC (red) membranes, calculated from the power function fits to the individual experimental data set for five liposome diameters.
Lipid shape is determined by the relation between the acyl chain and headgroup volume and is traditionally represented by three classes: cones, inverted cones, and cylinders (32) (Fig. 1 B). We first modulated lipid shape by employing liposomes in the SLiC assay produced from POPC (one saturated, one mono-unsaturated acyl chain, with relatively compact cylindrical lipid shape) or DOPC (two mono-unsaturated chains, with relatively less-compact inverted cone lipid shape) (33) (Fig. 1 B). To facilitate an easier comparison between the two systems, we converted the obtained absolute density data to normalized densities (by normalizing to a density of 1 for a liposome diameter of 400 nm) and plotted the normalized tN-Ras density on individual DOPC or POPC liposomes as a function of diameter (Fig. 1, C and D).
To quantify the correlation between membrane curvature and tN-Ras density, we fitted the data of Fig. 1, C and D, with an off-set power function as described in Larsen et al. (11) (and see Supporting Material). This allowed us to extract the recruitment ratio (R) as the fold-increase in tN-Ras density, when reducing the diameter by a factor of 10. We quantified R = 7.4 ± 0.4 for the DOPC system and R = 31.8 ± 6.2 for the POPC system (Fig. 1, C and D), whereas a negative control, binding streptavidin to biotinylated lipids, gave R = 1.01 ± 0.01 (Fig. S2). This demonstrated a significant selective recruitment of tN-Ras by membrane curvature for both the DOPC and POPC systems, relative to the negative control.
Thus our experiments revealed an approximately fourfold increase in the selective recruitment by membrane curvature for the POPC system compared to the DOPC system, illustrating that lipid shape can have a pronounced effect on the selective recruitment of tN-Ras by membrane curvature.
The ability of the SLiC assay to measure the absolute binding densities of tN-Ras allowed us to decipher if the change in the selective recruitment by membrane curvature originated from variations in the binding densities only on curved membranes, only on planar membranes, or both. For each individual dataset we calculated the absolute density from the off-set power function fit at five specific liposome sizes. We used these values to calculate the average absolute density on either DOPC or POPC membranes for the five different liposome diameters and presented these in Fig. 1 E, along with the mean ± SE from nDOPC = 9 and nPOPC = 8 independent experiments (see Supporting Material).
For liposomes larger than 90 nm, we found a higher tN-Ras density on the DOPC, as compared to the POPC, system. However, for smaller liposomes (<90 nm) the trend was reversed, resulting in a higher tN-Ras density on POPC liposomes. Thus, the increased selective recruitment by curvature quantified for the POPC system originated both from a decreased density on flat, and an increased density on curved, membranes compared to the DOPC system. This illustrates the complex interplay between membrane composition and curvature and how it is the combination of both properties that accounts for the absolute binding of lipidated proteins to membranes.
Molecular field calculations reveal that composition-dependent variations in the lipid packing density modulate the curvature-selective recruitment ability
Next we used a molecular field theory, as we have shown in Larsen et al. (11), to determine the thermodynamic and structural characteristics of the lipid bilayers, to provide a mechanistic understanding of how the selective tN-Ras recruitment by membrane curvature was influenced by bilayer lipid shape. The theory explicitly incorporates the conformational energy and entropy of the acyl chains of the bilayer lipids and tN-Ras anchor molecules. Together with terms accounting for the lipid headgroups and the translational mobility of the lipids, these contributions collectively form a density functional that encompasses fluctuations over all possible states of the systems.
The equilibrium state of a given system is calculated through functional minimization with respect to the system’s free variables, and is represented by the system’s thermodynamic free energy (34, 35). There is only one free parameter in the model. The relaxation ratio, which determines the amount of flip-flop between the leaflets as a function of curvature, is fit in the theory to match up with experimentally determined bending moduli for the lipid bilayers of interest in this article (Table S1). Our experimental data are then directly and quantitatively compared with the theoretical prediction of absorbed density; no further fitting is involved.
The resulting probability distributions are shown to have the same functional form for each chain in the hydrophobic region (here written with the explicit configuration dependence, αδ, and curvature dependence, (c). The probability of each chain being in a particular configuration within a membrane with a given curvature is
| (1) |
where ε(αδ) denotes the internal energy of the chains in configuration αδ; π(z,c) is the lateral pressure profile, determined self-consistently for each curved state of the bilayer; and νδ(z,αδ) is the contribution to the volume of the hydrophobic core at z (distance from the midplane of the bilayer) from the molecules in leaflet δ (inner or outer leaflet), existing in configuration αδ. The integration is performed over the entire hydrophobic region of the bilayer (–l < z < +l). The value qδ(c) is the single molecule partition function (normalization of the probability), defined as
| (2) |
When a bulk solution containing lipid anchors is in contact with a curved or planar lipid bilayer, thermodynamics requires that the chemical potential of the lipid anchors in the bulk solution, , must be equal to the chemical potential of the anchors bound to the external leaf of the bilayer in contact with the solution, . The chemical potential of the lipid anchors within this theoretical framework is
| (3) |
where qpal,E(c) and qfar,E(c) are the partition functions of the palmitoyl and farnesyl chain-anchors of tN-Ras, respectively. Because the lipid anchors are not explicitly present during the functional minimization that returns the lateral pressure fields of the curved lipid bilayers, we use Widom’s potential distribution theorem (34, 36) to obtain the chemical potential of the membrane-bound lipid anchors.
From this method, we may calculate the bound density of the lipid anchors in a curved bilayer, relative to that of the planar bilayer, in terms of the corresponding ratio of their partition functions:
| (4) |
The molecular model explicitly treats the packing interactions in the hydrophobic fatty-acid chain region of the bilayer. Whereas previous work with this molecular theory has included additional interactions including both the electrostatics of the headgroups (37) and attractive Maier-Saupe interactions to properly model the liquid-disorder/liquid-order phase transition (11), we decided to use the minimum required physics to capture the experimental observations. For example, note that there are systems where the additional complexity of attractive interactions is needed when studying curvature sensing in lipid bilayers in the liquid-order phase. It was recently shown that curvature effects are highly nonadditive in the liquid-order phase using simulation (38). We have recently published a similar finding when comparing experimental observations to theoretical methods that used the Maier-Saupe attractive interaction to study curvature sensing of tN-Ras binding to liquid-disorder and liquid-order phases (11). In this study, the lipid bilayers are all in the liquid-disorder phase, so the model did not include any additional attractive interactions. For more details, see Supporting Material (11, 34, 36). This theoretical approach allowed us to determine the structure and thermodynamics of various lipid systems to elucidate differences in the exact molecular structural organization of the membrane lipids when curving the bilayer and how this subsequently affected the localization of tN-Ras.
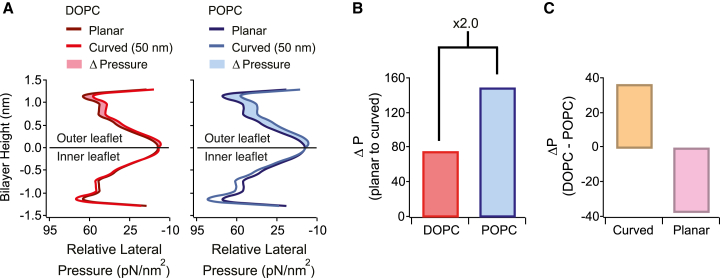
The theoretically calculated tN-Ras recruitment (black markers in Fig. 1, C and D) displayed a relative good agreement with the experimental data and recapitulated the more potent selective recruitment by membrane curvature for the POPC versus the DOPC system. Both theoretical (34, 35, 39) and experimental (11, 40, 41) studies have shown that protein partitioning to membranes depends explicitly on the lipid packing density, expressed in the molecular field theory as the area per lipid and the lateral membrane pressure.
We calculated very similar relative increases in the area per lipid when curving DOPC and POPC membranes to a diameter of 50 nm (8 and 7%, respectively) (Table 1). This suggests that membrane composition-dependent variations in the area per lipid are not driving the observed differences in the selective recruitment by membrane curvature, in agreement with previous studies on amphipathic helix insertion (42). Our calculations additionally reveal that curving membranes distorts the transbilayer symmetry of the lateral pressure profile, increasing the pressure in the inner leaflet and relieving the pressure in the outer leaflet (Fig. 2 A). Comparing the curvature-mediated relief in the lateral pressure of the outer leaflet (ΔP, shaded area in Fig. 2 A) revealed a 2.0-fold higher ΔP for the POPC system as compared to the DOPC system (Fig. 2 B; Table 1). Thus, we propose that the larger curvature-dependent decrease in the lateral pressure of the outer leaflet observed for POPC could drive the more potent selective recruitment by membrane curvature quantified for the POPC-versus-DOPC system.
Table 1.
Summary of Data from Molecular Field Theory Calculations
| Area per Lipid (nm2/Molecule) |
Lateral Pressure (pN/nm2) |
R Value | |||||
|---|---|---|---|---|---|---|---|
| Planar | 50 nm | % Change | Planar | 50 nm | ΔP | ||
| DOPC | 0.695 | 0.741 | 6.6 | 480 | 404 | 76 | 8.9 |
| POPC | 0.672 | 0.727 | 8.2 | 517 | 367 | 150 | 36.8 |
| 0% PE | 0.695 | 0.741 | 6.6 | 480 | 404 | 76 | 8.9 |
| 25% PE | 0.688 | 0.732 | 6.4 | 456 | 396 | 60 | 7.7 |
| 50% PE | 0.681 | 0.725 | 6.5 | 432 | 383 | 48 | 6.5 |
| DMPC | 0.621 | 0.661 | 6.4 | 442 | 334 | 108 | 17.7 |
| DLPC | 0.627 | 0.671 | 7.1 | 458 | 296 | 162 | 28.3 |
Figure 2.
Molecular field theory calculations reveal a greater curvature-mediated relief in lateral pressure for POPC versus DOPC. (A) Given here are theoretically calculated relative lateral pressure profiles along the bilayer normal for the hydrophobic region of (left, red) DOPC membranes or (right, blue) POPC membranes. The top part represents the outer monolayer (outer leaflet), the bottom represents the inner monolayer (inner leaflet) of the membrane, and the relative lateral pressure profile is depicted for either planar (dark line) or curved (pale line, 50 nm diameter liposome) membranes. The curvature-dependent relief in the relative lateral pressure of the outer monolayer, ΔP, is calculated as the total area between the curves (shaded area). (B) Given here is quantification of ΔP imposed by membrane curvature (planar to 50 nm) for either DOPC (red) or POPC (blue). (C) Given here is quantification of the compositional dependent change in the total integrated lateral pressure as compared for curved (50 nm, orange) or planar (pink) DOPC and POPC membranes.
To further elucidate the importance of the lateral pressure in governing the absolute tN-Ras membrane density, we compared the integrated lateral pressures between the outer leaflets of DOPC and POPC membranes in both planar and curved geometries (Fig. 2 C; Table 1). Intuitively we would predict that a higher integrated lateral pressure would increase the work of insertion and thus reduce tN-Ras binding. We calculated a higher integrated lateral pressure for planar POPC than DOPC (Fig. 2 C, pink), corresponding to augmented tN-Ras binding in planar DOPC membranes, in complete agreement with the experimental data. In contrast, for highly curved membranes we found the highest integrated lateral pressure in the DOPC membranes (Fig. 2 C, orange), again corroborating our experimental finding of the highest tN-Ras binding on curved POPC membranes. The nontrivial correlation between the curvature-dependent absolute binding densities of tN-Ras and compositionally mediated variations in the lateral pressure profile in the membrane’s external leaf, further support compositional and curvature-mediated variation of the lateral pressure as an underlying mechanism determining lipidated protein recruitment.
Lipid headgroup size modulates tN-Ras binding and selective recruitment by membrane curvature
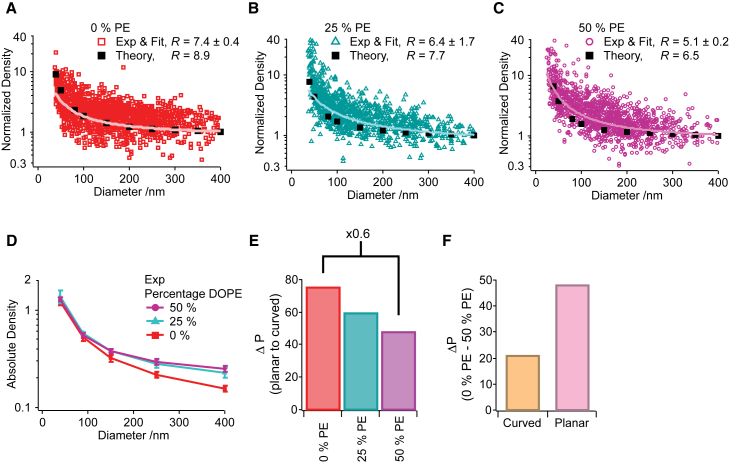
To further elucidate the mechanistic role of lipid shape-dependent variations in the curvature-mediated relief in lateral pressure, we modulated another lipid shape parameter, namely the lipid headgroup size. We employed DOPE lipids, which display a more pronounced inverted cone shape due to their small headgroup volume as compared to other phospholipids, e.g., DOPC lipids (32, 43) (Fig. 1 B). We prepared liposomes from lipid mixtures containing DOPC and increasing DOPE concentrations (0, 25, or 50 mol % DOPE) and quantified the selective recruitment of tN-Ras on all lipid formulations. R values decreased for higher mol % of DOPE, ranging from R = 7.4 ± 0.4, R = 6.4 ± 1.7 to R = 5.1 ± 0.2, respectively, for 0, 25, and 50 mol % DOPE (Fig. 3, A–C; Fig. S3). This constitutes a small, yet significant, reduction in the selective recruitment by membrane curvature of tN-Ras when increasing the DOPE content.
Figure 3.
Introducing lipids with reduced headgroup size increases the absolute tN-Ras binding and decreases recruitment by membrane curvature. Given here is the normalized tN-Ras density and experimental fit as a function of liposome diameter for liposomes prepared from DOPC lipids and containing 0% PE (A), 25% (B), or 50% PE (C) and the molecular field theory calculations depicting the equilibrium binding concentrations of tN-Ras on the three systems. R is reported as the average mean ± SE of n0% PE = 9, n25% PE = 3, and n50% PE = 6 independent experiments. (D) Shown here is the average absolute tN-Ras density on either 0% PE (red), 25% PE (turquoise), or 50% PE (purple) membranes, calculated for five liposome diameters. (E) Shown here is the quantification of ΔP imposed by membrane curvature for either 0% PE (red), 25% PE (turquoise), or 50% PE (purple) membranes of 50 nm in diameter. (F) Shown here is the quantification of the compositional dependent change in the total integrated lateral pressure as compared for curved (50 nm, orange) or planar (pink) 0 and 50% PE membranes.
Absolute tN-Ras density data on liposomes of different curvatures quantified an ∼88% increase in the density for a 400-nm liposome when increasing the DOPE content from 0 to 50 mol %, but a nonsignificant increase in density on smaller 40-nm liposomes for the same DOPE range (Fig. 3 D). This demonstrates that the reduced R values for higher DOPE concentrations originate from a DOPE-dependent increase in tN-Ras density on large liposomes.
The molecular field theory calculations showed a relatively good agreement with the experimental data and recapitulated the trend of decreasing R values for higher DOPE concentrations. Again, the difference in the curvature-mediated change in the area per lipid was found to be minimal (6.6 and 6.5% for 0 and 50% PE, respectively) (Table 1). In contrast, we calculated a 37% decrease in the curvature-dependent relief in the lateral pressure for increasing DOPE concentration (from 0 to 50% PE), again suggesting this to be a mechanism for the quantified reduction in the curvature-sensitivity of tN-Ras (Fig. 3 E; Tables 1 and S3).
We also calculated a notably larger reduction in the integrated lateral pressure between 0 and 50% PE in planar membranes than in curved membranes (Fig. 3 F; Table 1). This corroborated the experimental data demonstrating a larger increase in the tN-Ras binding density with rising PE concentration on the planar versus curved membrane systems. Overall, the experimental data and theoretical calculations further validate lipid shape-dependent variations in the curvature-mediated relief in lateral pressure as a molecular mechanism underlying differences in the selective membrane recruitment of lipidated proteins.
Decreasing lipid length increases the selective recruitment by membrane curvature
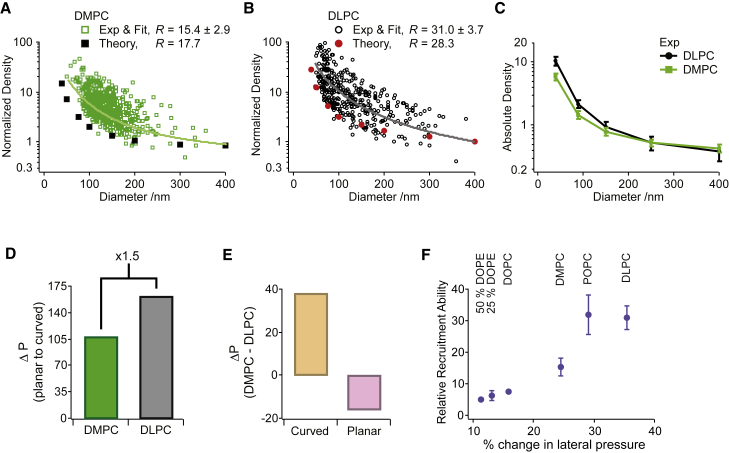
Varying the number of acyl groups, hereby affecting lipid length and membrane thickness, is the only remaining basic approach to changing lipid shape, a modulation that, to our knowledge, has yet to be examined for its potential role in regulating curvature-dependent protein recruitment (Fig. 1 B). We analyzed a relatively thicker membrane system, produced from DMPC lipids (diC14, 3.67 ± 0.07 nm) (44), and compared it to a thinner system produced from the short-tailed DLPC lipids (diC12, 3.26 ± 0.07 nm) (44). Although all other experiments were performed at 22°C, the comparison of DMPC and DLPC was performed at 30°C, where both lipid systems were in the fluid (liquid-disordered) phase and have previously been shown to display an ∼12% difference in bilayer thickness (44).
We quantified R = 15.4 ± 2.9 and R = 31.0 ± 3.7 for the DMPC and the DLPC systems, respectively (Fig. 4, A and B; Fig. S3). Hence, decreasing membrane thickness caused a twofold increase in the selective recruitment of tN-Ras by membrane curvature. The average absolute tN-Ras density plots demonstrated similar densities of tN-Ras on larger DMPC and DLPC liposomes, but significantly higher densities on highly curved DLPC versus DMPC liposomes, indicating differential binding on curved membranes to be the origin of the increased R value found for the DLPC system (Fig. 4 C).
Figure 4.
Thinner membranes show increased recruitment of tN-Ras by membrane curvature. Given here is the normalized tN-Ras density as a function of liposome diameter for liposomes prepared from longer DMPC lipids (A) or shorter DLPC lipids (B) and the molecular field theory calculations depicting the equilibrium binding concentrations of tN-Ras on the two systems. R is reported as the average mean ± SE of nDMPC = 7 and nDLPC = 6 independent experiments. (C) Shown here is the average absolute tN-Ras density on either DLPC (black) or DMPC (green), calculated for five liposome diameters. (D) Given here is the quantification of ΔP imposed by membrane curvature for either DMPC (green) or DLPC (black) membranes of 50 nm in diameter. (E) Given here is the quantification of the compositional-dependent change in the total integrated lateral pressure as compared for curved (50 nm, orange) or planar (pink) DLPC and DMPC membranes. (F) Shown here are experimentally determined R values versus the theoretically calculated curvature-dependent % change in the lateral pressure.
Molecular field calculations found a thickness difference of ∼11%, matching well the experimentally determined value of 12% (44), and fully captured the trend of increased selective tN-Ras recruitment for the DLPC as compared to the DMPC system, albeit while following the lower part of the experimental data (Fig. 4, A and B; Tables 1 and S1). The calculations quantified a 1.5-fold increase in ΔP for the DLPC versus the DMPC system and essentially no difference for the area per lipid (7 and 6%, respectively) (Figs. 4 D and S4; Table 1). We also calculated a markedly larger difference between the integrated lateral pressure of curved DMPC and DLPC membranes than in planar DMPC and DLPC membranes, all of which mirrors the experimental observations.
Here we show that changing lipid length can modulate both membrane binding and the selective recruitment by membrane curvature of membrane-anchored proteins. Finally, comparing all the experimentally collected R values versus the theoretically calculated curvature-dependent % change in lateral pressure displays an almost linear dependency (Figs. 4 F and S5). This very strong correlation further strengthens curvature-mediated relief in the lateral pressure as a molecular mechanism governing the curvature-sensitivity of lipidated proteins, and that any variation in lipid shape and membrane curvature synergistically tune the recruitment of such protein.
Discussion
Membrane-based sorting of lipidated proteins in general, and Ras proteins in particular, have traditionally been discussed in the context of partitioning between ordered membrane domains, previously referred to as “raft domains” (5, 45, 46, 47). Only recently was membrane curvature introduced as a generic regulator of lipidated protein localization (12) and suggested to work in synergy with ordered membrane domains to facilitate preferential tN-Ras up-concentration (11). This illustrates how the unique composition of complex lipid mixtures, which modulates collective membrane properties such as phase state, could synergize with membrane curvature in regulating the binding of lipidated proteins. Here we revealed how modulating individual properties affecting lipid shape also affect the curvature-selective recruitment of lipidated proteins and we have proposed a molecular mechanism.
Organelle-specific variations in lipid composition are tightly controlled and even small perturbations can elicit stress responses, triggering cell death (9). The Golgi represents the main sorting station for saturated lipids. Here, trafficking vesicles are believed to selectively transport more saturated lipid species to the plasma membrane, maintaining a gradient of increasing degree of acyl chain saturation along the secretory pathway (48, 49). These vesicle carriers have additionally been proposed to constitute the vesicular-based transport mechanism responsible for correct Ras localization to its main signaling compartment, the plasma membrane (6, 50, 51). Our findings suggest that the combination of high membrane curvature and increased levels of saturated lipids might constitute a potent molecular cue driving selective Ras localization to such trafficking vesicles, hereby efficiently delivering Ras to the plasma membrane.
Lipid length, and consequently membrane thickness, also differs between organelles, showing an ∼13% increase from the ER to the plasma membrane (52). Comparable to this, we modulated the lipid length in our model system by two acyl groups with a subsequent ∼12% change in thickness (44). Traditionally, such differences in membrane thickness have been proposed to regulate the sorting of integral membrane proteins based on hydrophobic mismatch between the transmembrane segment and the bilayer (53). However, the regulatory role of membrane thickness has, to our knowledge, not yet been considered in relation to the selective recruitment of membrane-anchored proteins by membrane curvature.
Here we showed that reducing lipid length increased the selective recruitment of tN-Ras by membrane curvature. This suggests a stronger selective recruitment to vesicular carriers with thinner membranes budding from the ER or Golgi as compared to carriers with thicker membrane budding from the plasma membrane. Such a mechanism might infer a selective driving force for localizing N-Ras on anterograde vesicular carriers, moving N-Ras to and retaining it at the plasma membrane where it primarily elicits its function (54).
Our finding that modulating acyl chain saturation or headgroup size affects the selective recruitment by membrane curvature of tN-Ras, is in line with previous observations for proteins anchored through shallow insertion of amphipathic helices (40, 42, 55, 56). This suggests that the interplay among lipid saturation state, headgroup size, and curvature might be a generic mechanism for regulating the localization of proteins anchored through hydrophobic insertion.
Previously, the biophysical mechanism relating how composition and curvature synergize to regulate selective protein recruitment has predominantly been discussed in the context of interfacial lipid packing defects (40, 42, 57, 58). This concept has been instrumental in understanding how composition and curvature regulate the selective recruitment of proteins anchored through shallowly inserted amphipathic helices. However, for lipidated proteins, which insert deeper into the outer membrane monolayer, a continuum model based on lipid packing density variations controlled by composition and curvature may be more appropriate (34, 35). Further evidence supporting this comes from coarse-grained MD simulation work describing the recruitment of various Ras peptides to the interface between phase-separated membranes (7, 59, 60, 61). Here a direct link was found between increased Ras clustering at the domain interface and changes in various structural membrane properties, including the lateral pressure profile, subsequently leading to introduction of membrane curvature. This work suggests an intimate relationship between how Ras peptides can be recruited by membrane curvature and how the recruitment can lead to curvature stabilization, potentially working as a positive feedback loop, driving Ras protein recruitment.
The theoretical calculations presented in this work suggest that, in mechanistic terms, the curvature- and composition-dependent relief in the lateral pressure of the outer leaflet will lead to a reduction in the work of insertion for tN-Ras, essentially driving the increased recruitment. An additional important corollary from the theoretical calculations is that any extrinsic parameters affecting membrane lateral pressure will lead to altered tN-Ras recruitment. This could be the binding and subsequent scaffolding of other peripheral proteins, the lateral segregation of lipids or transmembrane proteins, or interactions between the membrane and the cell cytoskeleton. All would work in synergy with composition and curvature and provide the cell with additional means to locally tune the density of lipidated proteins.
We have previously proposed curvature-mediated changes in lateral pressure as the molecular mechanism explaining how different complex lipid mixtures, regulating collective membrane properties like phase state, influence the selective recruitment by membrane curvature (11). Here we strengthened this hypothesis, demonstrating that modifying individual lipid properties, like degree of acyl chain saturation, headgroup area, or lipid length, will also lead to variations in the curvature-mediated relief in lateral pressure in the absence of changes in phase state.
Conclusions
We show that modulating the bilayer lipid shape can affect the selective recruitment by membrane curvature of the N-Ras lipid anchors. Molecular field theory calculations suggest lipid shape-dependent variation in the curvature-mediated relief in lateral pressure as an underlying biophysical parameter regulating the selective recruitment by membrane curvature. In cells, multiple factors contribute to the selective organization of lipidated proteins. Here, we propose that the compositional heterogeneity between compartments could allow the cell to promote or impede the curvature-selective recruitment of lipidated proteins, and thereby locally fine-tune protein localization.
Author Contributions
J.B.L., N.S.H., and D.S. designed research. J.B.L. performed research. C.K. and M.J.U. performed theoretical modeling. S.L.P. and K.J.J. contributed new reagents/analytic tools. J.B.L. and N.S.H. analyzed data. J.B.L., N.S.H., C.K., M.J.U., and D.S. wrote the paper. All authors discussed the results and commented on the manuscript at all stages.
Acknowledgments
This work was supported by the Lundbeck Foundation Center for Biomembranes in Nanomedicine, the Danish Councils for Independent and Strategic Research (1311-00002B), and the University of Copenhagen Programs of Excellence “Single Molecule Nanoscience”, “BioScaRT”, and “SYNBIO”. M.J.U. acknowledges support from the National Institutes of Health under grant P20GM103499.
Editor: Anne Kenworthy.
Footnotes
Jannik B. Larsen’s present address is Department of Micro- and Nanotechnology, Centre for Nanomedicine and Theranostics, DTU Nanotech, Technical University of Denmark, 2800 Kng. Lyngby, Denmark.
Søren L. Pedersen’s present address is Gubra Aps, 2970 Hørsholm, Denmark.
Nikos S. Hatzakis’s present address is Department of Chemistry, Nano-Science Center, University of Copenhagen, 2100 Copenhagen, Denmark.
Supporting Materials and Methods, one table, and five figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30701-4.
Contributor Information
Mark J. Uline, Email: uline@cec.sc.edu.
Dimitrios Stamou, Email: stamou@nano.ku.dk.
Supporting Citations
References (62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78) appear in the Supporting Material.
Supporting Material
References
- 1.Tate E.W., Kalesh K.A., Thinon E. Global profiling of protein lipidation using chemical proteomic technologies. Curr. Opin. Chem. Biol. 2015;24:48–57. doi: 10.1016/j.cbpa.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groves J.T., Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocks O., Peyker A., Bastiaens P.I.H. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 4.Resh M.D. Targeting protein lipidation in disease. Trends Mol. Med. 2012;18:206–214. doi: 10.1016/j.molmed.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prior I.A., Harding A., Hancock J.F. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 6.Simons K., Gerl M.J. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 7.Janosi L., Li Z., Gorfe A.A. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc. Natl. Acad. Sci. USA. 2012;109:8097–8102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holthuis J.C.M., Menon A.K. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 10.Bigay J., Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Larsen J.B., Jensen M.B., Stamou D. Membrane curvature enables N-ras lipid anchor sorting to liquid-ordered membrane phases. Nat. Chem. Biol. 2015;11:192–194. doi: 10.1038/nchembio.1733. [DOI] [PubMed] [Google Scholar]
- 12.Hatzakis N.S., Bhatia V.K., Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 13.Shibata Y., Hu J., Rapoport T.A. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 14.Lippincott-Schwartz J., Phair R.D. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu. Rev. Biophys. 2010;39:559–578. doi: 10.1146/annurev.biophys.093008.131357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prior I.A., Hancock J.F. Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatia V.K., Madsen K.L., Stamou D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen J., Hatzakis N.S., Stamou D. Observation of inhomogeneity in the lipid composition of individual nanoscale liposomes. J. Am. Chem. Soc. 2011;133:10685–10687. doi: 10.1021/ja203984j. [DOI] [PubMed] [Google Scholar]
- 19.Elizondo E., Larsen J., Ventosa N. Influence of the preparation route on the supramolecular organization of lipids in a vesicular system. J. Am. Chem. Soc. 2012;134:1918–1921. doi: 10.1021/ja2086678. [DOI] [PubMed] [Google Scholar]
- 20.Kunding A.H., Mortensen M.W., Stamou D. A fluorescence-based technique to construct size distributions from single-object measurements: application to the extrusion of lipid vesicles. Biophys. J. 2008;95:1176–1188. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatia V.K., Hatzakis N.S., Stamou D. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Semin. Cell Dev. Biol. 2010;21:381–390. doi: 10.1016/j.semcdb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Bendix P.M., Pedersen M.S., Stamou D. Quantification of nano-scale intermembrane contact areas by using fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA. 2009;106:12341–12346. doi: 10.1073/pnas.0903052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes L.D., Rawle R.J., Boxer S.G. Choose your label wisely: water-soluble fluorophores often interact with lipid bilayers. PLoS One. 2014;9:e87649. doi: 10.1371/journal.pone.0087649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonny B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 25.Cooke I.R., Deserno M. Coupling between lipid shape and membrane curvature. Biophys. J. 2006;91:487–495. doi: 10.1529/biophysj.105.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamal M.M., Mills D., Howard J. Measurement of the membrane curvature preference of phospholipids reveals only weak coupling between lipid shape and leaflet curvature. Proc. Natl. Acad. Sci. USA. 2009;106:22245–22250. doi: 10.1073/pnas.0907354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callan-Jones A., Sorre B., Bassereau P. Curvature-driven lipid sorting in biomembranes. Cold Spring Harb. Perspect. Biol. 2011;3:a004648. doi: 10.1101/cshperspect.a004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian A., Baumgart T. Sorting of lipids and proteins in membrane curvature gradients. Biophys. J. 2009;96:2676–2688. doi: 10.1016/j.bpj.2008.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorre B., Callan-Jones A., Bassereau P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. USA. 2009;106:5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh W.T., Hsu C.J., Baumgart T. Curvature sorting of peripheral proteins on solid-supported wavy membranes. Langmuir. 2012;28:12838–12843. doi: 10.1021/la302205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parthasarathy R., Yu C.H., Groves J.T. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir. 2006;22:5095–5099. doi: 10.1021/la060390o. [DOI] [PubMed] [Google Scholar]
- 32.Israelachvili J. 3rd Ed. Elsevier; London, UK: 1991. Intermolecular and Surface Forces. [Google Scholar]
- 33.Strandberg E., Tiltak D., Ulrich A.S. Lipid shape is a key factor for membrane interactions of amphipathic helical peptides. Biochim. Biophys. Acta. 2012;1818:1764–1776. doi: 10.1016/j.bbamem.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 34.Uline M.J., Longo G.S., Szleifer I. Calculating partition coefficients of chain anchors in liquid-ordered and liquid-disordered phases. Biophys. J. 2010;98:1883–1892. doi: 10.1016/j.bpj.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uline M.J., Szleifer I. Mode specific elastic constants for the gel, liquid-ordered, and liquid-disordered phases of DPPC/DOPC/cholesterol model lipid bilayers. Faraday Discuss. 2013;161:177–191. doi: 10.1039/c2fd20091k. discussion 273–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widom B. Potential-distribution theory and the statistical mechanics of fluids. J. Phys. Chem. 1982;86:869–872. [Google Scholar]
- 37.Grillo D., de la Cruz M.O., Szleifer I. Theoretical studies of the phase behavior of DPPC bilayers in the presence of macroions. Soft Matter. 2011;7:4672–4679. [Google Scholar]
- 38.Sodt A.J., Venable R.M., Pastor R.W. Nonadditive compositional curvature energetics of lipid bilayers. Phys. Rev. Lett. 2016;117:138104. doi: 10.1103/PhysRevLett.117.138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campelo F., Kozlov M.M. Sensing membrane stresses by protein insertions. PLOS Comput. Biol. 2014;10:e1003556. doi: 10.1371/journal.pcbi.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinot M., Vanni S., Barelli H. Lipid cell biology. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science. 2014;345:693–697. doi: 10.1126/science.1255288. [DOI] [PubMed] [Google Scholar]
- 41.Bigay J., Gounon P., Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 42.Vanni S., Hirose H., Gautier R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat. Commun. 2014;5:4916. doi: 10.1038/ncomms5916. [DOI] [PubMed] [Google Scholar]
- 43.Frolov V.A., Shnyrova A.V., Zimmerberg J. Lipid polymorphisms and membrane shape. Cold Spring Harbor Perspect. Biol. 2011;3:a004747. doi: 10.1101/cshperspect.a004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kucerka N., Nieh M.-P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim Biophys Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 45.Resh M.D. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 46.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 47.Veatch S.L., Keller S.L. Seeing spots: complex phase behavior in simple membranes. Biochim Biophys Acta. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 48.van Meer G., de Kroon A.I.P.M. Lipid map of the mammalian cell. J. Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 49.Sprong H., van der Sluijs P., van Meer G. How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2001;2:504–513. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- 50.Apolloni A., Prior I.A., Hancock J.F. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plowman S.J., Hancock J.F. Ras signaling from plasma membrane and endomembrane microdomains. Biochim. Biophys. Acta. 2005;1746:274–283. doi: 10.1016/j.bbamcr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Mitra K., Ubarretxena-Belandia I., Engelman D.M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andersen O.S., Koeppe R.E., 2nd Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 54.Hancock J.F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 55.Bigay J., Casella J.F., Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mesmin B., Drin G., Antonny B. Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry. 2007;46:1779–1790. doi: 10.1021/bi062288w. [DOI] [PubMed] [Google Scholar]
- 57.Vamparys L., Gautier R., Fuchs P.F.J. Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature. Biophys. J. 2013;104:585–593. doi: 10.1016/j.bpj.2012.11.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui H., Lyman E., Voth G.A. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys. J. 2011;100:1271–1279. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., Gorfe A.A. Membrane remodeling by surface-bound protein aggregates: insights from coarse-grained molecular dynamics simulation. J. Phys. Chem. Lett. 2014;5:1457–1462. doi: 10.1021/jz500451a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z., Gorfe A.A. Deformation of a two-domain lipid bilayer due to asymmetric insertion of lipid-modified Ras peptides. Soft Matter. 2013;9:11249–11256. doi: 10.1039/C3SM51388B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z., Gorfe A.A. Modulation of a small two-domain lipid vesicle by linactants. J. Phys. Chem. B. 2014;118:9028–9036. doi: 10.1021/jp5042525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McIntosh T.J., Simon S.A. Roles of bilayer material properties in function and distribution of membrane proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- 63.Kucerka N., Pencer J., Katsaras J. Curvature effect on the structure of phospholipid bilayers. Langmuir. 2007;23:1292–1299. doi: 10.1021/la062455t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kollmitzer B., Heftberger P., Pabst G. Monolayer spontaneous curvature of raft-forming membrane lipids. Soft Matter. 2013;9:10877–10884. doi: 10.1039/C3SM51829A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kooijman E.E., Chupin V., Rand P.R. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 2005;44:2097–2102. doi: 10.1021/bi0478502. [DOI] [PubMed] [Google Scholar]
- 66.Zimmerberg J., Kozlov M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 67.Orsi M., Michel J., Essex J.W. Coarse-grain modelling of DMPC and DOPC lipid bilayers. J. Phys. Condens. Matter. 2010;22:155106. doi: 10.1088/0953-8984/22/15/155106. [DOI] [PubMed] [Google Scholar]
- 68.Marsh D. Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys. J. 2007;93:3884–3899. doi: 10.1529/biophysj.107.107938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poger D., Mark A.E. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: a comparison with experiment. J. Chem. Theory Comput. 2010;6:325–336. doi: 10.1021/ct900487a. [DOI] [PubMed] [Google Scholar]
- 70.Flory P.J. Wiley-Interscience; New York, NY: 1969. Statistical Mechanics of Chain Molecules. [Google Scholar]
- 71.Rawicz W., Olbrich K.C., Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arriaga L.R., López-Montero I., Hellweg T. Stiffening effect of cholesterol on disordered lipid phases: a combined neutron spin echo + dynamic light scattering analysis of the bending elasticity of large unilamellar vesicles. Biophys. J. 2009;96:3629–3637. doi: 10.1016/j.bpj.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kucerka N., Liu Y., Nagle J.F. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using x-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 2005;88:2626A–2637A. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dimova R. Recent developments in the field of bending rigidity measurements on membranes. Adv. Colloid Interface Sci. 2014;208:225–234. doi: 10.1016/j.cis.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 75.Gullingsrud J., Schulten K. Lipid bilayer pressure profiles and mechanosensitive channel gating. Biophys. J. 2004;86:3496–3509. doi: 10.1529/biophysj.103.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Orsi M., Essex J.W. Physical properties of mixed bilayers containing lamellar and nonlamellar lipids: insights from coarse-grain molecular dynamics simulations. Faraday Discuss. 2013;161:249–272. doi: 10.1039/c2fd20110k. discussion 273–303. [DOI] [PubMed] [Google Scholar]
- 77.Rowlinson J.S., Widom B. Dover Publications; New York, NY: 1982. Molecular Theory of Capillarity. [Google Scholar]
- 78.Peter B.J., Kent H.M., McMahon H.T. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.